Abstract
Introduction
Alzheimer's disease (AD) is associated with altered metabolites. This study aimed to determine the validity of using circulating metabolites to differentiate AD from other dementias.
Methods
Blood metabolites were measured in three data sets. Data set 1 (controls, 27; AD, 28) was used for analyzing differential metabolites. Data set 2 (controls, 93; AD, 92) was used to establish a diagnostic AD model with use of a metabolite panel. The model was applied to Data set 3 (controls, 76; AD, 76; other dementias, 205) to verify its capacity for differentiating AD from other dementias.
Results
Data set 1 revealed 7 upregulated and 77 downregulated metabolites. In Data set 2, a panel of 11 metabolites was included in a model that could distinguish AD from controls. In Data set 3, this panel was used to successfully differentiate AD from other dementias.
Discussion
This study revealed an AD‐specific panel of 11 metabolites that may be used for AD diagnosis.
Keywords: Alzheimer's disease, dementia, diagnostic model, metabolite, neurodegenerative disease
1. INTRODUCTION
Alzheimer's disease (AD) is the most prevalent neurodegenerative dementia. AD is associated with a considerable social and economic burden in countries with aging populations. 1 Currently, biomarkers in cerebrospinal fluid (CSF) and neuroimaging are the most well‐validated AD biomarkers in routine clinical settings. 2 However, these diagnostic biomarkers are invasive, time‐consuming, and expensive, and are thus not ideal first‐line approaches for screening large numbers of candidate AD patients. In addition, AD and other types of dementia, such as vascular dementia (VaD), Parkinson disease dementia (PDD), behavioral variant frontotemporal dementia (bvFTD), and dementia with Lewy body (DLB) share overlapping clinical manifestations, pathology, and biomarkers, which often result in difficulties in clinical diagnosis. 3 Therefore, the development of blood‐based biomarkers that enable the identification and differentiation of AD from other types of dementia is required.
AD is increasingly recognized as a heterogeneous syndrome underpinned by genetic and environmental factors. Although the accumulation of amyloid beta (Aβ) and tau in the brain is considered the core pathologic hallmark of AD, 4 other pathophysiological pathways, such as oxidative stress, 5 inflammation, 6 lipid metabolism, 7 and mitochondrial dysfunction 8 have been implicated in AD pathogenesis. With anti‐amyloid antibody therapies failing to improve the cognitive function of patients with AD, 9 the identification of alternative and modifiable biological pathways is an urgent priority. Crucially, identifying novel biological pathways may reveal circulating biomarkers for risk prediction and early diagnosis. Therefore, an extensive screening of circulating biomarkers may further enhance our understanding of AD pathogenesis and risk prediction.
Metabolites are small molecules involved in biochemical pathways and include lipids, amino acids, peptides, nucleic acids, and organic acids. These metabolites are the products of complex biological processes in cells, tissues, or whole organisms, and are thus potential candidates that may reflect disease phenotypes. 10 Metabolomics is a profiling method for the collective quantification of metabolites, which has emerged as a powerful tool for the discovery of novel biomarkers and contributed to our understanding of the mechanistic pathways underpinning AD. Previous metabolomics studies have reported peripheral changes in metabolites linked to AD, highlighting the potential of blood metabolomics to elucidate the pathogenic mechanisms and identify novel biomarkers of AD. 11 , 12 , 13 Furthermore, metabolite panels consisting of multifactorial biochemical pathways are a promising tool for the accurate diagnosis of AD. 14 , 15 , 16 , 17 , 18 , 19 Although these studies highlight specific metabolic underpinnings of AD, some metabolomics findings have failed to be replicated.
To ensure the potential clinical application of our findings, we recruited participants in strict accordance with CSF biomarkers (phosphorylated tau (P‐tau)/Aβ42 and Aβ42), which enabled the maximization of diagnostic accuracy for AD and exclusion of preclinical AD participants from controls. In addition, we employed ultra‐high‐performance liquid chromatography‐high resolution mass spectrometry (UPLC‐HRMS) and recruited three separate data sets to validate the results. This study aimed to evaluate whether (1) blood‐based metabolites differed in AD, and (2) if they could be used to differentiate AD from cognitively normal controls and participants with other types of dementia.
RESEARCH IN CONTEXT
Systematic Review: We searched PubMed using the terms “Alzheimer's disease,” “dementia,” “metabolites,” and “biomarkers” since January 1, 1990. However, whether metabolites can differentiate Alzheimer's disease (AD) from other types of dementias has not been addressed.
Interpretation: AD and other dementias may have overlapping clinical manifestations, pathology, and biomarkers, often resulting in difficulties in clinical diagnosis. We aimed to determine metabolites as an AD‐specific biomarker. A panel of 11 metabolites successfully differentiated AD from controls and other dementias, such as vascular dementia (VaD), Parkinson disease dementia (PDD), behavioral variant frontotemporal dementia (bvFTD), and dementia with Lew body (DLB). Our finding may provide a minimally invasive and widely available AD‐specific biomarker.
Future Directions: The clinical application of the 11‐metabolite panel for screening AD will be strengthened by the prospective longitudinal studies. In addition, more samples in international multiple centers will provide powerful evidence before extensive clinic use.
2. METHODS
2.1. Subjects
Three data sets were acquired in this study. Data set 1 was used to analyze the differential metabolites in a Beijing center (n = 55; controls, 27; AD, 28). Data to confirm the differential metabolites and develop the diagnostic model (Data set 2) were collected from centers in the provinces of Shandong, Henan, and Guangxi (n = 185, controls, 92; AD, 93). Data for the application of the model (Data set 3) were acquired from a Beijing center (n = 357; control, 76; AD, 76; VaD, 50; PDD, 52; bvFTD, 52; DLB, 51). Diagnoses of AD were based on the criteria published by the National Institute on Aging and Alzheimer's Association (NIA‐AA). 2 A cutoff value of 0.14 for CSF P‐tau/Aβ42 was used to differentiate between subjects with AD and normal controls. This value was calculated based on our previously published data 20 and is consistent with that reported in other studies. 21 Based on the amyloid, tau, and neurodegeneration (ATN) framework, low CSF Aβ42 is the key “Alzheimer's pathological change.” 22 Therefore, we used a reported CSF Aβ42 cutoff of 500 pg/mL as another inclusion criterion. 23 Diagnoses of VaD, 24 PDD, 25 bvFTD, 26 and DLB 27 were based on previously published criteria. To avoid the influence of metabolic diseases, we excluded individuals with obesity, diabetes, and other metabolic diseases, such as hyperlipidemia, hyperhomocysteinemia, and abnormality of liver and renal function. Written informed consent was obtained from all participants or their legal guardians. This study was approved by the institutional ethics board of Xuanwu Hospital, Capital Medical University.
2.2. CSF collection and measurements
Aβ42, total tau (T‐tau), and P‐tau were measured in CSF to help diagnose AD. CSF samples were collected according to international guidelines. 28 Briefly, subjects were placed in the left lateral position in the morning following a 12‐h fast when the lumbar puncture was performed. The L3‐L5 intervertebral disc spaces were chosen as the site of puncture. Ten‐milliliter samples of CSF were collected with 20‐gauge atraumatic needles and centrifugated at 2000 × g for 10 min at room temperature. Aβ42, T‐tau, and P‐tau were measured according to a published protocol. 29
2.3. Blood sample collection and preparation
Blood samples of 10 mL volume were collected from all enrolled individuals in the morning after a 12‐hour fast. Each serum sample was divided into two parts to extract polar metabolites for untargeted metabolomic profiling analysis and non‐polar lipid molecules for untargeted lipidomic analysis. Briefly, 100 μL aliquots of polar metabolite extraction were deproteinized using 400 μL methanol‐acetonitrile solutions containing multiple isotope‐labeled compounds as extraction internal standards. The resultant supernatants were transferred into a polypropylene 96‐well plate and dried in a CentriVap Concentrator (Labconco Corporation, Kansas City, MO, USA). The extracts were then dissolved in a methanol‐water mixture for metabolomic analyses. The solution was divided into three fractions for different analytical profiling methods, as described below. For non‐polar lipid extraction, serum lipidomes were extracted using liquid‐liquid partition with methanol and methyl tert‐butyl ether from 50 μL aliquots of serum samples. 30 The upper layer extracts were transferred into a polypropylene tube and dried under a vacuum in a CentriVap Concentrator. The lipidome extracts were then redissolved in a solution containing acetonitrile and isopropanol for lipidomic analyses.
2.4. Metabolomic analysis
Samples were analyzed using the meta‐Phenotype high‐definition metabolomic platform, in which five complementary analytical methods based on UPLC‐HRMS were employed. Untargeted metabolomic analyses were conducted on an Ultimate 3000 UHPLC system coupled to a Q Exactive quadrupole Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA, USA). 31 Briefly, the first fraction of the polar metabolite extract was analyzed on an Acquity HSS C18 column (Waters Corporation, Milford, MA, USA) with 0.1% formic acid‐water and 0.1% formic acid‐acetonitrile as the binary mobile phase, and then detected under positive electrospray mode (Figure S1A). The second fraction was analyzed on an Acquity BEH C18 column (Waters Corporation) column and eluted with a mixture of water and acetonitrile/methanol containing 5 mM ammonium bicarbonate and detected under negative electrospray mode (Figure S1B). The third polar fraction was measured by hydrophilic interaction chromatography on an Acuity BEH Amide column (Waters Corporation) and detected in negative electrospray ionization mode (Figure S1C), in which binary mobile phases consisting of 10% water in acetonitrile and 50% acetonitrile in water with 10 mM ammonium acetate as buffer salt were employed. Untargeted lipidomic analyses were performed on the same analytical instrument, which was operated under positive/negative polarity switching mode for lipid molecule detection. Chromatographic separation of the lipidome was achieved on an Accucore C30 core–shell column (Thermo Scientific). A binary mobile phase consisting of 60% acetonitrile in water and 10% acetonitrile in isopropanol containing 10 mM ammonium formate and 0.1% formate was utilized to elute lipid molecules (Figure S1D and S1E). In all the employed profiling methods, full scan mass spectra data under 70,000 full width half maximum (FWHM) resolution and the top seven or 10 full‐scan data‐dependent MS/MS spectra data were acquired with XCalibur software (Thermo Scientific).
2.5. Metabolomic data processing
For polar metabolites, metabolic peak extraction was analyzed using Compound Discoverer 2.1 software (Thermo Scientific) for metabolite peak extraction, and further structural annotation through searching against a local Human Metabolome Database (HMDB) 32 and a local proprietary MS/MS spectrum library created using authentic standards as well as online mzCloud library (www.mzcloud.org). Multiple chemical details, such as the exact mass of precursors, isotopic pattern fit scores, MS/MS spectra similarity, and retention time were included in the structural annotation of metabolites. An untargeted lipidomic data process including peak picking and lipid identification was executed by LipidSearch software (Thermo Scientific), where the acquired MS/MS spectra were searched against in silico predicted spectra of diverse endogenous lipid classes. The proposed identification results were further manually checked individually to eliminate false positives. The peak areas under the curve were extracted as relative quantitative information for annotated metabolites and lipids using TraceFinder software (Thermo Scientific), and peak integration results were checked manually by well‐trained technicians to guarantee accuracy. Finally, the resultant data set from each measurement was normalized to total peak area sum, respectively, and then merged, and trimmed before statistical analysis. Multivariate analysis including principal component analysis and orthogonal partial least square‐discriminant analysis were conducted with SIMCA‐P software (Sartorius Umetrics, Germany). Other univariate analyses including independent sample t‐test, P‐value false discovery rate (FDR) adjustments, metabolic enrichment analysis, and pathway analysis were conducted on the MetaboAnalyst website. 33
2.6. Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics for Windows, version 22.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA). The data from each data set were analyzed independently. Group differences in categorical data, such as sex, clinical subgroups, and apolipoprotein E (APOE) ε4 carrier distributions, were analyzed using the χ2 test. Group differences in numerical data, such as biomarker levels, were analyzed using Welch t‐test. In Data sets 2 and 3, the predicted values were calculated using a binary logistic regression model with age, sex, education years, or APOE ε4 status as covariates, which were subsequently used for the receiver‐operating characteristic (ROC) curve analysis. The multicollinearity between each metabolite was estimated by analyzing tolerances, variance inflation factors (VIFs), eigenvalues, and condition indices. All tests were two tailed, and the level of statistical significance was set at P < .05.
3. RESULTS
3.1. Participant characteristics
Three independent data sets were included. Table 1 lists the characteristics of the study subjects. In all three data sets, no significant differences were observed in age and male/female ratio between the AD and control groups. Significant differences were observed in the percentage of APOE ε4, and Mini‐Mental State Examination (MMSE) scores, CSF Aβ42, T‐tau, and P‐tau between subjects with AD and the control groups in Data sets 1, 2, and 3 (P < .05). The percentage of APOE ε4 was also increased in patients with VaD in Dataset 3 (P < .05). Levels of Aβ42 and T‐tau in VaD, PDD, FTD, and DLB were slightly decreased or increased compared to controls (P < .05). The body mass index (BMI), fasting blood glucose, HbA1c, lipids, homocysteine (HCY), folic acid, and vitamin B12 levels; and liver and renal function were not significantly different among groups (Table S1, P > .05).
TABLE 1.
Characteristics of participants in data sets 1, 2, and 3
| A. Characteristics of participants in Data set 1 | |||
|---|---|---|---|
| Characteristic |
Total Sample (n = 55) |
Controls (n = 27) |
AD (n = 28) |
| Age, mean (SD) | 67.7 (5.8) | 67.3 (5.3) | 68.2 (6.3) |
| Education year, mean (SD), | 8.4 (2.6) | 8.6 (2.7) | 8.3 (2.5) |
| Women, no. (%) | 28 (59.1) | 14 (51.9) | 14 (50.0) |
| APOE ε4 positive (%) | 16 (29.1) | 5 (18.5) | 11 (39.3)* |
| MMSE score (SD) | 25.1 (4.6) | 29.2 (0.6) | 22.2 (3.2)* |
| Aβ42 (pg/ mL) | 545.8 (220.9) | 734.6 (144.5) | 363.7 (86.3)* |
| T‐tau (pg/ mL) | 501.5 (221.7) | 331.7 (95.9) | 665.3 (181.0)* |
| P‐tau (pg/ mL) | 88.8 (50.9) | 55.4 (23.3) | 121.1 (49.6)* |
| B. Characteristics of participants in Data set 2 | |||
|---|---|---|---|
| Characteristic |
Total sample (n = 185) |
Controls (n = 92) |
AD (n = 93) |
| Age, mean (SD) | 68.2 (7.1) | 67.5 (7.5) | 68.9 (6.8) |
| Education year, mean (SD) | 8.9 (2.3) | 9.3 (2.3) | 8.7 (2.3) |
| Women, no. (%) | 91 (49.2) | 45 (48.9) | 46 (49.5) |
| APOE ε4 positive (%) | 55 (29.7) | 17 (18.5) | 38 (40.9)* |
| MMSE score (SD) | 24.9 (4.5) | 29.0 (0.6) | 20.1 (2.7)* |
| Aβ42 (pg/ mL) | 540.1 (198.4) | 703.2 (140.2) | 387.8 (80.1)* |
| T‐tau (pg/mL) | 471.3 (208.8) | 326.5 (93.4) | 614.6 (191.9)* |
| P‐tau (pg/ mL) | 89.7 (53.7) | 53.4 (24.7) | 125.5 (50.4)* |
| C. Characteristics of participants in Data set 3 | |||||||
|---|---|---|---|---|---|---|---|
| Characteristic |
Total Sample (n = 357) |
Controls (n = 76) |
AD (n = 76) |
VaD (n = 50) |
PDD (n = 52) |
bvFTD (n = 52) |
DLB (n = 51) |
| Age, mean (SD) | 67.5 (5.9) | 68.0 (5.9) | 68.6 (5.1) | 67.2 (6.4) | 68.4 (6.8) | 66.5 (5.1) | 65.9 (5.9) |
| Education year, mean (SD) | 9.3 (2.2) | 9.5 (2.2) | 8.3 (2.4) | 9.9 (2.2) | 9.1 (1.8) | 10.2 (2.1) | 9.3 (1.9) |
| Women, no. (%) | 179 (50.1) | 39 (51.3) | 39 (51.3) | 23 (46.0) | 25 (48.1) | 27 (51.9) | 26 (51.0) |
| APOE ε4 positive (%) | 89 (24.9) | 13 (18.4) | 31 (40.8) * | 13 (26.0) * | 10 (19.2) | 11 (21.2) | 10 (19.6) |
| MMSE score (SD) | 22.3 (4.1) | 29.0 (0.6) | 21.1 (3.1) * | 19.8 (2.4) * | 19.9 (2.3) * | 20.4 (1.7) * | 20.8 (2.0) * |
| Aβ42 (pg/ mL) | 573.4 (192.7) | 712.3 (163.5) | 375.4 (80.1) * | 602.8 (158.9) * | 638.8 (185.3) * | 550.5 (158.2) * | 554.7 (156.7) * |
| T‐tau (pg/ mL) | 443.2 (166.6) | 330.7 (96.9) | 620.9 (205.7) * | 395.0 (89.9) * | 375.6 (88.6) * | 467.4 (112.4) * | 437.1 (127.7) * |
| P‐tau (pg/ mL) | 65.4 (39.4) | 47.0 (12.3) | 118.0 (54.5) * | 49.6 (10.1) | 47.5 (13.3) | 49.9 (12.3) | 48.3 (11.9) |
Note: The values of age, estimated year prior to onset and MMSE are shown as mean (SD).
Abbreviations: AD, Alzheimer's disease; VaD, vascular dementia; PDD, Parkinson disease dementia; bvFTD, behavioral variant frontotemporal dementia; DLB, dementia with Lewy body; APOE ε4, apolipoprotein ε4; MMSE, Mini‐Mental State Examination. PSEN, presenilin; SD, standard deviation
P < .05 compared with controls.
3.2. Differential analysis of metabolites
A differential analysis of metabolites was performed in Data set 1. The quality control experiments revealed that the metabolite data obtained in the project were of excellent quality and satisfied the technical requirements for further statistical analysis (Figure S2). The results revealed 847 metabolites in the blood of subjects with AD and controls, which could be classified into 23 categories (Figure S3A). Student t‐test with FDR‐controlling procedures revealed 77 downregulated and 7 upregulated metabolites in the AD group (Figure 1 and Figure S3B). The top 10 downregulated and 7 upregulated metabolites (downregulated: hexanoylcarnitine AcCa (6:0), 4‐decenoylcarnitine AcCa (10:1), propionylcarnitine AcCa (3:0), tetradecadiencarnitine AcCa (14:2), piperine, decanoylcarnitine AcCa (10:0), octanoylcarnitine AcCa (8:0), paraxanthine, L‐acetylcarnitine, and serotonin; 7 upregulated: glycerophosphocholine, aspartic acid, X236, LysoPI (18:0/0:0), LysoPI (0:0/18:0), hydroxypalmitic acid, and choline) were further analyzed in Data set 2 to establish a diagnostic panel of AD. The chemical classes of differential metabolites included 21 categories, such as acylcarnitine, lysophospholipid (LPC), triglycerides, amino acids, lysophosphatidylethanolamine (LPE), fatty acid, ceramide, phosphatidylethanolamine (PE), choline, lysophosphatidylinositol (LPI), vitamin, cholesterol ester, lysophosphatidylserine (LPS), peptide, phosphatidylglycerol (PG), phosphatidylinositol (PI), polyamine, phosphatidylserine (PS), and sphingosine (Figure S3C, Table S2). Using the method of gene set enrichment analysis (GSEA), metabolite set enrichment analysis was performed through quantitative enrichment analysis in different databases. Metabolic pathway analysis was also conducted using the over‐representation method. Our data revealed that differential metabolites were enriched in phospholipid biosynthesis, folate metabolism, fatty acid degradation, and taurine and hypotaurine metabolism (Figure S4A and S4B, Table S3 and S4). Location‐based metabolite set enrichment analysis revealed that the differential metabolites were enriched in the brain, neurons, and nerve cells (Figure S4C, Table S5). Pathway analysis revealed that differential metabolites were enriched in the pathways of nicotinate, nicotinamide, glycerophospholipid, alanine, aspartate, and glutamate metabolism (Figure S4D, Table S6).
FIGURE 1.

Heat map after hierarchical clustering of the 7 upregulated and 77 downregulated metabolites in Data set 1. Abbreviations: AD, Alzheimer's disease; NC, normal control. All P’s < .05 for each metabolite compared between AD and control
3.3. Diagnostic panel of metabolites
A relatively large sample (Data set 2) was recruited to further confirm the differential metabolites. All of the top 10 downregulated and 7 upregulated metabolites in Data set 1 were confirmed in Data set 2, which supported the significance of the differential metabolites. Logistic analysis was performed to estimate their ability to distinguish subjects with AD from controls. The top 10 downregulated and 7 upregulated metabolites were included as covariates with the diagnosis (AD or controls) as the dependent variable. After adjusting for age, sex, education years, and APOE ε4 status, a panel of 11 metabolites (downregulated: hexanoylcarnitine AcCa (6:0), 4‐decenoylcarnitine AcCa (10:1), tetradecadiencarnitine AcCa (14:2), piperine, decanoylcarnitine AcCa (10:0), L‐acetylcarnitine, and serotonin; upregulated: glycerophosphocholine, aspartic acid, hydroxypalmitic acid, and choline) were observed to be associated with AD (Figure 2A‐K). Age, sex, education years, and APOE ε4 status demonstrated a P‐value > .05 in the logistic model and were therefore excluded from further analysis. Analyses were performed to estimate the multicollinearity between the 11 metabolites in subjects with AD and controls. All tolerances were > 0.1, VIFs were < 10, eigenvalues were > 0, and condition indices were < 30, indicating that there was no significant multicollinearity among the 11 metabolites. Using the predictive values from the logistic analysis, the diagnostic capacity of the 11‐metabolite panel was assessed using ROC curve analysis. Our data revealed significantly high area under the curve (AUC) values (AUC = 0.97, P < .001, Figure 3A), indicating that the metabolite panel could successfully differentiate subjects with AD from controls. The diagnostic capacity of single metabolite was also assessed, revealing poor AUCs (0.63–0.73, Figure 3B) and indicating that a combination of the 11 metabolites was necessary to obtain an effective diagnosis.
FIGURE 2.
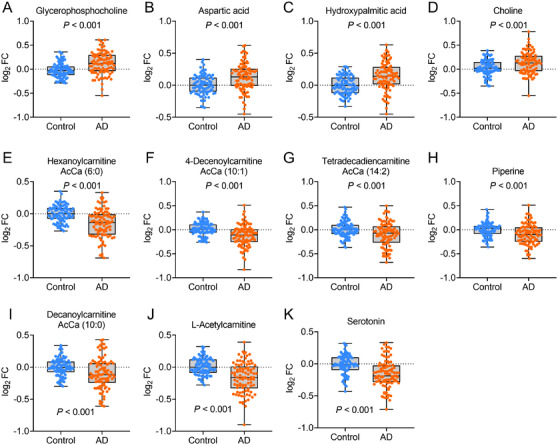
The measurements of metabolites in Data set 2. Glycerophosphocholine (A), aspartic acid (B), hydroxypalmitic acid (C), and choline (D), were increased; and hexanoylcarnitine AcCa (6:0) (E), 4‐decenoylcarnitine AcCa (10:1) (F), tetradecadiencarnitine AcCa (14:2) (G), piperine (H), decanoylcarnitine AcCa (10:0) (I), L‐acetylcarnitine (J), and serotonin (K) levels were decreased in AD patients. Abbreviations: AD, Alzheimer's disease; FC, fold‐change. All P’s < .001 for each metabolite compared between AD and control
FIGURE 3.
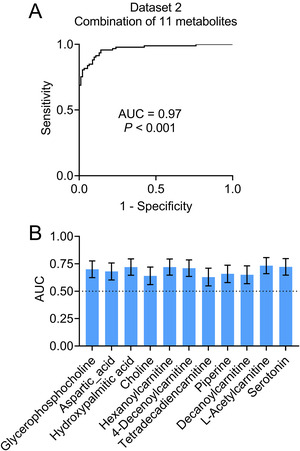
Establishment of diagnostic panel for Alzheimer's disease in Data set 2. Receiver‐operating characteristic (ROC) curve analyses were performed by combining the 11 metabolites (A). ROC analyses of each metabolite were performed (B). Abbreviations: AUC, area under the curve. P < .001 (A) or .01 (B)
In addition, to further examine the relationships between metabolite levels and cognitive decline in AD, we performed a linear correlation analysis between MMSE scores and each metabolite. Our results showed significant correlations between MMSE scores and the combination of the 11 metabolites (R2 = 0.62, P < .0001, Table S7), whereas single metabolites exhibited low correlations with MMSE (R2 = 0.05–0.14, P < .0001, Table S7).
3.4. Application of the predictive model
To assess the diagnostic capacity of the model when applied to subjects in clinical practice that may include controls, AD, and other types of dementia, such as VaD, PDD, bvFTD, and DLB, a third data set was used. Similar results to those of Data sets 1 and 2 were obtained. Levels of hexanoylcarnitine AcCa (6:0), 4‐decenoylcarnitine AcCa (10:1), tetradecadiencarnitine AcCa (14:2), piperine, decanoylcarnitine AcCa (10:0), L‐acetylcarnitine, and serotonin were decreased; whereas levels of glycerophosphocholine, aspartic acid, hydroxypalmitic acid, and choline were increased in patients with AD (P < .001; Figure 4A–K). Most of the 11 metabolites were not altered in patients diagnosed with VaD, PDD, bvFTD, and DLB (all P > .05), except for glycerophosphocholine in PDD (Figure 4A), aspartic acid in DLB (Figure 4B), choline in VaD (Figure 4D), and serotonin in bvFTD (Figure 4K). Further ROC analysis revealed very high AUCs (0.96–0.97, P < .001, Figure 5), indicating that the diagnostic panel was highly effective for distinguishing AD from controls (Figure 5A), non‐AD (combination of controls and other dementias) (Figure 5B), and other types of dementias (Figure 5C).
FIGURE 4.
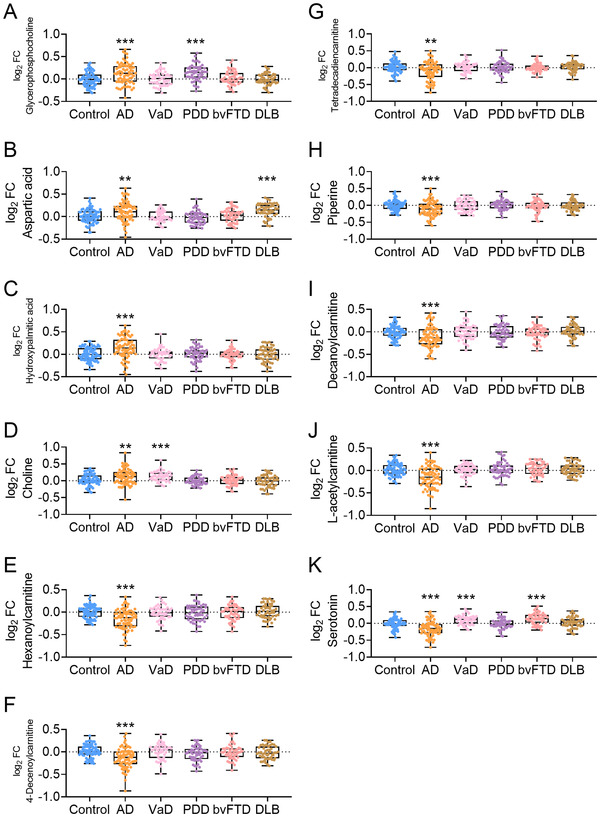
The measurements of metabolites in AD, VaD, PDD, bvFTD, and DLB in Data set 3. Glycerophosphocholine (A), aspartic acid (B), hydroxypalmitic acid (C), choline (D), hexanoylcarnitine AcCa (6:0) (E), 4‐decenoylcarnitine AcCa (10:1) (F), tetradecadiencarnitine AcCa (14:2) (G), piperine (H), decanoylcarnitine AcCa (10:0) (I), L‐acetylcarnitine (J), and serotonin (K) levels were measured. Abbreviations: AD, Alzheimer's disease; VaD, vascular dementia; PDD, Parkinson disease dementia; bvFTD, behavioral variant frontotemporal dementia; DLB, dementia with Lewy body; FC, fold‐change. **P < .01, ***P < .001
FIGURE 5.
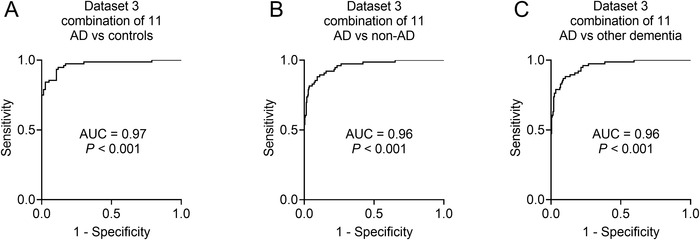
Receiver‐operating characteristic (ROC) curve analyses in Data set 3. The ROCs of AD versus controls (A), AD versus non‐AD (B), and AD versus other types of dementia (C). Non‐AD indicates a combination of controls and other types of dementia. Other types of dementia indicate a combination of VaD, Parkinson's disease dementia, behavioral variant frontotemporal dementia, and DLB. Abbreviations: AD, Alzheimer's disease; AUC, area under the curve; DLB, dementia with Lewy body; VaD, vascular dementia. All P’s < 0.001
The relationships between the combined metabolite levels and MMSE scores in participants with AD were statistically significant (R2 = 0.59, P < .0001, Table S8), whereas single metabolites exhibited low correlations with MMSE scores (R2 = 0.05–0.13, P < .0001, Table S8). There were no significant associations between single or combined metabolites in participants with VaD, PDD, bvFTD, and DLB (all P > .05).
4. DISCUSSION
In the current study, we identified a panel of metabolites that could differentiate AD from controls and other types of dementias, including VaD, PDD, bvFTD, and DLB. To the best of our knowledge, this is the first effort to screen differential metabolite signatures among these diseases. This approach is less invasive, antibody‐independent, and relatively low cost, which may facilitate its use in the screening of AD in the general elderly population.
Metabolomics is a powerful tool for detecting metabolites, which are downstream of genomic, transcriptomic, and proteomic processes. Aberrant upstream changes associated with certain diseases may eventually give rise to disease‐specific metabolic profiles. Metabolomics technology provides the opportunity to identify therapeutic targets and diagnostic biomarkers. Recent studies have reported metabolic differences in Parkinson disease, depression, and schizophrenia, 34 highlighting the promise of this approach to identify biomarkers and reveal disease pathogenesis. Notably, given the association of metabolic diseases such as diabetes and dyslipidemia with AD, and the impairments in metabolic enzymes in the glycolytic, tricarboxylic acid cycle, and oxidative phosphorylation pathways in AD, 35 identifying metabolic dysfunction is necessary to understand AD pathogenesis and identify relevant biomarkers.
Studies on sample sources from the central nervous system, such as brain tissues and CSF, demonstrated that metabolite changes are present in patients with AD. 36 In addition, metabolites in peripheral blood were reported to be altered in AD. 36 , 37 , 38 , 39 , 40 , 41 , 42 However, previous data are inconsistent. For example, several plasma phospholipids were reported to be associated with AD in some studies 36 , 37 , 38 but not others. 39 Data on sphingomyelin and docosahexaenoic acid are also inconsistent among studies. 36 , 38 , 40 , 41 , 42 Some of the relevant metabolites identified in this study were consistent with those reported previously. However, some positive metabolites in other studies were not identified in our study. For example, our data indicated that two lysophospholipids (lysoPC a C17:0 and lysoPC a C18:0) were altered in AD, which is consistent with other studies. 36 However, we did not observe any changes in phospholipids, which is consistent with some previous reports 39 but not others. 36 , 37 In addition, some positive metabolites in our study were also reported by others, such as glycerophosphocholine, 43 aspartic acid, 44 choline, 18 acetylcarnitine, 45 serotonin, and decanoylcarnitine AcCa (10:0). 45 Several of these metabolites were reported previously to be decreased in some studies but increased in others. For instance, choline was reported to be reduced in AD in some studies 46 but was reported to be increased in other studies. 12 , 18 This inconsistency may be due to the different inclusion criteria and testing methods used in the studies, which need to be further confirmed. In this study, we recruited participants in strict accordance with CSF biomarkers (P‐tau/Aβ42 and Aβ42) and screened the metabolites using UPLC‐HRMS. This method provides simultaneous global metabolic analysis and quantitative analysis of multiple analytes in full scan mode. 47 In addition, the top upregulated and downregulated metabolites were validated in three separate data sets, which supports the validity of our findings.
Biomarkers play an important role in AD diagnosis 2 and research. 22 Due to its minimal invasiveness and relatively low cost, the use of peripheral blood to diagnose AD has garnered increasing attention. The surge in research has revealed a series of promising blood biomarkers, including Aβ42, 48 neurofilament light protein, 49 P‐tau181, 50 P‐tau217, 50 exosomal Aβ42, T‐tau, P‐tau, and synaptic proteins. 20 , 29 In this study, we created a diagnostic panel of AD by detecting metabolites in the blood. Using three independent data sets, we confirmed that a combination of 11 metabolites was strongly associated with AD. We further compared metabolites between AD and VaD, PDD, bvFTD, and DLB, and confirmed that the diagnostic panel was AD specific. Although these degenerative diseases share certain clinical manifestations, such as cognitive impairment, AD is underpinned by unique pathological processes, which may underscore the AD‐specific changes in positive metabolites and enable the differentiation of AD from other dementia types.
The potential mechanisms by which the panel of 11 metabolites distinguishes AD from other dementias require further investigation. Among the 11 metabolites, hexanoylcarnitine AcCa (6:0), 4‐decenoylcarnitine AcCa (10:1), tetradecadiencarnitine AcCa (14:2), decanoylcarnitine AcCa (10:0), and L‐acetylcarnitine can be classified as acylcarnitines. Previous studies have shown that acylcarnitines are involved in mitochondrial energy metabolism, and serum levels of multiple acylcarnitines can identify the patients before the phenotype conversion to AD. 45 Serotonin and aspartic acid are amino acids. As amino acid neurotransmitters in the brain, increased aspartic acid impairs hippocampal neurons and induces synaptic plasticity and spatial memory decay, 51 , 52 whereas low serotonin levels are associated with high amounts of amyloid plaques. 53 Glycerophosphocholine and choline comprise membrane phospholipids, which breakdown during neurodegeneration. In the course of neurodegeneration, glycerophosphocholine and choline can be generated by hydrolysis of phospholipids. 43 Levels of glycerophosphocholine in the CSF of patients with AD were reportedly higher before. 43 In line with these data, our study showed that increased levels of glycerophosphocholine and choline in the blood are associated with AD. Hydroxypalmitic acid is a type of fatty acid. Multiple fatty acids are associated with AD neuropathology and cognitive function. 54 Fatty acids have roles in alpha‐secretase‐dependent amyloid precursor protein processing 55 and aging neuronal membranes. 56 However, the effects of elevated hydroxypalmitic acid levels in patients with AD need further study. Piperine is a kind of alkaloid. Studies showed that low levels of piperine in blood are associated with AD. Mechanically, piperine has been shown to perform as a β‐secretase inhibitor and acetylcholinesterase inhibitor, 57 , 58 which have potential therapeutical effects on AD. The above pathways are jointly involved in the pathogenesis of AD, so when these 11 metabolites change, the status of AD can be reflected. Therefore, the panel of 11 metabolites can be used to diagnose AD. However, the specific roles of these metabolites, their sequence in the progression of AD, and their upstream and downstream relationships are still unclear and need to be further studied.
Our study has several strengths. We used CSF Aβ42 and P‐tau /Aβ42 as diagnostic biomarkers, which ensured the accuracy of AD diagnosis. In addition, cognitively normal controls may include preclinical AD individuals with normal cognition, but their CSF Aβ42 and/or P‐tau/Aβ42 may have been abnormal. The use of biomarkers enabled the exclusion of preclinical AD from controls. Therefore, the diagnostic accuracy for participants enrolled in the current study was high. In addition, we validated differential metabolites in three separate data sets, which included patients with AD, VaD, PDD, bvFTD, and DLB, confirming that the diagnostic panel was clinically promising and AD specific.
This study was limited by its cross‐sectional design. Although we demonstrated that a panel of 11 metabolites could be used as a diagnostic biomarker for AD, a longitudinal design is more suitable for evaluating the performance of these biomarkers. Longitudinal studies on the relationship between biomarkers and cognitive decline in patients are necessary. A further limitation of this study was that it did not include patients with mild cognitive impairment that progressed to AD or stable amnestic mild cognitive impairment. Therefore, the applicability of our method to predict the progression from prodromal to probable AD is unclear. Finally, the detection of metabolites is a relatively quantitative method that cannot indicate the absolute levels of metabolites in the blood, which limits the comparison of absolute levels of metabolites between the current and future studies.
Here, to avoid the effects of other metabolic diseases, we used strict enrollment criteria for their exclusion. Therefore, all participants had normal BMI, HCY, triglyceride (TG), total cholesterol (TC), low‐density lipoprotein cholesterol (LDL‐C), high‐density lipoprotein cholesterol (HDL‐C), and glycosylated hemoglobin (HbA1c), and there was no difference in these values between the AD cases and cases of VD, FTD, or DLB. In the current data set, these metabolic parameters have no influence on the diagnostic model. Therefore, the 11‐metabolite panel did not consider metabolic disease effects. However, these conditions are often present in real clinical practice, which potentially limits the clinical application of our metabolite panel. In the future we plan to recruit patients with AD and other dementias with co‐morbid metabolic diseases and apply the model to these patients.
In addition, it has been demonstrated that many DLB mix with AD pathology. In the current study, we diagnosed DLB with clinical diagnostic criteria. In this context, DLB patients with AD pathology cannot be excluded. We may have included mixed DLB. Therefore, our diagnostic model may apply to mixed DLB, not pure DLB. However, we believe that such a sample has greater practical relevance because patients with DLB are seen in real clinical practice. The CSF biomarkers Aβ42, T‐tau, and P‐tau were not used here to diagnose DLB, but to identify controls and AD cases; DLB diagnoses were based on previously published criteria, which do not depend on CSF biomarkers. However, for consistency, in Data set 4 we present the CSF data for the AD cases and the controls, as well as for other dementias ie, VaD, PDD, bvFTD, and DLB).
In summary, the results of the present study indicate that a panel of 11 metabolites are potential blood biomarkers for AD. Specifically, the diagnostic panel can differentiate AD from other types of dementias, highlighting its potential clinical value. However, our findings require further validation in longitudinal studies.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
Longfei Jia had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Longfei Jia designed the study. Longfei Jia contributed to the acquisition, analysis, and interpretation of data. Longfei Jia performed the statistical analysis. Longfei Jia, Min Zhu and Jianwei Yang drafted the manuscript. Longfei Jia, Min Zhu, Jianwei Yang, Yana Pang, Qi Wang, Qin Wei, Ying Li, TingTing Li, Fangyu Li, Qigeng Wang, Yan Li, and Yiping Wei provided the administrative, technical, and material support. All authors critically reviewed the article and approved the final manuscript.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGMENTS
The authors thank Zeming Wu from iPhenome (Yunpukang) Biotechnology Inc. and BioMiao Biological Technology (Beijing) Company for technically supporting the experiments. This study was financially supported by Beijing Brain Initiative from Beijing Municipal Science & Technology Commission (Z201100005520016); National Natural Science Foundation of China (81870825, 82071194); and Beijing Municipal Natural Science Foundation (7202061).
Jia L, Yang J, Zhu M, et al. A metabolite panel that differentiates Alzheimer's disease from other dementia types. Alzheimer's Dement. 2022;18:1345–1356. 10.1002/alz.12484
REFERENCES
- 1. Jia L, Quan M, Fu Y, et al. Dementia in China: epidemiology, clinical management, and research advances. Lancet Neurol. 2020;19:81‐92. [DOI] [PubMed] [Google Scholar]
- 2. McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging‐Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kapasi A, DeCarli C, Schneider JA. Impact of multiple pathologies on the threshold for clinically overt dementia. Acta Neuropathol. 2017;134:171‐186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scheltens P, Blennow K, Breteler MM, et al. Alzheimer's disease. Lancet. 2016;388:505‐517. [DOI] [PubMed] [Google Scholar]
- 5. Chen Z, Zhong C. Oxidative stress in Alzheimer's disease. Neurosci Bull. 2014;30:271‐281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Calsolaro V, Edison P. Neuroinflammation in Alzheimer's disease: current evidence and future directions. Alzheimers Dement. 2016;12:719‐732. [DOI] [PubMed] [Google Scholar]
- 7. Zhu TB, Zhang Z, Luo P, Wang SS, Peng Y, Chu SF, et al. Lipid metabolism in Alzheimer's disease. Brain Res Bull. 2019;144:68‐74. [DOI] [PubMed] [Google Scholar]
- 8. Kerr JS, Adriaanse BA, Greig NH, et al. Mitophagy and Alzheimer's disease: cellular and molecular mechanisms. Trends Neurosci. 2017;40:151‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tolar M, Abushakra S, Sabbagh M. The path forward in Alzheimer's disease therapeutics: reevaluating the amyloid cascade hypothesis. Alzheimers Dement. 2020;16:1553‐1560. [DOI] [PubMed] [Google Scholar]
- 10. Wang JH, Byun J, Pennathur S. Analytical approaches to metabolomics and applications to systems biology. Semin Nephrol. 2010;30:500‐511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mapstone M, Lin F, Nalls MA. What success can teach us about failure: the plasma metabolome of older adults with superior memory and lessons for Alzheimer's disease. Neurobiol Aging. 2017;51:148‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gonzalez‐Dominguez R, Garcia A, Garcia‐Barrera T, Barbas C, Gomez‐Ariza JL. Metabolomic profiling of serum in the progression of Alzheimer's disease by capillary electrophoresis‐mass spectrometry. Electrophoresis. 2014;35:3321‐3330. [DOI] [PubMed] [Google Scholar]
- 13. de Leeuw FA, Peeters CFW, Kester MI, et al. Blood‐based metabolic signatures in Alzheimer's disease. Alzheimers Dement (AMST). 2017;8:196‐207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Olazaran J, Gil‐de‐Gomez L, Rodriguez‐Martin A, et al. A blood‐based, 7‐metabolite signature for the early diagnosis of Alzheimer's disease. J Alzheimers Dis. 2015;45:1157‐1173. [DOI] [PubMed] [Google Scholar]
- 15. Fiandaca MS, Zhong X, Cheema AK, et al. Plasma 24‐metabolite panel predicts preclinical transition to clinical stages of Alzheimer's disease. Front Neurol. 2015;6:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Figueira J, Adolfsson R, Nordin Adolfsson A, Nyberg L, Ohman A. Serum metabolite markers of dementia through quantitative NMR analysis: the importance of threonine‐linked metabolic pathways. J Alzheimers Dis. 2019;69:763‐774. [DOI] [PubMed] [Google Scholar]
- 17. Stamate D, Kim M, Proitsi P, et al. A metabolite‐based machine learning approach to diagnose Alzheimer‐type dementia in blood: results from the European Medical Information Framework for Alzheimer disease biomarker discovery cohort. Alzheimers Dement (N Y). 2019;5:933‐938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pena‐Bautista C, Baquero M, Vento M, Chafer‐Pericas C. Omics‐based biomarkers for the early Alzheimer disease diagnosis and reliable therapeutic targets development. Curr Neuropharmacol. 2019;17:630‐647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Casanova R, Varma S, Simpson B, et al. Blood metabolite markers of preclinical Alzheimer's disease in two longitudinally followed cohorts of older individuals. Alzheimers Dement. 2016;12:815‐822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jia L, Qiu Q, Zhang H, et al. Concordance between the assessment of Abeta42, T‐tau, and P‐T181‐tau in peripheral blood neuronal‐derived exosomes and cerebrospinal fluid. Alzheimers Dement. 2019;15:1071‐1080. [DOI] [PubMed] [Google Scholar]
- 21. Seeburger JL, Holder DJ, Combrinck M, et al. Cerebrospinal fluid biomarkers distinguish postmortem‐confirmed Alzheimer's disease from other dementias and healthy controls in the OPTIMA cohort. J Alzheimers Dis. 2015;44:525‐539. [DOI] [PubMed] [Google Scholar]
- 22. Jack CR Jr, Bennett DA, Blennow K, et al. NIA‐AA Research Framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14:535‐562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Humpel C. Identifying and validating biomarkers for Alzheimer's disease. Trends Biotechnol. 2011;29:26‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roman GC, Tatemichi TK, Erkinjuntti T, et al. Vascular dementia: diagnostic criteria for research studies. Report of the NINDS‐AIREN International Workshop. Neurology. 1993;43:250‐260. [DOI] [PubMed] [Google Scholar]
- 25. Goetz CG, Emre M, Dubois B. Parkinson's disease dementia: definitions, guidelines, and research perspectives in diagnosis. Ann Neurol. 2008;64(Suppl 2):S81‐92. [DOI] [PubMed] [Google Scholar]
- 26. Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, Neuhaus J, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134:2456‐2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McKeith IG, Boeve BF, Dickson DW, et al. Diagnosis and management of dementia with Lewy bodies: fourth consensus report of the DLB consortium. Neurology. 2017;89:88‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Teunissen CE, Petzold A, Bennett JL, et al. A consensus protocol for the standardization of cerebrospinal fluid collection and biobanking. Neurology. 2009;73:1914‐1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jia L, Zhu M, Kong C, et al. Blood neuro‐exosomal synaptic proteins predict Alzheimer's disease at the asymptomatic stage. Alzheimers Dement. 2020;17:49‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl‐tert‐butyl ether for high‐throughput lipidomics. J Lipid Res. 2008;49:1137‐1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ye G, Ding D, Gao H, et al. Comprehensive metabolic responses of HepG2 cells to fine particulate matter exposure: insights from an untargeted metabolomics. Sci Total Environ. 2019;691:874‐884. [DOI] [PubMed] [Google Scholar]
- 32. Wishart DS, Feunang YD, Marcu A, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608‐D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chong J, Soufan O, Li C, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486‐W94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu XH, Huang Y, Wang G, Chen SD. Metabolomics: a novel approach to identify potential diagnostic biomarkers and pathogenesis in Alzheimer's disease. Neurosci Bull. 2012;28:641‐648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Murray IV, Proza JF, Sohrabji F, Lawler JM. Vascular and metabolic dysfunction in Alzheimer's disease: a review. Exp Biol Med (Maywood). 2011;236:772‐782. [DOI] [PubMed] [Google Scholar]
- 36. Varma VR, Oommen AM, Varma S, et al. Brain and blood metabolite signatures of pathology and progression in Alzheimer disease: a targeted metabolomics study. PLoS Med. 2018;15:e1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mapstone M, Cheema AK, Fiandaca MS, et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med. 2014;20:415‐418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li D, Misialek JR, Boerwinkle E, et al. Plasma phospholipids and prevalence of mild cognitive impairment and/or dementia in the ARIC neurocognitive study (ARIC‐NCS). Alzheimers Dement (AMST). 2016;3:73‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tynkkynen J, Chouraki V, van der Lee SJ, et al. Association of branched‐chain amino acids and other circulating metabolites with risk of incident dementia and Alzheimer's disease: a prospective study in eight cohorts. Alzheimers Dement. 2018;14:723‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tan ZS, Harris WS, Beiser AS, et al. Red blood cell omega‐3 fatty acid levels and markers of accelerated brain aging. Neurology. 2012;78:658‐664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schaefer EJ, Bongard V, Beiser AS, et al. Plasma phosphatidylcholine docosahexaenoic acid content and risk of dementia and Alzheimer disease: the framingham heart study. Arch Neurol. 2006;63:1545‐1550. [DOI] [PubMed] [Google Scholar]
- 42. Mielke MM, Haughey NJ, Bandaru VV, et al. Plasma sphingomyelins are associated with cognitive progression in Alzheimer's disease. J Alzheimers Dis. 2011;27:259‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Walter A, Korth U, Hilgert M, et al. Glycerophosphocholine is elevated in cerebrospinal fluid of Alzheimer patients. Neurobiol Aging. 2004;25:1299‐1303. [DOI] [PubMed] [Google Scholar]
- 44. Kim M, Snowden S, Suvitaival T, et al. Primary fatty amides in plasma associated with brain amyloid burden, hippocampal volume, and memory in the European Medical Information Framework for Alzheimer's disease biomarker discovery cohort. Alzheimers Dement. 2019;15:817‐827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cristofano A, Sapere N, La Marca G, et al. Serum levels of acyl‐carnitines along the continuum from normal to Alzheimer's dementia. PLoS One. 2016;11:e0155694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Wijk N, Slot RER, Duits FH, et al. Nutrients required for phospholipid synthesis are lower in blood and cerebrospinal fluid in mild cognitive impairment and Alzheimer's disease dementia. Alzheimers Dement (AMST). 2017;8:139‐146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rochat B. From targeted quantification to untargeted metabolomics: why LC‐high‐resolution‐MS will become a key instrument in clinical labs. Trac‐Trend Anal Chem. 2016;84:151‐164. [Google Scholar]
- 48. Nakamura A, Kaneko N, Villemagne VL, et al. High performance plasma amyloid‐beta biomarkers for Alzheimer's disease. Nature. 2018;554:249‐254. [DOI] [PubMed] [Google Scholar]
- 49. Elahi FM, Casaletto KB, La Joie R, et al. Plasma biomarkers of astrocytic and neuronal dysfunction in early‐ and late‐onset Alzheimer's disease. Alzheimers Dement. 2020;16:681‐695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Karikari TK, Emersic A, Vrillon A, et al. Head‐to‐head comparison of clinical performance of CSF phospho‐tau T181 and T217 biomarkers for Alzheimer's disease diagnosis. Alzheimers Dement. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu R, Huang D, Tong J, Liao Q, Hu Z, Ouyang W. Aspartic acid in the hippocampus: a biomarker for postoperative cognitive dysfunction. Neural Regen Res. 2014;9:143‐152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Errico F, Nistico R, Napolitano F, et al. Persistent increase of D‐aspartate in D‐aspartate oxidase mutant mice induces a precocious hippocampal age‐dependent synaptic plasticity and spatial memory decay. Neurobiol Aging. 2011;32:2061‐2074. [DOI] [PubMed] [Google Scholar]
- 53. Larsson SC, Markus HS. Branched‐chain amino acids and Alzheimer's disease: a Mendelian randomization analysis. Sci Rep. 2017;7:13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Snowden SG, Ebshiana AA, Hye A, An Y, Pletnikova O, O'Brien R, et al. Association between fatty acid metabolism in the brain and Alzheimer disease neuropathology and cognitive performance: a nontargeted metabolomic study. PLoS Med. 2017;14:e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang X, Sheng W, Sun GY, Lee JC. Effects of fatty acid unsaturation numbers on membrane fluidity and alpha‐secretase‐dependent amyloid precursor protein processing. Neurochem Int. 2011;58:321‐329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yehuda S, Rabinovitz S, Carasso RL, Mostofsky DI. The role of polyunsaturated fatty acids in restoring the aging neuronal membrane. Neurobiol Aging. 2002;23:843‐853. [DOI] [PubMed] [Google Scholar]
- 57. Abdul Manap AS, Wei Tan AC, Leong WH, et al. Synergistic effects of curcumin and piperine as potent acetylcholine and amyloidogenic inhibitors with significant neuroprotective activity in SH‐SY5Y cells via computational molecular modeling and in vitro assay. Front Aging Neurosci. 2019;11:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Murata K, Matsumura S, Yoshioka Y, Ueno Y, Matsuda H. Screening of beta‐secretase and acetylcholinesterase inhibitors from plant resources. J Nat Med. 2015;69:123‐129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information


