Abstract
Porphyromonas gingivalis can use hemoglobin bound to haptoglobin and heme complexed to hemopexin as heme sources; however, the mechanism by which hemin is released from these proteins has not been defined. In the present study, using a variety of analytical methods, we demonstrate that lysine-specific cysteine proteinase of P. gingivalis (gingipain K, Kgp) can efficiently cleave hemoglobin, hemopexin, haptoglobin, and transferrin. Degradation of hemopexin and transferrin in human serum by Kgp was also detected; however, we did not observe extensive degradation of hemoglobin in serum by Kgp. Likewise the β-chain of haptoglobin was partially protected from degradation by Kgp in a haptoglobin-hemoglobin complex. Arginine-specific gingipains (gingipains R) were also found to degrade hemopexin and transferrin in serum; however, this was observed only at relatively high concentrations of these enzymes. Growth of P. gingivalis strain A7436 in a minimal media with normal human serum as a source of heme correlated not only with the ability of the organism to degrade hemoglobin, haptoglobin, hemopexin, and transferrin but also with an increase in gingipain K and gingipain R activity. The ability of gingipain K to cleave hemoglobin, haptoglobin, and hemopexin may provide P. gingivalis with a useable source of heme for growth and may contribute to the proliferation of P. gingivalis within periodontal pockets in which erythrocytes are abundant.
The ability of a pathogen to colonize and proliferate within a particular environmental niche in the host is essential for the initiation of an infection. Growth depends, in part, on the ability of a pathogen to scavenge essential nutrients including iron, which plays a crucial role in both the establishment of a pathogen and the progression of disease (21). Within the human host the majority of iron is found intracellularly in the form of hemoglobin, heme proteins, or ferritin (21). Following intracellular release, heme and hemoglobin are bound by serum proteins hemopexin and haptoglobin, respectively. Small quantities of extracellular iron are also complexed to iron-binding proteins, primarily transferrin, which is found in serum, and lactoferrin, which is present within mucosal surfaces.
To survive in the iron-limited environment of the host, microorganisms have developed diverse and elaborate systems to obtain this element, which is needed for growth. These include the production of low-molecular-weight iron-chelating compounds (siderophores) and iron-regulated outer membrane receptors that function to bind iron-containing compounds directly. A new class of heme- and hemoglobin-binding proteins (hemophores) recently described appears to function similarly to siderophores for the capture of heme (3, 9, 22, 29–31, 38). An extracellular heme- and hemoglobin-binding protein (Hbp) in a human pathogenic strain of Escherichia coli has also recently been described and has been proposed to function as a shuttle protein of a hemophore-dependent hemin acquisition system (43). In addition to binding hemoglobin, the E. coli Hbp can degrade hemoglobin and subsequently binds the released hemin.
Porphyromonas gingivalis, the etiological agent of adult periodontitis, requires iron for growth (5, 12, 18); however, the specific mechanisms utilized for the acquisition of iron are poorly understood. Hemoglobin bound to haptoglobin and hemin complexed to hemopexin can be used as iron sources by P. gingivalis (5), indicating that this bacterium has a mechanism for removing the hemin from these host iron-binding proteins. Several reports have described P. gingivalis iron-regulated outer membrane proteins, which could function to bind heme compounds directly (2, 6, 57, 58); however, conclusive evidence for the role of these proteins in heme binding and uptake has not been presented.
We recently identified a hemin and hemoglobin receptor in P. gingivalis strain A7436 (HmuR), which exhibits a high degree of homology to TonB-dependent outer membrane hemin and hemoglobin receptors from several different gram-negative bacteria (56). A P. gingivalis hmuR mutant exhibited diminished ability to bind hemoglobin and to grow with either this protein or hemin as the sole iron source. We also demonstrated that the recombinant HmuR protein bound hemoglobin, hemin, and several different metalloporphyrins in vitro (42, 56). Recombinant HmuR binds hemopexin and serum albumin complexed with hemin and haptoglobin complexed with hemoglobin, but with affinity lower than that for hemoglobin alone (42). Recent studies have described three P. gingivalis genes (hemR, tlr, and ihtA) which have homology to genes encoding TonB-dependent receptors (13, 26, 57). The role of the protein products of the hemR, tlr, and ihtA genes in binding and utilization of heme or iron from different serum proteins in P. gingivalis has not been delineated.
In addition to requiring outer membrane receptors, utilization of hemin from hemoglobin in P. gingivalis requires participation of the P. gingivalis cysteine proteases, collectively referred to as gingipains (11, 25, 47, 55). These bacterial proteases exhibit activity against a wide range of host proteins including immunoglobulins, extracellular matrix proteins, bactericidal proteins, collagen, fibronectin, fibrinogen, tumor necrosis factor, interleukin-8, and proteins involved in the complement, coagulation, and kallikrein/kinin cascades (20, 28, 33, 50). The arginine-specific gingipains (HRgpA and RgpB) are encoded by genes rgpA and rgpB, respectively, while a lysine-specific gingipain (Kgp) is encoded by gene kgp (1, 4, 10, 39, 40, 44, 45). The translated portions of the rgpA and kgp genes encode prepropeptide, catalytic, and hemagglutinin domains, and the initial polyprotein is subject to posttranslational processing by Kgp itself. RgpB is expressed as a prepropeptide missing the majority of the hemagglutinin domain but is otherwise closely related to the rgpA gene product (34, 36). Kgp, HRgpA, and RgpB can be found both associated with the outer membrane and in a soluble form.
Recent reports have documented the ability of the Kgp complex to bind hemoglobin, hemin, porphyrins, and metalloporphyrins (14, 17, 27, 37, 41, 42). Characterization of defined P. gingivalis mutants also supports a role for Kgp in hemin and hemoglobin binding and utilization (19, 41, 53, 55); W. Simpson and C. A. Genco, unpublished data). Furthermore, a recent report has documented that Kgp can cleave soluble hemoglobin (32). Collectively these studies indicate that Kgp may function to both bind and degrade hemoglobin, ultimately releasing heme, which could be utilized for growth. In this study we demonstrate that, in addition to its ability to bind and degrade hemoglobin, purified Kgp can degrade soluble haptoglobin, hemopexin, and transferrin present in normal human serum. Degradation of these serum proteins was observed during growth of P. gingivalis in a minimal medium supplemented with human serum as a sole source of heme, and cleavage of these proteins coincided with an increase in lysine- and arginine-specific cysteine proteinase activity in bacterial cultures.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. gingivalis strain A7436 was used in these studies and was maintained on anaerobic blood agar plates (Remel, Lenexa, Kans.). All P. gingivalis cultures were incubated at 37°C in an anaerobic chamber (Coy Laboratory Products, Inc., Ann Arbor, Mich.) with 85% N2, 5% H2, and 10% CO2 for 3 to 5 days. To examine the ability of P. gingivalis to degrade serum proteins, strain A7436 was first grown on anaerobe broth MIC (Difco, Detroit, Mich.) at 37°C for 24 h and then inoculated into a basal medium (BM; 10 g of Trypticase peptone, 0.2 g of tryptophan, 2.5 g of NaCl, 0.1 g of sodium sulfite, and 0.4 g of cysteine per liter). After another 24 h of cultivation under anaerobic conditions, the culture was inoculated into BM or BM supplemented with 10% normal human serum (Sigma, St. Louis, Mo.).
Purification of gingipains.
Kgp, HRgpA, and RgpB were purified from P. gingivalis strain HG66 cultures as previously described (46, 49). Approximately 5 mg of each gingipain from 1 liter of bacterial culture was purified to homogeneity as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the concentration of active gingipain in each batch was determined by active-site titration using specific inhibitors Z-Phe-Lys-2,4,6-trimethylbenzoyloxymethylketone and H-d-Phe-Phe-Arg-chloromethylketone (FFRck) (Bachem Biosciences Inc., King of Prussia, Pa.) for gingipain K and gingipains R, respectively (48).
Degradation of hemoglobin by purified gingipains.
Samples of hemoglobin (0.5 mg/ml, 7.5 μM) were incubated at 37°C with gingipains at 50 to 200 nM concentrations in 200 mM HEPES–150 mM NaCl–1 mM CaCl2, pH 7.6, supplemented with 10 mM cysteine. At specific times the reaction was stopped by the addition of FFRck (1.0 μM), and the mixture was subjected to SDS-PAGE analysis (52). SDS-PAGE gels were also subjected to laser densitometry, and hemoglobin degradation was monitored as the relative optical densities of the hemoglobin bands. The rate of hemoglobin degradation was also measured as a decrease of absorbance at 415 nm (the Soret band) during incubation of hemoglobin with gingipains. For these studies 7.5 μM hemoglobin and 100 to 200 nM gingipains were used. Degradation of hemoglobin by gingipains in human serum was also examined. For these studies hemoglobin (0.5 mg/ml) dissolved in 20 mM Tris–150 mM NaCl, pH 7.4, was added to human serum (0.5 mg/ml, final concentration) and the serum was incubated with Kgp (50 nM) in the presence of 10 mM cysteine at 37°C for 3 or 7 h. After SDS-PAGE, proteins were electrotransferred onto a nitrocellulose membrane and Western blot analysis was performed using antihemoglobin serum (see below).
Degradation of haptoglobin and haptoglobin-hemoglobin complexes by purified gingipains.
Purified haptoglobin (1 mg/ml) was incubated with RgpB (90 nM), HRgpA (90 nM), or Kgp (30 nM) for various times and examined by SDS-PAGE analysis as described above. For some studies purified haptoglobin (1 mg/ml) was preincubated with hemoglobin (2 mg/ml) and then incubated with the gingipains.
Degradation of hemoglobin, haptoglobin, hemopexin, and transferrin in normal human serum by gingipains.
Samples of human serum obtained from healthy volunteers (diluted fivefold) were incubated at 37°C with gingipains (RgpB, HRgpA, and Kgp) in the presence of 10 mM cysteine. Concentrations of gingipains for these studies ranged from 10 to 1,000 nM. At specific times, aliquots were withdrawn, the reaction was stopped by addition of FFRck (inhibits all three enzymes; 100 nM), and the mixture was subjected to SDS-PAGE analysis. Protein bands were visualized by Coomassie brilliant blue R-250 staining and by Western blot analysis (60) using antihemoglobin, antihaptoglobin, antihemopexin, and antitransferrin serum. Rabbit anti-human hemoglobin serum was purchased from DAKO (Glostrup, Denmark), goat anti-human haptoglobin serum and goat anti-human transferrin serum were purchased from Biomeda Co. (Foster City, Calif.), and rabbit anti-human hemopexin serum was purchased from Cortex Biochem (San Leadro, Calif.). Antihemoglobin serum was used at a 1:2,000 dilution, and antihaptoglobin, antihemopexin, and antitransferrin sera were used at 1:5,000 dilution. The appropriate secondary antibodies conjugated to alkaline phosphatase (Sigma) were added, and the blot was developed with a kit obtained from Bio-Rad according to manufacturer's protocol. For some studies hemoglobin (50 μg/ml to 10 mg/ml) or hemin (0.1 mg/ml) was added to serum samples and samples were incubated for 1 h at 37°C prior to the addition of gingipains.
Degradation of hemoglobin, haptoglobin, hemopexin, and transferrin in normal human serum during bacterial growth.
P. gingivalis A7436 was cultivated in BM or BM supplemented with 10% normal human serum (Sigma) as previously described (19). At specific times, samples were withdrawn, gingipains were inhibited by addition of FFRck (100 nM final concentration), and after centrifugation (10,000 × g, 10 min) the resulting supernatant was subjected to SDS-PAGE analysis. Protein bands corresponding to hemoglobin, haptoglobin, hemopexin, and transferrin were visualized by Western blot analysis as described above.
Lysine- and arginine-specific activities.
The amidolytic activities of P. gingivalis A7436 in whole cultures and in cell-free supernatant fractions were determined with either N-benzoyl-l-arginine-p-nitroanilide or Z-lysine-p-nitroanilide (Z-Lys-pNA). Samples were preincubated in 200 mM Tris-HCl–100 mM NaCl–5 mM CaCl2 (pH 7.6), supplemented with 10 mM cysteine, for 5 min at 37°C and assayed for amidase activity with 0.5 mM substrate. The formation of p-nitroanilide was monitored spectrophotometrically at 405 nm.
RESULTS
Kgp can degrade hemoglobin.
To investigate and compare the functions of P. gingivalis proteolytic enzymes in hemoglobin degradation and heme release, we incubated hemoglobin with active-site-titrated Kgp, HRgpA, or RgpB. The purity of the three enzymes used in this study was confirmed by SDS-PAGE analysis (Fig. 1). Following incubation of each purified gingipain with hemoglobin, we monitored the rate of hemoglobin degradation by SDS-PAGE analysis. Following a 3-h incubation of hemoglobin with Kgp we observed almost complete digestion of this protein (Fig. 2A, lane e). In contrast, at a 100 nM gingipain concentration hemoglobin was refractory to degradation by HRgpA and RgpB (Fig. 2A, lanes c and d). Degradation of the hemoglobin chains by Kgp occurred in a time-dependent manner and without accumulation of any discrete cleavage products (Fig. 2A, lanes h to n). Hemoglobin was completely degraded by Kgp following incubation for 360 min (Fig. 2A, lane n). For samples of hemoglobin incubated with Kgp, the decrease of the absorbance correlated with the disappearance of the globin chains as determined by laser densitometry analysis of SDS-PAGE gels (data not shown).
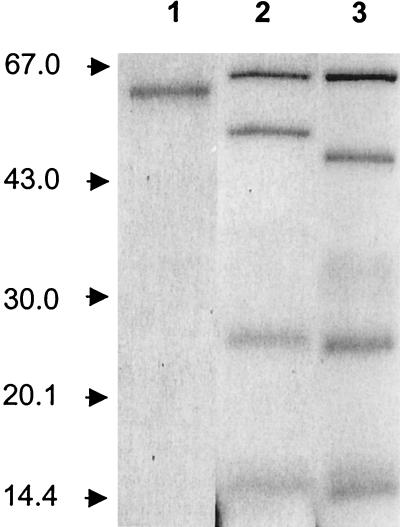
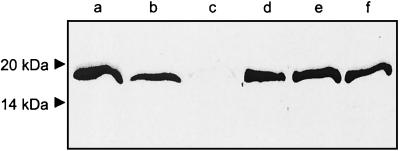
FIG. 1.
Purity of gingipains. RgpB (lane 1), HRgpA (lane 2), Kgp (lane 3) were purified from P. gingivalis strain HG66. Samples were boiled in reducing sample treatment buffer and analyzed by SDS-PAGE. Molecular weights are indicated on the left.
FIG. 2.
Degradation of hemoglobin. (A) Samples of hemoglobin (0.5 mg/ml, 7.5 μM) were incubated with gingipains RgpB, HRgpA, and Kgp (100 nM) at 37°C in the presence of 10 mM cysteine. At specific times, aliquots were withdrawn and treated with FFRck (100 nM, final concentration) to terminate the reaction. Samples were boiled in reducing sample treatment buffer and analyzed by SDS-PAGE. Lane a, molecular mass standards; lanes b and g, hemoglobin control incubated without gingipains; lanes c to e, hemoglobin incubated for 3 h with HRgpA, RgpB, and Kgp, respectively; lane f, Kgp alone; lanes h to n, hemoglobin incubated with Kgp for 30, 60, 90, 120, 150, 180, and 360 min, respectively. (B) Samples of hemoglobin (7.5 μM) were incubated with Kgp (100 [solid circles] or 200 nM [open circles]) as described above, and the optical density at 415 nm (OD415nm) was recorded every 10 min. Triangles, control samples containing all reagents except Kgp.
The release of heme during hemoglobin degradation by Kgp was examined by monitoring the decrease of absorbance at 415 nm (the Soret band). In agreement with the results obtained by SDS-PAGE analysis, incubation of hemoglobin with Kgp resulted in a time- and concentration-dependent decrease in the intensity of the Soret band of hemoglobin (Fig. 2B). Likewise, as observed in SDS-PAGE analysis, we did not observe a decrease in the Soret band following incubation of hemoglobin with HRgpA or RgpB (data not shown). Hemoglobin degradation by Kgp measured spectrophotometrically followed a typical Michaelis-Menten kinetic plot. The Km and kcat (catalytic constant) values were determined to be 2.9 ± 0.3 μM and 9.4 min−1, respectively, indicating the relatively high catalytic efficiency (kcat/Km = 3.2 × 106 M−1 min−1) of hemoglobin cleavage (data not shown).
Degradation of haptoglobin and the haptoglobin-hemoglobin complex by gingipains.
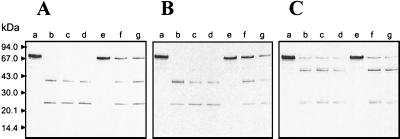
Under physiological conditions, hemoglobin released from erythrocytes is tightly bound to haptoglobin with a dissociation constant greater than 10−15 M (15, 21). Interestingly, the P. gingivalis hemoglobin receptor HmuR binds with very low affinity to haptoglobin complexed to hemoglobin and does not bind apo-haptoglobin (42). To utilize hemoglobin in vivo, P. gingivalis should thus also recognize and degrade haptoglobin and/or the hemoglobin-haptoglobin complex, releasing hemoglobin. Therefore, we examined the ability of gingipains to degrade haptoglobin. Three major phenotypic forms of human haptoglobin, designated Hp 1-1, Hp 2-2, and Hp 2-1 have been described; Hp 2-2 and 2-1 are polymerized forms of higher molecular mass (15). In vitro, Kgp (30 nM) efficiently cleaved the invariant heavy β-chain and both light α1- and α2-chains with purified Hp 1-2 (Fig. 3) and Hp 1-1 and Hp 2-2 (data not shown) phenotypes. This degradation was initially observed following a 15-min incubation period of haptoglobin with Kgp and was virtually complete by 60 min (Fig. 3). In contrast, HRgpA and RgpB (90 nM) degraded only the α-chains of haptoglobin, as first observed following a 60-min incubation (Fig. 3).
FIG. 3.
Degradation of soluble haptoglobin by gingipains. Haptoglobin (1 mg/ml) was incubated with RgpB (90 nM; lanes c to e), HRgpA (90 nM; lanes f to h), and Kgp (30 nM; lanes i to k) for 15 (lanes c, f, and i), 60 (lanes d, g, and j), and 180 min (lanes e, h, and k) in 20 mM HEPES–150 mM NaCl–1 mM CaCl2–10 mM cysteine, pH 7.6. The reaction was terminated with FFRck (10 μM, final concentration); samples were boiled in reducing treatment buffer and analyzed by SDS-PAGE. Haptoglobin incubated without gingipains for 180 min was loaded in lane b. Lane a, molecular mass standards.
The ability of gingipains to cleave haptoglobin in serum, where other proteins could inhibit the degradation of haptoglobin by these proteases, was confirmed. Human serum was incubated with gingipains at 37°C for various times and degradation of haptoglobin was monitored by Western blot analysis using rabbit antihaptoglobin serum. As observed above, HRgpA and RgpB (90 nM, final concentration) were found to cleave the haptoglobin α-chains, as detected following a 3-h incubation of these proteases with serum. Kgp at a lower concentration (30 nM) degraded both β- and α-chains of haptoglobin, as initially observed at 3 h (Fig. 4). However, the extent of haptoglobin susceptibility to degradation varied considerably among tested serum samples of the same haptoglobin phenotype (data not shown). This variation was at least partially due to differing degrees of erythrocyte hemolysis since saturation of susceptible samples with hemoglobin protected the α-chains and β-chain of haptoglobin from degradation by gingipains R and Kgp, respectively. However, we cannot exclude the possibility that Kgp and HRgpA bound the excess hemoglobin and that these proteases could not digest the haptoglobin.
FIG. 4.
Degradation of haptoglobin and the haptoglobin-hemoglobin complex in normal human serum. Human serum (lanes a to d) and serum saturated with hemoglobin (10 mg/ml; 1 h at 37°C) (lanes e to h) were incubated with HRgpA (90 nM) (A), RgpB (90 nM) (B), and Kgp (30 nM) (C) in the presence of 10 mM cysteine for 3 (lanes b and f), 7 (lanes c and g), and 15 h (lanes d and h) at 37°C. Lanes a and e, control serum samples incubated overnight under the same conditions but without gingipains. Following SDS-PAGE, Western blot analysis was performed using antihaptoglobin serum. Molecular mass standards are shown on the left. Arrowheads, α- and β-chains of haptoglobin.
The protective effect of hemoglobin on gingipain-mediated haptoglobin degradation was confirmed with isolated haptoglobin and with haptoglobin in complex with hemoglobin, as well as with human serum supplemented with increasing amounts of purified hemoglobin and incubated with Kgp. Some slight degradation of haptoglobin complexed with hemoglobin by Kgp was observed following a 180-min incubation; however, this was not as extensive as that of uncomplexed haptoglobin (data not shown). In the haptoglobin-hemoglobin complex, hemoglobin appeared to be completely refractory to degradation by Kgp. Human serum or purified haptoglobin was mixed with a saturating concentration of hemoglobin and incubated with Kgp and analyzed by Western blotting using antihemoglobin serum. Whereas free hemoglobin was readily degraded by Kgp in a time-dependent manner, as indicated by disappearance of the 16-kDa immunoreactive α- and β-hemoglobin chains, hemoglobin present in a complex in serum was resistant to digestion (Fig. 5). Similar results were obtained using Kgp at a final concentration of 200 nM (data not shown). These results indicate that in the haptoglobin-hemoglobin complex both hemoglobin and the β-chain of haptoglobin are protected from degradation by Kgp.
FIG. 5.
Degradation of hemoglobin in Tris-buffered saline and in normal human serum. Hemoglobin (0.5 mg/ml) dissolved in 20 mM Tris–150 mM NaCl, pH 7.4 (lanes a to c) or added to human serum (0.5 mg/ml, final concentration) (lanes d to f) was incubated with Kgp (50 nM) in the presence of 10 mM cysteine at 37°C for 3 (lanes b and e) or 7 h (lanes c and f). Lane a, hemoglobin in buffer; lane d, hemoglobin in serum without gingipains. After SDS-PAGE, proteins were electrotransferred onto a nitrocellulose membrane and Western blot analysis was performed using antihemoglobin serum. Molecular mass standards are shown on the left.
Hemopexin degradation by gingipains.
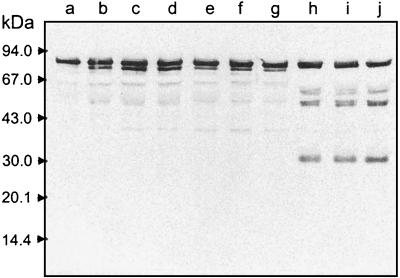
Heme is avidly scavenged by the serum heme-binding proteins hemopexin and albumin (21). The affinity of hemopexin for hemin is higher than that of hemoglobin and several orders of magnitude higher than that of albumin (35). Since P. gingivalis can utilize hemin complexed to hemopexin as an iron source (5), it must be able to capture hemin from this host hemin-binding protein. We have established that the P. gingivalis hemoglobin receptor HmuR binds hemin and, with lower affinity, hemopexin complexed with hemin, but not apo-hemopexin (42). Because hemopexin has such a high affinity for hemin, we reasoned that the ability of P. gingivalis to acquire hemin from this protein could involve a mechanism in which the serum protein was degraded by proteinases from this organism. To determine if the gingipains could degrade hemopexin, normal human serum was incubated with Kgp, HRgpA, or RgpB and degradation was monitored over time using Western blot analysis with antihemopexin serum. When Kgp (30 nM) was added to the serum samples, we observed rapid cleavage of hemopexin as indicated by the disappearance of the intact 70-kDa polypeptide and the appearance of degradation products (23 and 47 kDa) immunoreactive with antihemopexin serum (Fig. 6C). Serum hemopexin was also efficiently cleaved by HRgpA and RgpB, yielding two main proteolytic products of 23 and 40 kDa (Fig. 6A and B). However, degradation of hemopexin by HRgpA or RgpB was achieved only at higher concentrations (90 nM) of these enzymes (Fig. 6A and B).
FIG. 6.
Hemopexin degradation in human serum by gingipains. Serum (lanes b to d) and serum saturated with hemin (0.1 mg/ml, 30 min, 37°C) (lanes f and g) was incubated with HRgpA (90 nM) (A), RgpB (90 nM) (B), and Kgp (30 nM) (C) at 37°C for 3 (lanes b and f), 7 (lanes c), and 15 h (lanes d and g). Lanes a and e, serum samples incubated without gingipains. After SDS-PAGE, proteins were electrotransferred onto a nitrocellulose membrane and Western blot analysis was performed using antihemopexin serum. Molecular mass standards are shown on the left.
To determine if heme saturation of hemopexin could influence the susceptibility of the protein to proteolytic degradation by gingipains, human serum was saturated with hemin prior to the addition of gingipains. We found that the addition of hemin to human serum influenced the ability of Kgp to degrade hemopexin (Fig. 6C, lanes f and g). Similar results were observed in serum samples incubated with HRgpA and RgpB (Fig. 6A and B, lanes f and g).
Transferrin degradation.
In addition to hemin- and heme-containing proteins, P. gingivalis can utilize nonhemin iron source transferrin for growth (5, 7, 24, 54, 59). However, specific receptors for transferrin in this organism have not been described. To determine if transferrin in normal human serum is susceptible to cleavage by gingipains, we incubated serum with HRgpA, RgpB, or Kgp and monitored the degradation over time. Incubation of human serum with each of the three enzymes resulted in limited proteolysis of transferrin, as detected by Western blot analysis using antitransferrin serum. We found that the patterns of cleavage differed considerably between the arginine-specific gingipains and Kgp. While the addition of HRgpA and RgpB to human serum resulted in only a slight truncation of the transferrin polypeptide chain (Fig. 7, lanes b to g), the addition of Kgp resulted in the cleavage of transferrin into two fragments (30 and 50 kDa) (Fig. 7, lanes h to j).
FIG. 7.
Degradation of transferrin in human serum by gingipains. Normal human serum was diluted fivefold and incubated with a 50 nM concentration of RgpB (lanes b to d), HRgpA (lanes e to g), and Kgp (lanes h to j) for 3 (lanes b, e, and h), 7 (lanes c, f, and g), and 15 h (lanes d, g, and j). After SDS-PAGE, proteins were electrotransferred onto a nitrocellulose membrane and Western blot analysis was performed using antitransferrin sera. Lane a, serum sample incubated without gingipain. Molecular mass standards are shown on the left.
P. gingivalis can degrade hemoglobin, haptoglobin, hemopexin, and transferrin in human serum during bacterial growth.
To determine if P. gingivalis can cleave hemoglobin, haptoglobin, hemopexin, and transferrin in culture, we grew the organism in a minimal medium with human serum as the sole source of heme and iron. We monitored the degradation of serum proteins and the cell-associated and extracellular arginine- and lysine-specific activity during the growth period. We found that P. gingivalis strain A7436 exhibited a long lag phase in minimal media supplemented with normal human serum; this was followed by a rapid increase in growth (data not shown). The increase in bacterial growth correlated with a two- to threefold increase in both lysine- and arginine-specific proteinase activity (Table 1). The levels of lysine- and arginine-specific proteinase activity produced by P. gingivalis A7436 cultures were comparable to the levels of activity previously reported under similar growth conditions (19).
TABLE 1.
Arginine- and lysine-specific amidolytic activities of P. gingivalis culture grown in the presence of 10% human serum
| Time (h)b | Activitya
|
|
|---|---|---|
| Z-Lys-pNA | BApNA | |
| 0 | 0.09 | 0.98 |
| 6 | 0.18 | 2.3 |
| 24 | 0.38 | 2.65 |
| 30 | 0.34 | 2.41 |
| 48 | 0.37 | 1.08 |
| 54 | 1.29 | 0.77 |
Activity was determined from 1-μl samples of bacterial culture adjusted to the same optical density (OD) and expressed as milli-OD units per minute for each substrate. BApNA, N-benzoyl-l-arginine-p-nitroanilide; Z-Lys-pNA, Z-lysine-p-nitroanilide.
Time when samples were removed from the bacterial culture.
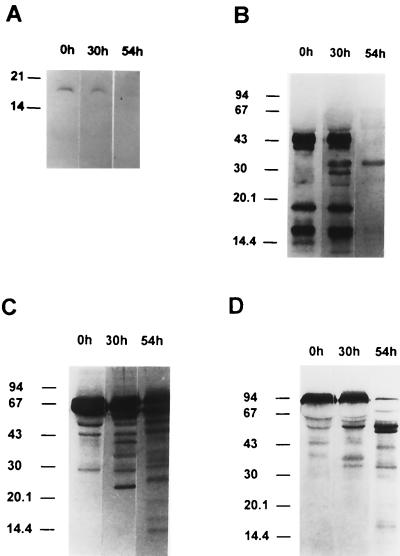
Bacterial growth and the expression of the lysine-specific proteinase activities in P. gingivalis A7436 were correlated with the degradation of serum proteins in supernatant fractions. We observed degradation of hemoglobin in supernatant fractions obtained from P. gingivalis A7436 cultures as demonstrated by the disappearance of the protein band immunoreactive with antihemoglobin serum, observed after 54 h of growth (Fig. 8). This correlated with high levels of Kgp activity at this time point (Table 1). We also observed degradation of haptoglobin in cultures at 30 h as determined by the appearance of haptoglobin degradation products (Fig. 8). By 54 h both the heavy and light chains of haptoglobin were degraded. We also observed degradation of hemopexin. By 54 h the hemopexin was almost completely degraded. P. gingivalis strain A7436 was also found to degrade transferrin following growth in normal human serum (Fig. 8). We did not observe degradation of hemoglobin, haptoglobin, hemopexin, or transferrin in BM containing serum that was incubated for 72 h without the addition of bacteria (data not shown).
FIG. 8.
Degradation of serum proteins during growth of P. gingivalis. P. gingivalis strain A7436 was grown in minimal media supplemented with 10% normal human serum as a heme source. Samples were removed from cultures grown with serum at the indicated times, supernatants were separated by SDS-PAGE, and the Western blot was developed using antisera to hemoglobin (A), haptoglobin (B), hemopexin (C), and transferrin (D). Results are from one experiment and are representative of three separate experiments.
DISCUSSION
P. gingivalis can utilize several different iron sources for bacterial growth including hemin, hemoglobin, and transferrin (5, 7, 18, 24, 54, 59). Hemoglobin has been reported to be more effective in supporting the growth of P. gingivalis than hemin or transferrin (53, 54). However, under physiological conditions, free hemoglobin or hemin may not be available since these compounds are efficiently scavenged by haptoglobin and hemopexin, respectively (15, 21, 35). P. gingivalis can utilize hemoglobin bound to haptoglobin and hemin bound to hemopexin (5); however, the mechanism(s) by which hemin is released from these proteins has not been defined. One effective way to release iron or hemin from these proteins may involve their proteolytic degradation, and in this study we have explored this possibility. We have demonstrated that soluble gingipains can degrade the host heme iron-binding proteins, hemoglobin, haptoglobin, hemopexin, and transferrin, with Kgp being the most effective. This enzyme at nanomolar concentrations degraded free hemoglobin apparently due to significant catalytic efficiency against this substrate, as reflected by the high value of the kcat/Km ratio (3.2 × 106 M−1 min−1). The kinetic value in this range indicates that this reaction can occur in vivo, especially in the absence of any endogenous inhibitors of Kgp (23, 51). In contrast to Kgp, gingipains R possessed weak hemoglobinase activity. Similarly, Kgp was more active than gingipains R in the degradation of haptoglobin, hemopexin, and transferrin in human serum. While arginine-specific gingipains HRgpA and RgpB also cleaved these serum proteins, the concentrations required for effective degradation were 5 to 10 times higher than that required by Kgp.
Our results fully confirm a recent study by Lewis et al. (32) on hemoglobin degradation by Kgp. Our study extends that report by providing kinetic data on hemoglobin degradation by Kgp as well as documenting the proteolysis of other heme iron-binding proteins by gingipains. Interestingly, in our study we did not observe extensive degradation of hemoglobin in human serum by soluble Kgp at the concentrations that we tested. Likewise the β-chain of haptoglobin was partially protected from degradation by Kgp in a haptoglobin-hemoglobin complex. However, we observed the time-dependent degradation of all of the serum proteins examined in culture media supplemented with human serum during P. gingivalis growth, which was correlated with an increase in arginine- and lysine-specific proteinase activities. The observation that host heme and iron serum proteins are degraded by P. gingivalis cultures is in agreement with previous studies in which a suspension of P. gingivalis cells were reported to degrade serum proteins (8, 16). Collectively, these observations may result from differences in the enzymatic activity of soluble versus membrane-associated gingipains. It was previously reported that the activities of the gingipains are dependent on the form in which they are purified (34). This is not an unexpected finding considering that, when gingipains are embedded in the membrane, the exposure of the catalytic and hemagglutinin portions would be expected to differ from that for the protein in its soluble form. Alternatively, it is also possible that unidentified P. gingivalis proteinases may be utilized for the cleavage of hemoglobin bound to haptoglobin in serum during bacterial growth.
The results presented in this study and in an accompanying paper (42), together with several other reports (7, 14, 17, 27, 32, 37, 41), have established that Kgp can bind and degrade hemoglobin and other serum heme and iron proteins. Characterization of P. gingivalis kgp mutants further supports a role for Kgp in hemoglobin binding and utilization in P. gingivalis (19, 41, 55). Furthermore, we have recently established that hemoglobin utilization in P. gingivalis A7436 requires both outer membrane receptor HmuR and Kgp (42, 55, 56; Simpson and Genco, unpublished data). We found that a P. gingivalis hmuR kgp mutant did not grow with either hemin or hemoglobin as the sole iron source (Simpson and Genco, unpublished data). We have also shown that HmuR can interact with Kgp (42). Based on these results, we propose that Kgp may facilitate hemoglobin binding and utilization in P. gingivalis by functioning as a bacterial heme-scavenging protein or hemoglobinase.
In E. coli extracellular hemoglobin-binding protein Hbp has been proposed to function as a bacterial hemophore or hemoglobinase (43). Similar to Kgp, E. coli Hbp appears to function as a hemoglobin-degrading proteinase and as a hemin-binding protein. Hbp, similar to P. gingivalis Kgp, is synthesized as a polyprotein precursor that is processed following export to the cell surface. However, it is not known if the E. coli hemoglobinase can degrade hemoglobin when it is complexed to haptoglobin or when it is present in human serum. We know, however, that any Kgp activity in human serum would be fully functional because of the absence of any Kgp-specific inhibitor that might restrict degradation of heme-containing proteins in serum (23). In contrast, gingipain R activity can be, at least partially, limited by α2-macroglobulin in human serum (23, 51).
The environmental conditions in the periodontal pocket during P. gingivalis infection are not precisely known. However, inflammation and tissue damage can result in an altered pattern of nutrients; when gingival crevicular fluid flow is increased and bleeding occurs, the availability of heme-containing proteins may increase, resulting in the enrichment of P. gingivalis. The ability to acquire hemin from hemoglobin released from erythrocytes appears to be of particular importance for the survival of P. gingivalis in vivo. In this study we have determined that lysine-specific cysteine proteinase Kgp can efficiently degrade soluble hemoglobin, as well as haptoglobin and hemopexin in serum. The proteinase-hemagglutinin complexes of Kgp may thus be important in the uptake of heme, via binding (42) and subsequent degradation of heme-containing proteins. The ability of P. gingivalis to efficiently bind and degrade host heme-containing proteins represents an additional mechanism adapted by this pathogen to ensure its successful growth and proliferation. Whether both soluble and cell-bound Kgps function in hemoglobin binding and degradation remains to be determined.
ACKNOWLEDGMENTS
This study was supported by Public Health Service grants from the National Institute of Dental and Craniofacial Research DE09161 (to C. A. Genco) and DE09761 (to J. Travis) and by grant 6 PO4A 047 17 from the State Committee of Scientific Research (KBN, Poland; to J. Potempa).
We acknowledge Dabney Dixon for critical review of the manuscript and Teresa Olczak for assistance with photography.
REFERENCES
- 1.Aduse-Opoku J, Muir J, Slaney J M, Rangarajan M, Curtis M A. Characterization, genetic analysis, and expression of a protease antigen (PrpRI) of Porphyromonas gingivalis W50. Infect Immun. 1995;63:4744–4754. doi: 10.1128/iai.63.12.4744-4754.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aduse-Opoku J, Slaney J, Rangarajan M, Muir J, Young K, Curtis M A. The Tla protein of Porphyromonas gingivalis W50: a homolog of the R1 protease precursor (PrpR1) is an outer membrane receptor required for growth on low levels of hemin. J Bacteriol. 1997;179:4778–4788. doi: 10.1128/jb.179.15.4778-4788.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnoux P, Haser R, Izadi N, Lecroisey A, Delepierre M, Wandersman C, Czjzek M. The crystal structure of HasA, a hemophore secreted by Serratia marcescens. Nat Struct Biol. 1999;6:516–520. doi: 10.1038/9281. [DOI] [PubMed] [Google Scholar]
- 4.Barkocy-Gallagher G A, Han N, Patti J M, Whitlock J, Progulske-Fox A, Lantz M S. Analysis of the prtP gene encoding porphypain, a cysteine proteinase of Porphyromonas gingivalis. J Bacteriol. 1996;178:2734–2741. doi: 10.1128/jb.178.10.2734-2741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bramanti T E, Holt S C. Roles of porphyrins and host iron transport proteins in regulation of growth of Porphyromonas gingivalis W50. J Bacteriol. 1991;173:7330–7339. doi: 10.1128/jb.173.22.7330-7339.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bramanti T E, Holt S C. Hemin uptake in Porphyromonas gingivalis: Omp 26 is a hemin-binding surface protein. J Bacteriol. 1993;175:7413–7420. doi: 10.1128/jb.175.22.7413-7420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brochu V, Grenier D, Nakayama K, Mayrand D. Acquisition of iron from human transferrin by Porphyromonas gingivalis: a role for Arg- and Lys-gingipain activities. Oral Microbiol Immunol. 2001;2:79–87. doi: 10.1034/j.1399-302x.2001.016002079.x. [DOI] [PubMed] [Google Scholar]
- 8.Carlsson J, Hofling J F, Sundqvist G K. Degradation of albumin, haemopexin, haptoglobin and transferrin by black-pigmented Bacteroides species. J Med Microbiol. 1984;18:39–46. doi: 10.1099/00222615-18-1-39. [DOI] [PubMed] [Google Scholar]
- 9.Cope L D, Thomas S E, Hrkal Z, Hansen E J. Binding of heme-hemopexin complexes by soluble HxuA protein allows utilization of this complexed heme by Haemophilus influenzae. Infect Immun. 1998;66:4511–4516. doi: 10.1128/iai.66.9.4511-4516.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis M A, Aduse-Opoku J, Slaney M, Rangarajan M, Booth C I, Sheperd P. Characterization of an adherence and antigenic determinant of the Arg1 protease of Porphyromonas gingivalis which is present on multiple gene products. Infect Immun. 1996;64:2532–2539. doi: 10.1128/iai.64.7.2532-2539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtis M A, Kuramitsu H K, Lantz M, Macrina F L, Nakayama K, Potempa J, Reynolds E C, Aduse-Opoku J. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J Periodontal Res. 2000;34:464–472. doi: 10.1111/j.1600-0765.1999.tb02282.x. [DOI] [PubMed] [Google Scholar]
- 12.Cutler C W, Kalmar J R, Genco C A. Pathogenic strategies of the oral anaerobe Porphyromonas gingivalis. Trends Microbiol. 1995;3:45–51. doi: 10.1016/s0966-842x(00)88874-5. [DOI] [PubMed] [Google Scholar]
- 13.Dashper S G, Hendtlass A, Slakeski N, Jackson C, Cross K J, Brownfield L, Hamilton R, Barr I, Reynolds E C. Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J Bacteriol. 2000;182:6456–6462. doi: 10.1128/jb.182.22.6456-6462.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeCarlo A A, Paramaesvaran M, Yun P L W, Collyer C, Hunter N. Porphyrin-mediated binding to hemoglobin by the HA2 domain of cysteine proteinases (gingipains) and hemagglutinins from the periodontal pathogen Porphyromonas gingivalis. J Bacteriol. 1999;181:3784–3791. doi: 10.1128/jb.181.12.3784-3791.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobryszycka W. Biological functions of haptoglobin—new pieces to an old puzzle. Eur J Clin Chem Clin Biochem. 1997;35:647–654. [PubMed] [Google Scholar]
- 16.Fishburn C S, Slaney J M, Carman R J, Curtis M A. Degradation of serum proteins by the trypsin-like enzyme of Porphyromonas gingivalis and inhibition of protease activity by a serine protease inhibitor of human serum. Oral Microbiol Immunol. 1991;6:209–215. doi: 10.1111/j.1399-302x.1991.tb00479.x. [DOI] [PubMed] [Google Scholar]
- 17.Fujimura S, Shibata Y, Hirai K, Nakamura T. Binding of hemoglobin to the envelope of Porphyromonas gingivalis and isolation of the hemoglobin-binding protein. Infect Immun. 1996;64:2339–2342. doi: 10.1128/iai.64.6.2339-2342.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genco C A, Odusanya B M, Brown G. Binding and accumulation of hemin in Porphyromonas gingivalis are induced by hemin. Infect Immun. 1994;62:2885–2892. doi: 10.1128/iai.62.7.2885-2892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genco C A, Simpson W, Forng R Y, Egal M, Odusanya B M. Characterization of a Tn4351-generated hemin uptake mutant of Porphyromonas gingivalis: evidence for the coordinate regulation of virulence factors by hemin. Infect Immun. 1995;63:2459–2466. doi: 10.1128/iai.63.7.2459-2466.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genco C A, Potempa J, Mikolajczyk-Pawlinska J, Travis J. Role of gingipains R in Porphyromonas gingivalis pathogenesis. Clin Infect Dis. 1999;28:456–465. doi: 10.1086/515156. [DOI] [PubMed] [Google Scholar]
- 21.Genco C A, Dixon D W. Emerging strategies in microbial heme capture. Mol Microbiol. 2001;39:1–11. doi: 10.1046/j.1365-2958.2001.02231.x. [DOI] [PubMed] [Google Scholar]
- 22.Ghigo M J, Letoffe S, Wandersman C. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J Bacteriol. 1997;179:3572–3579. doi: 10.1128/jb.179.11.3572-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gron H, Pike R, Potempa J, Travis J, Thogersen I B, Enghild J J, Pizzo S V. The potential role of alpha 2-macroglobulin in the control of cysteine proteinases (gingipains) from Porphyromonas gingivalis. J Periodontal Res. 1997;32:61–68. doi: 10.1111/j.1600-0765.1997.tb01383.x. [DOI] [PubMed] [Google Scholar]
- 24.Inoshita E, Iwakura K, Amano A, Tamagawa H, Shizukuishi S. Effect of transferrin on the growth of Porphyromonas gingivalis. J Dent Res. 1991;70:1258–1261. doi: 10.1177/00220345910700090501. [DOI] [PubMed] [Google Scholar]
- 25.Kadowaki T, Nakayama K, Okamoto K, Abe N, Baba A, Shi Y, Ratnayake D B, Yamamoto K. Porphyromonas gingivalis proteinases as virulence determinants in progression of periodontal diseases. J Biochem. 2000;128:153–159. doi: 10.1093/oxfordjournals.jbchem.a022735. [DOI] [PubMed] [Google Scholar]
- 26.Karunakaran T, Madden T, Kuramitsu K. Isolation and characterization of a hemin-regulated gene, hemR, from Porphyromonas gingivalis. J Bacteriol. 1997;179:1898–1908. doi: 10.1128/jb.179.6.1898-1908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuboniwa M, Amano A, Shizukuishi S. Hemoglobin-binding protein purified from Porphyromonas gingivalis is identical to lysine-specific cysteine proteinase (lys gingipain) Biochem Biophys Res Commun. 1998;249:38–43. doi: 10.1006/bbrc.1998.8958. [DOI] [PubMed] [Google Scholar]
- 28.Kuramitsu H K. Proteases of Porphyromonas gingivalis: what don't they do? Oral Microbiol Immunol. 1999;13:263–270. doi: 10.1111/j.1399-302x.1998.tb00706.x. [DOI] [PubMed] [Google Scholar]
- 29.Letoffe S, Ghigo J M, Wandersman C. Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc Natl Acad Sci USA. 1994;91:9876–9880. doi: 10.1073/pnas.91.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letoffe S, Redeker V, Wandersman C. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol Microbiol. 1998;28:1223–1234. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 31.Letoffe S, Nato F, Goldberg M E, Wandersman C. Interactions of HasA, a bacterial haemophore, with haemoglobin and with its outer membrane receptor HasR. Mol Microbiol. 1999;33:546–555. doi: 10.1046/j.1365-2958.1999.01499.x. [DOI] [PubMed] [Google Scholar]
- 32.Lewis J P, Dawson J A, Hannis J C, Muddiman D, Macrina F L. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J Bacteriol. 1999;181:4905–4913. doi: 10.1128/jb.181.16.4905-4913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikolajczyk-Pawlinska J, Travis J, Potempa J. Modulation of interleukin-8 activity by gingipains from Porphyromonas gingivalis: implications for pathogenicity of periodontal disease. FEBS Lett. 1998;440:282–286. doi: 10.1016/s0014-5793(98)01461-6. [DOI] [PubMed] [Google Scholar]
- 34.Mikolajczyk-Pawlinska J, Kordula T, Pavloff N, Pemberton P A, Chen W C, Travis J, Potempa J. Genetic variation of Porphyromonas gingivalis genes encoding gingipains, cysteine proteinases with arginine or lysine specificity. Biol Chem. 1998;379:205–211. doi: 10.1515/bchm.1998.379.2.205. [DOI] [PubMed] [Google Scholar]
- 35.Muller-Eberhard U. Hemopexin. Methods Enzymol. 1988;163:536–541. doi: 10.1016/0076-6879(88)63049-7. [DOI] [PubMed] [Google Scholar]
- 36.Nakayama K. Domain-specific rearrangement between the two Arg-gingipain-encoding genes in Porphyromonas gingivalis: possible involvement of nonreciprocal recombination. Microbiol Immunol. 1997;41:185–196. doi: 10.1111/j.1348-0421.1997.tb01189.x. [DOI] [PubMed] [Google Scholar]
- 37.Nakayama K, Ratnayake D B, Tsukuba T, Kadowaki T, Yamamoto K, Fujimura S. Hemoglobin receptor protein is intragenically encoded by the cysteine proteinase encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol Microbiol. 1998;27:51–61. doi: 10.1046/j.1365-2958.1998.00656.x. [DOI] [PubMed] [Google Scholar]
- 38.Ochsner U A, Johnson Z, Vasil M. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology. 2000;146:185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 39.Okamoto K, Misumi Y, Kadowaki T, Yoneda M, Yamamoto K, Ikehara Y. Structural characterization of arg gingipain, a novel arginine-specific cysteine proteinase, as a major periodontal pathogenic factor from Porphyromonas gingivalis. Arch Biochem Biophys. 1995;316:917–925. doi: 10.1006/abbi.1995.1123. [DOI] [PubMed] [Google Scholar]
- 40.Okamoto K, Kadowaki T, Nakayama K, Yamamoto K. Cloning and sequencing of the gene encoding a novel lysine-specific cysteine proteinase (Lys-gingipain) in Porphyromonas gingivalis: structural relationship with the arginine-specific cysteine proteinase (Arg-gingipain) J Biochem. 1996;120:398–406. doi: 10.1093/oxfordjournals.jbchem.a021426. [DOI] [PubMed] [Google Scholar]
- 41.Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake D B, Yamamoto K. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J Biol Chem. 1998;273:21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 42.Olczak T, Dixon D W, Genco C A. Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins. J Bacteriol. 2001;183:5599–5608. doi: 10.1128/JB.183.19.5599-5608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otto B R, van Dooren S J M, Nuijens J H, Lurink J, Oudega B. Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J Exp Med. 1998;188:1091–1103. doi: 10.1084/jem.188.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pavloff N, Potempa J, Pike R N, Prochazka V, Kiefer M C, Travis J, Barr P J. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase-adhesin polyprotein. J Biol Chem. 1995;270:1007–1010. doi: 10.1074/jbc.270.3.1007. [DOI] [PubMed] [Google Scholar]
- 45.Pavloff N, Pemberton P A, Potempa J, Chen W C, Pike R N, Prochazka V, Kiefer M C, Travis J, Barr P J. Molecular cloning and characterization of Porphyromonas gingivalis lysine-specific gingipain. A new member of an emerging family of pathogenic bacterial cysteine proteinases. J Biol Chem. 1997;272:1595–1600. doi: 10.1074/jbc.272.3.1595. [DOI] [PubMed] [Google Scholar]
- 46.Pike R, McGraw W, Potempa J, Travis J. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins. J Biol Chem. 1994;269:406–411. [PubMed] [Google Scholar]
- 47.Potempa J, Pavloff N, Travis J. Porphyromonas gingivalis: a proteinase/gene accounting audit. Trends Microbiol. 1995;3:430–434. doi: 10.1016/s0966-842x(00)88996-9. [DOI] [PubMed] [Google Scholar]
- 48.Potempa J, Pike R, Travis J. Titration and mapping of the active site of cysteine proteinases from Porphyromonas gingivalis (gingipains) using peptidyl chloromethanes. Biol Chem. 1997;378:223–230. doi: 10.1515/bchm.1997.378.3-4.223. [DOI] [PubMed] [Google Scholar]
- 49.Potempa J, Mikolajczyk-Pawlinska J, Brassell D, Nelson D, Thogersen I B, Enghild J J, Travis J. Comparative properties of two cysteine proteinases (gingipains R), the products of two related but individual genes of Porphyromonas gingivalis. J Biol Chem. 1998;273:21648–21657. doi: 10.1074/jbc.273.34.21648. [DOI] [PubMed] [Google Scholar]
- 50.Potempa J, Banbula A, Travis J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontology. 2000;24:153–192. doi: 10.1034/j.1600-0757.2000.2240108.x. [DOI] [PubMed] [Google Scholar]
- 51.Rangarajan M, Scragg M A, Curtis M A. Bait region cleavage and complex formation of human alpha2M with a Porphyromonas gingivalis W50 protease is not accompanied by enzyme inhibition. Biol Chem. 2000;381:57–65. doi: 10.1515/BC.2000.008. [DOI] [PubMed] [Google Scholar]
- 52.Schagger H, von Yagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 53.Shi Y, Ratnayake D B, Okamoto K, Abe N, Yamamoto K, Nakayama K. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. J Biol Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 54.Shizukuishi S, Tazaki K, Inoshita E, Kataoka K, Hanioka T, Amano A. Effect of concentration of compounds containing iron on the growth of Porphyromonas gingivalis. FEMS Microbiol Lett. 1995;131:313–317. doi: 10.1111/j.1574-6968.1995.tb07793.x. [DOI] [PubMed] [Google Scholar]
- 55.Simpson W, Wang C Y, Mikolajczyk-Pawlinska J, Potempa J, Travis J, Bond V C, Genco C A. Transposition of the endogenous insertion sequence element IS1126 modulates gingipain expression in Porphyromonas gingivalis. Infect Immun. 1999;67:5010–5020. doi: 10.1128/iai.67.10.5012-5020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simpson W, Olczak T, Genco C A. Characterization and expression of HmuR a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J Bacteriol. 2000;182:5737–5748. doi: 10.1128/jb.182.20.5737-5748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slakeski N, Dashper S G, Cook P, Poon C, Moore C, Reynolds E C. A Porphyromonas gingivalis genetic locus encoding a heme transport system. Oral Microbiol Immunol. 2000;15:388–392. doi: 10.1034/j.1399-302x.2000.150609.x. [DOI] [PubMed] [Google Scholar]
- 58.Smalley J W, Birss A J, McKee A S, Marsh P D. Haemin-binding proteins of Porphyromonas gingivalis W50 grown in a chemostat under haemin-limitation. J Gen Microbiol. 1993;139:2145–2150. doi: 10.1099/00221287-139-9-2145. [DOI] [PubMed] [Google Scholar]
- 59.Tazaki K, Inoshita E, Amano A, Hanioka T, Tamagawa H, Shizukuishi S. Interaction of Porphyromonas gingivalis with transferrin. FEMS Microbiol Lett. 1995;131:161–166. doi: 10.1016/0378-1097(95)00253-2. [DOI] [PubMed] [Google Scholar]
- 60.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]