Abstract
We have isolated a recombinant phage antibody (Phab) that binds a distinct epitope of the subclass of the ς54-dependent prokaryotic enhancer-binding proteins that respond directly to aromatic effectors, e.g., those that activate biodegradative operons of Pseudomonas spp. The DNA segments encoding the variable (V) domains of the immunoglobulins expressed by mice immunized with the C-terminal half of TouR (TouRΔA) of Pseudomonas stutzeri OX1 were amplified and rearranged in vitro as single-chain Fv (scFv) genes. An scFv library was thereby constructed, expressed in an M13 display system, and subjected to a panning procedure with TouR. One clone (named B7) was selected with high affinity for TouR and XylR (the regulator of the upper TOL operon of the pWW0 plasmid). The epitope recognized by this Phab was mapped to the peptide TPRAQATLLRVL, which seems to be characteristic of the group of enhancer-binding proteins to which TouR and XylR belong and which is located adjacent to the Walker B motif of the proteins. The Phab B7 was instrumental in measuring directly the intracellular levels of XylR expressed from its natural promoter in monocopy gene dosage in Pseudomonas putida under various conditions. Growth stage, the physical form of the protein produced (XylR or XylRΔA), and the presence or absence of aromatic inducers in the medium influenced the intracellular pool of these molecules. XylR oscillated from a minimum of ∼30 molecules (monomers) per cell during exponential phase to ∼140 molecules per cell at stationary phase. Activation of XylR by aromatic inducers decreased the intracellular concentration of the regulator. The levels of the constitutively active variant of XylR named XylRΔA were higher, fluctuating between ∼90 and ∼570 molecules per cell, depending on the growth stage. These results are compatible with the present model of transcriptional autoregulation of XylR and suggest the existence of mechanisms controlling the stability of XylR protein in vivo.
The regulators that belongs to the NtrC-family of prokaryotic enhancer-binding proteins activate transcription at a distance through the alternative sigma factor ς54 (8, 15, 26). A subclass of these proteins (e.g., XylR, DmpR, TouR, MopR, PhhR, Ph1R, TmbR, and PheR) specialize in the activation of catabolic operons involved in degradation of recalcitrant aromatic compounds (e.g., toluene, xylene, phenol, cresols, and other ring-containing hydrocarbons) (1, 4, 23, 24, 39). These proteins are activated upon association with cognate aromatic effectors (the substrates of the catabolic operons), and thus, they directly translate effector binding into transcriptional activation (38). These operons are commonly found in environmental isolates, especially in those belonging to Pseudomonas and Pseudomonas-like genera (40). For instance, the XylR protein was found in Pseudomonas putida mt-2, a strain capable of degrading toluene and meta- and p-xylene (1). Similarly, TouR regulates a catabolic pathway for degradation of o-xylene in P. stutzeri OX1 (although its actual effector is 2,3-dimethyl phenol, an intermediate of the o-xylene metabolic pathway) (4). Since XylR is an intensively studied specimen of such a group, we will refer hereafter to these proteins generically as members of the XylR class.
XylR and its related proteins have a common organization divided into four structural domains that also play different functional roles (22, 26). The N-terminal or A domain is involved in the recognition of the aromatic effector that triggers the activation of the protein (9, 28); the central or C domain has an ATPase activity and is responsible for the activation of the RNA polymerase-ς54 complex (29, 41). A short sequence (referred as to the B domain) connects the A and C domains (43). Although its function is still unclear, it may be involved in the intramolecular derepression of the protein after binding of the aromatic effector by the A domain (11, 27). Finally, the C-terminal or D domain contains a helix-turn-helix motif that is required for the binding-specific DNA sites located at the promoters of these catabolic operons (31). The C and D domains of XylR-like regulators have amino acid identities ranging from 60 to 70%. In general, the A domains are less conserved than the C domains, a fact that can be partially explained by their different specificities in recognition of aromatic effectors. In some cases, however, the similarity between two A domains responding to different effectors might be higher than that of two A domains responding to the same effector in different strains (37).
Monitoring XylR behavior in vivo requires specific tools able to reveal the number and the physical form of the protein in its natural host and stoichiometry (i.e., monocopy gene dosage). Although we have produced anti-XylR serum in the past (9), this material failed to detect adequately the protein expressed from its natural promoter in P. putida. This was likely to be caused by the very low concentration of intracellular XylR. We have thus resorted in this work to the production of a high-affinity single-chain antibody able to detect minute amounts of the protein in its natural state.
The technology for antibody production in Escherichia coli is based on the amplification of the V gene segments encoding the variable domains from the heavy (VH) and light (VL) chains of immunoglobulins (Igs) and their cloning into a filamentous phage or phagemid vector that displays the reconstructed Fv molecule in the phage particle (33, 42). The repertoires of VH and VL gene segments can be assembled in vitro as single-chain fragments (scFvs) by means of a linker encoding a flexible peptide. These pools of scFv-encoding genes are cloned in a phagemid vector that can be packaged in vivo into M13 phage particles that display the scFv library as hybrids with the minor coat protein III. The physical association within the same phage particle of the scFv fragment and its encoding gene allows the selective amplification of those clones binding a given antigen, a procedure known as panning (14). In this study, we have utilized this strategy for the selection of a high-affinity phage antibody (Phab) which specifically recognizes not only XylR but also the other members of the XylR class of regulators. With this antibody in hand, we have been able to visualize for the first time the fluctuations in intracellular XylR levels of P. putida in respect to growth phase and exposure to aromatic inducers.
MATERIALS AND METHODS
Bacteria, phages, growth, and induction conditions.
The E. coli strain XL-1 Blue (recA1 gyrA96 relA1 endA1 hsdR17 supE44 thi1 lac [F′ proAB lacIq lacZΔM15 Tn10] Tcr; Stratagene) was used as host for bacteriophages and phagemids. E. coli XL-1 Blue cells, harboring a phagemid encoding an scFv, were routinely grown at 30°C in 2× yeast extract-tryptone (YT) liquid medium or Luria-Bertani (LB) agar plates, containing glucose (2% [wt/vol]) for repressing the lac promoter, 10 μg of tetracycline (TET)/ml for F′ selection, and 150 μg of ampicillin (AMP)/ml for phagemid selection. For packaging of phagemids into M13 particles, these E. coli cells were infected with VCS-M13 helper phage (Kmr; Stratagene). Amplification of VCS-M13 helper phage was carried out in E. coli XL-1 Blue cells grown at 30°C in 2× YT medium containing 50 μg of kanamycin (KAN)/ml. E. coli strain BL21(DE3) (ompT hsdSB rB− mB− gal dcm λDE3; Novagen) transformed with plasmid pLysS was employed for the production of TouRΔA fragments encoded by pET derivatives (Novagen). The E. coli DH5α F′ [Δ(lacZYA-argF)U169 φ80(lacZΔM15) hsdR17 recA1 endA1 gyrA96 relA1 supE44 thi F′] was the host strain for construction and amplification of pET derivatives. E. coli BL21(DE3) and DH5α F′ strains were grown at 37°C in LB medium (21) containing the appropriate antibiotics. Chloramphenicol (CHL; 30 μg/ml) and AMP (150 μg/ml) were employed for selection of pLysS and pET derivatives, respectively. The production of TouRΔA fragments in E. coli BL21(DE3)(pLysS) cells, harboring a pET derivative, was induced by addition of 1 mM isopropyl-1-thio-β-d-galactoside (IPTG) to mid-log-phase (optical density at 600 nm [OD600], ∼0.5) cultures. After 4 h of induction, E. coli cells were harvested from the cultures and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting (see below). P. putida strains KT2442, MAD1, and MAD2 (9) and P. stutzeri OX1 (4) were grown at 30°C in LB medium. Activation of XylR in P. putida MAD1 was performed by adding 2 mM 3-methyl-benzyl-alcohol (3-MBA; from a 1 M stock in ethanol) directly to cultures. Following 1 to 3 h of incubation at 30°C, as indicated, the cells were harvested (5,000 × g, 10 min) and analyzed by SDS-PAGE and Western blotting (see below).
Phagemids, plasmids, and DNA constructs.
Standard methods were used to purify, analyze, manipulate, and amplify DNA (5). All oligonucleotides were synthesized by Isogen Bioscience BV and Cruachem. DNA constructs and phagemids were sequenced using the dideoxy method and an ABI-PRISM automated DNA sequencer (Perkin-Elmer). The phagemid pCANTAB-5E (Apr; Amersham Pharmacia Biotech) was utilized for the cloning of scFv genes. The phagemid pHen-MBP (Apr) (25) encodes an scFv against the E. coli maltose binding protein (MBP). To construct the pET vectors expressing truncations of TouRΔA, the DNA fragments encoding these deletions were amplified by PCR from plasmid pFP3038, carrying the wild-type touR gene (4), and cloned into the BamHI and HindIII sites of pET21d (Novagen). The primers used in these amplifications were the following: 5′-GGTCGGATCCGACTTGAGAAACAGCAG-3′ and 5′-GGCCGCAAGCTTGGTGGCGGCGGTTAC-3′ for fragment F1, 5′-GGTCGGATCCGAGAAGCGGAATTGTTT-3′ and 5′-GGCCGCAAGCTTGACTGCGAATAGGGA-3′ for fragment F2, and 5′-GGTCGGATCCGACTCAAGAAGTTCCAC-3′ and 5′-GGCCGCAAGCTTGGCTTCAGAAAAAATGCC-3′ for fragment F3.
Immunizations.
Three female BALB/c mice were immunized by intraperitoneal injection with the recombinant 6xhisTouRΔA protein, a truncated form of TouR in which the initial 225 N-terminal amino acids had been deleted and replaced by a six-histidine tag (3, 4). This protein was purified by immobilized metal affinity chromatography from overproducing E. coli cells as described previously for 6xhisXylRΔA (30). For immunizations, 6xhisTouRΔA protein was dialyzed against phosphate-buffered saline (PBS; 8 mM Na2HPO4, 1.5 mM KH2PO4, 3 mM KCl, 137 mM NaCl, pH 7.0.) and diluted at 0.5 mg/ml in PBS containing 0.1% (wt/vol) SDS. Just prior to injection, 0.3 ml of this protein stock was mixed with an identical volume of MPL+TDM adjuvant (Sigma) previously reconstituted in PBS at 1 mg/ml. For each mouse, 0.2 ml of this antigen-adjuvant emulsion (corresponding to 50 μg of 6xhisTouRΔA) was injected intraperitoneally (13). Three immunizations, at days 0, 21, and 42, were made. Ten days after the last boosting, an ∼100-μl blood sample was taken from each mouse, and the sera obtained were employed to determine the specific Ig response elicited against 6xhisTouRΔA by enzyme-linked immunosorbent assay (ELISA) (13).
scFv library construction.
The protocols described by McCafferty and Johnson (20) were basically followed with some modifications. The spleens from 6xhisTouRΔA-immunized mice were removed, placed into independent petri dishes containing sterile PBS, and finely disaggregated. Large clumps were discarded, and the individual cells were harvested by centrifugation (100 × g, 7 min). The erythrocytes were lysed by resuspension of the cell pellet in 5 ml of sterile EL solution (155 mM NH4Cl, 10 mM NaHCO3, 0.1 mM EDTA), and the splenocytes (mostly lymphocytes) were quickly harvested by centrifugation (100 × g, 7 min). The cell pellet was immediately lysed, and the total RNA was isolated according to the guanidinium isothiocyanate-acid phenol procedure (Ultraspec RNA isolation; Biotecx). The poly(A)+ mRNA was purified using an oligo(T) resin (OligoTex mRNA minikit; Qiagen) and then employed as template for a first-strand cDNA synthesis reaction (Amersham Pharmacia Biotech). The VH and VL gene segments were amplified from the cDNA samples by PCR (30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min). The sequences of the oligonucleotides used as primers, except those indicated, have been published previously (20). Typically, 1 μl of a 1:100 dilution of the cDNA synthesis was utilized as template in a 50-μl amplification reaction mixture (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 0.001% gelatin, 1.25 mM MgCl2, 250 μM [each] deoxynucleoside triphosphate [dNTP], 0.6 μM VH oligonucleotide mix, 0.6 μM VL oligonucleotide mix, and 1 U of Taq DNA polymerase). The VH oligonucleotide mix was an equimolar combination of oligonucleotides VH1FOR-2 and VH1BACK. The VL oligonucleotide mix was a combination of oligonucleotides VK2BACK-2 (5′-GACATTGAGCTCACCCAGTCTC-3′), MJK1FONX, MJK2FONX, MJK4FONX, and MJK5FONX in a molar ratio (4:1:1:1:1). As a negative control for amplification, the mock template used in the PCR was either 1 μl of the poly(A)+ mRNA or 1 μl of a cDNA synthesis mixture without added mRNA. Identical quantities of the V genes amplified from the three mice were pooled, and the ∼350-bp VH DNA fragments and the ∼320-bp VL DNA fragments were purified from agarose gels (Qiaex II kit; Qiagen). A DNA fragment of ∼100 bp was used as the linker for the assembly of the scFv genes. This DNA segment encoded a (Gly4Ser)3 sequence and contained regions of homology to the 3′ end of VH genes and the 5′ end of VL genes. The linker fragment was amplified (30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min) using 1 μl of Linker Primer mix (Amersham Pharmacia Biotech) as template DNA in a 50-μl PCR mixture (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 0.001% gelatin, 1.25 mM MgCl2, 250 μM [each] dNTP, 0.5 μM LINKBACK oligonucleotide, 0.5 μM LINKFOR5′-2 [5′-GAGACTGGGTGAGCTCAATGTC-3′], and 1 U of Taq DNA polymerase).
The scFv genes were assembled in a VH-linker-VL configuration by a homology-driven reaction without primers (25 cycles of 94°C for 1 min, 60°C for 2 min, and 72°C for 2 min) using a 25-μl reaction mixture that contained a Taq high-fidelity buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 0.001% gelatin, 4 mM MgCl2, 1 mM [each] dNTP), 30 ng of each DNA fragment (i.e., VH, VL, and linker), and Taq DNA polymerase (5 U). For optimal performance, this assembly reaction mixture was covered with mineral oil (Sigma) to minimize evaporation. Next, a standard amplification of the scFv genes was performed with oligonucleotides that incorporated SfiI and NotI restriction sites flanking the scFv gene (20). Typically, 1 μl of the assembly reaction mixture was included as DNA template in a 50-μl PCR mixture (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 0.001% gelatin, 1.25 mM MgCl2, 250 μM [each] dNTP, 0.5 μM VH1BACKSfi oligonucleotide, 0.5 μM JKNOT oligonucleotide mix, and 1 U of Taq DNA polymerase). The JKNOT oligonucleotide mix is an equimolar combination of oligonucleotides JK1NOT10, JK2NOT10, JK4NOT10, and JK5NOT10. After amplification, the ∼0.7-kb DNA fragments corresponding to the assembled scFv genes were digested with SfiI and NotI restriction enzymes, gel purified, and ligated into the same sites of pCANTAB-5E (Amersham Pharmacia Biotech). Finally, the products of different ligations were electroporated into E. coli XL-1 Blue cells, plated in 2× YT–glucose-TET-AMP medium (containing 2% glucose, 10 μg of TET/ml, and 150 μg of AMP/ml), and incubated at 30°C. At least 2 × 106 independent colonies were harvested from these plates in LB plus glycerol (15% [vol/vol]), pooled, and stored at −80°C. A control ligation of the SfiI/NotI pCANTAB-5E vector used gave ∼1% transformant colonies.
Rescue of phagemids.
Assembling of M13 particles displaying scFv-protein III hybrid (Phab production) was accomplished in E. coli XL-1 cells harboring a phagemid and infected with VCS-M13 (Kmr) helper phage under growth conditions that allow a weak expression of the lac promoter. For large-scale preparation of Phab(s), a single colony of E. coli XL-1 cells harboring a phagemid clone (or a mixture of E. coli cells representing the library or a subpopulation after panning) was inoculated in 40 ml of 2× YT–glucose-TET-AMP medium (containing 2% glucose, 10 μg of TET/ml, and 150 μg of AMP/ml) and incubated at 30°C until the OD600 was ∼0.2. At this point, ∼1010 PFU of VCS-M13 helper phage was added, and the cultures were incubated at 37°C for 1 h with gentle agitation. Then, E. coli cells were harvested by centrifugation (4,000 × g, 5 min) and resuspended in 400 ml of fresh 2× YT–TET-AMP-KAN medium. The absence of glucose guarantees a low level of expression of the scFv-gene III fusion, and the presence of KAN (50 μg/ml) allows the selection of the E. coli cells infected with the M13 helper phage. After 16 h of incubation at 30°C, the cultures were chilled on ice and centrifuged (8,000 × g, 10 min at 4°C) to remove the E. coli cells. To recover the M13 particles from the supernatant, 100 ml of polyethylene glycol-NaCl solution (20% [wt/vol] polyethylene glycol 8000; 2.5 M NaCl) was added, and the resulting mixture was kept on ice for an additional 45 min. The phage pellets obtained after centrifugation (8,000 × g, 20 min at 4 oC) were resuspended in 4 ml of TE (10 mM Tris-HCl, 1.0 mM EDTA, pH 8.0) and stored at −80°C. Phage stocks were titrated by serial 10-fold dilutions over E. coli XL-1 Blue cells which had been grown at 37°C in 2× YT liquid medium containing TET (10 μg/ml) until the OD600 was ∼0.2. The mixtures were incubated for 1 h at 37°C without agitation, plated over LB agar-glucose (2%) plates, containing either AMP (150 μg/ml) or KAN (50 μg/ml), and incubated at 30°C for 16 to 36 h. The number of Apr colonies represents the titer of the phagemid, whereas the number of Kmr colonies indicates the degree of contamination by VCS-M13 helper phage in the stock (usually <2%). The production of scFv-protein III fusions during viral packaging was tested by immunoblotting of E. coli whole-cell protein extracts (see below).
For rapid screening of TouR and XylR Phab binders, a small-scale rescue of phagemids was performed in cultures of E. coli cells grown in 96-well microtiter plates. Single colonies of E. coli XL-1 harboring phagemids were toothpicked into a 96-well flat-bottomed plate (Nunclon ΔSurface; Nunc) containing 100 μl of 2× YT–glucose-TET-AMP medium/well (see above). After overnight incubation at 30°C with shaking (inside a box with a water-saturated atmosphere), this master plate was used to inoculate (using a 96-well sterile transfer device) a round-bottomed 96-well plate (Nunclon ΔSurface) containing 150 μl of 2× YT–glucose-TET-AMP medium/well. The master plate was frozen at −80°C after addition of 50 μl of 60% (vol/vol) glycerol/well. This plate was kept for the recovery of positive clones. The replica plate was incubated at 37°C for 2 h (OD600, ∼0.4), and then ∼109 PFU of VCS-M13 was added per well. After 45 min of incubation at 37°C, the plate was centrifuged (585 × g, 10 min at room temperature), and the cell pellets were resuspended in 150 μl of 2× YT–TET-AMP-KAN medium without glucose (see above). After incubation overnight at 30°C, the plate was centrifuged (585 × g, 10 min at room temperature), and the supernatants (containing the Phabs) were used in ELISA (see below) to determine their specific binding to the antigen.
Panning of Phabs binding 6xhisTouRΔA.
All steps were performed at room temperature. Purified 6xhisTouRΔA (10 μg/ml) was adsorbed for 2 h to eight wells (50 μl/well) of a microtiter immunoplate (Maxisorb; Nunc) in 50 mM NaHCO3 (pH 9.0). The 6xhisTouRΔA solution was discarded, and the wells were blocked by adding 200 μl of MBT buffer (3% [wt/vol] skimmed milk, 1% [wt/vol] bovine serum albumin, 0.1% [vol/vol] Tween 20 in PBS)/well. After 2 h, the blocking solution was replaced by a total of 2 × 1011 PFU of Phabs in MBT buffer (50 μl/well at 5 × 1012 PFU/ml). Phabs were allowed to bind for 1 h, and the unbound Phabs were removed from the plates by 20 washes of 1 min employing 200 μl of washing solution (PBS, 0.1% [vol/vol] Tween 20)/well followed by an additional 20 washes of 1 min using PBS (200 μl/well). To collect the Phabs bound to 6xhisTouRΔA, the wells were incubated for 5 min with 0.1 M glycine, pH 2.5 (50 μl/well). The glycine solutions from the different wells were pooled in a sterile tube and immediately equilibrated by addition of 1 volume (400 μl) of 1 M Tris-HCl, pH 7.5. The titer of the Phab in this solution was determined over E. coli XL-1 Blue cells (see above) and referred to as bound phage. The bound Phabs were later used to infect E. coli XL-1 Blue cells and plated on 2× YT–glucose-AMP-TET medium. After 24 h of incubation at 30°C, the colonies grown on these plates were harvested as a pool and used for phagemid rescue. A new round of panning was performed over the new Phab sublibrary generated. Finally, individual Phab clones were rescued on a small scale, and their specific binding to 6xhisTouRΔA was determined by ELISA.
ELISAs.
The ELISAs were performed at room temperature. Purified 6xhisTouRΔA, 6xhisXylRΔA, 6xhisTouR, or ovalbumin (Sigma), as a negative control, was adsorbed for 2 h to 96-well immunoplates (Maxisorb; Nunc) at 10 μg/ml in 50 mM NaHCO3 (pH 9.0). Excess antigen was washed out, and the plates were blocked for 2 h using 200 μl of MBT buffer per well (see above regarding panning). The blocking solution was discarded, and the primary antibodies (Phabs or immune sera containing Igs) were added to the wells (50 μl of the dilution indicated in each case in MBT buffer). After 1 h of incubation, the unbound Phabs (or Igs) were removed by four 3-min washings of the wells with the same washing solution as used in panning. For detection of the bound Phab, the anti-M13 monoclonal antibody (MAb)-peroxidase conjugate (Amersham Pharmacia Biotech) was added at a 1:5,000 dilution in MBT buffer (50 μl/well). For detection of bound Igs, a goat anti-mouse IgG-peroxidase-horseradish peroxidase conjugate (Boehringer Mannheim; 0.03 U/ml) was used. After 1 h of incubation with the secondary antibody, the microtiter plates were washed as before and developed using a mixture of o-phenylenediamine (0.4 mg ml−1; Sigma) and H2O2 (0.012% [vol/vol]; Sigma) in phosphate-citrate buffer (103 mM dibasic sodium phosphate, 24 mM citric acid, pH 5.0; 80 μl per well). The reaction was allowed to proceed in the dark for 10 min and stopped with 0.6 N HCl (20 μl of 3 N HCl per well), and the OD490 of the plates was determined (Benchmark microplate reader; Bio-Rad Laboratories). Background binding to ovalbumin (usually at an OD490 of ≤0.05) was subtracted from the values of specific antigen binding obtained in all cases.
Gel electrophoresis and Western blotting.
SDS-PAGE was performed by standard protocols using the Miniprotean system (Bio-Rad). Whole-cell protein extracts from E. coli and Pseudomonas were prepared by harvesting the cells (10,000 × g, 5 min) from 1 ml of stationary-phase cultures (OD600 of ∼2.5; ∼2.5 × 109 CFU/ml), 10 ml of exponential cultures (OD600 of ∼0.5; ∼2.5 × 108 CFU/ml), or 5 ml of late-exponential cultures (OD600 of ∼1.1; ∼5 × 108 CFU/ml), as indicated, and resuspension of the cell pellet in 100 μl of 10 mM Tris-HCl, pH 7.5. Next, 100 μl of reducing 2× SDS sample buffer (120 mM Tris-HCl [pH 6.8], 2% [wt/vol] SDS, 10% [vol/vol] glycerol, 0.01% [wt/vol] bromophenol blue, 2% [vol/vol] 2-mercaptoethanol) was added to the samples, boiled for 10 min, sonicated briefly (∼5 s), and centrifuged (14,000 × g, 10 min) to eliminate the DNA viscosity and any insoluble material (i.e., peptidoglycan). When indicated, soluble protein extracts were employed instead of whole-cell protein extracts. To prepare soluble protein extracts, bacterial cells (20 ml of an overnight culture) were resuspended in 1 ml of buffer containing 20 mM Tris-HCl, 150 mM NaCl, and 1 mM EDTA (pH 7.5) and lysed by six pulses of sonication (30 s each) employing a LabSonic U instrument (B. Braun) with an output setting of −65. After a short spin to remove unbroken cells (3,000 × g, 5 min), the cell lysate was centrifuged at high speed (100,000 × g, 1 h) in a TL-100 centrifuge (Beckman), and the supernatant was collected as soluble protein extract. As described above, the protein samples were boiled in 2-mercapthoethanol–SDS sample buffer (1×) before electrophoresis. In all cases, the proteins were separated by SDS-PAGE (usually ∼10 μl was loaded per lane; ∼1.25 × 108 CFU per lane) and transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon P; Millipore) using a semidry electrophoresis transfer apparatus (Bio-Rad). The Kaleidoscope prestained standards (Bio-Rad) were used as markers of known molecular weights for the SDS-PAGE. After protein transfer, the membranes were blocked for 2 h at room temperature (or for 16 h at 4°C) using MBT buffer. For immunodetection of XylR-like proteins using Phabs, the membranes were incubated with 30 ml of MBT buffer containing 5 × 109 PFU of Phab B7/ml. The use of a large volume of MBT buffer during this step strongly diminished a background binding of the phages to the PVDF membranes. Unbound Phabs were eliminated by four washing steps of 5 min in 40 ml of PBS–0.1% (vol/vol) Tween 20. Next, anti-M13-peroxidase conjugate (1:5,000 in MBT buffer) was added to detect the bound Phab. For immunodetection of the E-tagged scFvs and scFv-protein 3 hybrids in whole-cell protein extracts of E. coli XL-1 cells (obtained after phage rescue), the membranes were incubated for 1 h with anti-E-tag MAb-peroxidase conjugate (1 μg/ml in MBT buffer; Amersham Pharmacia Biotech). For immunodetection of GroEL, a rabbit serum raised against the purified protein of E. coli (a gift of J. M. Valpuesta, Centro Nacional de Biotecnología) was employed in MBT buffer (1:5,000). After 1 h of incubation, the membranes were washed four times with PBS–0.1% (vol/vol) Tween 20 and further incubated for 1 h with protein A-peroxidase conjugate (Sigma; 1:5,000) in MBT buffer. In all cases, the membranes were washed four times with PBS–0.1% (vol/vol) Tween 20, and the bound peroxidase conjugates were developed by a chemiluminescence mixture of 1.25 mM luminol (Sigma) and 42 μM luciferin (Roche) in 100 mM Tris-HCl (pH 8.0). The membranes were soaked in this mixture, and H2O2 was added at 0.0075% (vol/vol). After 1 min of incubation in the dark, the PVDF membrane was exposed to an X-ray film (X-OMAT; Kodak) or to a Chemi Doc (Bio-Rad) luminometer. The intensity of the light in the protein bands was quantified in a Chemi Doc employing the Quantity One software (Bio-Rad). This standard procedure allowed the detection of ∼1 ng of XylR-like protein per lane. Higher sensitivity was obtained with an enhanced chemiluminescence mixture for detection of peroxidase (Roche).
Peptide synthesis.
Overlapping deca- and dodecapeptides from the TouR sequence (amino acids Glu301 to Thr372) were prepared as previously described (10, 36) by automated spot synthesis (Abimed, Langerfeld, Germany) onto an amino-derivatized cellulose membrane, immobilized by their C termini via a polyethylene glycol spacer, and N-terminally acetylated. Membranes were blocked in MBT buffer, incubated with Phab B7, and washed and developed as described for Western blotting.
RESULTS AND DISCUSSION
Selection of a phage antibody against regulators of the XylR class.
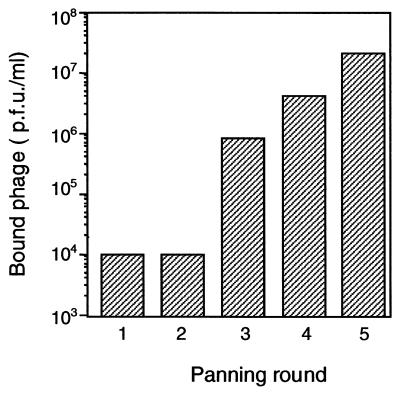
In order to obtain a phage library displaying scFvs specific against the XylR class of proteins, three BALB/c mice were immunized with 6xhisTouRΔA, a polypeptide containing the C and D domains of TouR, which are well conserved among proteins of the XylR class, and that was recently purified in our laboratory in high quantities from an overproducing E. coli strain (3). The splenocytes of these animals were employed for mRNA isolation and cDNA synthesis, and the VH and VL gene segments of the Igs were amplified by PCR (20). These V segments were assembled as scFv genes in a VH-linker-VL configuration and finally cloned into the SfiI and NotI sites of phagemid pCANTAB-5E. This vector accommodates the scFvs between an N-terminal signal peptide and protein III from M13. A library of ∼2 × 106 independent clones was obtained after transformation of E. coli XL-1 Blue cells, and the phagemids were rescued into M13 particles that display on their capsid the scFv library. These phage were utilized in a panning procedure to select clones binding to purified 6xhisTouRΔA. Five rounds of selection and amplification of the bound phage were performed. In each round, the input phage titers were kept uniformly at 2 × 1011 PFU, and after each selection, the number of phage bound to 6xhisTouRΔA was determined (Fig. 1). The results show that the titer of phage bound to 6xhisTouRΔA increased steadily from ∼1 × 104 PFU in the first and second rounds to ∼2 × 107 PFU in the fifth round.
FIG. 1.
Selection of Phabs binding to TouRΔA. The number of phage bound to ELISA plates coated with 6xhisTouRΔA after each panning round is indicated. The bound phage were eluted from the plates by incubation with 0.1 M glycine (pH 2.5) as explained in Materials and Methods. After recovery, the titers of these phage were determined on E. coli XL-1 Blue cells and selected for AMP resistance. In each panning round, the number of input phage was kept constant at 2 × 1011 PFU and the phage that did not bind 6xhisTouRΔA were removed by 40 washing steps with PBS. The increase in the number of bound phage after the second round of panning is indicative of a preferential amplification of Phab clones binding to 6xhisTouRΔA.
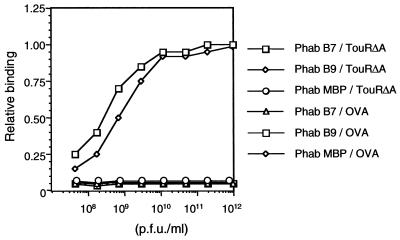
The above results suggested a selective amplification of Phab clones binding to 6xhisTouRΔA. To ascertain the binding properties of the amplified Phabs, 96 clones were individually rescued and the binding of the amplified Phab to 6xhisTouRΔA, 6xhisXylRΔA, and ovalbumin (as a negative control) was measured by ELISA (data not shown). Most of these clones (ca. 88) bound to 6xhisTouRΔA and, importantly, to 6xhisXylRΔA (OD490 ≥ 1.5), whereas no binding could be detected to ovalbumin (OD490 ≤ 0.1). The double-stranded phagemid DNA was isolated from the E. coli cells of 20 positive clones, and DNA sequences of their scFv genes were determined. Out of these 20 clones, 19 encoded the same scFv (hereinafter referred to as B7) and a single clone (named B9) coded for an scFv with five amino acid changes compared to B7 (Fig. 2). The high similarity of their amino acid sequence suggested that the two scFvs recognized the same epitope. Since the two scFvs have similar expression levels within E. coli cells (data not shown), the preferential amplification throughout the panning procedure of Phab B7 suggested a higher affinity of this clone for 6xhisTouRΔA. A direct comparison by ELISA of the binding properties of Phab B7 and Phab B9 also revealed a higher affinity of the B7 clone (Fig. 3). By ELISA, Phab B7 displayed identical affinities for 6xhisTouRΔA and for 6xhisXylRΔA (data not shown).
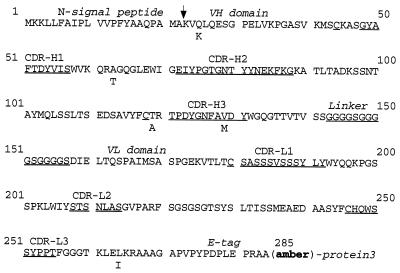
FIG. 2.
Amino acid sequence of scFv B7. The amino acid sequence of the scFv B7 polypeptide encoded by the phagemid is shown. The positions of the N-terminal signal peptide, the VH domain, the (Gly4Ser)3 linker peptide, the VL domain, and the E tag are indicated. The complementarity-determining regions (CDR) of the VH and VL domains are labeled and underlined. The site of cleavage of the bacterial signal peptidase is marked by an arrow. The five amino acid changes found in the scFv B9 are marked below the sequence of scFv B7. When produced in E. coli XL-1 Blue cells (supE), these scFvs are also synthesized as hybrids with protein 3 of M13. The location of the suppressed stop codon (amber), which is placed between the scFv and protein 3 coding sequences, is indicated.
FIG. 3.
Binding of Phabs B7 and B9 to TouRΔA. The binding of Phabs B7 and B9 and MBP (as a negative control [25]) to 6xhisTouRΔA or ovalbumin (OVA) as a specificity control antigen was determined by ELISA. Different dilutions of Phabs were incubated on ELISA plates coated with the antigens as indicated. After washing with PBS, the bound Phab was developed using anti-M13-peroxidase conjugate and the plates were read at OD490. The data shown are relative to the maximal OD490 obtained by Phab B7 at the higher titer employed (OD490 of ∼2.0).
Detection of XylR and TouR by Phab B7 in Western blots.
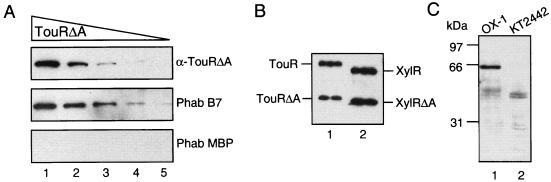
We investigated the ability of Phab B7 to detect XylR and TouR proteins after denaturing SDS-PAGE and Western blotting. As shown in Fig. 4A, Phab B7 allowed the detection of ∼5-fold-lower amounts of purified 6xhisTouRΔA than did the polyclonal serum obtained from the immunized mice. The detection limit of 6xhisTouRΔA determined for Phab B7 was ∼0.5 ng, whereas that of the polyclonal serum was ∼2.5 ng. As in the ELISA, Phab B7 recognized TouR, XylR, and their respective ΔA forms in Western blots (Fig. 4B). Next, we analyzed whether Phab B7 could specifically light up TouR out of complex protein mixtures from P. stutzeri OX1. To this end, soluble protein extracts were prepared from stationary cultures of P. stutzeri OX1 (the original strain from which touR was cloned [4]), and P. putida KT2442, a strain without known XylR-like regulators. As shown in Fig. 4C, Phab B7 clearly recognized a protein band in the extracts of P. stutzeri OX1 with an apparent molecular mass in SDS-PAGE of ∼65 kDa, in good agreement with the expected mass of TouR deduced from its amino acid sequence (4). Phab B7 did not reveal any band of the expected size range typical of XylR-like regulators in the soluble protein extracts of P. putida KT2442.
FIG. 4.
Recognition of XylR and TouR in Western blots by Phab B7. Different protein preparations were separated by denaturing SDS-PAGE (10%), transferred to a PVDF membrane, and probed with the antibodies and Phabs indicated. (A) Different amounts of purified 6xhisTouRΔA (50, 16.6, 5.5, 1.8, and 0.61 ng [lanes 1 to 5, respectively]) were probed with a serum obtained from immunized mice (α-TouRΔA; top panel), Phab B7 (middle panel), or Phab MBP (as a negative control; bottom panel). (B) A mixture of 10 ng of purified 6xhisTouRΔA and 6xhisTouR (lane 1) or 30 ng of 6xhisXylRΔA and 6xhisXylR (lane 2) was probed with Phab B7. (C) Whole-cell protein extracts from P. stutzeri OX1 (lane 1) or P. putida KT2442 (lane 2) were probed with Phab B7. In all cases, the bound antibodies or Phabs were developed with anti-mouse-peroxidase or anti-M13-peroxidase conjugates, respectively.
Mapping of the epitope recognized by Phab B7.
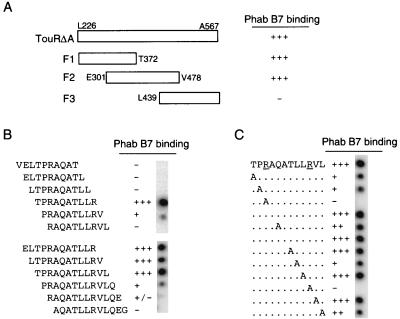
To define the protein region bound by Phab B7, three overlapping fragments of TouRΔA (F1, F2, and F3) were obtained by PCR and cloned into the BamHI and HindIII sites of vector pET21d (Fig. 5A). These fragments comprise the amino acids Leu226 to Thr372 (in F1), Glu301 to Val478 (in F2), and Leu439 to Ala567 (in F3) of the original TouR sequence. The resulting plasmids (pET-F1TouR, pET-F2TouR, and pET-F3TouR) encode their respective TouRΔA fragments under the control of the T7 RNA polymerase promoter and insert common N-terminal (T7 tag) and C-terminal (six-histidine tag) amino acid sequences into all the fragments. Induction with IPTG of E. coli BL21(DE3)(pLysS) cells harboring one of these plasmids led to the overproduction of the expected TouRΔA fragments. These were detected in the whole-cell protein extracts after Coomassie blue staining of denaturing SDS-polyacrylamide gels and by Western blots developed with a MAb specific for the six-histidine tag (data not shown). When Phab B7 was employed for detection of TouRΔA fragments in Western blots, only fragments F1 and F2 were recognized, indicating that the epitope bound by Phab B7 was restricted between amino acids Glu301 and Thr372 of TouR (data not shown). To accurately delimit the amino acid sequence bound by Phab B7, cellulose membranes containing deca- or dodecapeptides with one amino acid overlap of the TouR sequence between Glu301 and Thr372 were incubated with Phab B7 and developed as for Western blots. As summarized in Fig. 5B, these experiments revealed that the peptides having the sequence TPRAQATLLR were strongly bound by Phab B7. This sequence corresponds to the amino acid positions 340 to 349 of TouR, located downstream of the consensus Walker box B for ATP binding (30, 41).
FIG. 5.
Peptide mapping of the TouR epitope recognized by Phab B7. (A) Summary of the binding of Phab B7 to three fragments derived from TouRΔA (F1, Leu226-Thr372; F2, Glu301-Val478; and F3, Leu439-Ala567). Recognition of these fragments by Phab B7 (+) was determined by Western blotting of whole-cell protein extracts of E. coli BL21(DE3) strains overproducing each fragment in pET vectors under the control of the T7 promoter (see Materials and Methods). (B) The amino acid sequence of TouR between positions Glu301 and Thr372 was synthesized onto cellulose membranes as deca- and dodecapeptides with an overlap of one amino acid. These membranes were probed with Phab B7 and developed with anti-M13-peroxidase conjugate. The results of these experiments are summarized, showing only the peptides around the TPRAQATLLR sequence, which forms the core epitope required for Phab B7 recognition. (C) The dodecapeptide TPRAQATLLRVL and a collection of derivatives in which each of the amino acid positions was changed consecutively to alanine (as indicated) were synthesized onto cellulose membranes and probed with Phab B7 as described for panel A. The change of any of the Arg residues (underlined) to Ala completely abolished Phab B7 recognition.
To characterize the amino acid residues that are essential for Phab B7 recognition, a series of dodecapeptides were synthesized in which each of the positions of the peptide TPRAQATLLRVL was changed to alanine. The cellulose membrane containing this array of alanine-substituted peptides was incubated with Phab B7 and developed as before. The results of this experiment (Fig. 5C) revealed that the change of either of the two Arg residues of this sequence completely abolished Phab B7 binding. None of the other alanine substitutions had such a dramatic effect on Phab B7 binding, although the Pro-to-Ala change diminished recognition of the epitope.
Next, we examined the presence of the TPRAQATLLRVL sequence in the known members of the XylR-like family. The BLAST program (2) was employed to search for homologues of the TouRΔA sequence in the SwissProt and TrEMBL data banks. This analysis showed that all XylR-like regulators cloned from Pseudomonas strains contained the Phab B7 epitope TPRAQATLLRVL with an amino acid identity between 83 and 100% (Fig. 6). The conservation of this epitope is higher than the average identity found in the conserved C and D domains among these XylR-like regulators from Pseudomonas species (i.e., ∼63 to 69%). More crucial, the two Arg residues of the Phab B7 epitope that are essential for binding are conserved in the XylR-like regulators found in Pseudomonas (Fig. 6) but not in more distant homologues (e.g., from Ralstonia, Comamonas, Sphingomonas, and Burkholderia) or in other types of the ς54-dependent activators (e.g., NifA).
FIG. 6.
Conservation of the epitope bound by Phab B7 in XylR-like regulators. XylR-like sequences from data banks were aligned using BLAST (2), and the epitope bound by PhabB7 is shown. As indicated, XylR-like regulators from the Pseudomonas spp. (TouR, XylR, TmbR, Ph1R, PhhR, DmpR, and PheR) conserved all the residues required for Phab B7 recognition. The GenBank accession numbers of the XylR-like sequences shown are as follows: TouR, AJ005663; XylR, P06519; TmbR, U41301; Ph1R, X91145; PhhR, X79599; DmpR, X68033; PheR, D63814; MopR, Z69251; PoxR, AF026065; PhyR, AB031996; PhcR, AB024741; AphR, AB006480; PhnR, AF061751; TbuT, U72645; PhlR, AF065891; and NifA, P09570.
Quantification of the number of XylR molecules per cell in P. putida under various growth conditions.
Because of the intricate interplay between plasmid-encoded and chromosome-encoded factors, the regulation of the degradative operons of the TOL plasmid pWW0 of P. putida mt-2 is one outstanding paradigm of prokaryotic gene expression (35). One of the pillars of the present model is that the Pr promoter (which drives expression of the xylR gene) is autorepressed (7, 18). Because of this, the intracellular levels of the activator are predicted to remain approximately constant through all conditions, and thus, the only regulated event is the activation-deactivation of XylR with the aromatic effector. This view is based on experiments with transcriptional fusions (7) and with measurement of transcript production under various conditions (18). However, the actual levels of the protein in each case could not be directly quantitated. We have exploited the high affinity and specificity of Phab B7 described above to give an answer to this question.
The strains subjected to scrutiny were those termed P. putida MAD1 and P. putida MAD2. P. putida MAD1 bears a hybrid mini-Tn5 transposon which includes a tellurite resistance selection marker, the sequence of the wild-type xylR gene expressed through its native Pr promoter, and a transcriptional Pu-lacZ fusion (9). This ensures that all regulatory elements controlling expression from Pu are placed in one copy per chromosome. P. putida MAD2 bears an equivalent insertion which encodes the truncated protein XylRΔA with the deletion of amino acids 1 to 223 but expressed under the same translation initiation region and promoter (Pr) which drive expression of the native xylR-encoded protein in P. putida MAD1. Because of the activation mechanism for this type of regulator discussed above, XylRΔA can activate Pu without any inducer.
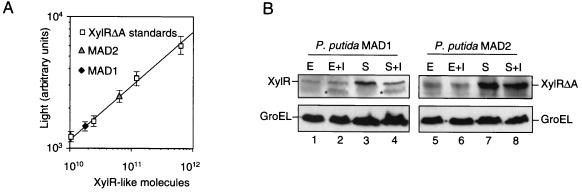
To estimate the number of XylR and XylRΔA proteins per cell at stationary phase and without effector, P. putida MAD1 and MAD2 cells were harvested from noninduced cultures grown in LB medium until stationary phase (OD600 of ∼2.5), and whole-cell protein extracts were prepared. The numbers of CFU per milliliter in these cultures were determined by plating serial 10-fold dilutions on LB agar plates. Then, the protein extracts were loaded onto SDS–10% polyacrylamide gels so that each lane contained the proteins from ∼1.25 × 108 CFU, and the extracts were subjected to Western blotting with Phab B7. Four serial fivefold dilutions of purified 6xhisXylRΔA as standards (starting at 40 ng) were also loaded in these gels. The intensity of light in the protein bands was quantified after chemiluminescence was developed. As shown in Fig. 7A, a relationship exists between the log10 of the number of molecules of 6xhisXylRΔA applied per lane in the gel and the log10 of light intensity of their corresponding protein bands. This standard curve was used to judge the number of XylR and XylRΔA molecules present in P. putida MAD1 and P. putida MAD2 by extrapolation with the light intensity values of their protein bands in these Western blots. Three independent experiments were made to carry out these estimations. Using this approach, the number of XylR molecules (monomers) per cell in P. putida MAD1 at stationary phase was calculated to be 142 ± 12, whereas that of XylRΔA in P. putida MAD2 was 575 ± 66. The ∼4-fold-higher intracellular level of XylRΔA could indicate a higher stability of the truncated protein in vivo or (more likely) a weaker down-regulation of its own promoter (6, 7, 18).
FIG. 7.
Quantification of XylR and XylRΔA in P. putida MAD1 and MAD2. (A) Intensity of light emission after chemiluminescence development of a Western blot containing four serial fivefold dilutions of purified 6xhisXylRΔA (40, 8, 1.6, and 0.3 ng) as standards and whole-cell protein extracts of P. putida MAD1 and MAD2, corresponding to ∼1.25 × 108 cells from stationary-phase cultures without inducer. A standard curve is shown employing the log of the number of molecules of purified 6xhisXylRΔA applied versus the log of light intensity of their corresponding protein bands. The Phab B7 and anti-M13-peroxidase MAb were used for detection (Materials and Methods) (B) P. putida MAD1 and MAD2 were grown in LB medium in the absence or presence of XylR inducer 3-MBA (+I). Bacterial cells were harvested from cultures induced by adding 2 mM 3-MBA either at exponential phase (OD600 of ∼0.5) with further incubation for 1 h (final OD600 of ∼1.1; E+I) or at early stationary phase (OD600 of ∼1.2) with further incubation for 3 h (final OD600 of ∼2.5; S+I). Bacterial cells were also harvested at exponential (E) and stationary (S) phases from aliquots of these cultures to which 3-MBA had not been added. Whole-cell protein extracts corresponding to identical numbers of P. putida MAD1 (lanes 1 to 4) and P. putida MAD2 (lanes 5 to 8) (∼1.25 × 108 CFU/lane) were loaded on an SDS–10% polyacrylamide gel, blotted onto a membrane, and probed with Phab B7 (for XylR and XylRΔA detection) or an anti-GroEL polyclonal serum as an internal control. The protein bands of XylR, XylRΔA, and GroEL are indicated. The appearance of a band of lower molecular weight in samples induced with 3-MBA and developed with Phab B7 is labeled with an asterisk.
Next, we investigated whether the levels of XylR and XylRΔA changed during growth and upon exposure of the cells to an aromatic inducer. To this end, cultures of P. putida MAD1 and P. putida MAD2 were grown in LB medium; 2 mM 3-MBA, an inducer of XylR, was added at an OD600 of ∼0.5; and the cultures were incubated further for 1 h (final OD600 of ∼1.1). Alternatively, the inducer was added at an OD600 of ∼1.3, and the cultures were further incubated for 3 h (final OD600 of ∼2.5). Cells were harvested from induced and noninduced cultures, at exponential and stationary phases, and whole-cell protein extracts were loaded onto SDS–10% polyacrylamide gels (∼1.25 × 108 CFU per lane), blotted onto a membrane, and probed with Phab B7. The intensity of light in the protein bands was quantified after chemiluminescence as described above. Three independent experiments were made that produced identical results. Figure 7B shows a representative Western blot of protein extracts of P. putida MAD1 (lanes 1 to 4) and MAD2 (lanes 5 to 8) developed with Phab B7 to detect XylR and XylRΔA (upper panel). A separate blot (lower panel) was probed with a rabbit serum against the stress-responsive protein GroEL (16, 17). This not only provided an internal control to verify that equivalent protein samples were being loaded in the gels but also verified that addition of the aromatic inducer did not cause a massive heat shock response which could distort the measurement of intracellular XylR and XylRΔA levels (19, 34).
The experiments of Fig. 7 showed that both XylR and XylRΔA increased by at least fivefold during the transition from exponential to stationary phase in the absence of effector (from ∼28 ± 5 to ∼142 ± 12 XylR molecules in P. putida MAD1 and from ∼90 ± 9 to ∼575 ± 66 XylR molecules in P. putida MAD2). Inducer addition also caused variations in the intracellular pool of XylR. As shown in Fig. 7, 3-MBA caused a significant (>2.5-fold) decrease of the intracellular XylR pool of P. putida MAD1 at stationary phase (∼58 ± 10 molecules) but not at exponential phase (∼27 ± 6). Furthermore, an extra band of a lower molecular mass than XylR (∼55 kDa) appeared in the induced samples of P. putida MAD1 at stationary phase (Fig. 7B). The levels of XylRΔA in P. putida MAD2 remained relatively constant (variation is below ∼1.5 times) in stationary phase regardless of 3-MBA addition, and no extra bands were detected.
Taken together, these experiments revealed that even during exponential growth phase, when produced from single gene copies per chromosome of P. putida, the number of available XylR or XylRΔA monomers per cell (∼28 ± 5 for XylR and 90 ± 9 for XylRΔA) exceeds by at least 1 order of magnitude the number of DNA targets (upstream activating sequences [UAS]) (12) in the cell. The intracellular XylR and XylRΔA levels further increase by fivefold in the stationary phase of growth, coinciding with the activity of the Pu promoter. A first inspection of these numbers indicates that intracellular levels of the activator in vivo are sufficient for activation of Pu under any of the conditions tested. However, these data alone cannot entirely rule out the possibility that XylR levels could be limiting for an optimal activation of the promoter during exponential growth, especially since an oligomeric complex needs to be formed at the UAS during the activation process (12, 29). Nonetheless, overexpression of XylR in P. putida did not alleviate the silencing of the promoter during the exponential phase (32).
The results presented in this work also show that the presence of an organic inducer (such as 3-MBA) causes a modulation in the intracellular concentration of XylR. It is possible that this modulation is mediated by a combination of (i) an enhanced autorepression of XylR at its own promoter, (ii) a proteolysis of XylR after effector recognition, and (iii) a shorter half-life of the protein (or a diminished protein synthesis) during exposure to the aromatic compounds. Further work is under way to characterize these possibilities in detail and their actual contribution to the physiological regulation of Pu activity in vivo.
ACKNOWLEDGMENTS
We are indebted to F. Arenghi for his kind gift of purified TouR and TouRΔA proteins.
This work was supported by EU contracts QLK3-CT2000-00170 and QLK3-CT1999-00041, by grant BIO98-0808 of the Spanish Comisión Interministerial de Ciencia y Tecnología (CICYT), and by the Strategic Research Groups Program of the Comunidad Autónoma de Madrid.
REFERENCES
- 1.Abril M A, Michan C, Timmis K N, Ramos J L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arenghi F L, Barbieri P, Bertoni G, de Lorenzo V. New insights into the activation of o-xylene biodegradation in Pseudomonas stutzeri OX1 by pathway substrates. EMBO Rep. 2001;2:409–414. doi: 10.1093/embo-reports/kve092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arenghi F L, Pinti M, Galli E, Barbieri P. Identification of the Pseudomonas stutzeri OX1 toluene–o-xylene monooxygenase regulatory gene (touR) and of its cognate promoter. Appl Environ Microbiol. 1999;65:4057–4063. doi: 10.1128/aem.65.9.4057-4063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1994. [Google Scholar]
- 6.Bertoni G, Marques S, de Lorenzo V. Activation of the toluene-responsive regulator XylR causes a transcriptional switch between sigma54 and sigma70 promoters at the divergent Pr/Ps region of the TOL plasmid. Mol Microbiol. 1998;27:651–659. doi: 10.1046/j.1365-2958.1998.00715.x. [DOI] [PubMed] [Google Scholar]
- 7.Bertoni G, Perez-Martin J, de Lorenzo V. Genetic evidence of separate repressor and activator activities of the XylR regulator of the TOL plasmid, pWW0, of Pseudomonas putida. Mol Microbiol. 1997;23:1221–1227. doi: 10.1046/j.1365-2958.1997.3091673.x. [DOI] [PubMed] [Google Scholar]
- 8.Buck M, Gallegos M T, Studholme D J, Guo Y, Gralla J D. The bacterial enhancer-dependent ς54 (ςN) transcription factor. J Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez S, de Lorenzo V, Perez-Martin J. Activation of the transcriptional regulator XylR of Pseudomonas putida by release of repression between functional domains. Mol Microbiol. 1995;16:205–213. doi: 10.1111/j.1365-2958.1995.tb02293.x. [DOI] [PubMed] [Google Scholar]
- 10.Frank R, Overwing H. Spot synthesis: epitope analysis with arrays of synthetic peptides prepared on cellulose membrane. Methods Mol Biol. 1996;66:149–169. doi: 10.1385/0-89603-375-9:149. [DOI] [PubMed] [Google Scholar]
- 11.Garmendia J, de Lorenzo V. The role of the interdomain B linker in the activation of the XylR protein of Pseudomonas putida. Mol Microbiol. 2000;38:401–410. doi: 10.1046/j.1365-2958.2000.02139.x. [DOI] [PubMed] [Google Scholar]
- 12.Garmendia J, de Lorenzo V. Visualization of DNA-protein intermediates during activation of the Pu promoter of the TOL plasmid of Pseudomonas putida. Microbiology. 2000;146:2555–2563. doi: 10.1099/00221287-146-10-2555. [DOI] [PubMed] [Google Scholar]
- 13.Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 14.Hoogenboom H R. Designing and optimizing library selection strategies for generating high-affinity antibodies. Trends Biotechnol. 1997;15:62–70. doi: 10.1016/S0167-7799(97)84205-9. [DOI] [PubMed] [Google Scholar]
- 15.Kustu S, North A K, Weiss D S. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem Sci. 1991;16:397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- 16.Llorca O, Galan A, Carrascosa J L, Muga A, Valpuesta J M. GroEL under heat-shock. Switching from a folding to a storing function. J Biol Chem. 1998;273:32587–32594. doi: 10.1074/jbc.273.49.32587. [DOI] [PubMed] [Google Scholar]
- 17.Lund P A. Microbial molecular chaperones. Adv Microb Physiol. 2001;44:93–140. doi: 10.1016/s0065-2911(01)44012-4. [DOI] [PubMed] [Google Scholar]
- 18.Marques S, Gallegos M T, Manzanera M, Holtel A, Timmis K N, Ramos J L. Activation and repression of transcription at the double tandem divergent promoters for the xylR and xylS genes of the TOL plasmid of Pseudomonas putida. J Bacteriol. 1998;180:2889–2894. doi: 10.1128/jb.180.11.2889-2894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marques S, Manzanera M, Gonzalez-Perez M M, Gallegos M T, Ramos J L. The XylS-dependent Pm promoter is transcribed in vivo by RNA polymerase with sigma 32 or sigma 38 depending on the growth phase. Mol Microbiol. 1999;31:1105–1113. doi: 10.1046/j.1365-2958.1999.01249.x. [DOI] [PubMed] [Google Scholar]
- 20.McCafferty J, Johnson K S. Construction and screening of antibody display libraries. In: Kay B K, Winter J, McCafferty J, editors. Phage display of peptides and proteins. San Diego, Calif: Academic Press, Inc.; 1996. pp. 79–111. [Google Scholar]
- 21.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 22.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller C, Petruschka L, Cuypers H, Burchhardt G, Herrmann H. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J Bacteriol. 1996;178:2030–2036. doi: 10.1128/jb.178.7.2030-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng L C, Poh C L, Shingler V. Aromatic effector activation of the NtrC-like transcriptional regulator PhhR limits the catabolic potential of the (methyl)phenol degradative pathway it controls. J Bacteriol. 1995;177:1485–1490. doi: 10.1128/jb.177.6.1485-1490.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nissim A, Hoogenboom H R, Tomlinson I M, Flynn G, Midgley C, Lane D, Winter G. Antibody fragments from a ‘single pot’ phage display library as immunochemical reagents. EMBO J. 1994;13:692–698. doi: 10.1002/j.1460-2075.1994.tb06308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.North A K, Klose K E, Stedman K M, Kustu S. Prokaryotic enhancer-binding proteins reflect eukaryote-like modularity: the puzzle of nitrogen regulatory protein C. J Bacteriol. 1993;175:4267–4273. doi: 10.1128/jb.175.14.4267-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Neill E, Wikstrom P, Shingler V. An active role for a structured B-linker in effector control of the sigma54-dependent regulator DmpR. EMBO J. 2001;20:819–827. doi: 10.1093/emboj/20.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Martin J, De Lorenzo V. The amino-terminal domain of the prokaryotic enhancer-binding protein XylR is a specific intramolecular repressor. Proc Natl Acad Sci USA. 1995;92:9392–9396. doi: 10.1073/pnas.92.20.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Martin J, de Lorenzo V. ATP binding to the sigma 54-dependent activator XylR triggers a protein multimerization cycle catalyzed by UAS DNA. Cell. 1996;86:331–339. doi: 10.1016/s0092-8674(00)80104-x. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Martin J, de Lorenzo V. In vitro activities of an N-terminal truncated form of XylR, a sigma 54-dependent transcriptional activator of Pseudomonas putida. J Mol Biol. 1996;258:575–587. doi: 10.1006/jmbi.1996.0270. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Martin J, de Lorenzo V. Physical and functional analysis of the prokaryotic enhancer of the sigma 54-promoters of the TOL plasmid of Pseudomonas putida. J Mol Biol. 1996;258:562–574. doi: 10.1006/jmbi.1996.0269. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Martin J, de Lorenzo V. VTR expression cassettes for engineering conditional phenotypes in Pseudomonas: activity of the Pu promoter of the TOL plasmid under limiting concentrations of the XylR activator protein. Gene. 1996;172:81–86. doi: 10.1016/0378-1119(96)00193-x. [DOI] [PubMed] [Google Scholar]
- 33.Plückthun A, Krebber C, Krebber U, Horn U, Knüpfer U, Wenderoth R, Nieba L, Proba K, Riesenberg D. Producing antibodies in Escherichia coli: from PCR to fermentation. In: McCafferty J, Hoogenboom H R, editors. Antibody engineering: a practical approach. Oxford, United Kingdom: IRL Press; 1996. pp. 203–252. [Google Scholar]
- 34.Ramos J L, Gallegos M, Marques S, Ramos-Gonzalez M, Espinosa-Urgel M, Segura A. Responses of Gram-negative bacteria to certain environmental stressors. Curr Opin Microbiol. 2001;4:166–171. doi: 10.1016/s1369-5274(00)00183-1. [DOI] [PubMed] [Google Scholar]
- 35.Ramos J L, Marques S, Timmis K N. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu Rev Microbiol. 1997;51:341–373. doi: 10.1146/annurev.micro.51.1.341. [DOI] [PubMed] [Google Scholar]
- 36.Reusch P, Arnold S, Heusser C, Wagner K, Weston B, Sebald W. Neutralizing monoclonal antibodies define two different functional sites in human IL-4. Eur J Biochem. 1994;222:491–499. doi: 10.1111/j.1432-1033.1994.tb18890.x. [DOI] [PubMed] [Google Scholar]
- 37.Schirmer F, Ehrt S, Hillen W. Expression, inducer spectrum, domain structure, and function of MopR, the regulator of phenol degradation in Acinetobacter calcoaceticus NCIB8250. J Bacteriol. 1997;179:1329–1336. doi: 10.1128/jb.179.4.1329-1336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shingler V. Signal sensing by sigma 54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–416. doi: 10.1046/j.1365-2958.1996.388920.x. [DOI] [PubMed] [Google Scholar]
- 39.Shingler V, Bartilson M, Moore T. Cloning and nucleotide sequence of the gene encoding the positive regulator (DmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J Bacteriol. 1993;175:1596–1604. doi: 10.1128/jb.175.6.1596-1604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Timmis K N, Steffan R J, Unterman R. Designing microorganisms for the treatment of toxic wastes. Annu Rev Microbiol. 1994;48:525–557. doi: 10.1146/annurev.mi.48.100194.002521. [DOI] [PubMed] [Google Scholar]
- 41.Weiss D S, Batut J, Klose K E, Keener J, Kustu S. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- 42.Winter G, Griffiths A D, Hawkins R E, Hoogenboom H R. Making antibodies by phage display technology. Annu Rev Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 43.Wootton J C, Drummond M H. The Q-linker: a class of interdomain sequences found in bacterial multidomain regulatory proteins. Protein Eng. 1989;2:535–543. doi: 10.1093/protein/2.7.535. [DOI] [PubMed] [Google Scholar]