Abstract
Mass spectrometry imaging (MSI) combines molecular and spatial information in a valuable tool for a wide range of applications. Matrix‐assisted laser desorption/ionization (MALDI) is at the forefront of MSI ionization due to its wide availability and increasing improvement in spatial resolution and analysis speed. However, ionization suppression, low concentrations, and endogenous and methodological interferences cause visualization problems for certain molecules. Chemical derivatization (CD) has proven a viable solution to these issues when applied in mass spectrometry platforms. Chemical tagging of target analytes with larger, precharged moieties aids ionization efficiency and removes analytes from areas of potential isobaric interferences. Here, we address the application of CD on tissue samples for MSI analysis, termed on‐tissue chemical derivatization (OTCD). MALDI MSI will remain the focus platform due to its popularity, however, alternative ionization techniques such as liquid extraction surface analysis and desorption electrospray ionization will also be recognized. OTCD reagent selection, application, and optimization methods will be discussed in detail. MSI with OTCD is a powerful tool to study the spatial distribution of poorly ionizable molecules within tissues. Most importantly, the use of OTCD−MSI facilitates the analysis of previously inaccessible biologically relevant molecules through the adaptation of existing CD methods. Though further experimental optimization steps are necessary, the benefits of this technique are extensive.
Keywords: chemical derivatization, mass spectrometry imaging, matrix‐assisted laser desorption ionization
Abbreviations
- CD
chemical derivatization
- DESI
desorption electrospray ionization
- LC‐MS
liquid chromatography‐mass spectrometry
- LESA
liquid extraction surface analysis
- MALDI
matrix‐assisted laser desorption ionization
- MS
mass spectrometry
- MSI
mass spectrometry imaging
- OTCD
on‐tissue chemical derivatization
- SIMS
secondary ion mass spectrometry
1. INTRODUCTION
Mass spectrometry imaging (MSI) is a powerful technique that allows untargeted assessment of analytes and their spatial distribution within the sample structure. Thousands of molecules, such as metabolites (Smith et al., 2020), lipids (Wu et al., 2016), peptides (Dilillo et al., 2017), and glycans (Holst et al., 2016) can be imaged in a single experiment without labeling. Spatial distributions of ions are visualized across sample sections, as unique mass spectra are generated from each individual pixel. Inks (Ifa et al., 2007), paint (Liu et al., 2010), synthetic fibers (van Geenen et al., 2017), and fingerprints (Francese et al., 2013) have been analyzed using MSI, along with an extensive list of biological tissues: Heart (Mezger et al., 2019), brain (Shariatgorji et al., 2014, 2015), kidney (Kuo et al., 2019), liver (Flinders et al., 2018), testes (Cobice et al., 2016), three‐dimensional (3D) cell cultures (Liu & Hummon, 2016), “living skin equivalents” (Russo et al., 2018), plant roots and seeds (Dueñas et al., 2019), and hair (Beasley et al., 2016).
MSI is a multi‐step process involving sample preparation, analyte desorption and ionization, mass analysis, and image coregistration to produce spatial molecular maps. To analyze the abundance and distribution of molecules directly from tissue sections both successfully and accurately, a reproducible and robust workflow is essential (Figure 1). Variations and fluctuations in the preparation or technical aspects can have detrimental effects at later stages of MSI throughput, leading to erroneous or misleading data (Goodwin et al., 2012). Briefly, tissue sections are prepared based on the optimal conditions for the target analyte, ionization method, and instrumentation. Molecules are ionized and desorbed from the surface and directed into the mass analyzer (Zimmerman et al., 2008). With ionization from known positions across a sample, collated mass spectral information can be used to generate distribution data from the molecules detected. The resulting ion distributions are typically presented as two‐dimensional ion images showing the relative abundance of selected mass‐to‐charge ratios. The sample‐processing pipeline for MSI, offers multiple stages for optimization and modification, as differing resolving power can lead to a range in complexity of data sets, corresponding to the type of analyzer selected. Consideration of spatial and spectral resolution for targeted or nontargeted (broadband) analysis needs to be addressed in preparation for MSI studies, as achieving high spatial resolution can mean sacrificing high spectral resolution.
Figure 1.
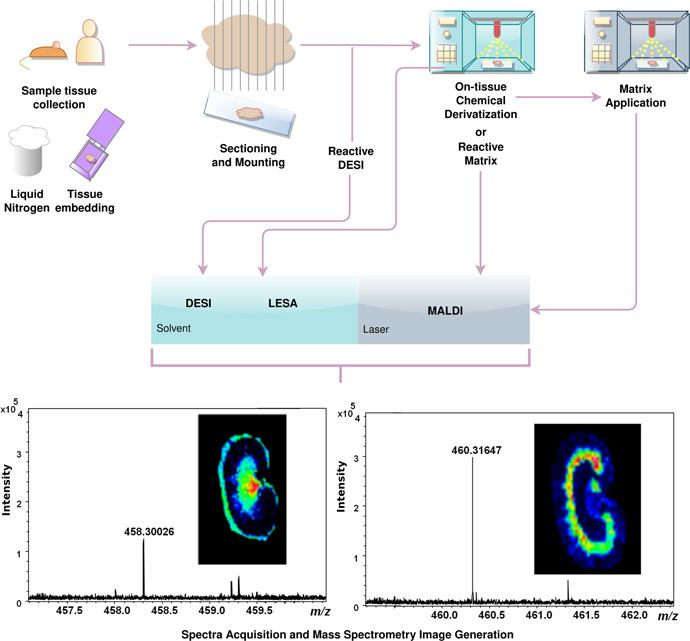
Mass spectrometry (MS) imaging workflow including methods of OTCD across MALDI, LESA, and DESI platforms. Following sample tissue collection, thin sections are mounted onto glass slides. From here, OTCD and analysis by reactive DESI do not require further preparation. In LESA, MALDI, and nonreactive DESI platforms, the derivatization reagent is sprayed directly onto the tissue. For MALDI, if the derivatization reagent also serves as a matrix (reactive matrix), no additional step is required. Alternatively, matrix must also be applied to the slide before analysis. Once sample preparation is complete, ionization and MS analysis are carried out by the appropriate instrument. DESI, desorption electrospray ionization; LESA, liquid extraction surface analysis; MALDI, matrix‐assisted laser desorption/ionization; OTCD, on‐tissue chemical derivatization [Color figure can be viewed at wileyonlinelibrary.com]
Critically, to effectively detect analytes directly from tissue sections by MSI, the molecule of interest must be ionized efficiently and must be present at concentrations relevant to MSI. Not all molecules are amenable to ionization, however, due to the absence of chargeable moieties or possessing intrinsically low ionization efficiency. In matrix‐assisted laser desorption/ionization (MALDI), prudent matrix selection and solvent composition can help achieve the desired sensitivity, nevertheless, some molecules still present a challenge and prove increasingly difficult to detect by mass spectrometry (MS). In desorption electrospray ionization (DESI), modifications to the solvent composition, such as organic solvent content (Green et al., 2010) and introducing surfactants to reduce surface tension (Badu‐Tawiah & Cooks, 2010) have been shown to improve ionization efficiency.
Similarly, in liquid extraction surface analysis (LESA) ionization, alterations to the solvent composition and solvent dwell time can increase ion production yields. For example, in LESA MSI analysis of drugs in liver tissue, an acetonitrile‐based solvent was found to increase signal intensity when compared to isopropanol and methanol‐based solutions. In addition, the signal increased in tandem with the solvent on‐tissue dwell time from 1 to 3 s, but fell at 4 and 5 s (Swales et al., 2016). In secondary ionization mass spectrometry (SIMS), initially a “hard” ionization technique, the use of softer ion clusters for penetration and sample ionization such as or Ar+ (Chang et al., 2012) and (Kaya et al., 2018) induce less fragmentation and retain the integrity of large biomolecules. Sample coating with gold or silver has been shown to increase the yield of large organic molecular ions, termed “metal‐assisted‐SIMS or MetA‐SIMS” (Altelaar et al., 2006; Delcorte & Bertrand, 2004; Nygren, Johansson, et al., 2004; Nygren, Malmberg, et al., 2004).
Chemical derivatization (CD) has been extensively used as a strategy for improving the detection of poorly ionizable molecules by liquid chromatography‐mass spectrometry (LC‐MS), for both atmospheric pressure chemical ionization (APCI) and electrospray ionization analysis (ESI) (Higashi et al., 2005; Quirke & van Berkel, 2001; Zaikin & Halket, 2006).
Carefully selected derivatization reagents react with functional groups located in target molecules, increasing ionization efficiency and sensitivity for small molecule detection (Chacon et al., 2011) and minimizing ion suppression, for example by endogenous metabolites or MALDI matrix cluster peaks (Manier et al., 2011). Within the field of MSI, on‐tissue chemical derivatization (OTCD) is still in its infancy but is constantly growing with promising results targeting key biological molecules: amino acids (Manier et al., 2014), hormones (H. Enomoto, Sensu, et al., 2018), N‐glycans (Holst et al., 2016), steroids (Cobice et al., 2013), fatty acids (Wu et al., 2016), neurotransmitters (Shariatgorji et al., 2019), thiols (Fülöp et al., 2020), and vitamin metabolites (Smith et al., 2020). Successful applications in drug localization demonstrate the value of OTCD in the pharmacological field also (Barré et al., 2016; Chacon et al., 2011; Flinders et al., 2015) (Table 1). The number of publications in this area has visibly increased over the last 5 years and is currently on a rising trend (Figure 2). This review focuses on “OTCD” methods for MALDI‐MSI techniques, however, strategies for LESA, DESI, and SIMS MSI will also be mentioned briefly.
Table 1.
Derivatization reagents used for OTCD in MSI applications
| Reagent | Structure | Sample | Target | Platform | Application | Reference |
|---|---|---|---|---|---|---|
| Aliphatic alcohols, phenols, and thiols | ||||||
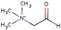
| Betaine aldehyde |
|
Atherosclerotic plaque (human) | Cholesterol | Reactive DESI‐ MSI | Lipidomics | Manicke et al. (2009) |
| Brain (rat) | Cholesterol | Reactive DESI‐ MSI | Lipidomics | C. Wu et al. (2009) | ||
| Adrenal gland (pig) | Cholesterol | Reactive DESI‐ MSI | Lipidomics | Wu et al. (2010) | ||
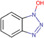
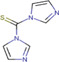
| 1,1‐thiocarbonyldiimidazole (TCDI) |
|
Liver and kidney (mouse) | 3‐methoxysalicylamine(3‐MoSA) (hydroxyl group) | MALDI‐MSI | Pharmacology | Chacon et al. (2011) |
| 2‐fluoro‐1‐methylpyridinium p‐tolunesolfonate (FMTPS) |
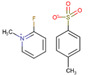
|
Hair (human) | Cannabinoids (hydroxyl group) | MALDI‐MSI | Illicit Drug Use Testing/Forensics | Beasley et al. (2016) |
| (E)‐2‐cyano‐N‐(2‐(2,5‐dioxo‐2,5‐dihydro‐1H‐pyrrol‐1‐yl) ethyl)‐3‐(4‐hydroxyphenyl) acrylamide (CHC‐Mal) |

|
Xenograft tumors (mouse) Liver and Pancreas (pig) |
sulfhydryl groups | MALDI‐MSI (reactive matrix) | Pharmacology | Fülöp et al. (2020) |
| Carboxylic acids | ||||||
| 2‐picolamine (PA) |
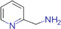
|
Brain (rat) | Fatty acids (carboxyl group) | MALDI‐ MSI | Lipidomics | Q. Wu et al. (2016) |
| Seeds and leaves (maize plant) | Carboxylic acids | MALDI‐MSI | Plant metabolomics | Dueñas et al. (2019) | ||
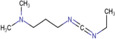
| N,N‐dimethylpiperazine iodide (DMPI) |
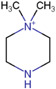
|
Thyroid carcinoma (human) | Fatty acids (carboxyl group) | MALDI‐MSI | Cancer metabolomics | S. S. Wang et al. (2019) |
| N,N,N‐trimethyl‐2‐(piperazin‐1‐yl)ethan‐1‐aminium iodide (TMPA) |
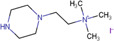
|
Kidney and brain (rat) | Carboxyl‐Containing Metabolites | MALDI‐MSI | Metabolomics | Sun et al. (2020) |
| 4‐(N‐methyl)‐pyridinium boronic acid |
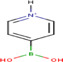
|
Adrenal gland (pig) | Catecholamines |
LDI‐MSI SIMS‐MSI |
Endocrinology | Kaya et al. (2018) |
|
Ethyl esterification and dimethylamidation Two‐step reaction: Step 1: |
|
Leiomyosarcoma and colon carcinoma (human) | N‐glycans (linkage‐specific sialic acid) | MALDI‐MSI | Cancer metabolomics | Holst et al. (2016) |
| ||||||
| Step 2: | ||||||
| ||||||
| Unsaturated systems (alkenes) | ||||||
| Paternò–Büchi reaction |
|
Brain (mouse) | C═C double‐bond (db) positional isomers | MALDI‐MSI | Lipidomics | Bednařík et al. (2018) |
| ||||||
| Brain (mouse) | C═C double‐bond (db) positional isomers | MALDI‐MSI | Lipidomics | Wäldchen et al. (2019) | ||
| Ozone |
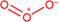
|
Brain (rat) | C═C double‐bond (db) positional isomers | MALDI‐MSI | Lipidomics | Paine et al. (2018) |
|
Brain (mouse) Colon (human) |
C═C double‐bond (db) positional isomers | MALDI‐MSI | Lipidomics | Bednařík et al. (2020) | ||
| meta‐Chloroperoxybenzoic acid (mCPBA) |
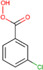
|
Kidney (mouse) Metastatic lung (mouse) |
C═C double‐bond (db) positional isomers | Reactive DESI‐MSI | Lipidomics | Kuo et al., 2019) |
| Amines | ||||||
| p‐N, N, N trimethylammonioanilyl N′‐hydroxysuccinimidyl carbamate iodide (TAHS) |
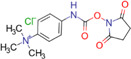
|
Metastatic liver (mouse) | Amino acids | MALDI‐MSI | Cancer metabolomics | Toue et al. (2014) |
| Brain (mouse) | Amino neurotransmitters | MALDI‐MSI | Neuroscience | Esteve et al. (2016) | ||
| Liver (mouse) | Amino acids | MALDI‐MSI | Proteomics | Arts et al. (2017) | ||
| Adrenal gland with Pheochromocytoma (human) | Catecholamine | MALDI ‐MSI | Cancer metabolomics | Takeo et al. (2019) | ||
| 4‐Hydroxy‐3‐methoxycinna‐maldehyde (CA) |

|
Lung (rabbit) | Isoniazid (antituberculosis drug) | MALDI‐MSI | Pharmacology | Manier et al. (2011) |
|
Adrenal gland (pig) Brian (rat) |
Amine metabolites and neurotransmitters | MALDI‐MSI | Proteomics/Neurosciences | Manier et al. (2014) | ||
| Brain (mouse) | Amino neurotransmitters | MALDI‐MSI | Neuroscience | Esteve et al. (2016) | ||
| Brain (rat) | Amino Acids | MALDI‐MSI | Metabolomics | Guo et al. (2019) | ||
| Roots and leaves (maize plant) | Amino acids | MALDI‐MSI | Plant metabolomics | Dueñas et al. (2019) | ||
| Root (maize plant) | Amino acids | MALDI‐MSI | Plant metabolomics | O'Neill and Lee (2020) | ||
| 2,4‐diphenyl‐pyrylium (DPP) |
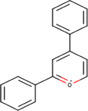
|
Brain (mouse) | Amino neurotransmitters | MALDI‐MSI | Neuroscience | Sugiyama et al. (2019) |
| 2,4‐diphenyl‐pyranylium tetrafluoroborate (DPP‐TFB) |
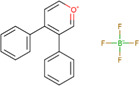
|
Brain (rat, mouse, rhesus monkey) | Amino neurotransmitters | MALDI‐MSI (reactive matrix) | Neuroscience | Shariatgorji et al. (2014) |
| Brain (rat, mouse) | Amino neurotransmitters | MALDI‐MSI (reactive matrix) | Neuroscience | Shariatgorji et al. (2015) | ||
| Brain (rat, mouse) | Amino neurotransmitters | DESI‐MSI | Neuroscience | Shariatgorji et al. (2016) | ||
| Brain (mouse) | Amino neurotransmitters | MALDI‐MSI | Neuroscience | Esteve et al. (2016) | ||
| Brain glioma (mouse) | Amino neurotransmitters/metabolites | MALDI‐MSI | Cancer metabolomics | Dilillo et al., 2017) | ||
| Whole head (Drosophila) | γ‐ aminobutyric acid (GABA) | MALDI‐MSI | Neuroscience | Y. Enomoto, Phan, et al. (2018) | ||
| Brain (crustacean) | Amino neurotransmitters | MALDI‐MSI | Neuroscience | Cao et al. (2019) | ||
| Brain (mouse) | Amino neurotransmitters | DESI‐MSI | Neuroscience | Davison et al. (2019) | ||
| Pyrilium salts (general) |
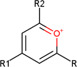
|
Brain (rat, mouse) | Amino neurotransmitters | MALDI‐MSI (reactive matrix) | Neuroscience | Shariatgorji et al. (2015) |
| Fluoromethylpyridinium (FMP) |
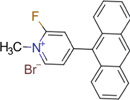
|
Brain (rat, rhesus monkey, human) | Amino transmitters and their metabolites (phenolic hydroxyl groups) | MALDI–MSI (reactive matrix) | Neuroscience | Shariatgorji et al. (2019) |
| 2‐(4‐bromo‐phenyl)‐4,6‐ diphenyl‐pyranylium (Br‐TPP) |
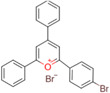
|
Brain (rat, mouse) | Amino neurotransmitters |
MALDI–MSI (reactive matrix) DESI‐MSI |
Neuroscience | Shariatgorji et al. (2020) |
| Mass differential tags for relative and absolute quantification (mTRAQ®)Δ0 |
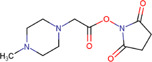
|
Brain (rat) | Amino acids and Neurotransmitters | (MALDI ‐MSI) | Neuroscience | Ito and Hiramoto (2019) |
| N‐succinimidyloxycarbony methyl) tris(2,4,6 trimethoxy phenyl) phosphonium bromide (TMPP) |
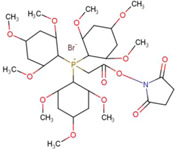
|
Brain (rat) | Peptides and Proteins | MALDI‐MSI | Proteomics | Franck et al. (2009) |
| 3‐Sulfobenzoic acid succinimidyl ester (3‐SBASE) |
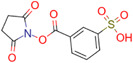
|
|||||
| 4‐Sulfophenyl isothiocyanate (4‐SPITC) |
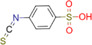
|
|||||
| Ketones/Aldehydes | ||||||
| Girard's reagent T (GT) |
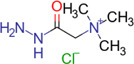
|
Brain (rat, mouse) Adrenal gland (rat, mouse) | Corticsteroids | MALDI‐MSI | Intracrinology | Cobice et al. (2013) |
| Cartilage (human) | Triamcinolone acetonide (osteoarthritis drug) | MALDI‐MSI | Pharmacology | Barré et al. (2016) | ||
| Testes (mouse) | Androgens | MALDI‐MSI | Endocrinology | Cobice et al. (2016) | ||
| Testes (mouse) | Testosterone | MALDI‐MSI | Intracrinology | Shimma et al. (2016) | ||
| Brain (mouse) | Corticsteroids | MALDI‐MSI | Pharmacology | Cobice et al. (2018) | ||
| Bean seeds | Acids | MALDI‐MSI | Plant metabolomics | H. Enomoto, Sensu, et al. (2018) | ||
| Adrenal gland (rat, human) | Corticosteroids | MALDI‐MSI | Intracrinology | Sugiura et al. (2018) | ||
| Brain (rat) | Steroids | MALDI ‐MSI | Intracrinology | Guo et al. (2019) | ||
| Adrenal gland (rat, human) | Steroids | MALDI ‐MSI | Cancer metabolomics | Takeo et al. (2019) | ||
| Girard's reagent P (GP) |
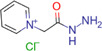
|
Brain (mouse) | Sterols and Oxysterols | MALDI‐MSI | Metabolomics | Yutuc et al. (2020) |
| 2,4‐Dinitrophenylhydrazine (DNPH) |
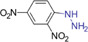
|
Spinal cord (rat) | Malondialdehyde | DESI‐MSI (reactive) | Lipidomics (peroxidation) | Girod et al. (2012) |
| Lung (rat) | Fluticasone proprionate (asthma drug‐steroid) | MADLI‐MSI (Reactive matrix) | Pharmacology | Flinders et al. (2015) | ||
| 4‐dimethylamino‐6‐(4‐methoxy‐1‐naphthyl)‐1,3,5‐triazine‐2‐hydrazine (DMNTH) |
|
Lung (rat) | Fluticasone proprionate (asthma drug‐steroid) | MADLI‐MSI (Reactive matrix) | Pharmacology | Flinders et al. (2015) |
| Platinum‐containing compounds | ||||||
| Diethyldithiocarbamate (DDTC) |
|
Three‐dimensional multicellular tumor spheroids (MCTS) | Oxaliplatin (chemotherapy drug) | MALDI‐MSI | Pharmacology | Liu and Hummon (2016) |
| Vitamin‐D metabolites | ||||||
| Amplifex™ |
|
Kidney (mouse) | Vitamin‐D metabolites |
MALDI‐MSI DESI−MSI |
Metabolomics | Smith et al. (2020) |
Abbreviations: DESI, desorption electrospray ionization; LDI, laser desorption ionization; MALDI, matrix‐assisted laser desorption/ionization; MSI, mass spectrometry imaging; OTCD, on‐tissue chemical derivatization.
Figure 2.
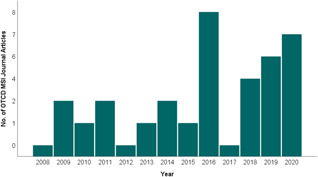
Frequency of OTCD/MSI publications since 2008. This figure is based on the average number of relevant results from the Pubmed and EBSCO (Medline) databases with the following search terms (((On tissue derivatization) AND (On tissue derivatization)) AND (Mass spectrometry)) AND (Imaging)) from years 2008–2020. Review articles were not counted. Only those which described OTCD in tandem with MSI analysis as a method used in their analysis. A sharp increase in 2016 is seen followed by a dip in 2017. However, it can be seen that since 2018, the number of papers, which include OTCD/MSI, is on a rising trend. MSI, mass spectrometry imaging; OTCD, on‐tissue chemical derivatization [Color figure can be viewed at wileyonlinelibrary.com]
2. IONIZATION TECHNIQUES
2.1. Matrix‐assisted laser desorption ionization
Brought to attention in the field of MSI by Caprioli et al. (1997), MALDI has become the most widely used ionization source, overtaking secondary ionization mass spectrometry (Pacholski & Winograd, 1999) due to its availability and capabilities imaging large biomolecules at a high spatial resolution. In practice, frozen sections of tissue are typically thaw mounted onto electro‐conductive slides, commonly coated with indium tin oxide (Figure 1). The slides are then coated with a matrix, which cocrystallizes with sample molecules. Surface ablation (or irradiation) with an ultraviolet (UV) laser (335‐ to 349‐nm wavelength) leads to desorption of charged matrix/analyte clusters. The matrix absorbs the high energy of the laser, preserving the integrity of analyte molecules and aiding ionization. Primary ions are generated from neutral molecules in the samples through photoionization. Secondary ions are generated in the resulting “plume” of acidic matrix ions, typically by proton or electron‐transfer or cationization in the gas phase (Dreisewerd & Hillenkamp, 1995; Dreisewerd, 2003; Zenobi & Knochenmuss, 1998).
The selection and deposition of matrix must be considered for each analysis as it has a significant effect on the quality of results. It must have the ability to absorb the wavelength applied by the laser, be stable under vacuum conditions and ensure ionization of the analyte while preventing cluster formation (Dreisewerd, 2003; Leopold et al., 2018). Commonly used matrices include organic acids such as alpha‐cyano‐4‐hydroxycinnamic acid (CHCA), 2,5‐dihydroxybenzoic acid (DHB), and sinapinic acid (SA), these matrices possess an aromatic ring to absorb ultraviolet energy and an acidic side chain to act as a proton donor and facilitate ionization of the analyte in positive mode. To promote ionization in negative mode, 9‐aminoacridine (9‐AA) and norharmane are commonly used as they have aromatic moieties and an amine group to act as a proton acceptor. To further enhance the efficacy of targeted ionization, reactive matrices have been developed which incorporate CD with matrix within a single molecule, thereby increasing analyte sensitivity and eliminating the need for a matrix application step (Flinders et al., 2015; Fülöp et al., 2020; Kaya et al., 2018; Shariatgorji et al., 2014, 2015, 2019, 2020). This concept is further discussed in Section 8. MALDI remains the most prominent ionization source in OTCD‐MSI as seen in the examples listed in Table 1. However, ambient ionization methods are growing in popularity as improvements in spatial resolution and analysis speed have been made in recent years (Duncan et al., 2017; Yin et al., 2019).
2.2. Desorption electrospray ionization
Developed by the Cooks research laboratory in 2004, DESI offers the advantage of analyzing surfaces under atmospheric conditions in situ, with little‐to‐no sample preparation. Ionization of target analytes is achieved by angling a high velocity charged solvent spray onto the sample surface (Takáts et al., 2004). The solvent is passed through a charged capillary assisted by a nebulizing gas (nitrogen/compressed air/argon) and bombardment of the sample dissolves and accelerates analyte/solvent droplets away from the surface in a “splash” motion. Desorbed ions travel into a mass analyzer via an inlet capillary close to the sample surface. Post‐desorption ionization mechanisms are similar to conventional ESI, in the desolvation of the charged particle droplets, before resulting ions are propelled into the heated inlet of the analyzer by the gas stream (Takáts et al., 2004; Venter et al., 2006; Wiseman et al., 2008). Coulombic repulsion between ions overcomes the surface tension of the droplet, leading to droplet fission. Gaseous ions are formed via direct emission from the droplet surface (ion evaporation model) or by repeated fission until no solvent remains (charge release model) (Cole, 2000; Enke, 1997; Kebarle, 2000).
In contrast to MALDI, where spatial resolution is associated with the size of the focal laser spot, in DESI, data is acquired in line scans, rather than spots, which means the definition of spatial resolution is altered. It can be measured as the distance in which features show an increase in relative signal intensity from 20% to 80% (Luxembourg et al., 2004; Yin et al., 2019). Optimization of geometrical settings such as the angle and distance of the solvent and MS inlet capillaries to the sample surface can serve to drastically improve the spatial resolution and reproducibility of DESI‐MSI experiments (Campbell et al., 2012; Green et al., 2010; Takats et al., 2017). In addition, the solvent capillary itself was identified as a problematic source of variability and was rectified by stabilizing a machine‐tapered fused silica capillary, within a stainless steel gas nozzle, thereby enabling a spatial resolution of 40–60 µm with 30 times faster acquisition time (Tillner et al., 2016, 2017). Solvent choice and adjustment of the aforementioned geometric features can optimize ionization of target analytes (Campbell et al., 2012). DESI also lends itself to OTCD via two routes; first, application of derivatization reagent onto tissue before analysis or incorporation of the derivatization reagent into a solvent delivery system (Cotte‐Rodriguez et al., 2005). The latter is termed “reactive DESI” and is further discussed in Section 8.
The development of nano‐DESI drastically improved the spatial resolution to less than 12 µm in on‐tissue surface sampling (Laskin et al., 2012; Roach et al., 2010). The modification allowed formation of a constant‐flow liquid bridge between two capillaries, which then meets the sample surface. One capillary ensures solvent delivery to the sample, the second carries analytes and solvent to the mass analyzer through generation of a nanoelectrospray. A recent modification includes the addition of a nebulizer to the inlet capillary to improve analyte sensitivity via more complete desolvation and reduced ion suppression (Duncan et al., 2017). A key parameter affecting the efficacy of this technique is the distance between the nano‐DESI probe and the sample surface. Implementation of automated sample platforms (Lanekoff et al., 2012) and, more recently, the use of shear force microscopy serve to maintain a constant probe‐to‐sample distance. Spatial resolutions of better than 10 µm were obtained when shear force microscopy was used to optimize nano‐probe distance (Nguyen et al., 2018; Yin et al., 2018, 2019). Continuing improvements in spatial resolution and analysis speed in DESI and nano‐DESI coupled with minimal sample preparation are increasing their popularity in MSI applications.
2.3. Secondary ion mass spectrometry
SIMS predates both MALDI and DESI for use in imaging applications and due to its nature offers the highest spatial resolution capabilities. However, excessive ion fragmentation limits its functionality in MSI, where detection and identification of intact biomolecules are often desired. A beam of primary ions bombards the sample surface and uses kinetic energy to release surface ions and molecules either singly or in clusters (Altelaar et al., 2005, 2006). High energy at the impact site can cause fragmentation of ionized moieties before entry to the mass analyzer. SIMS offers the advantage of achieving higher spatial resolution (~50–100 nm) than that of MALDI (~20–50 µm), as no matrix is required for analyte ionization, reducing the potential for diffusion and analyte delocalization (Hammond, 2010). As ion beams can be focused with greater precision than lasers, the high spatial resolution of SIMS offers a unique tool for MSI of abundant inorganic elements and small organic molecules in cellular organelles (Altelaar et al., 2007). Furthermore, the depth of SIMS imaging can be refined to nm as opposed to μm layers (Chandra et al., 2000). The wide introduction of polyatomic ion beams such as C60 +, Ar+ (Chang et al., 2012), (CO2)6k + (Kaya et al., 2018), and (H2O) n + cluster ions (n = 500–10,000) has had an even more dramatic effect on molecular SIMS (Sheraz née Rabbani et al., 2015). Not only did these beams increase ion yield, but it was also discovered that, for many materials, but the degree of observable bombardment‐induced damage was also significantly reduced so that analysis could be carried out well beyond the static limit; indeed, in some cases, the whole sample could be sputtered during analysis and the chemical information was not compromised. This was a revolutionary development for molecular SIMS because, in principle, it allowed molecular depth profiling of organic materials (Fletcher & Szakal, 2013). However, the use of SIMS as an ionization source is limited due to most of the instruments being built in‐house at a high expense, which vastly reduces the accessibility of these systems.
2.4. Liquid extraction surface analysis
A recent development in the context of MSI ionization is LESA (Kertesz & van Berkel, 2010), which utilizes the formation of a liquid junction between a conductive pipette tip and the tissue sample surface to extract and ionize analytes of interest. The LESA‐MSI approach does not require matrix, eliminating further sample preparation. The ionization process is soft, potentially revealing labile molecules or metabolites and it allows analysis of negatively or positively charged analytes by a simple exchange of the modifier used in the extraction solvent. However, LESA is a new analysis tool available for ambient surface analysis and its extraction process on thin tissue sections is not well characterized or understood. Its sensitivity might be compound‐dependent and affected by ion suppression of co‐extracted biological matrix molecules. Differences in extraction and ion suppression from different tissues might also limit the ability for relative quantification across a thin section (Kertesz & van Berkel, 2010). Optimization for target analytes includes adjustment of solvent composition, organic content, solvent dwell time, and volume dispensed on the tissue to improve dissolution and ionization of analytes (Swales et al., 2016). Another limiting factor of LESA is spatial resolution, as it can only be optimized to approximately 1 mm, which is not suitable for high‐resolution tissue imaging (Eikel et al., 2011). The development of µLESA has enabled a higher spatial resolution of around 400 µm (Yutuc et al., 2020). LESA has been effective in MSI of protein from mouse and rat tissue surfaces coupled with high field asymmetric waveform ion mobility spectrometry (FAIMS) (Griffiths et al., 2016), cylindrical FAIMS (Griffiths et al., 2020), and native LESA traveling wave ion mobility spectrometry (TWIMS) (Hale et al., 2020). Quantitative MSI of ubiquitin in rat and mouse brain tissue was reported using a stable isotope‐labeled mimetic tissue and LESA MSI (Havlikova et al., 2019). However, these techniques did not require the use of OTCD and so, are out of the scope of this review.
2.5. Additional ambient sources
The use of ambient ionization sources in the field of mass spectrometry imaging has grown significantly in recent years. Comprehensive reviews on the many different modes of ambient ionization for MSI applications are available (Perez et al., 2019; Wu et al., 2013). Most are liquid‐based such as, liquid ablation electrospray ionization (LAESI), which combines laser desorption of the analyte with aided ionization via electrospray and has been applied in the imaging of synthetic fibers and biological tissues (van Geenen et al., 2017; Nemes & Vertes, 2007; Román et al., 2018). Another example is the use of a liquid surface micro junction surface sampling probe (LMJ‐SSP) which creates a liquid junction between solvent delivery and aspiration tubes on the sample surface. Dissolved analytes are subsequently ionized via an atmospheric pressure source like ESI (van Berkel et al., 2008; Kertesz & van Berkel, 2010).
Plasma‐based techniques are another classification of ambient ionization applied in MSI (Ding & Duan, 2015; Smoluch et al., 2016). The combination of a multi‐wavelength laser and a heated metastable plasma achieved a spatial resolution of 60 µm when imaging the ink of printed Chinese characters on a vermilion inkpad and the molecular structure of the traditional Chinese medicine Radix Scutellariae (Feng et al., 2014). Low‐temperature plasma ionization sources are nondestructive to the sample which is a requirement in certain applications, for example examining the ink profiles of artwork without damage (Liu et al., 2010). Coupling of laser ablation to flowing atmospheric‐pressure afterglow source was also applied to caffeine deposits on paper and in celery sections and to lidocaine spotted onto turkey tissue (Shelley et al., 2008). To the best of our knowledge, there are not yet examples of OTCD‐MSI using these sources, which is why they will not be discussed in any further detail here. However, there is considerable potential for OTCD using these sources, which could serve to enhance their performance with certain analytes.
3. ION SUPPRESSION
Ion suppression occurs when ions of interest are masked by interfering ions in the mass spectrum. Source can be endogenous, that is, from the sample itself, or exogenous, introduced to the sample through preparation processes (Antignac et al., 2005). The concept was first introduced when experiments demonstrated the reduced intensity of a target organic base as the concentration of another organic base increased (Kebarle & Tang, 1993). In further experiments, the phenomenon of ion suppression was identified and proven separate to insufficient extraction methods which were thought to be causative of observed lower signal intensity (Buhrman et al., 1996). Ion suppression leads to reduced sensitivity and repeatability of experiments and spectral changes may inhibit the identification of target molecules from a database (Antignac et al., 2005). There are many sources for ion suppression in MSI, including MALDI matrices cluster ions (for MALDI‐MSI), endogenous tissue metabolites, exogenous substances, and contaminants from tissue embedding and fixation. Certain factors make a compound a prime candidate for inducing ion suppression; for example, concentration, molecular weight, and physicochemical properties such as basicity and hydrophobicity (Goodwin et al., 2012). Attempts to ameliorate ion suppression effects by endogenous metabolites involve the addition of a washing step (ethanol and chloroform) after tissue sectioning and mounting to remove interfering salts and lipids (Figure 3) (Dilillo et al., 2017; Franck et al., 2009; Holst et al., 2016; Shariatgorji et al., 2012; Sun et al., 2019). In lipidomics, pH‐controlled ammonium acetate (Bednařík et al., 2020) and ammonium formate (Wäldchen et al., 2019) washes increased the signal of specific lipids by removal of interfering ions. Though these may provide some assistance, the risk of delocalization and even loss of analyte is cause for concern mainly for small drug‐like molecules. CD has the advantage of being selective as only the group or moiety of interest is targeted.
Figure 3.
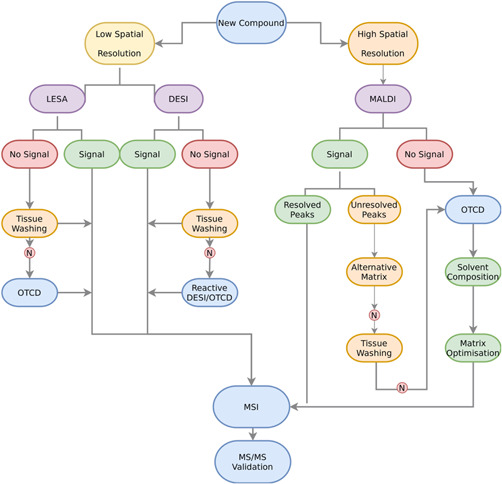
Mass spectrometry imaging (MSI) strategy workflow for analysis of a new compound. Spatial resolution and availability of instrument may govern the chosen platform. When there is no signal for the chosen analyte or unresolved peaks, tissue washing steps or alternative matrices (matrix‐assisted laser desorption/ionization [MALDI]) may help but if unsuccessful, on‐tissue chemical derivatization (OTCD) provides a targeted effective method to increase signal before MSI analysis and validation [Color figure can be viewed at wileyonlinelibrary.com]
4. CHEMICAL DERIVATIZATION
Mass spectrometry analysis requires the ionization of analytes to allow detection. Challenging analytes with low ionization efficiencies are not easily detected and therefore, are not visualized by MSI and are also prone to masking effects by intrinsic ion suppression. In addition, certain mass analyzers have limitations at lower m/z values creating issues for small molecule detection, which is particularly an issue with MALDI‐time‐of‐flight mass spectrometry (MALDI‐TOF‐MS). Including a charged (or easily ionized) moiety in the design of a target‐specific CD reagent counteracts low ionization efficiency problems. Ion suppression in a certain mass range can be avoided by the reaction with another molecule, which increases the analyte m/z value and minimizes spectral isobaric interferences (Manier et al., 2014). Targeted derivatization is achieved through a reaction between the potentially active functional groups of the analytes and the CD reagent used. It has been demonstrated across a range of mass spectrometry platforms and been transferred to imaging platforms such as MALDI (Cobice et al., 2018), DESI (Girod et al., 2012), LESA (Yutuc et al., 2020), and SIMS (Kaya et al., 2018).
5. ON‐TISSUE CHEMICAL DERIVATIZATION
OTCD denotes the transfer of the principles of CD to biological tissue surfaces. Though effective, there are specific challenges associated with this change including analyte delocalization. Increased sample preparation stages may jeopardize spatial resolution if the sample surface is “wetted” with the derivatization reagent. This is particularly true in MALDI where derivatization and matrix deposition can be separate procedures. Longer incubation/reaction times can affect delocalization and if increased temperatures are involved, tissue integrity may be compromised. Consequently, the choice of reagent, matrix compatibility, and application method must be optimized to prevent adverse effects from this preparation step.
Figure 3 shows a strategy for MSI and the situational preconditions for OTCD depending on ionization source and initial signal output. When faced with analysis of a new compound in MSI, requirements concerning spatial resolution must first be considered. A higher spatial resolution may require MALDI as opposed to LESA or DESI. With all platforms, the ionization efficiency of the compound of interest must first be tested. In MALDI, matrix screening is carried out alongside to investigate the most compatible choice for the analysis. An absence of signal following initial screening may indicate the need for OTCD.
Initial presence of signal on‐tissue with no interferences is a promising result for MSI and the method can be further developed for the analysis. However, if there are interferences which cannot be resolved by alternative matrices, tissue washing, or the assistance of higher resolution platforms such as Fourier transform ion cyclotron resonance mass spectrometry (FT‐ICR‐MS), OTCD should be considered. Derivatization reagents should be selected based on the target functional group while considering parameters such as; cocrystallization with matrix, deposition method, solvent composition, reagent concentration, which are discussed in subsequent sections of this review. Though CD can introduce further complications, such as identification of a suitable reagent (further detailed in this section) and optimization of reaction and ionization conditions, the advantages in ionization efficiency and analyte detection can drastically outweigh potential issues.
5.1. Reagent selection
OTCD targets many functional groups of endogenous metabolites as well as synthetic drugs to investigate their distribution within biological tissues. It can also lead to specific fragmentation patterns, which help with molecule identification (Manier et al., 2014; Toue et al., 2014). The following sections detail CD reagents successfully applied to specific functional groups in OTCD. It should be noted that only derivatization methods in OTCD, mainly focused on MALDI‐MSI have been described here. There are many other derivatization reagents targeting a variety of drugs and biological molecules in mass spectrometry platforms that have not been translated to MSI and thus outside the scope of this review.
5.2. Aliphatic alcohols, phenols, and thiols
An early example of OTCD by Chacon et al., 2011, targeted the hydroxyl group of the levuglandin scavenger 3‐methoxysalicylamine with 1,1′‐thiocarbonyldiimidazole, enhancing the sensitivity and spatial distribution of the drug in murine tissue sections. Another interesting application of OTCD is seen in the analysis of cannabinoids in single hairs (Beasley et al., 2016). The use of 2‐fluoro‐1‐methylpyridinium p‐tolunesulfonate to target hydroxyl groups forming N‐methylpyridinium derivatives, improved the ionization efficiencies of Δ9‐tetrahydrocannabinol, cannabinol, cannabidiol, and other metabolites, demonstrating the potential for application in illicit drug testing. A recent paper established the use of the first derivatization reagent for sulfhydryl, ((E)‐2‐cyano‐N‐(2‐(2,5‐dioxo‐2,5‐dihydro‐1H‐pyrrol‐1‐yl)ethyl)‐3 (4hydroxyphenyl)acrylamide) (Fülöp et al., 2020). Measurement of thiols can be indicative of various reactive oxidative species‐related metabolic changes and important in the development of diseases such as cancer. The derivatization reagent enabled the detection of low mass metabolites in porcine tissues and mouse xenograft tissue.
5.3. Carboxylic acids
Formation of an amide bond with the carboxyl group of target free fatty acids (FFAs) and phospholipids (PL) has been demonstrated using 2‐picolamine (PA) (Dueñas et al., 2019; Q. Wu et al., 2016) in the presence of 2,2‐dipyridyl disulfide and triphenylphosphine (Figure 4B). Using the same mechanism, N,N‐dimethylpiperazine iodide, was designed to derivatize FFAs and PLs simultaneously in thyroid carcinomas. This method showed improved signal intensities and on‐target sensitivity in comparison to PA (S. S. Wang et al., 2019). In addition, it allowed FFAs and PLs to be analyzed in the same experiment as they ionize in different modes. Recently, a novel reagent, N,N,N‐trimethyl‐2‐(piperazin‐1‐yl)ethan‐1‐aminium iodide (TMPA) was synthesized for the analysis of carboxyl‐containing groups and allowed tricarboxylic acid cycle intermediates, fatty acids, and bile acids and their metabolites to be detected simultaneously in rat kidney tissues (Sun et al., 2020).
Figure 4.
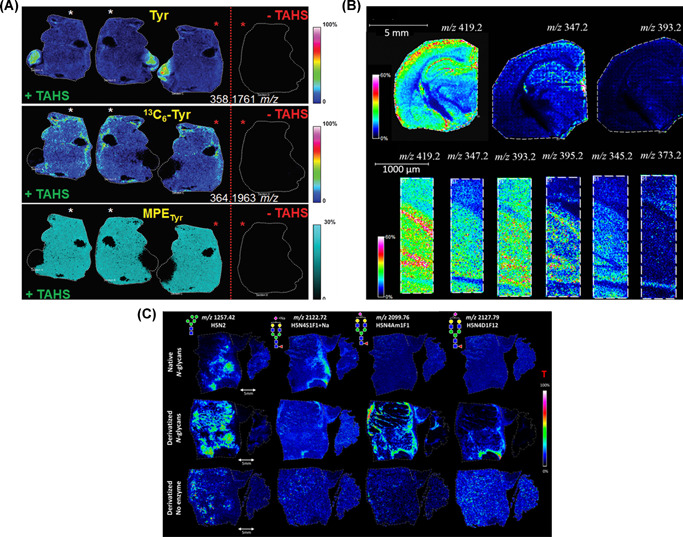
Examples of OTCD with MALDI MSI in proteomics, lipidomics and glycomics. (A) Proteomics: Murine liver sections derivatized with TAHS (+TAHS) (left) showing the direct localization of tyrosine (Tyr) (top), 13C6‐labeled l‐phenylalanine (middle), and the molar percentage excess (MPE) of tyrosine (bottom) compared to underivatized tissue (−TAHS) (right). Consecutive sections with identical intersections, rotated 180°, are denoted by similarly colored asterisks. Adapted from Arts et al. (2017) with permission. (B) Lipidomics: MSI ion maps of fatty acids obtained using OTCD of rat cerebrum with 2‐picolylamine, 100‐μm spatial resolution (above), and from selected subregions, 20‐μm spatial resolution images (below). Adapted from Q. Wu et al. (2016) with permission. (C) Glycomics: MSI of colon carcinoma showing the expression of several N‐glycans; native (top), derivatized by dimethylamidation of sialic acid residues (middle), and negative control sample (bottom). Green circle = mannose, yellow circle = galactose, blue square = N‐acetylglucosamine, yellow square = N‐acetylgalactosamine, white square = N‐acetylhexosamine, red triangle = fucose, purple diamond = N‐acetylneuraminic acid, T = total ion current normalization. Adapted from Holst et al. (2016), with permission. MALDI, matrix‐assisted laser desorption/ionization; MSI, mass spectrometry imaging; OTCD, on‐tissue chemical derivatization [Color figure can be viewed at wileyonlinelibrary.com]
5.4. Unsaturated systems (alkenes)
The positioning of carbon–carbon double bonds (db) and stereospecific numbering (sn) are important factors in determining lipid functionality; however, fragmentation of larger lipids during analysis causes the loss of this information. Derivatization of carbon–carbon db has been employed as a method to overcome this issue and to facilitate characteristic fragmentation for detailed analysis. One such method is the use of the Paternò–Büchi reaction that has been applied in mass spectrometry platforms but more recently in MSI (Bednařík et al., 2018; Wäldchen et al., 2019). Photochemical reaction of a carbonyl group in an excited state with carbon–carbon db form oxetane rings. Fragmentation of this four‐ring moiety by collision‐induced dissociation (CID) results in characteristic fragments revealing positional information of C═C bonds. Typically, acetone acts as the carbonyl moiety source but can cause analyte delocalization. Benzaldehyde was shown to be a practical alternative (Bednařík et al., 2018). To increase availability of this reaction to researchers without access to specialized equipment, a reactive matrix (benzophenone) was developed (Wäldchen et al., 2019). This reagent absorbs at MALDI standard UV laser wavelength (337–355 nm) and once energized, reacts with unsaturated lipids in the same way. This double functioning system was effective with localization of phosphatidylcholines in mouse cerebellum at 15‐µm pixel size (Wäldchen et al., 2019). An alternative method utilizes meta‐chloroperoxybenzoic acid (mCPBA), in an epoxidation reaction with the C═C bonds (Kuo et al., 2019). This strategy, termed “MELDI” (mCPBA epoxidation for lipid db identification) facilitated spatial imaging of tumor‐associated unsaturated lipid changes in metastatic tissue sections. A further method of saturated isomer identification is ozone‐induced dissociation (Bednařík et al., 2020; Paine et al., 2018). Oxygen reacts with carbon db forming ozonide derivatives and reveals positional information through fragmentation. This has been introduced as an on‐tissue derivatization process investigating phosphatidylcholines (PCs) in mouse brain and in human colon samples (Bednařík et al., 2020). Ultraviolet photodissociation (UVPD) (193 nm) has been coupled with DESI to differentiate isomeric lipids from a variety of human tissue samples (Klein et al., 2018). This application of high‐energy photons to lipid ions generated by DESI allowed for characteristic fragmentation and localization of db in PCs by promoting cleavage of adjacent carbon–carbon bonds (Brodbelt et al., 2020). This method identified notable changes in abundance of PC isomers between white and gray matter of mouse and human brain tissues and between cancerous and healthy portions of human lymph node. Recently, ion–ion reactions were applied to image PC structures in rat brain by gas‐phase charge inversion (Specker et al., 2020). Briefly, the mass analyzer was modified to trap lipid cations generated from the tissue surface by MALDI within the collision cell. Radical cations generated by ESI were reacted with cations in the gas‐phase to alter the singly charged initial cation to a charge inverted, anion. Collision‐induced dissociation of the resulting anion allowed structural information of fatty acid tails to be obtained which would not have been possible with a precursor PC cation.
5.5. Amines
Franck et al., (2009) demonstrated improvement in peptide identification aided by the derivatization of basic N‐terminals. Following trypsin digestion, 4‐sulfophenyl isothiocyanate (4‐SPITC) and 3‐sulfobenzoic acid succinimidyl ester (3‐SBASE) facilitated derivatization by bonding of the negatively charged group at the basic peptide N‐terminus. This greatly improved spectral interpretation for peptide identification. 3‐BASE was found to be more efficient, owing to higher peak intensities and reaction yield. An additional reagent, TMPP, also proved efficient and did not depend on the presence of a basic N‐terminus (Franck et al., 2009). Typically, in amine derivatization, a primary amine group undergoes a reaction with larger, commonly aromatic compounds. An example of such is 4‐hydroxy‐3‐methoxycinnamaldehyde (CA). The carbonyl group in this molecule forms a stable Schiff base with target amine groups (Dueñas et al., 2019; Esteve et al., 2016). Its use has been applied in localization of an antituberculosis drug Isoniazid in rabbit lung tissue (Manier et al., 2011). Derivatization with CA resulted in minimal interference and localization within tissue; whereas, without OTCD, it was impossible to distinguish control tissue from dosed due to endogenous signal interferences. This reagent has also been applied in the analysis of amino acids and neurotransmitters in plant‐based tissues (Dueñas et al., 2019), pig adrenal gland (Manier et al., 2014), and rodent brain (Esteve et al., 2016; Manier et al., 2014). Alternatively, the reagent p‐N,N,N trimethylammonioanilyl N‐hydroxysuccinimidyl carbamate iodide (TAHS) reacts with a primary amine, the oxygen next to the carbonyl group is removed and a ureide bond (R–NH–(C═O)–NH–R′ where R is from the amino‐compound and R’ is the charged reagent moiety) is formed with a permanent cationic charge. This approach has been applied in the investigation of amino acid alteration in cancer (Toue et al., 2014), to obtain pathological information in liver disease (Arts et al., 2017) (Figure 4A) and to investigate the distribution of catecholamines to differentiate between tumors of the adrenal gland (Takeo et al., 2019). This reaction offers the benefit of creating a permanent charge, aiding ionization.
Similarly, the use of compounds containing a pyrilium group (C5H5O+) (Table 1) reacts specifically with primary amines to form pyridinium cations (C5H5NH+). The molecule 2,4‐diphenyl‐pyranylium (DPP) (Sugiyama et al., 2019) and 2,4‐diphenylpyranylium tetrafluoroborate (DPP‐TFB) has been found to be effective in the localization of neurotransmitters in murine and crab brain sections (Cao et al., 2019; Davison et al., 2019; Esteve et al., 2016) and in the heads of Drosophila species (Y. Enomoto, Phan, et al., 2018). It was also effective at showing neurotransmitter alterations in a Parkinson's disease model, promoted by injection of the neurotoxin 6‐hydroxydopamine (6‐OHDA) (Figure 5C) (Shariatgorji et al., 2014, 2015). An added advantage of this method is that the aromatic moiety of the pyrylium salts absorbs at the wavelength of standard MALDI lasers and, therefore, acts as a reactive matrix, which means it does not require further matrix addition (discussed further in Section 8). Commercial stable isotope‐labeled derivatization reagents, termed mTRAQ® reagents, were applied in an analysis to compare levels of the neurotransmitter gamma‐aminobutyric acid (GABA) in the brains of different rat models (Ito & Hiramoto, 2019). These probing reagents were designed specifically for use in multiple reaction monitoring MS experiments for peptide analysis and involve the substitution of an N‐hydroxysuccinamide with the amine group of targeted analytes. In these experiments, mTRAQΔ0 solution was sprayed directly onto sample tissue to derivatize endogenous amines. Second, its stable isotope‐labeled analog mTRAQΔ4 was used to derivatize a GABA standard and applied to the tissue as an internal standard for normalization purposes. Third, the DHB matrix was applied completing the “triple‐spray method.” The method was successful in detecting statistically significant differences in GABA levels between stroke‐prone spontaneous‐hypertensive rats and Wistar‐Kyoto rats.
Figure 5.
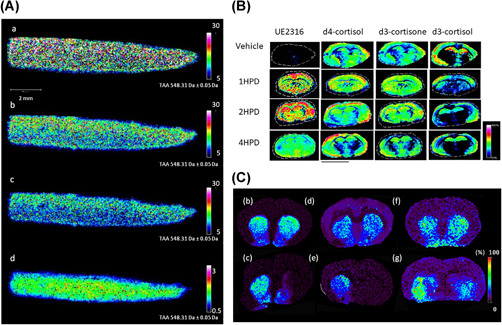
Examples of OTCD with MALDI MSI in pharmacology, intracrinology, and neurosciences. (A) Pharmacology: MALDI‐MSI visualization of the distribution of triamcinolone acetonide (TAA) with OTCD using Girard's T reagent on osteoarthritis cartilage tissue sections using different normalization methods: (a) nonnormalization, (b) median normalization, (c) TIC normalization, (d) normalization against the labeled analog of the drug. Adapted from Barré et al. (2016) with permission. (B) Intracrinology: Evaluation of changes in abundances of UE2316, d4‐cortisol (d4F), d3‐cortisone (d3E), and d3‐cortisol (d3F) at varying time points (HPD) in murine brain regions, measured following OTCD using Girard's T reagent. Adapted from Cobice et al. (2018) with permission. (C) Neurosciences: MALDI‐MSI images of dopamine (DA) in coronal tissue sections of control (b, d, f) and unilaterally 6‐OHDA lesioned (c, e, g) rat brains after derivatization with DPP (b, c), PBDPP (d, e) or TMP (f, g). Signal intensity is indicated using a rainbow scale. Scale bar = 2 mm; spatial resolution = 120 μm. Adapted from Shariatgorji et al. (2015), with permission. DPP, 2,4‐diphenyl‐pyranylium; HPD, hours postdosing; MALDI, matrix‐assisted laser desorption/ionization; MSI, mass spectrometry imaging; OTCD, on‐tissue chemical derivatization [Color figure can be viewed at wileyonlinelibrary.com]
5.6. Ketones/Aldehydes
Localization of keto‐containing molecules has been targeted using hydrazine‐based reagent via Schiff‐base (hydrazones) formation. Hydrazone formation by Girard reagents (e.g., Girard's T reagent [GT]) and Girard's P reagent [GP]) is a two‐step reaction that involves condensation, normally achieved by using protic solvent in a weak acid media as catalyst. It contains a positively charged trimethylamine (GT) or pyridine (GP) and reactive acetyl hydrazine, which forms stable hydrazones with carbonyl groups. The first application of GT in OTCD was in the detection of the substrate and product of glucocorticoid‐amplifying enzyme 11β‐HSD1 in rat adrenal gland and mouse brain (Cobice et al., 2013). This technique even allowed pharmacokinetic and pharmacodynamics effects of an11β‐HSD1 inhibitor which demonstrated potential in drug screening for the treatment of neurological disorders (Cobice et al., 2018) (Figure 5b). Further to this, GT enabled the localization of androgens in mouse testes (Cobice et al., 2016; Shimma et al., 2016). Use of on‐tissue GT derivatization aided the separation of steroidal structural isomers with characteristic fragmentations in ion trap MS/tandem MS (MS3) analysis in human and rat adrenal glands (Takeo et al., 2019). Another study utilized GT to determine the depth penetration of an osteoarthritis drug triamcinolone acetonide in human cartilage (Figure 5A) (Barré et al., 2016). This study found a higher signal intensity observed from GT reagent when compared with an alternative carbonyl derivatizing reagent 2,4‐dinitrophenylhydrazine (DNPH). GT has also been applied to the derivatization of abscisic acid and 12‐oxo phytodienoic acid and enhanced visualization in seed sections (H. Enomoto, Sensu, et al., 2018). Sterols and oxysterols are important molecules in the study of neurological disorders but present ionization issues during MS analysis. To overcome this issue, an enzyme‐assisted derivatization technique has been developed which uses a bacterial cholesterol oxidase enzyme to convert sterols to carbonyl‐possessing molecules so they can then be derivatized by Girard's P reagent and form stable hydrazones (Yutuc et al., 2020). This method proved successful for the spatial localization of oxysterols in mouse brain by µLESA‐LC‐MS and work is currently underway to translate this technique to a MALDI MSI platform. DNPH and 4‐dimethylamino‐6‐(4‐methoxy‐1‐naphthyl)‐1, 3, 5‐triazine‐2‐hydrazine (DMNTH) were used to derivatize the corticosteroid fluticasone propionate in a similar, hydrazone‐forming mechanism and demonstrated the distribution of spotted standards on rat lung tissue (Flinders et al., 2015).
6. REAGENT‐MATRIX CO‐CRYSTALLIZATION
In MALDI, the analyte of interest must be soluble in the matrix solvent to achieve a homogeneous cocrystallization (analyte inclusion in the matrix crystal structure), hence, a successful desorption and ionization process. In the case of CD, the reagent used must also have the same properties. Matrix crystallization should be as small and homogenous as possible to achieve higher spatial resolution and avoid “hot‐spots” or artifacts. Solvent composition influences crystal formation; the pH and organic to aqueous ratio must be optimized to allow maximum analyte dissolution and inclusion without loss of spatial resolution. Increasing organic solvent percentage reduces the size of matrix crystals, however, may have a negative impact on analyte dissolution. On the contrary, if composition and coverage are too wet, analyte delocalization may occur. The concentration and density of the matrix applied should not be too high to mask the analyte and the application method used should be considered (Section 7). On the contrary, sufficient matrix coating is needed to keep the ionization “soft,” particularly for biological applications where fragmentation of larger biomolecules is undesirable. The composition of the tissue itself can cause interference with the quality of analyte‐matrix crystals. High concentrations of lipids and salts can interfere and suppress the analyte within matrix crystals. Washing of the tissue sample with ethanol or chloroform to remove abundant PLs (Dilillo et al., 2017; Franck et al., 2009; Holst et al., 2016) or ammonium acetate/formate to remove salts and alkali metal ions (Bednařík et al., 2020; Wäldchen et al., 2019) have alleviated these effects. Microscopic observation of matrix crystals can reveal the best conditions for an experiment and give valuable information on the nature of the crystals formed. Comparison of a specially designed reactive matrix to detect neurotransmitters in brain tissue was compared to the commonly used DHB matrix (Shariatgorji et al., 2019). Crystals of 15‐ to 20‐µm size were formed with the reactive matrix in a mosaic pattern, which was favorable compared to larger DHB crystals. The addition of derivatization reagent (N,N‐dimethylpiperazine iodide) to the tissue before DHB matrix by electrospray was investigated by scanning electron microscope during method development (S. S. Wang et al., 2019) and crystal formation was unchanged by this step. The consideration of the change in crystal formation upon addition of another reagent to the tissue is an often‐overlooked parameter and only a small number of authors have reported microscopic imaging of this phenomenon to investigate its effect.
7. REAGENT DEPOSITION AND ANALYTE DIFFUSION
The effectiveness of OTCD and MSI analysis depends highly on the methods used for reagent and matrix (MALDI‐MSI) deposition. A balance must be reached which allows the reagent to penetrate the sample biomatrix and react with analytes of interest without causing delocalization (Table 2). Uniformity of both reagent and matrix application are important parameters to allow homogenous derivatization of target molecules. In practice, the derivatization reagent and matrix are applied to sample tissues using similar methods, that is, hand‐held or automated/pneumatic sprayers.
Table 2.
Chemical derivatization reagent deposition techniques
| Technique | Advantages | Disadvantages |
|---|---|---|
| Acoustic multi‐spotter | Uniform, fast, good reproducibility | Droplet application |
| Electrospray deposition | Homogenous | Limited time for analyte–matrix interaction |
| Pneumatic Sprayer | Homogenous | Droplet size not constant |
| ImagePrep™ | Controlled conditions, automated, homogenous | Droplet size not constant, expensive |
| Dry‐coating | Cheap, high‐purity matrix | Poor analyte–matrix extraction |
| Sublimation | Homogenous, reproducible, fast | Poor analyte–matrix extraction |
| Desktop inkjet printer | Uniform droplets (multichannel) | Slow, solvent compatibility, clogging |
Reagent concentration, reaction time, and the coverage density must all be optimized for the experiment, along with physical parameters of the deposition such as ambient temperature, spray temperature, flow rate, velocity, and distance of source from tissue. For example, increase in flow rate can improve signal intensity but also results in larger droplets, compromising analyte localization (Q. Wu et al., 2016). A balance must be struck for optimal spatial information production. Simple artistic airbrushes and thin layer chromatography sprayers have been used to apply derivatization reagents (H. Enomoto, Sensu, et al., 2018; Sugiura et al., 2018, Takeo et al., 2019), but cause issues with uniformity and reproducibility (Smith et al., 2020). High‐intensity hot spots have been generated on tissue with this method in comparison to an automated system (Q. Wu et al., 2016). Effective use of this method relies heavily on operator experience and environmental factors such as temperature and humidity are not controlled. The development of automated pneumatic sprayer systems allows for a controlled, reproducible application and there are many options. The HTX TM‐Sprayer™ (HTXImaging) (Davidson et al., 2019; Dueñas et al., 2019; Kaya et al., 2018) and the SunCollect™ (SunChrom) (Barré et al., 2016; Fülöp et al., 2020) are commonly used for both derivatization and matrix application. They have similar mechanisms in that the reagent solvent is sprayed onto the slide in a controlled environment with programmable parameters (e.g., velocity and flow rate) to achieve homogeneous layer depositions across the tissue. Both systems are rapid, however, blockages can arise depending on the nature of the solvent used.
The ImagePrep™ (Bruker Daltonics) deposits an aerosol created by vibrational vaporization onto a glass slide (Shariatgorji et al., 2015; Smith et al., 2020). A major difference between this and other spray systems is that reagent sprays into the tightly controlled chamber from the side, rather than overhead allowing droplets to settle gently onto the surface. A series of incubation periods allow for adequate reaction and drying in between spraying, reducing the likelihood of delocalization. This system also includes a sensor to detect matrix density and dryness, permitting increased uniformity and repeatability between samples. The ImagePrep™ has the advantage of a closed chamber, which fills with nitrogen gas before the deposition begins, creating an inert environment for the process. Increased control and uniformity of external factors in this way, greatly improve experimental reproducibility.
Homemade electrospray systems have enabled simultaneous detection of fatty acids and PLs at 20‐µm spatial resolution in rat cerebrum, in comparison to airbrush application, which generated random nonhomogenous high‐intensity spots (Q. Wu et al., 2016). Electrospray systems have also generated smaller matrix crystals and achieved higher sensitivity and spatial resolution when compared to the ImagePrep (Li et al., 2016). In addition, scanning electron microscope images of DHB crystals created with this system have shown that the addition of the derivatization reagent does not negatively affect crystal uniformity (S. S. Wang et al., 2019). The application of an electric field to the slide itself was shown to increase the sensitivity of lipid and protein detection (X. Wang et al., 2015). Recently, modified 3D printers have been developed for solvent/reagent deposition on tissues (Tucker et al., 2018; Smith et al., 2020). They are inexpensive compared to commercial systems while allowing a controlled spray with programmable parameters. Spotting applicators apply reagent in spots at uniform distances rather than covering the tissue in a spray, for example, the Portrait™ 630 Reagent Multi‐Spotter (Labcyte Inc.) (Flinders et al., 2015) and inkjet printers, such as the ChIP‐1000 (Shimadzu) (Franck et al., 2009; Ljungdahl et al., 2011). Though this has analyte delocalization within the spots deposited, it also ensures the spatial resolution of the analysis is now limited to the spot size. Solvent‐free application methods, such as matrix dusting (Ferguson et al., 2013; Goodwin et al., 2010) or sublimation (Gemperline et al., 2014) removes the effects of solvent delocalization but, may also limit tissue penetration leaving several analytes undetected. Transfer to a humid environment (Goodwin et al., 2010; Gemperline et al., 2014), or subsequent solvent spray onto the sample surface (Ferguson et al., 2013; Lauzon et al., 2015) have been necessary to enhance analyte extraction. Coating the slide with derivatization reagent and matrix, before thaw mounting of tissue, simplifies preparation and is thought to reduce delocalization (Manier et al., 2014). However, as thaw‐mounting of the sample is relied on to resolubilize the reagents and analyte dissolution may be limited. An interesting new OTCD method called laser‐assisted tissue transfer (LATT) enhances the effects of OTCD (Guo et al., 2019). The technique follows tissue treatment with derivatization reagent and matrix and allows a thin film of tissue to be transferred to a clean slide using localized heat from a blue laser beam, which is then analyzed. The thinner section reduced ion suppression effects, derivatization time, and delocalization, led to increased intensities of target derivatives when compared to standard derivatization and matrix application protocols and even allowed visualization of several new peaks in the LATT‐treated sample.
As shown, there are many application systems available, each with its own benefits and limitations. For this reason, it is extremely important to optimize this step in OTCD and MALDI MSI to achieve the best results possible.
8. REACTIVE MATRICES AND REACTIVE‐DESI
In MALDI, recent developments include the use of a reagent that combines the properties of derivatization and matrix into one. This advantageously reduces the risk of undesired effects, such as longer sample preparation time and further “wetting” or incubation of sample tissue, which could cause analyte delocalization or degradation. For this to be successful, in addition to facilitating ionization of target analytes, the reagent in question must have the ability to absorb UV radiation at the same wavelength as the ablation laser and be stable under vacuum conditions. Aromatic structures present in the derivatization reagents DNPH and DMNTH facilitate laser absorption, whereas amine functional groups target carbonyl groups for derivatization (Flinders et al., 2015). When applied to detection of fluticasone propionate in rat lung tissue, both reagents were successful in facilitating selective detection of the target protonated hydrazone derivative without matrix addition, however, DMNTH demonstrated superior linearity when spotted standards were analyzed (Flinders et al., 2015). Kaya et al., (2018) designed a reactive matrix, 4‑(N‑methyl) pyridinium boronic acid, for the detection of catecholamines in the porcine adrenal gland. In this reaction, boronic acid reacts with the catechol group of the target analytes forming a boranate ester (boronic acid–diol reaction). The inclusion of boron added the advantage of a characteristic isotopic pattern to identify the derivatives. In addition, the reaction allowed absorption at the laser wavelength, and the permanent positive charged molecule aided ionization. The use of this reagent enhanced the sensitivity of detection for catecholamines using both SIMS and MALDI platforms. There are several examples of reactive matrices based on pyrylium salts designed to react with primary amine groups forming pyridium cations (Davison et al., 2019; Dilillo et al., 2017; Y. Enomoto, Phan, et al., 2018; Esteve et al., 2016; Sugiyama et al., 2019). One group recognized the potential for pyrilium salts to be used as reactive matrices as initial experiments revealed DPP absorbed at the 335‐nm wavelength of the standard MALDI lasers that are used, and formed small crystals (Shariatgorji et al., 2014, 2015). This reagent enabled pmol/mg level detection of l‐BMAA in rat sagittal tissue sections without the addition of a common MALDI matrix. Subsequent modifications to the structure facilitated simultaneous targeting of primary and secondary amines and subsequent phenolic metabolites of catecholamines (Shariatgorji et al., 2019). This reaction is based on a nucleophilic substitution of target functional groups with an electrophilic fluoropyridinium moiety, which has a chromophore entity to facilitate laser absorption. This reactive matrix improved the detection limit of dopamine by a factor of 500 in comparison to 2,5‐dihydroxybenzoic acid. In addition, incorporation of bromine to the pyrilium structure provided isotopically characteristic derivative spectra and improved the limit of detection of neurotransmitters in brain tissue (Shariatgorji et al., 2020). A novel reactive matrix was used to analyze acidity in Ponkan fruit (Ma et al., 2018). Glycosyl‐3‐aminoquinioline (Gly‐3AQ) undergoes catalysis under acidic conditions, therefore, facilitating the association between a change in ion m/z ratio and the level of acidity in the fruit during different developmental stages. This method is interesting as it is not targeting a specific molecule but rather reacting to the environment within the tissue itself.
Inorganic matrices have been successful in on‐tissue analyses, by removing interfering ions. The image quality of low molecular weight molecules in mouse brain sections was improved using graphene oxide (Zhou et al., 2017). Metal nanoparticles such as silver carry a charge and absorb the wavelength of the UV laser, fulfilling matrix requirements. The deposition of silver (Ag) nanoparticles in place of liquid AgNO3 avoided delocalization and proved more effective in the analysis. For example, silver nanoparticles resulted in lower limits of detection than liquid in the analysis of cholesterol in human fibroblast monolayers (Xu et al., 2015). Electrophilic silver ions are selective for carbon–carbon db, forming charge‐transfer complexes, allowing their use to lend itself the title of “reactive matrix.” This feature is effectively demonstrated in the detection of unsaturated lipid molecules and has been utilized on‐tissue to detect hydrophobic lipids in kidneys (Muller et al., 2015), brain (Guan et al., 2018), and heart (Jackson et al., 2014) tissues. Another advantage is a characteristic isotope pattern (107Ag and 109Ag) which assists in db localization (Xu et al., 2015).
Though this review focuses on the application of OTCD for MALDI platforms, it is important to also briefly mention the use of reactive DESI. The introduction of targeted derivatization reagents to the DESI solvent has allowed analysis of challenging analytes. The technique has developed alongside DESI itself with the use of a lysozyme substrate to identify the enzyme–substrate complex from sample on a solid surface reported by Takáts et al. (2004). Early examples of samples spotted on solid surfaces include the analysis of steroids in urine by a reaction of hydroxylamine with the steroid carbonyl groups (Huang et al., 2007). Use of phenylboronic acid (Chen et al., 2006) to target cis‐diols in spotted samples and later in urine and serum samples (Zhang & Chen, 2010). On‐tissue transfer of this technique has also proven effective. Though the method does include optimization of reagent concentration, solvent choice, and so forth, the risk of analyte delocalization is potentially lower as sample “wetting” takes place in a much more controlled way during analysis. In addition, the reaction can occur in tandem with ionization and under ambient conditions. The technique has been applied in the analysis of steroids (Girod et al., 2012), peptides (Shin & Cha, 2018), and lipids (Lostun et al., 2015; C. Wu et al., 2009, 2010). The amino group of dinitrophenylhydrazine targets steroid carbonyl groups, and doubled the intensity of the signal from the lipid peroxidation product malondialdehyde in traumatized rat spinal cord, giving information on the state of oxidative stress and delineation damage (Girod et al., 2012). Free cholesterol was targeted by incorporating charge‐tagged betaine aldehyde into the DESI solution which reacted with cholesterol hydroxyl groups. This has been applied on several tissue types such as rat brain (C. Wu et al., 2009) and rabbit and porcine adrenal gland (C. Wu et al., 2010), and human atherosclerotic plaque tissue (Manicke et al., 2009). Further lipid analysis employed dications to charge‐tag negative lipids for analysis in positive mode. This was achieved using synthetic 2‐ring aromatic compounds, linked with a hydrocarbon chain each having a charged nitrogen. This improved the signal‐to‐noise ratio of low molecular weight lipids and phosphoethanolamines (PE) in rat brain and zebrafish tissue (Lostun et al., 2015). A DESI method to detect citrullinated peptides using selective reaction with phenylglyoxal was successfully developed by Shin and Cha (2018) but was not applied on the tissue. The use of reactive matrices and reactive DESI eliminates the developmental issues associated with the introduction of a derivatization reagent in a separate sample preparation step. Rapid analysis is achieved by halving optimization experiments while incorporating selective and enhancing the performance of OTCD.
9. APPLICATIONS OF OTCD
9.1. Neurosciences
Understanding of the distribution of neurotransmitters and their metabolites would divulge insightful information into behavior, sleep, cognitive function, and diseases such as Parkinson's and Alzheimer's. The largest portion of examples mentioned throughout this review has made possible the localization and tracing of previously undetectable neurotransmitters in animal tissue (Davison et al., 2019; Esteve et al., 2016; Ito & Hiramoto, 2019; Manier et al., 2014; Shariatgorji et al., 2014, 2015, 2016, 2019, 2020; Sugiyama et al., 2019). OTCD of amino‐neurotransmitters with TAHS and CA reagents to target the amine group have been successful in localizing neurotransmitters in mice brain tissue (Esteve et al., 2016). However, pyrilium salts have overtaken as the gold standard for this application, with the added advantage that they can be used as reactive matrices for MALDI‐MSI (Shariatgorji et al., 2014). The reagent DPP‐TFB has been most commonly used; however, modifications to these structures have enhanced the efficacy and scope of their use. The highly electrophilic cation, 2‐fluoro‐1‐methylpyridinium, reacts with primary amines and metabolite hydroxyl groups in a nucleophilic aromatic substitution reaction, allowing detection of not only neurotransmitters but subsequent metabolites (Shariatgorji et al., 2019). Most recently, the synthesis of 2‐(4‐bromo‐phenyl)‐4,6‐diphenyl‐pyranylium, incorporating bromine into the pyrilium structure generated characteristic isotopic spectra of derivatives (Shariatgorji et al., 2020). Application in rat, mice, and primate models of neurological disease states has been effective. The use of a combination of three reagents (TAHS, CA, and DPP‐TFB) detected neurotransmitters in mouse brain sagittal sections and to investigate changes when a cortical spreading depression was induced in one hemisphere (Esteve et al., 2016). Pyrilium‐based reagents were used to derivatize dopamine in control rat brains in a Parkinson's disease model, promoted by injection of the neurotoxin 6‐OHDA (Shariatgorji et al., 2014, 2015) (Figure 5C). This group moved forward to simultaneously detect neurotransmitters and their metabolites in rat, rhesus monkey, and human brain tissue by synthesizing a derivatization reagent targeting primary amines and hydroxyl groups (Shariatgorji et al., 2019) and easing identification of target derivatives with the addition of bromine to generate characteristic isotopic spectra (Shariatgorji et al., 2020). The application of OTCD in neurosciences is significant with huge potential for future contribution to pathological understanding of neurotransmitter behaviors in disease model tissues.
9.2. Lipidomics
Lipids are widely researched in MSI as targets for information on disease progression, biomarker analysis, cellular membrane components, and signaling molecules. Identification of lipid changes is extremely useful but problematic to carry out. The use of derivatization has been applied to mass spectrometry platforms for lipid analysis. Not all methods have translated to MSI however; though there are several that show promising results. Problems in the detection of lower molecular weight lipid metabolites can be aided with OTCD. Cholesterol targeting using betaine aldehyde was an early example of lipid detection using OTCD (Manicke, et al., 2009; C. Wu et al., 2009, 2010). Malondialdehyde, a low‐mass reactive aldehyde resulting from lipid peroxidation was detected when derivatized by DNPH in reactive DESI‐MSI; giving information on lipid metabolism in injured rat spinal cord tissue (Girod et al., 2012). Investigating lipid metabolism in brain tissues was possible by targeting the carboxyl group in low molecular weight fatty acids with PA (Q. Wu et al., 2016) and oxo‐containing sterols with GT (Yutuc et al., 2020).
A widespread problem in the field of lipidomics is the inability to dissociate between isomeric fragments of lipids. The positioning of carbon–carbon db and sn are key factors in determining functionality but when larger molecules are fragmented, this information becomes lost. Derivatization of carbon–carbon db has been employed as a method to overcome this issue and to facilitate specific fragmentation for analysis. The Paternò–Büchi reaction has been employed in this manner and allowed for the identification of db positional isomers in mouse brain tissues (Bednařík et al., 2018; Wäldchen et al., 2019). In addition, derivatization of C═C bonds into epoxides identified tumor‐specific isomeric changes in metastatic tissues (Kuo et al., 2019). Addressing the same issue is the method of ozone‐induced dissociation, which is based on the formation of ozonide derivatives when oxygen reacts with the lipid db, revealing positional information (Paine et al., 2018). Initially, this was developed to introduce ozone gas into the collision cell with the collision gas but has since been introduced as an on‐tissue derivatization process investigating phosphatidylcholines (PCs) in mouse brain and in human colon samples (Bednařík et al., 2020).
Ultraviolet photodissociation (UVPD) (193 nm) coupled with DESI differentiated isomeric lipids from white and gray matter of mouse and human brain tissues and between cancerous and healthy portions of human lymph node (Klein et al., 2018). The reaction between high‐energy photons and lipid ions released from tissue allowed for a characteristic dissociation and localization of structural isomeric features. Charge inversion of lipid cations with ion–ion reactions in the gas phase revealed structural information that would not have been observed by dissociation of the original cation (Specker et al., 2020).
Issues of low molecular weight and inability to distinguish structures which only differ in the position of the C═C bonds have been remedied by OTCD. The potential for the previously unattainable understanding of lipid changes in disease states is vast.
9.3. Proteomics
An early example of peptide derivatization utilized sulfonates and TMPP to aid protein identification in a bottom‐up imaging strategy (Franck et al., 2009). Although these reagents were not used in any subsequent analyses identified during the course of this review, the concept of peptide derivatization to aid protein identification was successful. Disease‐specific peptide alterations and amino acid metabolites have been identified by OTCD offering a platform to obtain vital pathological information in liver disease (Arts et al., 2017) (Figure 4A), cancer (Dilillo et al., 2017; Toue et al., 2014), and neurological conditions such as cortical spreading depression (Esteve et al., 2016), and Alzheimer's and Parkinson's (Guo et al., 2019). Derivatization of amino acid metabolites in maize roots and seeds enhanced signal intensity (Dueñas, et al., 2019) and distinguished amino acid genotypic differences (O'Neill & Lee, 2020). Protein behavior is a core aspect of disease pathology and knowledge in this area is vital to progression in understanding and treatment. OTCD of small peptides and metabolites has proven a useful tool in this endeavor.
9.4. Endocrinology/Intracrinology
Localization of steroidal compounds by OTCD has been very effectively demonstrated in recent years (Cobice et al., 2013, 2018; Dueñas et al., 2019; Sugiura et al., 2018). GT has allowed the localization of steroidal hormones in mouse testes (Cobice et al., 2016; Shimma et al., 2016) and adrenal glands (Cobice et al., 2018; Sugiura et al., 2018). This reagent targeted ketones in the androgen structures, enhancing the sensitivity of detection within target tissues. A novel reagent, 4‐(N‐methyl)‐pyridinium boronic acid, allowed for targeted derivatization of catecholamines in the adrenal gland incorporating boron in its structure for generation of characteristic spectra (Kaya et al., 2018). Used as a reactive matrix and across multiple platforms, it shows great promise for future analysis in this area. Spatial localization of hormones will increase knowledge of their roles in disease states; not only in diseases with hormonal causes such as primary aldosteronism (Sugiura et al., 2018) but those with lesser‐known hormonal involvement such as dementia and various cancers.
9.5. Alternative metabolites
Outside of proteomics, lipidomics and endocrinology, OTCD can be applied to many other metabolic targets. N‐glycans are important biological molecules and due to their availability for posttranslational modifications and presence on cell surfaces, they have enormous potential as biomarkers of disease. The presence of sialic acid on N‐glycans hinders the ability to detect and localize them in tissues. The derivatization of sialic acid itself not only increases the ionization efficiency of the molecules but also can distinguish alternative linkages which are important in disease progression, including cancer (Figure 4C) (Holst et al., 2016). Vitamin D plays a vital role in calcium homeostasis, immune function, and has effects on intracrine pathways (Smith et al., 2019). Visualization of vitamin‐D precursor and product metabolites in mouse kidneys was made possible by OTCD using Amplifex™ reagent, which targeted structural cis‐diene bonds (Smith et al., 2020). Fatty acids, bile salts, and other carboxyl‐containing metabolites were visualized in rat kidney and brain after OTCD with a novel reagent, TMPA (Sun et al., 2020). The ability to measure such molecules and provide information on complex metabolic processes is enhanced with OTCD.
9.6. Pharmacology
The spatial distribution of drugs in tissue is important in drug development, and MALDI MSI has become a vital tool in this field. Depth penetration of the osteoarthritis drug triamcinolone acetonide in cartilage tissue is important in understanding the effectiveness of the treatment (Barré et al., 2016). Low ionization efficiency meant that OTCD was required for visualization (Figure 4A). Enhanced effectiveness of MSI in tissue for small molecule drugs (Chacon et al., 2011) and for compounds with isobaric interferences, such as the amine‐containing drug Isoniazid in rabbit lung tissue (Manier et al., 2011) was achieved with OTCD. Increased understanding of pharmacokinetics and pharmacodynamics of a dementia treatment drug in mice liver and brain tissue (Figure 5B) (Cobice et al., 2018) and targeted analysis of drug and metabolites simultaneously has been made possible (Liu & Hummon, 2016). Analysis of human hair by MSI creates another possibility of applications for this technique (Beasley et al., 2016). The detection of low abundance cannabinoids assisted with OTCD demonstrates the possibility of applications in illicit drug testing or forensics. MSI is at the forefront of pharmacological research and with the assistance of OTCD for problematic targets, the opportunities for advancement in this area are vast.
10. CONCLUSION
CD greatly enhances the detection of target analytes in mass spectrometry. When applied in MSI, spatial localization enables the generation of visual information that would have been previously unattainable, and the nature of OTCD means that it can be tailored for use in many applications. Suitable reagents can be synthesized to suit the target, the ionization platform, and even to generate characteristic spectra for ease of detection. Though MALDI remains the standard ionization platform for MSI, recent developments in the speed and spatial resolution capabilities of ambient ionization methods suggest a rise in their use. OTCD has proven effective with ambient sources also, and so is not limited to the ionization platform used. Pathophysiological understanding of a range of diseases can be improved with OTCD; changes in the abundance or localization of neurotransmitters, fatty acids, amino acids, and other metabolites provide indispensable information which may not have been possible otherwise. The changes in metabolic profiles in diseased tissues, such as Alzheimer's, Parkinson's, and cancers, for example, are essential to advancing diagnosis and treatment. Testing the efficacy of drug treatments with OTCD MSI and the ability to view distributions in tissue are important to determining the correct treatment and preventing the continued use of those which do not perform correctly.
Applications in plant tissues allowed for a deeper understanding of developmental stages and metabolic processes involved. There are only a few examples of this but there is huge potential for growth in this area.
Challenging analytes have been specifically targeted and amplified using OTCD, supplying vital spatial information within sample tissues in a wide variety of applications and research. Careful reagent design suits specialized experiments and precise requirements of the analysis and are adaptable over a range of ionization platforms. Though optimization must be considered, the benefits of OTCD in a variety of scientific fields are clear.
11. FUTURE PERSPECTIVES
The development of increasing spatial resolution, while providing valuable molecular information in biological tissues, reduces ion yield per pixel as they become smaller. The introduction of the post‐ionization technique MALDI 2 (Soltwisch et al., 2015), has been shown to increase ion intensity and sensitivity by several orders of magnitude (Ellis et al., 2017; Niehaus et al., 2019; Soltwisch et al., 2020; Spivey et al., 2019). In this approach, another laser irradiates the plume of ions from the initial surface ablation, facilitating secondary ionization reactions. Used in tandem with CD, this could be a very promising and effective strategy for molecules which are difficult to ionize. It could offer a method of exploration for molecules not yet tested by MSI. However, the infancy of this technique means availability may be limited; therefore, CD presents a cheaper alternative with no necessary additional instrumentation needed. In addition, improvements in spatial resolution and speed of ambient ionization methods such as DESI and nanoDESI suggest their use in MSI may increase. Consequently, so too may the use of OTCD in these platforms. Ambient ionization reduces sample preparation time and handling, which is advantageous in biological applications for maintaining tissue structure. Similarly, the development of reactive MALDI matrices is encouraging, removing extra sample preparation steps and the risk of delocalization. The requirements of an effective MALDI matrix are well known as are the availability of suitable CD reagents, paving the way for promising future results in this field.
There are many derivatization reagents which have been successful in mass spectrometry platforms, such as LC‐MS that have not been adapted to OTCD. There is immense potential for introduction of new derivatization reagents which would widen the scope of the technique and the fields of implementation.
Although the examples presented in this review are biological in nature, there are many nonbiological applications of MSI which may benefit from OTCD. The detection of cannabinoids in human hair (Beasley et al., 2016) demonstrates potential in forensic applications. The detection of low abundance molecules in hair paves the way for detection of a variety of compounds in such samples providing vital evidence in criminal cases. MSI has been applied to the characterization of surface and polymer layers of synthetic fibers (van Geenen et al., 2017). OTCD could be applied in this way to detect trace compounds. Detection of trace amounts of illicit drugs (Groeneveld et al., 2015), condom lubricants (Bradshaw et al., 2013), and blood (Bradshaw et al., 2014) in fingerprints has been demonstrated. OTCD may allow for analysis of previously undetectable compounds and add intelligence to lawful investigation of criminal cases.
In summary, OTCD applications in MSI are growing and expected to continue. Previously unionizable molecules are being detected with MALDI and DESI platforms. Ionization techniques are improving drastically and new techniques are being introduced, which OTCD can be tailored to suit. Although the investigation of tissue metabolomics, disease pathology, and pharmacological analyses will continue to benefit from OTCD, we hope the potential in other fields will be recognized.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
Biographies
Carla Harkin will complete her PhD degree in Bioanalytical Mass Spectrometry from Ulster University, Coleraine in 2021. She received her 2‐year MSc by research in Letterkenny Institute of Technology in 2014 as part of a larger global project. She then worked as a Research Assistant and Microbiology Technician before taking up employment as a QC Analyst at Randox Teoranta where she was offered a place in the Randox‐Ulster University‐Industrial PhD Academy. Her current research area focuses on the identification of potential biomarkers for diabetic kidney disease using mass spectrometry imaging (MSI) among other platforms.

Karl W. Smith received his BSc (Hons) in Biology from Ulster University in 2016 and PhD in biomedical sciences in 2020 from Ulster University, where he studied the effects of vitamin D in prostate cancer using mass spectrometry imaging. Currently, Dr. Smith is a postdoctoral researcher with the National High Magnetic Field Laboratory at Florida State University. His research interests include advancing instrumentation and imaging capabilities at ultra‐high mass resolution in FT‐ICR MS, and how this can be applied to tissue sections relevant to disease or biochemical pathways.

Faye L. Cruickshank, PhD, completed her MChem in 2011 at the University of Edinburgh working with Prof. Perdita Barran. She then obtained her PhD in 2015 from the University of Edinburgh, working with Prof. Alison Hulme on drug repurposing for cancer therapeutics. Since completing her Ph.D., she has worked at SIRCAMS at the University of Edinburgh.

Dr. Logan MacKay, PhD, obtained his PhD in physical chemistry in 2003 from the University of Edinburgh on mass spectrometric techniques for single‐cell analysis. After 2 years of post‐doctoral work under Alan Marshall at the National High Magnetic Field Lab in Tallahassee, Florida, he returned to the University of Edinburgh to work at SIRCAMS to continue working with FT‐ICR instrumentation, taking over as facility manager in 2008. His academic research has focused on the development of ultra‐high resolution mass spectrometry techniques, with applications including complex mixture analysis, mass spectrometry imaging, and hydrogen‐deuterium exchange.

Bryn Flinders, PhD, obtained his bachelor's degree in forensic and analytical science and later PhD from Sheffield Hallam University. His PhD focused on the application of MALDI‐MS imaging to monitor the distribution of pharmaceuticals in the lungs. He then moved to the Netherlands, to do post‐doctoral research at FOM‐Institute AMOLF in Amsterdam with Prof. Ron M.A. Heeren on the application of mass spectrometry imaging techniques to monitor the distribution of drugs of abuse in hair. He then moved to Maastricht, to work at the Maastricht Multimodal Molecular Imaging (M4I) Institute to research the application of mass spectrometry imaging techniques to monitor the distribution of pharmaceutical compounds in tissues. Currently, he works for Hair Diagnostics, part of the Dutch Screening Group as a researcher developing new methods for the commercial application of mass spectrometry imaging for hair testing.

Prof. Ron M. A. Heeren obtained a PhD degree in technical physics in 1992 at the University of Amsterdam on plasma‐surface interactions. He was the research group leader at FOM‐AMOLF for macromolecular ion physics and biomolecular imaging mass spectrometry in the period 1995–2015. In 2001 he was appointed professor at the chemistry faculty of Utrecht University lecturing on the physical aspects of biomolecular mass spectrometry. In 2014 he was appointed as distinguished professor and Limburg Chair at the University of Maastricht. He is the scientific director of M4I, the Maastricht MultiModal Molecular Imaging institute and heads the division of imaging MS. In 2019 he received the Physics Valorization prize from the Dutch national science foundation. He is the 2020 recipient of the Thomson medal of the international mass spectrometry foundation. His academic research interests are mass spectrometry‐based personalized medicine, translational molecular imaging research, high‐throughput bioinformatics, and the development and validation of new mass spectrometry‐based “omics” imaging techniques for the life sciences.

Tara Moore, Professor of Personalized Medicine, holds a chair at Ulster University in Northern Ireland and concurrently is Chief of Research and Innovation for Avellino, USA. Subsequent to her return home to Northern Ireland from Research fellowship training in Harvard Medical School, Tara has undertaken research sabbaticals in Mount Sinai Hospital New York, Dundee Wellcome Trust Laboratories, and The Stem Cell Institute at the University of Modena and Reggio. As a result of intensive training to obtain skills and expertise Tara has a wealth of experience of working in Universities and industrial laboratories focusing on the development of partner diagnostics and treatment for genetic eye disease. The research featuring in this edition is part of the Ulster Randox Industrial PhD Academy that Tara spearheaded as a mechanism of aligning the University's strategic research investment with the needs of industry. Tara's research team concentrates on identifying genes which cause disease and in developing gene editing and gene silencing treatments for autosomal dominantly inherited eye diseases. Her work is disseminated worldwide through her contribution to guest lectures and workshops and internationally co‐authored peer‐reviewed publications. The culmination of Tara's contribution to health, well‐being and safety and improving people's lives whilst inspiring others to see the opportunities to make a difference through science was recognized with the prestigious WISE award presented by HRH Princess Anne, THE Outstanding Research Supervisor of the year award and US‐Ireland Research Innovation Award. Tara is a Fellow of Royal Society of Biology, United Kingdom, Royal Society of Arts, United Kingdom, Royal Society of Medicine, United Kingdom, Senior Associate of Royal Society of Medicine and Honorary Fellow of Royal College of Physicians (London) FRCP, FFLM.

Simon Brockband, PhD, has approximately 20 years' experience in medical/clinical research activities with particular emphasis on biomarker discovery and the role proteins and posttranslational modifications play in pathology. Having gained his PhD in 2000, Simon embarked on postdoctoral studies at Queen's University Belfast employing proteomic strategies to characterize novel retinal peptides and proteins associated with rod and cone photoreceptor survival and pathology. This was followed by a further postdoctoral project aimed at investigating the expression and toxicity of α‐synuclein and its metabolites in Parkinson's patients. In April 2006, he was appointed as Lecturer in the School of Medicine & Dentistry, at Queens and investigated the phenomenon of aspirin nonresponsiveness in cardiovascular disease and then the pathobiology of AGE protein modification in the diabetic retina. Simon joined Randox Laboratories Limited in 2011 and currently focuses on innovative clinical diagnostic tests to aid in the prediction, diagnosis, and prognosis of disease and monitoring of treatment strategies/therapeutic interventions. He specializes in the development of relevant multiplexed immunoassays and partakes in the identification and definition of new research projects within Randox.

Dr. Diego Cobice, PhD, has been contributing in the field of Mass Spectrometry (MS) for more than 20 years. He obtained a Medicinal Chemistry degree in Buenos Aires, Argentina. He was Head of the Mass Spectrometry Laboratory at Boehringer Ingelheim Pharmaceuticals R&D Center in Argentina for five years. After 7 years in the pharmaceutical sector, he moved to Edinburgh to pursue an MSc in applied MS to cardiovascular disease using “Mass Spectrometry Imaging” (MSI) platforms at the University of Edinburgh. After completing his MSc, he obtained his PhD at the University of Edinburgh under the supervision of Prof. Brian Walker studying the effects of 11‐βHSD1 manipulation on glucocorticoids turnover using a novel MSI platform. This research led to the first publication that achieved spatial distribution and quantification of glucocorticoids in a mouse brain by using a novel on‐tissue chemical derivatization platform. After completing his PhD, he worked as a senior research scientist in the MSI DMPK Oncology Department at AstraZeneca, Cambridge, United Kingdom. Finally, he was appointed Director of the Mass Spectrometry Centre at the Biomedical Research Institute at Ulster University in Northern Ireland. His research interest is mainly focused on the development of novel MSI platforms for the identification of novel molecular signatures in several endocrine degenerative diseases to unravel new biological mechanisms that can lead to new therapeutic targets. He is a member of the Royal Society of Chemistry (RSC) and American Society for Mass Spectrometry, International Mass Spectrometry Foundation (affiliate delegate member), European Society of Molecular Imaging, and member of the Advisory Board Committee of the Irish Mass Spectrometry Society.

Harkin C, Smith KW, Cruickshank FL, et al. On‐tissue chemical derivatization in mass spectrometry imaging. Mass Spec Rev. 2022;41:662‐694. 10.1002/mas.21680
REFERENCES
- Altelaar AFM, Klinkert I, Jalink K, De Lange RPJ, Adan RAH, Heeren RMA, Piersma SR. Gold‐enhanced biomolecular surface imaging of cells and tissue by SIMS and MALDI mass spectrometry. Anal Chem. 2006;78:734–742. [DOI] [PubMed] [Google Scholar]
- Altelaar AFM, Luxembourg SF, Mc Donnell LA, Piersma SR, Heeren RMA. Imaging mass spectrometry at cellular length scales. Nature Protoc. 2007;2(5):1185–1196. [DOI] [PubMed] [Google Scholar]
- Altelaar AFM, Van Minnen J, Jiménez CR, Heeren RMA. Direct molecular imaging of Lymnaea stagnalis nervous tissue at subcellular spatial resolution by mass spectrometry. Anal Chem. 2005;77:735–741. [DOI] [PubMed] [Google Scholar]
- Antignac JP, de Wasch K, Monteau F, de Brabander H, Andre F, Le Bizec B. The ion suppression phenomenon in liquid chromatography‐mass spectrometry and its consequences in the field of residue analysis. Anal Chim Acta. 2005;529(1–2 Spec. Iss.):129–136. [Google Scholar]
- Arts M, Soons Z, Ellis SR, Pierzchalski KA, Balluff B, Eijkel GB, Dubois LJ, Lieuwes NG, Agten SM, Hackeng TM, van Loon LJC, Heeren RMA, Olde Damink SWM. Detection of localized hepatocellular amino acid kinetics by using mass spectrometry imaging of stable isotopes. Angew Chemie Int Ed. 2017;56(25):7146–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badu‐Tawiah A, Cooks RG. Enhanced ion signals in desorption electrospray ionization using surfactant spray solutions. J Am Soc Mass Spectrom. 2010;21(8):1423–1431. [DOI] [PubMed] [Google Scholar]
- Barré FPY, Flinders B, Garcia João P, Imke J, Huizing Lennart RS, Porta T, Creemers LB, Heeren RMA, Berta CP. Derivatization strategies for the detection of triamcinolone acetonide in cartilage by using matrix‐assisted laser desorption/ionization mass spectrometry imaging. Anal Chem. 2016;88(24):12051–12059. [DOI] [PubMed] [Google Scholar]
- Beasley E, Francese S, Bassindale T. Detection and mapping of cannabinoids in single hair samples through rapid derivatization and matrix‐assisted laser desorption ionization mass spectrometry. Anal Chem. 2016;88(20):10328–10334. [DOI] [PubMed] [Google Scholar]
- Bednařík A, Bölsker S, Soltwisch J, Dreisewerd K. An on‐tissue Paternò‐Büchi reaction for localization of carbon‐carbon double bonds in phospholipids and glycolipids by matrix‐assisted laser‐desorption‐ionization mass‐spectrometry imaging. Angew Chemie. 2018;130(37):12268–12272. [DOI] [PubMed] [Google Scholar]
- Bednařík A, Preisler J, Bezdeková D, Machálková M, Hendrych M, Navrátilová J, Knopfová L, Moskovets E, Soltwisch J, Dreisewerd K. Ozonization of tissue sections for MALDI MS imaging of carbon–carbon double bond positional isomers of phospholipids. Anal Chem. 2020;92(9):6245–6250. [DOI] [PubMed] [Google Scholar]
- Bradshaw R, Bleay S, Clench MR, Francese S. Direct detection of blood in fingermarks by MALDI MS profiling and imaging. Sci Justice. 2014;54(2):110–117. [DOI] [PubMed] [Google Scholar]
- Bradshaw R, Wolstenholme R, Ferguson LS, Sammon C, Mader K, Claude E, Blackledge RD, Clench MR, Francese S. Spectroscopic imaging based approach for condom identification in condom contaminated fingermarks. Analyst. 2013;138(9):2546–2557. [DOI] [PubMed] [Google Scholar]
- Brodbelt JS, Morrison LJ, Santos I. Ultraviolet photodissociation mass spectrometry for analysis of biological molecules. Chem Rev. 2020;120(7):3328–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhrman DL, Price PI, Rudewicz PJ. Quantitation of SR 27417 in human plasma using electrospray liquid chromatography‐tandem mass spectrometry: A study of ion suppression. J Am Soc Mass Spectrom. 1996;7(11):1099–1105. [DOI] [PubMed] [Google Scholar]
- Campbell DI, Ferreira CR, Eberlin LS, Cooks RG. Improved spatial resolution in the imaging of biological tissue using desorption electrospray ionization. Anal Bioanal Chem. 2012;404(2):389–398. [DOI] [PubMed] [Google Scholar]
- Cao Q, Wang Y, Chen B, Ma F, Hao L, Li G, Ouyang C, Li L. Visualization and identification of neurotransmitters in crustacean brain via multifaceted mass spectrometric approaches. ACS Chem Neurosci. 2019;10(3):1222–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI‐TOF MS. Anal Chem. 1997;69(23):4751–4760. [DOI] [PubMed] [Google Scholar]
- Chacon A, Zagol‐Ikapitte I, Amarnath V, Reyzer ML, Oates JA, Caprioli RM, Boutaud O. On‐tissue chemical derivatization of 3‐methoxysalicylamine for MALDI‐imaging mass spectrometry. J Mass Spectrom. 2011;46(8):840–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, Smith DR, Morrison GH. Peer reviewed: A subcellular imaging by dynamic SIMS ion microscopy. Anal Chem. 2000;72:104A–114A. [DOI] [PubMed] [Google Scholar]
- Chang CJ, Chang HY, You YW, Liao HY, Kuo YT, Kao WL, Yen GJ, Tsai MH, Shyue JJ. Parallel detection, quantification, and depth profiling of peptides with dynamic‐secondary ion mass spectrometry (D‐SIMS) ionized by C 60+‐Ar+ co‐sputtering. Anal Chim Acta. 2012;718:64–69. [DOI] [PubMed] [Google Scholar]
- Chen H, Cotte‐Rodríguez I, Cooks RG. cis‐Diol functional group recognition by reactive desorption electrospray ionization (DESI). Chem Commun. 2006; 597–599. [DOI] [PubMed] [Google Scholar]
- Cobice DF, Livingstone DEW, MacKay CL, Goodwin RJA, Smith LB, Walker BR, Andrew R. Spatial localization and quantitation of androgens in mouse testis by mass spectrometry imaging. Anal Chem. 2016;88(21):10362–10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobice DF, Livingstone DEW, McBride A, MacKay CL, Walker BR, Webster SP, Andrew R. Quantification of 11β‐hydroxysteroid dehydrogenase 1 kinetics and pharmacodynamic effects of inhibitors in brain using mass spectrometry imaging and stable‐isotope tracers in mice. Biochem Pharmacol. 2018;148:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobice DF, Mackay CL, Goodwin RJA, McBride A, Langridge‐Smith PR, Webster SP, Walker BR, Andrew R. Mass spectrometry imaging for dissecting steroid intracrinology within target tissues. Anal Chem. 2013;85(23):11576–11584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RB. Some tenets pertaining to electrospray ionization mass spectrometry. J Mass Spectrom. 2000;35(7):763–772. [DOI] [PubMed] [Google Scholar]
- Cotte‐Rodríguez I, Takáts Z, Talaty N, Chen H, Cooks RG. Desorption electrospray ionization of explosives on surfaces: Sensitivity and selectivity enhancement by reactive desorption electrospray. Anal Chem. 2005;77(21):6755–6764. [DOI] [PubMed] [Google Scholar]
- Davison AS, Strittmatter N, Sutherland H, Hughes AT, Hughes J, Bou‐Gharios G, Milan AM, Goodwin RJA, Ranganath LR, Gallagher JA. Assessing the effect of nitisinone induced hypertyrosinaemia on monoamine neurotransmitters in brain tissue from a murine model of alkaptonuria using mass spectrometry imaging. Metabolomics. 2019;15(5):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcorte A, Bertrand P. Interest of silver and gold metallization for molecular SIMS and SIMS imaging. Appl Surf Sci. 2004;231–232:250–255. [Google Scholar]
- Dilillo M, Ait‐Belkacem R, Esteve C, Pellegrini D, Nicolardi S, Costa M, Vannini E, de Graaf EL, Caleo M, McDonnell LA. Ultra‐high mass resolution MALDI imaging mass spectrometry of proteins and metabolites in a mouse model of glioblastoma. Sci Rep. 2017;7(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Duan Y. Plasma‐based ambient mass spectrometry techniques: The current status and future prospective. Mass Spectrom Rev. 2015;34(4):449–473. [DOI] [PubMed] [Google Scholar]
- Dreisewerd K. The desorption process in MALDI. Chem Rev. 2003;103(2):395–425. [DOI] [PubMed] [Google Scholar]
- Dreisewerd K, Hillenkamp F. Influence of the laser intensity and spot size on the desorption of molecules and ions in matrix‐assisted laser desorption/ionization with a uniform beam profile. Int J Mass Spectrom Ion Process. 1995;141:127–148. [Google Scholar]
- Dueñas ME, Larson EA, Lee YJ. Toward mass spectrometry imaging in the metabolomics scale: Increasing metabolic coverage through multiple on‐tissue chemical modifications. Front Plant Sci. 2019;10(July):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan KD, Bergman HM, Lanekoff I. A pneumatically assisted nanospray desorption electrospray ionization source for increased solvent versatility and enhanced metabolite detection from tissue. Analyst. 2017;142(18):3424–3431. [DOI] [PubMed] [Google Scholar]
- Eikel D, Vavrek M, Smith S, Bason C, Yeh S, Korfmacher WA, Henion JD. Liquid extraction surface analysis mass spectrometry (LESA‐MS) as a novel profiling tool for drug distribution and metabolism analysis: The terfenadine example. Rapid Commun Mass Spectrom. 2011;23(July):3587–3596. [DOI] [PubMed] [Google Scholar]
- Ellis SR, Soltwisch J, Paine MRL, Dreisewerd K, Heeren RMA. Laser post‐ionisation combined with a high resolving power orbitrap mass spectrometer for enhanced MALDI‐MS imaging of lipids. Chem Commun. 2017;53(53):7246–7249. [DOI] [PubMed] [Google Scholar]
- Enke CG. A predictive model for matrix and analyte effects in electrospray ionization of singly‐charged ionic analytes. Anal Chem. 1997;69(23):4885–4893. [DOI] [PubMed] [Google Scholar]
- Enomoto H, Sensu T, Yumoto E, Yokota T, Yamane H. Derivatization for detection of abscisic acid and 12‐oxo‐phytodienoic acid using matrix‐assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun Mass Spectrom. 2018;32(17):1565–1572. [DOI] [PubMed] [Google Scholar]
- Enomoto Y, Phan ANT, Yamaguchi M, Fukusaki E, Shimma S. Mass spectrometric imaging of GABA in the Drosophila melanogaster adult head. Anal Sci. 2018;34(9):1055–1059. [DOI] [PubMed] [Google Scholar]
- Esteve C, Tolner EA, Shyti R, van den Maagdenberg AMJM, McDonnell LA. Mass spectrometry imaging of amino neurotransmitters: A comparison of derivatization methods and application in mouse brain tissue. Metabolomics. 2016;12(2):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Zhang J, Chang C, Li L, Li M, Xiong X, Guo C, Tang F, Bai Y, Liu H. Ambient mass spectrometry imaging: Plasma assisted laser desorption ionization mass spectrometry imaging and its applications. Anal Chem. 2014;86(9):4164–4169. [DOI] [PubMed] [Google Scholar]
- Ferguson LS, Creasey S, Wolstenholme R, Clench MR, Francese S. Efficiency of the dry‐wet method for the MALDI‐MSI analysis of latent fingermarks. J Mass Spectrom. 2013;48(6):677–684. [DOI] [PubMed] [Google Scholar]
- Fletcher JS, Szakal C. Cluster and polyatomic primary ion beams. In: Vickerman JC, Briggs D, eds. TOF‐SIMS: Materials Analysis by Mass Spectrometry. 2nd Ed. UK: IM Publications LLP & SurfaceSpectra Ltd.; 2013:291–310. [Google Scholar]
- Flinders B, Huizing LRS, van Heerden M, Cuyckens F, Neumann UP, van der Laan LJW, Damink SWMO, Heeren RMA, Schaap FG, Vreeken RJ. Cross‐species molecular imaging of bile salts and lipids in liver: Identification of molecular structural markers in health and disease. Anal Chem. 2018;90:11835−11846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinders B, Morrell J, Marshall PS, Ranshaw LE, Clench MR. The use of hydrazine‐based derivatization reagents for improved sensitivity and detection of carbonyl containing compounds using MALDI‐MSI. Anal Bioanal Chem. 2015;407(8):2085–2094. [DOI] [PubMed] [Google Scholar]
- Francese S, Bradshaw R, Ferguson LS, Wolstenholme R, Clench MR, Bleay S. Beyond the ridge pattern: Multi‐informative analysis of latent fingermarks by MALDI mass spectrometry. Analyst. 2013;138(15):4215–4228. [DOI] [PubMed] [Google Scholar]
- Franck J, Arafah K, Barnes A, Wisztorski M, Salzet M, Fournier I. Improving tissue preparation for matrix‐assisted laser desorption ionization mass spectrometry imaging. Part 1: Using microspotting. Anal Chem. 2009;81(19):8193–8202. [DOI] [PubMed] [Google Scholar]
- Fülöp A, Bausbacher T, Rizzo S, Zhou Q, Gillandt H, Hopf C, Rittner M. New derivatization reagent for detection of free thiol‐groups in metabolites and proteins in matrix‐assisted laser desorption/ionization mass spectrometry imaging. Anal Chem. 2020;92(9):6224–6228. [DOI] [PubMed] [Google Scholar]
- Gemperline E, Rawson S, Li L. Optimization and comparison of multiple MALDI matrix application methods for small molecule mass spectrometric imaging. Anal Chem. 2014;86(20):10030–10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod M, Shi Y, Cheng JX, Cooks RG. mapping lipid alterations in traumatically injured rat spinal cord by desorption electrospray ionization imaging mass spectrometry. Anal Chem. 2012;83(1):207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin RJ, MacIntyre L, Watson DG, Scullion SP, Pitt AR. A solvent‐free matrix application method for matrix‐assisted laser desorption/ionization imaging of small molecules. Rapid Commun Mass Spectrom. 2010;24:1682–1686. [DOI] [PubMed] [Google Scholar]
- Goodwin RJA, Iverson SL, Andren PE. The significance of ambient‐temperature on pharmaceutical and endogenous compound abundance and distribution in tissues sections when analyzed by matrix‐assisted laser desorption/ionization mass spectrometry imaging. Rapid Commun Mass Spectrom. 2012;26(5):494–498. [DOI] [PubMed] [Google Scholar]
- Green FM, Salter TL, Gilmore IS, Stokes P, Connor GO. The effect of electrospray solvent composition on desorption electrospray ionisation (DESI) efficiency and spatial resolution. Analyst. 2010;135:731–737. [DOI] [PubMed] [Google Scholar]
- Griffiths RL, Creese AJ, Race AM, Bunch J, Cooper HJ. LESA FAIMS mass spectrometry for the spatial profiling of proteins from tissue. Anal Chem. 2016;88:6758–6766. [DOI] [PubMed] [Google Scholar]
- Griffiths RL, Hughes JW, Abbatiello SE, Belford MW, Styles IB, Cooper HJ. Comprehensive LESA mass spectrometry imaging of intact proteins by integration of cylindrical FAIMS. Anal Chem. 2020;92(4):2885–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeneveld G, de Puit M, Bleay S, Bradshaw R, Francese S. Detection and mapping of illicit drugs and their metabolites in fingermarks by MALDI MS and compatibility with forensic techniques. Sci Rep. 2015;5:0–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan M, Zhang Z, Li S, Liu J, Liu L, Yang H, Zhang Y, Wang T, Zhao Z. Silver nanoparticles as matrix for MALDI FTICR MS profiling and imaging of diverse lipids in brain. Talanta. 2018;179(December 2017):624–631. [DOI] [PubMed] [Google Scholar]
- Guo S, Tang W, Hu Y, Chen Y, Gordon A, Li B, Li P. Enhancement of on‐tissue chemical derivatization by laser‐assisted tissue transfer for MALDI MS imaging. Anal Chem. 2019;92:1431−1438. [DOI] [PubMed] [Google Scholar]
- Hale OJ, Sisley EK, Griffiths RL, Styles IB, Cooper HJ. Native LESA TWIMS‐MSI: Spatial, conformational, and mass analysis of proteins and protein complexes. J Am Soc Mass Spectrom. 2020;31(4):873–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JS. Comparison of SIMS and MALDI for Mass Spectrometric Imaging. In: Setou M, ed. Imaging Mass Spectrometry. Tokyo: Springer; 2010:235–257. [Google Scholar]
- Havlikova J, Randall EC, Griffiths RL, Swales JG, Goodwin RJA, Bunch J, Styles IB, Cooper HJ. Quantitative imaging of proteins in tissue by stable isotope‐labeled mimetic liquid extraction surface analysis mass spectrometry. Anal Chem. 2019;91:14198−14202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi T, Yamauchi A, Shimada K. 2‐Hydrazino‐1‐methylpyridine: A highly sensitive derivatization reagent for oxosteroids in liquid chromatography‐electrospray ionization‐mass spectrometry. J Chromatogr B: Anal Technol Biomed Life Sci. 2005;825(2):214–222. [DOI] [PubMed] [Google Scholar]
- Holst S, Heijs B, de Haan N, van Zeijl RJM, Briaire‐De Bruijn IH, van Pelt GW, Mehta AS, Angel PM, Mesker WE, Tollenaar RA, Drake RR, Bovée JVMG, McDonnell LA, Wuhrer M. Linkage‐specific in situ sialic acid derivatization for N‐glycan mass spectrometry imaging of formalin‐fixed paraffin‐embedded tissues. Anal Chem. 2016;88(11):5904–5913. [DOI] [PubMed] [Google Scholar]
- Huang G, Chen H, Zhang X, Cooks RG, Ouyang Z. Rapid screening of anabolic steroids in urine by reactive desorption electrospray ionization achieved by combining reactive desorption electrospray. Anal Chem. 2007;79(21):8327–8332. [DOI] [PubMed] [Google Scholar]
- Ifa DR, Wiseman JM, Song Q, Cooks RG. Development of capabilities for imaging mass spectrometry under ambient conditions with desorption electrospray ionization (DESI). Int J Mass Spectrom. 2007;259(1–3):8–15. [Google Scholar]
- Ito T, Hiramoto M. Use of mTRAQ derivatization reagents on tissues for imaging neurotransmitters by MALDI imaging mass spectrometry: The triple spray method. Anal Bioanal Chem. 2019;411(26):6847–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SN, Baldwin K, Muller L, Womack VM, Schultz JA, Balaban C, Woods AS. Imaging of lipids in rat heart by MALDI‐MS with silver nanoparticles. Anal Bioanal Chem. 2014;406(5):1377–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya I, Brülls SM, Dunevall J, Jennische E, Lange S, Martensson J, Ewing AG, Malmberg P, Fletcher JS. On‐tissue chemical derivatization of catecholamines using 4‐(N‐methyl)pyridinium boronic acid for ToF‐SIMS and LDI‐ToF mass spectrometry imaging. Anal Chem. 2018;90(22):13580–13590. [DOI] [PubMed] [Google Scholar]
- Kebarle P. A brief overview of the present status of the mechanisms involved in electrospray mass spectrometry. J Mass Spectrom. 2000;35:804–817. [DOI] [PubMed] [Google Scholar]
- Kebarle P, Tang L. From ions in solution to ions in the gas phase. Anal Chem. 1993;65(22):972A–986A. [Google Scholar]
- Kertesz V, van Berkel GJ. Liquid microjunction surface sampling coupled with high‐pressure liquid chromatography‐electrospray ionization‐mass spectrometry for analysis of drugs and metabolites in whole‐body thin tissue sections. Anal Chem. 2010;82(14):5917–5921. [DOI] [PubMed] [Google Scholar]
- Klein DR, Feider CL, Garza KY, Lin JQ, Eberlin LS, Brodbelt JS. Desorption electrospray ionization coupled with ultraviolet photodissociation for characterization of phospholipid isomers in tissue sections. Anal Chem. 2018;90(17):10100–10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo TH, Chung HH, Chang HY, Lin CW, Wang MY, Shen TL, Hsu CC. Deep lipidomics and molecular imaging of unsaturated lipid isomers: A universal strategy initiated by mCPBA epoxidation. Anal Chem. 2019;91(18):11905–11915. [DOI] [PubMed] [Google Scholar]
- Lanekoff I, Heath BS, Liyu A, Thomas M, Carson JP, Laskin J. Automated platform for high‐resolution tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2012;84(19):8351–8356. [DOI] [PubMed] [Google Scholar]
- Laskin J, Heath, BS , Roach, PJ , Cazares L, Semmes, JO . Tissue imaging using nanospray DESI. Anal Chem. 2012;84(1):141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauzon N, Dufresne M, Chauhan V, Chaurand P. Development of laser desorption imaging mass spectrometry methods to investigate the molecular composition of latent fingermarks. J Am Soc Mass Spectrom. 2015;26(6):878–886. [DOI] [PubMed] [Google Scholar]
- Leopold J, Popkova Y, Engel KM, Schiller J. Recent developments of useful MALDI matrices for the mass spectrometric characterization of lipids. Biomolecules. 2018;8(4):173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang Y, Liu J, Han J, Guan M, Yang H, Lin Y, Xiong S, Zhao Z. Electrospray deposition device used to precisely control the matrix crystal to improve the performance of MALDI MSI. Sci Rep. 2016;6(June):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hummon AB. Chemical imaging of platinum‐based drugs and their metabolites. Sci Rep. 2016;6(November):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ma X, Lin Z, He M, Han G, Yang C, Xing Z, Zhang S, Zhang X. Imaging mass spectrometry with a low‐temperature plasma probe for the analysis of works of art. Angew Chemie Int Ed. 2010;49(26):4435–4437. [DOI] [PubMed] [Google Scholar]
- Ljungdahl A, Hanrieder J, Fälth M, Bergquist J, Andersson M. Imaging mass spectrometry reveals elevated nigral levels of dynorphin neuropeptides in L‐DOPA‐induced dyskinesia in rat model of Parkinson's disease. PLOS One. 2011;6(9):e25653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lostun D, Perez CJ, Licence P, Barrett DA, Ifa DR. Reactive DESI‐MS imaging of biological tissues with dicationic ion‐pairing compounds. Anal Chem. 2015;87:3286−3293 [DOI] [PubMed] [Google Scholar]
- Luxembourg SL, Mize TH, McDonnell LA, Heeren RMA. High‐spatial resolution mass spectrometric imaging of peptide and protein distributions on a surface. Anal Chem. 2004;76(18):5339–5344. [DOI] [PubMed] [Google Scholar]
- Ma G, Zhao X, Guo C, Li S, Liu Y, He Q, Chen K, Pan Y. Glycosylamines‐based reactive matrix designed for imaging acidity in Ponkan fruit using matrix assisted laser desorption/ionization mass spectrometry imaging. Anal Chim Acta. 2018;1041:78–86. [DOI] [PubMed] [Google Scholar]
- Manicke NE, Nefliu M, Chunping W, Woods JW, Reiser V, Hendrickson RC, Cooks RG. Imaging of lipids in atheroma by desorption electrospray ionization mass spectrometry. Anal Chem. 2009;81(21):8702–8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manier ML, Reyzer ML, Goh A, Dartois V, Via LE, Barry CE, Caprioli RM. Reagent precoated targets for rapid in‐tissue derivatization of the anti‐tuberculosis drug isoniazid followed by MALDI imaging mass spectrometry. J Am Soc Mass Spectrom. 2011;22(8):1409–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manier ML, Spraggins JM, Reyzer ML, Norris JL, Caprioli RM. A derivatization and validation strategy for determining the spatial localization of endogenous amine metabolites in tissues using MALDI imaging mass spectrometry. J Mass Spectrom. 2014;49(8):665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezger STP, Mingels AMA, Bekers O, Cillero‐Pastor B, Heeren RMA. Trends in mass spectrometry imaging for cardiovascular diseases. Anal Bioanal Chem. 2019;411(17):3709–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller L, Kailas A, Jackson SN, Roux A, Barbacci D, Schultz JA, Balaban C, Woods AS. Lipid imaging within the normal rat kidney using silver nanoparticles by matrix‐assisted laser desorption/ionization mass spectrometry. Kidney Int. 2015;88(1):186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem. 2007;79(21):8098–8106. [DOI] [PubMed] [Google Scholar]
- Nguyen SN, Sontag RL, Carson JP, Corley RA, Ansong C, Laskin J. Towards high‐resolution tissue imaging using nanospray desorption electrospray ionization mass spectrometry coupled to shear force microscopy. J Am Soc Mass Spectrom. 2018;29(2):316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus M, Soltwisch J, Belov ME, Dreisewerd K. Transmission‐mode MALDI‐2 mass spectrometry imaging of cells and tissues at subcellular resolution. Nat Methods. 2019;16(9):925–931. [DOI] [PubMed] [Google Scholar]
- Nygren H, Johansson BR, Malmberg P. Bioimaging TOF‐SIMS of tissues by gold ion bombardment of a silver‐coated thin section. Microsc Res Technol. 2004;65(6):282–286. [DOI] [PubMed] [Google Scholar]
- Nygren H, Malmberg P, Kriegeskotte C, Arlinghaus HF. Bioimaging TOF‐SIMS: Localization of cholesterol in rat kidney sections. FEBS Lett. 2004;566(1–3):291–293. [DOI] [PubMed] [Google Scholar]
- O'Neill KC, Lee YJ. Visualizing genotypic and developmental differences of free amino acids in maize roots with mass spectrometry imaging. Front Plant Sci. 2020;11(May):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacholski ML, Winograd N. Imaging with mass spectrometry. Chem Rev. 1999;99:2977–3005. [DOI] [PubMed] [Google Scholar]
- Paine MRL, Poad BLJ, Eijkel GB, Marshall DL, Blanksby SJ, Heeren RMA, Ellis SR. Mass spectrometry imaging with isomeric resolution enabled by ozone‐induced dissociation. Angew Chemie Int Ed. 2018;57(33):10530–10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez CJ, Bagga AK, Prova SS, Yousefi TM, Ifa DR. Review and perspectives on the applications of mass spectrometry imaging under ambient conditions. Rapid Commun Mass Spectrom. 2019;33(S3):27–53. [DOI] [PubMed] [Google Scholar]
- Quirke JME, van Berkel GJ. Electrospray tandem mass spectrometric study of alkyl 1‐methylpyridinium ether derivatives of alcohols. J Mass Spectrom. 2001;36(12):1294–1300. [DOI] [PubMed] [Google Scholar]
- Roach PJ, Laskin J, Laskin A. Nanospray desorption electrospray ionization: An ambient method for liquid‐extraction surface sampling in mass spectrometry. Analyst. 2010;135(9):2233–2236. [DOI] [PubMed] [Google Scholar]
- Román JK, Walsh CM, Oh J, Dana CE, Hong S, Jo KD, Alleyne M, Miljkovic N, Cropek DM. Spatially resolved chemical analysis of cicada wings using laser‐ablation electrospray ionization (LAESI) imaging mass spectrometry (IMS). Anal Bioanal Chem. 2018;410(7):1911–1921. [DOI] [PubMed] [Google Scholar]
- Russo C, Lewis EEL, Flint L, Clench MR. Mass spectrometry imaging of 3D tissue models. Proteomics. 2018;18(14):1–4. [DOI] [PubMed] [Google Scholar]
- Shariatgorji M, Källback P, Gustavsson L, Schintu N, Svenningsson P, Goodwin RJA, Andren PE. Controlled‐pH tissue cleanup protocol for signal enhancement of small molecule drugs analyzed by MALDI‐MS imaging. Anal Chem. 2012;84(10):4603–4607. [DOI] [PubMed] [Google Scholar]
- Shariatgorji M, Nilsson A, Fridjonsdottir E, Vallianatou T, Källback P, Katan L, Sävmarker J, Mantas I, Zhang X, Bezard E, Svenningsson P, Odell LR, Andrén PE. Comprehensive mapping of neurotransmitter networks by MALDI–MS imaging. Nat Methods. 2019;16(10):1021–1028. [DOI] [PubMed] [Google Scholar]
- Shariatgorji M, Nilsson A, Goodwin RJA, Källback P, Schintu N, Zhang X, Crossman AR, Bezard E, Svenningsson P, Andren PE. Direct targeted quantitative molecular imaging of neurotransmitters in brain tissue sections. Neuron. 2014;84(4):697–707. [DOI] [PubMed] [Google Scholar]
- Shariatgorji M, Nilsson A, Källback P, Karlsson O, Zhang X, Svenningsson P, Andren PE. Pyrylium salts as reactive matrices for MALDI‐MS imaging of biologically active primary amines. J Am Soc Mass Spectrom. 2015;26(6):934–939. [DOI] [PubMed] [Google Scholar]
- Shariatgorji R, Nilsson A, Strittmatter N, Vallianatou T, Zhang X, Svenningsson P, Goodwin RJA, Andren PE, Bromopyrylium derivatization facilitates identification by mass spectrometry imaging of monoamine neurotransmitters and small molecule neuroactive compounds. J Am Soc Mass Spectrom. 2020;31:2553‐2557 [DOI] [PubMed] [Google Scholar]
- Shariatgorji M, Strittmatter N, Nilsson A, Källback P, Alvarsson A, Zhang X, Vallianatou T, Svenningsson P, Goodwin RJA, Andren PE. Simultaneous imaging of multiple neurotransmitters and neuroactive substances in the brain by desorption electrospray ionization mass spectrometry. NeuroImage. 2016;136:129–138. [DOI] [PubMed] [Google Scholar]
- Shelley JT, Ray SJ, Hieftje GM. Laser ablation coupled to a flowing atmospheric pressure afterglow for ambient mass spectral imaging. Anal Chem. 2008;80:8308–8313. [DOI] [PubMed] [Google Scholar]
- Sheraz née Rabbani S, Razo IB, Kohn T, Lockyer NP, Vickerman JC. Enhancing ion yields in time‐of‐flight‐secondary ion mass spectrometry: A comparative study of argon and water cluster primary beams. Anal Chem. 2015;87:2367–2374. [DOI] [PubMed] [Google Scholar]
- Shimma S, Kumada HO, Taniguchi H, Konno A, Yao I, Furuta K, Matsuda T, Ito S. Microscopic visualization of testosterone in mouse testis by use of imaging mass spectrometry. Anal Bioanal Chem. 2016;408(27):7607–7615. [DOI] [PubMed] [Google Scholar]
- Shin E, Cha S. In situ probing citrullinated sites in a peptide by reactive desorption electrospray ionization mass spectrometry. Bull Korean Chem Soc. 2018;39:40–44. [Google Scholar]
- Smith KW, Flinders B, Thompson PD, Cruickshank FL, Mackay CL, Heeren RMA, Cobice DF. Spatial localization of vitamin D metabolites in mouse kidney by mass spectrometry imaging. ACS Omega. 2020;5(22):13430–13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KW, Thompson PD, Rodriguez EP, Mackay CL, Cobice DF. Effects of vitamin D as a regulator of androgen intracrinology in LNCAP prostate cancer cells. Biochem Biophys Res Commun. 2019;519(3):579–584. [DOI] [PubMed] [Google Scholar]
- Smoluch M, Mielczarek P, Silberring J. Plasma‐based ambient ionization mass spectrometry in bioanalytical sciences. Mass Spectrom Rev. 2016;35(1):22–34. [DOI] [PubMed] [Google Scholar]
- Soltwisch J, Heijs B, Koch A, Vens‐Cappell S, Höhndorf J, Dreisewerd K. MALDI‐2 on a trapped ion mobility quadrupole time‐of‐flight instrument for rapid mass spectrometry imaging and ion mobility separation of complex lipid profiles. Anal Chem. 2020;92:8697−8703. [DOI] [PubMed] [Google Scholar]
- Soltwisch J, Kettling H, Vens‐Cappell S, Wiegelmann M, Muthing J, Dreisewerd K. Mass spectrometry imaging with laser‐induced postionization. Science. 2015;348(6231):211–215. [DOI] [PubMed] [Google Scholar]
- Specker JT, van Orden SL, Ridgeway ME, Prentice BM. Identification of phosphatidylcholine isomers in imaging mass spectrometry using gas‐phase charge inversion ion/ion reactions. Anal Chem. 2020;92(19):13192–13201. [DOI] [PubMed] [Google Scholar]
- Spivey EC, McMillen JC, Ryan DJ, Spraggins JM, Caprioli RM. Combining MALDI‐2 and transmission geometry laser optics to achieve high sensitivity for ultra‐high spatial resolution surface analysis. J Mass Spectrom. 2019;54(4):366–370. [DOI] [PubMed] [Google Scholar]
- Sugiura Y, Takeo E, Shimma S, Yokota M, Higashi T, Seki T, Mizuno Y, Oya M, Kosaka T, Omura M, Nishikawa T, Suematsu M, Nishimoto K. Aldosterone and 18‐oxocortisol coaccumulation in aldosterone‐producing lesions. Hypertension. 2018;72(6):1345–1354. [DOI] [PubMed] [Google Scholar]
- Sugiyama E, Guerrini MM, Honda K, Hattori Y, Abe M, Källback P, Andrén PE, Tanaka KF, Setou M, Fagarasan S, Suematsu M, Sugiura Y. Detection of a high‐turnover serotonin circuit in the mouse brain using mass spectrometry imaging. iScience. 2019;20:359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Li Z, Ma C, Zang Q, Li J, Liu W, Zhao H, Wang X. Acetone immersion enhanced MALDI‐MS imaging of small molecule metabolites in biological tissues. J Pharm Biomed Anal. 2019;176:112797. [DOI] [PubMed] [Google Scholar]
- Sun C, Liu W, Geng Y, Wang X. On‐tissue derivatization strategy for mass spectrometry imaging of carboxyl‐containing metabolites in biological tissues. Anal Chem. 2020;92(18):12126–12131. [DOI] [PubMed] [Google Scholar]
- Swales JG, Strittmatter N, Tucker JW, Clench MR, Webborn PJH, Goodwin RJA. Spatial quantitation of drugs in tissues using liquid extraction surface analysis mass spectrometry imaging. Nat Publ Gr. 2016;6(June):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts Z, Strittmatter N, McKenzie JS. Ambient Mass Spectrometry in Cancer Research. 1st ed. 134, Advances in Cancer Research. Elsevier Inc. 2017: p. 231– 256 [DOI] [PubMed] [Google Scholar]
- Takáts Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306(5695):471–473. [DOI] [PubMed] [Google Scholar]
- Takeo E, Sugiura Y, Uemura T, Nishimoto K, Yasuda M, Sugiyama E, Ohtsuki S, Higashi T, Nishikawa T, Suematsu M, Fukusaki E, Shimma S. Tandem mass spectrometry imaging reveals distinct accumulation patterns of steroid structural isomers in human adrenal glands. Anal Chem. 2019;91(14):8918–8925. [DOI] [PubMed] [Google Scholar]
- Tillner J, McKenzie JS, Jones EA, Speller AVM, Walsh JL, Veselkov KA, Bunch J, Takats Z, Gilmore IS. Investigation of the impact of desorption electrospray ionization sprayer geometry on its performance in imaging of biological tissue. Anal Chem. 2016;88(9):4808–4816. [DOI] [PubMed] [Google Scholar]
- Tillner J, Wu V, Jones EA, Pringle SD, Karancsi T, Dannhorn A, Veselkov K, McKenzie JS, Takáts Z. Faster, more reproducible DESI‐MS for biological tissue imaging. J Am Soc Mass Spectrom. 2017;28(10):2090–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toue S, Sugiura Y, Kubo A, Ohmura M, Karakawa S, Mizukoshi T, Yoneda J, Miyano H, Noguchi Y, Kobayashi T, Kabe Y, Suematsu M. Microscopic imaging mass spectrometry assisted by on‐tissue chemical derivatization for visualizing multiple amino acids in human colon cancer xenografts. Proteomics. 2014;14(7–8):810–819. [DOI] [PubMed] [Google Scholar]
- Tucker LH, Conde‐González A, Cobice DF, Hamm GR, Goodwin RJA, Campbell CJ, Clarke DJ, Mackay CL. MALDI matrix application utilizing a modified 3D printer for accessible high resolution mass spectrometry imaging. Anal Chem. 2018;90(15):8742–8749. [DOI] [PubMed] [Google Scholar]
- van Berkel GJ, Kertesz V, Koeplinger K, Vavrek M, Ah‐Ng TK. Liquid microjunction surface sampling probe electrospray mass spectrometry for detection of drugs and metabolites in thin tissue sections. J Mass Spectrom. 2008;43:500–508. [DOI] [PubMed] [Google Scholar]
- van Geenen FAMG, Franssen MCR, Schotman AHM, Zuilhof H, Nielen MWF. Ambient characterization of synthetic fibers by laser ablation electrospray ionization mass spectrometry. Anal Chem. 2017;89(7):4031–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter A, Sojka PE, Cooks RG. Droplet dynamics and ionization mechanisms in desorption electrospray ionization mass spectrometry. Anal Chem. 2006;78(24):8549–8555. [DOI] [PubMed] [Google Scholar]
- Wäldchen F, Spengler B, Heiles S. Reactive matrix‐assisted laser desorption/ionization mass spectrometry imaging using an intrinsically photoreactive paternò‐büchi matrix for double‐bond localization in isomeric phospholipids. J Am Chem Soc. 2019;141(30):11816–11820. [DOI] [PubMed] [Google Scholar]
- Wang SS, Wang YJ, Zhang J, Sun TQ, Guo YL. Derivatization strategy for simultaneous molecular imaging of phospholipids and low‐abundance free fatty acids in thyroid cancer tissue sections. Anal Chem. 2019;91(6):4070–4076. [DOI] [PubMed] [Google Scholar]
- Wang X, Han J, Yang J, Pan J, Borchers CH. Matrix coating assisted by an electric field (MCAEF) for enhanced tissue imaging by MALDI‐MS. Chem Sci. 2015;6(1):729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman JM, Ifa DR, Zhu Y, Kissinger CB, Manicke NE, Kissingera PT, Cooks RG. Desorption electrospray ionization mass spectrometry: Imaging drugs and metabolites in tissues. Proc Natl Acad Sci USA. 2008;105(47):18120–18125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Dill AL, Eberlin LS, Cooks RG, Ifa DR. Mass spectrometry imaging under ambient conditions. Mass Spectrom Rev. 2013;32(3):218–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Ifa DR, Manicke NE, Cooks RG. Rapid, direct analysis of cholesterol by charge labeling in reactive desorption electrospray ionization. Anal Chem. 2009;81(18):7618–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Ifa DR, Manicke NE, Cooks RG. Molecular imaging of adrenal gland by desorption electrospray ionization mass spectrometry. Analyst. 2010;135(1):28–32. [DOI] [PubMed] [Google Scholar]
- Wu Q, Comi TJ, Li B, Rubakhin SS, Sweedler JV. On‐tissue derivatization via electrospray deposition for matrix‐assisted laser desorption/ionization mass spectrometry imaging of endogenous fatty acids in rat brain tissues. Anal Chem. 2016;88(11):5988–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Kliman M, Forsythe JG, Korade Z, Hmelo AB, Porter NA, McLean JA. Profiling and imaging ion mobility‐mass spectrometry analysis of cholesterol and 7‐dehydrocholesterol in cells via sputtered silver MALDI. J Am Soc Mass Spectrom. 2015;26(6):924–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Burnum‐Johnson KE, Sun X, Dey SK, Laskin J. High spatial resolution imaging of biological tissues using nanospray desorption electrospray ionization mass spectrometry. Nat Protoc. 2019;14(12):3445–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Kyle J, Burnum‐Johnson K, Bloodsworth K, Sussel L, Ansong C, Laskin J. High spatial resolution imaging of mouse pancreatic islets using nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2018;90(11):6548–6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutuc E, Angelini R, Baumert M, Mast N, Pikuleva I, Newton J, Clench MR, Skibinski DOF, Howell OW, Wang Y, Griffiths WJ. Localization of sterols and oxysterols in mouse brain reveals distinct spatial cholesterol metabolism. Proc Natl Acad Sci USA. 2020;117(11):5749–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaikin VG, Halket JM. Derivatization in mass spectrometry—8. Soft ionization mass spectrometry of small molecules. Eur J Mass Spectrom. 2006;12(2):79–115. [DOI] [PubMed] [Google Scholar]
- Zenobi R, Knochenmuss R. Ion formation in maldi mass spectrometry. Mass Spectrom Rev. 1998;17(5):337–366. [Google Scholar]
- Zhang Y, Chen H. Detection of saccharides by reactive desorption electrospray ionization (DESI) using modified phenylboronic acids. Int J Mass Spectrom. 2010;289:98–107. [Google Scholar]
- Zhou D, Guo S, Zhang M, Liu Y, Chen T, Li Z. Mass spectrometry imaging of small molecules in biological tissues using graphene oxide as a matrix. Anal Chim Acta. 2017;962(January):52–59. [DOI] [PubMed] [Google Scholar]
- Zimmerman TA, Monroe EB, Sweedler JV. Adapting the stretched sample method from tissue profiling to imaging.. Proteomics. 2008;8(18):3809–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]