Abstract
Aim
To determine, using a mouse model of obesity, whether low‐dose hydralazine prevents obesity‐related chronic kidney disease (CKD).
Methods
From 8 weeks of age, male C57BL/6 mice received a high‐fat diet (HFD) or chow, with or without low‐dose hydralazine (25 mg/L) in drinking water, for 24 weeks. Biometric and metabolic variables, renal function and structural changes, renal global DNA methylation, DNA methylation profile and markers of renal fibrosis, injury, inflammation and oxidative stress were assessed.
Results
The HFD‐fed mice developed obesity, with glucose intolerance, hyperinsulinaemia and dyslipidaemia. Obesity increased albuminuria and glomerulosclerosis, which were significantly ameliorated by low‐dose hydralazine in the absence of a blood pressure‐lowering effect. Obesity increased renal global DNA methylation and this was attenuated by low‐dose hydralazine. HFD‐induced changes in methylation of individual loci were also significantly reversed by low‐dose hydralazine. Obese mice demonstrated increased markers of kidney fibrosis, inflammation and oxidative stress, but these markers were not significantly improved by hydralazine.
Conclusion
Low‐dose hydralazine ameliorated HFD‐induced albuminuria and glomerulosclerosis, independent of alterations in biometric and metabolic variables or blood pressure regulation. Although the precise mechanism of renoprotection in obesity is unclear, an epigenetic basis may be implicated. These data support repurposing hydralazine as a novel therapy to prevent CKD progression in obese patients.
Keywords: chronic kidney disease, kidney fibrosis, methylation, Obesity
1. INTRODUCTION
Obesity is a major public health concern, and its prevalence globally is projected to increase by 40% over the next decade. 1 Obesity is increasingly recognized as an important risk factor in the development of chronic kidney disease (CKD) and progression towards end‐stage kidney disease. 1 , 2 , 3 , 4 Obesity can lead to CKD through mechanisms including altered renal haemodynamics, induction of albuminuria, activation of complex neuroendocrine pathways, inflammation and oxidative stress. 1 , 5 , 6 , 7
There is compelling evidence of associations between CKD and epigenetic mechanisms, particularly DNA methylation. 8 In blood and kidney tissue, differentially methylated genes have been identified between patients with CKD and healthy controls, 9 , 10 and in patients with renal fibrosis and rapidly progressive CKD. 11 , 12 Furthermore, increased global DNA methylation has been observed in the blood of end‐stage kidney disease patients with features of inflammation, 13 , 14 whereas global hypomethylation of DNA has been noted in uraemic dialysis patients. 15
Drugs that modify epigenetic factors could potentially prevent CKD progression. The antihypertensive drug, hydralazine, has been shown to have DNA demethylating actions, when used at low doses. 16 , 17 Hydralazine has been used clinically since the 1950s, mainly for the management of resistant hypertension and hypertensive disorders of pregnancy. 18 In animal models of CKD involving unilateral ureteral obstruction and folic acid nephropathy, low‐dose hydralazine has been shown to attenuate renal fibrosis through epigenetic mechanisms. 19 , 20
The objective of this study was to discover if low‐dose hydralazine can prevent obesity‐related CKD and determine the underlying mechanisms. We also aimed to determine whether hydralazine alters DNA methylation profile in the kidney.
2. MATERIALS AND METHODS
2.1. Animal experiments
The study, which was approved by the Animal Ethics Committee of the Northern Sydney Local Health District (Resp/18/102) and complied with the Australian code for the care and use of animals for scientific purposes, 21 took place at Kearns Animal Facility. From 8 weeks of age, male C57BL/6 mice were fed either a high‐fat diet (HFD; 20 kJ/g, 43% of total energy from fat [Specialty Feeds, Glen Forrest, WA, Australia]), or standard rodent chow diet (11 kJ/g [Gordon′s Specialty Stockfeeds, Yanderra, NSW, Australia]). At the same time, they were either treated with low‐dose hydralazine (Alphapharm, Millers Point, NSW, Australia; 25 mg/L in the drinking water), or received normal drinking water. The dose of hydralazine was determined following a pilot study which demonstrated that 25 mg/L had no effect on blood pressure regulation (Figure S1). Male mice were used because previous experimental models of CKD have generally shown that disease progression is more rapid in males than in females. 22 There were four animal groups (n = 8–15): chow/water; HFD/water; chow/hydralazine; and HFD/hydralazine. Hydralazine drinking water was changed twice weekly.
In the week prior to the endpoint assessment, blood pressure was measured via a noninvasive CODA® tail vein cuff method (Kent Scientific, Torrington, Connecticut), and intraperitoneal glucose tolerance testing was conducted as we have described previously. 23 , 24
At 32 weeks of age, the mice were killed whilst anaesthetized (2% isoflurane), and tissues were harvested under fasting conditions. Blood was collected via cardiac puncture and urine was collected by bladder puncture. The body was perfused with phosphate‐buffered saline, and kidneys, liver and fat were weighed, either fixed in 10% formalin solution for histological analysis, or snapped frozen for the analysis of protein, RNA and DNA.
2.2. Serum measures
Measurement of serum insulin was performed (n = 5) using the Ultra‐Sensitive Mouse Insulin ELISA Kit (Crystal Chem Inc., Elk Grove Village, Illinois). Serum creatinine was measured (n = 8) using a colorimetric assay kit (Cayman Chemical, Ann Arbor, Michigan). Colorimetric assays were used to measure triglycerides and non‐esterified fatty acids (NEFA) (n = 6–8). 23
2.3. Urine measures
Urinary albumin: creatinine ratio was measured at endpoint as previously described. 23 Kidney injury molecule‐1 (KIM‐1, n = 6) was determined using a KIM‐1 ELISA Kit (Abcam, Cambridge, UK), and expressed as urinary KIM‐1: creatinine ratio.
2.4. Global DNA methylation analysis
Renal genomic DNA (n = 5) was extracted from cortical tissue using the ISOLATE II Genomic DNA Kit (Bioline, London, UK). 5‐methylcytosine (5mC; as a percentage of total DNA) was determined using the Methylated DNA Quantification Kit (Abcam).
2.5. Reduced representation bisulphite sequencing
DNA library preparation and reduced representation bisulphite sequencing (RRBS) were performed using the Illumina NovaSeq sequencing platform by the Australian Genome Research Facility (AGRF, Melbourne, VIC, Australia). Four mice were sequenced in each of the relevant treatment groups (chow/water, HFD/water and HFD/hydralazine). No pooling of samples occurred. The RRBS library was produced using the NuGEN Ovation® RRBS Methyl‐Seq system. Reads (30 million, 100 bp single end run) were assessed using FastQC v.0.11.8, 25 then trimmed using Trim Galore v0.5.0. 26 Further trimming was performed to remove NuGEN′s RRBS‐specific primer. Bismark v0.21.0 27 allowed mapping to GRCm38 builds for the mouse genome. Alignments were performed using Bowtie 2 v2.3.4, 28 with default parameters allowing 0 mismatch in a 20‐bp seed. Alignments were then deduplicated using a barcoded RRBS mode of Bismark. As RRBS tends to yield data with much lower sequencing coverage (the total read counts of both methylated and unmethylated intensity) compared to array‐based sequencing, in order to obtain meaningful data, library sizes were limited to loci with coverages ≥8, then beta and M values were calculated as previously described. 29 The offset value (α) was set at 0.1 to avoid zero and infinite values in M‐value computation. P values and adjusted P values (false discovery rate) of each pair‐wise comparison were calculated based on M values using the limma package. 30 Beta values, which reflect methylation level, were used for data presentation. A CpG site was deemed to be differentially methylated if the false discovery rate was less than 0.05 and the difference in methylation levels (deltaB) was greater than 5%. Pathway analysis was performed using g: Profiler (https://biit.cs.ut.ee/gprofiler/gost).
2.6. Renal structural changes
Formalin‐fixed kidneys (n = 6), sectioned at 2‐μm thickness, underwent periodic acid Schiff staining and were assessed using a light microscope (Leica, Wetzlar, Germany). Glomerulosclerosis was assessed in 20 glomeruli per slide by two independent, blinded investigators. Picrosirius red staining was used to determine renal expression of collagens I and III. Four non‐overlapping images per sample were analysed by Image J software (National Institutes of Health, Bethesda, Maryland). Slides were also assessed for tubulointerstitial fibrosis, tubular vacuolation and tubular dilatation, as we described previously. 6
2.7. Immunohistochemistry
Renal immunohistochemistry was assessed on 4‐μm thick sections as we have described previously, 23 using primary antibodies against 8‐hydroxydeoxyguanosine (8‐OHdG; 1:750, Bioss, Woburn, Massachusetts), and nitrotyrosine (1:500; Merck Millipore Ltd, Darmstadt, Germany). Six non‐overlapping images per slide were photographed at 200× magnification (Leica Application Suite, Leica, Germany) and were quantified by two independent investigators using Image J software.
2.8. Western blotting
Protein was extracted from frozen kidney tissue (n = 6) and was quantitated using the Pierce BCA Protein Kit (Thermo Fisher Scientific, Melbourne, VIC, Australia). Then, 20 μg of each protein sample was loaded onto an Invitrogen Bolt 4% to 12% Bis‐Tris Plus Gel (Thermo Fisher Scientific), and underwent Western blotting 31 using the following primary antibodies: DNA methyltransferase (DNMT)‐1 (dilution 1:1000; Abcam), DNMT3a (1:1000; Abcam), Ten‐eleven translocation‐3 (Tet3; 1:1000; Abcam), fibronectin (1:2000; Thermo Fisher Scientific), collagen I (1:2000; Abcam), collagen IV (1:5000; Abcam), glutathione peroxidase‐1 (GPx‐1; 1:500; R&D Systems, Minneapolis, Minnesota), manganese superoxide dismutase (MnSOD; 1:2000 [Merck Millipore]), nitrotyrosine (1:1000; Merck Millipore), COX‐IV (1:1000; Abcam) and β‐actin (Santa Cruz, Dallas, Texas). Protein level was assessed using GelPro Analyser software (Informer Technologies Inc.) and expressed as a proportion relative to control. β‐actin was used as the internal control for DNMT1, DNMT3a, Tet3, fibronectin, collagen I, collagen IV, GPx‐1 and nitrotyrosine, whereas COX‐IV was used as the internal control for MnSOD.
2.9. Quantitative real‐time PCR
RNA was extracted (n = 6) using the RNeasy Plus Mini Kit (Qiagen, Valencia, California). RT‐PCR was performed using the QuantStudio 12K Flex Real‐Time PCR System (Thermo Fisher Scientific), with QuantiNova SYBR Green (Qiagen) mastermixes and the PCR primers listed in Table S1. Results are expressed as fold change after normalization to either 18S or β‐actin.
2.10. Statistical analysis
Analysis of DNA methylation data is described above. All other results are presented as mean ± SEM. GraphPad Prism version 8.3.1 (San Diego, California) was used to perform statistical analysis. Analyses were performed using two‐way analysis of variance (ANOVA) followed by Fisher′s least significant difference test. P values of <0.05 were considered statistically significant.
3. RESULTS
3.1. Biometric and metabolic variables
As shown in Table 1, HFD‐fed mice displayed an obese phenotype by 32 weeks, with significantly increased body weight compared to chow‐fed mice (P < 0.0001). Hydralazine did not alter body weight. Kidney weight/body weight percentage was significantly reduced in obese mice (P < 0.01), irrespective of treatment with hydralazine. As a percentage of total body weight, liver, retroperitoneal and epididymal weights were significantly elevated in obese animals (P < 0.01, P < 0.001 and P < 0.0001, respectively). There were no differences in mean blood pressure between groups. As expected, low‐dose hydralazine did not exert a blood pressure‐lowering effect. Glucose intolerance was significantly higher in HFD‐fed mice, compared to control (P < 0.01), and this was not affected by hydralazine. There was a trend towards increased fasting blood glucose level in obese animals compared to controls. Obese animals had significantly higher serum insulin concentration compared to controls (P < 0.05); this remained unchanged by hydralazine. Serum triglycerides and NEFA were significantly elevated in obese mice compared to controls (P < 0.01 and P < 0.05, respectively), although these serum measures were unaffected by hydralazine. There were no differences in serum creatinine between groups. Mice fed the HFD developed significantly increased albuminuria compared to chow‐fed mice (P < 0.01; HFD/water vs. chow/water). Administration of hydralazine to obese mice markedly reduced HFD‐induced albuminuria (P < 0.01 vs. HFD/water), to the level observed in control animals.
TABLE 1.
Biometric and metabolic parameters
| Chow/water | HFD/water | Chow/hydralazine | HFD/hydralazine | |
|---|---|---|---|---|
| Body weight (g) | 31.91 ± 0.73 | 45.05 ± 0.86**** | 33.16 ± 0.88 | 46.63 ± 0.40#### |
| Kidney weight/body weight (%) | 0.72 ± 0.04 | 0.57 ± 0.02** | 0.71 ± 0.04 | 0.53 ± 0.03### |
| Liver weight/ body weight (%) | 5.34 ± 0.56 | 7.11 ± 0.45** | 4.79 ± 0.26 | 7.27 ± 0.39### |
| Retroperitoneal weight/ body weight (%) | 0.60 ± 0.12 | 1.20 ± 0.11*** | 0.54 ± 0.11 | 1.06 ± 0.10## |
| Epididymal fat weight/ body weight (%) | 2.70 ± 0.36 | 5.07 ± 0.24**** | 2.23 ± 0.40 | 4.10 ± 0.22#### |
| Mean BP (mmHg) | 83.74 ± 7.00 | 76.87 ± 2.91 | 83.78 ± 2.88 | 83.61 ±2.85 |
| IPGTT AUC | 1721 ± 245 | 2452 ± 106** | 1484 ± 140 | 2345 ± 197### |
| Glucose (mmol/L) | 12.98 ± 0.44 | 16.30 ± 1.56 | 11.16 ± 1.62 | 14.74 ± 2.29 |
| Serum insulin (ng/mL) | 0.58 ± 0.05 | 2.27 ± 0.73* | 0.54 ± 0.05 | 2.60 ± 0.53## |
| Serum triglycerides (mg/mL) | 0.57 ± 0.06 | 0.82 ± 0.03** | 0.54 ± 0.05 | 0.68 ± 0.06 |
| NEFA (mmol/L) | 0.62 ± 0.07 | 0.80 ± 0.05* | 0.64 ± 0.05 | 0.78 ± 0.04 |
| Serum creatinine (μmol/L) | 43.93 ± 2.92 | 50.60 ± 2.37 | 45.42 ± 4.51 | 51.58 ± 2.19 |
| Urinary albumin:creatinine ratio (μg/mg) | 16.93 ± 8.15 | 58.19 ± 13.19** | 15.44 ± 3.02 | 26.40 ± 4.20§§ |
Notes: Results expressed as mean ± SEM, n = 8‐15.
Chow/water vs HFD/water: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
Chow/hydralazine vs HFD/hydralazine: ##P < 0.01, ###P < 0.001, ####P < 0.0001.
HFD/water vs HFD/hydralazine: §§ p < 0.01.
3.2. DNA methylation
There was a significantly increased 5mC percentage of total DNA in obese mice versus lean mice (P < 0.05; Figure 1A). There was no difference in global DNA methylation between control mice and hydralazine‐treated lean mice. Interestingly, there was a marked reduction in 5mC percentage of total DNA in hydralazine‐treated obese mice compared to HFD/water mice (P < 0.0001; Figure 1A), suggesting that the DNA demethylating effect of hydralazine occurs only in the presence of an HFD, or alternatively, that hydralazine only reverses pathological methylation back to normal levels.
FIGURE 1.
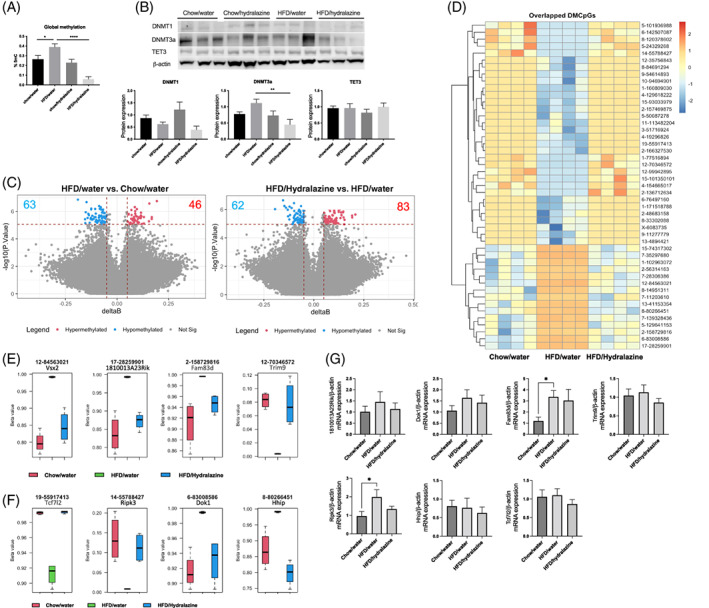
DNA methylation markers and differential methylation by a high‐fat diet (HFD) and hydralazine in mouse kidney. A, Global methylation expressed as 5‐methylcytosine percentage (%5mC) of total DNA. Results expressed as mean ± SEM. n = 5‐6, *P < 0.05, ****P < 0.0001. B, Western blotting of DNA methyltransferase (DNMT)1, DNMT3a and ten‐eleven translocation‐3 (Tet3). Results expressed as mean ± SEM. n = 6, **P < 0.01. C, Volcano plots of significant differentially methylated CpGs between conditions. D, Heatmap of common differentially methylated CpGs by HFD and Hydralazine in mouse kidney. E, Top four differentially methylated genes. F, Differentially methylated genes with known association with inflammation and fibrosis. G, Renal mRNA expression of genes of interest
There were no changes in the protein expression of DNMT1 in HFD/water compared to control (Figure 1B). There was a trend of increased DNMT3a protein expression in HFD/water versus chow/water, although this did not reach statistical significance (Figure 1B). Interestingly, hydralazine significantly reduced the level of DNMT3a in the HFD/hydralazine group compared to HFD/water group (P < 0.01; Figure 1B). Tet3 protein expression was similar between groups (Figure 1B).
3.3. Gene methylation profiling
In line with the changes in the global DNA methylation levels, we identified 63 hypomethylated and 46 hypermethylated CpGs in the kidneys of HFD‐fed mice compared with chow‐fed mice, as well as 62 hypomethylated and 83 hypermethylated CpGs in the kidney of HFD‐fed mice treated with hydralazine compared to mice treated with vehicle control (Figure 1C, Tables S2 and S3). Pathway analysis revealed enrichments of the cation transmembrane transport pathway due to HFD and cell junction organization due to hydralazine treatment (Figure S2A). Hydralazine also affected cell response to growth factor and presynapse assembly pathways.
Among the CpGs that were significantly hypo‐ or hypermethylated due to either HFD or HFD/hydralazine, 47 loci were common (Figure S2B). Strikingly, for each of these 47 CpGs, methylation levels were completely normalized by hydralazine (Figure 1D, Table S4). The top four significant CpGs are associated with genes including visual system homeobox 2 (Vsx2), RIKEN cDNA 1810013A23 gene (1810013A23Rik), family with sequence similarity 83 member D (Fam83d) and tripartite motif‐containing 9 (Trim9; Figure 1E). Among 47 differentially methylated genes, we identified genes implicated in inflammation, cell death and fibrosis. These include transcription factor 7 like 2 (Tcf7l2), receptor‐interacting serine–threonine kinase 3 (Ripk3), docking protein 1 (Dok1) and Hedgehog‐interacting protein (Hhip; Figure 1F). The mRNA expression of Fam83d was significantly increased in the HFD/water group compared to control (P < 0.05; Figure 1G). Similarly, the mRNA level of Ripk3 significantly increased in HFD/water compared to control (P < 0.05; Figure 1G). Interestingly, this significance was lost in the presence of hydralazine. There were no significant differences in the mRNA expression of other genes (Figure 1G). Vsx2 was not expressed in kidney at transcriptional levels.
3.4. Renal structural changes
Increased glomerulosclerosis was observed in HFD‐fed versus chow‐fed mice (P < 0.05) and this was ameliorated by hydralazine (P < 0.05; Figure 2A,B). Obese mice had more pronounced renal picrosirius red staining compared to lean mice (P < 0.01). This was unaffected by hydralazine (Figure 2C,D). Similarly, there was more tubulointerstitial fibrosis, tubular vacuolation and tubular dilatation in kidneys of obese mice compared to lean controls (P < 0.001, P < 0.0001 and P < 0.05, respectively; Figure 2E‐G). However, hydralazine did not improve these markers.
FIGURE 2.
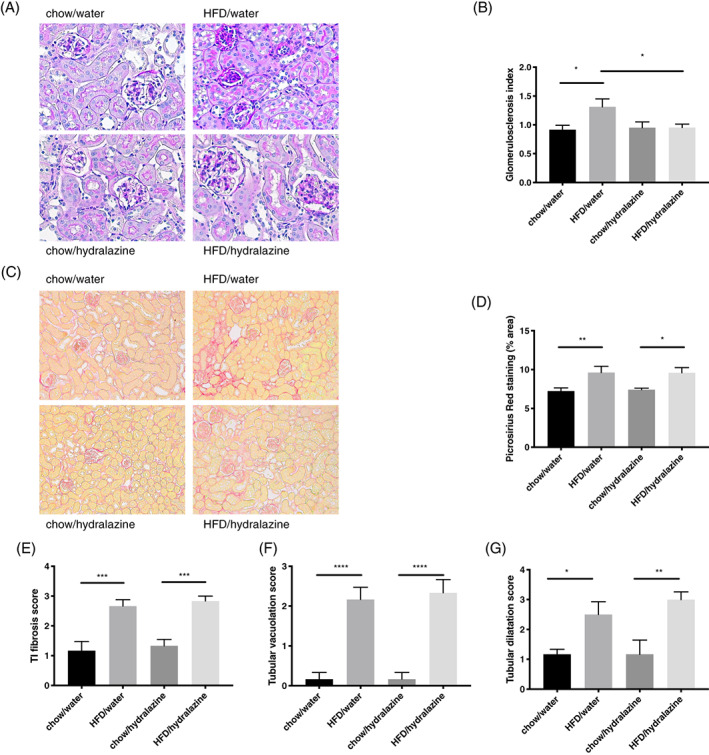
Renal structural changes. A, Representative images of periodic acid Schiff staining for glomerulosclerosis at 400× magnification. B, Glomerulosclerosis index. C, D, Picrosirius red staining representative images at 200× magnification and quantitation. E, Tubulointerstitial (TI) fibrosis. F, Tubular vacuolation. G, Tubular dilatation. Results expressed as mean ± SEM, n = 6, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. HFD, high‐fat diet
3.5. Renal fibrosis
Kidneys of obese mice demonstrated significantly increased fibronectin mRNA expression compared to controls (P < 0.05), however, this was not ameliorated by hydralazine (Figure 3A). There was a trend towards increased fibronectin protein expression in HFD/water compared to chow/water (Figure 3B). Hydralazine significantly reduced fibronectin expression in obese mice (P < 0.05; HFD/hydralazine vs. HFD/water [Figure 3B]).
FIGURE 3.
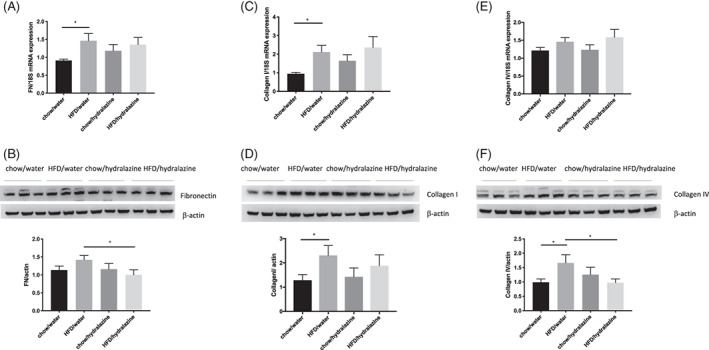
Renal fibrotic changes as measured by fibronectin, collagen I and collagen IV. mRNA expression and Western blotting of fibronectin (FN; A, B), collagen I (C, D) and collagen IV (E, F). Representative Western blots are shown for n = 3 per group. Results expressed as mean ± SEM, n = 6, *P < 0.05. HFD, high‐fat diet
Obese mice had significantly elevated renal collagen I mRNA and protein expression compared with lean mice (P < 0.05), yet this was not affected by hydralazine (Figure 3C,D). There were no differences in collagen IV mRNA expression between groups (Figure 3E), although collagen IV protein expression was significantly increased by obesity (P < 0.05; Figure 3F). Collagen IV protein expression was significantly attenuated by hydralazine, (P < 0.05; HFD/hydralazine vs. HFD/water [Figure 3F]).
3.6. Kidney injury and inflammation
Urinary KIM‐1:creatinine ratio was significantly elevated in obesity (P < 0.05), however, no protective effect was seen for hydralazine (Figure 4A). HFD increased renal KIM‐1 mRNA expression compared to control (P < 0.001), and this effect was ameliorated by hydralazine (P < 0.05; HFD/hydralazine vs. HFD/water [Figure 4B]).
FIGURE 4.
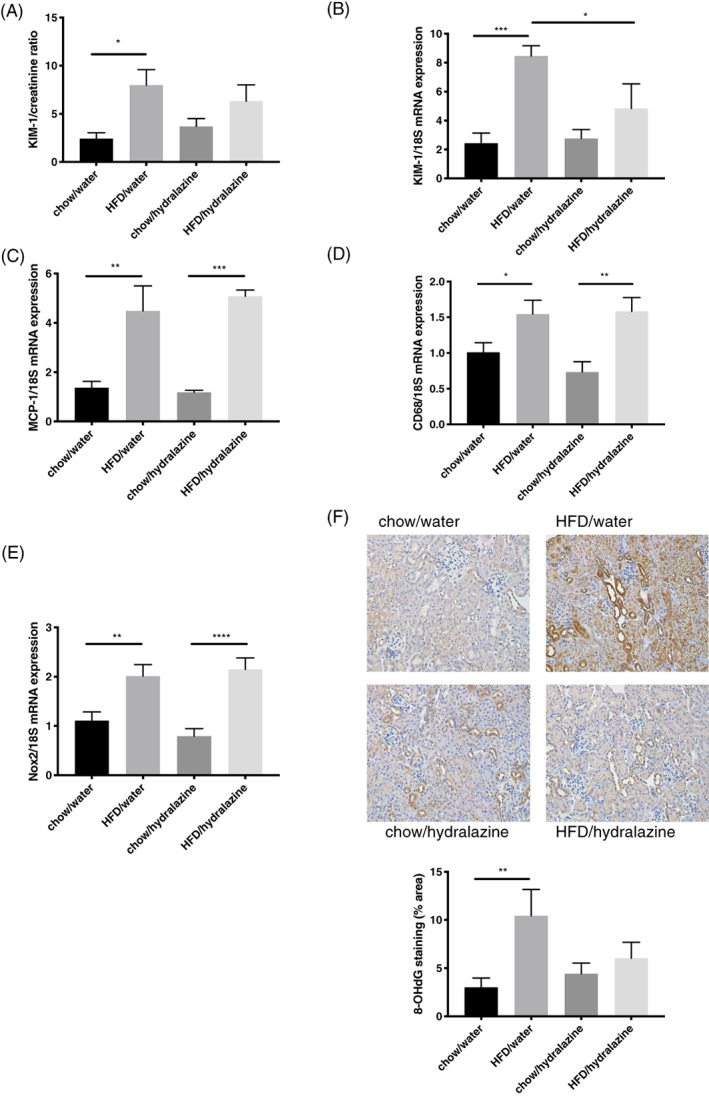
Kidney injury, inflammation and oxidative stress. A, Urinary kidney injury molecule‐1 (KIM‐1): creatinine ratio. B, KIM‐1 mRNA expression in kidney tissue. C, Renal monocyte chemoattractant protein‐1 (MCP‐1) mRNA expression. D, Renal CD68 mRNA expression. E, Renal NADPH oxidase 2 (Nox2) mRNA expression. F, Representative images of 8‐OHdG staining at 200× magnification and quantitation of immunohistochemistry staining for 8‐OHdG. Results expressed as mean ± SEM, n = 6, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. 8‐hydroxy‐2′‐deoxyguanosine (8‐OHdG)
The HFD increased renal mRNA expression of monocyte chemoattractant protein‐1 (MCP‐1) compared to control (P < 0.01; Figure 4C); this was not altered by hydralazine. Additionally, kidney mRNA expression of the macrophage marker CD68 was significantly elevated in obese mice compared to lean mice (P < 0.05) and hydralazine was not protective (Figure 4D).
3.7. Renal oxidative stress
Obesity increased the renal mRNA expression of NADPH oxidase 2 (Nox2), and protein expression of 8‐hydroxy‐2′‐deoxyguanosine (8‐OHdG), a marker of oxidative damage of DNA (P < 0.01 for both, HFD/water vs. chow/water [Figure 4E,F]). Hydralazine tended to reduce 8‐OHdG expression due to obesity, albeit without reaching statistical significance (P = 0.08; Figure 4F). There were no significant changes in the levels of other renal oxidative stress markers including glutathione peroxidase‐1 (GPx‐1), nitrotyrosine, manganese superoxide dismutase (MnSOD), catalase, Nox4, superoxide dismutase‐1 (SOD1), and inducible nitric oxide synthase (iNOS; Figure S3).
4. DISCUSSION
In this study, we observed that administration of low‐dose hydralazine to obese mice reduced albuminuria, glomerulosclerosis and renal KIM‐1, independent of metabolic effects or blood pressure regulation. This was associated with significant suppression of renal global DNA methylation and normalization of the DNA methylation profile.
As we have previously described, HFD consumption resulted in an obese phenotype with several features of the metabolic syndrome, including increased adiposity, reduced glucose tolerance, hyperinsulinaemia and dyslipidaemia. 24 Several deleterious renal consequences of obesity were also observed, including albuminuria, glomerulosclerosis, tubulointerstitial fibrosis, tubular dilatation, and tubular vacuolation. Increased renal markers of injury, inflammation, fibrosis and oxidative stress were also demonstrated in obese mice.
Albuminuria is a marker of kidney damage and is one of the earliest signs of asymptomatic renal injury in diabetes, hypertension and several other primary kidney diseases. 32 It is also a significant predictor of progressive renal dysfunction in CKD. 33 Therefore, lowering albuminuria represents a valid renoprotective strategy. 32 Hence, the fact that hydralazine ameliorates albuminuria, glomerulosclerosis and renal KIM‐1 levels in obese mice supports the renoprotective nature of the drug in our obesity‐related CKD model.
Obesity and hydralazine did not affect blood pressure, confirming that the renoprotective effects of low‐dose hydralazine occur independently of blood pressure regulation. The potential impact of blood pressure was an important confounding factor to exclude, given that intraglomerular hypertension injures the glomerular filtration barrier and can contribute to albuminuria. 1 Although we could not exclude the possibility that local renal haemodynamic effects may have contributed to reduced albuminuria, this appears unlikely given that hydralazine at higher doses predominantly induces afferent arteriolar dilatation, 34 which would increase intraglomerular pressure and potentially exacerbate albuminuria.
The role of DNA methylation was explored in this study, which may provide further insight into the potential therapeutic role of hydralazine in obesity‐induced CKD. We observed increased renal global DNA methylation in obese mice compared to lean mice. This was significantly reduced in the presence of hydralazine. The protein expression level of DNMT1, an enzyme responsible for maintenance of DNA methylation, was not significantly different between obese and lean mice. However, obesity was associated with a near‐significant increase in the protein expression of DNMT3a, an enzyme responsible for de novo DNA methylation, which may explain the increase in renal global DNA methylation in that group. Hydralazine significantly decreased DNMT3a expression in obese mice, which may explain the reduction in renal global DNA methylation in that group, supporting hydralazine′s role as a DNA demethylating agent.
To provide greater understanding of the effect of hydralazine on epigenetic mechanisms, beyond global DNA methylation analysis, we have assessed the methylation status of individual genes by RRBS. Our data revealed significant changes in the DNA methylation pattern in the kidney of HFD‐fed animals, which were also significantly reversed by the administration of hydralazine. Among these differentially methylated genes are genes that have been shown to play important roles in inflammation, cell death and fibrosis, which may contribute to the development and/or resolution of CKD. TCF7L2 variants have been associated with CKD progression and renal function in population‐based cohorts. 35 Ripk3 is an important mediator of cell death 36 and its expression was shown to increase in response to oxidative stress and inflammation. 37 Our recent study also demonstrated that Ripk3 blockade can attenuate tubulointerstitial fibrosis in a mouse model of diabetic nephropathy. 38 This is consistent with our finding that Ripk3 gene expression is significantly upregulated by an HFD. Similarly, Hhip has been shown to be upregulated by high glucose in vitro and promotes fibrosis and apoptosis in glomerular endothelial cells in in vivo models of diabetes. 39 The direct role of Dok1 in CKD is unknown; however, it has been implicated in the effect of HFD to induce adipocyte hypertrophy and obesity. 40
It is known that DNA methylation does not always lead to gene silencing, and instead may result in gene activation when it inhibits factors known to reduce gene transcription. 8 , 41 In our study, we have confirmed that only Ripk3 and Fam83d were regulated at a gene level in obesity. Given that low‐dose hydralazine normalized Ripk3 expression and attenuated albuminuria, renal kidney injury markers and glomerulosclerosis, this suggests that the renoprotective effect of hydralazine might involve epigenetic modification of Ripk3. This novel mechanism for Ripk3 requires further validation. In this study, we only examined the gene expression of eight out of 47 differentially methylated genes which are commonly regulated by HFD and hydralazine. Therefore, it is highly likely that other genes may be involved in hydralazine′s effect on obesity‐related CKD. This could be assessed in future studies.
Hydralazine did not protect obese mice against tubulointerstitial fibrosis, although there were significant reductions in renal fibronectin and collagen IV protein expression. Tampe et al. previously demonstrated attenuation of renal fibrosis by hydralazine in mouse models of unilateral ureteral obstruction and folic acid nephropathy. 19 , 20 This difference most likely relates to the model of CKD employed in our study. Unilateral ureteral obstruction and folic acid nephropathy models produce features of overt renal fibrosis. 42 , 43 Our HFD‐induced model of CKD resulted in more subtle histological changes, and although we did see evidence of increased renal fibrosis markers, the changes were more consistent with those of kidney injury rather than severe renal fibrosis.
Inflammation and oxidative stress are known to be important contributing factors which act synergistically in the initiation of renal fibrosis and the progression of CKD. 6 , 44 , 45 , 46 In the present study, there was evidence of increased renal inflammation in obese mice, with significantly increased mRNA expression of MCP‐1 and CD68. Hydralazine was not protective against obesity‐induced kidney inflammation. We also observed an increase in oxidative stress markers in the context of obesity. Renal Nox2 mRNA expression was significantly elevated in obese mice, and this remained unchanged by hydralazine treatment. There is some evidence that hydralazine may have antioxidant properties, either by directly inhibiting NADPH oxidase to reduce superoxide production, or by scavenging free radicals. 47 , 48 It is possible that the failure of our model to demonstrate such an inhibitory effect on Nox2 could relate to the low dose of hydralazine used. Interestingly, administration of hydralazine prevented the significant upregulation of 8‐OHdG induced by HFD. Whether hydralazine has any effect on DNA copy number is unclear, and this could be a focus of additional studies.
In summary, we observed that low‐dose hydralazine ameliorates albuminuria and glomerulosclerosis in a mouse model of obesity‐induced CKD, and so we propose that hydralazine may have a therapeutic role in renoprotection in obesity. These effects occur independently of alterations in metabolic variables and blood pressure. Hydralazine did not attenuate HFD‐induced renal fibrosis or inflammation. The exact mechanism of hydralazine′s renoprotective actions remains unclear, however, demethylation of DNA or reduced DNA oxidation may be involved, as hydralazine not only suppressed the increased global DNA methylation but also normalized the abnormal methylation pattern induced by the HFD in the kidney. These data support the repurposing of hydralazine, an antihypertensive drug used clinically for many decades, as a novel therapy to prevent the progression of obesity‐related CKD.
AUTHOR CONTRIBUTIONS
Benjamin P. Larkin: design, conduct/data collection, analysis, writing manuscript, writing—review and editing. Long T. Nguyen: conduct/data collection, analysis, writing manuscript, writing—review and editing. Miao Hou: conduct/data collection, writing—review and editing. Sarah J. Glastras: writing—review and editing. Hui Chen: writing—review and editing. Alen Faiz: analysis, writing—review and editing. Jason Chen: conduct/data collection, writing—review and editing. Rosy Wang: conduct/data collection, writing—review and editing. Carol A. Pollock: design, analysis, writing—review and editing. Sonia Saad: design, conduct/data collection, analysis, writing—review and editing.
CONFLICT OF INTEREST
The authors declare that no competing interests exist.
Supporting information
Figure S1 Pilot study to determine the optimal hydralazine dose. Mean blood pressure according to hydralazine concentration in the drinking water. Results expressed as mean ± SEM, n = 4, ***P < 0.001.
Figure S2 A, Pathway analysis showing enriched biological processes (BP) as per gene ontology (GO). B, Common CpGs that were regulated by both HFD and hydralazine.
Figure S3 Markers of renal oxidative stress at 32 weeks of age. (A, B) Renal nitrotyrosine expression as measured on immunohistochemistry and western blotting, (C) renal SOD1 mRNA expression, (D) renal iNOS mRNA expression, (E) renal Nox4 mRNA expression, (F) renal catalase mRNA expression, (G) renal GPx‐1 expression measured by western blotting, (H) renal mRNA expression of GPx‐1 and (I) renal expression of MnSOD on western blotting. Results expressed as mean ± SEM. n = 6.
Table S1 Mouse‐specific primers used in quantitative real‐time PCR
Table S2 Differential methylated CpGs in the kidney of HFD‐fed mice
Table S3 Differential methylated CpGs in the kidney of HFD‐fed mice treated with hydralazine
Table S4 Common differential methylated CpGs induced by HFD and HFD + Hydralazine
ACKNOWLEDGMENTS
B. Larkin is the recipient of a Jacquot Research Entry Scholarship in Nephrology from the Royal Australasian College of Physicians. This study was partly funded by grants from the Diabetes Australia Research Trust (DART) and the Rebecca L. Cooper Medical Research Foundation. Open access publishing facilitated by The University of Sydney, as part of the Wiley ‐ The University of Sydney agreement via the Council of Australian University Librarians.
Larkin BP, Nguyen LT, Hou M, et al. Low‐dose hydralazine reduces albuminuria and glomerulosclerosis in a mouse model of obesity‐related chronic kidney disease. Diabetes Obes Metab. 2022;24(10):1939‐1949. doi: 10.1111/dom.14778
Funding information This study was funded by grants from the Diabetes Australia Research Trust (DART) and the Rebecca L. Cooper Medical Research Foundation.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Kovesdy CP, Furth SL, Zoccali C. Obesity and kidney disease: hidden consequences of the epidemic. Am J Nephrol. 2017;45(3):283‐291. [DOI] [PubMed] [Google Scholar]
- 2. Ejerblad E, Fored CM, Lindblad P, Fryzek J, McLaughlin JK, Nyren O. Obesity and risk for chronic renal failure. J Am Soc Nephrol. 2006;17(6):1695‐1702. [DOI] [PubMed] [Google Scholar]
- 3. Stenvinkel P, Zoccali C, Ikizler TA. Obesity in CKD—what should nephrologists know? J Am Soc Nephrol. 2013;24(11):1727‐1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rhee CM, Ahmadi SF, Kalantar‐Zadeh K. The dual roles of obesity in chronic kidney disease: a review of the current literature. Curr Opin Nephrol Hypertens. 2016;25(3):208‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Glastras SJ, Chen H, McGrath RT, et al. Effect of GLP‐1 receptor activation on offspring kidney health in a rat model of maternal obesity. Sci Rep. 2016;6:23525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glastras SJ, Chen H, Tsang M, et al. The renal consequences of maternal obesity in offspring are overwhelmed by postnatal high fat diet. PLoS One. 2017;12(2):e0172644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Whaley‐Connell A, Sowers JR. Obesity and kidney disease: from population to basic science and the search for new therapeutic targets. Kidney Int. 2017;92(2):313‐323. [DOI] [PubMed] [Google Scholar]
- 8. Smyth LJ, Duffy S, Maxwell AP, McKnight AJ. Genetic and epigenetic factors influencing chronic kidney disease. Am J Renal Physiol. 2014;307(7):F757‐F776. [DOI] [PubMed] [Google Scholar]
- 9. Smyth LJ, McKay GJ, Maxwell AP, McKnight AJ. DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics. 2014;9(3):366‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ko YA, Mohtat D, Suzuki M, et al. Cytosine methylation changes in enhancer regions of core pro‐fibrotic genes characterize kidney fibrosis development. Genome Biol. 2013;14(R108):R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chu AY, Tin A, Schlosser P, et al. Epigenome‐wide association studies identify DNA methylation associated with kidney function. Nat Commun. 2017;8(1):1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wing MR, Devaney JM, Joffe MM, et al. DNA methylation profile associated with rapid decline in kidney function: findings from the CRIC study. Nephrol Dial Transplant. 2014;29(4):864‐872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stenvinkel P, Karimi M, Johansson S, et al. Impact of inflammation on epigenetic DNA methylation—a novel risk factor for cardiovascular disease? J Intern Med. 2007;261(5):488‐499. [DOI] [PubMed] [Google Scholar]
- 14. Kato S, Lindholm B, Stenvinkel P, et al. DNA hypermethylation and inflammatory markers in incident Japanese dialysis patients. Nephron Extra. 2012;2(1):159‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ingrosso D, Cimmino A, Perna AF, et al. Folate treatment and unbalanced methylation and changes of allelic expression induced by hyperhomocysteinaemia in patients with uraemia. Lancet. 2003;361(9370):1693‐1699. [DOI] [PubMed] [Google Scholar]
- 16. Tampe B, Steinle U, Tampe D, et al. Low‐dose hydralazine prevents fibrosis in a murine model of acute kidney injury–to–chronic kidney disease progression. Kidney Int. 2017;91(1):157‐176. [DOI] [PubMed] [Google Scholar]
- 17. Zeisberg EM, Zeisberg MA. A rationale for epigenetic repurposing of hydralazine in chronic heart and kidney failure. J Clin Epigenet. 2016;2(1):2472‐1158. [Google Scholar]
- 18. Kandler MR, Mah GT, Tejani AM, Stabler SN, Salzwedel DM. Hydralazine for essential hypertension. Cochrane Database Syst Rev. 2011 Nov 9;(11):CD004934. [DOI] [PubMed] [Google Scholar]
- 19. Tampe B, Tampe D, Muller CA, et al. Tet3‐mediated Hydroxymethylation of epigenetically silenced genes contributes to bone Morphogenic protein 7‐induced reversal of kidney fibrosis. J Am Soc Nephrol. 2014;25(5):905‐912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tampe B, Tampe D, Zeisberg EM, et al. Induction of Tet3‐dependent epigenetic remodeling by low‐dose hydralazine attenuates progression of chronic kidney disease. EBioMedicine. 2015;2(1):19‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Council NHaMR . Australian Code for the Care and Use of Animals for Scientifc Purposes. 8th ed. Canberra: National Health and Medical Research Council; 2013. [Google Scholar]
- 22. Neugarten J, Golestaneh L. Gender and the prevalence and progression of renal disease. Adv Chronic Kidney Dis. 2013;20(5):390‐395. [DOI] [PubMed] [Google Scholar]
- 23. Larkin BP, Nguyen LT, Hou M, et al. Novel role of gestational hydralazine in limiting maternal and dietary obesity‐related chronic kidney disease. Front Cell Dev Biol. 2021;9:705263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Glastras SJ, Chen H, Teh R, et al. Mouse models of diabetes, obesity and related kidney disease. PLoS One. 2016;11(8):e0162131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andrews S. FastQC: A quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 26. Krueger F. Trim Galore!: A Wrapper Tool Around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files, Babraham Institute; 2015. [Google Scholar]
- 27. Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for bisulfite‐Seq applications. Bioinformatics. 2011;27(11):1571‐1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langmead B, Salzberg SL. Fast gapped‐read alignment with bowtie 2. Nat Methods. 2012;9:357‐359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weinhold L, Wahl S, Pechlivanis S, Hoffmann P, Schmid M. A statistical model for the analysis of beta values in DNA methylation studies. BMC Bioinform. 2016;17(1):480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Larkin BP, Saad S, Glastras SJ, et al. Low‐dose hydralazine during gestation reduces renal fibrosis in rodent offspring exposed to maternal high fat diet. PLoS One. 2021;16(3):e0248854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lambers Heerspink HJ, Gansevoort RT. Albuminuria is an appropriate therapeutic target in patients with CKD: the pro view. Clin J Am Soc Nephrol. 2015;10(6):1079‐1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cravedi P, Remuzzi G. Pathophysiology of proteinuria and its value as an outcome measure in chronic kidney disease. Br J Clin Pharmacol. 2013;76(4):516‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wolf S, Risler T. Are all antihypertensive drugs renoprotective? Herz. 2004;29(3):248‐254. [DOI] [PubMed] [Google Scholar]
- 35. Köttgen A, Hwang SJ, Rampersaud E, et al. TCF7L2 variants associate with CKD progression and renal function in population‐based cohorts. J Am Soc Nephrol. 2008;19(10):1989‐1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Orozco S, Oberst A. RIPK3 in cell death and inflammation: the good, the bad, and the ugly. Immunol Rev. 2017;277(1):102‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Imamura M, Moon J‐S, Chung K‐P, et al. RIPK3 promotes kidney fibrosis via AKT‐dependent ATP citrate lyase. JCI insight. 2018;3(3):e94979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi Y, Huang C, Zhao Y, et al. RIPK3 blockade attenuates tubulointerstitial fibrosis in a mouse model of diabetic nephropathy. Sci Rep. 2020;10(1):10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao XP, Chang SY, Liao MC, et al. Hedgehog interacting protein promotes fibrosis and apoptosis in glomerular endothelial cells in murine diabetes. Sci Rep. 2018;8(1):5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hosooka T, Noguchi T, Kotani K, et al. Dok1 mediates high‐fat diet‐induced adipocyte hypertrophy and obesity through modulation of PPAR‐gamma phosphorylation. Nat Med. 2008;14(2):188‐193. [DOI] [PubMed] [Google Scholar]
- 41. Larkin BP, Glastras SJ, Chen H, Pollock CA, Saad S. DNA methylation and the potential role of demethylating agents in prevention of progressive chronic kidney disease. FASEB J. 2018;32(10):5215‐5226. [DOI] [PubMed] [Google Scholar]
- 42. Martinez‐Klimova E, Aparicio‐Trejo OE, Tapia E, Pedraza‐Chaverri J. Unilateral ureteral obstruction as a model to investigate fibrosis‐attenuating treatments. Biomolecules. 2019;9(4):141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eddy AA, Lopez‐Guisa JM, Okamura DM, Yamaguchi I. Investigating mechanisms of chronic kidney disease in mouse models. Pediatr Nephrol. 2012;27(8):1233‐1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Small DM, Coombes JS, Bennett N, Johnson DW, Gobe GC. Oxidative stress, anti‐oxidant therapies and chronic kidney disease. Nephrol Ther. 2012;17:311‐321. [DOI] [PubMed] [Google Scholar]
- 45. Daenen K, Andries A, Mekahli D, Van Schepdael A, Jouret F, Bammens B. Oxidative stress in chronic kidney disease. Pediatr Nephrol. 2019;34(6):975‐991. [DOI] [PubMed] [Google Scholar]
- 46. Sung CC, Hsu YC, Chen CC, Lin YF, Wu CC. Oxidative stress and nucleic acid oxidation in patients with chronic kidney disease. Oxidative Med Cell Longev 2013;2013:301982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Daiber A, Mülsch A, Hink U, et al. The oxidative stress concept of nitrate tolerance and the antioxidant properties of hydralazine. Am J Cardiol. 2005;96(7):25‐36. [DOI] [PubMed] [Google Scholar]
- 48. Münzel TKS, Rajagopalan S, Thoenes M, et al. Hydralazine prevents nitroglycerin tolerance by inhibiting activation of a membrane‐bound NADH oxidase. J Clin Invest. 1996;98(6):1465‐1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Pilot study to determine the optimal hydralazine dose. Mean blood pressure according to hydralazine concentration in the drinking water. Results expressed as mean ± SEM, n = 4, ***P < 0.001.
Figure S2 A, Pathway analysis showing enriched biological processes (BP) as per gene ontology (GO). B, Common CpGs that were regulated by both HFD and hydralazine.
Figure S3 Markers of renal oxidative stress at 32 weeks of age. (A, B) Renal nitrotyrosine expression as measured on immunohistochemistry and western blotting, (C) renal SOD1 mRNA expression, (D) renal iNOS mRNA expression, (E) renal Nox4 mRNA expression, (F) renal catalase mRNA expression, (G) renal GPx‐1 expression measured by western blotting, (H) renal mRNA expression of GPx‐1 and (I) renal expression of MnSOD on western blotting. Results expressed as mean ± SEM. n = 6.
Table S1 Mouse‐specific primers used in quantitative real‐time PCR
Table S2 Differential methylated CpGs in the kidney of HFD‐fed mice
Table S3 Differential methylated CpGs in the kidney of HFD‐fed mice treated with hydralazine
Table S4 Common differential methylated CpGs induced by HFD and HFD + Hydralazine
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


