Abstract
Both Salmonella enterica serovar Typhimurium and Escherichia coli contain the cspH gene encoding CspH, one of the cold shock proteins (CSPs). In this study, we investigated the expression of cspH in S. enterica serovar Typhimurium and found that it was induced in response to a temperature downshift during exponential phase. The cspH promoter was activated at 37°C, and its mRNA was more stable than the other csp mRNAs at 37°C. Moreover, lacZ expression of the translational cspH-lacZ fusion was induced at that temperature. Interestingly, the cspH mRNA had a much shorter 5′-untranslated region than those in the other cold-shock-inducible genes, and the promoter sequence, which was only 55 bp, was sufficient for cspH expression. The 14-base downstream box located 12 bases downstream of the initiation codon of cspH mRNA was essential for its cold shock activation.
Most bacteria react against a sudden decrease in temperature, a cold shock response (from 30 to 10°C), in what is called an acclimation phase (16). During this phase, cold shock proteins (CSPs) are synthesized, and subsequently the cells resume growth (19, 23, 24). CSP genes from Salmonella spp., cspA, cspB, cspC, cspE, and cspH, have been sequenced, and the cold-shock inducibility of cspA and cspB has been reported (6, 19, 20), although their functions have not yet been clearly elucidated. Previously, it was reported that the expression of cspA and cspB in Salmonella spp. was slightly different from that in Escherichia coli (19, 20). CSPs in E. coli have been extensively studied, and CspA has been identified as a major CSP (15). Other CSPs, CspB through CspI, which are collectively known as the CspA family, are known to be present in E. coli (17). All nine CSPs in E. coli are not inducible by a temperature downshift (37, 39). For cspH, neither its expression nor its regulation in E. coli and Salmonella enterica serovar Typhimurium has been investigated.
Cold shock genes, such as cspA, cspB, cspG, and cspI, have a common feature: an unusually long 5′-untranslated region (5′-UTR) (13, 21, 28, 36). In S. enterica serovar Typhimurium, the 5′-UTR of cspA is 145 nucleotides and that of cspB is 162 nucleotides. For E. coli, cspA has 159 nucleotides, cspB has 161 nucleotides, cspG has 156 nucleotides, and cspI has 145 bp. In E. coli, the 5′-UTRs are known to be involved in the regulation of their expression (36). The 5′-UTRs of the cspA and cspI mRNAs have a negative effect on the expression of the genes at 37°C, while they have a positive effect at cold temperatures (21, 36). In S. enterica serovar Typhimurium, the role of the 5′-UTR has not been yet characterized.
The downstream box (DB) located in the downstream of AUG initiation codon enhances translation of several bacterial and phage mRNAs, and it is also required for efficient translation of CSPs during cold shock (9, 10, 28). It has been proposed that the DB enhances translation by base pairing transiently to bases 1469 to 1483 of 16S rRNA, the so-called anti-DB, during the initiation phase of translation (33). Contrary to this, O'Connor et al. (30) reported that the enhancement of translation by the DB did not involve base pairing of mRNA with the penultimate stem sequence of 16S rRNA. However, a mechanism of translational activation by the DB is not known.
Several promoters in E. coli and S. enterica serovar Typhimurium have a sequence of seven consecutive G158C base pairs (GCGCCNC; GC-rich discriminator) starting from 2 bp downstream of the putative −10 region (34). The GC-rich discriminator renders promoters sensitive to regulation by stringent response (5, 34, 35).
We investigated, in this work, the expression of cspH and characterized the transcriptional start site and promoter of cspH in S. enterica serovar Typhimurium. The 5′-UTR of its mRNA was very short, and its mRNA was stable at 37°C.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are shown in Table 1. Luria-Bertani broth (LB) media were used for growth. When necessary, ampicillin was added at the final concentration of 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relative genotype or properties | Reference or source |
|---|---|---|
| Strains | ||
| S. enterica serovar Typhimurium | ||
| SF530 (χ3761) | Wild-type UK-1 | 7 |
| SF586 (JR501) | hsdSA29 hsdSB121 hsdL6 metE551 trpC2 ilv452 rpL120 galE719 H1-6 H2-c, n, x nml fla-66 | 4 |
| YH1 | UK-1 containing the pYH1; Apr | This study |
| YH2 | UK-1 containing the pYH2; Apr | This study |
| YH3 | UK-1 containing the pYH3; Apr | This study |
| YHD | UK-1 containing the pYHD; Apr | This study |
| YHS | UK-1 containing the pYHS; Apr | This study |
| E. coli | ||
| DH5α | F−endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 gvrA96 relA1 φ80dlacZM15 Δ(lacZYA-argF)U169 | Gibco-BRL, Gaithersburg, Md. |
| EYH5 | DH5α containing pYH5; Apr | This study |
| M182 | Δ (laclPOZYA)74 | 40 |
| MYH1 | M182 containing pYH1; Apr | This study |
| Plasmids | ||
| pRS414 | Plasmid for translational lacZ fusion; Apr | 32 |
| pRS415 | Plasmid for transcriptional lacZ fusion; Apr | 32 |
| pGEM-T-easy | PCR cloning vector; Apr | Promega Co. |
| pYH1 | pRS414 derivative carrying cspH::lacZ (from −363 to +118 of cspH); Apr | This study |
| pYH2 | pRS415 derivative carrying cspH::lacZ (from −363 to +118 of cspH); Apr | This study |
| pYH3 | pRS414 derivative carrying cspH::lacZ (from −55 to +118 of cspH); Apr | This study |
| pYH5 | pGEM-T-easy vector derivative carrying a partial pagD fragment and complete cspH; Apr | This study |
| pYHD | pYH1 derivative; a deletion of DB fragment; Apr | This study |
| pYHS | pYH1 derivative; a deletion of SD fragment; Apr | This study |
General techniques.
DNA cloning was carried out by a method described previously (31). PCR amplification was carried out as described in the manufacturer's manual (Takara Co., Kyoto, Japan) with 30 cycles of amplification steps, each 30 s at 95°C, 30 s at 51°C, and 1 min at 72°C. Restriction enzymes were purchased from Boehringer GmbH, Mannheim, Germany.
Plasmid constructions.
To construct the translational and transcriptional cspH-lacZ fusions, the upper portion of cspH was amplified by PCR with oligonucleotides proL8064 (5′-aagaattcTCATGCTTTTATGCTTTGG-3′) and proR8065 (5′-ggggatccACCTGAACATCTTTGCG-3′), where the 5′ tails are in lowercase and the cleavage sites of EcoRI and BamHI, respectively, are underlined. The template for PCR was obtained from wild-type strain UK-1. The PCR product was digested with BamHI and EcoRI and then cloned into pRS414 and pRS415 for construction of translational and transcriptional lacZ fusions, respectively. These fusion plasmids were designated pYH1 and pYH2, respectively. To construct a cspH-lacZ translational fusion containing only −55 (pYH3), we used proL8066 (5′-aagaattcTGATTGCCATCAAATATAGC-3′) and proR8065.
For DB site deletion construct pYHD, primers proL8064, DBL (5′-aagcttTTTACGAGACAAACAAATTCC-3′), DBR (5′-aagcttAAAACCTTTGATTGTAAGAGC-3′), and proR8065 were used. To prepare the Shine-Dalgarno (SD) fragment deletion construct (pYHS), primers proL8064, SDL (5′-aagcttAATGGGGATAACGAGGCG-3′), SDR (5′-aagcttTTTGTTTGTCTCGTAAAATGA-3′), and proR8065 were used. The underlined sequence indicates the HindIII cleavage site. Each PCR product was cloned into the T-easy vector, and two clones were digested with HindIII and SphI (the T-easy vector does not contain a HindIII site). The two digested fragments were ligated by T4 DNA ligase. The constructs on the PCR cloning vector were digested with EcoRI and BamHI and then cloned into pRS414 (translational lacZ fusion vector). All pRS414-cspH constructs were cloned in DH5α as previously described (31), and those constructs were introduced into UK-1 by electroporation. The DNA sequences of all the constructs were confirmed by DNA sequencing with a sequencing kit (ABI Prism; part no. 4303152) and an ABI model 310 system (version 3.0).
Synthetic oligonucleotide primers P0424 (5′-AAATACTGATAAAAAGCAAGCC-3′) and P0425 (5′-CACAGGACTGATTAGCAAAATA-3′) were used for constructing the template for sequencing, yielding pYH5.
Assay for β-galactosidase activity.
To measure the cold shock activation of cspH, cells were grown in LB at 37°C to mid-exponential (optical density at 600 nm [OD600], 0.6) or stationary phase (OD600, ≥1.3) and transferred to 15°C. Cell samples were taken immediately after cold shock and at 1, 2, and 3 h after cold shock, and β-galactosidase activity was measured. For the measurement of growth-dependent expression, cells were grown to each growth phase. As the density of early-exponential-phase cells is not sufficient (OD600, 0.15) for the β-galactosidase assay, the cells were centrifuged and the pellet was suspended in fresh LB medium (OD600, ∼0.4). The stationary-phase cells were immediately diluted to an OD600 of 0.4 in fresh LB medium, and β-galactosidase activity was measured as described by Miller (27). The assay was done at least in duplicate at each time point.
Isolation of total RNA and Northern blotting analysis.
Strain UK-1 was grown to different growth phases at 37°C or incubated to mid-log and stationary phases and then transferred to 15°C in LB medium. Then, the cells were collected and total RNAs were extracted with hot phenol as described by Laodie and Ullman (26). The extracted RNA was electrophoresed in a denaturing formaldehyde-agarose gel and transferred to a nylon membrane (Amersham Co.). The membrane was blotted overnight at 54°C. The high-sodium dodecyl sulfate buffer with 50% formamide (see the DIG manual of Boehringer) was added as hybridization buffer. The transferred membrane was probed with PCR products of cspH30 (5′-CTGGCATTATTTTTCCTGAC-3′ [forward primer]) and cspH264 (5′-AGGCCATTAATACGACAAAA-3′ [reverse primer]). The 16S rDNA probe (5′-AGAGTTTGATCATGGCCCA-3′ [forward primer] and 5′-GGTTACCTTGGTTACCTTTGTTACGACTT-3′ [reverse primer]) was used as the control. The probes were primed at 37°C with a DIG labeling kit (Boehringer) for 3 h. For washing and detection procedures, we followed manufacturer's protocol (DIG detection kit; Boehringer).
Primer extension and sequencing.
The RNA was extracted from UK-1 after cold shock (15°C) for 1 h at mid-log phase as described above. Primers CS1 (5′-ATGCTGAAATGTGGACCTGAA-3′) and CS2 (5′-TTACGAGACAAACAAATTCC TTA-3′) for extension were labeled with [32γ-P]ATP by using T4 polynucleotide kinase (Boehringer). RNA (5 μg) and labeled primers were incubated at 72°C for 2 min and slowly transferred to room temperature for 40 min to make RNA bound to the primer. Primer extension was carried out at 37°C for 90 min in a total volume of 20 μl, which contained 50 mM Tris-HCl (pH 8.5), 8 mM MgCl2, 30 mM KCl, 1 mM dithiothreitol, 0.4 pmol of 32P-labeled primer, 0.5 mM dATP, 0.5 mM dGTP, 0.5 mM dCTP, 0.5 mM dTTP, 10 U of RNase inhibitor (Boehringer Mannheim), and 10 U of avian myeloblastosis virus reverse transcriptase (Promega Co). Only half of the primer extension mixture (20 μl) was loaded on the gel.
For the mRNA stability experiments at 37°C during the exponential period, cells were incubated to an OD600 of 0.15. Then, rifampin was added at a final concentration of 200 μg/ml to stop transcription, and a 10-ml culture was taken at each point. To measure the mRNA stability at 15°C, cells were cultured to an OD600 of 0.15 at 37°C, transferred to 15°C, and incubated for 1 h. At that point, rifampin was added at the same concentration as for 37°C, and a 10-ml culture was taken at each point. The products were analyzed on a denatured polyacrylamide gel and quantified by using a PhosphorImager (Bio-Rad).
For sequencing, CS1 and CS2 were used as primers and the reaction was performed with an alkali-denatured double-stranded pYH5 plasmid with Sequenase, version 2.0 (Amersham). The products of primer extension and sequencing were analyzed on an 8% polyacrylamide gel under denaturing conditions.
Nucleotide sequence accession number.
The nucleotide sequence of the cspH gene has been deposited in the EMBL, GenBank, and DDBJ databases under accession no. AF191799.
RESULTS
Sequence comparison between E. coli and S. enterica serovar Typhimurium cspH genes.
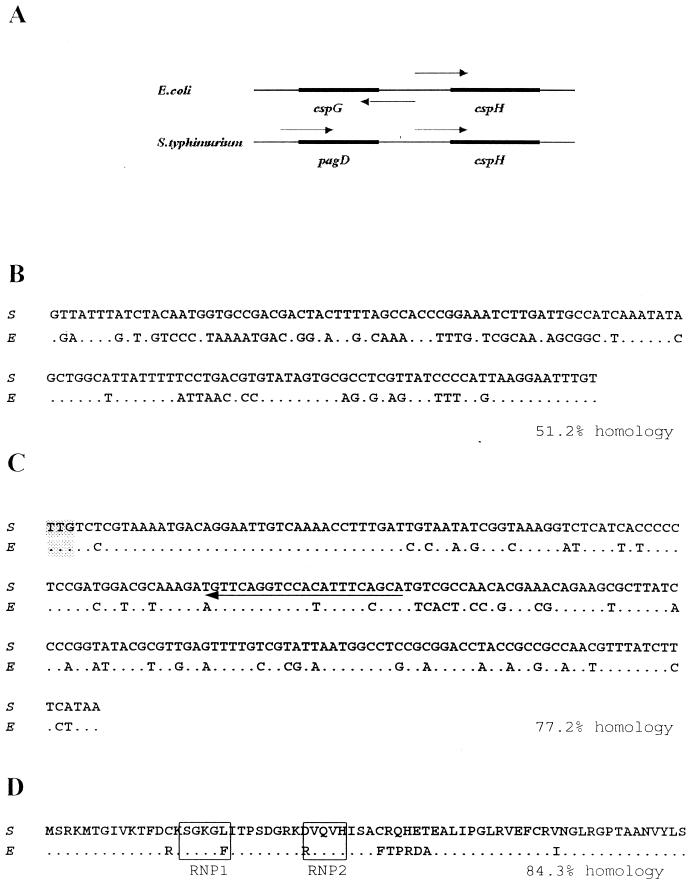
The organization of cspH genes in E. coli and S. enterica serovar Typhimurium is known to be as follows: the initiation codons of E. coli cspG and cspH are separated by 285 bp and are arranged divergently, and both genes are likely to have a common upstream promoter region (38). Extensive studies have revealed that cspG is a cold-shock-inducible gene (29), but how cspH is induced is not known. For S. enterica serovar Typhimurium, the pagD gene is present upstream of cspH (Fig. 1A) (18). The cspH gene from S. enterica serovar Typhimurium UK-1 was sequenced, and the region upstream of the initiation codon, the structural gene, and the amino acid sequence were compared with those determined from the E. coli cspH sequence (Fig. 1B to D). The cspH promoter regions of S. enterica serovar Typhimurium and E. coli have only a 52% nucleotide identity (Fig. 1B). This difference in promoter sequence indicates that the regulatory expression of S. enterica serovar Typhimurium cspH might be quite different from that of E. coli cspH. However, Fig. 1C and D show that the structural gene and amino acid sequences derived from the two species are similar to each other, suggesting that CspH in S. enterica serovar Typhimurium might have a mode of action similar to that of E. coli. Unlike other CSPs, CspH in E. coli and S. enterica serovar Typhimurium contained few aromatic amino acid residues (Fig. 1D) (38), indicating that the CspH of S. enterica serovar Typhimurium can bind to DNA rather than RNA (see Discussion).
FIG. 1.
Comparison of E. coli cspH (E) and S. enterica serovar Typhimurium UK-1 cspH (S). (A) Relative gene compositions near cspH on the chromosome. Arrows, directions of transcription of the open reading frames. A comparison of putative cspH promoters (B), structure genes (C), and amino acid sequences (D) between E. coli and S. enterica serovar Typhimurium is also shown. Boxes, RNP1and RNP2 RNA-binding motifs, which also exist in other cold shock genes. The translational start codon is shaded. Arrow (C), primer used for primer extension (CS1) (see Materials and Methods). The nucleotide and amino acid sequences in E. coli and S. enterica serovar Typhimurium have database accession no. AF003591 and AF191799, respectively.
The cspH of S. enterica serovar Typhimurium is induced after temperature downshift.
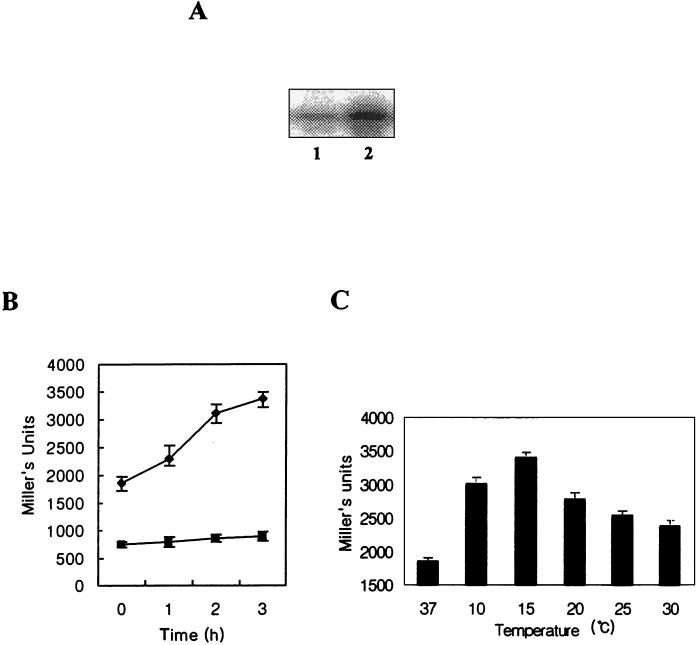
We examined whether the expression of cspH in S. enterica serovar Typhimurium was cold shock inducible. Figure 2A showed that cspH mRNA expression was increased dramatically 1 h after the temperature downshift (15°C) during exponential phase. Its level upon cold shock was four- to fivefold that at normal temperature (data not shown). However, it was hardly inducible during stationary phase (data not shown). To confirm this result, a cspH-lacZ translational fusion (pYH1) was constructed (see Fig. 6A) as described in Materials and Methods. β-Galactosidase activity of YH1 (UK-1 strain containing pYH1) was increased following cold shock during log phase but hardly increased during stationary phase (Fig. 2B). These results are in good agreement with the primer extension analysis (Fig. 2A), suggesting that cspH might be expressed not only at the transcriptional level but also at the translational level.
FIG. 2.
(A) Cold shock activation of cspH mRNA in wild-type UK-1 by primer extension analysis. Total RNAs were extracted from cells incubated at mid-log phase under the following conditions: no cold shock (lane 1) and cold shock at 15°C for 1 h (lane 2). (B) β-Galactosidase activity expressed in UK-1 (YH1) carrying a translational cspH-lacZ fusion after cold shock. Time zero, no shock after incubation. ●, mid-log phase; ▪, stationary phase. (C) At the indicated temperatures, β-galactosidase activity of YH1 was measured after cold shock for 3 h. Error bars, standard errors.
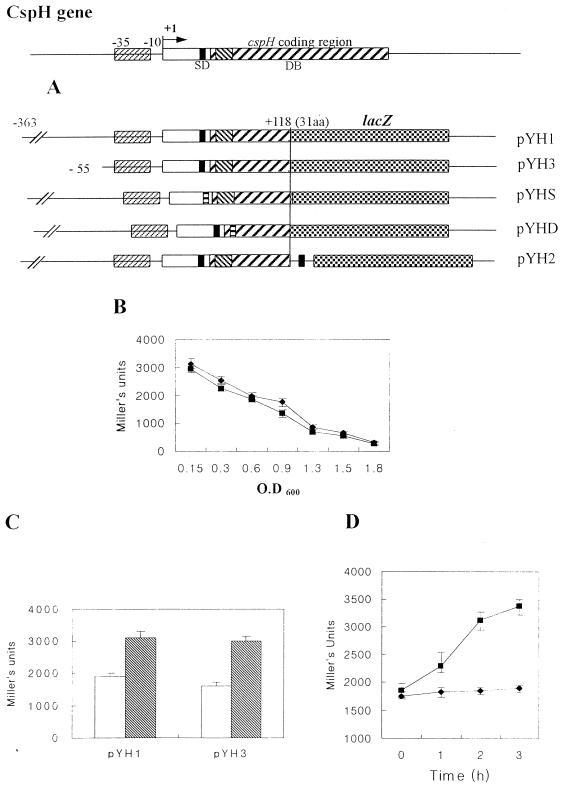
FIG. 6.
(A) Construction of each cspH-lacZ fusion. (B) Expression pattern of β-galactosidase of YH1 (⧫) and YH3 (▪) at 37°C. (C) Cold shock activation of YH1 and YH3. Open bars, expression at 37°C in exponential phase; hatched bars, cold shock activation at 15°C for 3 h. (D) Comparison of expression patterns by cold shock of YH1 (▪) and YHD (●) at exponential phase. Error bars, standard errors.
A transcriptional lacZ fusion (pYH2) was also constructed (see Fig. 6A), and the β-galactosidase activity of the construct was induced by cold shock as was done for the translational fusion. However, the cold induction was less efficient for the lacZ fusion than for the translational fusion (data not shown). Because β-galactosidase activities from both pYH1 and pYH2 were increased after cold shock, it is likely that the expression of cspH on cold shock might be regulated during both transcription and translation.
Since not all E. coli csp genes are inducible optimally at 15°C (11, 29, 36), a β-galactosidase assay with YH1 was carried out after temperature downshift at each temperature between 10 and 30°C for 3 h (Fig. 2C). CspH of S. enterica serovar Typhimurium was induced optimally at 15°C.
cspH has a short 5′-UTR.
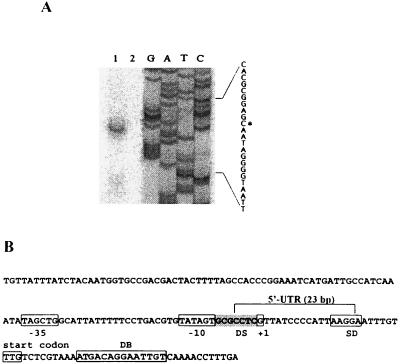
To find a transcription start site of cspH, a primer extension analysis and sequencing were carried out. A putative promoter region, −35 and −10 regions (TAGCTG and TATAGT), and the transcription start site are indicated in Fig. 3A and B. All genes which are inducible upon cold shock, including cspA and cspB of Salmonella, have a long 5′-UTR, which contains at least 145 bp (6, 36, 38). To our surprise, however, the cspH mRNA contained a very short 5′-UTR consisting of only 23 bases (Fig. 3B).
FIG. 3.
(A) Primer extension analysis of the cspH mRNA and sequencing. RNA was extracted from S. enterica serovar Typhimurium UK-1 after cold shock for 1 h at an OD600 of 0.6 as described in Materials and Methods. Primer extension analysis was carried out with (lane 1) and without (lane 2) RNA extracted from UK-1. The primer extension product was analyzed on an 8% polyacrylamide gel under denaturing conditions (lane1) with a sequencing ladder. The sequence is shown at the right; asterisk, transcription start site. (B) Nucleotide sequence of cspH. DS, discriminator.
Within the coding sequence of cspH, there was a 14-base DB sequence. The DB is known to play a critical role in the cold induction of CSPs (9, 10, 28). CspH also contained a well-conserved GC-rich discriminator sequence (GCGCCTC) at the −7 to −1 region (Fig. 3B) (34). It has been proposed that the discriminator renders the promoter sensitive to a stringent response (5).
cspH expression is also dependent on growth phase, with maximal level at early log phase.
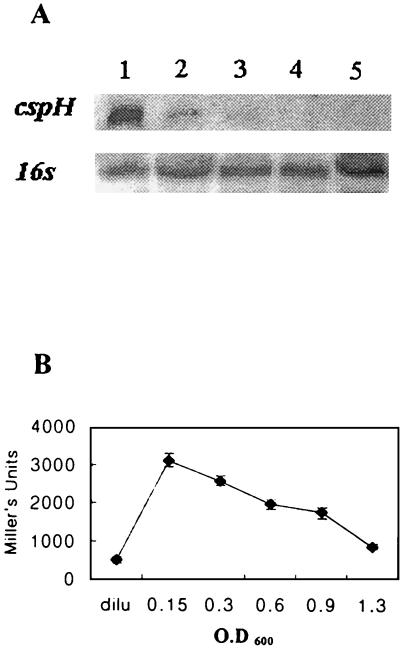
As shown in Fig. 2A (lane 1) and 2B (at zero time), cspH was expressed even at 37°C during early exponential phase but was hardly inducible during stationary phase. From these results, we speculated that cspH might be induced without cold shock and that its expression might be dependent on growth phase. To substantiate this assumption, the cells were grown to different phases at 37°C and the total RNAs were extracted: As shown in Fig. 4A, the expression of cspH at 37°C was induced to maximal level at early exponential phase (OD600, 0.15) but was repressed as cells entered into the stationary phase, indicating that the cspH promoter was under growth phase control. Although the cspH mRNA was inducible at 37°C, it might not have been translated. Therefore, we carried out a β-galactosidase assay at 37°C with YH1. As shown in Fig. 4B, its expression also showed growth phase dependence.
FIG. 4.
Growth-dependent expression of cspH at 37°C. (A) Northern hybridization with cspH-specific probe at OD600s of 0.15 (lane 1), 0.3 (lane 2), 0.6 (lane 3), 0.9 (lane 4), and 1.3 (lane 5). Northern hybridization of 16S rRNA was shown as a control for equal amounts of total RNA. (B) β-Galactosidase with YH1 at each growth phase. YH1 was diluted after overnight culture with fresh LB medium and cultured until each growth phase. At each interval, β-galactosidase activities were measured. dilu, β-galactosidase activity at the point of dilution with fresh medium after overnight culture. Error bars, standard errors.
Stability of cspH mRNA at 37 and 15°C.
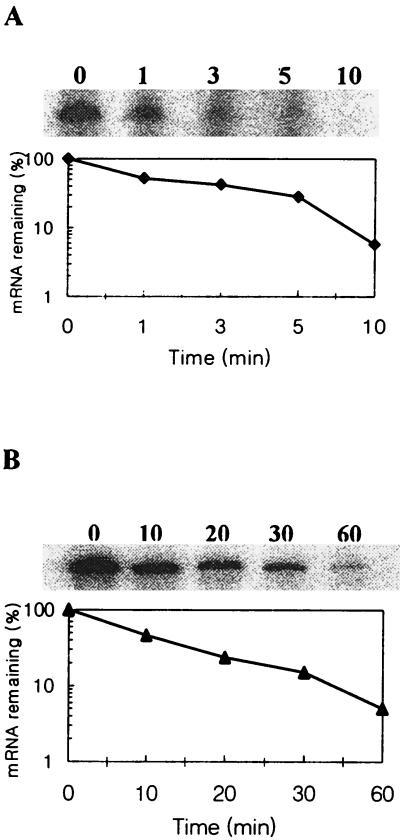
Since the 5′-UTR of cspH was very short, we expected that cspH mRNA might be more stable at 37°C than the other csp mRNAs. Therefore, the stability of cspH mRNA at both 37 and 15°C was measured. As shown in Fig. 5A, the mRNA was relatively stable at 37°C, with a half-life of approximately 70 s, while the half-life of cspA mRNA was estimated to be less than 20 s (2, 12, 14). Interestingly, more than 40% of cspH mRNA remained stable after 3 min, while less than 10% of other csp mRNAs remained stable after 2 min (36). One hour after the temperature downshift, the cspH mRNA remained stable with a half-life of approximately 10 min (Fig. 5B), while the cspA mRNA was estimated to be stable longer than 10 min (36). However, the stability of cspH mRNA was generally similar to that of cspA mRNA during cold shock.
FIG. 5.
mRNA stability of cspH. (A) Measurement of the mRNA stability at 37°C. A culture of strain UK-1 (wild type) was grown to early exponential phase at 37°C, and then rifampin was immediately added to the culture to a final concentration of 200 μg/ml. RNA was extracted at the indicated times after the addition of rifampin. Also shown is the radioactivity of primer extension products, with the product at zero time set to 100%. (B) Measurement of mRNA stability at 15°C. A culture of strain UK-1 was grown to early exponential phase at 37°C, transferred to 15°C, and incubated for 1 h. Then, rifampin was added, and RNA was extracted at each time indicated.
A promoter sequence of only 55 bp is sufficient to generate the cspH expression pattern.
To detect an upstream promoter site required for the expression of cspH, we constructed a cspH-lacZ translational fusion containing only −55 (pYH3) (Fig. 6A). As shown in Fig. 6B and C, pYH3 was sufficient for the characteristic expression of cspH.
The DB mediates cold shock activation of cspH
DB is known to play a critical role in cold shock activation of CSPs (8, 9). We constructed a DB-deleted cspH-lacZ plasmid (pYHD) (Fig. 6A) to understand whether the DB affects the cold induction of cspH. As shown in Fig. 6D, without DB, CspH was not inducible by cold shock. But LacZ expression of pYHD at 37°C was not affected (data not shown). We also constructed a Shine-Dalgarno (SD) sequence-deleted cspH-lacZ fusion (pYHS) (see Materials and Methods) and found that the construct was expressed only at the basal level at 37°C (data not shown).
DISCUSSION
In the present study, we revealed that the cspH from S. enterica serovar Typhimurium was another cold-shock-inducible gene (Fig. 2) and that the promoter of cspH was active during exponential phase at 37°C (Fig. 4A). Using the translational cspH-lacZ fusion, pYH1 (Fig. 6A), we also showed that CspH might be expressed during cold shock and exponential phase at 37°C (Fig. 4B). Interestingly, the expression mode of cspH was very similar to that of cspA in E. coli: cold shock activation and growth-dependent expression at 37°C (3). However, in stationary phase Salmonella cspH was not expressed at 37°C or at lower temperatures; on the other hand, E. coli cspA was highly induced in stationary-phase cells. It was also found that the cspH mRNA at 37°C was more stable than other csp mRNAs (Fig. 5A). The results from assays of translational fusion and mRNA stability indicated the possible presence of CspH at 37°C, although they were not conclusive. To confirm the presence of CspH at 37°C, use of antibodies is warranted.
Cold-inducible genes in E. coli and S. enterica serovar Typhimurium have unusually long 5′-UTRs. In E. coli, it has been proposed that the 5′-UTRs in their mRNAs play an important role in their expression at low temperature (2, 13, 21, 28) but have a negative effect on expression at 37°C (36). In this study, we have shown that the 5′-UTR of cspH mRNA was unusually short (Fig. 3A), implying that CspH in S. enterica serovar Typhimurium is regulated without the conserved sequence at the 5′-UTR during cold shock. Also, it is considered that this short 5′-UTR might be a reason for the stability of cspH mRNA because of a low susceptibility to cellular RNase.
To study whether Salmonella cspH could be induced in E. coli, we transformed pYH1 into E. coli strain M182 and named the resulting strain MYH1. The β-galactosidase activity of MYH1 was measured during cold shock and at normal temperature. The expression of MYH1 was similar to that of YH1 at normal temperature. However, the β-galactosidase activity of MYH1 was not induced during cold shock (data not shown). From the above result, it appears that the cspH promoter, which contains the short 5′-UTR, may be active in E. coli at normal temperature but not during cold shock. Salmonella cspH contains a conserved DB but does not have a long 5′-UTR, which is known to play an essential role in cold shock activation in E. coli (21, 36). Therefore, it is speculated that either the long 5′-UTR is important for cold-shock activation of E. coli csp or Salmonella cspH is inducible upon cold shock without the long 5′-UTR.
The result that cspH required a promoter sequence of only 55 bp for its growth-dependent and cold-shock-inducible expression (Fig. 6B and C) suggests that the cspH promoter may contain the regulatory fragments within −55. To analyze if a second promoter and possibly another start point exist, we performed primer extension with primer CS2 instead of CS1 (see Materials and Methods). CS1 is from +112 to +132, and CS2 is from +12 to +34. Only one band was observed in primer extension by CS2, and sequencing with CS2 revealed that the band was at the same point as that found by primer extension with CS1 (data not shown). Therefore, it is tempting to speculate that cspH contains only one promoter both at normal temperature and during cold shock.
For E. coli cspA, it was proposed that Fis regulated the expression of cspA at 37°C (3). Here, we found putative Fis binding sequences at −54 and −30 (data not shown). The relationship between cspH and fis in Salmonella enterica serovar Typhimurium should be further investigated.
CspA, the major CSP of E. coli, is known to be an RNA chaperone, which was thought to facilitate translation at low temperature by destabilizing mRNA structures (22). Recently, it was proposed that CspA, CspC, and CspE in E. coli play a role in the transcription antiterminator (1). In the present study, we have shown that CspH in S. enterica serovar Typhimurium did not contain the well-conserved aromatic residues (Fig. 1D). In E. coli, all the CspA homologues except CspF and CspH have eight aromatic residues (six Phe, one Tyr, and one Trp) on their amino acid sequences and these residues are known to be essential for the binding to RNA and single-stranded (38). As CspF and CspH of E. coli have only three and four aromatic residues, respectively, Yamanaka et al. (38) suggested that those might bind to DNA rather than to RNA. Therefore, it is likely that S. enterica serovar Typhimurium CspH might bind to DNA, since it contains only two aromatic residues (Phe and Tyr) (Fig. 1D).
As shown in Fig. 3B, the cspH promoter has a well-conserved discriminator sequence (between −10 and +1; GCGCCTC). It is known that the discriminator sequence plays a role in stringent control and in growth rate control (25). Therefore, we suggest that the GC-rich discriminator may affect the growth-dependent expression of cspH at 37°C.
We demonstrated that cspH had a DB in its structural gene like other cold-shock-inducible genes (Fig. 3B). The DB is known to be required for efficient translation during cold shock (10, 28). In this study, a DB-deleted cspH-lacZ translational fusion (pYHD) showed that cspH required the DB to carry out an efficient translation upon cold shock (Fig. 6D) but that the DB did not affect cspH expression at 37°C (data not shown). From these results, it can be inferred that the DB mediates efficient translation of S. enterica serovar Typhimurium cspH upon cold shock, although a precise mechanism of activation by the DB is not known. We also constructed an SD sequence-deleted cspH-lacZ fusion (Fig. 6A) and showed that, in the absence of the SD sequence, cspH was not induced at 15 or 37°C (data not shown), indicating that the DB of cspH is operational only in the presence of the SD sequence.
In E. coli, Csp family members have their respective optimal temperature ranges. The temperature dependence of CspA induction is broad (30 to 10°C), while that of CspB and CspG is restricted to lower temperatures and to a narrower temperature range (20 to 10°C) (11). CspI induction occurs over the narrowest and lowest temperature range (15 to 10°C) (36). The present study has shown that CspH induction in S. enterica serovar Typhimurium occurred over the broad temperature range (30 to 10°C) (Fig. 2C). This indicates that CspH expression in S. enterica serovar Typhimurium is due to small changes in the temperature downshift.
In conclusion, the present findings indicate that cspH from S. enterica serovar Typhimurium is another cold shock gene and that its product, CspH, might be present at 37°C. Also, the short 5′-UTR, in contrast to those of mRNAs of other cold-inducible genes, may provide a new insight into the molecular mechanism during cold-shock induction. Certainly, the precise function of CspH and the mode of its regulation during the temperature downshift and under normal condition are worth further investigation.
ACKNOWLEDGMENTS
We are grateful to Sangduk Kim (Graduate School of Biotechnology, Korea) for her advice and comments. We are also grateful to Jin Lee (Graduate School of Biotechnology, Korea) for her technical assistance in this work.
This work was supported by Korea Research Foundation grants (KRF-1998-019-D00058 and KRF-2000-015-DP0316).
REFERENCES
- 1.Bae W, Xia B, Inouye M, Severinov K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc Natl Acad Sci USA. 2000;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandi A, Pietroni P, Gualerzi C O, Pon C L. Post-transcriptional regulation of CspA expression in Escherichia coli. Mol Microbiol. 1996;19:231–240. doi: 10.1046/j.1365-2958.1996.362897.x. [DOI] [PubMed] [Google Scholar]
- 3.Brandi A, Spurio R, Gualerzi C O, Pon C L. Massive presence of the Escherichia coli ‘major cold shock protein’ CspA under non-stress conditions. EMBO J. 1999;18:1653–1659. doi: 10.1093/emboj/18.6.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullas L R, Ryu J I. Salmonella typhimurium LT2 strains which are r− m+ for all three chromosomally located systems for DNA restriction and modification. J Bacteriol. 1983;156:471–474. doi: 10.1128/jb.156.1.471-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa X J D, Stanley W A. Mutations that render the promoter of the histidine operon of Salmonella typhimurium insensitive to nutrient-rich medium repression and amino acid downshift. J Bacteriol. 1997;179:5211–5217. doi: 10.1128/jb.179.16.5211-5217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig J E, Boyle D, Fransis K P, Gallagher M P. Expression of the cold-shock gene cspB in Salmonella typhimurium occurs below a threshold temperature. Microbiology. 1998;144:697–704. doi: 10.1099/00221287-144-3-697. [DOI] [PubMed] [Google Scholar]
- 7.Curtiss R, III, Porter S B, Munson M, Tinge S A, Hassan J O, Gentry-Weeks C, Kelly S M. Nonrecombinant and recombinant avirulent Salmonella vaccines for poultry. In: Blankenship L C, Bailey J H S, Cox N A, Stern N J, Meinersmann R J, editors. Colonization control of human bacterial enteropathogens in poultry. New York, N.Y: Academic Press; 1991. pp. 169–198. [Google Scholar]
- 8.Etchegaray J P, Inouye M. CspA, CspB, and CspG, major cold shock proteins of Escherichia coli, are induced at low temperature under conditions that completely block protein synthesis. J Bacteriol. 1999;181:1827–1830. doi: 10.1128/jb.181.6.1827-1830.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Etchegaray J P, Inouye M. A sequence downstream of the initiation codon is essential for cold shock induction of cspB of Escherichia coli. J Bacteriol. 1999;181:5852–5854. doi: 10.1128/jb.181.18.5852-5854.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Etchegaray J P, Inouye M. Translational enhancement by an element downstream of the initiation codon in Escherichia coli. J Biol Chem. 1999;274:10079–10085. doi: 10.1074/jbc.274.15.10079. [DOI] [PubMed] [Google Scholar]
- 11.Etchegaray J P, Jones P G, Inouye M. Differential thermoregulation of two highly homologous cold-shock genes, cspA and cspB, of Escherichia coli. Genes Cells. 1996;1:171–178. doi: 10.1046/j.1365-2443.1996.d01-231.x. [DOI] [PubMed] [Google Scholar]
- 12.Fang L, Jiang W, Bae W, Inouye M. Promoter-independent cold-shock induction of cspA and its derepression at 37°C by mRNA stabilization. Mol Microbiol. 1997;23:355–364. doi: 10.1046/j.1365-2958.1997.2351592.x. [DOI] [PubMed] [Google Scholar]
- 13.Fang L, Hou T, Inouye M. Role of the cold-box region in the 5′ untranslated region of the cspA mRNA in its transient expression at low temperature in Escherichia coli. J Bacteriol. 1998;180:90–95. doi: 10.1128/jb.180.1.90-95.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldenberg D, Azar I, Oppenheim A B. Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol Microbiol. 1996;19:241–248. doi: 10.1046/j.1365-2958.1996.363898.x. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein J, Pollitt N S, Inouye M. Major cold shock proteins of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graumann P, Marahiel M A. Some like it cold: response of microorganism to cold shock. Arch Microbiol. 1996;166:293–300. doi: 10.1007/s002030050386. [DOI] [PubMed] [Google Scholar]
- 17.Graumann P, Marahiel M A. A superfamily of proteins that contain the cold-shock domain. Trends Biochem Sci. 1998;23:286–290. doi: 10.1016/s0968-0004(98)01255-9. [DOI] [PubMed] [Google Scholar]
- 18.Gunn J S, Aranda C M A, Loomis W P, Belden W J, Miller S I. Characterization of the Salmonella typhimurium pagD/pagD chromosomal region. J Bacteriol. 1995;177:5040–5047. doi: 10.1128/jb.177.17.5040-5047.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton A J, Hak K M, Steffan R J, Foster J W, Bej A K. Adaptive response to cold temperatures and characterization of cspA in Salmonella typhimurium LT2. Antonie Leeuwenhoek. 2000;77:13–20. doi: 10.1023/a:1002055719798. [DOI] [PubMed] [Google Scholar]
- 20.Jeffreys A G, Hak K M, Steffan R J, Foster J W, Bej A K. Growth, survival and characterization of cspA in Salmonella enteritidis following cold shock. Curr Microbiol. 1998;36:29–35. doi: 10.1007/s002849900275. [DOI] [PubMed] [Google Scholar]
- 21.Jiang W, Fang L, Inouye M. The role of 5′-end untranslated region of the mRNA for CspA, the major cold shock protein of Escherichia coli, in cold-shock adaptation. J Bacteriol. 1996;178:4919–4925. doi: 10.1128/jb.178.16.4919-4925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 23.Jones P G, Inouye M. RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol Microbiol. 1996;21:1207–1218. doi: 10.1111/j.1365-2958.1996.tb02582.x. [DOI] [PubMed] [Google Scholar]
- 24.Jones P G, VanBogelen R A, Neidhardt F C. Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol. 1987;169:2092–2095. doi: 10.1128/jb.169.5.2092-2095.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Josaitis C A, Gaal T, Gourse R L. Stringent control and growth-rate-dependent control have nonidentical promoter sequence requirements. Proc Natl Acad Sci USA. 1995;92:1117–1121. doi: 10.1073/pnas.92.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laodie B M, Ullmann A. Virulence dependent and independent regulation of Bordetella pertussis cya operon. EMBO J. 1990;9:999–1005. doi: 10.1002/j.1460-2075.1990.tb08202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 28.Mitta M, Fang L, Inouye M. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold shock induction. Mol Microbiol. 1997;26:321–335. doi: 10.1046/j.1365-2958.1997.5771943.x. [DOI] [PubMed] [Google Scholar]
- 29.Nakashima K, Kanamaru K, Mizuno T, Horikoshi K. A novel member of the cspA family of genes that is induced by cold shock in Escherichia coli. J Bacteriol. 1996;178:2994–2997. doi: 10.1128/jb.178.10.2994-2997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Connor M, Asai T, Squires C L, Dahlberg A E. Enhancement of translation by the downstream box does not involve base pairing of mRNA with the penultimate stem sequence of 16S rRNA. Proc Natl Acad Sci USA. 1999;96:8973–8978. doi: 10.1073/pnas.96.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 33.Sprengart M L, Fuchs E, Porter A G. The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J. 1996;15:665–674. [PMC free article] [PubMed] [Google Scholar]
- 34.Travers A A. Conserved features of coordinately regulated E. coli promoters. Nucleic Acids Res. 1984;12:2605–2618. doi: 10.1093/nar/12.6.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker K A, Atkins C L, Osuna R. Functional determinants of the Escherichia coli fis promoter: roles of −35, −10, and transcription initiation regions in the response to stringent control and growth phase-dependent regulation. J Bacteriol. 1999;181:1269–1280. doi: 10.1128/jb.181.4.1269-1280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang N, Yamanaka K, Inouye M. CspI, ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J Bacteriol. 1999;181:1603–1609. doi: 10.1128/jb.181.5.1603-1609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamanaka K, Inouye M. Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J Bacteriol. 1997;179:5126–5130. doi: 10.1128/jb.179.16.5126-5130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamanaka K, Fang L, Inouye M. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]
- 39.Yamanaka K, Mitani T, Ogura T, Niki H, Hiraga S. Cloning, sequencing and characterization of a mukB mutation in Escherichia coli. Mol Microbiol. 1994;13:301–312. doi: 10.1111/j.1365-2958.1994.tb00424.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang A, Rimsky S, Reaban M E, Buc H, Belfort M. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]