Abstract
Gene arrays containing all currently known open reading frames of Bacillus subtilis were used to examine the general stress response of Bacillus. By proteomics, transcriptional analysis, transposon mutagenesis, and consensus promoter-based screening, 75 genes had previously been described as ςB-dependent general stress genes. The present gene array-based analysis confirmed 62 of these already known general stress genes and detected 63 additional genes subject to control by the stress sigma factor ςB. At least 24 of these 125 ςB-dependent genes seemed to be subject to a second, ςB-independent stress induction mechanism. Therefore, this transcriptional profiling revealed almost four times as many regulon members as the proteomic approach, but failure of confirmation of all known members of the ςB regulon indicates that even this approach has not yet elucidated the entire regulon. Most of the ςB-dependent general stress proteins are probably located in the cytoplasm, but 25 contain at least one membrane-spanning domain, and at least 6 proteins appear to be secreted. The functions of most of the newly described genes are still unknown. However, their classification as ςB-dependent stress genes argues that their products most likely perform functions in stress management and help to provide the nongrowing cell with multiple stress resistance. A comprehensive screening program analyzing the multiple stress resistance of mutants with mutations in single stress genes is in progress. The first results of this program, showing the diminished salt resistance of yjbC and yjbD mutants compared to that of the wild type, are presented. Only a few new ςB-dependent proteins with already known functions were found, among them SodA, encoding a superoxide dismutase. In addition to analysis of the ςB-dependent general stress regulon, a comprehensive list of genes induced by heat, salt, or ethanol stress in a ςB-independent manner is presented. Perhaps the most interesting of the ςB-independent stress phenomena was the induction of the extracytoplasmic function sigma factor ςW and its entire regulon by salt shock.
Almost 15 years ago we began to analyze the response of Bacillus subtilis cells to stress and starvation because these unfavorable conditions are the rule in natural ecosystems and adaptation to stress and starvation is crucial for survival in nature. We used the highly sensitive two-dimensional gel electrophoresis technique to visualize global changes in the gene expression pattern (24, 25, 38). These studies revealed a large group of stress proteins that seemed to be induced together by physical stress such as heat, salt, ethanol, or acid stress, as well as by glucose, oxygen, or phosphate starvation. This complex induction profile encouraged us to suggest that these proteins may have a rather nonspecific, but nevertheless very essential, protective function in response to stress or starvation, regardless of the specific stress stimulus. Therefore, the proteins were called nonspecific or general stress proteins (24, 25, 38).
Subsequently, stress induction of this protein group was shown to be mediated by the alternative sigma factor ςB, the general stress sigma factor of gram-positive bacteria. W. G. Haldenwang and R. Losick discovered ςB more than 20 years ago (22), but its role and physiological function remained matters of speculation for more than a decade. In the early 1990s the laboratories of W. G. Haldenwang and C. W. Price independently discovered that the sigB operon was induced by the same stimuli as the general stress proteins, namely, either heat, ethanol, or salt stress or entry into the stationary-growth phase, and that this induction was achieved by ςB itself (6, 7, 9, 11). These findings strongly suggested that the genes encoding the general stress proteins belong to the ςB regulon. Identification of numerous general stress proteins by N-terminal sequencing or matrix-assisted laser desorption ionization–time-of-flight mass spectrometry and subsequent detailed analysis of gene regulation proved that ςB indeed controls induction of the general stress genes. By use of transposon mutagenesis C. W. Price and coworkers investigated the effects of ςB on transcription and identified eight ςB-dependent genes (for reviews see references 26 and 37).
Finally, analysis of the B. subtilis genome for ςB-dependent promoters was used to identify additional members of the ςB regulon. This computer-aided identification of new general stress genes became feasible because of the highly conserved and distinct consensus sequence of ςB-dependent promoters. Screening of the potential target genes by oligonucleotide hybridization revealed more than 20 new genes that are probably under ςB control (36). The three approaches described above and additional genetic and transcriptional studies have led thus far to the identification of 75 ςB-dependent general stress genes. The number of genes identified by each of these approaches is given in Table 3.
TABLE 3.
Analysis of the frequency of retrieving known ςB regulon members on gene arrays
| Identification strategy | No. known | No. verified by DNA array analysisa |
|---|---|---|
| Proteome analysis | 34 | 31 (+3) |
| Promoter- and oligonucleotide-based screening | 24 | 12 (+2) |
| Transposon mutagenesis | 8 | 6 |
| Other | 19 | 10 (+6) |
| Total | 75 | 51 (+11) |
Number in parentheses is the number of genes subject to additional ςB-independent stress induction.
Many of the general stress genes display basal level transcription from vegetative ςA-dependent promoters. However, activation of ςB activity following metabolic or environmental stress dramatically increases the transcription of the general stress genes. As a result of this massive induction of the regulon, the fraction of total translational capacity utilized for production of general stress proteins rises from approximately 1% in growing cells to 20% or even more in starved or stressed bacteria (8). During exponential growth ςB is kept in an inactive complex by binding to its anti-sigma factor, RsbW (5). Activation of ςB requires the dephosphorylation of an antagonist protein, RsbV, which then forms a complex with RsbW and releases ςB from its inhibition (1, 17). During exponential growth RsbV is phosphorylated and inactivated by RsbW (17, 52), but after the imposition of stress or starvation two specific PP2C type phosphatases, RsbU and RsbP, can shift the equilibrium from RsbV∼P to RsbV and consequently trigger stress gene activation (48, 55).
Comparative phenotypic studies of sigB mutants and wild-type bacteria have meanwhile proven that high-level expression of the general stress regulon provides stressed or starved cells with multiple, nonspecific, prospective stress resistance in anticipation of “future stress” (18, 19, 51). This protective function is particularly important for cells that are no longer able to grow (51). Therefore, the general stress response might be an essential alternative for all resting Bacillus cells that do not sporulate efficiently either because the cell density is too low (21) or because stress conditions (e.g., osmotic stress, oxygen limitation) do not allow sporulation (28, 39).
Analysis of the precise function of the general stress regulon in stress management will undoubtedly profit from a comprehensive description of all ςB-dependent genes. Therefore, we decided to use DNA macroarrays for transcriptional profiling of stress adaptation in B. subtilis to detect the still missing members of the ςB regulon. By this approach more than 60 new ςB-dependent genes were discovered. The screening was also utilized for the characterization of ςB-independent stress gene induction. Interestingly, these studies showed salt shock induction of the regulon of the extracytoplasmic function (ECF) sigma factor ςW.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The following B. subtilis strains were used: 168 (trpC2), BSA46 (trpC2 SPβ ctc::lacZ), ML6 (trpC2 sigB::ΔHindIII-EcoRV::cat), BSA272 (trpC2 sigB::ΔHindIII-EcoRV::cat), and BSA386 (trpC2 rsbX::spc sup20a SPβ ctc::lacZ; obtained from W. G. Haldenwang). The yjbC (BFA2841) and yjbD (BFA2842) mutants were constructed by inserting the nonreplicative plasmid pMUTIN4, carrying fragments of the yjbC and yjbD structural genes, respectively, lacking the ribosome binding site and the first N-terminal codons, into the corresponding genes via a Campbell type single-crossover event (41). Strains were grown with vigorous agitation at 37°C in a synthetic medium with 0.2% (wt/vol) glucose as the carbon source (strains 168, ML6, BFA2841, and BFA2842) (44) or in Luria-Bertani medium (strains BSA46, BSA272, and BSA386). Ethanol or osmotic stress was imposed by adding ethanol or NaCl to exponentially growing cells to a final concentration of 4% (vol/vol or wt/vol, respectively). For heat stress, the temperature was shifted from 37 to 48°C.
Survival of growth-preventing salt stress was examined by transferring exponentially growing cultures into minimal medium containing an initial NaCl concentration of 4% (wt/vol). After a preadaptation period of 30 min, this was raised to a final NaCl concentration of 10% (wt/vol).
Cell lysis and RNA isolation.
RNA was isolated either according to the acid phenol method of Majumdar et al. (34), with the modifications previously described (49), or after mechanical disruption of the cells as described by Hauser et al. (23). In the latter case sedimented cells were resuspended in 200 μl of growth medium and immediately frozen in a small Teflon vessel of a grinding mill (B. Braun Biotec Int., Melsungen, Germany) in liquid nitrogen. After addition of a tungsten carbide bead, the frozen drops were mechanically broken for 2 min at top speed. The frozen powder was instantly taken up in guanidine thiocyanate buffer (4 M guanidine thiocyanate, 25 mM sodium acetate [pH 5.2], 0.5% [wt/vol] N-lauroylsarcosinate) and was extracted three times with 1 volume of acid phenol-chloroform-isoamyl alcohol solution (25:24:1, vol/vol/vol), and twice with chloroform-isoamyl alcohol (24:1, vol/vol). After ethanol precipitation and washing with 70% ethanol, the RNA pellet was dried and dissolved in diethyl pyrocarbonate-treated distilled water.
Preparation of labeled cDNA, array hybridization, and DNA macroarray regeneration.
Prior to cDNA synthesis, the quality of the RNA was routinely verified by standard Northern blot analysis with digoxigenin-labeled antisense RNA probes specific for known general stress genes. For cDNA synthesis, 2 μg of total RNA was mixed with 4 μl of a commercially available primer mix (Sigma-Genosys Ltd.) and 3 μl of 10× hybridization buffer (100 mM Tris [pH 7.9], 10 mM EDTA, 2.5 M KCl) in a total volume of 30 μl. The primer mix consisted of 4,107 specific oligonucleotide primers complementary to the 3′ ends of all B. subtilis mRNAs (Sigma-Genosys Ltd.). The sample was heated to 95°C for 10 min and subsequently cooled to 42°C for primer annealing. Reverse transcription was performed in a total volume of 60 μl with SuperScript II reverse transcriptase and [α-33P]dCTP in the appropriate buffer for 1 h (Life Technologies, GmbH, Karlsruhe, Germany). After addition of 2 μl of 1% sodium dodecyl sulfate (SDS), 2 μl of 0.5 M EDTA (pH 8.0), and 6 μl of 3 M NaOH, the remaining RNA was hydrolyzed by incubation at 65°C for 30 min and at room temperature for 15 min. Prior to ethanol precipitation, the cDNA solution was neutralized with 20 μl of 1 M Tris (pH 8.0) and 6 μl of 2 N HCl. After a wash with 70% ethanol the pellet was carefully dried and resolved in 100 μl of distilled water. Labeling efficiency was determined with a liquid scintillation counter. This study was performed with Panorama B. subtilis gene arrays from Sigma-Genosys Ltd., which carry duplicate spots of PCR products representing 4,107 currently known B. subtilis genes. cDNA denaturation, probe hybridization, and washing of filters were performed as described by Hauser et al. (23).
The arrays were exposed for 2 and 4 days to storage phosphor screens (Molecular Dynamics, Sunnyvale, Calif.) and scanned with a Storm 840/860 PhosphorImager (Molecular Dynamics) at a resolution of 50 μm and a color depth of 16 bits.
Bound cDNA was stripped off the membranes by a short (1-min) washing step with 250 ml of boiling buffer (5 mM sodium phosphate [pH 7.5], 0.1% SDS), incubation in 250 ml of fresh buffer at 95°C for 20 min, and two additional wash steps with fresh boiling buffer.
Data analysis.
Hybridization signals were quantified with ArrayVision software (Imaging Research Inc.) after direct import of the PhosphorImager files. After subtraction of the background, which was defined as the median of signals surrounding the entire spot fields, the overall spot normalization function of ArrayVision was used to calculate the normalized intensity values of individual spots, thus facilitating the comparison of results from different hybridizations and filters. Briefly, this procedure involved two steps: (i) calculation of the intensity of an average spot by dividing the sum of the intensities of all PCR product specific signals on the array by the total number of spots and (ii) dividing the intensity of the individual spot by the intensity of this average spot.
For each growth condition mRNA was prepared from two independent cultivations and then used for independent cDNA synthesis and DNA array hybridizations. For exponentially growing bacteria and ethanol treatment, three entirely independent replicates were processed. In total, 32 array hybridizations were performed. For each gene the average of the normalized intensity values from all the replicate experiments was calculated. To avoid extreme intensity ratios for genes close to or below the detection limit, the average normalized intensity for these low values was arbitrarily set to a value corresponding to a signal-to-noise ratio of 2. These average values were then used to calculate expression ratios for the following comparisons: (i) stressed (ethanol, salt, or heat shock applied for 10 min) versus exponentially growing wild-type strains, (ii) stressed (ethanol, salt, or heat shock applied for 10 min) versus exponentially growing sigB mutant cells, (iii) stressed wild-type cells versus stressed sigB mutant cells (both treated with ethanol, salt, or heat shock for 10 min), and (iv) the rsbX sup20a hyperexpression mutant versus the sigB mutant 60 min after ethanol addition. rsbX mutants lack an essential negative regulator of the ςB regulatory cascade, fail to restrict the ςB response, and therefore display artificially high and extended ςB activity (50). In the suppressor mutant (rsbX sup20a) artificially high ςB activity is compatible with growth (W. G. Haldenwang, unpublished data).
Experiments involving ethanol, salt, or heat stress (10 min) were performed with the isogenic B. subtilis strain pair (168 and ML6). In order to substantiate the results and to minimize the number of false positives, experiments involving treatment with ethanol for 10 min were also performed with an independent strain pair, BSA46 and BSA272, and Panorama B. subtilis gene arrays (Sigma-Genosys Ltd.) from a different batch. Due to the different strain pair and the different array batches, hybridizations involving this strain pair did not produce exactly the same induction ratios but confirmed the candidates identified in this study with the strain pair 168 and ML6.
The ratios of the expression levels obtained from the averaged normalized intensities of all replicate experiments were imported into GeneSpring 3.2.12 software (Silicon Genetics, Redwood City, Calif.) and used to find additional ςB-dependent and ςB-independent stress genes. Seventy-five genes have previously been assigned to the ςB regulon. Twelve of these (clpC, ctsR, opuE, sms, trxA, yacH, yacI, yacK, ytxG, ytxH, ytxJ, and yvyD) exhibit an additional stress induction mechanism, but the remaining 63 (aldY, bmr, bmrR, bmrU, bofC, clpP, csbA, csbB, csbD, csbX, ctc, dps, gsiB, gspA, gtaB, katB, katX, nadE, rsbV, rsbW, rsbX, sigB, yacL, ycdF, ydaD, ydaE, ydaG, ydaP, ydaS, ydaT, ydbD, ydbP, ydhK, yfhK, yfkM, yflA, yflT, yhdF, yhdN, yhxD, yjbC, yjgB, yjgC, ykgA, ykzA, yocK, yotK, yoxA, yoxC, ypuB, yqhA, yqhQ, yqxL, yrvD, ysnF, ytkL, yugU, yvrE, yxaB, yxbG, yxcC, yxkO, and yycD) should display clear ςB-dependent induction in response to all three stresses. These genes were also used to obtain a consensus sequence of ςB-dependent promoters.
The DNA sequences preceding genes with potential ςB-dependent stress induction were subsequently inspected for the occurrence of the characteristic −35 and −10 boxes recognized by the RNA holoenzyme carrying ςB with the MotivFinder program (Decodon GmbH, Greifswald, Germany).
World Wide Web access.
The complete data sets for all the growth conditions investigated are available online (http:///www.uni-marburg.de/mpi/voelker/functional -genomics).
RESULTS
Identification of ςB-dependent general stress genes.
DNA macroarrays that contain all currently known genomic open reading frames of B. subtilis were used to record the comparative transcriptional profiles of exponentially growing cells and cultures exposed to mild ethanol, heat, or salt stress. Bacteria were exposed to the stresses for 10 min in order to achieve maximal transcriptional induction.
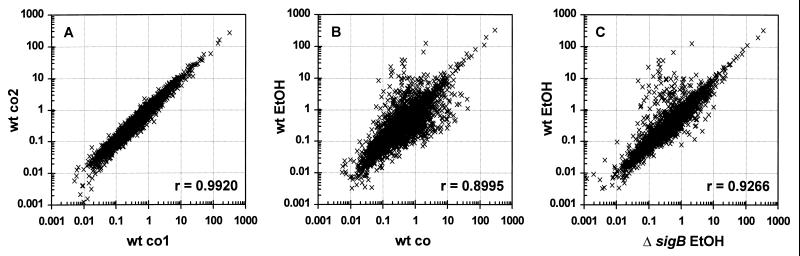
Prior to quantification of the array data, the reproducibility of the array experiments was estimated by comparing the normalized spot intensities in scatter diagrams (Fig. 1). Array data from hybridizations of independent samples from the same cultivation condition always yielded high Pearson correlation coefficients (see Fig. 1A for an example; r = 0.992). As expected, Pearson correlation coefficients calculated for comparison of untreated and stressed samples of the wild type or for comparison of ethanol-stressed samples from a wild-type strain and from the corresponding sigB mutant were lower (r = 0.8995 and r = 0.9266, respectively), and the scatter diagrams displayed genes induced and genes repressed by ethanol stress (Fig. 1B and C).
FIG. 1.
Scatter diagrams of normalized spot intensities. (A) Spot intensities of two array hybridizations with two different unstressed samples from the wild-type strain 168 (wt co1 versus wt co2). (B) Spot intensities of array hybridizations from a nonstressed probe of strain 168 (wt co) and an ethanol-stressed sample of the same strain (wt EtOH). (C) Comparison of spot intensities of filters hybridized with probes of wild-type strain 168 (wt EtOH) and its isogenic sigB mutant ML6 (ΔsigB EtOH), both treated with ethanol. For the presentation, spot intensities of the 4,107 genes have been normalized and the duplicate spots on the filter have been averaged as described in Materials and Methods. r, Pearson correlation coefficient.
In this study a gene was considered to require ςB for stress induction when it complied with all of the following three criteria. (i) Expression of the gene had to be induced more than twofold by at least two of the three stresses in the wild type. (ii) The ratio of induction had to be at least 2 in three of the four mutant comparisons employed, i.e., wild type versus sigB mutant after heat, salt, or ethanol stress and expression in the rsbX sup20a mutant versus expression in the sigB mutant 60 min after exposure to ethanol stress. (iii) A ςB-dependent promoter had to be located in front of the gene or the transcriptional unit to which the gene belonged. These criteria clearly differentiated between specific and general ςB-dependent stress genes.
Table 1 lists the 101 genes that met these selection criteria. Fifty-one genes belong to the group of 75 ςB-dependent genes already described in the literature, and 50 genes correspond to potential new members of the ςB regulon.
TABLE 1.
Summary of ςB-dependent general stress genes
| Genea | Function or nearest homolog (E value)b | Regulatory region or potential promoter sequencec | Operon structure
|
Induction ratiof
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Predictedd | Validatede | EtOH, 10 min/co | EtOH, wt/sigB | Heat, wt/sigB | Salt, wt/sigB | EtOH, rsbX/sigB | |||
| yaaH | Cortical fragment-lytic enzyme SleL, Bacillus cereus (e-120) | Potential yaaIH operon | 2nd | 4.3 | 3.3 | 1.9 | 3.0 | 7.4 | |
| yaaI | Isochorismatase (EntB) homolog, A. fulgidus (1e-21) | CAAAGTTTTTTCATTGCCTAAAAAGGCTACATAT-N33-TTG | 1st | 16 | 15 | 2.1 | 3.5 | 1.9 | |
| ctc | Probable ribosomal protein L25, Pseudomonas aeruginosa (1e-18) | CGAGGTTTAAATCCTTATCGTTATGGGTATTGTTTGTAAT(A)GG | 1st | + | 8.2 | 27 | 9.1 | 8.4 | 21 |
| spoVC | Peptidyl-tRNA hydrolase (Pth), stage V sporulation protein C (1e-106) | ctc spoVC operon | 2nd | + | 8.4 | 8.4 | 2.1 | 3.2 | 2.7 |
| ybdT | Fatty acid beta-hydroxylating cytochrome P450 (CypC) (<1e-180) | GTCGGAAGATTAACGTGAAAATAGAGGGTAAAAAG-N17-ATG | m | 6.5 | 6.3 | 4.2 | 4.1 | 39 | |
| ybyB | Unknown conserved protein BH2666, B. halodurans (7e-08) | ACAGGTTTAGCAATTT.CCAAAACGGGAATGATA-N38-ATG | m | 40 | 120 | 53 | 1.7 | 273 | |
| ycbK | Probable integral membrane protein CJ0860, Campylobacter jejuni (2e-07) | AAAGGTGTTATTCTGACTGCTCAAGGATATACGC-N55-ATG | m | 8.0 | 6.3 | 2.5 | 6.3 | 1.2 | |
| ycbP | Conserved hypothetical protein YndM, B. subtilis (6e-11) | TAAGGTTTAACTTTTT.ACATTTGAGGAATTATA-N34-ATG | m | 20 | 31 | 14 | 23 | 12 | |
| ycdF | Glucose 1-dehydrogenase II, B. megaterium (2e-66) | TGCTGTTTTCAACTCGGAAAAACAGGGTATTTTC-N35-ATG | 1st | 16 | 15 | 6.3 | 21 | 191 | |
| ycdG | Oligo-1,6-glucosidase (EC 3.2.1.10), B. thermoglucosidasius (<1e-180) | Potential ycdFG operon | 2nd | 7.4 | 5.2 | 2.1 | 3.2 | 22 | |
| nadE | NH3-dependent NAD+ synthetase (EC 6.3.5.1) | TCATGATTCATTTTCATTGATTTAGGGAAATGATCAGT(A)ATA | m | + | 3.2 | 2.3 | 1.4 | 8.7 | 3.1 |
| ydaD | Hypothetical oxidoreductase YhdF, B. subtilis (e-113) | CCATGTTTATCCAAAGAGTGTGTGAGGTACACAACAAA(TA)GA | 1st | + | 132 | 110 | 16 | 59 | 168 |
| ydaE | No similarity | ydaDE operon | 2nd | + | 66 | 52 | 9.9 | 36 | 7.3 |
| ydaG | Probable general stress protein 26, D. radiodurans (7e-04) | TCTTGTTTAAATCTTCCCCGGATGTGGAAAAGTAACAG(C)GGA | m | + | 6.4 | 9.0 | 3.4 | 15 | 59 |
| ydaP | Pyruvate dehydrogenase-related protein, T. acidophilum (e-103) | CCGGGTTTTAAAGCCTTTCTCCTGTGGTATTGAAAAAA(GG)AA | m | + | 22 | 30 | 9.8 | 18 | 155 |
| ydaS | Conserved hypothetical membrane protein YwzA, B. subtilis (5e-06) | AAGTGTTTCGAAATGATCAGGAGCGGTTATTGAT-N29-ATG | m | 5.3 | 3.2 | 1.4 | 2.7 | 6.7 | |
| ydaT | No similarity | ATGCGTTTTTATTTTT.CACCTGCGGGTACCATT-N27-ATG | m | 3.9 | 3.6 | 2.4 | 4.4 | 9.6 | |
| gsiB | Embryonic abundant protein Em1, Arabidopsis thaliana (4e-18) | GTTTGTTTAAAAGAAT.TGTGAGCGGGAATACAACAAC(CA)AC | m | + | 61 | 64 | 16 | 44 | 263 |
| ydbD | Manganese-containing catalase BH2190, B. halodurans (e-102) | TTTCGTTTATCTTTCT.ATCGATCGGAAATATAA-N31-ATG | m | 83 | 75 | 9.8 | 18 | 454 | |
| rsbV | Anti-anti-sigma factor of SigB | AAAGGTTTAACGTCTGTCAGACGAGGGTATAAAGCCAACT(A)G | 1st | + | 4.8 | 6.0 | 4.6 | 15 | 12 |
| rsbW | Switch protein/serine kinase, anti-sigma factor of SigB | rsbVW sigB rsbX operon | 2nd | + | 7.1 | 8.6 | 3.8 | 13 | 18 |
| sigB | RNA polymerase general stress sigma factor | rsbVW sigB rsbX operon | 3rd | + | 7.1 | 15 | 7.3 | 8.7 | 51 |
| rsbX | PP2C type serine phosphatase | rsbVW sigB rsbX operon | 4th | + | 5.8 | 9.9 | 6.2 | 8.2 | 16 |
| yfnI | Anion-binding protein homolog, YflE, B. subtilis (<e-180) | TGTTGTGTGATGCCTTGATTTTGTTTGGAAAAGAAG-N34-ATG | m | 5.6 | 12 | 2.3 | 6.8 | 14 | |
| yflT | No similarity | ATCTGTTTCAGGTACA.GACGATCGGGTATGAAA-N33-ATG | m | 105 | 202 | 58 | 283 | 646 | |
| yflH | Unknown conserved protein BH2390, B. halodurans (5e-08) | GCAGGTTTAACCTCCTCCAATTGCAGGTAAAGCA-N28-ATG | m | 13 | 5.5 | 2.2 | 2.5 | 11 | |
| yflA | Amino acid transporter BH4033, B. halodurans (8e-68) | AAAGGTTTATGTTTTTCCATCTATGGGAAATGAT-N25-ATG | m | 3.3 | 5.2 | 2.7 | 7.4 | 3.6 | |
| yfkM | General stress protein BH3025, B. halodurans (3e-64) | GAATGTTTATTTTAGT.GTGGCTGGGGTAGAGTG-N36-ATG | m | 24 | 26 | 5.9 | 17 | 171 | |
| yfkJ | Protein-tyrosine-phosphatase BH2238, B. halodurans (5e-46) | GAAGGTTTCTTTTTAGAGAAATAGGGGCAAAGAA-N17-ATG | 1st | 11 | 8.7 | 2.1 | 14 | 6.0 | |
| yfkI | No similarity | Potential yfkJIH operon | 2nd | 21 | 14 | 3.0 | 38 | 11 | |
| yfkH | Hypothetical protein YfkH, B. cereus (5e-59) | Potential yfkJIH operon | 3rd | 4.3 | 3.3 | 1.3 | 3.7 | 8.5 | |
| yfhD | No similarity | Potential yfhFED operon | 3rd | 6.8 | 4.9 | 3.2 | 7.1 | 9.2 | |
| yfhE | No similarity | Potential yfhFED operon | 2nd | 2.3 | 2.9 | 1.5 | 2.2 | 4.9 | |
| yfhF | Cell division inhibitor SLR1223, Synechocystis sp. (6e-54) | ACGCGTTTTCTTTTAT.TACAATGAGGTAAAGTA-N30-ATG | 1st | 4.0 | 2.8 | 1.9 | 2.7 | 3.9 | |
| yfhK | Conserved hypothetical protein YwsB, B. subtilis (1e-22) | TTATGTTTGGCTTTGCAAACAAAGGGGAATAGGA-N84-TTG | 1st | 8.5 | 11 | 7.2 | 52 | 43 | |
| yfhLg | Hypothetical protein YvaZ, B. subtilis (5e-05) | Potential yfhKLM operon | 2nd | 5.3 | 3.4 | 2.0 | 1.8 | 1.2 | |
| yfhM | Epoxide hydrolase-related protein DR2549, D. radiodurans (9e-61) | Potential yfhKLM operon | 3rd | 9.8 | 3.4 | 2.7 | 2.1 | 1.5 | |
| yhcM | ORF DG1040 (fragment), D. discoideum (4e-06) | TAACGGTTAATTTGTCTAACGAGGGGGAAAATAT-N23-ATG | m | 8.9 | 7.3 | 2.1 | 5.6 | 52 | |
| ygxB | CmpX, Pseudomonas fluorescens (6e-10) | ATCTGTTTGCTTATTG.GATAAGTGGGTAAACAC-N31-TTG | m | 27 | 21 | 6 | 18 | 90 | |
| yhdF | Oxidoreductase (short chain DH/Red.) BH1511, B. halodurans (e-104) | TGGCGTTTATTCATTTCTGTCGTGTGGTAACGTTCAGT(A)TC | m | 11 | 5.1 | 2.1 | 2.4 | 0.9 | |
| yhdN | Probable oxidoreductase PA1127, P. aeruginosa (e-112) | AAAGGTTTAACATTTTTTCCAGAGGGGAAAAGAT-N25-ATG | m | + | 2.9 | 8.8 | 4.9 | 9.4 | 32 |
| yheK | YxiE, B. subtilis (5e-12) | AAAGGTTAATTGTGCT.CAAATTCGGGTAGTAGT-N30-ATG | m | 8.1 | 13 | 5.1 | 28 | 33 | |
| yhxD | Ribitol dehydrogenase, Z. mobilis (6e-66) | ACATGTTTTTCTGCTTATGCTCAGGGGTACACAT-N35-ATG | m | 4.8 | 7.9 | 2.4 | 1.9 | 15 | |
| yitT | Unknown conserved protein BH2916, B. halodurans (6e-99) | TCGTGTTCAAATATTT.GTTTTAAGGGAAAACAT-N30-ATG | m | 6.2 | 4.9 | 1.2 | 5.1 | 7.9 | |
| yjcE | No similarity | TGCCGTTTTACAAGAA....ACACGGGTATCGCG-N122-ATG | m | 3.2 | 6.3 | 3.8 | 36 | 5.7 | |
| yjgB | No similarity | GGCTGTTTAAACAAGA.AGAAATGGGGTATATCT-N38-ATG | m | 37 | 38 | 4.8 | 12 | 36 | |
| yjgC | Formate dehydrogenase homolog YrhE, B. subtilis (<e-180) | GTATGTTTTATTGAGTTGTTGTAAGGGAACTGAA-N40-ATG | 1st | 34 | 24 | 5.2 | 17 | 47 | |
| yjgDh | Conserved hypothetical protein YrhD, B. subtilis (4e-11) | yjgCD operon | 2nd | 83 | 88 | 18 | 64 | 120 | |
| ykgA | Hypothetical protein BH1779, B. halodurans (3e-64) | AATGGTTTAATGATTTTCATGATGAGGGAATAATA-N33-ATG | m | 20 | 26 | 3.8 | 11 | 6.7 | |
| ykzA | Organic hydroperoxide resistance protein Ohr, P. aeruginosa (2e-21) | GCATGTTTAAAAAGAT..CAGAAAGGGAAATATAACAACT(A)G | m | + | 79 | 147 | 36 | 162 | 238 |
| yknA | Putative deoxycytidylate deaminase, C. arietinum (1e-27) | GAATGTTCTTTGCATT..CTTTTCGGCTATACTA-N29-ATG | m | 4.1 | 4.1 | 1.4 | 2.0 | 2.1 | |
| ykuT | Unknown conserved protein BH2666, B. halodurans (4e-72) | AAACGTTTAACATGGTCATGTACAGGGTAACTAG-N38-ATG | m | 2.0 | 1.7 | 3.2 | 9.9 | 2.5 | |
| ykzI | Unknown conserved protein BH2636, B. halodurans (2e-06) | GCGCGTTTGAAGTAAGGAGAAATGTGGTAATAAA-N139-ATG | m | 13 | 24 | 6.5 | 38 | 17 | |
| ylxP | Unknown conserved protein BH2412, B. halodurans (2e-21) | AGAAGTTTCACAAGGCTATGAATGTGGTATTACA-N71-GTG | m | 4.6 | 10 | 1.6 | 2.3 | 18 | |
| ymzB | Unknown conserved protein BH1784, B. halodurans (5e-04) | TCCCGTGTAAAAGATGCAGCAATTGGGAATAGTA-N24-TTG | m | 6.1 | 9.2 | 1.4 | 2.7 | 23 | |
| yoxC | No similarity | TTCTGATTAAAAAAAC.GGATACAGGGTAATGAC-N20-ATG | m | 16 | 26 | 3.4 | 10 | 14 | |
| yocB | No similarity | TCAGGTTTGATCGTTT.TTAAGAGAGGAAAAAGA-N30-ATG | m | 29 | 31 | 12 | 7.4 | 60 | |
| yocK | DnaK suppressor DksA, Chlamydia pneumoniae (3e-16) | CCATGTTTGACAGAAG.GCAAAACGGGAAACAGG-N20-ATG | m | 14 | 36 | 5.3 | 3.0 | 27 | |
| bmrU | Multidrug resistance protein | CTTCGTTTACTGCTTACAGAAAAAGGGGATTATATAACC(A)GA | 1st | + | 5.8 | 3.8 | 1.2 | 4.4 | 3.3 |
| bmrg | Multidrug efflux transporter | bmrU bmr bmrR operon | 2nd | + | 1.1 | 1.1 | 1.1 | 2.7 | 0.7 |
| bmrRg | Transcriptional regulator (MerR family) | bmrU bmr bmrR operon | 3rd | + | 4.3 | 1.5 | 1.1 | 1.5 | 3.4 |
| yqxL | Mg2+ and Co2+ transport protein CorA, A. aeolicus (2e-11) | ACTGGTTTAGTGACGC.GGTTATTGGGCAATTAA-N33-ATG | m | 8.0 | 11 | 3.6 | 12 | 25 | |
| yqhB | Putative membrane protein with hemolysin domain, C. jejuni (3e-68) | ACATGTTTTATGAGCATTTTCAGGTGGTATGGAA-N29-ATG | m | 5.2 | 6.7 | 3.0 | 6.0 | 12 | |
| yqgZh | Arsenate reductase BH3485, B. halodurans (3e-16) | AATGGTTTAAATGAAAAATGATCCGGGTAGTTAT-N37-ATG | m | 5.4 | 101 | 76 | 257 | 337 | |
| bofC | Forespore regulator of the sigma K checkpoint | Potential csbX bofC operon | 2nd | + | 5.2 | 6.4 | 2.7 | 3.6 | 60 |
| csbX | α-Ketoglutarate permease | ACAGGATTACAATTCAGCAAGCTTGGGTATATACTCCATT(G)A | 1st | + | 7.9 | 8.9 | 4.0 | 4.3 | 5.1 |
| ysnF | Hypothetical protein DR1314, D. radiodurans (1e-13) | TTTTGTTTAATTCAAA.GAACAGCGGGAATTACA-N46-ATG | m | + | 116 | 194 | 16 | 40 | 398 |
| ytxJ | General stress protein BH3013, B. halodurans (7e-11) | ytxGHJ operon | 3rd | + | 8.8 | 5.6 | 5.4 | 23 | 5.2 |
| ytxH | Seed maturation protein PM30, Glycine max (6e-04) | ytxGHJ operon | 2nd | + | 7.7 | 4.4 | 7.4 | 26 | 6.8 |
| ytxG | General stress protein BH3245, B. halodurans (2e-19) | ACATGTTTATGATTGA.AGAAAACGGGTAAACAGCAG(T)ATAT | 1st | + | 7.7 | 5.0 | 10 | 38 | 5.7 |
| dps | DNA-protecting protein | AGAGGTTTTAGCGTAG.ATATTAAGGGTATACATAGTCAT(A)T | m | + | 20 | 47 | 11 | 111 | 31 |
| ytiA | Ribosomal protein L31, B. halodurans (9e-30) | TGAGGTTTACGATGTG.AAACAGAGGGAAGGATA-N75-ATG | m | 13 | 25 | 6.1 | 16 | 1.0 | |
| ytaB | ORF168 protein, A. rubrum (6e-07) | GGGGGTTTGATATTTATAAGATAAAGGGTAATTAA-N21-ATG | m | 9.3 | 12 | 2.6 | 12 | 28 | |
| yugU | Unknown conserved protein BH3498, B. halodurans (2e-48) | TCGGGTTTATGAGAGCGGTTTAACAGGAAAAAAAA-N21-ATG | m | 3.0 | 3.2 | 1.9 | 3.5 | 3.8 | |
| yuzA | Hypothetical conserved protein BH3345, B. halodurans (7e-22) | GATGGTTTTATTTTGCAAGGTGCTGGGAAAGAAG-N113-ATG | m | 13 | 11 | 2.6 | 3.4 | 1.6 | |
| yvrE | Senescence marker protein-30 (SMP30-fam.), Xenopus aevis (2e-50) | AGTGGTTTGGACACCT.CTTTGCCGGGAATAACA-N32-ATG | m | 12 | 12 | 4.0 | 14 | 33 | |
| yvgO | Hypothetical 19.6-kDa protein, B. amyloliquefaciens (1e-53) | TTGAGATTACAAATACATTGAGCAGGGTATGCCT-N36-TTG | m | 10 | 7.0 | 3.8 | 5.1 | 0.9 | |
| yvaA | Hypothetical oxidoreductase YdgJ, Escherichia coli (2e-76) | TTAGGTTTTACCATTTGATCAGGAGGGTATATAC-N35-GTG | m | 10 | 12 | 6.3 | 13 | 20 | |
| yvaJ | 3′-to-5′ exoribonuclease RNase R | yvaK yvaJ operon | 2nd | 7.1 | 5.1 | 1.7 | 3.0 | 12 | |
| yvaK | Carboxylesterase precursor (EC 3.1.1.1), B. stearothermophilus (2-107) | AAACGTTTTTTTCTGATTAAACTGTGGAAAACTA-N28-ATG | 1st | 6.6 | 7.8 | 1.5 | 3.5 | 10 | |
| gtaB | UTP-glucose-1-phosphate uridylyltransferase (EC 2.7.7.9) | AAATGTGTAAAAACATATTGAAAAGGGTAAATGTGCTGT(A)GT | m | + | 5.2 | 4.4 | 2.0 | 6.1 | 4.6 |
| ywtG | d-Xylose-proton symporter XylT, L. breyis (e-113) | AAAGGTTTAATGGCCGGAAAAAGAGGCTAAAAGA-N60-ATG | m | 4.7 | 4.5 | 1.8 | 3.5 | 79 | |
| ywsB | Hypothetical protein YfhK, B. subtilis (2e-22) | TGATGTTTAGGAACCTGCGATAACGTGAATAGAG-N42-ATG | m | 18 | 61 | 20 | 159 | 60 | |
| csbD | No similarity | GAATGTTTATTGCCTC.TCAGATCGGGAAGTTAA-N34-ATG | m | 60 | 240 | 66 | 245 | 21 | |
| ywmE | No similarity | ATTGGTTTAAAAACAG.TTTGGGCGGGAATGATA-N21-ATG | m | 8.4 | 6.5 | 3.1 | 4.1 | 6.3 | |
| ywjC | No similarity | ATAGGTTTACGACTTGTCAGCTTTGGGAACTTAG-N36-ATG | m | 7.2 | 16 | 10 | 13 | 69 | |
| ywiE | Cardiolipin synthetase BH2858, B. halodurans (e-115) | CAAGGTTTATCGATTAGAAAAAAGAGGTAATACA-N32-ATG | m | 2.4 | 2.3 | 1.3 | 2.6 | 2.6 | |
| ywzA | Conserved hypothetical protein YdaS, B. subtilis (2e-06) | AGTTGTTTATCTTATACAAAAAAGAGGAATGATA-N130-ATG | m | 88 | 140 | 29 | 208 | 459 | |
| gspA | Putative glycosyl transferase LgtC, P. multocida (6e-30) | ACGTGTTTATTTTTTT.GAAAAAGGGTATGTAACTTGT(A)CA | m | + | 42 | 88 | 33 | 57 | 395 |
| yxzF | No similarity | Potential yxlJ yxzF operon | 2nd | 2.6 | 2.3 | 1.6 | 2.0 | 13 | |
| yxlJ | N-Methylpurine-DNA glycosylase, Mus musculus (3-e21) | AGCCGTTTTTTTTGAT.CTGCTTCGGGAATGGAT-N36-GTG | 1st | 4.2 | 3.2 | 1.6 | 3.0 | 5.3 | |
| katX | Catalase X (EC 1.11.1.6) | GGCTGTTTTAAAATCTTTCCATTCAGGGAATATTGTTAC(C)GT | m | + | 5.2 | 3.8 | 1.6 | 2.0 | 11 |
| yxkO | Hypothetical protein TM0922, T. maritima (2-e30) | TTTTGTTTGAAAAAGAAAAGGGACAGGAAAAATA-N27-ATG | m | 3.9 | 2.4 | 2.0 | 2.1 | 37 | |
| yxiS | No similarity | katB yxiS operon | 2nd | + | 3.6 | 4.0 | 3.0 | 7.9 | 9.2 |
| katB | Catalase 2 (EC 1.11.1.6) | AGCAGTTTATATGAAGAACGCCACGGGTAAATGTGCTGT(A)GA | 1st | + | 2.2 | 2.1 | 1.3 | 2.6 | 11 |
| csbC | Metabolite transport protein homolog YwtG, B. subtilis (e-115) | AAATGTTTCAAATGAGATAGGAAATGGGTACTAATCT(A)TTAA | m | + | 4.3 | 3.6 | 2.2 | 6.6 | 73 |
| yxbG | Probable short-chain dehydrogenase PA1649, P. aeruginosa (1e-30) | GCATGTTTATCACTGC.ACATAGCGGGAAGACAA-N23-ATG | m | 2.7 | 2.0 | 1.4 | 3.0 | 8.7 | |
| yxnA | Glucose 1-dehydrogenase II (EC 1.1.1.47), B. megaterium (4e-24) | AGGGGTAAGACCCTTC..CGGATGGGGTAATGTA-N29-ATG | m | 4.8 | 3.1 | 1.6 | 3.1 | 23 | |
| yxaB | EpsL, S. thermophilus (4e-32) | CAATGCATAGCCATCCTTCTTTTTGGGTAGAGAC-N45-ATG | m | 8.5 | 8.5 | 1.9 | 3.6 | 7.1 | |
| yycD | No similarity | GATCGTTTCGGACAGTAACAAGGCGGGAAAAATG-N27-ATG | m | 33 | 34 | 8.3 | 14 | 104 | |
Genes are listed according to their order in the B. subtilis genome. Genes in bold letters were known from the literature to be ςB dependent prior to the DNA macroarray analysis.
The function of the protein encoded is given if it has been supported by genetic or biochemical data. Otherwise the nearest homolog of the protein encoded is listed. The expect values (E values) given in parentheses were obtained with BLAST 2.0 searches using the BLOSUM 62 matrix against the SWALL-none redundant protein database. “No similarity” indicates that the E value was above e-03. In cases where the best homolog has not been characterized yet, the next possible homolog with known function is given instead.
The presumed −35 and −10 regions of potential ςB-dependent promoters and the start codon of the coding region are shown in boldface. The experimentally determined 5′ ends of the mRNAs are represented by bold letters in parentheses. If the gene is not the first gene in an operon, the operon is shown instead of the regulatory region.
Potential operon structures derived from the B. subtilis genome sequence. m, monocistronic; ordinal numbers indicate the position in the potential operon.
Plus sign indicates verification of operon structure by Northern blot analysis.
Calculated by dividing the normalized hybridization signals of individual genes in the induced sample or wild type (wt) by the relative intensities of the untreated controls. Values reflect averages of at least two independent array hybridizations. EtOH 10 min/co, RNA was prepared from a wild-type strain either immediately before or 10 min after treatment with 4% (vol/vol) ethanol; EtOH, heat, or salt, wt/sigB, ratios were obtained with RNAs prepared from the wild-type strain and the sigB mutant treated for 10 min with 4% (vol/vol) ethanol, 48°C, or 4% (wt/vol) NaCl; EtOH, rsbX/sigB, the ratios were obtained with RNA prepared from an rsbX suppressor mutant (BSA386) and the sigB mutant (BSA272) 60 min after addition of ethanol to a final concentration of 4% (vol/vol).
Operon internal genes that do not fulfill the stringent expression criteria are included here if the flanking genes display ςB-dependent expression or if the operon structure has been proven by Northern blot analysis.
The expression pattern has been verified by Northern blot analysis.
However, it was apparent that we preferentially missed ςB-dependent genes subject to an additional ςB-independent stress induction mechanism, because those genes would not always display much stronger induction in the wild type than in the sigB mutant. Therefore, we applied a modified two-step discrimination protocol to the same set of data to hunt for this class of genes. Table 2 displays the results of this search, which required twofold stress induction by at least two of the three stress factors in the wild type and the sigB mutant. Potential candidates passing this first test were subsequently screened for the presence of the conserved ςB-dependent promoter. This search strategy identified 24 additional genes, 11 of which had previously been described as general stress genes. The latter group includes yvyD, which remained inducible by ethanol stress in the sigB mutant at the ςH-dependent promoter (16), as well as the clpC operon and clpP, both of which remained stress inducible in a sigB mutant at a ςA-dependent promoter after inactivation of the CtsR repressor by stress (20, 32). trxA also belongs to this group, but the mechanism for stress induction in the sigB mutant has not been clarified yet (40). Assigning the other 13 genes to the ςB regulon is more complicated because all of them still displayed stress induction in a sigB mutant (Table 2). In the case of the presumed yceCDEFGH operon this additional stress induction mechanism seems to involve the ECF sigma factor ςW (see below). Detailed transcriptional analysis will be necessary to confirm ςB dependency for each single gene listed in Table 2.
TABLE 2.
Summary of putative ςB-dependent genes subject to additional ςB-independent stress inductiona
| Gene | Function or nearest homolog (E value) | Regulatory region or potential promoter sequences | Operon structure
|
Induction ratiob
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| wt
|
sigB
|
|||||||||
| Predicted | Validated | EtOH | Heat | Salt | EtOH | Heat | Salt | |||
| ctsR | Negative transcriptional regulator of class III stress genes | TCAGGTTTTGTGGACCGGGAAAATGGAAATAATGAAGG(A)T(A) | 1st | + | 9.0 | 10 | 4.2 | 6.5 | 8.1 | 2.8 |
| mcsA | Modulator of CtsR repression | ctsR mcsAB clpC sms yacK operon | 2nd | + | 3.3 | 5.4 | 1.9 | 4.3 | 4.4 | 1.5 |
| mcsB | Modulator of CtsR repression | ctsR mcsAB clpC sms yacK operon | 3rd | + | 10 | 16 | 5.4 | 14 | 15 | 3.4 |
| clpC | Class III stress response-related ATPase | ctsR yacHI clpC sms yacK operon | 4th | + | 5.8 | 19 | 5.0 | 7.4 | 13 | 2.3 |
| sms | DNA repair protein homolog BH0104, B. halodurans (e-178) | ctsR yacHI clpC sms yacK operon | 5th | + | 6.2 | 4.6 | 4.2 | 4.5 | 3.0 | 3.2 |
| yacK | Putative DNA-binding protein SCE9403, Streptomyces coelicolor (1e-80) | ctsR yacHI clpC sms yacK operon | 6th | + | 3.5 | 3.2 | 3.7 | 4.5 | 2.2 | 3.3 |
| yacL | Putative glutamyl-tRNA-transferase, Listeria monocytogenes (e-108) | TTTCGGTTAAAACCTTATGAATACGGGTATATTAATGTT(G)GTT | m | 2.4 | 3.2 | 2.5 | 2.5 | 2.2 | 1.9 | |
| yceC | Resistance protein CdrC, C. acetobutylicum (2e-43) | AAACGTATATATTAGT.AATTTACGGCTTATTTT-N61-GTG | 1st | 2.9 | 1.9 | 9.6 | 6.1 | 2.2 | 12 | |
| yceD | Resistance protein CdrC, C. acetobutylicum (7e-63) | Potential yceCDEFGH operon | 2nd | 3.2 | 1.8 | 6.8 | 5.7 | 1.7 | 8.8 | |
| yceE | Resistance protein CdrC, C. acetobytylicum (1e-74) | Potential yceCDEFGH operon | 3rd | 2.7 | 1.4 | 6.2 | 8.3 | 2.5 | 12 | |
| yceF | Toxic anion resistance protein YkoY, B. subtilis (4e-28) | Potential yceCDEFGH operon | 4th | 2.1 | 1.6 | 5.2 | 4.0 | 1.6 | 7.3 | |
| yceG | No similarity | Potential yceCDEFGH operon | 5th | 2.7 | 3.2 | 6.8 | 4.1 | 3.0 | 7.9 | |
| yceH | Tellurite resistance protein TelA, R. sphaeroides (1e-30) | Potential yceCDEFGH operon | 6th | 1.7 | 2.5 | 4.9 | 3.6 | 3.1 | 9.6 | |
| ycnHc | Succinate-semialdehyde DH GabD, B. halodurans (e-141) | AACGGATTACTTTTGC.TGACAGCGGGAATTAACGGTA(A)TATC | m | 13 | 6.3 | 4.5 | 11 | 4.0 | 2.0 | |
| yjbC | Conserved protein BH2863, B. halodurans (8e-21) | GGCTGTTTAAACAAGA.AGAAATGGGGTATATCTAAAAGT(A)TG | 1st | + | 8.9 | 3.8 | 9.2 | 18 | 3.2 | 18 |
| yjbD | Conserved protein BH2861, B. halodurans (1e-56) | yjbCD operon | 2nd | + | 4.8 | 2.3 | 2.7 | 5.6 | 2.4 | 4.1 |
| ypuD | No similarity | TACGGTTTTTTATTCATGAAAAAAAGGAATAACT-N40-ATG | m | 2.2 | 2.3 | 1.6 | 3.4 | 1.8 | 2.5 | |
| yqjL | Probable hydrolase PA2934, P. aeruginosa (2e-05) | CCCTGTTCAATACGAA.AGAGGCTGGTAATGCCC-N104-ATG | m | 5.1 | 5.6 | 2.0 | 9.4 | 5.2 | 2.8 | |
| sodA | Superoxide dismutase (EC 1.15.1.1) | GCAGGTTTAATGGGGCAGATTATCGGTTAAAGTG-N132-ATG | m | 5.3 | 3.1 | 3.0 | 5.4 | 3.7 | 1.3 | |
| yraA | Intracellular proteinase I PfpI, P. furiosus (4e-28) | GATTGATATTTCCACATTGTAATAGGTTAGTCCT-N118-ATG | m | 6.9 | 3.6 | 3.5 | 9.1 | 3.2 | 2.0 | |
| trxA | Thioredoxin | TCAGGTTTTAAAACAGCTCCGGCAGGGCATGGTAAAGTAC(A)TG | m | + | 6.9 | 3.9 | 3.9 | 4.8 | 2.9 | 1.7 |
| yvgN | Putative morphine dehydrogenase YtbE, B. subtilis (e-116) | AAGCGTATTATTGGTATCGGCTGAGAGGAATGTGA-N44-GTG | m | 3.1 | 6.1 | 2.8 | 7.6 | 7.4 | 2.4 | |
| clpP | ATP-dependent Clp protease proteolytic subunit (EC 3.4.21.92) | GATGGTTTGAACCCCTGTATTTTTGGGGAAAATGGGAAAA(AG)A | m | + | 2.6 | 17 | 5.1 | 4.9 | 18 | 4.1 |
| yvyD | Ribosomal protein S30AE family, B. halodurans (7e-62) | TTATGTTTCAGCAGGAATTGTAAAGGGTAAAAGAGAAAT(A)GA | m | + | 4.6 | 2.1 | 2.0 | 8.5 | 0.6 | 0.5 |
For details on the data presentation, see Table 1, footnotes a through e.
Calculated by dividing the relative hybridization signals of individual genes in the induced sample of the wild type (wt) or the sigB mutant by the respective control values. For experimental details, see Materials and Methods. EtOH, ethanol.
Expression pattern has been verified by Northern blot analysis.
In total this DNA array analysis revealed 125 ςB-dependent genes, 24 of which seem to be subject to a second, ςB-independent stress induction. We confirmed 62 of the 75 ςB-dependent genes known from the literature. Most notably, all the ςB-dependent genes identified by the proteomics approach were confirmed by this array analysis (Table 3). This observation is not surprising, because the proteomics approach should have mainly detected genes displaying strong expression as well as clear transcriptional induction. Also, most of the genes identified by transcriptional studies (27, 37) or by the transposon-based approach of Price and coworkers (10, 12) were confirmed (Table 3). However, only 14 of 24 genes newly described as ςB dependent by a promoter consensus search (36) were validated by DNA macroarrays. The reason for the surprisingly low validation rate of this group is not yet known. The list of genes already described in the literature as ςB dependent but not confirmed by this DNA macroarray analysis includes aldY, csbA, csbB, opuE, ydbP, ydhK, yotK, yoxA, ypuB, yqhA, yqhQ, yrvD, and ytkL. We cannot exclude the possibility that a few of these genes constitute false positives that were described in earlier studies. However, we suspect that in some cases the apparent lack of detection by this DNA array approach might also be an artifact due to differences in the amount or quality of the PCR products on the membrane or in the primers utilized for the cDNA synthesis. One member of the latter group is certainly opuE, whose ςB dependency has been unambiguously demonstrated (43).
Induction of ςB-dependent genes ranged from twofold to several hundredfold. Such extreme induction ratios might be explained by the fact that ςB, which is almost inactive during exponential growth, exclusively controls some ςB-dependent genes. Frequently, general stress genes contain additional promoters, in most cases ςA-dependent promoters, that allow significant basal expression level during growth. Accordingly, the stress induction ratios of genes in the latter group are lower than those for the former.
When the data were analyzed, it was apparent that salt and ethanol triggered much stronger induction of the ςB regulon than heat stress, although heat stress was effective in inducing heat-specific stress proteins (see below). The reason for this difference is not clear but might be related to the influence of the stresses on growth and consequently their stringency. Both ethanol and salt reduced the growth rate slightly at the concentration used (final concentration, 4%), whereas a temperature shift from 37 to 48°C still stimulated growth.
Sometimes not all genes of an operon met the stringent criteria applied. If the missing genes were flanked by genes displaying a clear induction pattern (e.g., yfhL) or if the operon structure had been experimentally proven (e.g., the bmrU bmr bmrR operon), the genes were added to the table, since the failure of detection was most likely caused by the limitations of the array analysis described above. Applying stringent criteria to the searches will certainly minimize the detection of false-positive candidates, but at the risk of producing false negatives. Besides the genes listed in Tables 1 and 2, we uncovered a group of genes with either a less conserved ςB-dependent promoter or a stress induction pattern just failing to fulfill the requirements outlined above. These 38 genes (aldY, aroI, mtrA, purK, rbfA, spoIIQ, yabK, yacO, yazC, ycsE, ydcF, yddS, yeaC, yerD, yfjB, yfkC, yfkT, yfmG, yfmK, ygxA, yhdE, ykrS, ykrT, ykyB, ykzC, yrrU, ysdB, ytzB, ytzE, yumB, yusD, yusS, yutK, yvaM, ywdJ, ywdL, ywlB, and ywmF) are currently the subject of detailed Northern blot analysis to clarify their potential ςB dependence.
Locations and functions of new general stress proteins.
The proteomic approach almost exclusively identified general stress proteins that were localized in the cytoplasm. The transposon mutagenesis as well as the promoter search- and oligonucleotide screening-based approaches have already indicated that the synthesis of membrane proteins is also subject to ςB control, leading to the assumption that ςB also contributes to the maintenance of the integrity of the cell envelope during stress (19, 36). Inspection of the ςB-dependent gene products described in this study for membrane-spanning helices (MSH) revealed that 25 of them contain at least one potential MSH (Table 4). Furthermore, at least six of the general stress proteins seemed to contain signal sequences indicating an extracellular location. Four of those proteins are potential lipoproteins and are most likely attached to the outside of the cytoplasmic membrane (Table 5).
TABLE 4.
Membrane-localized specific and general stress proteins
| No. of MSHa |
Proteinsb |
|---|---|
| 1 | YbbM, YbyB, YdjG, YpuD, YqfB, YtxG, YtxH, YuaG, YvlC, YxzE |
| 2 | YdbS, YdjH, YfhL, YjcE, YobJ, YqfA, YqxL, YrvD, YteJ, YuzA, YvlA, YxiS |
| 3 | YdaS, YkuT, YrkA, YuaF, YvlD, YvqI, YwrE |
| 4 | MrpB, YcbP, YflA, YknZ, YqeZ, YqhB, YwoA |
| 5 | OpuBB, OpuCB, OpuCD, YdbT, YfnI, YitT, YknW, YtaB |
| 6 | YfkH |
| 7 | YvgW |
| 8 | YceF, YthQ |
| 11 | YdaR, YgxB, YhaU, YhfA, YwtG |
| 12 | Bmr, CsbC, CsbX |
Determined by using either the dense alignment surface method of Cserzo et al. (15) or a hidden Markov model-based method introduced by Sonnhammer et al. (42). Proteins containing only one or two potential transmembrane helices (MSH) were classified as potential membrane proteins only if the MSH were predicted by both algorithms.
Proteins displaying SigB-dependent stress-induced synthesis are shown in bold.
TABLE 5.
Extracellular specific and general stress proteinsa
| Protein class | Proteins |
|---|---|
| Predicted lipoproteins | SacC, YfhK, YobJ, YoxC, YpuA, YvgO, YwsB, YvlA |
| Predicted extracellular proteins | BofC, OpuAC, OpuBC, OpuCC, YjgB, YknX |
All stress-induced genes were analyzed for the occurrence of a potential signal sequence with the SignalP algorithm of Nielsen et al. (35), and potential candidates were subsequently compared with a recent genome-based analysis of the secretome of B. subtilis by Tjalsma et al. (45). Proteins displaying ςB-dependent stress-induced synthesis are shown in boldface.
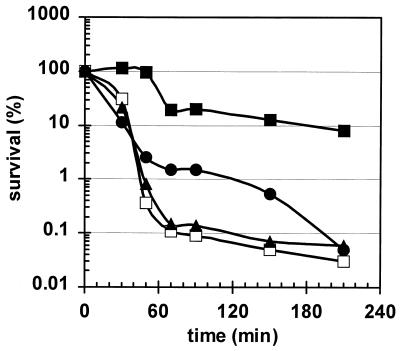
The functions of most of these newly described ςB-dependent genes are still unknown. Probably the proteins encoded by those genes are involved, like the proteins already known, in the development of nonspecific multiple stress resistance in starving cells or in growing cells subject to harsh stress. In order to define the kind of stress resistance in which the individual genes are involved, a comprehensive screening program analyzing the stress resistance of mutants with mutations in single stress genes is in progress (41). This screening has already revealed a number of stress genes that have dramatic effects on resistance to specific stress factors. Figure 2 displays the sensitivities of two selected mutants to growth-preventing salt stress (see reference 51). The newly described yjbCD operon, which is at least in part under ςB control (see Table 2), therefore codes for proteins that are somehow involved in salt resistance.
FIG. 2.
Survival of B. subtilis strains during growth-preventing salt stress. The wild-type strain 168 (solid squares) and its isogenic mutants with mutations in sigB (ML6) (open squares), yjbC (BFA2841) (solid triangles), and yjbD (BFA2842) (solid circles) were grown in a synthetic medium and exposed to salt stress. Survival was determined by plating appropriate dilutions on Luria-Bertani agar plates. Cultures were pretreated with a mild salt stress of 4% NaCl for 30 min at time zero before the sodium chloride concentration was raised to 10% (wt/vol).
Identification of ςB-independent stress genes.
The DNA array data had thus far been used only for the identification of genes strictly requiring ςB for stress induction or possessing a ςB-dependent stress induction component. However, the same array data also provide a comprehensive picture of genes inducible by ethanol, salt, or heat stress independently of ςB. Table 6 lists genes displaying significant induction by only one stress or a combination of two or all three stresses. In those cases stress induction is very likely ςB independent, because similar or even stronger induction was observed in the sigB mutant and the corresponding transcriptional units seemed to lack ςB-dependent promoters. Seven genes (murG, sacC, yugJ, yutG, ywaC, ywnF, and ywoA) seemed to lack a potential ςB promoter and displayed at least twofold induction in the wild type and the sigB mutant under all stress conditions tested. For most of these genes, the precise biochemical functions of the products have not yet been determined.
TABLE 6.
ςB-independent stress gene induction in B. subtilisa
| Geneb | Function or nearest homolog (E value) | Regulatory protein or sigma factor | Operon structure
|
Induction ratio
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| wt
|
sigB
|
|||||||||
| Predicted | Validated | EtOH | Heat | Salt | EtOH | Heat | Salt | |||
| Induction by EtOH, heat, and salt shock | ||||||||||
| murG | Undecaprenyl-PP-MURNAC-pentapeptide-UDPGlCNAc GlCNAc transferase | m | 2.5 | 2.7 | 2.3 | 2.9 | 2.2 | 2.5 | ||
| sacC | Levanase (EC 3.2.1.65) | CcpA, LevR | m | 3.2 | 4.4 | 3.2 | 4.1 | 4.4 | 4.9 | |
| yugJ | Butanol dehydrogenase (EC 1.1.1.-), C. acetobutylicum (1e-97) | m | 3.7 | 6.7 | 5.1 | 6.3 | 5.0 | 2.7 | ||
| yutG | Low temperature requirement C protein, B. halodurans (4e-51) | m | 2.2 | 8.4 | 3.9 | 3.0 | 7.6 | 2.4 | ||
| ywoA | Probable bacitracin transport permease BcrC, B. licheniformis (4e-17) | m | 2.5 | 3.1 | 2.0 | 4.1 | 2.5 | 2.4 | ||
| ywnF | Unknown conserved protein, B. halodurans (6e-05) | m | 4.7 | 5.0 | 5.9 | 6.0 | 3.5 | 3.1 | ||
| ywaC | GTP pyrophosphokinase, B. halodurans (4e-36) | m | 2.4 | 3.2 | 2.3 | 4.3 | 2.9 | 5.1 | ||
| Induction by EtOH and heat shock | ||||||||||
| groES | Class I heat shock protein (chaperonin) | HrcA | 1st | + | 7.2 | 11 | 1.0 | 16 | 16 | 1.6 |
| groEL | Class I heat shock protein (chaperonin) | 2nd | + | 5.0 | 14 | 1.3 | 11 | 18 | 1.9 | |
| yhfA | Unknown conserved protein, B. halodurans (1e-41) | 2nd | 2.5 | 3.7 | 1.3 | 2.8 | 3.4 | 1.3 | ||
| yhfI | Unknown conserved protein, B. halodurans (2e-59) | 1st | 2.6 | 2.7 | 1.1 | 3.4 | 2.6 | 0.8 | ||
| yhfJ | Lipoate-protein ligase, B. halodurans (e-136) | 2nd | 2.8 | 3.2 | 1.2 | 3.3 | 2.8 | 0.8 | ||
| yhfK | Unknown conserved protein BH1520, B. halodurans (3e-32) | 3rd | 3.3 | 3.0 | 1.9 | 5.2 | 2.8 | 1.3 | ||
| yitW | dTDP-4-keto-l-rhamnose reductase RmlD, S. mutans (3e-31) | m | 3.2 | 2.9 | 1.2 | 4.0 | 2.7 | 1.2 | ||
| yjbG | Oligopeptidase F homolog, B. subtilis (<3e-180) | m | 4.3 | 4.1 | 1.8 | 4.6 | 2.8 | 1.2 | ||
| ykuV | Unknown conserved protein, B. halodurans (1e-59) | m | 2.6 | 3.1 | 1.3 | 2.5 | 2.6 | 0.8 | ||
| yoeB | No similarity | m | 7.4 | 20 | 1.5 | 17 | 6.0 | 0.7 | ||
| yqkF | Oxidoreductase BH1011, B. halodurans (e-102) | m | 6.7 | 4.0 | 1.8 | 6.9 | 2.8 | 1.1 | ||
| yqiG | NADH oxidase, T. brockii (2e-45) | m | 2.5 | 4.6 | 1.7 | 5.1 | 5.4 | 1.6 | ||
| hrcAc | Transcriptional repressor of class I heat shock genes | HrcA | 1st | + | 5.2 | 7.8 | 5 | 6.8 | 7.6 | 8.6 |
| grpE | Heat shock protein (HSP-70 cofactor) | 2nd | + | 2.8 | 3.9 | 1.8 | 4.9 | 4.3 | 2.8 | |
| dnaKc | Molecular chaperone | 3rd | + | 1.4 | 5.5 | 1.8 | 3.2 | 6.4 | 3.7 | |
| yvrD | 3-Hydroxybutyrate dehydrogenase, S. meliloti (1e-30) | m | 3.1 | 4.8 | 2.4 | 4.0 | 4.3 | 1.5 | ||
| iolS | Ion channel homolog YccK, B. subtilis (3e-94) | m | 2.6 | 3.1 | 1.8 | 3.9 | 3.2 | 1.0 | ||
| Induction by heat shock | ||||||||||
| ycnD | NADPH-flavin oxidoreductase, B. halodurans (4e-49) | m | 1.4 | 4.1 | 2.1 | 1.9 | 4.1 | 1.6 | ||
| yjeA | Peptidoglycan GlcNAc deacetylase, Streptococcus pneumoniae (1e-48) | m | 0.6 | 6.2 | 0.6 | 0.9 | 6.6 | 0.7 | ||
| clpE | ATP-dependent Clp protease-like (class III stress gene) | CtsR | m | + | 1.0 | 12 | 1.2 | 2.3 | 15 | 4.4 |
| yqjM | NADH oxidase, B. halodurans (e-126) | m | 1.6 | 6.5 | 1.4 | 2.3 | 5.4 | 1.1 | ||
| yrkF | ORF H0532, Halobacterium sp. (5e-13) | 3rd | 1.2 | 9.0 | 2.8 | 5.7 | 6.1 | 1.8 | ||
| yrkE | Hypothetical protein, Staphylococcus aureus (5e-36) | 2nd | 1.0 | 4.7 | 2.2 | 3.1 | 3.5 | 1.2 | ||
| yrkD | Hypothetical protein, S. aureus (5e-14) | 1st | 0.8 | 3.5 | 1.5 | 2.8 | 3.4 | 1.1 | ||
| Induction by EtOH shock | ||||||||||
| ygaC | Unknown conserved protein BH3193, B. halodurans (6e-28) | m | 3.1 | 1.2 | 2.3 | 3.2 | 1.2 | 2.8 | ||
| yhcX | Nitrilase-related protein, D. radiodurans (1e-32) | 2nd | 3.8 | 2.5 | 2.2 | 3.9 | 1.8 | 1.2 | ||
| yhgB | No similarity | 1st | 3.4 | 1.1 | 2.0 | 3.7 | 1.0 | 1.7 | ||
| ykgB | Hypothetical protein YadB, L. lactis (1e-60) | m | 4.9 | 1.9 | 1.6 | 3.8 | 1.8 | 0.7 | ||
| yktC | myo-Inositol-1(or 4)-monophosphatase homolog, B. subtilis (e-153) | 1st | 3.4 | 1.6 | 2.7 | 3.4 | 1.3 | 1.1 | ||
| ypiA | α-Acetolactate synthase Als, L. lactis (9e-34) | 1st | 4.2 | 2.2 | 1.9 | 6.7 | 1.0 | 1.8 | ||
| yrzF | No similarity | 1st | 3.2 | 1.9 | 2.3 | 3.7 | 1.8 | 2.1 | ||
| yrzG | No similarity | 2nd | 3.1 | 1.6 | 2.6 | 5.6 | 1.7 | 2.4 | ||
| ytkL | No similarity | m | 3.8 | 2.4 | 2.0 | 3.1 | 1.3 | 0.9 | ||
| ytxK | Unknown conserved protein BH3193, B. halodurans (3e-76) | m | 3.6 | 1.2 | 0.9 | 5.9 | 1.3 | 0.7 | ||
| yuaE | No similarity | m | 4.3 | 2.5 | 2.2 | 6.5 | 1.9 | 1.5 | ||
| yvqH | Phage shock protein A, E. coli (8e-07) | 2nd | 3.6 | 1.1 | 0.6 | 23 | 1.2 | 0.9 | ||
| yvqI | No similarity | 1st | 3.4 | 1.6 | 1.3 | 16 | 1.5 | 0.9 | ||
| yvgW | Cation-transporting ATPase, P type (PacS) PAB0626, P. abyssi | m | 3.5 | 1.4 | 1.0 | 4.0 | 1.6 | 0.8 | ||
| ureB | Urease (gamma subunit) (EC 3.5.1.5) | SigA, SigH | 1st | 3.8 | 0.8 | 0.8 | 3.2 | 0.7 | 0.6 | |
| ureA | Urease (beta subunit) (EC 3.5.1.5) | Cod, GlnR | 2nd | 4.4 | 1.3 | 1.4 | 3.5 | 1.0 | 0.9 | |
| Induction by salt shock | ||||||||||
| yaaN | Tellurite resistance protein, R. sphaeroides (2e-50) | SigW | 2nd | 1.4 | 1.3 | 4.7 | 2.2 | 1.2 | 5.7 | |
| sigW | RNA polymerase ECF type sigma factor | SigW | 1st | 1.3 | 0.8 | 3.3 | 1.4 | 0.7 | 3.7 | |
| ybbM | Anti-ς factor of SigW | 2nd | 1.2 | 0.6 | 4.2 | 2.7 | 1.2 | 8.6 | ||
| ybfA | Ribosomal protein S18 alanine acetyltransferase homolog, A. fulgidus (1e-06) | m | 2.2 | 1.9 | 3.7 | 3.8 | 2.1 | 6.0 | ||
| opuAAc | Glycine betaine ABC transporter (ATP-binding protein) | SigA | 1st | + | 0.3 | 0.4 | 2.2 | 0.6 | 0.4 | 3.6 |
| opuABc | Glycine betaine ABC transporter (permease) | 2nd | + | 0.8 | 0.9 | 1.9 | 0.7 | 0.7 | 2.9 | |
| opuAC | Glycine betaine ABC transporter (glycine betaine-binding protein) | 3rd | + | 0.3 | 0.6 | 3.2 | 0.6 | 0.5 | 4.0 | |
| ydbS | Unknown conserved protein, B. halodurans (3e-28) | SigW | 1st | 2.1 | 1.3 | 6.7 | 2.8 | 1.3 | 5.3 | |
| ydbT | Unknown conserved protein BH1721, B. halodurans (9e-68) | 2nd | 1.4 | 0.8 | 7.1 | 2.2 | 0.7 | 4.9 | ||
| ydjF | Phage shock protein A, D. radiodurans (4e-20) | SigW | m | 2.3 | 0.9 | 9.1 | 3.9 | 1.1 | 14 | |
| ydjG | Unknown conserved protein, B. halodurans (6e-74) | SigW | 1st | 1.2 | 1.1 | 5.5 | 2.4 | 1.3 | 9.6 | |
| ydjH | Unknown conserved protein, B. halodurans (2e-11) | 2nd | 1.5 | 1.5 | 3.0 | 1.8 | 1.3 | 4.5 | ||
| ydjI | Unknown conserved protein, B. halodurans (4e-5) | 3rd | 0.9 | 1.0 | 4.1 | 1.7 | 1.0 | 7.8 | ||
| ydjO | No similarity | SigW | 3rd | 1.0 | 0.7 | 4.2 | 1.0 | 0.7 | 5.3 | |
| ydjP | Bromide peroxidase, S. aureofaciens (1e-18) | 2nd | 1.8 | 1.5 | 5.9 | 1.4 | 1.2 | 5.9 | ||
| yeaA | No similarity | 1st | 1.4 | 1.2 | 4.8 | 1.2 | 1.1 | 4.8 | ||
| spoOM | Sporulation control protein | SigH | m | 3.7 | 0.8 | 6.1 | 4.2 | 0.9 | 6.6 | |
| yhaU | Putative transmembrane transport protein, S. coelicolor (8e-65) | 3rd | 0.9 | 0.9 | 7.3 | 0.8 | 0.7 | 4.5 | ||
| yhaTc | Hypothetical protein YrvC, B. subtilis (4e-49) | 2nd | 1.1 | 1.0 | 3.7 | 1.2 | 1.2 | 2.6 | ||
| yhaS | No similarity | 1st | 1.6 | 1.6 | 20 | 1.7 | 1.5 | 12 | ||
| yhgD | Transcriptional regulator (TetR/AcrR family), B. halodurans (8e-32) | m | 1.1 | 1.6 | 5.3 | 0.6 | 1.4 | 11 | ||
| yjoB | FtsH, Helicobacter pylori (2e-20) | SigW | m | 3.6 | 1.3 | 15 | 2.2 | 1.0 | 13 | |
| ykrL | Probable protease HtpX, E. coli (2e-51) | m | 1.2 | 1.1 | 6.9 | 2.4 | 1.5 | 12 | ||
| yknW | Unknown conserved protein, B. halodurans (4e-20) | SigW | 1st | 1.0 | 0.6 | 7.5 | 2.4 | 0.8 | 11 | |
| yknXc | ATP-binding cassette transporter-like protein TptB, S. cristatus (5e-23) | 2nd | 0.4 | 0.7 | 0.7 | 0.9 | 0.9 | 0.6 | ||
| yknY | Putative ABC transporter YvrO, B. subtilis (3e-75) | 3rd | 1.2 | 0.6 | 3.4 | 3.2 | 0.8 | 5.5 | ||
| yndN | Glutathione transferase FosB (EC 2.5.1.18), S. epidermidis (8e-48) | Potential SigW | m | 1.3 | 1.1 | 4.5 | 2.2 | 1.1 | 6.4 | |
| proJ | Glutamate 5-kinase | SigA | 2nd | + | 1.3 | 1.3 | 4.9 | 1.2 | 1.2 | 4.4 |
| proH | Pyrroline-5-carboxylate reductase | 1st | + | 1.4 | 1.2 | 4.6 | 1.1 | 0.9 | 4.1 | |
| yoaF | No similarity | Potential SigW | m | 0.3 | 0.5 | 3.2 | 0.7 | 0.7 | 6.4 | |
| yobJ | No similarity | Sig W | m | 1.2 | 0.5 | 5.2 | 3.9 | 0.9 | 10 | |
| yocL | No similarity | 2nd | 1.0 | 0.8 | 3.3 | 1.0 | 1.0 | 6.3 | ||
| yocM | Small heat shock protein HspC, B. japonicum (2e-9) | 1st | 0.8 | 1.1 | 3.6 | 0.8 | 1.0 | 5.7 | ||
| yozO | Hypothetical protein YjqA, B. subtilis (7e-10) | Potential SigW | m | 0.6 | 0.6 | 6.1 | 0.6 | 0.6 | 6.4 | |
| yqfB | No similarity | Potential SigW | 3rd | 1.2 | 1.0 | 3.9 | 1.6 | 1.2 | 6.2 | |
| yqfA | Protein of unknown function ORF1, B. megaterium (1e-151) | 2nd | 1.7 | 0.9 | 24 | 2.8 | 1.1 | 22 | ||
| yqeZ | No similarity | 1st | 1.3 | 0.9 | 17 | 1.8 | 1.0 | 17 | ||
| yrkA | Integral membrane protein with hemolysin domain, C. jejuni (1e-73) | m | 0.9 | 0.8 | 9.4 | 1.1 | 1.1 | 16 | ||
| yteJ | Unknown conserved protein, B. halodurans (3e-31) | SigW | 2nd | 1.3 | 1.0 | 3.4 | 4.1 | 1.0 | 5.3 | |
| yteI | Proteinase IV, A. aeolicus (1e-37) | 1st | 2.4 | 0.9 | 3.6 | 5.3 | 0.8 | 4.7 | ||
| ythQ | ABC transporter (permease), B. halodurans (6e-05) | Potential SigW | 2nd | 1.5 | 1.1 | 3.4 | 2.3 | 1.2 | 4.7 | |
| ytgB | ABC transporter, ATP-binding protein (TroB), T. pallidum (6e-79) | 2nd | 0.8 | 0.8 | 3.7 | 0.9 | 1.1 | 10 | ||
| ytgA | ABC transporter, periplasmic binding protein (TroA), T. pallidum (9e-63) | 1st | 0.9 | 1.0 | 4.2 | 0.9 | 1.1 | 10 | ||
| yuaI | Probable acetyltransferase, D. radiodurans (1e-15) | SigW | 3rd | 2.5 | 0.8 | 34 | 7.0 | 2.1 | 62 | |
| yuaG | Epidermal surface antigen, B. halodurans (4e-78) | 2nd | 3.6 | 0.9 | 57 | 7.2 | 1.9 | 68 | ||
| yuaF | No similarity | 1st | 1.7 | 1.0 | 14 | 2.7 | 1.2 | 20 | ||
| gbsBg | Alcohol dehydrogenase | SigA | 2nd | + | 1.0 | 1.1 | 2.0 | 1.0 | 1.2 | 3.8 |
| gbsA | Glycine betaine aldehyde dehydrogenase | 1st | + | 1.0 | 1.7 | 4.6 | 1.2 | 2.1 | 15 | |
| mrpB | Na+/H+ antiporter BH1318, B. halodurans (2e-26) | 2nd | + | 0.8 | 1.7 | 3.1 | 0.7 | 1.6 | 6.9 | |
| opuBDc | Choline ABC transporter (membrane protein) | SigA | 4th | + | 1.2 | 1.2 | 2.6 | 0.9 | 1.2 | 4.2 |
| opuBCc | Choline ABC transporter (choline-binding protein) | 3rd | + | 0.6 | 0.7 | 2.9 | 0.8 | 1.0 | 9.4 | |
| opuBB | Choline ABC transporter (membrane protein) | 2nd | + | 0.9 | 1.0 | 3.0 | 1.0 | 1.0 | 5.3 | |
| opuBA | Choline ABC transporter (ATP-binding protein) | 1st | + | 0.5 | 0.7 | 3.3 | 0.7 | 1.0 | 12 | |
| opuCDc | Glycine betaine/carnitine/choline ABC transporter (membrane protein) | SigA | 4th | + | 4.3 | 0.8 | 2.6 | 0.3 | 0.8 | 3.2 |
| opuCC | Glycine betaine/carnitine/choline ABC transporter (binding protein) | 3rd | + | 0.3 | 0.7 | 15 | 0.5 | 1.0 | 28 | |
| opuCB | Glycine betaine/carnitine/choline ABC transporter (membrane protein) | 2nd | + | 0.7 | 0.9 | 9.8 | 1.2 | 1.1 | 16 | |
| opuCA | Glycine betaine/carnitine/choline ABC transporter (ATP-binding protein) | 1st | + | 0.4 | 0.9 | 13 | 0.4 | 0.9 | 15 | |
| yvlD | Unknown conserved protein, B. halodurans (4e-23) | SigW | 4th | 1.5 | 1.2 | 6.0 | 2.7 | 1.3 | 9.3 | |
| yvlCc | Unknown conserved protein BH3592, B. halodurans (3e-11) | 3rd | 0.9 | 1.3 | 2.3 | 1.2 | 1.1 | 2.9 | ||
| yvlB | Unknown conserved protein, B. halodurans (4e-63) | 2nd | 2.7 | 1.3 | 7.2 | 3.4 | 1.2 | 11 | ||
| yvlA | No similarity | 1st | 2.0 | 1.3 | 6.1 | 3.1 | 1.3 | 9.2 | ||
| yxjI | Hypothetical protein SCGD3.06, S. coelicolor (2e-05) | SigW | m | 1.1 | 0.5 | 3.2 | 1.2 | 0.6 | 4.8 | |
| ahpF | Alkyl hydroperoxide reductase (large subunit) (EC 1.6.99.3) | 2nd | + | 0.9 | 1.7 | 3.7 | 1.0 | 1.6 | 3.6 | |
| ahpCc | Alkyl hydroperoxide reductase (small subunit) | SigA | 1st | + | 0.7 | 2.1 | 2.8 | 1.0 | 2.0 | 3.4 |
See Table 1, footnotes b through e, and Table 2, footnote b, for explanations of data presentation.
Genes are sorted according to their order in the B. subtilis genome. Genes previously known to be induced by the stimulus are shown in bold. EtOH, ethanol.
Operon internal genes that do not meet the stringent expression criteria are included here if the flanking genes display ςB-dependent expression or if the operon structure has been proven by Northern blot analysis.
In addition to ςB-independent stress genes induced by all three stresses, there are proteins induced at least 2.5-fold by heat alone or heat plus ethanol (Table 6). Because ethanol may induce cellular signals similar to those induced by heat stress, genes induced by ethanol and heat stress might actually prove to encode specific heat stress proteins. The well-known members of the HrcA regulon belong to this group. Induction of the HrcA regulon by ethanol was more pronounced in the sigB mutant, most likely because ςB-dependent stress gene induction did not compete for the limiting RNA polymerase core enzyme in this mutant.
The group of genes preferentially induced at least threefold by ethanol stress alone is somewhat surprising, since it was not recognized in the proteomic studies. For most of those genes neither an induction mechanism nor their functions in adaptation to ethanol can be inferred from the currently available data. Induction of the ureAB operon is most likely accomplished via ςH, which has already been shown to be involved in stress induction of the ytxGHJ operon and the yvyD gene (16, 47).
Activation of the ECF sigma factor ςW following salt shock.
Investigation of the genes specifically induced after imposition of salt stress revealed 64 genes that were at least threefold induced in the wild type and the sigB mutant or that belonged to an operon fulfilling this criterion (Table 6). In accordance with expectations, screening for genes specifically induced by salt stress revealed four (opuA, opuB, opuC, and proHJ) of the already known osmoregulated operons of B. subtilis (13). Interestingly, the gbsAB operon, encoding proteins for the conversion of choline to the osmoprotectant glycine betaine (13), also displayed salt shock induction. Quite surprisingly, the gene coding for the ECF sigma factor ςW was clearly induced following salt shock. Frequently, the genes of ECF sigma factors, sigW included, are subject to autoregulation (29). Consequently, the list of salt-induced genes included 23 genes previously described as ςW dependent (30). Screening of the regulatory regions of the remaining salt-induced genes revealed that 11 of them either possessed a putative ςW promoter or belonged to a potential ςW-dependent transcriptional unit. Further extending this analysis, we inspected (i) the salt induction pattern of other previously described members of the ςW regulon (30) for their response to salt shock and (ii) genes displaying stress induction via ςB and at least one other mechanism (Table 2) for putative ςW promoters. This approach suggested 14 more genes that seemed to belong to a ςW-dependent transcriptional unit displaying salt shock induction. An overview of the induction of the ςW regulon by salt shock is presented in Table 7. Examination of the data indicates that many of these potential ςW-dependent genes were also—although to a lesser extent—induced by ethanol (Tables 6 and 7), an effect that was more pronounced in the sigB mutant than in the wild type strain.
TABLE 7.
Induction of the ςW regulon in B. subtilis by salt or ethanol stressa
| Gene | Function or nearest homolog (E value) | Regulatory region or potential promoter sequences | Induction ratio
|
|||
|---|---|---|---|---|---|---|
| wt
|
sigB
|
|||||
| EtOH | Salt | EtOH | Salt | |||
| xpaC | 5-Bromo-4-chloro-indolyl phosphate hydrolysis protein | AAGATGAAACTTGTTTAAGGATTGAACGTAGTAG-N41-ATG | 1.4 | 2.3 | 1.6 | 2.3 |
| yaaN | Tellurite resistance protein TelA, R. sphaeroides (2e-50) | xpaC yaaN operon | 1.4 | 4.7 | 2.2 | 5.7 |
| sigW | RNA polymerase ECF type sigma factor | AAATTGAAACCTTTTGAAACGAAGCTCGTATACATACA(GA)C | 1.3 | 3.3 | 1.4 | 3.7 |
| ybbM | Anti-ς factor | sigW ybbM operon | 1.2 | 4.2 | 2.7 | 8.6 |
| yceC | Resistance protein CdrC, C. acetobutylicum (2e-43) | TTTACGAAACTTTGATATAATAACAAACGTATATA-N83-GTG | 2.9 | 9.6 | 6.1 | 1.2 |
| yceD | Resistance protein CdrC, C. acetobutylicum (7e-63) | Potential yceCDEFGH operon | 3.2 | 6.8 | 5.7 | 8.8 |
| yceE | Resistance protein CdrC, C. acetobutylicum (1e-74) | Potential yceCDEFGH operon | 2.7 | 6.2 | 8.3 | 1.2 |
| yceF | Toxic anion resistance protein YkoY, B. subtilis (4e-28) | Potential yceCDEFGH operon | 2.1 | 5.2 | 4.0 | 7.3 |
| yceG | No similarity | Potential yceCDEFGH operon | 2.7 | 6.8 | 4.1 | 7.9 |
| yceH | Tellurite resistance protein TelA, R. sphaeroides (1e-30) | Potential yceCDEFGH operon | 1.7 | 4.9 | 3.6 | 9.6 |
| ydbS | Conserved protein, B. halodurans (3e-28) | AGAATGAAACCTTTCTGTAAAAGAGACGTATAAATAA(CG)A | 2.1 | 6.7 | 2.8 | 5.3 |
| ydbT | Conserved protein BH1721, B. halodurans (9e-68) | ydbST operon | 1.4 | 7.1 | 2.2 | 4.9 |
| ydjF | Phage shock protein A DR1473, D. radiodurans (4e-20) | AAAGTGAAACTTTTAACGATAATAAATAGTATATG-N40-ATG | 2.3 | 9.1 | 3.9 | 14 |
| ydjG | Unknown protein BH1806, B. halodurans (6e-74) | ydjFGHI operon | 1.2 | 5.5 | 2.4 | 9.6 |
| ydjH | Unknown protein BH1807, B. halodurans (2e-11) | ydjFGHI operon | 1.5 | 3.0 | 1.8 | 4.5 |
| ydjI | Unknown protein BH1805, B. halodurans (4e-5) | ydjFGHI operon | 0.9 | 4.1 | 1.7 | 7.8 |
| ydjO | No similarity | yeaA ydjPO operon | 1.0 | 4.2 | 1.0 | 5.3 |
| ydjP | Bromide peroxidase, S. aureofaciens (1e-18) | yeaA ydjPO operon | 1.8 | 5.9 | 1.4 | 5.9 |
| yeaA | No similarity | TTTATGAAACCTTTGGCCCTATTTATCGTATTACGT(A)AAA | 1.4 | 4.8 | 1.2 | 4.8 |
| yfhL | Hypothetical protein YvaZ, B. subtilis (5e-05) | TGCATGAAACATTTCTTCTTTCTGCACGTAACAATGA(GA)A | 5.3 | 3.9 | 1.6 | 2.0 |
| yfhM | Epoxide hydrolase-related protein, D. radiodurans (9e-61) | yfhLM operon | 9.8 | 7.5 | 3.8 | 4.1 |
| yjoB | FtsH, H. pylori (2e-20) | GGGATGAAACAAAATGCTATGTCAATCGTATATATAAC(G)T | 3.6 | 15 | 2.2 | 13 |
| yknW | Unknown conserved protein, B. halodurans (4e-20) | AACATGAAACTTTTTGATATCCTTCCCGTACTATTTGT(T)A | 1.0 | 7.5 | 2.4 | 11 |
| yknXb | ABC-transporter-like protein TptB, S. cristatus (5e-23) | yknWXYZ operon | 0.4 | 0.7 | 0.9 | 0.6 |
| yknY | Putative ABC transporter YvrO, B. subtilis (3e-75) | yknWXYZ operon | 1.2 | 3.4 | 3.2 | 5.5 |
| yknZ | ABC transporter–ATP-binding protein NMB0549, Neisseria meningitidis (6e-52) | yknWXYZ operon | 1.2 | 2.8 | 2.3 | 3.8 |
| yndN | Glutathione transferase FosB (EC 2.5.1.18), S. epidermidis (8e-48) | TGTATGAAACTTTCTTATGAAAAAAGTCGTATATG-N32-GTG | 1.3 | 4.5 | 2.2 | 6.4 |
| yoaF | No similarity | ATAATGAAACCCGGAGTATGCCAAGCCCGTATAAC-N28-ATG | 0.3 | 3.2 | 0.7 | 6.4 |
| yobJ | No similarity | TATATGAAACCTTTTTTATTTTAGCCCGTATTAAAAGT(A)A | 1.2 | 5.2 | 3.9 | 10 |
| yozO | Hypothetical protein YjqA, B. subtilis (7e-10) | ATATTGAAACTTTTTTCTCTATATGTGCGTATTAC-N171-TTG | 0.6 | 6.1 | 0.6 | 6.4 |
| yqfB | No similarity | yqeZ yqfAB operon | 1.2 | 3.9 | 1.6 | 6.2 |
| yqfA | ORF1, B. megaterium (1e-151) | yqeZ yqfAB operon | 1.7 | 24 | 2.8 | 22 |
| yqeZ | Conserved protein BH1356, B. halodurans (e-103) | AAAATGAAACCTTTGATACATTTGTTACGTATGAA-N42-TTG | 1.3 | 17 | 1.8 | 17 |
| yteI | Proteinase IV, A. aeolicus (1e-37) | GAAGTGAAACATTTTTCATATTGAATCGTATAATGAG(AG)A | 2.4 | 3.6 | 5.3 | 4.7 |
| ythQ | ABC transporter (permease) EcsB, B. halodurans (6e-05) | ythPQ operon | 1.5 | 3.4 | 2.3 | 4.7 |
| ythP | ABC transporter–ATP-binding protein EcsA, B. subtilis (2e-45) | TTAAAGAAACTTTTTTTATTCTATTTCGTAGTAA-N20-TTG | 1.3 | 2.5 | 1.3 | 2.6 |
| yuaI | Probable acetyltransferase, D. radiodurans (1e-15) | yuaFGI operon | 2.5 | 34 | 7.0 | 62 |
| yuaG | Epidermal surface antigen, B. halodurans (4e-78) | yuaFGI operon | 3.6 | 57 | 7.2 | 68 |
| yuaF | No similarity | ATTTTGAAACTTTTCCCGAGGTGTCTCGTATAAATGGT(A)A | 1.7 | 14 | 2.7 | 20 |
| yvlD | Unknown conserved protein, B. halodurans (4e-23) | yvlABCD operon | 1.5 | 6.0 | 2.7 | 9.3 |
| yvlC | Conserved protein BH3592, B. halodurans (3e-11) | yvlABCD operon | 0.9 | 2.3 | 1.2 | 2.9 |
| yvlB | Unknown conserved protein, B. halodurans (4e-63) | yvlABCD operon | 2.7 | 7.2 | 3.4 | 11 |
| yvlA | No similarity | AATTTGAAACCTGAAGAGATTTTAAACGTATAAATAA(GT)A | 2.0 | 6.1 | 3.1 | 9.2 |
| ywrE | No similarity | TTTATGAAACGTTTTTCCTTTTTCTTCGTATAAAGGTA(GA) | 0.7 | 2.4 | 0.7 | 3.5 |
See Table 1, footnotes a through e, and Table 2, footnote b, for explanations of data presentation.
Operon internal genes that do not meet our stringent expression criteria are included here if the flanking genes display ςB-dependent expression or if the operon structure has been proven by Northern blot analysis.
Twenty-three of the salt-induced proteins contain at least one putative MSH, and five seem to be exported or attached to the membrane as lipoproteins. These proteins are involved either in the acquisition of compatible solutes (the Opu-class of proteins) (13) or in the compensation of constraints imposed by salt stress on the membrane or the cell wall.
Because other stress stimuli such as oxidative, alkaline, or acid stress were not considered in this study, care should be taken in the classification of genes as stress specific from these data alone. AhpC and AhpF, for instance, should be considered oxidative stress proteins, because both are particularly induced by peroxide (3, 14). In this case induction by salt stress most likely reflects a secondary oxidative stress.
DISCUSSION
The ςB-dependent general stress regulon is one of the largest regulons of B. subtilis. The discovery and functional characterization of almost all ςB-dependent genes will be necessary for a comprehensive understanding of the physiological role of this huge regulon. Therefore, the DNA array technique was used to detect the candidates not yet found by proteomics, transcriptional analysis, consensus promoter-based transcriptional screening, or transposon mutagenesis (2, 8, 10, 12, 26, 36, 37). The DNA array induction pattern of the previously published ςB-dependent genes (see Materials and Methods for a comprehensive list) was utilized to formulate the following criteria for identifying the remaining members of the regulon: (i) induction in the wild type by at least two of the three stresses analyzed (heat shock, salt stress, and ethanol stress), (ii) ςB dependency of stress induction, that is, absence in the sigB mutant and/or presence for a prolonged time in an RsbX− suppressor mutant that displayed prolonged and increased ςB activity following stress, and (iii) presence of a putative ςB-dependent promoter in front of the gene or operon. This approach is validated by the fact that it detected 51 of the 64 genes already known to be strictly ςB dependent. In addition to this large group, 50 new genes, all subject to the control of a putative ςB-dependent promoter, were identified. In order to also facilitate the recognition of genes with an additional ςB-independent stress induction component, target genes displaying stress induction in the wild type and the sigB mutant were screened for the presence of the typical ςB promoter structure in the regulatory region. This adjustment of the data analysis revealed 11 already known ςB-dependent genes for which complex regulation had been described previously as well as 13 new candidates.
In total we describe 125 genes that belong to the ςB regulon in this study. For the new members of the regulon detected in this study, ςB dependency is highly probable but has to be confirmed by additional transcriptional studies in each case. Northern blot hybridizations have been conducted and confirmed ςB dependency for ycnH, yjgD, and yqgZ.
However, a few genes described as ςB dependent in earlier studies, such as aldY, csbA, csbB, opuE, ydbP, ydhK, yotK, yoxA, ypuB, yqhA, yqhQ, yrvD, and ytkL, were not detected by our approach. The majority of these belong to a group of genes that had been found to be ςB dependent by a consensus promoter-directed slot blot hybridization screening (36). Of the 24 new candidates identified by this strategy, only 14 could be confirmed in the present investigation. Possible reasons for this failure include (i) the complex control of genes, which could blur the ςB dependency, especially if it was combined with a weak ςB promoter that showed only a low induction rate, and, alternatively, (ii) false-positive candidates described in earlier studies. We suspect (iii) that some genes were not confirmed because of artifacts due to either the quality or quantity (or both) of the PCR product on the membrane or the quality of the primers utilized for synthesis of the labeled cDNA. opuE is a clear example of this class, because its ςB-dependent stress induction has been unequivocally demonstrated (43, 53).
Therefore, it should be stressed that the real number of ςB-dependent genes might be even higher. Thirty-eight genes displayed ςB dependency but failed to comply with all the criteria applied in this study. Those genes had to be listed separately either because they did not display induction by multiple stresses or because they lacked the well-conserved ςB-dependent promoter, although they exhibited much stronger induction in the wild type than in the sigB mutant. Failure to display an obvious ςB promoter, for a gene that shows clear ςB-dependent induction, might reflect indirect control, probably via a transcriptional regulator subject to ςB-dependent induction. However, this hypothesis remains to be substantiated by experimental data. Besides additional ςB-dependent stress genes, this list of 38 genes probably also contains some false-positive candidates. Detailed transcriptional analysis of each single gene or operon is currently being performed so that a final decision on their ςB dependency can be made.
Although the limitations of the DNA array hybridization at best allow a semiquantitative comparison of the expression profiles of different genes, the variations in the expression level of the ςB-dependent genes were striking. Most of the ςB-dependent genes displaying the strongest induced signals on the DNA macroarrays have already been found by the proteomic approach, which should preferentially identify the strongly expressed genes. Examples of this group are ctc, gsiB, clpP, ydaD, yflT, and ykzA. Three other strongly expressed ςB-dependent genes have not yet been identified on two-dimensional gels. This is not surprising, because one of them encodes a membrane protein (ytxG), and the other two most likely escaped detection by the proteomic approach because their small products have a very basic pI (csbD and ywzA). The reasons for the strong expression of these genes are not immediately apparent because most of their promoters do not show what we currently believe to be perfect −35 (GTTTAA) and −10 (GGGWAW) boxes. In the case of gsiB the strong ribosome binding site, leading to high stability of the mRNA, seems to be an additional factor contributing to a high expression rate (31). For the other genes the factors determining strong expression still need to be elucidated.
The complete description of all members of a regulon is only a prerequisite for a full understanding of its physiological role. Detailed biochemical and physiological studies must now follow to obtain substantially new information on the physiological role of the ςB regulon. Previous studies showed that ςB-dependent stress proteins provide the starved or stressed cell with oxidative, pH, salt, and heat stress resistance (3, 18, 19, 51). So far only Dps has been shown to be required for oxidative stress resistance (4), and the ςB-dependent proteins essential for salt, heat, and acid resistance are not known. Because the clpC operon or the clpP gene remains heat inducible in a sigB mutant, a limiting amount of ClpC or ClpP should not be the main reason for the impaired heat stress resistance of a sigB mutant (for reviews see references 26 and 27). The newly identified ςB-dependent genes do not immediately help to answer this question, because most of them encode proteins of thus far undefined function. However, many membrane proteins belong to this group, indicating an essential role in the maintenance of cell envelope or transport capacity, as already discussed by C. W. Price (19, 37). Experimental evidence for this suggestion has been provided by studies by E. Bremer's group, who showed that some genes encoding proteins involved in the uptake of compatible solutes are at least partly under ςB control (43, 53). A few of the new ςB-dependent genes seem to encode proteins with interesting functions. YfhF, a probable cell division inhibitor, might prevent division under conditions of severe stress, giving cells time to recover. Some of the products might be involved in detoxification, such as the products of the yceCDEFGH operon, which seems to encode toxic anion resistance proteins, or that of yqgZ, which encodes a potential arsenate reductase. Other proteins seem to perform functions in maintaining the redox balance of the cell, including the products of yxnA, ycnH, and yvaA, encoding a glucose-1-dehydrogenase, a potential succinate semialdehyde dehydrogenase, and a hypothetical oxidoreductase, respectively. The superoxide dismutase SodA is certainly required for detoxification of superoxide, whereas the potential intracellular proteinase YraA might be required to degrade proteins that cannot be repaired. However, detailed functional analysis will be necessary to ascertain the precise functions of these proteins in stress management.
One of the interesting findings of this study is the surprisingly large number of genes with unknown functions that belong to the ςB regulon (see Tables 1 and 2). Of the 4,100 B. subtilis genes, about 1,700 code for proteins with still unknown functions. Elucidation of the functions of all these proteins is a great challenge for future research. Allocating unknown proteins to their regulation groups is a useful approach for a preliminary prediction of their functions. This approach indicates that almost 100 ςB-dependent proteins with still unknown functions are probably involved in the development of multiple prospective stress resistance in cells entering the stationary-growth phase or in the development of heat or osmotic stress resistance. However, detailed phenotypic screening of mutants is necessary to assign each protein to a single facet of stress resistance as a first step and to uncover its exact function by detailed biochemical experiments. Such experiments are in progress in order to gain a more comprehensive picture of the physiological role of this huge regulon in stress adaptation. The first results of this screening are presented in Fig. 2. Obviously, inactivation of yjbC renders B. subtilis almost as sensitive to growth-preventing salt stress as a sigB null mutant. In the yjbD mutant, too, stress resistance is significantly diminished from that in the wild type. It is noteworthy that yjbD, the downstream gene of an operon (yjbCD) that is at least partially ςB dependent, encodes a protein (YjbD) whose Lactococcus lactis homolog seems to affect degradation of nonnative proteins and thereby stress tolerance (H. Ingmer, personal communication). Further studies are in progress to analyze the precise physiological role of both B. subtilis proteins in more detail.
Despite some restrictions discussed above, this study clearly shows that the DNA array technique is a very useful approach for defining the structure and function of already known or still unknown regulons. Fewer than 30% of the regulon members had been identified thus far by a proteomics approach, and low-abundance proteins or proteins with membrane-spanning domains had been missed entirely. Even though the proteomics approach will still have wide application because the final and active products of gene expression, as well as information on posttranslational modification or protein sorting, can be visualized only by proteomics, it will be replaced by the DNA array technologies for the purpose of defining regulon structure. The latter approach is easier to handle and is certainly more comprehensive, but even such a sophisticated screening will require detailed follow-up experiments to validate the data, including quantification of the results.
Among ςB-independent stress induction phenomena, the salt shock induction of the ςW regulon was certainly the most interesting. ςW is one of seven ECF type sigma factors of B. subtilis, the functions of all of which are still not well understood. In general this class of sigma factors controls uptake or secretion of specific molecules and ions or responses to a variety of stresses (33). ςW in particular has been implicated in detoxification responses and the production of antimicrobial compounds (30, 46). Although a sigW mutant displays altered resistance to cell wall biosynthesis inhibitors (46), no difference in the zone of inhibition was observed when a sigW mutant and its corresponding wild type were exposed to HCl, NaOH, NaCl, EDTA, dithiothreitol, 2-mercaptoethanol, lysozyme, SDS, hydrogen peroxide, methyl viologen, metal ions, and various antibiotics (30). In this study we provide evidence that the ςW regulon is induced by salt shock. This is most likely not an osmotic but an ionic effect, because none of the known osmoregulated genes of B. subtilis possesses a ςW promoter. Probably salt shock interferes with the cell envelope or the transport capacity of the cell and thereby triggers induction of the ςW regulon. Sensing of transport processes has already been implicated in triggering ςW activity (46). Salt shock does not seem to be the only stress inducing the ςW regulon. Recently, Schumann and coworkers discovered alkaline shock induction of the ςW regulon (54). Alkaline shock could also interfere with the transport capacity of the cell and thus release ςW from its inhibition by its anti-sigma factor RsiW.
ACKNOWLEDGMENTS
A. Petersohn and M. Brigulla contributed equally to this work.
We are grateful to W. G. Haldenwang for providing strain BSA386 and to Jeff Errington for construction of the BFA2841 and BFA2842 mutants. We thank N. Hauser, M. Scheideler, and T. Beißbarth for initial help with the gene array hybridizations and analysis of the data.
This work was supported by grants from the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, the BMBF, the European Union (GLG2-CT-1999-01455) and the Max-Planck-Gesellschaft to J.H., U.V., and M.H.
REFERENCES
- 1.Alper S, Dufour A, Garsin D A, Duncan L, Losick R. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J Mol Biol. 1996;260:165–177. doi: 10.1006/jmbi.1996.0390. [DOI] [PubMed] [Google Scholar]
- 2.Antelmann H, Bernhardt J, Schmid R, Mach H, Völker U, Hecker M. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis. 1997;18:1451–1463. doi: 10.1002/elps.1150180820. [DOI] [PubMed] [Google Scholar]
- 3.Antelmann H, Engelmann S, Schmid R, Hecker M. General and oxidative stress responses in Bacillus subtilis—cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J Bacteriol. 1996;178:6571–6578. doi: 10.1128/jb.178.22.6571-6578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antelmann H, Engelmann S, Schmid R, Sorokin A, Lapidus A, Hecker M. Expression of a stress- and starvation-induced dps/pexB homologous gene is controlled by the alternative sigma factor ςB in Bacillus subtilis. J Bacteriol. 1997;179:7251–7256. doi: 10.1128/jb.179.23.7251-7256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson A K, Haldenwang W G. Bacillus subtilis ςB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci USA. 1993;90:2330–2334. doi: 10.1073/pnas.90.6.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson A K, Haldenwang W G. Regulation of ςB levels and activity in Bacillus subtilis. J Bacteriol. 1993;175:2347–2356. doi: 10.1128/jb.175.8.2347-2356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson A K, Haldenwang W G. The ςB-dependent promoter of the Bacillus subtilis sigB operon is induced by heat shock. J Bacteriol. 1993;175:1929–1935. doi: 10.1128/jb.175.7.1929-1935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernhardt J, Völker U, Völker A, Antelmann H, Schmid R, Mach H, Hecker M. Specific and general stress proteins in Bacillus subtilis—a two-dimensional protein electrophoresis study. Microbiology. 1997;143:999–1017. doi: 10.1099/00221287-143-3-999. [DOI] [PubMed] [Google Scholar]
- 9.Boylan S A, Redfield A R, Brody M S, Price C W. Stress-induced activation of the ςB transcription factor of Bacillus subtilis. J Bacteriol. 1993;175:7931–7937. doi: 10.1128/jb.175.24.7931-7937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boylan S A, Redfield A R, Price C W. Transcription factor ςB of Bacillus subtilis controls a large stationary-phase regulon. J Bacteriol. 1993;175:3957–3963. doi: 10.1128/jb.175.13.3957-3963.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boylan S A, Rutherford A, Thomas S M, Price C W. Activation of Bacillus subtilis transcription factor ςB by a regulatory pathway responsive to stationary-phase signals. J Bacteriol. 1992;174:3695–3706. doi: 10.1128/jb.174.11.3695-3706.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boylan S A, Thomas M D, Price C W. Genetic method to identify regulons controlled by nonessential elements: isolation of a gene dependent on alternate transcription factor ςB of Bacillus subtilis. J Bacteriol. 1991;173:7856–7866. doi: 10.1128/jb.173.24.7856-7866.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bremer E, Krämer R. Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes in bacteria. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 79–97. [Google Scholar]
- 14.Chen L, Keramati L, Helmann J D. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc Natl Acad Sci USA. 1995;92:8190–8194. doi: 10.1073/pnas.92.18.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 16.Drzewiecki K, Eymann C, Mittenhuber G, Hecker M. The yvyD gene of Bacillus subtilis is under dual control of ςB and ςH. J Bacteriol. 1998;180:6674–6680. doi: 10.1128/jb.180.24.6674-6680.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dufour A, Haldenwang W G. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV) J Bacteriol. 1994;176:1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelmann S, Hecker M. Impaired oxidative stress resistance of Bacillus subtilis sigB mutants and the role of katA and katE. FEMS Microbiol Lett. 1996;145:63–69. doi: 10.1111/j.1574-6968.1996.tb08557.x. [DOI] [PubMed] [Google Scholar]
- 19.Gaidenko T A, Price C W. General stress transcription factor ςB and sporulation transcription factor ςH each contribute to survival of Bacillus subtilis under extreme growth conditions. J Bacteriol. 1998;180:3730–3733. doi: 10.1128/jb.180.14.3730-3733.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerth U, Krüger E, Derre I, Msadek T, Hecker M. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol Microbiol. 1998;28:787–802. doi: 10.1046/j.1365-2958.1998.00840.x. [DOI] [PubMed] [Google Scholar]
- 21.Grossman A, Losick R. Extracellular control of spore formation in Bacillus subtilis. Proc Natl Acad Sci USA. 1988;85:4369–4373. doi: 10.1073/pnas.85.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haldenwang W G, Losick R. A modified RNA polymerase transcribes a cloned gene under sporulation control in Bacillus subtilis. Nature. 1979;282:256–260. doi: 10.1038/282256a0. [DOI] [PubMed] [Google Scholar]
- 23.Hauser N C, Vingron M, Scheideler M, Krems B, Hellmuth K, Entian K D, Hoheisel J D. Transcriptional profiling on all open reading frames of Saccharomyces cerevisiae. Yeast. 1998;14:1209–1221. doi: 10.1002/(SICI)1097-0061(19980930)14:13<1209::AID-YEA311>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 24.Hecker M, Heim C, Völker U, Wölfel L. Induction of stress proteins by sodium chloride treatment in Bacillus subtilis. Arch Microbiol. 1988;150:564–566. doi: 10.1007/BF00408250. [DOI] [PubMed] [Google Scholar]
- 25.Hecker M, Völker U. General stress proteins in Bacillus subtilis. FEMS Microbiol Ecol. 1990;74:197–213. [Google Scholar]
- 26.Hecker M, Völker U. General stress response of Bacillus subtilis and other bacteria. Adv Microb Physiol. 2001;44:35–91. doi: 10.1016/s0065-2911(01)44011-2. [DOI] [PubMed] [Google Scholar]
- 27.Hecker M, Völker U. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the ςB regulon. Mol Microbiol. 1998;29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann T, Troup B, Szabo A, Hungerer C, Jahn D. The anaerobic life of Bacillus subtilis: cloning of the genes encoding the respiratory nitrate reductase system. FEMS Microbiol Lett. 1995;131:219–225. doi: 10.1111/j.1574-6968.1995.tb07780.x. [DOI] [PubMed] [Google Scholar]
- 29.Huang X, Fredrick K L, Helmann J D. Promoter recognition by Bacillus subtilis ςW: autoregulation and partial overlap with the ςX regulon. J Bacteriol. 1998;180:3765–3770. doi: 10.1128/jb.180.15.3765-3770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang X J, Gaballa A, Cao M, Helmann J D. Identification of target promoters for the Bacillus subtilis extracytoplasmic function ς factor, ςW. Mol Microbiol. 1999;31:361–371. doi: 10.1046/j.1365-2958.1999.01180.x. [DOI] [PubMed] [Google Scholar]
- 31.Jürgen B, Schweder T, Hecker M. The stability of mRNA from the gsiB gene of Bacillus subtilis is dependent on the presence of a strong ribosome binding site. Mol Gen Genet. 1998;258:538–545. doi: 10.1007/s004380050765. [DOI] [PubMed] [Google Scholar]
- 32.Krüger E, Msadek T, Hecker M. Alternate promoters direct stress-induced transcription of the Bacillus subtilis clpC operon. Mol Microbiol. 1996;20:713–723. doi: 10.1111/j.1365-2958.1996.tb02511.x. [DOI] [PubMed] [Google Scholar]
- 33.Lonetto M A, Brown K L, Rudd K E, Buttner M J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase ς factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majumdar D, Avissar Y J, Wyche J H. Simultaneous and rapid isolation of bacterial and eukaryotic DNA and RNA: a new approach for isolating DNA. BioTechniques. 1991;11:94–101. [PubMed] [Google Scholar]
- 35.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Petersohn A, Bernhardt J, Gerth U, Höper D, Koburger T, Völker U, Hecker M. Identification of ςB-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J Bacteriol. 1999;181:5718–5724. doi: 10.1128/jb.181.18.5718-5724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price C W. Protective function and regulation of the general stress response in Bacillus subtilis and related gram-positive bacteria. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 179–197. [Google Scholar]
- 38.Richter A, Hecker M. Heat-shock proteins in Bacillus subtilis: a two-dimensional gel electrophoresis study. FEMS Microbiol Lett. 1986;36:69–71. [Google Scholar]
- 39.Ruzal S M, Lopez C, Rivas E, Sanchez-Rivas C. Osmotic strength blocks sporulation at stage II by impeding activation of early sigma factors in Bacillus subtilis. Curr Microbiol. 1998;36:75–79. doi: 10.1007/s002849900282. [DOI] [PubMed] [Google Scholar]
- 40.Scharf C, Riethdorf R, Ernst H, Engelmann S, Völker U, Hecker M. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J Bacteriol. 1998;180:1869–1877. doi: 10.1128/jb.180.7.1869-1877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schumann W, Ehrlich D S, Ogasawara N. Functional analysis of bacterial genes: a practical manual. Chichester, United Kingdom: John Wiley & Sons, Ltd.; 2001. [Google Scholar]
- 42.Sonnhammer E L L, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. In: Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C, editors. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. Menlo Park, Calif: AAAI Press; 1998. pp. 175–182. [PubMed] [Google Scholar]
- 43.Spiegelhalter F, Bremer E. Osmoregulation of the opuE proline transport gene from Bacillus subtilis—contributions of the ςA- and ςB-dependent stress-responsive promoters. Mol Microbiol. 1998;29:285–296. doi: 10.1046/j.1365-2958.1998.00929.x. [DOI] [PubMed] [Google Scholar]
- 44.Stülke J, Hanschke R, Hecker M. Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J Gen Microbiol. 1993;139:2041–2045. doi: 10.1099/00221287-139-9-2041. [DOI] [PubMed] [Google Scholar]
- 45.Tjalsma H, Bolhuis A, Jongbloed J D, Bron S, van Dijl J M. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol Mol Biol Rev. 2000;64:515–547. doi: 10.1128/mmbr.64.3.515-547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner M S, Helmann J D. Mutations in multidrug efflux homologs, sugar isomerases, and antimicrobial biosynthesis genes differentially elevate activity of the ςX and ςW factors in Bacillus subtilis. J Bacteriol. 2000;182:5202–5210. doi: 10.1128/jb.182.18.5202-5210.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varon D, Brody M S, Price C W. Bacillus subtilis operon under the dual control of the general stress transcription factor ςB and the sporulation transcription factor ςH. Mol Microbiol. 1996;20:339–350. doi: 10.1111/j.1365-2958.1996.tb02621.x. [DOI] [PubMed] [Google Scholar]
- 48.Vijay K, Brody M S, Fredlund E, Price C W. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the ςB transcription factor of Bacillus subtilis. Mol Microbiol. 2000;35:180–188. doi: 10.1046/j.1365-2958.2000.01697.x. [DOI] [PubMed] [Google Scholar]
- 49.Völker U, Engelmann S, Maul B, Riethdorf S, Völker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 50.Völker U, Luo T Q, Smirnova N, Haldenwang W G. Stress activation of Bacillus subtilis ςB can occur in the absence of the ςB negative regulator RsbX. J Bacteriol. 1997;179:1980–1984. doi: 10.1128/jb.179.6.1980-1984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Völker U, Maul B, Hecker M. Expression of the ςB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J Bacteriol. 1999;181:3942–3948. doi: 10.1128/jb.181.13.3942-3948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Völker U, Völker A, Haldenwang W G. Reactivation of the Bacillus subtilis anti-ςB antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J Bacteriol. 1996;178:5456–5463. doi: 10.1128/jb.178.18.5456-5463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Blohn C, Kempf B, Kappes R M, Bremer E. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol Microbiol. 1997;25:175–187. doi: 10.1046/j.1365-2958.1997.4441809.x. [DOI] [PubMed] [Google Scholar]
- 54.Wiegert T, Homuth G, Versteeg S, Schumann W. Alkaline shock induces the Bacillus subtilis ςW regulon. Mol Microbiol. 2001;41:59–71. doi: 10.1046/j.1365-2958.2001.02489.x. [DOI] [PubMed] [Google Scholar]
- 55.Yang X F, Kang C M, Brody M S, Price C W. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996;10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]