Abstract
Fitness‐enhancing adaptations of protein expression and its regulation are an important aspect of bacterial evolution. A key question is whether evolution has led to optimal protein expression that maximizes immediate growth rate (short‐term fitness) in a robust manner (consistently across diverse conditions). Alternatively, they could display suboptimal short‐term fitness, because they cannot do better or because they instead strive for long‐term fitness maximization by, for instance, preparing for future conditions. To address this question, we focus on the ATP‐producing enzyme F1F0 H+‐ATPase, which is an abundant enzyme and ubiquitously expressed across conditions. Its expression is highly regulated and dependent on growth rate and nutrient conditions. For instance, during growth on sugars, when metabolism is overflowing acetate, glycolysis supplies most ATP, while H+‐ATPase is the main source of ATP synthesis during growth on acetate. We tested the optimality of H+‐ATPase expression in Escherichia coli across different nutrient conditions. In all tested conditions, wild‐type E. coli expresses its H+‐ATPase remarkably close (within a few per cent) to optimal concentrations that maximize immediate growth rate. This work indicates that bacteria can indeed achieve robust optimal protein expression for immediate growth‐rate maximization.
Keywords: ATP synthase, Escherichia coli, fitness maximization, maximal growth rate, protein expression
To experimentally validate whether Escherichia coli maximizes its growth rate by optimal expression of its ATP synthase, we titrated the expression level of this enzyme in a mutant strain, using an isopropyl β‐ d‐1‐thiogalactopyranoside‐titratable construct. We confirmed that: i. an optimal expression level exists at which the growth rate is maximal, and ii. the wildtype strain achieves this same maximal growth rate across conditions.
Abbreviations
- ATP synthase
F1F0 H+‐ATPase
- IPTG
isopropyl β‐ d‐1‐thiogalactopyranoside
Introduction
According to the accepted view on fitness, the fittest microorganisms produce most viable offspring over a given period of time and (dynamic) conditions [1, 2]. One long‐term fitness strategy is to always aim for maximization of immediate growth rate (short‐term fitness). Another strategy is to prepare for future conditions, leading to a suboptimal short‐term fitness, but a more competitive long‐term fitness; for instance, by ensuring that adaptation times are generally short or by anticipatory expression of stress proteins [3, 4, 5, 6]. Which strategy is preferred depends on the evolutionary history of the organism [7, 8] and, likely, also on the optimization capabilities of the regulatory circuitry of cells [9].
Fitter microbes express proteins at better adapted concentrations or ones with better adapted (kinetic) properties. In principle, a cell can enhance its net biosynthetic rates, and hereby its immediate growth rate (short‐term fitness), by increasing concentrations of needed metabolic enzymes – since the rate of an enzyme is generally proportional to its concentration [10]. However, bounds exist on the total allowable protein concentrations in a cell [11, 12]. For instance, because proteins occupy space and finite transcriptional and translation resources limit biosynthetic processes [11, 13, 14, 15]. In addition, enzymes have per capita finite activities [10]. Thus, the immediate growth rate of a microorganism has a maximum. One possible perspective on bacterial protein‐expression regulation is that it resembles a constrained “optimization” problem to optimally allocate their limited biosynthetic resources over all (growth rate supporting) proteins in the cell [9, 11, 13, 16]. Examples of limited biosynthetic resources are, for instance, RNA polymerases, ribonucleic acids, ribosomes, amino acids, free‐energy carriers, membrane space, and cytosolic space [11, 16]. All of this occurs in the presence of complicating relationships between protein activities such as their reliance and influence on cellular pH.
That biosynthetic resources are limited in a bacterial cell is indicated by the fact that the expression of unneeded proteins reduces immediate growth rate, as their synthesis reduces concentrations of needed growth‐supporting proteins [12, 16, 17, 18]. This was, for instance, illustrated by expressing beta‐galactosidase during growth on a minimal medium supplemented with glucose rather than lactose [12].
Tuning the concentrations of needed proteins (e.g. for growth or stress relief) to optimal levels for the current environment can maximize immediate growth rate, as then no biosynthetic resources are wasted [19, 20, 21, 22]. Alternatively, a needed protein can be over‐ or underexpressed, relative to its optimal level, because the organism cannot perform better or because it is preparing itself for future conditions [4, 23]. In the latter cases, the expression level of the needed protein is suboptimal for immediate growth rate.
An optimal level appears to exist for any needed protein. Imagine that we slowly increase the concentration of a needed enzyme. When its concentration is increased, starting from zero, the growth rate increases from zero too. The growth rate continues to rise as long as the enzyme is “underexpressed” [19, 20, 21, 22]. Eventually, the growth rate reduces when the enzyme is increased even further, i.e. when it is “overexpressed” [12, 19, 20, 21, 22]. In between the under‐ and overexpression levels the growth rate reaches its maximum at the optimal expression level [19, 20, 21, 22]. In the overexpression regime, the growth rate reduces because, amongst other effects, too many limited biosynthetic resources are allocated to an enzyme that is no longer growth limiting [12, 17, 18]. Prevention of protein overexpression is therefore a requirement for maximization of the immediate growth rate (short‐term fitness). Since expression costs generally rise linearly with the expression level and the benefits maximize at a certain expression level (when other proteins become growth limiting), an optimum is found at intermediate expression levels where the difference between protein benefit and cost is maximal [19, 24]. The actual value of the growth rate of a cell, and its dependency on concentrations of proteins, is partially determined by kinetic effects, including regulatory mechanisms and cellular pH.
Optimal expression levels have been found experimentally for a number of different proteins across microorganisms. For instance, for glycolytic proteins in Lactococcus lactis [25, 26, 27]; enzymes of the PTS system of Salmonella typhimurium [28]; citrate synthase [29], catabolic genes regulated by CRP [6], H+‐ATPase [30, 31, 32], and the lac operon of Escherichia coli [19]; and dozens of proteins of Saccharomyces cerevisiae, involved in various processes [20]. These studies provide strong evidence for protein‐expression optimization by microorganisms. Natural selection for growth rate at constant conditions, i.e. for immediate growth rate, should also in principle lead to evolution of protein expression towards its optimum [16]. Also this has been shown experimentally [19, 33].
Whether a microorganism expresses its proteins to an optimal level to maximize growth rate is nontrivial, especially if it is subjected to a dynamic environment. It could well be that an organism has adapted its expression level to circumstances it most often encounters (e.g. glucose as carbon source), but performs sub‐optimally in more unusual circumstances (e.g. mixed carbon/nitrogen sources). Moreover, the exact regulatory circuits remain unknown for many proteins and pathways, which hampers the prediction of the extent to which they are able to robustly tune expression levels throughout different environments. Experimental studies are therefore needed to assess the optimality of protein expression across many conditions. However, current studies have so far focused on the optimality of expression of proteins in only a few environments. Therefore, we address whether E. coli is able to robustly optimize the expression level of a key protein through phenotypic adaptation (on short timescales, excluding adaptation through mutation and selection); thus, over a diverse set of nutrient conditions.
Results
Optimal expression of F1F0 H+‐ATPase across conditions
We chose to study the optimality of the expression of F1F0 H+‐ATPase (ATPase). It catalyses ATP synthesis, a vital cellular process. This membrane‐embedded protein is composed of eight subunits, each expressed from a single operon, i.e. atpIBEFHAGDC, with a single promoter [34, 35, 36]. ATPase has a total mass of ~ 545 kDa, making it about 15 times as large as the average bacterial protein [35 kDa (Bionumbers)]. Its size is 20% of the size of the largest protein, the ribosome (2700 kDa). ATPase generates vast amounts of ATP under aerobic conditions at the expense of the proton motive force that is maintained by an electron transport chain [35]. While glycolysis alone generates ~ 2 ATP molecules per glucose molecule, the activity of ATPase adds to that 28–38 ATP molecules per glucose molecule [23, 24, 25]. This amounts to a high ATP synthesis rate by ATPase and, as a consequence, about 10% of the membrane proteins can be ATPase complexes [37]; indicating that it is one of the top competitors for membrane space. When E. coli grows aerobically on other carbon sources than sugars (e.g. on acetate or pyruvate), ATPase is the only source of ATP and is, therefore, an essential enzyme. Also under non‐respiratory conditions, ATPase confers a benefit: it can hydrolyse ATP to generate a proton gradient to drive other cellular processes (like membrane transport) and maintain a physiological intracellular pH. ATPase is, therefore, a key enzyme for E. coli’s fitness, independent of environmental conditions.
The expression level of ATPase in E. coli has been measured using various techniques (including LC‐MS/MS, microarray, ribosome profiling and expression of a reporter protein), showing that the expression level differs depending on the growth conditions (i.e. medium composition, aeration, dilution rate, presence of stress conditions) [38, 39, 40, 41, 42, 43]. It remains unclear whether these expression levels are optimal or not, i.e. whether they indeed maximize the growth rate across those conditions. We therefore tested ATPase‐expression optimality across 26 different carbon sources, each supplemented to M9 minimal medium, and once in complex medium (Luria‐Bertani Broth). We exploited a previously characterized E. coli mutant (LM3113) and its wildtype, a K12‐derived strain (LM3118) [30, 31, 32, 44, 45]. The mutant has an isopropyl β‐ d‐1‐thiogalactopyranoside (IPTG)‐titratable atpIBEFHAGDC operon. Jensen et al. confirmed that IPTG titration indeed leads to changes in enzyme synthesis and confirmed that growth rate vs IPTG plots indeed report optimality in accordance with growth rate vs ATPase concentration plots (by measuring the c subunit of ATPase as a function of IPTG) [30, 31, 32]. In the 27 different conditions, we grew the IPTG‐titratable strain at a range of IPTG concentrations to determine the optimal ATPase‐expression level that maximizes the growth rate of the titratable mutant. We also grew the wildtype in this manner to be able to compare its growth rate to that of the mutant. An overview of the experimental approach is shown in Fig. 1. All the used nutrients were sole carbon sources, except for i. arginine, asparagine, glutamine, glycine, and ornithine, which were mixed carbon and nitrogen sources (in those cases, ammonium was not added to the medium) and ii. cytosine, alanine and glucosamine, which were used as sole nitrogen sources (glucose was added in these cases as the carbon source). The outcome of our experiments is shown in Fig. 2.
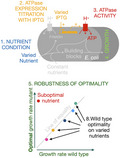
Fig. 1.
Experimental approach for identification of robust optimal expression of ATPase. Escherichia coli wildtype and an ATPase‐titratable strain were grown on minimal medium in different nutrient conditions. In a single condition, the ATPase concentration was titrated with IPTG and the growth rates of the wildtype and the mutant were measured. The optimal growth rate of the mutant strain was plotted versus the growth rate of the wildtype. If those values lie on the line of optimality for all carbon sources, then E. coli displays robust, optimal expression of its ATPase that maximizes the immediate growth rate. Alternatively, for a given nutrient source, the growth rate of the wildtype could be lower than that at the optimal value of the titrated strain, indicating suboptimal expression levels.
Fig. 2.
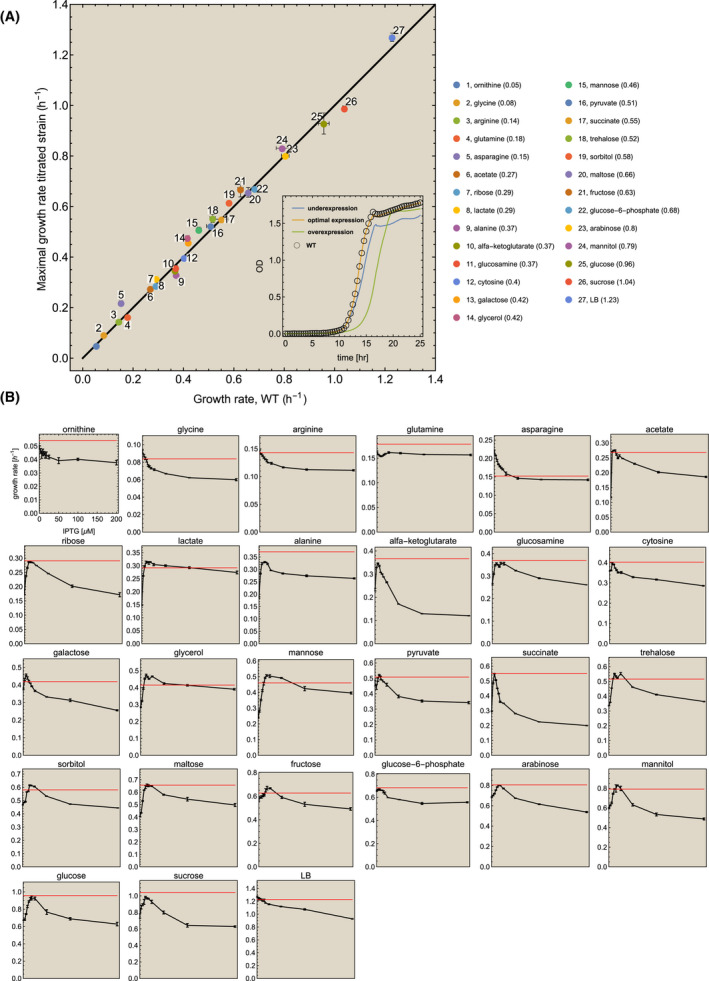
Robust, growth rate maximizing expression of ATPase. The wildtype and the ATPase titratable strain were grown in 27 different conditions. (A) At each condition, the optimal growth rate of the titratable strain was plotted as function of the growth rate of the wildtype. Error bars indicate the standard deviation. The growth rates of the wildtype and the mutant were compared at the optimal IPTG concentration. The growth rate of the wildtype strain is indicated in the legend after the name of the nutrient and indicated by the red line in the small plots below. The inset shows the growth profiles on glucose of the mutant strain, grown at underexpression, optimal expression and overexpression conditions (as lines), and the wildtype as open circles. The profile of the wildtype overlaps with that of the mutant at optimal conditions. (B) For each condition, a plot of the growth rate of the mutant strain (± SEM) as function of the IPTG concentration is shown (the axes are only shown once). The red line indicates the mean growth rate of the wildtype (at the optimal IPTG concentration). Note that the growth rate does not go to zero at high IPTG concentrations. There, the promoter is at maximal activity and ATPase cannot be increased more in concentration. Only with a stronger promoter or multiple gene copies would a higher reduction in growth rate become achievable. Also, the promoter is a bit leaky, leading to some expression at zero IPTG. We only show the growth rate values and IPTG concentrations for ornithine, the growth rate range of the other nutrients are different.
Figure 2 indicates that ATPase expression by wildtype E. coli is generally very close to optimal behaviour. The average absolute deviation from optimality is 6.1 ± 6.1%, indicating that optimal expression of ATPase is surprisingly robust. In a few cases, we found that the wildtype grew faster than the mutant. Note that, due to the experimental setup with a discrete range of IPTG concentrations, the possibility exists that the exact optimal concentration was not included.
A full overview of the growth rates at the optimum and of the wildtype, including the deviation of the wildtype from the optimum, can be found in Table 1. The absolute deviation values per carbon source are shown in Fig. 3. Four carbon sources had a statistically significant deviation (t‐test, P‐value < 0.05) of more than 10%: asparagine (30.3%), glycerol (12.7%), alanine (12.5%) and ornithine (10.2%). Of these 4, for ornithine the absolute values of the growth rates only differ by 0.005 h−1, and for alanine the wildtype outperforms the apparent optimum (most likely, as stated above, due to the discrete range of IPTG concentrations measured) – pointing towards experimental limitations in these 2 cases. For glycerol, it is interesting to note that an earlier study has also found that the wildtype grows sub‐optimally, and can improve after adaptive evolution [33]. In the case of asparagine, we cannot exclude that the wildtype also shows sub‐optimal behaviour that can be improved by evolution. When we exclude those four carbon sources, the mean deviation from optimality is 4.4 ± 3.0%. An additional statistical analysis is shown in Fig. 4.
Table 1.
Overview for all nutrient sources of wildtype strain (wt) and ATPase titratable strain at the optimum (opt). Values displayed are the IPTG concentration at the optimum for the titratable strain; the average growth rate (µ) in 1/h and standard error (SE) of the wildtype and titratable strain at the optimum; the deviation of the growth rate of the wildtype from the optimum; and the P‐value of the corresponding Student’s t‐test.
| Nutrient source | [IPTG] in microM at optimum |
Mean µmax wt (1/h) |
SE µmax wt |
Mean µmax optimum (1/h) |
SE µmax opt |
Deviation wt from opt (1/h) |
t‐test (P‐value) |
|---|---|---|---|---|---|---|---|
| Acetate | 9 | 0.268 | 0.002 | 0.275 | 0.002 | 0.02 | 0.0832 |
| Alanine | 12 | 0.371 | 0.002 | 0.330 | 0.002 | −0.13 | 0.0000 |
| Alfa‐ketoglutarate | 6 | 0.367 | 0.003 | 0.347 | 0.001 | −0.06 | 0.0043 |
| Arabinose | 15 | 0.804 | 0.008 | 0.801 | 0.003 | 0.00 | 0.7097 |
| Arginine | 0 | 0.143 | 0.000 | 0.145 | 0.003 | 0.01 | 0.5879 |
| Asparagine | 0 | 0.152 | 0.003 | 0.218 | 0.004 | 0.30 | 0.0001 |
| Cytosine | 6 | 0.403 | 0.002 | 0.397 | 0.003 | −0.01 | 0.2073 |
| Fructose | 18 | 0.626 | 0.006 | 0.668 | 0.015 | 0.06 | 0.0642 |
| Galactose | 6 | 0.419 | 0.003 | 0.458 | 0.005 | 0.08 | 0.0009 |
| Glucosamine | 18 | 0.369 | 0.004 | 0.357 | 0.005 | −0.03 | 0.0886 |
| Glucose‐6‐phosphate | 6 | 0.682 | 0.000 | 0.670 | 0.003 | −0.02 | 0.0317 |
| Glucose | 18 | 0.957 | 0.011 | 0.929 | 0.021 | −0.03 | 0.2869 |
| Glutamine | 0 | 0.178 | 0.001 | 0.163 | 0.002 | −0.09 | 0.0003 |
| Glycerol | 12 | 0.415 | 0.002 | 0.476 | 0.005 | 0.13 | 0.0001 |
| Glycine | 0 | 0.084 | 0.002 | 0.092 | 0.002 | 0.09 | 0.0312 |
| Lactate | 18 | 0.292 | 0.001 | 0.314 | 0.002 | 0.07 | 0.0001 |
| Lb | 0 | 1.228 | 0.005 | 1.270 | 0.008 | 0.03 | 0.0082 |
| Maltose | 15 | 0.657 | 0.003 | 0.656 | 0.010 | 0.00 | 0.9399 |
| Mannitol | 18 | 0.792 | 0.012 | 0.831 | 0.004 | 0.05 | 0.0412 |
| Mannose | 18 | 0.461 | 0.005 | 0.508 | 0.005 | 0.09 | 0.0007 |
| Ornithine | 0 | 0.054 | 0.000 | 0.049 | 0.001 | −0.10 | 0.0191 |
| Pyruvate | 9 | 0.508 | 0.008 | 0.523 | 0.002 | 0.03 | 0.1723 |
| Ribose | 9 | 0.291 | 0.001 | 0.286 | 0.001 | −0.02 | 0.0683 |
| Sorbitol | 15 | 0.581 | 0.004 | 0.615 | 0.003 | 0.06 | 0.0004 |
| Succinate | 6 | 0.551 | 0.014 | 0.550 | 0.002 | 0.00 | 0.9305 |
| Sucrose | 12 | 1.039 | 0.003 | 0.988 | 0.006 | −0.05 | 0.0036 |
| Trehalose | 25 | 0.516 | 0.006 | 0.553 | 0.008 | 0.07 | 0.0110 |
Fig. 3.
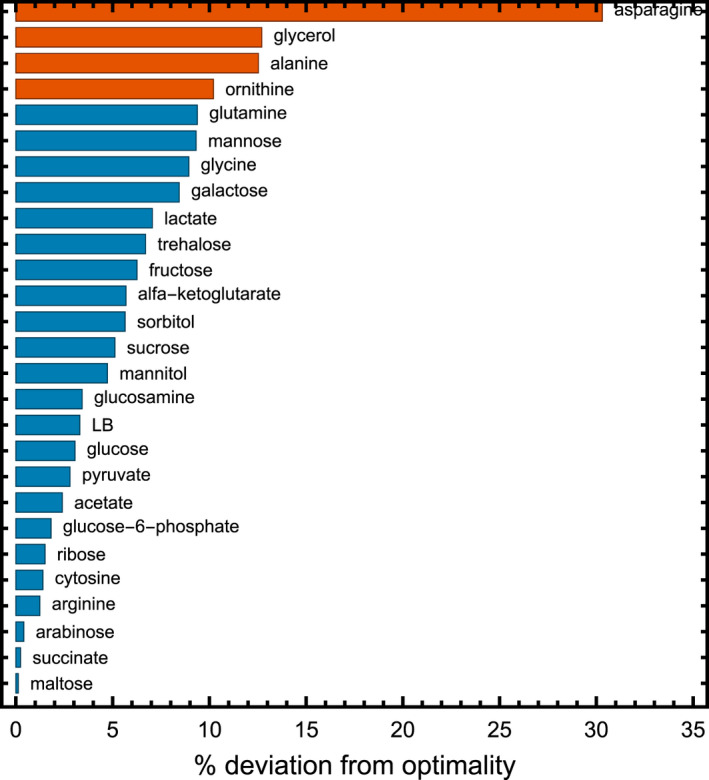
Percentage absolute deviation from optimal protein expression per carbon source. Only four carbon sources show a percentage deviation of more than 10%: ornithine (10.2%), glycerol (12.7%), alanine (12.3%) and asparagine (30.3%). Mean absolute deviation is 6.1 ± 6.1% and with those four carbon sources removed 4.3 ± 3.0%. The growth rates of the wildtype and the mutant were compared at the optimal IPTG concentration.
Fig. 4.
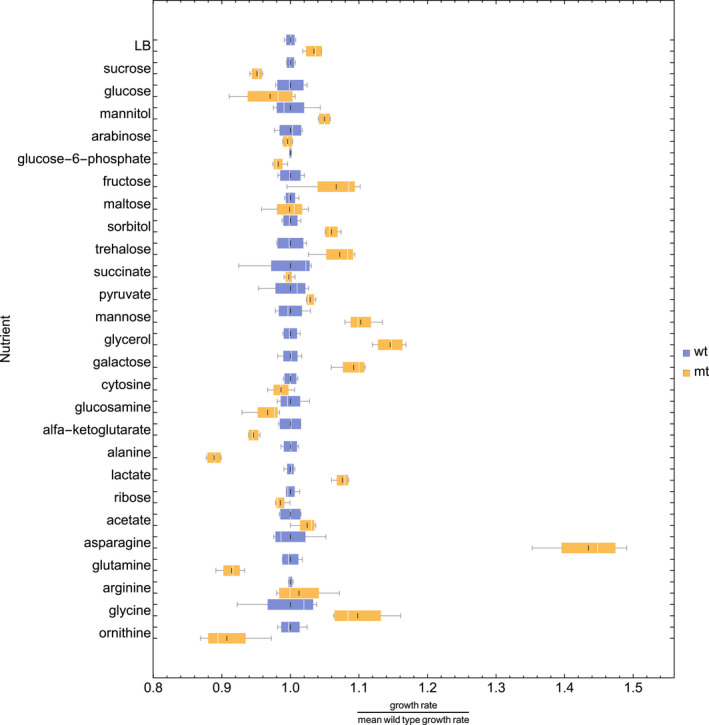
Box whisker plot of the titratable strain at optimum (mt, yellow) and wildtype (wt, blue) data. Each nutrient has two boxes, one blue the other yellow, indicating its wild and mutant growth rate values, respectively. The white line indicates the median value, the black line the mean value, the lower end of the box marks the 25% quantile and the upper the 75% quantile.
Since our main research aim is to get an impression of the robustness of the optimality of expression, we performed a regression analysis to evaluate to what extend the dataset as a whole indicates that the wildtype robustly optimises the expression of ATPase to maximise immediate growth rate. We found a regression coefficient (R 2) of 0.988, indicating that the wildtype robustly steers ATPase expression to its optimal level throughout the tested conditions.
Discussion
Studies that focus on growth rate effects of protein expression raise a number of fundamental questions at the interface of microbiology and evolutionary biology. [16, 21, 22] Many of them indicate direct [16, 19, 20, 25, 26, 27, 28, 29, 30] or indirect [11, 13, 15, 46, 47] evidence of optimal expression behaviour. Together, they suggest that microbial physiology may be predictable and understandable from an evolutionary perspective where selection for maximal growth is constrained by physicochemical and cellular limits. These limits bound cellular protein concentrations and, therefore, the maximally‐attainable growth rate [16]. Optimality of protein expression studies suggests that this optimal state is reached for dozens of key metabolic proteins [16, 19, 20, 25, 26, 27, 28, 29, 30]. In yeast, this was shown for over 70 proteins [20]. Here, we show for the first time that optimal expression of a key metabolic protein can be robust across a large range of diverse conditions.
Our findings indicate that the expression of the operon atpIBEFHAGDC is optimally regulated by its associated molecular‐control system across conditions. Many genes involved in respiration and metabolism are regulated by ArcA and Fnr [48, 49], but knocking out these proteins did not affect atp expression [38]. Regrettably, how the regulation of the atp operon molecularly works is still poorly understood. We note that theoretical studies have shown that optimal gene expression can be achieved by simple circuits composed of basic biochemical interactions, suggesting that even a single transcription factor and a promoter with a single binding site can suffice for optimal steering [6, 9, 13, 50]. One of the general caveats in our understanding of expression regulation of metabolic genes is that even if we know the identity of the associated transcription factors, we rarely know which metabolites regulate their DNA‐binding affinities by binding and stabilizing particular transcription‐factor conformations.
Optimal expression of ATPase for immediate growth‐rate maximization does not necessarily imply that E. coli does not carry out a long‐term fitness maximization strategy [6, 51]. In fact, evidence exists for long‐term fitness maximization too. It has, for instance, been shown that E. coli prepares for adverse conditions at slow growth [52, 53]. Nevertheless, a 1% overexpression of an abundant and relatively costly enzyme, like ATPase, represents a significant waste of resources and reduces the immediate growth rate. Thus, evolution under constant conditions can be expected to lead to a loss of preparatory overexpression.
We showed here that optimal expression of ATPase is preserved across a wide range of nutrient conditions in E. coli. This indicates that the expression regulation of a ubiquitous enzyme, central to growth and survival (i.e. fitness), can be understood in terms of growth‐rate maximization. Since optimal regulation of gene expression is achievable with simple circuits [9, 13, 15, 19, 50], optimization of metabolic protein levels for maximal growth rate might be widespread in microbiology. A quite universal principle may therefore exist for the regulation of (metabolic) gene expression by microorganisms.
Materials and methods
Media
For all experiments, M9 minimal medium [54] was freshly made every experiment day, to avoid degradation. It was supplemented with 1 μg·mL−1 thiamine, as well as the desired carbon source (concentration adjusted to carbon content or nitrogen content; 20 mm for C6‐carbohydrates, 18.7 mm for N1). During preculturing, 10 μm of IPTG was added to the LM3113 strain, to reduce the selective advantage of tacI mutants. For the experiments with mixed carbon and nitrogen sources (arginine, asparagine, glutamine, glycine, ornithine) NH4Cl and another carbon source were omitted from the medium; in the cases of cytosine, alanine and glucosamine, NH4Cl was omitted but glucose was added to the medium.
Strains and growth experiments
Derivative strains of E. coli K‐12 were used in this study, kindly provided by P. R. Jensen (Table 2). All strains have their lactose permease eliminated, making them reliant on passive diffusion of IPTG. Since lactose permease can act as a costly IPTG‐import system its deletion is preferred.
Table 2.
Overview of the used strains.
The obtained strains were sent on LB medium. A single colony was picked and grown on M9 medium + 1 µg·mL−1 thiamine + 20 mm glucose, then stored in aliquots at −80 °C in glycerol stock. For each experiment, an aliquot of the same glycerol stock was used to inoculate the cultures on M9 medium + thiamine + glucose, and grown overnight at 30 °C, shaking at 200 RPM. The next morning the OD600 was measured, and the cultures propagated to fresh medium containing the desired carbon source at 0.01 < OD < 0.05, followed by a few more hours of growth (depending on the growth rate) to ensure exponential growth of the cultures.
For the microplate experiments, M9 medium + thiamine + nutrient source was prepared containing a range of IPTG concentrations (0, 3, 6, 9, 12, 15, 18, 25, 50, 100, 200 μm). The OD600 of the propagated cultures was measured, and the cells were propagated to the (IPTG containing) media at OD = 0.0005, and put in microplate in quadruplicate per treatment. Every microplate also contained LM3118 wildtype on glucose without IPTG as an internal control to confirm proper growth conditions. The spaces in between wells were filled with 0.9% NaCl solution, and the plate sealed at the edges with parafilm to minimize evaporation. The OD600 was measured at 30 °C every 5 min using the Spectramax 384 plus, with shaking in between reads. The data were calibrated based on a dilution series of different ODs measured in a cuvette spectrometer and the microplate reader. The growth rate was then calculated during the exponential growth phase, from the linear ln(OD) range using an R script.
Statistics
The comparison between the growth rates at the optimal IPTG concentration of wildtype and the titratable strain on all separate carbon sources was tested using a two‐tailed, heteroscedastic Student's t‐test. The regression analysis on the overall dataset was done by calculating the total sum of squares with the growth rate of the wildtype at the optimal [IPTG] on nutrient source i for Yi , and the average growth rate of the wildtype throughout conditions for Y mean; the residual sum of squares with the growth rate of the titratable strain at the optimum for fi ; and the coefficient of determination as .
Conflict of interest
The authors declare no conflict of interest.
Author contributions
IR and FJB designed the experiments and analysed the data. IR performed the experiments. IR and FJB wrote the manuscript.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1111/febs.16401.
Acknowledgements
We thank Peter Jensen for providing us with the E. coli strains used in this study. We thank Dirk Bald, Greg Bokinsky, Johan van Heerden, Bas Teusink, Bob Planqué and Hans Westerhoff for in depth discussions. IR and FJB acknowledge funding from NWO‐VIDI Project 864.11.011.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Sæther BE, Engen S. The concept of fitness in fluctuating environments. Trends Ecol Evol. 2015;30:273–81. 10.1016/j.tree.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 2. Orr HA. Fitness and its role in evolutionary genetics. Nat Rev Genet. 2009;10:531–9. 10.1038/nrg2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. New AM, Cerulus B, Govers SK, Perez‐Samper G, Zhu B, Boogmans S, et al. Different levels of catabolite repression optimize growth in stable and variable environments. PLoS Biol. 2014;12:e1001764. 10.1371/journal.pbio.1001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Venturelli OS, Zuleta I, Murray RM, El‐Samad H. Population diversification in a yeast metabolic program promotes anticipation of environmental shifts. PLoS Biol. 2015;13:e1002042. 10.1371/journal.pbio.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang J, Atolia E, Hua B, Savir Y, Escalante‐Chong R, Springer M. Natural variation in preparation for nutrient depletion reveals a cost‐benefit tradeoff. PLoS Biol. 2015;13:e1002041. 10.1371/journal.pbio.1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Towbin BD, Korem Y, Bren A, Doron S, Sorek R, Alon U. Optimality and sub‐optimality in a bacterial growth law. Nat Commun. 2017;8:14123. 10.1038/ncomms14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haccou P, Iwasa Y. Optimal mixed strategies in stochastic environments. Theor Popul Biol. 1995;47:212–43. 10.1006/tpbi.1995.1009. [DOI] [Google Scholar]
- 8. Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–8. 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 9. Planqué R, Hulshof J, Teusink B, Hendriks JC, Bruggeman FJ. Maintaining maximal metabolic flux by gene expression control. PLoS Comput Biol. 2018;14:e1006412. 10.1371/journal.pcbi.1006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cleland W. The kinetics of enzyme‐catalyzed reactions with two or more substrates or products. Biochim Biophys Acta Spec Sect Enzymol Subj. 1963;6569:104–37. 10.1016/0926-6569(63)90211-6. [DOI] [PubMed] [Google Scholar]
- 11. Molenaar D, Van Berlo R, De Ridder D, Teusink B. Shifts in growth strategies reflect tradeoffs in cellular economics. Mol Syst Biol. 2009;5:323. 10.1038/msb.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science (80‐.). 2010;330:1099–102. 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- 13. Scott M, Klumpp S, Mateescu EM, Hwa T. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol Syst Biol. 2014;10:747. 10.15252/msb.20145379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ehrenberg M, Kurland CG. Costs of accuracy determined by a maximal growth rate constraint. Q Rev Biophys. 1984;17:45–82. 10.1017/S0033583500005254. [DOI] [PubMed] [Google Scholar]
- 15. Bosdriesz E, Molenaar D, Teusink B, Bruggeman FJ. How fast‐growing bacteria robustly tune their ribosome concentration to approximate growth‐rate maximization. FEBS J. 2015;282:2029–44. 10.1111/febs.13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bruggeman FJ, Planqué R, Molenaar D, Teusink B. Searching for principles of microbial physiology. FEMS Microbiol Rev. 2020;44:821–44. 10.1093/femsre/fuaa034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stoebel DM, Dean AM, Dykhuizen DE. The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics. 2008;178:1653–60. 10.1534/genetics.107.085399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kurland CG, Dong H. Bacterial growth inhibition by overproduction of protein. Mol Microbiol. 1996;21:1–4. 10.1046/j.1365-2958.1996.5901313.x. [DOI] [PubMed] [Google Scholar]
- 19. Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436:588–92. 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- 20. Keren L, Hausser J, Lotan‐Pompan M, Vainberg Slutskin I, Alisar H, Kaminski S, et al. Massively parallel interrogation of the effects of gene expression levels on fitness. Cell. 2016;166:1282–94.e18. 10.1016/j.cell.2016.07.024. [DOI] [PubMed] [Google Scholar]
- 21. Dykhuizen DE, Dean AM, Hartl DL. Metabolic flux and fitness. Genetics. 1987;115:25–31. 10.1093/genetics/115.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dykhuizen DE, Dean AM. Enzyme activity and fitness: evolution in solution. Trends Ecol Evol. 1990;5:257–62. 10.1016/0169-5347(90)90067-N. [DOI] [PubMed] [Google Scholar]
- 23. Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, et al. Adaptive prediction of environmental changes by microorganisms. Nature. 2009;460:220–4. 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- 24. Berkhout J, Bosdriesz E, Nikerel E, Molenaar D, de Ridder D, Teusink B, et al. How biochemical constraints of cellular growth shape evolutionary adaptations in metabolism. Genetics. 2013;194:505–12. 10.1534/genetics.113.150631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Solem C, Koebmann BJ, Jensen PR. Glyceraldehyde‐3‐phosphate dehydrogenase has no control over glycolytic flux in Lactococcus lactis MG1363. J Bacteriol. 2003;185:1564–71. 10.1128/JB.185.5.1564-1571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Solem C, Koebmann B, Yang F, Jensen PR. The las enzymes control pyruvate metabolism in Lactococcus lactis during growth on maltose. J Bacteriol. 2007;189:6727–30. 10.1128/JB.00902-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koebmann B, Solem C, Jensen PR. Control analysis as a tool to understand the formation of the las operon in Lactococcus lactis . FEBS J. 2005;272:2292–303. 10.1111/j.1742-4658.2005.04656.x. [DOI] [PubMed] [Google Scholar]
- 28. Vlag J, Hof R, Dam K, Postma PW. Control of glucose metabolism by the enzymes of the glucose phosphotransferase system in Salmonella typhimurium . Eur J Biochem. 1995;230:170–82. 10.1111/j.1432-1033.1995.0170i.x [DOI] [PubMed] [Google Scholar]
- 29. Walsh K, Koshland DE. Characterization of rate‐controlling steps in vivo by use of an adjustable expression vector. Proc Natl Acad Sci USA. 1985;82:3577–81. 10.1073/pnas.82.11.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jensen PR, Michelsen O, Westerhoff HV. Control analysis of the dependence of Escherichia coli physiology on the H+‐ATPase. Proc Natl Acad Sci USA. 1993;90:8068–72. 10.1073/pnas.90.17.8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jensen PR, Westerhoff HV, Michelsen O. Excess capacity of H+‐ATPase and inverse respiratory control in Escherichia coli . EMBO J. 1993;12:1277–82. 10.1002/j.1460-2075.1993.tb05772.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jensen PR, Michelsen O, Westerhoff HV. Experimental determination of control by the H+‐ATPase in Escherichia coli . J Bioenerg Biomembr. 1995;27:543–54. 10.1007/BF02111653. [DOI] [PubMed] [Google Scholar]
- 33. Ibarra RU, Edwards JS, Palsson BØ. Escherichia coli K‐12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:186–9. [DOI] [PubMed] [Google Scholar]
- 34. Kanazawa H, Mabuchi K, Futai M. Nucleotide sequence of the promoter region of the gene cluster for proton‐translocating ATPase from Escherichia coli and identification of the active promoter. Biochem Biophys Res Commun. 1982;107:568–75. 10.1016/0006-291X(82)91529-7. [DOI] [PubMed] [Google Scholar]
- 35. Senior AE. The proton‐translocating ATPase of Escherichia coli . Ann Rev Biophys Biophys Chem. 1990;19:7–41. 10.1146/annurev.bb.19.060190.000255. [DOI] [PubMed] [Google Scholar]
- 36. McCarthy JEG. Expression of the unc genes in Escherichia coli . J Bioenerg Biomembr. 1988;20:19–39. 10.1007/BF00762136. [DOI] [PubMed] [Google Scholar]
- 37. Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, et al. Protein abundance profiling of the Escherichia coli cytosol. BMC Genom. 2008;9:102. 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kasimoglu E, Park SJ, Malek J, Tseng CP, Gunsalus RP. Transcriptional regulation of the proton‐translocating ATPase (atpIBEFHAGDC) operon of Escherichia coli: control by cell growth rate. J Bacteriol. 1996;178:5563–7. 10.1128/jb.178.19.5563-5567.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Unden G, Bongaerts J. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta Bioenerg. 1997;1320:217–34. 10.1016/S0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 40. Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–35. 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schmidt A, Kochanowski K, Vedelaar S, Ahrné E, Volkmer B, Callipo L, et al. The quantitative and condition‐dependent Escherichia coli proteome. Nat Biotechnol. 2016;34:104–10. 10.1038/nbt.3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peebo K, Valgepea K, Maser A, Nahku R, Adamberg K, Vilu R. Proteome reallocation in Escherichia coli with increasing specific growth rate. Mol Biosyst. 2015;11:1184–93. 10.1039/C4MB00721B [DOI] [PubMed] [Google Scholar]
- 43. Valgepea K, Adamberg K, Seiman A, Vilu R. Escherichia coli achieves faster growth by increasing catalytic and translation rates of proteins. Mol Biosyst. 2013;9:2344. 10.1039/c3mb70119k. [DOI] [PubMed] [Google Scholar]
- 44. Jensen PR, Westerhoff HV, Michelsen O. The use of lac‐type promoters in control analysis. Eur J Biochem. 1993;211:181–91. [DOI] [PubMed] [Google Scholar]
- 45. Jensen PR, Michelsen O. Carbon and energy metabolism of atp mutants of Escherichia coli . J Bacteriol. 1992;174:7635–41. 10.1128/jb.174.23.7635-7641.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. De Groot DH, Van Boxtel C, Planqué R, Bruggeman FJ, Teusink B. The number of active metabolic pathways is bounded by the number of cellular constraints at maximal metabolic rates. PLoS Comput Biol. 2019;15:e1006858. 10.1371/journal.pcbi.1006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. O’Brien EJ, Lerman JA, Chang RL, Hyduke DR, Palsson B. Genome‐scale models of metabolism and gene expression extend and refine growth phenotype prediction. Mol Syst Biol. 2013;9:693. 10.1038/msb.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gunsalus RP, Park SJ. Aerobic‐anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res Microbiol. 1994;145(5‐6):437–50. 10.1016/0923-2508(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 49. Iuchi S, Lin ECC. Adaptation of Escherichia coli to redox environments by gene expression. Mol Microbiol. 1993;9:9–15. 10.1111/j.1365-2958.1993.tb01664.x. [DOI] [PubMed] [Google Scholar]
- 50. Berkhout J, Teusink B, Bruggeman FJ. Gene network requirements for regulation of metabolic gene expression to a desired state. Sci Rep. 2013;3:1417. 10.1038/srep01417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dai X, Zhu M, Warren M, Balakrishnan R, Patsalo V, Okano H, et al. Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat Microbiol. 2016;2:16231. 10.1038/nmicrobiol.2016.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ihssen J, Egli T. Specific growth rate and not cell density controls the general stress response in Escherichia coli . Microbiology. 2004;150:1637–48. 10.1099/mic.0.26849-0. [DOI] [PubMed] [Google Scholar]
- 53. Ihssen J, Egli T. Global physiological analysis of carbon‐ and energy‐limited growing Escherichia coli confirms a high degree of catabolic flexibility and preparedness for mixed substrate utilization. Environ Microbiol. 2005;7:1568–81. 10.1111/j.1462-2920.2005.00846.x. [DOI] [PubMed] [Google Scholar]
- 54. LaCroix RA, Sandberg TE, O'Brien EJ, Utrilla J, Ebrahim A, Guzman GI, et al. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K‐12 MG1655 on glucose minimal medium. Appl Environ Microbiol. 2015;81:17–30. 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


