Abstract
Previous genetic and biochemical studies have confirmed that hemoglobin and hemin utilization in Porphyromonas gingivalis is mediated by the outer membrane hemoglobin and heme receptor HmuR, as well as gingipain K (Kgp), a lysine-specific cysteine protease, and gingipain R1 (HRgpA), one of two arginine-specific cysteine proteases. In this study we report on the binding specificity of the recombinant P. gingivalis HmuR protein and native gingipains for hemoglobin, hemin, various porphyrins, and metalloporphyrins as assessed by spectrophotometric assays, by affinity chromatography, and by enzyme-linked immunosorbent assay. Protoporphyrin, mesoporphyrin, deuteroporphyrin, hematoporphyrin, and some of their iron, copper, and zinc derivatives were examined to evaluate the role of both the central metal ion and the peripheral substituents on binding to recombinant HmuR and soluble gingipains. Scatchard analysis of hemin binding to Escherichia coli cells expressing recombinant membrane-associated six-His-tagged HmuR yielded a linear plot with a binding affinity of 2.4 × 10−5 M. Recombinant E. coli cells bound the iron, copper, and zinc derivatives of protoporphyrin IX (PPIX) with similar affinities, and approximately four times more tightly than PPIX itself, which suggests that the active site of HmuR contains a histidine that binds the metal ion in the porphyrin ring. Furthermore, we found that recombinant HmuR prefers the ethyl and vinyl side chains of the PPIX molecule to either the larger hydroxyethyl or smaller hydrogen side chains. Kgp and HRgpA were demonstrated to bind various porphyrins and metalloporphyrins with affinities similar to those for hemin, indicating that the binding of Kgp and HRgpA to these porphyrins does not require a metal within the porphyrin ring. We did not detect the binding of RgpB, the arginine-specific cysteine protease that lacks a C-terminal hemagglutinin domain, to hemoglobin, porphyrins, or metalloporphyrins. Kgp and HRgpA, but not RgpB, were demonstrated to bind directly to soluble recombinant six-His-tagged HmuR. Several possible mechanisms for the cooperation between outer membrane receptor HmuR and proteases Kgp and HRgpA in hemin and hemoglobin binding and utilization are discussed.
Passive heme uptake through the outer membranes of gram-negative bacteria is not a significant route of heme entry (24, 33), and most bacteria possess specific heme uptake systems to use this compound as either an iron or iron-porphyrin source (reviewed in reference 20). In most gram-negative bacteria heme utilization is mediated by specific outer membrane receptors that bind directly to host heme-sequestering proteins. Several gram-negative bacteria also produce extracellular heme-binding proteins (hemophores). These secreted proteins extract heme from hemoglobin and deliver it to an outer membrane-associated protein, which transports heme into the cell. The best-characterized system is that of Serratia marcescens-secreted protein HasA, which captures heme and hemoglobin and delivers it to outer membrane receptor HasR (21, 28).
Porphyromonas gingivalis, the etiological agent of adult periodontal disease, requires hemin for growth (17, 46). The binding and utilization of hemoglobin (2, 16, 25, 52) and hemin (5, 17, 54) have been demonstrated in P. gingivalis and related species. P. gingivalis expresses several outer membrane proteins in response to iron and heme limitation (5, 51); however, the role of these proteins in heme transport is not well defined. Several reports have also described P. gingivalis genes hemR, ihtA, and tlr, which exhibit homology to genes encoding TonB-dependent receptors (13, 23, 49). A role for the protein products of the P. gingivalis hemR, ihtA, and tlr genes has not been delineated because the respective P. gingivalis mutants have not been isolated.
We have described a P. gingivalis heme and hemoglobin receptor (heme/hemoglobin receptor; HmuR) which has homology with TonB-dependent outer membrane hemoglobin/heme receptors (50). P. gingivalis hmuR mutant cells bound less hemoglobin and hemin than did the parental strain and exhibited diminished growth with hemoglobin or hemin (50). Furthermore, we demonstrated that recombinant HmuR expressed in E. coli bound hemin and hemoglobin (50). Amino acid comparisons of the conserved motifs of several different hemoglobin/heme receptors and the P. gingivalis HmuR protein revealed that HmuR contains highly conserved domains containing invariant histidine residues (His95 and His434), glutamic acid residues (Glu448 and Glu458), and the FRAP (in HmuR YRAP) and NPNL (in HmuR NPDL) amino acid boxes, which may be involved in hemoglobin and heme binding (4, 50). It was previously shown that the hemR gene is identical with the hmuR gene in the N-terminal portion but that these two genes differ in their C termini (23, 50). Despite the fact that previous studies have determined that hemR is present in strains 53977, 381, and W50, we were unable to amplify the hemR gene from P. gingivalis A7436, suggesting that in this strain hemin transport can occur independently of HemR.
In addition to conventional outer membrane receptors, heme and hemoglobin utilization in P. gingivalis also requires participation of the cysteine proteases referred to as gingipains (12, 19, 26). The gingipains exhibit proteolytic enzymatic activity against a range of host proteins including host proteinase inhibitors, immunoglobulins, iron-sequestering proteins, extracellular matrix proteins, bactericidal proteins and peptides, and proteins involved in the coagulation, complement, and kallikrein/kinin cascades (15, 22, 31, 45). P. gingivalis lysine-specific gingipain K (Kgp) and arginine-specific gingipain R1 (HRgpA) are purified as noncovalent complexes of the catalytic domain associated with four polypeptide chains derived from the hemagglutinin domain (3, 11, 36, 37, 40, 41, 42). These gingipains occur either in extracellular soluble or in membrane-associated forms (40, 41). In contrast to Kgp and HRgpA, a second arginine-specific gingipain, R2 (RgpB), contains only a catalytic domain (44) and is not required for hemoglobin and heme utilization in P. gingivalis. Previous studies have reported that different portions of Kgp and HRgpA can bind hemoglobin, hemin, and protoporphyrin IX (PPIX) (14, 25, 35, 38, 48). Kgp has also recently been demonstrated to degrade hemoglobin (29, 53), hemopexin, haptoglobin (53), and transferrin (6). Genetic analysis has confirmed a role for Kgp in hemoglobin and hemin utilization in P. gingivalis (18, 51). It has recently been proposed that soluble Kgp and outer membrane receptor HmuR function together for the transport of hemin from hemoglobin in P. gingivalis (20).
In this study we report on the binding specificity of recombinant P. gingivalis HmuR and native soluble gingipains for hemin, hemoglobin, porphyrins, and metalloporphyrins. A series of porphyrins and metalloporphyrins were chosen to evaluate the role of both the central metal ion and the peripheral substituents in porphyrin binding to recombinant HmuR and native gingipains. Our results demonstrate that outer membrane receptor HmuR binds heme and related metalloporphyrins more tightly than hemoglobin. The iron, copper, and zinc derivatives of PPIX bound to recombinant HmuR with similar affinities, and more tightly than PPIX itself, suggesting that the active site of HmuR has a histidine that binds to the metal ion present in the porphyrin ring. Native Kgp and HRgpA bound selected porphyrins approximately as well as the corresponding metalloporphyrins, indicating that the binding of Kgp and HRgpA to these compounds does not require a metal present in the porphyrin ring. RgpB, which is missing the C-terminal hemagglutinin domain present in Kgp and HRgpA, did not bind to these compounds. Finally, we demonstrate that soluble Kgp and HRgpA, but not RgpB, bind directly to recombinant HmuR.
MATERIALS AND METHODS
Construction of HmuR expression plasmids.
The hmuR gene was amplified from total genomic DNA of P. gingivalis strain A7436 as previously reported (50). Briefly, the forward primer (Table 1) was designed to produce hmuR either without (F1) or with (F2) its native signal peptide sequence. The reverse primer was designed to remove the native stop codon (R1) to preserve the reading frame through the C-terminal tag. To obtain recombinant HmuR without the tag consisting of the V5 epitope and six histidines (V5–six-His tag), the stop codon was included (R2). The amplified products were purified and cloned into vector pCRT7/CT-TOPO (Invitrogen, Carlsbad, Calif.), coding for the V5 epitope and polyhistidine (six-His) regions. The resulting plasmids (pTO1, pTO2, pTO3, and pTO4; Table 2) were then transformed into Escherichia coli strain TOP10F′ (Invitrogen). Transformants were selected on Luria-Bertani plates containing 100 μg of ampicillin/ml, and the insertion was confirmed by restriction enzyme analysis, PCR, and sequence analysis.
TABLE 1.
Primers used in this study
| Primera (sequence) | Description |
|---|---|
| F1 (ATGGCCAACCCTCCGGCCCAACCTA) | Amplifies the hmuR gene without the signal peptide sequence and without the stop codon |
| R1 (GAAAGTGATCCGAACCAACCCGTAT) | |
| F2 (ATGAAAAGTCTAGTAACAAAGCAGG) | Amplifies the hmuR gene with the signal peptide sequence and without the stop codon |
| R1 (GAAAGTGATCCGAACCAACCCGTAT) | |
| F1 (ATGGCCAACCCTCCGGCCCAACCTA) | Amplifies the hmuR gene without the signal peptide sequence and with the stop codon |
| R2 (TTAGAAAGTGATCCGAACCAACCCGTAT) | |
| F2 (ATGAAAAGTCTAGTAACAAAGCAGG) | Amplifies the hmuR gene with the signal peptide sequence and with the stop codon |
| R2 (TTAGAAAGTGATCCGAACCAACCCGTAT) |
F, forward; R, reverse.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| P. gingivalis A7436 | Wild type | Laboratory collection |
| E. coli TOP10F′ | F′ {lacIq/Tn10 (Tetr)} mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| E. coli BL21 (DE3)pLysE | F−ompT hsdSB (rB− mB−) gal dcm (DE3)pLysE(Camr) | Invitrogen |
| E. coli BL21 (DE3)pLysS | F−ompT hsdSB (rB− mB−) gal dcm (DE3)pLysS(Camr) | Invitrogen |
| pCRT7/CT-TOPO | Ampr | Invitrogen |
| pTO1 | pCRT7/CT-TOPO containing the hmuR gene without the signal peptide sequence and with the fusion tag | 50 |
| pTO2 | pCRT7/CT-TOPO containing the hmuR gene with the signal peptide sequence and with the fusion tag | 50 |
| pTO3 | pCRT7/CT-TOPO containing the hmuR gene with the signal peptide sequence and without the fusion tag | This study |
| pTO4 | pCRT7/CT-TOPO containing the hmuR gene without the signal peptide sequence and without the fusion tag | This study |
Expression and purification of recombinant HmuR.
E. coli strains BL21(DE3)pLysS and BL21(DE3)pLysE (Invitrogen) were transformed with either pTO1, pTO2, pTO3, pTO4, or pCRT7/CT-TOPO and grown overnight at 37°C in minimal medium (M9) supplemented with 100 μg of ampicillin and 34 μg of chloramphenicol/ml. Overnight bacterial cultures were inoculated into fresh M9 medium and grown at 37°C to an optical density at 600 nm (OD600) of 0.5 to 0.6. Expression of the P. gingivalis hmuR gene was induced by the addition of 0.5 to 1 mM isopropyl β-d-thiogalactopyranoside (IPTG; Sigma, St. Louis, Mo.), followed by a 3- to 5-h growth period. After IPTG induction, E. coli BL21(DE3)pLysS and BL21(DE3)pLysE cells expressing TO1 were harvested by centrifugation for 20 min at 8,000 × g. Recombinant HmuR containing the V5–six-His tag (rHmuR-6His) and lacking the signal sequence was purified from inclusion bodies using Ni-chelate chromatography under denaturing conditions as described previously (50). The purified protein was dialyzed to decreasing concentrations of urea and finally to 0.5 M urea made in 20 mM phosphate buffer, pH 7.4, containing 0.14 M NaCl (phosphate-buffered saline [PBS]) and 0.1% n-octyl-β-d-glucopyranoside (OG; Sigma). After dialysis less than 10% of the rHmuR-6His was susceptible to renaturation and present in the soluble fraction, as determined by protein concentration. The remainder of the rHmuR-6His protein obtained after the final dialysis was present in the denatured nonsoluble fraction.
To localize recombinant HmuR, total membrane fractions were isolated from E. coli cells (adjusted to an OD600 of 1.0) harboring pTO2, pTO3, or the vector alone after centrifugation (70,000 × g, 1 h) of the supernatant remaining after the first centrifugation at 8,000 × g for 20 min. Total-membrane fractions were solubilized in 0.5% sarcosyl (Sigma) in PBS containing protease inhibitors, and after centrifugation (100,000 × g, 1 h) outer membrane fractions were collected. Detection of the recombinant HmuR was performed after polyacrylamide gel electrophoresis (PAGE) in the presence of sodium dodecyl sulfate (SDS) using Coomassie brilliant blue G-250 (CBB; Invitrogen) staining or after transfer onto nitrocellulose membranes by probing with monoclonal anti-V5 antibodies (Invitrogen) or polyclonal anti-HmuR antibodies (Lampire Biological Laboratories, Pipersville, Pa.). The latter were raised to the rHmuR-6His purified from inclusion bodies. The immunoglobulin G (IgG) fraction was isolated from the resulting antiserum using a HiTrap protein A affinity column (Pharmacia Biotech, Piscataway, N.J.). After incubation with a secondary antibody conjugated with horseradish peroxidase (HRP; Sigma) chemiluminescence staining was used to detect the complexes formed (50).
UV-Vis spectrophotometric analysis.
Heme binding to rHmuR-6His purified from inclusion bodies was monitored by UV-visual (UV-Vis) absorption analysis on a Beckman DU 7500 spectrophotometer scanning from 200 to 800 nm. The final mixture of 5 μM rHmuR-6His (renatured by dialysis against PBS, pH 7.4, containing 0.1% OG and 0.5 M urea) with 20 μM hemin (dissolved in dimethyl sulfoxide [DMSO]; final DMSO concentration in the mixture was 1 to 10%) was placed in a cuvette (0.5-cm cell length), and spectra were recorded immediately after mixing (time zero) and after 5, 15, 30, and 60 min of incubation at room temperature (RT). Nonrenatured rHmuR-6His, present after dialysis in the denatured nonsoluble fraction, was dissolved in PBS containing 0.1% OG and 8 M urea and was also examined by UV-Vis spectrophotometric analysis for hemin binding. The UV-Vis analyses of serum protein complexes with hemoglobin or hemin were performed under similar conditions as described above, with the exception that PBS was used to dissolve and dilute all proteins.
Binding of E. coli cells expressing recombinant HmuR to hemoglobin, porphyrins, and metalloporphyrins.
E. coli BL21(DE3)pLysS and BL21(DE3)pLysE cells expressing rHmuR-6His (containing the V5–six-His tag) or rHmuR (V5–six-His tag not produced) deposited in inclusion bodies (lacking the signal sequence) or membrane associated (containing the signal sequence) and cells containing the vector alone were grown in M9 medium and harvested before and after IPTG induction. The cells were washed with PBS and adjusted to an OD600 of 1.0, and 0.8-ml aliquots of the cell suspension in PBS were mixed with 0.2 ml of hemoglobin (5 μM) or hemin (10 μM) or other porphyrins or metalloporphyrins (10 μM). To reduce self-aggregation of hemin and other metalloporphyrins or porphyrins, DMSO (1 to 10%) was included in all assays (10, 20). PPIX and mesoporphyrin (MPIX) were obtained from Aldrich Biochemicals (Milwaukee, Wis.); iron(III) α,β,γ,δ-tetraphenylporphine tetrasulfonic acid (FeTPPS4), hematoporphyrin (HPIX), deuteroporphyrin (DPIX), the copper derivative of MPIX (CuMPIX), and the zinc and copper derivatives of PPIX (ZnPPIX, and CuPPIX, respectively) were obtained from Porphyrin Products (Logan, Utah); the iron and zinc derivatives of MPIX (FeMPIX and ZnMPIX, respectively) were obtained from Midcentury (Posen, Ill.); hemoglobin and hemin were obtained from Sigma. Samples were incubated at RT for 1 h and centrifuged, and the OD400 of the supernatant was measured. Compounds diluted in PBS were incubated under the same conditions and served as appropriate controls. The binding of all compounds was determined by the decrease of absorbance of the supernatant compared to those for control samples, which were set as 100%. For the saturation of hemin binding to rHmuR-6His, nonspecific hemin binding to E. coli cells harboring the vector alone was subtracted from hemin binding to E. coli cells expressing rHmuR-6His.
Purification of gingipains.
Kgp, HRgpA, and RgpB were purified from P. gingivalis strain HG66 cultures as previously described (42, 44) and were kindly provided by Jan Potempa (Jagiellonian University, Cracow, Poland). Approximately 5 mg of each gingipain from 1 liter of bacterial culture was purified to homogeneity as determined by SDS-PAGE and CBB staining and, after transfer onto a nitrocellulose membrane, by reactivity with the appropriate antibodies. In addition, the concentration of active gingipain in each batch was determined by active-site titration using specific inhibitors Z-Phe-Lys-2,4,6-trimethylbenzoyloxymethylketone (FKck) and H-d-Phe-Phe-Arg-chloromethylketone (FFRck) (Bachem Biosciences Inc., King of Prussia, Pa.) for gingipain K and gingipain R, respectively (43). Antibodies to each purified gingipain (Lampire Biological Laboratories) were produced as previously described (19). IgG fractions were purified from the antisera using a HiTrap protein A affinity column (Pharmacia Biotech), and their specificities and activities were confirmed.
Binding of recombinant HmuR to hemoglobin, gingipains, and serum proteins.
Binding of rHmuR-6His to hemoglobin, gingipains, and serum proteins was examined by an enzyme-linked immunosorbent assay (ELISA). Six-His-tagged rHmuR purified from inclusion bodies was immobilized (0.05 nmol per well) onto the surface of Ni-nitrilotriacetic acid (NTA) HisSorb wells (Qiagen, Valencia, Calif.) and incubated overnight at 4°C. This was followed by the addition of hemoglobin, haptoglobin (Sigma), transferrin saturated with iron (Sigma), human serum albumin (HSA; Sigma), hemopexin (Sigma), Kgp, HRgpA, or RgpB (0.005 to 1.0 nmol per well). We also prepared complexes of hemoglobin with haptoglobin, hemopexin with hemin, and HSA with hemin by incubation of serum proteins with hemoglobin or hemin in a molar ratio of 1:1 for 1 h at 37°C. The formation of these complexes was confirmed by UV-Vis analysis at 200 to 800 nm. Free hemoglobin or hemin was not observed in these preparations. The complexes formed between rHmuR-6His and the proteins were detected by probing with specific antibodies to each serum protein (goat antihemoglobin, antihaptoglobin, antihemopexin, rabbit anti-HSA, and antitransferrin diluted 1:5,000, 1:10,000, 1:10,000, 1:5,000, and 1:5,000, respectively; Biomeda, Foster City, Calif., or Sigma) or gingipain (rabbit anti-Kgp, -HRgpA, and -RgpB diluted 1:30,000, 1:20,000, and 1:10,000, respectively), followed by appropriate secondary antibodies conjugated with HRP. Substrate o-phenylenediamine (Sigma) was added to wells and incubated at RT for 15 to 30 min. The reactions were stopped by addition of 50 μl of 12.5% sulfuric acid, and the absorbance was measured at 490 nm.
Binding of recombinant HmuR and gingipains to hemoglobin- or heme-agarose.
Hemoglobin- and heme-agarose beads (Sigma) were washed with PBS or PBS containing 0.1% OG, resuspended in 20 μl of the buffer, and incubated with agitation overnight at 4°C with rHmuR-6His purified from inclusion bodies (0.01 nmol of rHmuR-6His per sample) and Kgp, HRgpA, or RgpB (0.01 nmol of Kgp and HRgpA and 0.02 nmol of RgpB; gingipains contained FKck and FFRck as inhibitors) in a total volume of 50 μl. Hemoglobin-agarose contained 6.1 to 12.3 nmol of hemoglobin per sample (0.02 to 0.05 μmol of heme in a hemoglobin sample), and heme-agarose contained 0.08 to 0.16 μmol of heme per sample. After centrifugation, the supernatant fluids were collected and the conjugated agarose beads were washed three times with PBS or PBS containing 0.1% OG. All samples were then boiled in Laemmli sample buffer and after centrifugation were subjected to SDS-PAGE. Gels were either stained with CBB or transferred onto nitrocellulose membranes. Recombinant HmuR was detected on blots with an anti-V5-HRP antibody (Invitrogen), and the other proteins were detected with the appropriate antibodies. Chemiluminescence detection was performed as previously described (50).
Binding of gingipains to hemoglobin and various porphyrins and metalloporphyrins.
Binding of Kgp, HRgpA, and RgpB to hemoglobin, hemin, porphyrins, and metalloporphyrins was studied using an ELISA. Polystyrene plates (Dynex, Chantilly, Va.) were coated overnight at 4°C with 100 μl of 5 μM hemoglobin or 10 μM porphyrins or metalloporphyrins per well. Human hemoglobin was dissolved in PBS, whereas all other compounds were dissolved in DMSO and then diluted with PBS (final DMSO concentration of 1 to 10%). Nonspecific binding sites were blocked overnight at 4°C with 1% bovine serum albumin (BSA)-PBS, followed by addition of 0.01 to 2 nmol of Kgp, HRgpA, or RgpB (containing the appropriate inhibitors) in 100 μl of 1% BSA-PBS per well. Bound gingipains were detected by rabbit anti-Kgp, -HRgpA, or -RgpB antibodies diluted 1:30,000, 1:20,000, and 1:10,000, respectively, and by goat anti-rabbit IgG antibodies conjugated with HRP, diluted 1:10,000. All reactions were performed for 1 h at 37°C. Washing was performed with PBS or PBS containing 0.1% Tween 20. The color was developed as described above.
Statistical analysis.
Data expressed as means ± standard deviations (SD) were analyzed using Student's t test; P values below 0.05 were considered significant.
RESULTS
Recombinant HmuR-hemin spectra and saturation of hemin binding to membrane-associated rHmuR-6His.
In a previous study we demonstrated that recombinant HmuR containing its native signal peptide sequence and the V5–six-His tag (rHmuR-6His) was exported and associated with the outer membrane of E. coli cells (50). We also demonstrated that E. coli cells expressing membrane-bound rHmuR-6His bound hemoglobin and hemin (50). However, the binding specificity of rHmuR-6His for these ligands has not been examined in detail.
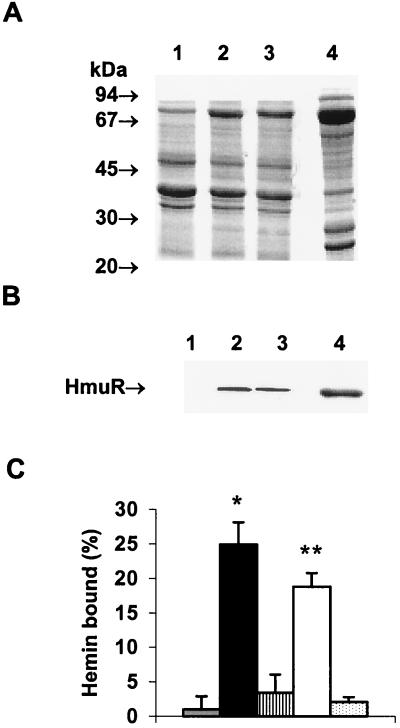
We examined the localization of the recombinant HmuR lacking the V5–six-His tag (rHmuR) and compared its expression and localization with those of rHmuR-6His. As shown in Fig. 1A and B the association of rHmuR and that of rHmuR-6His with the outer membranes of E. coli cells and expression levels of both proteins were comparable. We also determined that the addition of the His tag did not significantly affect the ability of E. coli cells expressing recombinant HmuR to bind hemin (Fig. 1C). As expected, only E. coli cells expressing membrane-associated rHmuR-6His (containing the His tag) or rHmuR (lacking the His tag) were found to bind hemin. In contrast, E. coli cells in which rHmuR-6His and rHmuR both lacked the signal sequences and after expression were deposited in inclusion bodies bound small amounts of hemin (Fig. 1C). These results indicate that E. coli cells expressing either rHmuR-6His or rHmuR can bind hemin and that the His tag did not significantly affect this binding.
FIG. 1.
E. coli cells expressing rHmuR-6His (containing the signal sequence and the V5–six-His tag) and rHmuR (containing the signal sequence but lacking the V5–six-His tag) bind hemin. (A and B) Localization of recombinant HmuR in E. coli cells. Membrane fractions of E. coli cells harboring the vector alone (lane 1), expressing rHmuR (lane 2), expressing rHmuR-6His (lane 3), and expressing rHmuR-6His lacking the signal sequence and deposited in inclusion bodies (lane 4) were adjusted to an OD600 of 1.0, and outer membranes (lanes 1 to 3) and inclusion bodies (lane 4) were isolated. (A) SDS–12.5% PAGE gel stained with CBB (10-μl portions of the samples were loaded onto the gel). (B) Western blot developed by probing with anti-HmuR antibodies raised to the recombinant six-His-tagged protein (5-μl portions of the samples were loaded onto the gel). (C) Hemin binding to E. coli cells expressing rHmuR. E coli cells were resuspended in PBS, adjusted to an OD600 of 1.0, and incubated for 1 h at RT with 10 μM hemin. Binding was determined by the decrease in the absorbance of the supernatant at 400 nm and was recorded as the percentage of the input hemin. Three independent experiments were performed in duplicate. Data are means ± SD. Asterisk, P < 0.001 for E. coli expressing membrane-associated rHmuR-6His (black bar) versus E. coli harboring the vector alone (grey bar); double asterisks, P < 0.05 for E. coli expressing membrane-associated rHmuR (open bar) versus E. coli harboring the vector alone. E. coli expressing rHmuR-6His (striped bar) and rHmuR (dotted bar) lacking the signal sequence, both of which were deposited in inclusion bodies, is also shown.
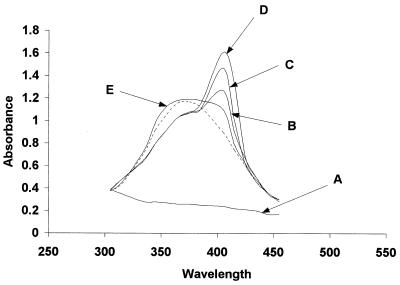
The ability of rHmuR-6His purified from inclusion bodies to bind heme was examined by incubating the soluble protein with hemin and measuring the absorbance of hemin at 200 to 800 nm. Following the addition of rHmuR-6His (5 μM) to a freshly prepared hemin solution (20 μM) the absorption spectrum of heme in the Soret region changed from the spectrum of free heme (380 nm) to the typical spectrum of a heme-protein complex (408 nm) (Fig. 2). Spectral changes of rHmuR-6His with hemin were essentially complete after a 15-min incubation. This shift in the wavelength of the absorbance reflects a modification of the heme absorption spectrum caused by the binding of hemin to rHmuR-6His. We also found that rHmuR-6His purified from inclusion bodies but nonrenatured and dissolved in 8 M urea did not bind hemin (Fig. 2).
FIG. 2.
Absorption spectra of hemin–rHmuR-6His complex. The absorption spectra in the Soret region of rHmuR-6His (5 μM) purified and renatured from inclusion bodies alone (A) or with added hemin (20 μM) were recorded immediately (B) and 5 (C) and 15 min (D) after mixing. (E) Absorption spectrum of hemin (20 μM) added to nonrenatured rHmuR-6His (5 μM) purified from inclusion bodies and dissolved in 8 M urea (recorded after 15 min of incubation with hemin). Dotted line, 20 μM hemin alone.
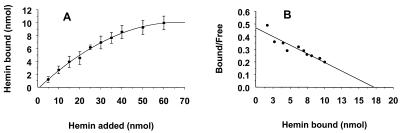
The saturation of hemin binding by membrane-associated rHmuR-6His expressed in E. coli cells was further examined by a spectrophotometric assay. Binding of hemin to whole E. coli cells was expressed as a decrease in the absorbance of the supernatant samples at 400 nm compared to that of samples containing only hemin (Fig. 3A). Scatchard analysis of hemin binding to rHmuR-6His yielded a linear plot with a binding affinity (Kd) of 2.4 × 10−5 M (Fig. 3B). E. coli cells containing the vector alone bound hemin nonspecifically with very low efficiency. This binding did not show saturation, and the transformed data clustered around the origin (data not shown). These results demonstrate that E. coli cells with recombinantly expressed HmuR can bind hemin.
FIG. 3.
Saturation of hemin binding to E. coli cells expressing membrane-associated rHmuR-6His. Saturability of hemin binding (A) and Scatchard plot analysis (B) of E. coli expressing recombinant HmuR are shown. E coli cells were resuspended in PBS and incubated for 1 h at RT with hemin. Binding was determined by the decrease of absorbance of the supernatant at 400 nm and recorded as the percentage of the input hemin. Nonspecific hemin binding to E. coli cells harboring the vector alone was subtracted from hemin binding to E. coli cells expressing rHmuR-6His. Two independent experiments were performed in duplicate. Data are means ± SD.
Binding of rHmuR-6His to hemin and hemoglobin immobilized on agarose.
The binding of recombinant HmuR to hemoglobin and hemin was also assessed using hemoglobin and heme immobilized on agarose beads. The amounts of hemoglobin and heme bound to agarose exceeded the amounts of rHmuR-6His. We found that rHmuR-6His bound to heme-agarose (Fig. 4). When rHmuR-6His was incubated with hemoglobin-agarose, a larger amount of nonbound recombinant protein was present in the flowthrough fraction than in the heme-agarose flowthrough fraction (Fig. 4). These results confirmed that rHmuR-6His purified from inclusion bodies can bind to hemoglobin and hemin and suggested that binding to hemin can be more efficient than binding to hemoglobin. Similar results were obtained using membrane-associated rHmuR-6His. E. coli cells expressing rHmuR-6His bound lower levels of hemoglobin than of hemin (see Fig. 6).
FIG. 4.
Binding of rHmuR-6His to hemoglobin or heme immobilized on agarose. Recombinant HmuR-6His purified from inclusion bodies (0.01 nmol per sample) was incubated with 20 μl of hemoglobin-agarose (6.1 to 12.3 nmol of hemoglobin per sample; 0.02 to 0.05 μmol of heme in hemoglobin sample) or heme-agarose (0.08 to 0.16 μmol of heme per sample). Samples were then separated by SDS-PAGE and detected using CBB staining (lane 1) or with the anti-V5 antibody (lanes 2 to 5). Lane 1, soluble rHmuR-6His purified and renatured from inclusion bodies; lanes 2 and 4, rHmuR-6His bound to hemoglobin (lane 2) or heme (lane 4); lanes 3 and 5, flowthrough fractions from hemoglobin-agarose (lane 3) and heme-agarose (lane 5).
FIG. 6.
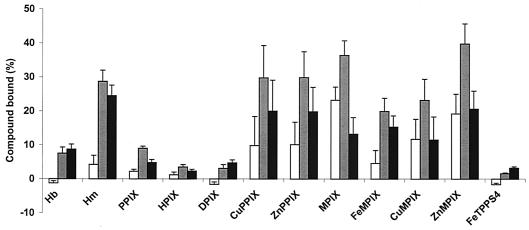
E. coli cells expressing membrane-associated rHmuR-6His bind porphyrins and metalloporphyrins. Binding to E. coli expressing 6His-rHmuR (grey bars), and to control E. coli harboring the vector alone (open bars) is shown. Black bars, differences between the two data sets. E. coli cells were resuspended in PBS and incubated for 1 h at RT with hemoglobin (5 μM) or porphyrins or metalloporphyrins (10 μM). Binding was determined by the decrease of absorbance at 400 nm of the supernatant and recorded as the percentage of the input porphyrins. Data are means ± SD from three independent experiments performed in duplicate. Hb, hemoglobin; Hm, hemin.
E. coli cells expressing membrane-associated rHmuR-6His bind porphyrins and metalloporphyrins.
We used a spectrophotometric assay to assess the ability of whole E. coli cells expressing membrane-associated rHmuR-6His to bind hemoglobin, porphyrins, and metalloporphyrins. We compared the binding of hemin with the corresponding zinc and copper metalloporphyrins (ZnPPIX and CuPPIX), as well as PPIX itself (Fig. 5). The same series of metals were evaluated for MPIX, in which the vinyl groups on the heme periphery at the 2 and 4 positions are replaced by ethyl groups (Fig. 5). To determine if alterations at the 2 and 4 positions of the heme substantially affect binding, we utilized DPIX and HPIX, which have H- and CHOHCH3, respectively, at the 2 and 4 positions (Fig. 5). E. coli cells expressing recombinant HmuR containing the His tag were used for these studies; in initial studies we found that the addition of the His tag did not significantly affect the ability of recombinant HmuR to bind hemin (Fig. 1C) and other metalloporphyrins (data not shown). The binding of all compounds was determined by the decrease in the absorbance of the supernatant compared to those for control samples, which were set as 100% (Fig. 6). Some of the porphyrins and metalloporphyrins showed substantial nonspecific binding to the control E. coli cells harboring the vector alone (Fig. 6). To account for this, the data were replotted for each compound in Fig. 6 as the difference between the two data sets. The discussion below refers to this difference for each compound.
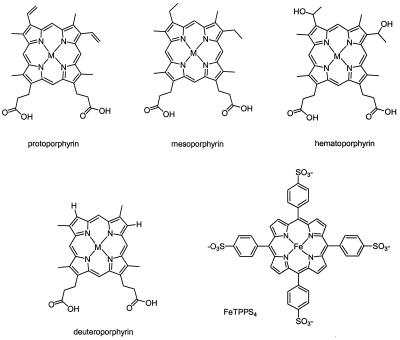
FIG. 5.
Structures of the porphyrins and metalloporphyrins used. The compounds utilized to examine the binding specificity of HmuR and gingipains for heme are depicted. M, metal (Fe, Zn, or Cu) within the PPIX ring.
The porphyrins themselves bound to E. coli expressing membrane-associated rHmuR-6His in the following order of affinity: MPIX > PPIX > DPIX ≈ HPIX. In the metalloprotoporphyrin series, E. coli expressing rHmuR-6His bound hemin, CuPPIX, and ZnPPIX with almost equal affinities, which were approximately four times higher than that for PPIX itself. The same series of metalloporphyrins with the MPIX skeleton was studied. In the MPIX series, there was substantial nonspecific binding; when this background was subtracted, all of the derivatives bound with the same affinity within experimental error.
We also examined FeTPPS4, as it was previously reported to support the growth of Vibrio vulnificus as a single iron source (34). E. coli cells expressing rHmuR-6His bound FeTPPS4 with very low efficiency; this binding was comparable to the nonspecific binding observed in control cells containing the vector alone (Fig. 6). Thus, the tetraphenylporphyrin structure makes this compound less accessible for HmuR than the other metalloporphyrins based on the natural porphyrin skeleton (Fig. 5).
These results demonstrate that E. coli cells expressing membrane-associated rHmuR-6His bind iron, copper, and zinc derivatives of PPIX more tightly than PPIX. This presumably suggests that the active site of HmuR has a histidine, which binds to the metal present in the porphyrin ring.
Binding of rHmuR-6His to serum proteins.
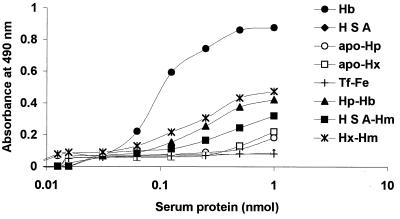
Serum hemoglobin released from erythrocytes is tightly bound to haptoglobin, and heme is bound to hemopexin or albumin (20). Hemoglobin bound to haptoglobin and hemin complexed to hemopexin can be used as iron sources by P. gingivalis, indicating that this microorganism has a mechanism for removing the hemin from these host iron-binding proteins (29, 53). To determine if recombinant HmuR could bind haptoglobin, hemopexin, or albumin, we examined the ability of rHmuR-6His purified from inclusion bodies to bind these proteins by ELISA. As a negative control, we utilized iron-saturated transferrin. We did not detect binding of rHmuR-6His to serum apo-haptoglobin, albumin, apo-hemopexin, or transferrin saturated with iron (Fig. 7). As expected, binding of rHmuR-6His to hemoglobin was observed in this assay. Recombinant HmuR was also demonstrated to bind complexes of haptoglobin with hemoglobin, hemopexin with hemin, and albumin with hemin; however, the affinities were lower than that of hemoglobin.
FIG. 7.
Binding of rHmuR-6His to serum proteins. Ni-NTA plates were coated with rHmuR-6His purified and renatured from inclusion bodies (0.05 nmol per well) and incubated with human hemoglobin (Hb), haptoglobin (Hp), human serum albumin (HSA), hemopexin (Hx), transferrin saturated with iron (Tf-Fe), and complexes of Hp-Hb, Hx-Hm, and HSA-Hm (0.005 to 1.0 nmol of serum protein per well). The binding was detected using antibodies to the appropriate protein. Values are representative of two separate experiments performed in triplicate.
Binding of gingipains to hemoglobin, porphyrins, and metalloporphyrins.
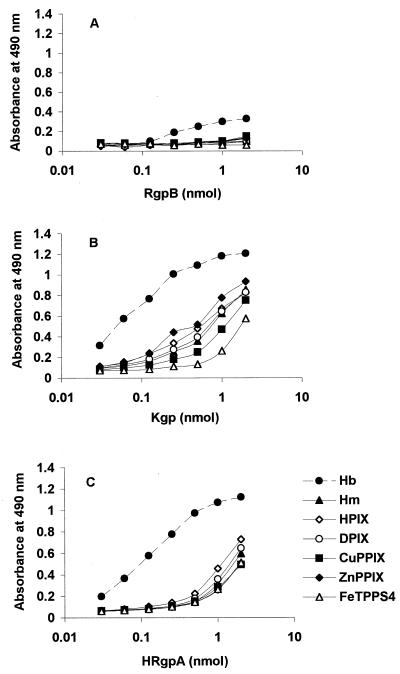
We next examined the ability of gingipains to bind hemoglobin, porphyrins, and metalloporphyrins using ELISA, as described above for rHmuR-6His. We found that both Kgp and HRgpA bound to the porphyrins and metalloporphyrins tested with affinities similar to those for hemin (Fig. 8). The ability of these gingipains to bind metalloporphyrins and porphyrins similarly confirms that the binding of Kgp and HRgpA to these porphyrins does not require a metal present within the porphyrin ring. Interestingly, we observed greater binding of Kgp and HRgpA to hemoglobin than to the tested porphyrins and metalloporphyrins (Fig. 8). These observations suggest that substantial portions of the recognition sites of Kgp and HRgpA for hemoglobin are due to protein-protein interactions. In general, Kgp bound hemoglobin and the porphyrins and metalloporphyrins more efficiently than HRgpA. RgpB, which lacks the hemagglutinin domain, showed little or no binding to hemoglobin, hemin, and the porphyrins and metalloporphyrins (Fig. 8).
FIG. 8.
Binding of gingipains to porphyrins and metalloporphyrins. Plates were coated with hemoglobin (Hb; 5 μM) or porphyrins or metalloporphyrins (10 μM). After incubation with RgpB (A), Kgp (B), or HRgpA (C) (0.01 to 1 nmol per well), binding was detected using antibodies to the appropriate protein. Values are representative of experiments performed in triplicate on three separate occasions. Hm, hemin.
Gingipain binding to heme- and hemoglobin-agarose.
To further examine the binding efficiency of the gingipains to hemoglobin and hemin, we utilized heme- and hemoglobin-agarose as described above for rHmuR-6His. We observed that Kgp and HRgpA bound to hemoglobin and heme immobilized on agarose (data not shown), confirming the results obtained by ELISA (Fig. 8). RgpB demonstrated little ability to bind hemoglobin or heme, since the majority of the protein incubated with heme- or hemoglobin-agarose was present in flowthrough fractions (data not shown). These results further confirm that hemoglobin and heme binding to Kgp and HRgpA is mediated via the hemagglutinin domains of these proteins. The low binding of RgpB to hemoglobin and heme is most likely caused by the nonspecific tendency of heme to bind to most molecules and surfaces.
Interactions of gingipains with recombinant HmuR.
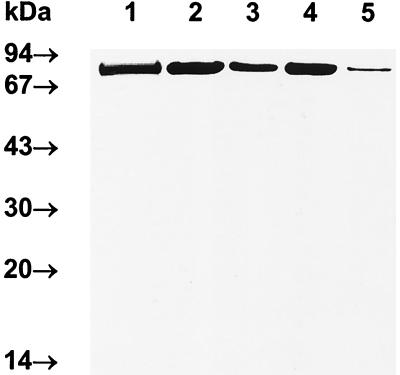
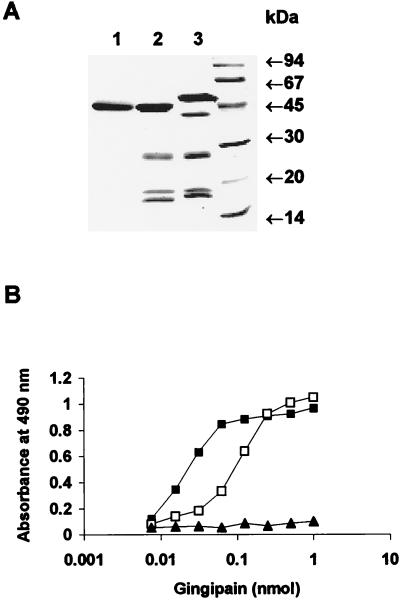
In addition to binding hemoglobin, Kgp can degrade hemoglobin, as well as hemopexin, haptoglobin, and transferrin (6, 29, 53). Based on these results and the results presented here, we reasoned that in a soluble form Kgp could function as a heme-scavenging and hemoglobinase protein. The concept that Kgp could function in a multifactor manner in heme transport in P. gingivalis is supported by studies of the recently described E. coli hemoglobinase, Hbp (39). In addition to binding hemoglobin, Hbp can degrade hemoglobin and subsequently binds the released hemin. In a similar fashion, we postulate that Kgp could bind and degrade hemoglobin, and the released hemin could then be delivered to outer membrane receptor HmuR. This would require a direct interaction of Kgp with outer membrane receptor HmuR. To examine the possible association between Kgp or HRgpA and HmuR, we examined this interaction by an ELISA. The purity of rHmuR-6His preparation is shown in Fig. 4. Our analysis indicated that the gingipain preparations were pure and showed the same banding pattern after SDS-PAGE and CBB staining (Fig. 9A) as previously described (42, 44). This was also confirmed after transfer onto nitrocellulose membranes by probing with the appropriate antibodies raised to each gingipain (data not shown). As shown in Fig. 9B, rHmuR-6His was found to bind to Kgp. We also observed binding of rHmuR-6His to HRgpA at higher concentrations than those at which binding to Kgp was observed. We did not detect binding of rHmuR-6His to RgpB (Fig. 9B). We also found that gingipain domains cleaved by boiling in the presence of SDS and then separated by SDS-PAGE and transferred onto nitrocellulose membrane failed to bind rHmuR-6His (data not shown). This suggests that the interaction between Kgp or HRgpA and HmuR requires the intact gingipain molecule but not denatured and separated gingipain domains.
FIG. 9.
Interaction between rHmuR-6His and gingipains. (A) SDS–12.5% PAGE gel stained with CBB showing the purity of gingipains RgpB (lane 1), HRgpA (lane 2), and Kgp (lane 3); molecular mass standards are shown on the right. (B) Reactivity of gingipains with soluble rHmuR-6His. Ni-NTA plates were coated with rHmuR-6His purified and renatured from inclusion bodies (0.05 nmol per well) and incubated with Kgp (▪), HRgpA (□), or RgpB (▴) (0.005 to 1.0 nmol of gingipains per well). Binding was detected using antibodies to the appropriate gingipain. Values are representative of two separate experiments performed in triplicate.
DISCUSSION
In this study we assessed the binding specificities of the P. gingivalis outer membrane hemoglobin/heme receptor, HmuR, and cysteine proteases Kgp, HRgpA, and RgpB for hemoglobin, hemin, serum proteins, various porphyrins, and metalloporphyrins. While rHmuR-6His purified from inclusion bodies bound to hemin and hemoglobin and also, with lower affinity, to complexes of haptoglobin-hemoglobin, hemopexin-hemin, and albumin-hemin, we did not detect the binding of recombinant HmuR to apo-haptoglobin, albumin, apo-hemopexin, or transferrin saturated with iron. We also found that E. coli cells expressing rHmuR-6His bound not only hemin but also CuPPIX and ZnPPIX with similar affinities. These were bound approximately four times more tightly than PPIX itself. All three metals that we examined in this study (iron, copper, and zinc) can bind to the histidine within the active site in heme-proteins (1, 47, 55). The observation that these metalloprotoporphyrins bind to HmuR somewhat better than PPIX itself is consistent with a histidine in HmuR serving as the axial ligand, which binds to the metal ion present in the porphyrin ring. For the porphyrins themselves, we found that the order of affinity for HmuR-6His binding was as follows: MPIX > PPIX > DPIX ∼ HPIX. This indicated that HmuR has a preference for ethyl or vinyl side chains of heme. In total, our results suggest that the HmuR binding site for heme has an axial histidine and accommodates a porphyrin structure with a periphery approximating that of the natural substrate hemin at the 2 and 4 positions. It should also be noted that differential-reconstitution experiments are subject to the issues that the porphyrins and metalloporphyrins may bind to other components in the mixture (20), that proteins and lipids interacting with one another may change the constant for binding to any individual component (8, 32), and that porphyrins and metalloporphyrins can aggregate in aqueous solution (7, 9, 27).
We demonstrated that soluble Kgp and HRgpA bound to the various porphyrins and metalloporphyrins with similar affinities. This indicates that any interaction between Kgp and HRgpA and the metal of the metalloporphyrin does not contribute significantly to recognition of the metalloporphyrin. Kgp and HRgpA bound hemoglobin more than 1 order of magnitude more tightly on a per-heme basis than any of the porphyrins and metalloporphyrins investigated. This is in agreement with a study by DeCarlo et al. (14), which reported that a recombinant polypeptide of the Kgp complex binds hemoglobin more efficiently than hemin. In that report the binding of the 19-kDa protein was inhibited by PPIX, indicating that hemin forms part of the recognition site for Kgp and hemoglobin. The results obtained in our study suggest that the recognition by Kgp and HRgpA of hemoglobin is mediated significantly via protein-protein interactions. Together these two studies point to roles for both heme-protein and protein-protein interactions in Kgp hemoglobin binding. Based on studies of two-component protein-protein complexes, it appears that heme itself will form only a minor portion of the recognition site, probably less than 20% (20, 30). The rest of the recognition could be due to the hemoglobin protein itself.
The dissociation constant of hemin binding to E. coli cells expressing recombinant HmuR is lower than Kds for hemin receptors in P. gingivalis described by Tompkins et al. (54). According to this work P. gingivalis whole cells had both low- and high-affinity binding sites for hemin (Kd = 2.6 × 10−7 to 6.5 × 10−8 M and 3.6 × 10−11 to 9.6 × 10−11 M, respectively). Our Kd for HmuR expressed in E. coli (2.4 × 10−5 M) is similar to that found for the S. marcescens TonB-dependent hemoglobin/heme receptor, HasR (Kd = 10−4 to 10−6 M) (21). In this study we have shown that P. gingivalis outer membrane receptor HmuR interacts with soluble gingipains Kgp and HRgpA. This suggests that Kgp and HRgpA might function as heme-scavenging proteins, cooperating with HmuR in hemoglobin and heme utilization in P. gingivalis. By analogy with the HasR-HasA system from S. marcescens, it is possible that HmuR requires a hemophore-like protein (Kgp or HRgpA) to increase hemoglobin and/or hemin binding to HmuR. Ghigo et al. (21) have shown that HasR alone is sufficient for hemoglobin and heme utilization but that E. coli more efficiently utilizes heme from hemoglobin via HasR-HasA cooperation (Kd < 10−8 M). The HmuR system in P. gingivalis may function in an analogous manner with Kgp and HRgpA.
Okamoto et al. (38) previously reported that P. gingivalis kgp mutants are nonpigmented and are decreased in their ability to bind hemoglobin. The phenotype of the kgp mutants described by these investigators is similar to the phenotype of P. gingivalis kgp mutants, which we previously constructed and characterized (18; W. Simpson and C. A. Genco, unpublished data). These P. gingivalis mutant cells bind reduced levels of hemoglobin and hemin and exhibit a delayed growth with hemoglobin compared to the parental strain. We conclude from these results that Kgp may not be absolutely required for hemoglobin utilization in P. gingivalis but may make the process of hemoglobin utilization more efficient. We have previously reported that a P. gingivalis hmuR mutant does not grow with hemoglobin or hemin as the sole iron source, indicating that HmuR is required for hemoglobin utilization in P. gingivalis (50). We also observed that, following prolonged growth on blood agar plates, a P. gingivalis hmuR mutant is characterized by higher pigmentation capacity than wild-type P. gingivalis and nonpigmented kgp mutants (50; Simpson and Genco, unpublished data). This may be due to excessive heme storage, possibly through Kgp, and an inability to internalize heme due to the absence of HmuR. Furthermore, we found that a P. gingivalis hmuR kgp mutant is nonpigmented (Simpson and Genco, unpublished data), suggesting that this mutant cannot store and/or use heme due to the absence of HmuR and Kgp.
Overall our results indicate that outer membrane receptor HmuR exhibits higher specificity for heme than either Kgp or HRgpA. This finding would be expected if indeed soluble Kgp and HRgpA function more as heme scavenger proteins than as outer membrane receptors, which transport heme into the cell. It is noteworthy that the amino acid sequences of Kgp and HRgpA exhibit no similarity to those of TonB-dependent outer membrane proteins, further suggesting that these proteins may function as extracellular heme-scavenging proteins rather than classical outer membrane receptors.
ACKNOWLEDGMENTS
This study was supported by Public Health Service grant DE09161 from the National Institute of Dental and Craniofacial Research (C. A. Genco) and by grant AI45883 from the National Institute of Allergy and Infectious Diseases (D. W. Dixon).
We acknowledge Frank Gibson, Jan Potempa, and Waltena Simpson for scientific discussions and critical review of the manuscript.
REFERENCES
- 1.Alston K, Storm C B. Copper (II) protoporphyrin IX as a reporter group for the heme environment in myoglobin. Biochemistry. 1979;18:4292–4300. doi: 10.1021/bi00587a006. [DOI] [PubMed] [Google Scholar]
- 2.Amano A, Kuboniwa M, Kataoka K, Tazaki K, Inoshita E, Nagata H, Tamagawa H, Shizukuishi S. Binding of hemoglobin by Porphyromonas gingivalis. FEMS Microbiol Lett. 1995;134:63–67. doi: 10.1111/j.1574-6968.1995.tb07915.x. [DOI] [PubMed] [Google Scholar]
- 3.Barkocy-Gallagher G A, Han N, Patti J M, Whitlock J, Progulske-Fox A, Lantz M S. Analysis of the prep gene encoding porphypain, a cysteine proteinase of Porphyromonas gingivalis. J Bacteriol. 1996;178:2734–2741. doi: 10.1128/jb.178.10.2734-2741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bracken C S, Baer M T, Abdur-Rashid A, Helms W, Stojiljkovic I. Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function. J Bacteriol. 1999;181:6063–6072. doi: 10.1128/jb.181.19.6063-6072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bramanti T E, Holt S C. Hemin uptake in Porphyromonas gingivalis: Omp26 is a hemin-binding surface protein. J Bacteriol. 1993;175:7413–7420. doi: 10.1128/jb.175.22.7413-7420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brochu V, Grenier D, Nakayama K, Mayrand D. Acquisition of iron from human transferrin by Porphyromonas gingivalis: a role for Arg- and Lys-gingipain activities. Oral Microbiol Immunol. 2001;16:79–87. doi: 10.1034/j.1399-302x.2001.016002079.x. [DOI] [PubMed] [Google Scholar]
- 7.Brown S B, Hazikonstantinou H, Herries D. The structure of porphyrins and haems in aqueous solution. Int J Biochem. 1980;12:701–707. doi: 10.1016/0020-711x(80)90147-0. [DOI] [PubMed] [Google Scholar]
- 8.Camejo G, Halberg C, Manschik-Lundin A, Hurt-Camejo E, Rosengren B, Olsson H, Olsson G I, Forsberg G B, Ylhen B. Hemin binding and oxidation of lipoproteins in serum: mechanisms and effect on the interaction of LDL with human macrophages. J Lipid Res. 1998;39:755–766. [PubMed] [Google Scholar]
- 9.Cohen S, Margalit R. Binding of porphyrin to human serum albumin. Structure-activity relationships. Biochem J. 1990;270:325–330. doi: 10.1042/bj2700325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collier G S, Pratt J M, DeWet C R, Tshabalala C F. Studies on haemin in dimethyl sulfoxide/water mixtures. Biochem J. 1979;179:281–289. doi: 10.1042/bj1790281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtis M A, Aduse-Opoku J, Slaney M, Rangarajan M, Booth C I, Sheperd P. Characterization of an adherence and antigenic determinant of the Arg1 protease of Porphyromonas gingivalis which is present on multiple gene products. Infect Immun. 1996;64:2532–2539. doi: 10.1128/iai.64.7.2532-2539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curtis M A, Kuramitsu H K, Lantz M, Macrina F L, Nakayama K, Potempa J, Reynolds E C, Aduse-Opoku J. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J Periodontal Res. 2000;34:464–472. doi: 10.1111/j.1600-0765.1999.tb02282.x. [DOI] [PubMed] [Google Scholar]
- 13.Dashper S G, Hendtlass A, Slakeski N, Jackson C, Cross K J, Brownfield L, Hamilton R, Barr I, Reynolds E C. Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J Bacteriol. 2000;182:6456–6462. doi: 10.1128/jb.182.22.6456-6462.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeCarlo A A, Paramaesvaran M, Yun P L W, Collyer C, Hunter N. Porphyrin-mediated binding to hemoglobin by the HA2 domain of cysteine proteinases (gingipains) and hemagglutinins from the periodontal pathogen Porphyromonas gingivalis. J Bacteriol. 1999;181:3784–3791. doi: 10.1128/jb.181.12.3784-3791.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fishburn C S, Slaney J M, Carman R J, Curtis M A. Degradation of plasma proteins by the trypsin-like enzyme of Porphyromonas gingivalis and inhibition of protease activity by a serine protease inhibitor of human plasma. Oral Microbiol Immunol. 1991;6:209–215. doi: 10.1111/j.1399-302x.1991.tb00479.x. [DOI] [PubMed] [Google Scholar]
- 16.Fujimura S, Shibata Y, Hirai K, Nakamura T. Binding of hemoglobin to the envelope of Porphyromonas gingivalis and isolation of the hemoglobin-binding protein. Infect Immun. 1996;64:2339–2342. doi: 10.1128/iai.64.6.2339-2342.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genco C A, Odusanya B M, Brown G. Binding and accumulation of hemin in Porphyromonas gingivalis are induced by hemin. Infect Immun. 1994;62:2885–2892. doi: 10.1128/iai.62.7.2885-2892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genco C A, Simpson W, Forng R Y, Egal M, Odusanya B M. Characterization of a Tn4351-generated hemin uptake mutant of Porphyromonas gingivalis: evidence for the coordinate regulation of virulence factors by hemin. Infect Immun. 1995;63:2459–2466. doi: 10.1128/iai.63.7.2459-2466.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genco C A, Potempa J, Mikolajczyk-Pawlinska J, Travis J. Role of gingipains R in Porphyromonas gingivalis-mediated periodontal disease. Clin Infect Dis. 1999;28:456–465. doi: 10.1086/515156. [DOI] [PubMed] [Google Scholar]
- 20.Genco C A, Dixon D W. Emerging strategies in microbial heme capture. Mol Microbiol. 2001;39:1–11. doi: 10.1046/j.1365-2958.2001.02231.x. [DOI] [PubMed] [Google Scholar]
- 21.Ghigo M J, Letoffe S, Wandersman C. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherchia coli. J Bacteriol. 1997;179:3572–3579. doi: 10.1128/jb.179.11.3572-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadowaki T, Nakayama K, Okamoto K, Abe N, Baba A, Shi Y, Ratnayake D B, Yamamoto K. Porphyromonas gingivalis proteinases as virulence determinants in progression of periodontal diseases. J Biochem. 2000;128:153–159. doi: 10.1093/oxfordjournals.jbchem.a022735. [DOI] [PubMed] [Google Scholar]
- 23.Karunakaran T, Madden T, Kuramitsu K. Isolation and characterization of a hemin-regulated gene, hemR, from Porphyromonas gingivalis. J Bacteriol. 1997;179:1898–1908. doi: 10.1128/jb.179.6.1898-1908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi Y, Nakae T. The permeability property of the outer membrane of Bacteroides fragilis, a strictly anaerobic opportunistic pathogen. Biochem Biophys Res Commun. 1986;141:292–298. doi: 10.1016/s0006-291x(86)80367-9. [DOI] [PubMed] [Google Scholar]
- 25.Kuboniwa M, Amano A, Shizukuishi S. Hemoglobin-binding protein purified from Porphyromonas gingivalis is identical to lysine-specific cysteine proteinase (Lys gingipain) Biochem Biophys Res Commun. 1998;249:38–43. doi: 10.1006/bbrc.1998.8958. [DOI] [PubMed] [Google Scholar]
- 26.Kuramitsu H K. Proteases of Porphyromonas gingivalis: what don't they do? Oral Microbiol Immunol. 1999;13:263–270. doi: 10.1111/j.1399-302x.1998.tb00706.x. [DOI] [PubMed] [Google Scholar]
- 27.Kuzelova K, Mrhalova M, Hrkal Z. Kinetics of heme interaction with heme-binding proteins: the effect of heme aggregation state. Biochem Biophys Acta. 1997;1336:497–501. doi: 10.1016/s0304-4165(97)00062-7. [DOI] [PubMed] [Google Scholar]
- 28.Letoffe S, Nato F, Goldberg M E, Wandersman C. Interactions of HasA, a bacterial hemophore, with hemoglobin and with its outer membrane receptor HasR. Mol Microbiol. 1999;33:546–555. doi: 10.1046/j.1365-2958.1999.01499.x. [DOI] [PubMed] [Google Scholar]
- 29.Lewis J P, Dawson J A, Hannis J C, Muddiman D, Macrina F L. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J Bacteriol. 1999;181:4905–4913. doi: 10.1128/jb.181.16.4905-4913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 31.Mikolajczyk-Pawlinska J, Travis J, Potempa J. Modulation of interleukin-8 activity by gingipains from Porphyromonas gingivalis: implications for pathogenicity of periodontal disease. FEBS Lett. 1998;440:282–286. doi: 10.1016/s0014-5793(98)01461-6. [DOI] [PubMed] [Google Scholar]
- 32.Miller Y I, Shaklai N. Kinetics of hemin distribution in plasma reveals its role in lipoprotein oxidation. Biochim Biophys Acta. 1999;1454:153–164. doi: 10.1016/s0925-4439(99)00027-7. [DOI] [PubMed] [Google Scholar]
- 33.Minnock A, Vernon D I, Schofield J, Griffiths J, Parish J H, Brown S B. Mechanism of uptake of a cationic water-soluble pyridinium zinc phthalocyanine across the outer membrane of Escherichia coli. Antimicrob Agents Chemother. 2000;44:522–527. doi: 10.1128/aac.44.3.522-527.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyoshi S, Kamei T, Inami Y, Ota Y, Yamamoto S, Tomochika K, Shinoda S. The ability of Vibrio vulnificus to use a synthetic hydrophilic heme compound, Fe-TPPS4, as a single iron source. FEMS Microbiol Lett. 1999;172:73–77. doi: 10.1111/j.1574-6968.1999.tb13452.x. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama K, Ratnayake D B, Tsukuba T, Kadowaki T, Yamamoto K, Fujimura S. Hemoglobin receptor protein is intragenically encoded by the cysteine proteinase-encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol Microbiol. 1998;27:51–61. doi: 10.1046/j.1365-2958.1998.00656.x. [DOI] [PubMed] [Google Scholar]
- 36.Okamoto K, Misumi Y, Kadowaki T, Yoneda M, Yamamoto K, Ikehara Y. Structural characterization of arg gingipain, a novel arginine-specific cysteine proteinase, as a major periodontal pathogenic factor from Porphyromonas gingivalis. Arch Biochem Biophys. 1995;316:917–925. doi: 10.1006/abbi.1995.1123. [DOI] [PubMed] [Google Scholar]
- 37.Okamoto K, Kadowaki T, Nakayama K, Yamamoto K. Cloning and sequencing of the gene encoding a novel lysine-specific cysteine proteinase (Lys-gingipain) in Porphyromonas gingivalis: structural relationship with the arginine-specific cysteine proteinase (Arg-gingipain) J Biochem. 1996;120:398–406. doi: 10.1093/oxfordjournals.jbchem.a021426. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake D B, Yamamoto K. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J Biol Chem. 1998;273:21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 39.Otto B R, van Dooren S J M, Nuijens J H, Lurink J, Oudega B. Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J Exp Med. 1998;188:1091–1103. doi: 10.1084/jem.188.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pavloff N, Potempa J, Pike R N, Prochazka V, Kiefer M C, Travis J, Barr P J. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase-adhesion polyprotein. J Biol Chem. 1995;270:1007–1010. doi: 10.1074/jbc.270.3.1007. [DOI] [PubMed] [Google Scholar]
- 41.Pavloff N, Pemberton P A, Potempa J, Chen W C A, Pike R N, Prochazka V, Kiefer M C, Travis J, Barr P J. Molecular cloning and characterization of Porphyromonas gingivalis Lys-gingipain. A new member of an emerging family of pathogenic bacterial cysteine proteinases. J Biol Chem. 1997;272:1595–1600. doi: 10.1074/jbc.272.3.1595. [DOI] [PubMed] [Google Scholar]
- 42.Pike R N, McGraw W, Potempa J, Travis J. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization and evidence for the existence of complexes with hemagglutinins. J Biol Chem. 1994;269:406–411. [PubMed] [Google Scholar]
- 43.Potempa J, Pike R, Travis J. Titration and mapping of the active site of cysteine proteinases from Porphyromonas gingivalis (gingipains) using peptidyl chloromethanes. Biol Chem. 1997;378:223–230. doi: 10.1515/bchm.1997.378.3-4.223. [DOI] [PubMed] [Google Scholar]
- 44.Potempa J, Mikolajczyk-Pawlinska J, Brassell D, Nelson D, Thogersen I B, Enghild J J, Travis J. Comparative properties of two cysteine proteinases (gingipains R), the products of two related but individual genes of Porphyromonas gingivalis. J Biol Chem. 1998;273:21648–21657. doi: 10.1074/jbc.273.34.21648. [DOI] [PubMed] [Google Scholar]
- 45.Potempa J, Banbula A, Travis J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol 2000. 2000;24:153–192. doi: 10.1034/j.1600-0757.2000.2240108.x. [DOI] [PubMed] [Google Scholar]
- 46.Shah H N, Bonnett R, Mateen B, Williams R A D. The porphyrin pigmentation of subspecies of Bacteroides melaninogenicus. Biochem J. 1979;180:45–50. doi: 10.1042/bj1800045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharp R E, Diers J R, Bocian D F, Dutton P L. Differential binding of iron(III) and zinc(II) protoporphyrin IX to synthetic four-helix bundles. J Am Chem Soc. 1998;120:7103–7104. [Google Scholar]
- 48.Shi Y, Ratnayake D B, Okamoto K, Abe N, Yamamoto K, Nakayama K. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. J Biol Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 49.Slakeski N, Dashper S G, Cook P, Poon C, Moore C, Reynolds E C. A Porphyromonas gingivalis genetic locus encoding a heme transport system. Oral Microbiol Immunol. 2000;15:388–392. doi: 10.1034/j.1399-302x.2000.150609.x. [DOI] [PubMed] [Google Scholar]
- 50.Simpson W, Olczak T, Genco C A. Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J Bacteriol. 2000;182:5737–5748. doi: 10.1128/jb.182.20.5737-5748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smalley J W, Birss A, McKee A S, Marsh P D. Haemin-binding proteins of Porphyromonas gingivalis W50 grown in a chemostat under haemin-limitation. J Gen Microbiol. 1993;139:2145–2150. doi: 10.1099/00221287-139-9-2145. [DOI] [PubMed] [Google Scholar]
- 52.Smalley J W, Birss A J, McKee A S, Marsh P D. Haemin regulation of haemoglobin binding by Porphyromonas gingivalis. Curr Microbiol. 1998;36:102–106. doi: 10.1007/s002849900287. [DOI] [PubMed] [Google Scholar]
- 53.Sroka A, Sztukowska M, Potempa J, Travis J, Genco C A. Degradation of host heme proteins by the lysine- and arginine-specific cysteine proteinases (gingipains) of Porphyromonas gingivalis. J Bacteriol. 2001;183:5609–5616. doi: 10.1128/JB.183.19.5609-5616.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tompkins G R, Wood D P, Birchmeher K R. Detection and comparison of specific hemin binding by Porphyromonas gingivalis and Prevotella intermedia. J Bacteriol. 1997;179:620–626. doi: 10.1128/jb.179.3.620-626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye S Y, Shen C Y, Cotton T M, Kostic N M. Characterization of zinc-substituted cytochrome c by circular dichroism and resonance Raman spectroscopic methods. J Inorgan Biochem. 1997;65:219–226. doi: 10.1016/s0162-0134(97)00001-9. [DOI] [PubMed] [Google Scholar]