Abstract
In Pseudomonas syringae strains, the hrp-hrc pathogenicity island consists of an HrpL-dependent regulon that encodes a type III protein translocation complex and translocated effector proteins required for pathogenesis. HrpR and HrpS function as positive regulatory factors for the hrpL promoter, but their mechanism of action has not been established. Both HrpR and HrpS are structurally related to enhancer-binding proteins, but they lack receiver domains and do not appear to require a cognate protein kinase for activity. hrpR and hrpS were shown to be expressed as an operon: a promoter was identified 5′ to hrpR, and reverse transcriptase PCR detected the presence of an hrpRS transcript. The hrpR promoter and coding sequence were conserved among P. syringae strains. The coding sequences for hrpR and hrpS were cloned into compatible expression vectors, and their activities were monitored in Escherichia coli transformants carrying an hrpL′-lacZ fusion. HrpS could function as a weak activator of the hrpL promoter, but the activity was only 2.5% of the activity detected when both HrpR and HrpS were expressed in the reporter strain. This finding is consistent with a requirement for both HrpR and HrpS in the activation of the hrpL promoter. By using a yeast two-hybrid assay, an interaction between HrpR and HrpS was detected, suggestive of the formation of a heteromeric complex. Physical interaction of HrpR and HrpS was confirmed by column-binding experiments. The results show that HrpR and HrpS physically interact to regulate the ς54-dependent hrpL promoter in P. syringae strains.
Pseudomonas syringae is a causal agent of leaf blights and related diseases in many plant species (19). When introduced into tissue of a susceptible plant, the bacterium colonizes the intercellular spaces of parenchymatous tissue, remaining external to plant cell walls. Colonizing bacteria produce extracellular polysaccharides, derivatized peptide toxins, and plant hormones that lead to altered ion fluxes across cellular membranes and slowly developing tissue necroses typical of leaf blights. Although P. syringae is capable of causing disease in most economically important plant species, a single strain usually causes disease only in a specific subset of plant species or in some cases specific genetic lines of a single plant species. When introduced into plants other than the susceptible host, a P. syringae strain elicits an active defense response that culminates in a rapid programmed cell death. This programmed cell death, also known as the hypersensitive response, and the associated defense responses prevent further colonization of the tissue and are thought to be major factors in the determination of a strain's host range (8, 23).
The colonization of plant tissue and elicitation of active defense responses by P. syringae strains have both been linked to a pathogenicity island (PAI) called the hrp gene cluster (for a recent review, see reference 8). The P. syringae hrp gene cluster encodes a type III protein export complex (PEC) similar to those encoded by PAIs of mammalian pathogens (8, 22, 24), such as Yersinia, Salmonella, Shigella, enteropathogenic and enterohemorrhagic Escherichia coli, and P. aeruginosa strains and, more recently, Chlamydia spp. (11, 20) and Bordetella pertussis (66). The gene products conserved among all known type III PECs are present in the hrp gene cluster and are required for activity. Like other type III PECs, the hrp-encoded type III PEC functions in the translocation of effector proteins into the cytoplasm of host cells (13, 39, 43), most likely through the HrpA pilus (57).
Effector proteins translocated by the hrp-encoded type III PEC are postulated to function as pathogenicity determinants to allow colonization of susceptible plant hosts (5, 26, 46). Some of these translocated effector proteins, identified as products of avr (avirulence) genes, can also elicit the aforementioned active defense responses in resistant plants through a host protein-mediated recognition process (see references in references23, 32, and 39). In addition, differential regulation of the hrp regulon in P. syringae strains may also affect host range by altering when pathogenicity and host range factors are secreted during pathogenesis.
Like many other type III PECs, expression of most P. syringae hrp and hrc genes is environmentally regulated. hrp, hrc, and avr expression is low during growth in most rich media containing broad-spectrum amino acid sources and is induced during pathogenesis or by culture in an acidic minimal salts medium (45, 56, 65). The acidic minimal salts medium is thought to mimic conditions found in planta. It is unclear at present whether hrp and avr genes are regulated by host cell contact similar to that postulated to occur during pathogenesis by Yersinia spp. (45, 48) or whether they are regulated via nutritional or physiological signals related to the growth conditions.
Several transcriptional factors that mediate the environmental regulation of the P. syringae hrp, hrc, and avr genes have been identified. The primary transcriptional factor controlling expression of most hrp, hrc, and avr genes is the alternative sigma factor HrpL (63), a member of the extracytoplasmic function (ECF) family of sigma factors (34). An HrpL-dependent promoter consensus sequence that is present in all known HrpL-dependent promoters (24) was identified (64) and is a required cis-acting element associated with transcription initiation (25, 49, 51). The operons carrying the hrp and hrc genes, which encode structural components of the type III PEC, as well as the genes for secreted effector proteins, such as the avr and hop genes, form the hrp regulon, which is dependent upon HrpL for expression (24). Related sigma factors controlling the type III PEC have been identified in Erwinia strains carrying closely related group I hrp clusters (HrpL) (59) and in Bordetella (Trs) (66).
Because HrpL is the primary transcription factor controlling expression of hrp regulon genes, regulation of hrpL transcription may in part control the environmental regulation of the hrp regulon. HrpR and HrpS have been reported to be positively acting regulators of hrpL expression (63). Both HrpR and HrpS are unusual members of the enhancer-binding family of proteins (9, 15, 16, 63) that normally function as response regulators of two-component regulatory systems (41). Most enhancer-binding proteins are typically modular, consisting of a large regulatory receiver (AB) domain, a central domain (C) involved in the interaction with ς54, and an enhancer- or upstream activating sequence-binding domain (D) (37, 41). Similar to other enhancer-binding proteins, HrpR and HrpS retain the ς54 interaction (C) and DNA binding domains (D) (37, 40). HrpR and HrpS differ from most enhancer-binding factors that function in two-component regulatory systems by the apparent absence of a receiver domain that functions in phosphorylation-dependent modulation of response regulator activity (see reference 52). Thus, HrpR and HrpS are similar to the stress response regulator PspF, which also lacks these domains (27, 28).
The mechanism by which HrpR and HrpS regulate hrpL promoter activity has not been established. Xiao et al. (63) reported that hrpL promoter activity in Escherichia coli transformants was dependent upon the expression of both hrpR and hrpS and suggested that an interaction between the two proteins may be required to activate expression of the ς54-dependent hrpL promoter. Grimm et al. (15) reported that hrpS expressed from a plasmid-borne construct could rescue the ability of an hrpR::Tn5 mutant of P. syringae NPS3121 to elicit the hypersensitive response in tobacco leaves. An apparent hrpS transcript was detected that appeared to initiate near a minimal ς54 promoter consensus sequence internal to the hrpR coding sequence. HrpS was thus proposed to function independently of HrpR to activate expression of the hrp regulon in P. syringae strains (15). Other bacteria carrying closely related group I hrp PAIs found in Erwinia strains carry an apparent HrpS homolog (30, 58) but not an HrpR homolog. Since other aspects of type III secretion in Erwinia strains appear to be similar to those of P. syringae, the role of hrpR in the regulation of group I hrp PAIs is unclear (22, 58).
The purpose of our experiments was to elucidate the role of HrpR and HrpS in the regulation of the P. syringae hrp regulon. Here we report that hrpR and hrpS are expressed as a single operon and that the gene products function together as a positive transcriptional factor for the hrpL promoter. For maximal activity of the hrpL promoter, both HrpR and HrpS were required. Physical interaction of HrpR and HrpS was detected by yeast two-hybrid analysis and confirmed biochemically in column-binding experiments. The results indicate that HrpR and HrpS form a stable heteromeric complex to regulate the ς54-dependent hrpL promoter in P. syringae strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains, plasmids, and primers used in this work are listed in Table 1. E. coli strains were grown at 37°C in King's B broth (1) unless otherwise noted. P. syringae strains were grown in King's B broth or M63 minimal salts medium supplemented with glucose, fructose, and/or 1% Casamino Acids as indicated in the text. Yeast (Saccharomyces cerevisiae) strains were maintained on defined media (7). The following antibiotics were included where indicated below at the indicated concentrations (in micrograms per milliliter): ampicillin, 200; kanamycin, 50; spectinomycin, 100; tetracycline, 25; and nalidixic acid, 50.
TABLE 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| P. syringae | ||
| DC3000 | Tomato and Arabidopsis pathogen | 60 |
| Pss61 | Weak bean pathogen | 2 |
| E. coli | ||
| DH5α | (rK− mK−) recA1 relA1 Δ(argF-lacZYA)U169 φ80dlacZDM15 | Invitrogen BRL |
| MC4100 | F′ Δ(argF-lacZYA)U169 | 4 |
| S. cerevisiae L40 | MATahis3D200 trp1-901 leu2-3, 112 ade2 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ gal80 | S. Hollenberg |
| Plasmids | ||
| pBTM116 | oriColE1ori2μm TRP1 Padh::LexA′-BD::mcs | 3 |
| pBTM-lamin | Lamin gene cloned into pBTM116 to create a LexA BD-lamin fusion | 3 |
| pDRR1R | 690-bp BstYI fragment cloned into pRG970 to create PhrpR-lacZ | This report |
| pDSK519 | incQ Knr | 29 |
| pDSK600 | incQ Spr, triple lacUV5 promoter, mcs | 38 |
| pDWR3BTM | 0.9-kb PCR product cloned into pBTM116, LexA′-′HrpR | This report |
| pDWR3GAD | 0.9-kb PCR product cloned into pGAD424, GAL4′-′HrpR | This report |
| pDWS4BTM | 0.9-kb PCR product cloned into pBTM116, LexA′-′HrpS | This report |
| pDWS4GAD | 0.9-kb PCR product cloned into pGAD424, GAL4′-′HrpS | This report |
| pFLAG-CTC | C-terminal FLAG epitope expression vector | Sigma |
| pGAD424 | oriColE1ori2μ-m LEU2 Padh::GAL4′-AD::mcs | 3 |
| pHIR11 | hrp PAI from Pss61 cloned into pLAFR3 | 21 |
| pJBR6R | 697-bp PCR product amplified from DC3000 and cloned into pRG970 to create PhrpR-lacZ | This report |
| pJBR7R | 1,087-bp PCR product amplified from DC3000 and cloned into pRG970 to create PhrpS-lacZ | This report |
| pLAFR3 | incP-1 Tcr cosmid vector | 54 |
| pMLB1034 | pBR322 derivative carrying ′lacZ for constructing translational fusions | 53 |
| pNTRS3D | 2-kb PCR product amplified from Pss61 genomic DNA using Pwo polymerase and ligated as an XbaI-HindIII fragment in to pDSK519 | This work |
| pQE30 | N-terminal six-His expression vector | Qiagen |
| pREP4 | Supplies LacI, Knr | Qiagen |
| pRG970 | incP Spr, promoterless ′lacZYA for constructing transcriptional fusions | 55 |
| pSGL4MS | 340-bp fragment from pYXL1R cloned into pMLB1034 as a BamHI-EcoRI fragment, PhrpL-lacZ | This report |
| pSHS23Q30 | 0.9-kb PCR product cloned into pQE30, Plac–six-His–hrpS | This report |
| pSJR2L | PCR-amplified 1-kb fragment carrying hrpR cloned into pLAFR3, Plac-hrpR | This report |
| pSJS3DS | PCR-amplified 1-kb fragment carrying hrpS cloned into pDSK600 as an XbaI-BamHI fragment, PlacUV5-hrpS | This report |
| pSJR2S3L | hrpS fragment from pSJS3DS cloned into pSJR2L as a BamHI-HindIII fragment, Plac-hrpRΔ30bp hrpS | This report |
| pTSR4GAD | 899-bp PCR product cloned into pGAD424, GAL4′-′HrpRΔN | This report |
| pTSR4R | 1,087-bp PCR product amplified from Pss61 cloned into pRG970 to create PhrpS-lacZ | This report |
| pTSR5GAD | 730-bp PCR product cloned into pGAD424, GAL4′-′HrpRΔC′ | This report |
| pTSR6GAD | 691-bp PCR product cloned into pGAD424, GAL4′-′HrpRΔNΔC′ | This report |
| pTSR8CTC | 0.9-kb PCR product cloned into pFLAG-CTC, Plac-hrpR-FLAG fusion | This report |
| pYXL1R | 340-bp PCR product cloned into pRG970, PhrpL-lacZ | 63 |
| pYXRS1B | BamHI-BglII fragment carrying hrpRS cloned into pBluescript SK(+) | 63 |
| pYXRS1D | BamHI-BglII fragment carrying hrpRS cloned into pDSK519 | 63 |
| Primers | ||
| DC715 | CGGATATCGACCGCTTTGCCAGTATCC | |
| DC985 | CGGATATCTCCGCTTGCCACCCACCA | |
| DC1412 | CGGGATCCTGATGACCCGCTGATAATGC | |
| DC2045 | CGGGATCCCTCGTCCAGATCATCCTCAA | |
| DC24200 | GGAGGCGATCACGCAGATA | |
| DC24901 | CAGCAAGGGGAAGCCGAG | |
| DC25619 | GATAAGGACGTCCGAGAGTG | |
| P23740 | GCGCAAGCTTTCAGATGCTCAATTCCTTGATGCGA | |
| P24200 | AGCCGACGCAATCACACAG | |
| P24607 | GCAGATCTCATCCAGGTCATCGTCAAAC | |
| P24634 | GATGATGAGTTTGACCATGA | |
| P24640 | GCGGATCCAATCTCGATGATGAGTTTGACGA | |
| P24692 | GCGAATTCGGACTCGGCTGCCAGGTC | |
| P24901 | TCACAGCAACGGCAATCCGAGG | |
| P25591 | GTAACTGCATTATCGGCTGG | |
| P25630R | CCTCAAGCTTAGCACAGACATTGATAACGA | |
| P25677R | TGTCCCGGGCGTGGTATTAATATTCCGCCT | |
| RC | GCGGATCCTCAGGACTCGGCTGCCAGGTCGA | |
| RN | GCGAATTCAGCACAGACATTGATAACGACG | |
| RΔC | GCGGATCCCAGCAACGGCAATCCGAGGAC | |
| RΔN | GCGAATTCGTAACTGCATTATCGGCTGGT | |
| SC | GCGGATCCTCAGATGCTCAATTCCTTGAT | |
| SN | GCGAATTCAATCTCGATGATGAGTTTGAC |
General DNA manipulations.
Restriction enzymes were purchased from Invitrogen BRL (Bethesda, Md.), and T4 DNA ligase was purchased from New England Biolabs (Beverly, Mass.) and used according to the manufacturer's recommendations. Basic manipulations were done using standard procedures (50). PCRs were performed using a Hybaid PCRSprint thermal cycler with 50-μl reaction volumes. Unless indicated otherwise, Pwo polymerase (Boehringer Mannheim) was used for amplifying fragments for cloning.
Construction of hrpR′-lacZ and hrpS′-lacZ promoter fusions.
The hrpR promoter region was isolated from pYXRS1B as a 690-bp BstYI fragment (Fig. 1). This fragment was cloned into BamHI-digested pRG970 to create pDRR1R. The fusion of the hrpR promoter to the vector's promoterless ′lacZYA cassette was confirmed by sequence analysis. The potential hrpS promoter was amplified from pHIR11 using the tailed primers P24607 and P25677R. The resulting 1,087-bp fragment encompassed the region extending from 44 bp 5′ of the hrpR coding sequence to 34 bp inside the hrpS coding sequence. The BglII- and XmaI-digested fragment was ligated into BamHI- and XmaI-digested pRG970 to construct pTSR4R. Inserts were confirmed by sequence analysis. To construct the equivalent constructs using DC3000 sequences, the primers DC715 and DC1412 (R6 fragment) and DC985 and DC2045 (R7 fragment) were used to amplify fragments from DC3000 genomic DNA that were subsequently ligated into SmaI- and BamHI-digested pRG970 as EcoRV-BamHI fragments to create pJBR6R carrying the hrpR promoter and pJBR7R carrying the putative hrpS promoter.
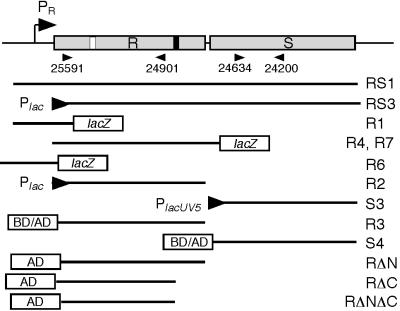
FIG. 1.
Features of the hrpRS region and primers and constructs used in experiments. A map of the hrpRS region is shown at the top. Shaded boxes represent deduced coding sequences for hrpR (R) and hrpS (S) (9, 63). The location of the hrpR promoter (Pr) is shown by the bent arrow. The HrpR box (white bar) and hrpS promoter (black bar) are positioned as proposed by Grimm et al. (15). The locations and orientations of the primers used in the RT-PCR experiments are indicated by the labeled arrowheads immediately below the map. Fragments used in other experiments are indicated by the labeled lines that represent the portion of the mapped region carried by the fragment. Promoter fusions are shown by the arrowheads, with the left side representing the left end of the fragment. Transcriptional fusions to lacZ are indicated by the labeled box. Translational fusions constructed to the LexA′ BD or GAL4′ AD are also shown. Not shown is the S23 fragment that is equivalent to the S4 fragment except that it has tails with different restriction sites; this fragment was used to construct a His-tagged derivative as described in Materials and Methods. The R8 fragment is equivalent to the R3 fragment but lacks the C-terminal stop codon and was used to make a FLAG-tagged derivative. Designations for plasmids carrying the above-described fragments represent the constructor's initials, the fragment designation shown above, and a suffix indicating the host vector. Vector codes are as follows: B, pBluescript SK+; BTM, pBTM116; CTC, pFLAG-CTC; D, pDSK519; DS, pDSK600; GAD, pGAD424; L, pLAFR3; MS, pMLB1034; Q30, pQE30; R, pRG970.
RNA extraction from P. syringae cells.
RNA was isolated from cells grown for 3 h in M63 medium (pH 5.5) containing fructose as the carbon source using hot Trizol (Gibco BRL) extraction as recommended by the manufacturer.
RT-PCR.
Extracted RNA was DNase I treated and used for cDNA synthesis with Superscript II reverse transcriptase (RT; Gibco BRL) and with the P24200 primer by using the manufacturer's protocol for DNA synthesis from high-GC-content RNA templates. A control reaction lacking RT (−RT) was run in parallel with cDNA synthesis. The cDNA preparation and the −RT control were treated with RNase H and used as templates for PCR. The PCR amplification, using Taq polymerase (Gibco BRL) and the primers indicated below, involved 35 cycles of 94°C for 10 s, 50°C for 10 s, and a 1.5-min extension at 72°C followed by a 4-min fill-in reaction at 72°C. Primers used to amplify Pss61 sequences were P24200, P24634, P24901, and P25591 (Fig. 1). For DC3000, the corresponding primer sequences were DC24200, DC24901, and DC25619.
β-Galactosidase assays.
β-Galactosidase activity in bacterial cells was estimated by the procedures of Miller (36).
Yeast two-hybrid analysis.
′hrpR (codons 2 to 314) was amplified by PCR using the tailed primers RN and RC from pYXRS1D. Similarly, ′hrpS (codons 2 to 302) was amplified using the tailed primers SN and SC. The resulting fragments were cloned as EcoRI-BamHI fragments into pBTM116 to create pDWR3BTM (LexA′-HrpR fusion) and pDWS4BTM (LexA′-HrpS fusion) or into pGAD424 to generate pDWR3GAD (GAL4′-HrpR fusion) and pDWS4GAD (GAL4′-HrpS fusion). Translational fusions to the LexA′ binding domain (BD) of pBTM116 or the GAL4′ activating domain (AD) of pGAD424 were confirmed by sequence analysis. Truncated derivatives of hrpR were constructed using the primers RΔN and RΔC and corresponding RN or RC primers. The resulting constructs were transformed into S. cerevisiae L40 singly or in combination with pBTM-lamin, pGAD424, and the corresponding pBTM116 or pGAD424 construct by using the Li acetate-polyethylene glycol one-step transformation protocol (12). Transformants were selected on defined media by complementation of Trp and/or Leu auxotrophy. The resulting transformants were initially screened for β-galactosidase activity by filter lift assay employing liquid N2-lysed cells floated on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-containing phosphate buffer (7). β-Galactosidase activity in liquid N2-lysed cells was quantitatively estimated by the procedures of Clark et al. (7).
Construction of epitope-tagged HrpR and HrpS.
To construct an N-terminal His-tagged HrpS, a fragment carrying ′hrpS (codons 2 to 302) was amplified using the primers P24640 and P23740 and Pwo polymerase. The resulting fragment was cloned into BamHI- and HindIII-digested pQE30 to create pSHS23Q30. To create a C-terminal FLAG-tagged HrpR, hrpR′ (codons 1 to 313) was amplified using the primers P25630R and P24692 and Pwo polymerase. The hrpR fragment was ligated as a HindIII-EcoRI fragment into pFLAG-CTC to generate pTSR8CTC.
Column binding assay.
DH5α(pREP4)(pSHS23Q30) or DH5α(pTSR8CTC) cultures (optical density at 600 nm [OD600] = 0.5) were induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and grown for an additional 5 h. Cells from 50-ml cultures were harvested and stored at −20°C until use. Frozen cells were thawed on ice in 5 ml of lysis buffer (50 mM NaHPO4 [pH 8.0], 100 mM KCl, 10 mM imidazole), and 5 mg of lysozyme was added. After 30 min of incubation, cells were lysed by sonication and fractionated by centrifugation at 10,000 × g for 30 min. The supernatant was collected and clarified by centrifugation in a microcentrifuge for 10 min at 4°C.
The clarified lysate of DH5α(pREP4)(pSHS23Q30) was mixed with 1.5 ml of Ni-nitrilotriacetic acid (NTA) slurry (Qiagen, Valencia, Calif.) and incubated for 2 h at 4°C with shaking. Resin was collected in a 6-ml polypropylene column and washed with 2 column volumes of lysis buffer, 2 column volumes of wash buffer (lysis buffer supplemented with 20 mM imidazole), and 1 column volume of lysis buffer. The clarified lysate of DH5α(pTSR8CTC) was applied to the column, and the column was washed with 1 column volume of lysis buffer, two column volumes of wash buffer, and two column volumes of lysis buffer. Bound proteins were eluted in elution buffer (lysis buffer supplemented with 250 mM imidazole).
Immunoblotting.
Proteins in samples were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12% polyacrylamide gels. Proteins were electroblotted onto polyvinylidene difluoride membranes in Tris-glycine buffer (pH 8.3) containing 20% methanol. Membranes were then blocked with 5% dry milk in phosphate-buffered saline (PBS)–0.05% Tween 20 (PBST) and incubated with anti-His antibody (Bio-Rad, Hercules, Calif.) and/or anti-FLAG M2 antibody (Sigma) in 3% bovine serum albumin in PBST for 1 h at room temperature. Membranes were washed three times in PBST and incubated with a 1:3,000 dilution of anti-mouse immunoglobulin G-horseradish peroxidase conjugant (Bio-Rad) in 5% dry milk in PBST for 1 h at room temperature. Membranes were washed three times in PBST and once in PBS, and immunoreactive proteins were detected by using an enhanced-chemiluminescence detection kit (Amersham Pharmacia Biotech).
RESULTS
hrpR and hrpS are expressed as an operon.
To determine whether promoters were associated with hrpR and/or hrpS, fragments 5′ to the hrpR and to the hrpS coding sequences from P. syringae Pss61 (R1 and R4) (Fig. 1) or P. syringae DC3000 (R6 and R7) genomic DNA were cloned into the low-copy-number plasmid pRG970 (Table 1) as described in Materials and Methods to create transcriptional fusions to ′lacZYA. The resulting constructs were confirmed by sequence analysis and transformed into P. syringae Pss61 or P. syringae DC3000. Cells were assayed for β-galactosidase activity during mid-log-phase growth in the inductive M63 fructose medium. Promoter activity was detected from the R1 construct carrying 259 bp upstream of the hrpR coding sequence, irrespective of the host bacterium (Table 2). Strains carrying the R1 construct exhibited >30-fold more β-galactosidase activity than the background. In contrast, little promoter activity was detected from the 1,070-bp R4 and R7 constructs, which include the predicted HrpR-dependent regulatory site (HrpR box) and potential hrpS promoter (HrpS box) (15), the hrpR-hrpS intergenic region, and the coding sequence for the first 13 amino acids (aa) of hrpS. β-Galactosidase levels in strains carrying these constructs expressed less than 40 Miller units of β-galactosidase activity. Similar results were obtained when these constructs were tested in E. coli MC4100. The R4 construct exhibited minimal if any promoter activity, as was true for the R7 construct (data not shown).
TABLE 2.
Activities of hrpR, hrpS, and hrpL promoter constructs in P. syringae Pss61, P. syringae DC3000, and E. coli MC4100
| Strain | hrpRS expresseda | β-Galactosidase activity from promoter fusionb:
|
|||
|---|---|---|---|---|---|
| Nonec | hrpRd | hrpSe | hrpLf | ||
| Pss61 | − | 3 ± 1 | 417 ± 16 | 31 ± 1 | 348 ± 16 |
| + | 3 ± 1 | 615 ± 38 | 40 ± 1 | 3,469 ± 184 | |
| DC3000 | − | 4 ± 1 | 126 ± 2 | 7 ± 3 | 57 ± 4 |
| + | 6 ± 1 | 147 ± 4 | 9 ± 1 | 216 ± 8 | |
| MC4100 | − | 1 ± 1 | 439 ± 23 | 6 ± 1 | 5 ± 1 |
| + | 1 ± 1 | 364 ± 19 | 5 ± 1 | 271 ± 5 | |
The indicated strains carried pDSK519 (−) or pNTR3D to ectopically express hrpRS (+).
Strains carrying the indicated reporter construct were grown overnight to an OD600 of 1.0, harvested, washed and used to inoculate inductive M63 fructose (pH 5.5) medium for P. syringae strains or King's B medium for E. coli MC4100. After 6 h of growth, β-galactosidase activity was determined by the procedures of Miller (36). The data reported are the means of values from a single experiment done in triplicate. Errors represent standard deviations. Each experiment was repeated at least three times with similar results.
Indicated strain carrying pRG970.
Pss61 carrying pDRR1R, DC3000 carrying pJBR6R, or MC4100 carrying pDRR1R.
Pss61 carrying pTSR4R, DC3000 carrying pJBR7R, or MC4100 carrying pTSR4R.
Indicated strain carrying pYXL1R.
Although the activity of the R1 construct indicates that hrpR was expressed in P. syringae strains under the conditions employed in the preceding experiments, additional experiments were performed using a plasmid-borne hrpR expression system. Consistent with previous results (63), vector-directed expression of hrpRS in Pss61 or MC4100 caused at least a 45-fold increase in hrpL promoter activity (Table 2), showing that the hrpRS expression system was functioning under the experimental conditions employed. Promoter activity of neither the hrpR promoter carried by the R1 construct nor the putative hrpS promoter included in the R4 construct, however, was substantially affected by the presence of the hrpRS expression system (Table 2). Levels of activity were similar irrespective of the presence of the hrpRS construct. Similar results were obtained when an hrpR construct was employed (data not shown). The absence of promoter activity in strains carrying the R4 construct even in the presence of expressed hrpR argues that the postulated HrpR-dependent hrpS promoter may be only weakly active in Pss61 and DC3000 and is inactive in E. coli strains, irrespective of the expression of hrpR.
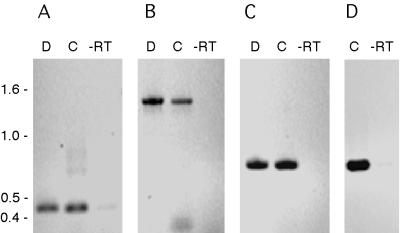
To determine if a transcript extends from hrpR into hrpS, RT-PCR was performed on total RNA extracted from P. syringae Pss61 cells. cDNA synthesis was initiated using the P24200 primer, which is complementary to a specific sequence internal to the hrpS coding sequence (Fig. 1). As shown in Fig. 2A, a 0.44-kb fragment was amplified from the P24200-primed cDNA preparation by using hrpS-specific primers. This product was identical in size to the expected product amplified from the native DNA template and absent from the control reaction mixture using just the DNase I-treated RNA preparation as a template (−RT). This PCR product confirms that cDNA synthesis occurred and that the RNA preparation was largely free of genomic DNA. When primers were employed to amplify a region encompassing both hrpR and hrpS, the predicted 1.4-kb fragment indicative of an hrpRS transcript was amplified from the same cDNA preparation (Fig. 2B). Since each of the preceding PCRs employed a primer that was also used in the cDNA synthesis, a third PCR was performed to confirm the apparent transcriptional linkage of hrpR and hrpS. The cDNA preparation generated using the hrpS P24200 primer was used as a template for PCR employing the P25591 and P24901 primers, specific to hrpR. As shown in Fig. 2C, a 0.7-kb fragment that was indistinguishable from the expected 0.7-kb fragment amplified from genomic DNA was amplified from the cDNA preparation but was absent from the −RT control. Similar results were obtained during parallel experiments with RNA extracted from P. syringae strain DC3000. A 0.7-kb hrpR fragment could also be amplified from the cDNA preparation generated from DNase-treated DC3000 RNA and DC3000-specific primers equivalent to P24200, P24901, and P25591 (Fig. 2D). The ability to detect the 1.4- and 0.7-kb products after RT-PCR of total RNA extracts of P. syringae indicates that a transcript that contains the coding sequences for both hrpR and hrpS is produced by these P. syringae strains.
FIG. 2.
RT-PCR of the hrpRS transcript. Total RNA was extracted using Trizol and precipitated, and 2 μg was treated with DNase I. The DNase I-treated RNA preparation was used for cDNA synthesis using the P24200 primer (Fig. 1) and for a −RT control. Genomic DNA (lanes D), the cDNA preparation (lanes C), and the −RT control (lanes −RT) were used as templates for PCR employing primers P24200 and P24634 to amplify an hrpS region (A), primers P24200 and P25591 to amplify an hrpRS fragment (B), or primers P24901 and P25591 to amplify an hrpR fragment (C). Panel D is similar to panel C except that DC3000 RNA and DC3000-specific primers were employed. Molecular sizes shown on the left in kilobases were estimated using a 1-kb ladder obtained from Invitrogen.
Conservation of the hrpRS region in P. syringae strains.
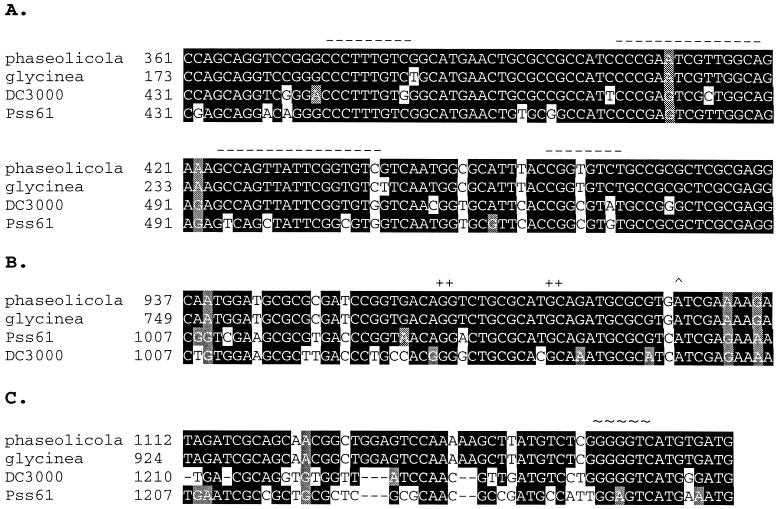
To determine if the hrpRS regulatory sequences might be unique to the strains examined above, the nucleotide sequences of the hrpRS regions of several P. syringae strains were compared. Within the region carried by the R4 construct, the Pss61 hrpR coding sequence exhibited 84% identity at the nucleotide level with the P. syringae pv. phaseolicola hrpR sequence (data not shown). For comparison, the hrpS coding sequence retained 81% identity. The postulated HrpR box (Fig. 3A) and the predicted hrpS promoter regions (Fig. 3B) were also conserved in all strains examined. Highest divergence was detected in the noncoding intergenic region between hrpR and hrpS (Fig. 3C). This 45- to 50-bp region, although large for an intergenic region, lacked motifs known to function as transcriptional terminators. The retention of major features of the region and the absence of significant sequence divergence in the region argue that the means of regulating and expressing hrpRS are likely to be similar in all P. syringae strains, irrespective of their host range.
FIG. 3.
Conservation of the hrpR promoter and coding sequence. Sequences were aligned using the CLUSTAL W version 1.8 program (61) and data from GenBank accession numbers AF2322004, U03853, U03852, X77638, and AF069650. (A) Proposed HrpR-responsive hrpS promoter element. The dashed lines represent the HrpR box proposed by Grimm et al. (15). (B) Postulated ς54-dependent hrpS promoter element. Conserved promoter elements (+) and the transcriptional initiation site (^) identified by Grimm et al. (15) are shown. (C) hrpRS intergenic region. The deduced ribosome binding site for hrpS is indicated by the ∼ symbols.
Maximal activation of the hrpL promoter requires both HrpR and HrpS.
As demonstrated above, HrpR and HrpS function as positively acting regulatory factors for the hrpL promoter. To determine whether HrpR or HrpS could function as an independent activator of the hrpL promoter, coding sequences for hrpR and hrpS together with their native ribosome binding sites were amplified from pHIR11 by PCR and cloned individually into the IncQ plasmid pDSK600 (pSJS3DS) (Fig. 1) or the IncP-1 plasmid pLAFR3 (pSJR2L) such that the cloned genes were expressed from vector Plac promoters (Fig. 1). To reconstruct the hrpRS operon from the individually cloned fragments and inactivate a potential open reading frame identified on the opposite strand, the hrpS fragment was ligated as a 1-kb fragment into pSJR2L to create pSJR2S3L. The resulting construct carried a 30-bp deletion in the intergenic region between hrpR and hrpS but retained the native ribosome binding site for hrpS. The above-named constructs were then transformed individually or in combination into MC4100(pSGL4MS) carrying a PhrpL-lacZ fusion and assayed for β-galactosidase activity.
The hrpL promoter construct carried by pSGL4MS was inactive in MC4100 in the absence of hrpRS (Table 3) as observed above. Vector-directed expression of hrpR by the pSJR2L construct alone had no detectable effect on the activity of the hrpL promoter. Consistent with the observations of Grimm et al. (15), expression of hrpS partially activated hrpL promoter activity. MC4100(pSGL4MS)(pSJS3DS) expressing just hrpS, however, accumulated only 83 Miller units of β-galactosidase activity during logarithmic growth. Significantly higher levels of hrpL promoter activity were detected in strains expressing (i) hrpR and hrpS from separate plasmids [MC4100(pSGL4MS)(pSJR3L)(pSJS2D)], (ii) the reconstructed hrpRS operon [MC4100 (pSGL4MS)(pSJR3S2L)], and (iii) hrpRS as a native construct [MC4100 (pSGL4MS)(pYXRS1D)]. Transformants expressing hrpR and hrpS in trans on separate plasmids expressed greater than 4,700 Miller units of β-galactosidase activity. In these constructs, the proposed hrpS promoter and regulatory sequences were physically separated from the hrpS coding sequence. The reconstructed hrpRS operon produced a similar level of activation. Transformants carrying the native RS construct exhibited greater than 13,000 Miller units of activity. As these activities were all more than 37-fold higher than the activity induced by hrpS alone, these results indicate that maximal activation of the hrpL promoter requires expression of both hrpR and hrpS.
TABLE 3.
Activation of the hrpL promoter in E. coli MC4100 by cloned hrpR and hrpS
| MC4100 (pSGL4MS) transformant plasmid(s)a | Expressed gene(s) | Promoter activityb |
|---|---|---|
| pDSK519 | None | 1 ± 1 |
| pYXRS1D | Native hrpRS | 13,162 ± 745 |
| pSJR2L | hrpR | 1 ± 1 |
| pSJS3DS | hrpS | 83 ± 9 |
| pSJR2L, pSJS3DS | hrpR + hrpS in trans | 4,725 ± 524 |
| pSJR2S3L | Reconstructed hrpRS operon | 3,077 ± 111 |
The indicated plasmids are described in Table 1.
Promoter activity as estimated by β-galactosidase activity (in Miller units) expressed in strains carrying the indicated plasmids as described in Materials and Methods. Values are means ± standard deviations.
Physical interaction of HrpR and HrpS is detected by yeast two-hybrid studies.
One possible interpretation for the requirement of both HrpR and HrpS in the activation of the hrpL promoter is that the two proteins physically interact. A yeast two-hybrid assay (3) was used to determine if HrpR and HrpS could interact. Fragments carrying codons 2 to 314 of hrpR and codons 2 to 302 of hrpS (Fig. 1) were amplified by PCR from pHIR11 and cloned as EcoRI-BamHI fragments into pBTM116 and pGAD424 to create translational fusions with the LexA′ DNA BD or the GAL4′ AD carried by each respective plasmid. The resulting constructs were verified by sequencing and transformed into the yeast reporter strain L40.
Yeast L40 transformants carrying either the LexA′ BD-HrpR or LexA′ BD-HrpS fusion alone did not exhibit a β-galactosidase-positive phenotype, indicating that neither protein alone could activate the reporter construct in yeast (Table 4). None of the HrpR or HrpS fusions exhibited an interaction with the GAL4′ AD expressed by pGAD424 or with a LexA′-lamin fusion routinely used to detect nonspecific interactions (3). A weak interaction was detected in strains carrying both HrpS constructs, but this activity was about 10% of the activity observed when both translational fusions were expressed in the yeast indicator strain. Strong interactions were detected when HrpR and HrpS fusions were expressed in the same strain, irrespective of the fusion domain. Lysed colony lifts floated on X-Gal solutions exhibited a positive phenotype within 15 min, and quantitative analyses detected relatively high levels of β-galactosidase activity. Twenty-five to 40 U of β-galactosidase activity was routinely detected in strains carrying these constructs.
TABLE 4.
Interaction of HrpR and HrpS detected by yeast two-hybrid analysis
| L40 transformant plasmid(s) | LexA′-BD fusion | GAL4′-AD fusion | β-Galactosidase activitya |
|---|---|---|---|
| pDWR3BTM | HrpR | Absent | <1 |
| pDWS4BTM | HrpS | Absent | <1 |
| pBTM-lamin, pDWR3GAD | Lamin | HrpR | <1 |
| pBTM-lamin, pDWS4GAD | Lamin | HrpS | <1 |
| pDWR3BTM, pGAD424 | HrpR | None | <1 |
| pDWS4BTM, pGAD424 | HrpS | None | 1.4 ± 0.6 |
| pDWR3BTM, pDWR3GAD | HrpR | HrpR | <1 |
| pDWS4BTM, pDWS4GAD | HrpS | HrpS | 3 ± 0.3 |
| pDWR3BTM, pDWS4GAD | HrpR | HrpS | 29 ± 10 |
| pDWS4BTM, pDWR3GAD | HrpS | HrpR | 28 ± 10 |
| pDWS4BTM, pTSR4GAD | HrpS | HrpRΔN | 26 ± 3 |
| pDWS4BTM, pTSR5GAD | HrpS | HrpRΔC | 1.4 ± 0.5 |
| pDWS4BTM, pTSR6GAD | HrpS | HrpRΔNΔC | 1.0 ± 0.2 |
The indicated S. cerevisiae L40 transformants were grown overnight at 25°C in defined medium lacking Leu and/or Trp (as required to maintain selection for plasmids) to an OD600 of 1.0. Cells were harvested and lysed by three freezing cycles in liquid N2, and β-galactosidase activity was determined as indicated in Materials and Methods.
To determine whether a specific domain was involved in the interaction between HrpR and HrpS, truncated derivatives of HrpR lacking the 15 N-terminal aa and/or the 70 C-terminal aa (includes the DNA BD [37]) were constructed in pGAD424. Such constructs have been stable in other systems (47). The resulting constructs were transformed into the yeast reporter strain carrying the LexA BD′-HrpS construct in pBTM116. Yeast transformants carrying the HrpR derivative lacking just the 15 N-terminal amino acids (RΔN) exhibited a β-galactosidase-positive phenotype (Table 3), whereas the derivatives lacking the C terminus (RΔC, RΔNΔC) exhibited little or no β-galactosidase activity. These results suggest that a C-terminal domain may control the interaction between HrpR and HrpS or that the C-terminal deletion renders the fusion protein unstable.
Copurification of His-tagged HrpS and FLAG-tagged HrpR in column-binding experiments.
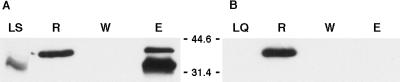
The requirement for both hrpR and hrpS in the activation of the hrpL promoter and the yeast two-hybrid experiments are highly suggestive of a physical interaction between HrpR and HrpS. To confirm the apparent interaction of HrpR and HrpS, column-binding experiments were performed. An N-terminal six-His fusion to HrpS was constructed in pQE30 (pSHS23Q30), and a C-terminal FLAG-tagged HrpR was constructed in pFLAG-CTC (pTSR8CTC) (Fig. 1). The His-tagged HrpS was collected from lysates of DH5α(pREP4)(pSHS23Q30) on Ni+-agarose matrix, washed, and allowed to interact with clarified lysates of DH5α(pTSR8CTC) containing the C-terminal FLAG-tagged HrpR. The matrix was washed, and bound proteins were eluted using 250 mM imidazole. As shown in Fig. 4, both His-tagged HrpS, which migrated as a 34-kDa protein, and FLAG-tagged HrpR, detected as a 40-kDa protein, were eluted from the column with the imidazole wash. Although the apparent 40-kDa mass of HrpR was larger than expected for the FLAG-tagged derivative (35 kDa), His-tagged HrpR also migrated as a 40-kDa protein (data not shown). The reason for this aberrant behavior of HrpR during SDS-PAGE has not been established. FLAG-tagged HrpR did not bind to an Ni+-agarose matrix loaded with lysates of DH5α(pREP4)(pQE30). The copurification of HrpS and HrpR under these conditions is consistent with the strong physical interaction detected in the yeast two-hybrid analyses.
FIG. 4.
Interaction of HrpR and HrpS detected by column-binding experiments. Clarified lysate from an IPTG-induced DH5α(pREP4)(pSHS23Q30) culture expressing HrpS was loaded onto an Ni+-nitrilotriacetic acid column (Qiagen), and the column was washed as described in Materials and Methods to release weakly bound proteins. A clarified lysate from an IPTG-induced DH5α(pTSR8CTC) culture was then applied to the column, and the column was washed with sufficient buffer to produce an HrpR-free elutant. Bound proteins were eluted using 250 mM imidazole in lysis buffer. Proteins in the indicated samples were fractionated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and probed using anti-His and anti-FLAG antibodies simultaneously as described in Materials and Methods. (A) HrpR eluted from an HrpS-loaded column. Lane LS, DH5α(pREP4)(pSHS23Q30) lysate applied to the column (contrast enhanced); lane R, DH5α(pTSR8CTC) lysate applied to the column; lane W, final wash prior to elution; lane E; imidazole-eluted proteins. (B) HrpR binding to an E. coli protein-loaded column. Lane LQ, DH5α(pREP4)(pQE30) lysate applied to the column; lane R, DH5α(pTSR8CTC) lysate; lane W, final wash; lane E, imidazole-eluted proteins. Molecular masses shown in kilodaltons were estimated using Kaleidoscope prestained markers (Bio-Rad).
DISCUSSION
The hrp-encoded type III PEC is central to the pathogenicity of P. syringae strains. Although the characterization of the regulatory system controlling assembly of the hrp-encoded PEC is still incomplete, it is clear that expression of hrp genes in P. syringae is coordinated by the activity of HrpL. HrpL is an alternative sigma factor required for transcription of the operons encoding structural elements of the PEC as well as the genes for the secreted effector proteins (24). As the only factor currently thought to affect HrpL activity is protein turnover, expression of hrpL is likely to be critical to the assembly of the hrp-encoded PEC of P. syringae. The results presented above indicate that the expression of hrpL is controlled in part at the transcriptional level by the interaction of two unusual enhancer-binding proteins, HrpR and HrpS.
HrpR and HrpS retain most of the structural features conserved in other members of the enhancer-binding protein family that function in transcriptional regulation of ς54-dependent promoters (37, 40). Consistent with these features, HrpR and HrpS activated the ς54-dependent hrpL promoter (63). This promoter contains a ς54 promoter consensus sequence (63), and transcription of hrpL initiates 12 bp downstream of this promoter motif (S. Heu and S. Hutcheson, unpublished results). hrpL expression in P. syringae pv. maculicola was recently reported to be dependent upon rpoN (17, 18).
In contrast to other known enhancer-binding proteins, both HrpR and HrpS were required for maximal activation of the hrpL promoter. HrpS expressed from a strong promoter on a multicopy plasmid could function only as a weak activator of hrpL promoter activity. This activity was less than 2.5% of the activity detected when both hrpR and hrpS were expressed in a cell, irrespective of the promoter construct used to drive expression. As the proposed HrpR-linked hrpS regulatory sequences internal to hrpR (15) were physically separated from hrpS in these experiments, it appears unlikely that HrpR directly influences transcription of hrpS in these constructs. The simplest explanation for these results is that both proteins are required to fully activate the hrpL promoter. The observation that HrpS can act as a weak activator of the hrpL promoter provides an explanation for the reported plant response-positive phenotype of a P. syringae hrpR mutant carrying an hrpS expression construct (15). Relatively little hrp expression appears to be necessary to assemble the hrp-encoded PEC (65). Ectopic expression of hrpS would have induced at least some expression of the hrp regulon and thus allowed the hrp-encoded PEC to be assembled.
Consistent with the requirement for both proteins in the activation of hrpL expression, hrpR and hrpS were shown to be expressed as an operon. The only fragment from the hrpRS region with significant promoter activity was 5′ to hrpR, and a transcript encompassing both hrpR and hrpS was detected by RT-PCR analysis. Although some sequence divergence was detected in the hrpRS region, most involved silent codon substitutions. The conservation of the hrpRS region argues that hrpR and hrpS are transcribed as an operon in all P. syringae strains. Regulation of hrpRS expression in distinct strains, however, may be different, as sequences upstream of the hrpR promoter were not well conserved.
A requirement for both HrpR and HrpS in the activation of the hrpL promoter may indicate that HrpR either activates HrpS or forms a stable complex with HrpS. In either model, HrpR and HrpS would be expected to physically interact. The yeast two-hybrid experiments demonstrated that a physical interaction between HrpR and HrpS can occur. This apparent strong interaction was confirmed in column-binding experiments. A FLAG-tagged HrpR derivative in crude cell lysates was retained by immobilized His-tagged HrpS. This complex formed under the relatively high-salt conditions (100 mM KCl) of the lysis buffer and was stable during washes exceeding 5 column volumes. Another enhancer-binding protein, NtrC, has been proposed to form a homodimer that upon phosphorylation assembles into a larger oligomeric activator complex (62). Dimerization involves the C terminus of the protein (31). The ability of HrpR and HrpS to form a stable complex during column-binding experiments in the absence of a target promoter suggests that these proteins form a heteromeric complex prior to activation of the hrpL promoter. The yeast two-hybrid assay results suggest that the formation of this complex may involve the C-terminal domain of HrpR, as reported for NtrC.
HrpR and HrpS lack the 130-aa receiver (AB) domain that is typically found in most other members of the protein family (37, 41). The receiver domain has been proposed to be a repressor of ATP hydrolysis in the absence of kinase-mediated phosphorylation or binding of a regulatory effector molecule (52). The absence of the receiver domains argues that HrpR and HrpS do not require posttranslational modification, such as phosphorylation or the binding of an effector molecule, to activate the target promoter. Consistent with this hypothesis, vector-directed expression of hrpRS as a minimal coding sequence produced a functional activator complex in E. coli transformants. HrpR and HrpS are thus functionally similar to E. coli PspF (27, 28) or truncated derivatives of DctD (33) and XylR (42). These proteins lack the AB receiver domain and are also constitutively active. The activity of HrpR and HrpS thus appears to be independent of a direct posttranslational modification mechanism, such as phosphorylation, but posttranslational modification by a broadly conserved mechanism cannot be fully excluded at this time.
Regulation of the P. syringae hrp PAI shares some similarities to the regulatory system controlling flagellar biosynthesis. Flagellar biosynthesis has been proposed to be a form of a type III PEC, and three classes of promoters have been identified for genes involved in the assembly of flagella (6). At the top of the regulatory system is the class 1 promoter for flhCD. FlhC and FlhD, once expressed, interact to form an FlhD-FlhC complex that then activates expression of class 2 promoters. FliA, expressed from a class 2 promoter, functions as an alternative sigma factor to direct expression of class 3 promoters. Like FlhD-FlhC, HrpR and HrpS are expressed as an operon and form a complex. There is, however, little if any sequence similarity between FlhD-FlhC and HrpR-HrpS. At present the only known target for the HrpR-HrpS complex appears to be the hrpL promoter, but other HrpR-HrpS-dependent promoters may exist in cells. HrpL is a sigma factor related to FliA that directs expression of the HrpL-dependent regulon. The HrpL-dependent promoters are analogous to the class 3 promoters of flagellar biosynthesis. Although the HrpR- or HrpS-HrpL regulatory system is superficially similar to the FlhD- or FlhC-FliA regulatory system, the genes controlled at each level of these regulatory systems are distinct. In flagellar biosynthesis, the genes encoding the type III PEC are considered to be class 2 operons, although there is some influence of FliA on their expression (35), whereas the hrp counterparts could be considered to be equivalent to the class 3 operons.
The HrpR-HrpS regulatory system also shares some similarity to the RcsB-RcsA system regulating capsular biosynthesis in several bacterial species (14). RcsB interacts with RcsA to regulate cps expression. RcsB is part of a two-component regulatory system involving RcsC. The RcsB-RcsC system can activate low-level expression of the cps genes but acts synergistically with RcsA. RcsA is present at limiting levels in which RcsA levels are regulated by turnover mediated by Lon protease. HrpS appears to be able to activate low-level expression of the hrp regulon but requires HrpR for maximal activity. A similar situation occurs in the regulation of Erwinia amylovora hrp genes. The Erwinia HrpS can initiate expression of the hrpL promoter but requires HrpX for maximal activity (57). HrpX is an enhancer-binding protein that is part of a classic two-component regulatory system involving a phosphorelay. As mentioned above, there is no evidence at present to indicate that HrpR or HrpS functions as part of a two-component regulatory system, and in contrast to rcsA and rcsBC, hrpR and hrpS are expressed as an operon.
Unresolved at present is the mechanism by which environmental signals generated during pathogenesis are transduced to alter hrp expression. The proposed regulatory system appears to represent a regulatory cascade in which expression of the Hrp regulon may be controlled by the expression of hrpRS in a manner analogous to that of PspF in the regulation of stress genes in E. coli (28) and flhCD in flagellar biosynthesis (6). hrpS transcript levels have been reported to be repressed in DC3000 during growth in repressive media (57). Other results suggest that hrpRS expression is constitutive in several strains because hrpRS transcripts could be detected by primer extension and RT-PCR, irrespective of the growth conditions (J. Bretz and S. Hutcheson, unpublished results). Interestingly, significant differences in hrpR promoters were observed between P. syringae strains as described above. This opens the possibility of strain-specific regulation of hrpRS expression. In contrast to the hrpR promoter, the hrpL promoter was observed to be environmentally regulated (24). This finding argues that additional factors must mediate the environmental regulation of the hrp cluster in addition to HrpR-HrpS and HrpL. One such candidate is HrpV, which has been reported to function as a negative regulator of hrp expression (44), and preliminary results suggest that it functions analogously to PspA (10) in the regulation of HrpR-HrpS activity (T. Sussan, X. Wei, and S. Hutcheson, unpublished results).
ACKNOWLEDGMENTS
This work was supported by grant MCB9729524 from the National Science Foundation.
The assistance of Sunggi Heu, Don Weaver, and Dan Rowley in the construction of several of the plasmids used in these experiments and the advice of Rick Stewart and Lisa Simpson in the yeast two-hybrid experiments are gratefully acknowledged.
REFERENCES
- 1.Atlas R M. Handbook of microbiological media. Boca Raton, Fla: CRC Press; 1993. [Google Scholar]
- 2.Baker C J, Atkinson M M, Collmer A. Concurrent loss in Tn5 mutants of the ability to induce the HR and host plasma membrane K+/H+ exchange in tobacco. Phytopathology. 1987;77:1268–1272. [Google Scholar]
- 3.Bartel P I, Chien C T, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley D A, editor. Cellular interactions in development: a practical approach. Oxford, United Kingdom: Oxford University Press; 1994. pp. 153–179. [Google Scholar]
- 4.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z, Kloek A P, Boch J, Katagiri F, Kunkel B N. The Pseudomonas syringae avrRpt2 gene product promotes pathogen virulence from inside plant cells. Mol Plant-Microbe Interact. 2000;13:1312–1321. doi: 10.1094/MPMI.2000.13.12.1312. [DOI] [PubMed] [Google Scholar]
- 6.Chilcott G S, Hughes K T. Coupling flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark K L, Larsen P B, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA. 1998;95:5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collmer A, Badel J L, Charkowski A O, Deng W-L, Fouts D E, Ramos A R, Rehm A H, Anderson D M, Schneewind O, van Dijk K, Alfano J R. Pseudomonas syringae Hrp type III secretion system and effector proteins. Proc Natl Acad Sci USA. 2000;97:8770–8777. doi: 10.1073/pnas.97.16.8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng W L, Preston G, Collmer A, Chang C-J, Huang H-C. Characterization of the hrpC and hrpRS operons of Pseudomonas syringae pathovars syringae, tomato, and glycinea and analysis of the ability of hrpF, hrpG, hrcC, hrpT, and hrpV mutants to elicit the hypersensitive response and disease in plants. J Bacteriol. 1998;180:4523–4531. doi: 10.1128/jb.180.17.4523-4531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dworkin J, Jovanovic G, Model P. The PspA protein of Escherichia coli is a negative regulator of ς54-dependent transcription. J Bacteriol. 2000;182:311–319. doi: 10.1128/jb.182.2.311-319.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields K A, Hackstadt T. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol Microbiol. 2000;38:1048–1060. doi: 10.1046/j.1365-2958.2000.02212.x. [DOI] [PubMed] [Google Scholar]
- 12.Gietz R D, Woods R A. High efficiency transformation in yeast. In: Johnson J A, editor. Molecular genetics of yeast: practical approaches. Oxford, United Kingdom: Oxford University Press; 1994. pp. 121–134. [Google Scholar]
- 13.Gopalan S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottesman S. Regulation of capsule synthesis: modification of the two-component paradigm by an accessory unstable regulator. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 253–262. [Google Scholar]
- 15.Grimm C, Aufsatz W, Panopoulos N J. The hrpRS locus of Pseudomonas syringae pv. phaseolicola constitutes a complex regulatory unit. Mol Microbiol. 1995;15:155–165. doi: 10.1111/j.1365-2958.1995.tb02230.x. [DOI] [PubMed] [Google Scholar]
- 16.Grimm C, Panopoulos N J. The predicted protein product of a pathogenicity locus from Pseudomonas syringae pv. phaseolicola is homologous to a highly conserved domain of several prokaryotic regulatory proteins. J Bacteriol. 1989;171:5031–5038. doi: 10.1128/jb.171.9.5031-5038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrickson E L, Guevera P, Ausubel F M. The alternative sigma factor RpoN is required for hrp activity in Pseudomonas syringae pv. maculicola and acts at the level of hrpL transcription. J Bacteriol. 2000;182:3508–3516. doi: 10.1128/jb.182.12.3508-3516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrickson E L, Guevera P, Penaloza-Vazquez A, Shao J, Bender C, Ausubel F M. Virulence of the phytopathogen Pseudomonas syringae pv. maculicola is rpoN dependent. J Bacteriol. 2000;182:3498–3507. doi: 10.1128/jb.182.12.3498-3507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirano S S, Upper C D. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae: a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev. 2000;64:624–653. doi: 10.1128/mmbr.64.3.624-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsia R, Pannekoek Y, Ingerowski E, Bavoil P. Type III secretion genes identify a putative virulence locus of Chlamydia. Mol Microbiol. 1997;25:351–359. doi: 10.1046/j.1365-2958.1997.4701834.x. [DOI] [PubMed] [Google Scholar]
- 21.Huang H C, Schuurink R, Denny T P, Atkinson M M, Baker C J, Yucel I, Hutcheson S W, Collmer A. Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco. J Bacteriol. 1988;170:4748–4756. doi: 10.1128/jb.170.10.4748-4756.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutcheson S W. Currents concepts of active defense in plants. Annu Rev Phytopathol. 1998;36:59–90. doi: 10.1146/annurev.phyto.36.1.59. [DOI] [PubMed] [Google Scholar]
- 24.Hutcheson S W. The hrp cluster of Pseudomonas syringae: a pathogenicity island encoding a type III protein translocation complex? In: Kaper J B, Harker J, editors. Pathogenicity islands and other mobile virulence elements. Washington, D.C.: American Society for Microbiology; 1999. pp. 309–329. [Google Scholar]
- 25.Innes R W, Bent A F, Kunkel B N, Bisgrove S R, Staskawicz B J. Molecular analysis of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syringae avirulence genes. J Bacteriol. 1993;175:4859–4869. doi: 10.1128/jb.175.15.4859-4869.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson R W, Athanassopoulos E, Tsiamis G, Mansfield J W, Sema A, Arnold D L, Gibbon M J, Murillo J, Taylor J D, Vivian A. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pv. phaseolicola. Proc Natl Acad Sci USA. 1999;96:10875–10880. doi: 10.1073/pnas.96.19.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jovanovic G, Dworkin J, Model P. Autogenous control of PspF, a constitutively active enhancer-binding protein of Escherichia coli. J Bacteriol. 1997;179:5232–5237. doi: 10.1128/jb.179.16.5232-5237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jovanovic G, Weiner L, Model P. Identification, nucleotide sequence, and characterization of PspF, the transcriptional activator of the Escherichia coli stress-induced psp operon. J Bacteriol. 1996;178:1936–1945. doi: 10.1128/jb.178.7.1936-1945.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keen N, Tamaki J, Kobayashi D, Trollinger D. Improved broad host range plasmids for DNA cloning in gram negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 30.Kim J F, Ham J H, Bauer D W, Collmer A, Beer S V. The hrcC and hrpN operons of Erwinia chrysanthemi EC16 are flanked by plcA and homologs of hemolysin/adhesion genes and accompanying activator/transporter genes. Mol Plant-Microbe Interact. 1998;11:563–567. doi: 10.1094/MPMI.1998.11.6.563. [DOI] [PubMed] [Google Scholar]
- 31.Klose K E, North A, Stedman K M, Kustu S. The major dimerization determinants of the nitrogen regulatory protein NtrC from enteric bacteria lies at its carboxy-terminal domain. J Mol Biol. 1994;241:233–245. doi: 10.1006/jmbi.1994.1492. [DOI] [PubMed] [Google Scholar]
- 32.Leach J E, White F F. Bacterial avirulence genes. Annu Rev Phytopathol. 1996;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]
- 33.Lee J L, Scholl D, Nixon B T, Hoover T. Constitutive ATP hydrolysis and transcription activation by a stable, truncated form of Rhizobium meliloti DctD, a ς54-dependent transcriptional activator. J Biol Chem. 1994;269:20401–20409. [PubMed] [Google Scholar]
- 34.Lonetto M A, Brown K L, Rudd K E, Bittner M J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase ς factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 36.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1971. [Google Scholar]
- 37.Morett E, Segovia L. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murillo J, Shen H, Gerhold D, Sharma A, Cooksey D A, Keen N T. Characterization of pPT23B, the plasmid involved in syringolide production by Pseudomonas syringae pv. tomato PT23. Plasmid. 1994;31:275–287. doi: 10.1006/plas.1994.1029. [DOI] [PubMed] [Google Scholar]
- 39.Nimchuk Z, Marios E, Kjemtrup S, Leister R, Katagiri F, Dangl J. Eukaryotic fatty acid acylation drives plasma membrane targeting and function of several type III effector proteins from Pseudomonas syringae. Cell. 2000;101:353–363. doi: 10.1016/s0092-8674(00)80846-6. [DOI] [PubMed] [Google Scholar]
- 40.Osuna J, Soberon X, Morett E. A proposed architecture for the central domain of the bacterial enhancer-binding proteins based on secondary structure prediction and fold recognition. Protein Sci. 1997;6:543–555. doi: 10.1002/pro.5560060304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 42.Periz-Martin J, deLorenzo V. The N-terminal domain of the prokaryotic enhancer-binding protein XylR is a specific intramolecular repressor. Proc Natl Acad Sci USA. 1995;92:9392–9396. doi: 10.1073/pnas.92.20.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pirhonen M U, Lidell M C, Rowley D, Lee S W, Silverstone S, Liang Y, Keen N T, Hutcheson S W. Phenotypic expression of Pseudomonas syringae avr genes in E. coli is linked to the activities of the hrp-encoded secretion system. Mol Plant-Microbe Interact. 1996;9:252–260. doi: 10.1094/mpmi-9-0252. [DOI] [PubMed] [Google Scholar]
- 44.Preston G, Deng W-L, Huang H-C, Collmer A. Negative regulation of hrp genes in Pseudomonas syringae by HrpV. J Bacteriol. 1998;180:4532–4537. doi: 10.1128/jb.180.17.4532-4537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahme L G, Mindronos M N, Panopoulos N J. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1992;174:3499–3507. doi: 10.1128/jb.174.11.3499-3507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ritter C, Dangl J L. The avrRpm1 gene of Pseudomonas syringae pv. maculicola is required for virulence on Arabidopsis. Mol Plant-Microbe Interact. 1995;8:444–453. doi: 10.1094/mpmi-8-0444. [DOI] [PubMed] [Google Scholar]
- 47.Ronson C W, Nixon B T, Ausubel F M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987;49:579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- 48.Rosqvist R, Magnusson K E, Wolf-Watz H. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salmeron J M, Staskawicz B J. Molecular characterization and hrp-dependence of the avirulence gene avrPto from Pseudomonas syringae pv. tomato. Mol Gen Genet. 1993;239:6–10. doi: 10.1007/BF00281595. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 51.Shen H, Keen N T. Characterization of the promoter of avirulence gene D from Pseudomonas syringae pv. tomato. J Bacteriol. 1993;175:5916–5924. doi: 10.1128/jb.175.18.5916-5924.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shingler V. Signal sensing by ς54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–416. doi: 10.1046/j.1365-2958.1996.388920.x. [DOI] [PubMed] [Google Scholar]
- 53.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 54.Staskawicz B J, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van den Eede G, Deblaere R, Goethals K, Van Montagu M, Holsters M. Broad host range and promoter selection vectors for bacteria that interact with plants. Mol Plant-Microbe Interact. 1992;5:228–234. doi: 10.1094/mpmi-5-228. [DOI] [PubMed] [Google Scholar]
- 56.van Dijk K, Fouts D E, Rehm A H, Hill A R, Collmer A, Alfano J R. The Avr (effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the Hrp (type III) protein secretion system in a temperature- and pH-sensitive manner. J Bacteriol. 1999;181:4790–4797. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei W, Plovanich-Jones A, Deng W-L, Jin Q-L, Collmer A, Huang H C, He S Y. The gene coding for the Hrp pilus structural protein is required for type III secretion of Hrp and Avr proteins in Pseudomonas syringae pv. tomato. Proc Natl Acad Sci USA. 2000;97:2247–2252. doi: 10.1073/pnas.040570097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei Z, Kim J F, Beer S V. Regulation of hrp genes and type III protein secretion in Erwinia amylovora by HrpX/HrpY, a novel two component system, and HrpS. Mol Plant-Microbe Interact. 2000;13:1251–1262. doi: 10.1094/MPMI.2000.13.11.1251. [DOI] [PubMed] [Google Scholar]
- 59.Wei Z M, Beer S V. hrpL activates Erwinia amylovora hrp gene transcription and is a member of the ECF subfamily of sigma factors. J Bacteriol. 1995;177:6201–6210. doi: 10.1128/jb.177.21.6201-6210.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whalen M C, Innes R W, Bent A F, Staskawicz B J. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Aribidopsis and soybean. Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Worley K, Culpepper P, Wiese B, Smith R. BEAUTY-X: enhanced BLAST searches for DNA queries. Bioinformatics. 1998;14:890–891. doi: 10.1093/bioinformatics/14.10.890. [DOI] [PubMed] [Google Scholar]
- 62.Wyman C, Rombel I, North A K, Bustamante C, Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 63.Xiao Y, Heu S, Yi J, Lu Y, Hutcheson S W. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J Bacteriol. 1994;176:1025–1036. doi: 10.1128/jb.176.4.1025-1036.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao Y, Hutcheson S W. A single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae. J Bacteriol. 1994;176:3089–3091. doi: 10.1128/jb.176.10.3089-3091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao Y, Lu Y, Heu S, Hutcheson S W. Organization and environmental regulation of the Pseudomonas syringae pv. syringae 61 hrp cluster. J Bacteriol. 1992;174:1734–1741. doi: 10.1128/jb.174.6.1734-1741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuk M H, Harvill E T, Cotter P A, Miller J F. Modulation of host immune responses, induction of apoptosis and inhibition of NF-κB activation by Bordetella type III secretion system. Mol Microbiol. 2000;35:991–1004. doi: 10.1046/j.1365-2958.2000.01785.x. [DOI] [PubMed] [Google Scholar]