Summary
The pentatricopeptide repeat protein GENOMES UNCOUPLED1 (GUN1) is required for chloroplast‐to‐nucleus signalling when plastid translation becomes inhibited during chloroplast development in Arabidopsis thaliana, but its exact molecular function remains unknown.
We analysed GUN1 sequences in land plants and streptophyte algae. We tested functional conservation by complementation of the Arabidopsis gun1 mutant with GUN1 genes from the streptophyte alga Coleochate orbicularis or the liverwort Marchantia polymorpha. We also analysed the transcriptomes of M. polymorpha gun1 knockout mutant lines during chloroplast development.
GUN1 evolved within the streptophyte algal ancestors of land plants and is highly conserved among land plants but missing from the Rafflesiaceae that lack chloroplast genomes. GUN1 genes from C. orbicularis and M. polymorpha suppress the cold‐sensitive phenotype of the Arabidopsis gun1 mutant and restore typical retrograde responses to treatments with inhibitors of plastid translation, even though M. polymorpha responds very differently to such treatments.
Our findings suggest that GUN1 is an ancient protein that evolved within the streptophyte algal ancestors of land plants before the first plants colonized land more than 470 million years ago. Its primary role is likely to be in chloroplast gene expression and its role in chloroplast retrograde signalling probably evolved more recently.
Keywords: chloroplast, evolution, gene expression, GUN1, retrograde signalling, streptophyte
Introduction
Plant chloroplasts evolved from endosymbiotic cyanobacteria. While chloroplasts retain their own genomes, the majority of the cyanobacterial genes were lost or have moved into the nuclear genome during evolution (Timmis et al., 2004). Consequently, many chloroplast protein complexes contain both nucleus‐ and chloroplast‐encoded components (Rolland et al., 2012). Therefore, expression of the chloroplast and nuclear genes needs to be tightly coordinated to ensure optimal chloroplast function during development and under changing environmental conditions (Chan et al., 2016). This coordination of chloroplast and nuclear gene expression requires both nucleus‐to‐chloroplast signals (anterograde signals) and chloroplast‐to‐nucleus signals (retrograde signals). Chloroplast‐to‐nucleus retrograde signalling mechanisms fall into two categories: biogenic retrograde signals that operate during early chloroplast development, and operational retrograde signals that are emitted by mature chloroplasts (reviewed in Pogson et al., 2008; Chan et al., 2016; Hernández‐Verdeja & Strand, 2018; Wu & Bock, 2021).
Chloroplasts develop from small, undifferentiated plastids called proplastids. In Arabidopsis, proplastid‐to‐chloroplast differentiation takes place in young cotyledons and in specific cells of the shoot apical meristem (Charuvi et al., 2012; Wu et al., 2018). Inhibition of chloroplast gene expression during proplastid‐to‐chloroplast differentiation results in strong transcriptional repression of photosynthesis‐associated nuclear genes (PhANGs), including genes coding for light‐harvesting Chl a/b binding (LHCB) proteins and rubisco small subunit (RbcS) proteins (Harpster et al., 1984; Mayfield & Taylor, 1984).
While the exact identity of the chloroplast‐emitted retrograde signal or signals is still being debated (reviewed in Hernández‐Verdeja & Strand, 2018; Wu & Bock, 2021), the retrograde signalling mechanism that operates during proplastid‐to‐chloroplast differentiation in Arabidopsis involves the GENOMES UNCOUPLED (GUN) genes. The genomes uncoupled mutants were isolated based on their inability to repress transcription of the nuclear LIGHT HARVESTING CHLOROPHYLL A/B‐BINDING PROTEIN 1.2 (LHCB1.2) gene in response to inhibition of chloroplast translation imposed by norflurazon treatment (Susek et al., 1993; Mochizuki et al., 2001; Woodson et al., 2011). Norflurazon inhibits carotenoid biosynthesis, which results in oxidative stress to the chloroplasts and loss of chloroplast ribosomes (Bartels & Watson, 1978; Breitenbach et al., 2001). While five out of the six GUN genes (GUN2–GUN6) code for proteins involved in tetrapyrrole metabolism, the molecular mechanism of GUN1 function has remained unresolved. GUN1 is unique among the other GUN genes in that it is also required for chloroplast‐to‐nucleus signalling in response to treatments with chemicals that specifically inhibit plastid gene expression (Susek et al., 1993; Koussevitzky et al., 2007).
GUN1 codes for a chloroplast‐localized pentatricopeptide peptide repeat (PPR) protein with a C‐terminal small MutS‐related (SMR) domain (Koussevitzky et al., 2007). PPR proteins constitute one of the largest protein families in seed plants, with 450 members in Arabidopsis (Barkan & Small, 2014). PPR proteins are characterized by tandem arrays of 31–36 amino‐acid‐long PPR motifs (Lurin et al., 2004). Most previously characterized PPR proteins function as sequence‐specific RNA binding proteins. PPR proteins are encoded by the nuclear genome and almost exclusively localize to chloroplasts or mitochondria, where they bind their target RNAs. PPR proteins have been described to alter the expression of their target RNAs in a variety of ways, including RNA cleavage, splicing, editing, stabilization and translational activation (Pfalz et al., 2009; Prikryl et al., 2011; Wu et al., 2016; Aryamanesh et al., 2017; Zhou et al., 2017; Rojas et al., 2018; Lee et al., 2019).
A subset of land plant PPR proteins, the PPR‐SMR proteins, contain a C‐terminal SMR domain. The SMR domain was first characterized in the MutS2 protein of the cyanobacterium Synechocystis, where it was reported to have DNA endonuclease activity (Moreira & Philippe, 1999; Fukui et al., 2007). SMR‐domain‐containing proteins are commonly found in both prokaryotic and eukaryotic organisms, although PPR proteins with SMR domains are (apart from a few exceptions) restricted to land plants and green algae (Liu et al., 2013). The Arabidopsis genome encodes for eight PPR‐SMR proteins, five of which locate to plastids. Three of these, PLASTID TRANSCRIPTIONALLY ACTIVE2 (pTAC2), SUPPRESSOR OF THYLAKOID FORMATION1 (SOT1) and SUPPRESSOR OF VARIEGATION7 (SVR7), are among the most abundant PPR proteins in plastids (Liu et al., 2013; Mergner et al., 2020). pTAC2 is an essential component of the plastid‐encoded RNA polymerase complex and ptac2 mutants are only able to grow on sucrose‐supplemented media (Pfalz et al., 2006). SOT1 facilitates proper assembly and maturation of the plastid ribosome by processing the plastid 23S–4.5S ribosomal RNA (rRNA) precursor (Wu et al., 2016; Zhou et al., 2017). svr7 mutants also possess plastid rRNA processing and gene expression defects, most notably impaired expression of the plastid ATP synthase (Liu et al., 2010; Zoschke et al., 2013). Therefore, PPR‐SMR proteins are required for appropriate plastid gene expression.
In contrast to the other chloroplast‐localized Arabidopsis PPR‐SMR proteins, the GUN1 protein only accumulates in specific tissues (Mergner et al., 2020). While the GUN1 transcript is abundantly expressed, GUN1 protein accumulation coincides with proplastid‐to‐chloroplast differentiation in the cotyledons during early seedling development and in specific cells of the shoot apical meristem, based on GUN1‐green fluorescent protein (GFP) fusion signal (Wu et al., 2018). In addition, the GUN1‐GFP protein accumulates in response to inhibitors that induce plastid retrograde signalling (Wu et al., 2018). Several recent studies have proposed roles for the Arabidopsis GUN1 protein based on its interactions with other chloroplast‐localized proteins. These include interaction with the chaperone cpHSC70‐1 to control chloroplast protein import (Tadini et al., 2016; Wu et al., 2019), interaction with nucleus‐encoded plastid RNA polymerase (NEP) to control the accumulation of NEP‐encoded transcripts (Tadini et al., 2020), interaction with multiple tetrapyrrole biosynthesis enzymes, haem and porphyrins to control the flux through the tetrapyrrole biosynthesis pathway (Shimizu et al., 2019), and interaction with MULTIPLE ORGANELLAR RNA EDITING FACTOR2 (MORF2) to control RNA editing (Zhao et al., 2019).
The numerous proposed GUN1 interaction partners and mechanisms of function are intriguing but confusing. Current data on GUN1 function are limited to Arabidopsis, so we set out to investigate the origin and evolution of the land plant GUN1 proteins. Here we identify GUN1 as an ancient protein that is highly conserved across land plants. The pattern of amino acid conservation along the GUN1 protein is consistent with the hypothesis that GUN1, like other characterized PPR proteins, codes for a nucleic acid binding protein. Finally, the retrograde signalling cascade that functions downstream of the conserved core GUN1 mechanism is not conserved and may have evolved more recently.
Materials and Methods
Identification of GUN1 sequences
Arabidopsis and Marchantia GUN1 sequences were retrieved from TAIR (https://www.arabidopsis.org/) and MarpoIBase (https://marchantia.info/), respectively. Full‐length GUN1 sequences were obtained from a representative set of land plants by Blast searches using the Arabidopsis sequence to search GenBank and selected translated sequence sets (whole genome shotgun or transcriptome shotgun assemblies) via the NCBI Sequence Set Browser (https://www.ncbi.nlm.nih.gov/Traces/wgs/). A set of 76 phylogenetically diverse GUN1 sequences (including representatives from algae, bryophytes, lycophytes, ferns, gymnosperms and angiosperms) were aligned using the G‐INS‐i algorithm in Mafft v.7 (Katoh & Standley, 2013). The most highly conserved region of this alignment (876 positions) was used to generate a GUN1 sequence profile with hmmbuild from the hmmer package (v.3.3.1) (http://hmmer.org; Eddy, 2011), which in turn was used to search for GUN1 sequences (using hmmsearch with default parameters) in translations of various transcriptome datasets, most notably putative PPR protein sequences compiled by Gutmann et al. (2020) from the 1KP dataset (Carpenter et al., 2019; Leebens‐Mack et al., 2019). The 1KP transcriptomes were filtered to remove those encoding fewer than 10 000 distinct proteins to avoid trivial false negatives due to low coverage and those from organisms other than green algae and land plants. This resulted in 1128 analysable samples from 894 plant species. Specific searches were also made in datasets of particular interest, or where GUN1 could not be found in the corresponding 1KP samples. These additional datasets included whole genome shotgun data from Sapria himalayana (Cai et al., 2021) and whole transcriptome data from Rafflesia cantleyi (Lee et al., 2016), both holoparasites from the Rafflesiaceae.
Plant lines and growth conditions
Arabidopsis thaliana wild‐type Columbia (Col‐0) and the Atgun1 T‐DNA insertion line gun1‐102 SAIL_290_D09 (Sessions et al., 2002; Tadini et al., 2016) were used in this study. Arabidopsis seeds were sterilized with chlorine gas for 2–4 h and grown on sterile half‐strength Murashige & Skoog medium (½MS medium; PhytoTech Labs, Lenexa, KS, USA) under long‐day conditions (16 h : 8 h, light : dark) at 100 μmol m−2 s−1 at 22°C unless otherwise mentioned.
Marchantia polymorpha wild‐type accessions Takaragaike‐1 (Tak‐1) male and Tagarakaike‐2 (Tak‐2) female were kindly provided by Prof. John Bowman, Monash University. Marchantia polymorpha CRISPR knockout lines Mpgun1‐1 and Mpgun1‐2 were generated in this study. Marchantia polymorpha plants were grown on sterile half‐strength Gamborg's medium (Duchefa Biochemie, Haarlem, the Netherlands) supplemented with 1.2% agar under long‐day conditions (16 h : 8 h, light : dark) unless otherwise mentioned. Crossing and spore sterilization were carried out as described in Supporting Information Methods S1.
Generation of A. thaliana gun1 mutant complementation lines
All primer sequences are provided in Table S1. Annotated sequences of the complementation constructs are provided in Notes S1. AtGUN1, MpGUN1 and CoGUN1 protein coding sequences were aligned using Geneious (Kearse et al., 2012). The region containing chloroplast transit peptides was estimated based on lack of conservation between the protein sequences (Fig. S1). The MpGUN1 coding sequence excluding the putative chloroplast transit peptide‐encoding region (amino acids 1–114) was amplified from M. polymorpha cDNA using PrimeSTAR HS DNA polymerase (Takara Bio, Shiga, Japan) with primers CDS_MpGUN1_At_gib_F and CDS_MpGUN1_At_gib_R. The CoGUN1 coding sequence excluding the chloroplast transit peptide‐encoding region (amino acids 1–145) was ordered as a gene synthesis fragment from Integrated DNA Technologies (IDT, Coralville, IA, USA). A fragment containing the AtGUN1 promoter, 5′ untranslated region and chloroplast transit peptide (amino acids 1–135) was amplified from Arabidopsis Col‐0 genomic DNA with primers pro_AtGUN1_gib_F and tp_AtGUN1_Mp_gib_R for the MpGUN1 construct, and primers pro_AtGUN1_gib_F and tp_AtGUN1_Co_gib_R for the CoGUN1 construct. The AtGUN1 terminator fragment was amplified with primers term_AtGUN1_Mp_gib_F and term_AtGUN1_gib_R for the MpGUN1 construct and with primers term_AtGUN1_Co_gib_F and term_AtGUN1_gib_R for the CoGUN1 construct. The three fragments for each construct were assembled on a XbaI‐ and SacI‐digested gel‐purified MpGWB101 vector (Ishizaki et al., 2015) using Gibson assembly. To create a control construct for complementation of the Atgun1 mutant with AtGUN1 genomic fragment, the wild‐type AtGUN1 gene sequence was amplified from Col‐0 DNA using primers pro_AtGUN1_gib_F and term_AtGUN1_gib_R and cloned onto the XbaI‐ and SacI‐linearized MpGWB101 vector backbone.
Arabidopsis thaliana wild‐type and gun1 mutant plants were transformed using the floral dip method (Zhang et al., 2006) with binary vectors described above using Agrobacterium strain GV3101. Seeds were surface‐sterilized and sown on sterile plates supplemented with 15 μg ml−1 hygromycin. After 1 wk, hygromycin‐resistant plants were transferred to soil. The genotype of Arabidopsis wild‐type, gun1 and gun1 complemented with AtGUN1, MpGUN1 and CoGUN1 was PCR‐verified using the Phire plant direct PCR kit (Thermo Fisher, Waltham, MA, USA) as recommended by the manufacturer. The presence of the T‐DNA insertion was verified using primers LBb1.3 and SAIL_280_D09_LP, whereas the presence/absence of intact wild‐type GUN1 sequence was verified with primers SAIL_280_D09_LP and SAIL_280_D09_RP. Seeds from 10 to 15 independent transformant lines were observed for the initial phenotypic assessment. All final phenotype and gene expression analyses were carried out on nonsegregating T2 or T3 seeds of three independent homozygous transformant lines for each construct.
Marchantia polymorpha transformation and generation of transgenic Mpgun1 CRISPR/Cas9 knockout lines
Marchantia polymorpha transformation and generation or transgenic Mpgun1 CRISPR/Cas9 knockout lines are described in Methods S1 and S2.
Plant growth experiments
Lincomycin treatment of A. thaliana
Arabidopsis seeds were sterilized and plated on sterile ½MS medium pH 5.7, 2% sucrose and 0.8% agar. For the lincomycin treatment, the growth medium was supplemented with 220 μg ml−1 lincomycin (Sigma‐Aldrich). The seeds were stratified at 4°C for 2 d, after which the plants were grown at 22°C under continuous light at 100 μmol m−2 s−1 for 5 d before imaging or harvesting the tissue for RNA extraction.
Combined low light and lincomycin treatment of A. thaliana
Arabidopsis seeds were sterilized and plated on sterile ½MS medium pH 5.7 (without sucrose) and 0.8% agar. For the lincomycin treatment the growth medium was supplemented with 220 μg ml−1 lincomycin (Sigma‐Aldrich). The seeds were stratified at 4°C for 4 d, and then grown at 22°C under continuous light at 1 μmol m−2 s−1 for 3 d before imaging.
Arabidopsis thaliana cold growth experiment
Arabidopsis seeds were sterilized and plated on ½MS medium pH 5.7 (without sucrose) and 0.8% agar. The plates were placed directly at 4°C in a growth room under long‐day conditions (8 h : 16 h, dark : light) at 100 μmol m−2 s−1. Plants were imaged after 7 wk.
Spectinomycin and norflurazon treatments of M. polymorpha spores
Marchantia polymorpha spores were sterilized and plated on ½ Gamborg's medium (Duchefa Biochemie) supplemented with 1.2% agar and 500 μg ml−1 spectinomycin or 5 μM norflurazon. The plates were placed under long‐day conditions for 48 h, after which the spores were resuspended in 1 ml of sterile water, transferred to a microcentrifuge tube and spun down at 3400 g for 1 min. The water was removed, and the spore pellet wasflash‐frozen in liquid nitrogen.
RNA extraction
Extraction of total RNA from Arabidopsis seedlings and Marchantia spores was carried out using the Direct‐Zol RNA MINIprep kit (Zymo Research, Irvine, CA, USA) following the manufacturer's protocol. Three independent biological replicates were extracted for each line. RNA was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific) and diluted to 250 ng μl−1 before DNase treatment using Turbo DNase (Ambion, Austin, TX, USA) as recommended by the manufacturer.
cDNA synthesis and quantitative real‐time PCR (qPCR)
One microgram of DNase‐treated RNA was used as a template for cDNA synthesis. cDNA was generated with oligo dT18 primer using the Protoscript II reverse transcriptase (NEB, Ipswich, MA, USA) in the presence of Murine RNase inhibitor (NEB). cDNA was diluted 1 : 5 before qPCR. qPCR was carried out using SYBR Premix Ex Taq II (Takara Bio) qPCR reagent. Each 5 μl reaction contained 1 μl primer mix, 1.5 μl cDNA dilution and 2.5 μl Sybr II master mix. Each biological replicate sample was run in three technical replicates. Amplification was carried out in a LightCycler480 instrument (Roche) using the following cycling conditions: initial denaturation for 1 min at 95, then 40 cycles of 10 s at 95°C, 10 s at 60°C and 20 s at 72°C. For primer sequences see Table S1. The data were analysed using LinRegPCR (Ruijter et al., 2009) v.2017.1. The expression of each gene of interest was first separately normalized against the reference genes AtPP2AA3 and AtUBQ10 for Arabidopsis and MpEF1α and MpACT for M. polymorpha. The geometric mean of the two normalized values was then recorded as the expression level.
RNA sequencing
Transcriptome libraries of M. polymorpha wild‐type and Mpgun1 mutant spores were prepared using 200 ng of DNase‐treated total RNA as a template for the TruSeq Stranded Total RNA kit with Ribo‐Zero Plant (Illumina, San Diego, CA, USA). The libraries were sequenced on an Illumina HiSeq 4000 platform (150 nt paired‐end reads) at Novogene, Hong Kong. At least 5.7 GB of raw data was obtained for each replicate library. Sequencing read data were deposited at the Short Read Archive database at the National Center for Biotechnology Information under project nos. PRJNA800059 and PRJNA838206.
Optical duplicate reads were first removed with clumpify (parameters: dedupe optical dist = 40) from the bbmap package (https://sourceforge.net/projects/bbmap/) and adapters were trimmed with bbduk (parameters: ktrim = r k = 23 mink = 11 hdist = 1 tpe tbo ftm = 5). The reads were then assigned to transcripts using Salmon v.1.3.0 (Patro et al., 2017) (parameters: ‐l A ‐‐validateMappings) against an index prepared with the M. polymorpha MpTak_v.6.1 reference genome and cDNA assemblies (https://marchantia.info/). Differential expression analyses were carried out using DESeq2 (Love et al., 2014). Functional annotations for the MpTak_v.5.1 genome release were used to annotate differentially expressed genes (log2 fold‐change > 1 or < −1 and P adj < 0.01) and to identify M. polymorpha photosynthesis‐associated nuclear genes. Gene Ontology (GO) enrichment analyses were performed on the Dicots Plaza 4.5 platform (Van Bel et al., 2018) using standard settings with differentially expressed genes showing log2 fold‐change > 1 or < −1 and P adj < 0.01 as an input.
Microscopy and image analysis
Arabidopsis seedlings and M. polymorpha gemmae were imaged using an Olympus Camedia C 7070 camera mounted on a Leica SZ61 dissecting microscope or with an Apple iPad (7th generation). Transmitted light micrographs of M. polymorpha spores were obtained using an Olympus Camedia C 7070 camera mounted on a Olympus BX51 microscope. Image analysis to quantify phenotypic differences was carried out using Fiji (Schindelin et al., 2012). Images were adjusted using Adobe Photoshop 2020.
Accession numbers
The following Arabidopsis genes were mentioned in this article: AtGUN1, AT2G31400; AtLHCB1.2, AT1G29910; AtLHCB2.2, AT2G05070; AtCA1, AT3G01500; and AtCP12, AT3G62410.
The following M. polymorpha genes were mentioned in this article: MpGUN1, Mp1g08430; MpLHCB2, Mp7g06790; MpRBCS, Mp4g10850; and MpFBPase, Mp6g13790.
Results
GUN1 evolved within the streptophyte algae and is conserved among land plants
To discover when GUN1 arose and its distribution within the plant lineage, we searched for GUN1‐like sequences using the Arabidopsis GUN1 (AtGUN1) sequence as a query in Blastp (Altschul et al., 1990) searches of the GenBank nonredundant protein database. Probable GUN1 sequences were verified by alignment to AtGUN1, by reciprocal Blastp to Arabidopsis protein sequences and by identification of the expected PPR motifs and C‐terminal SMR domain. Land plants form a monophyletic group that evolved from the streptophyte algae (Wickett et al., 2014). We identified putative GUN1 sequences in the streptophyte algal groups most closely related to land plants (Zygnematales, Coleochaetales and Charales), but not in other streptophyte algal groups (Klebsormidiales, Mesostigmatales and Chlorokybales) nor in other green algae (Chlamydomonadales) (Fig. S2) This suggests the GUN1 protein is ancient and evolved in the streptophyte algal ancestors of land plants before the first plants colonized land.
To systematically assess the conservation of GUN1 among plants, we selected 76 phylogenetically diverse full‐length GUN1 sequences from a representative set of land plants and streptophyte algae (Figs 1, S2), aligned them and retained the most conserved region of the alignment (which included all of the PPR motifs and the SMR domain). From this alignment we developed a sequence profile using hmmbuild (http://hmmer.org; Eddy, 2011) and used it to screen c. 500 000 PPR sequences derived from the 1KP plant transcriptome dataset (Carpenter et al., 2019; Leebens‐Mack et al., 2019; Gutmann et al., 2020) with hmmsearch. Putative GUN1 orthologues could be distinguished from other PPR‐SMR sequences based on the domain score reported for each match by hmmsearch (Fig. S3; Table S2). GUN1 transcripts were detected in 824 of the 894 species analysed. We identified conserved GUN1 sequences in 345 of the 366 plant families represented in this dataset. Notably, we found that transcriptomes of nonphotosynthetic parasitic plants with highly reduced plastid genomes, such as Pilostyles thunbergii (Bellot & Renner, 2016), Balanophora fungosa (Su et al., 2019) and Conopholis americana (Wicke et al., 2013), encode GUN1. Of the 21 families apparently lacking GUN1, 14 are green algal families outside the streptophyte clades closely related to land plants, and thus not expected to contain GUN1. One of the remaining seven is represented in the original set of 76 sequences used to build the GUN1 profile, and thus does contain GUN1, even though no GUN1 transcripts were detected in the 1KP sample. This left six land plant families expected to contain GUN1 (Agapanthaceae, Cyrillaceae, Hymenophyllaceae, Juglandaceae, Monocleaceae, Thelypteridaceae) for which the data so far did not provide clear evidence of GUN1 sequences (Table S2). After searching in additional whole transcriptome or whole genome shotgun sequencing datasets, we were able to identify putative GUN1 sequences from Agapanthus, Hymenophyllum and Juglans. For the three remaining families, each represented by only a single sample in our 1KP dataset, we were unable to find additional data to search. We also searched whole transcriptome shotgun sequences from R. cantleyi (Lee et al., 2016) and whole genome shotgun sequences from S. himalayana (Cai et al., 2021) (both Rafflesiaceae). These two closely related genera are reported to lack a plastid genome. We found no putative GUN1 sequences in either dataset. Sapria himalayana is the only embryophyte for which we were unable to find a putative GUN1 gene in its genome, and R. cantleyi one of the few for which we were unable to find putative GUN1 transcripts in its transcriptome.
Fig. 1.
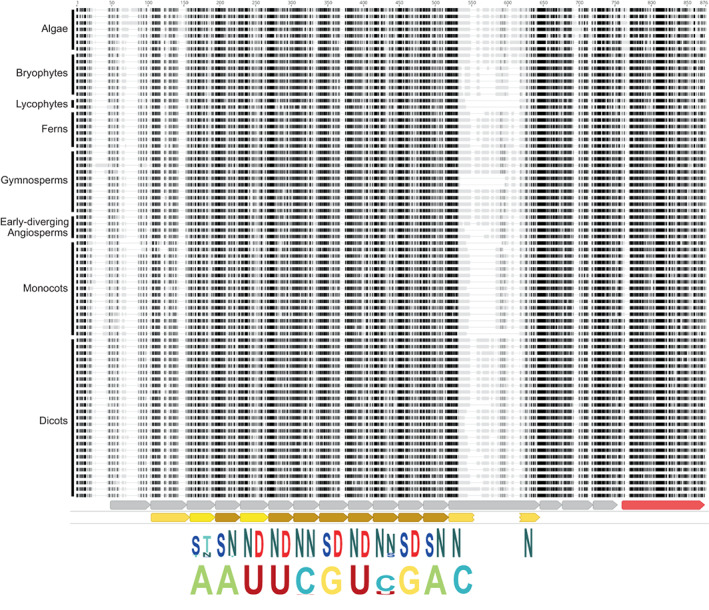
GUN1 is conserved in most streptophytes. The top part of the figure represents a multiple alignment of 76 GUN1 protein sequences from diverse streptophyte algae and land plants. The alignment was constructed using Mafft and visualized with Geneious. Only the central, most conserved region of the alignment is shown (corresponding to positions 135–888 in AtGUN1). Darker shading indicates higher similarity to the consensus. The sequences are grouped (approximately) by phylogenetic relationship as indicated; for the species names and amino acid sequences, see the alignment in Supporting Information Fig. S2. Below the alignment, the annotation tracks show helix–turn–helix motifs (grey arrows) and SMR domain (red arrow) predicted by AlphaFold and PPR motifs (brown arrows) predicted by hmmsearch using a P‐type PPR motif HMM. Dark brown motifs have higher confidence. The final PPR motif is interrupted by a long insertion but alphafold predicts that the N‐ and C‐terminal helices nevertheless interact together as in a typical PPR motif. Below these motif predictions, the fifth and last amino acids of the conserved PPR motifs are indicated as sequence logos generated by Skylign (Wheeler et al., 2014) (letter height represents information content) from the alignment of 893 GUN1 sequences (the 76 sequences shown here plus 817 sequences from the 1KP PPR dataset). Finally, at the bottom of the figure, the expected RNA binding specificities of the conserved PPR motifs are indicated, again as a Skylign sequence logo. The HMM profile and full alignment of all 893 identified GUN1 sequences are obtainable from Dryad (10.5061/dryad.x0k6djhmk).
The predicted binding specificity of GUN1 proteins is conserved
Conservation of protein function is likely to depend on the level of sequence conservation in the functionally important regions of the protein. The GUN1 protein consists of 12–16 PPR motifs (depending on the approach used to recognize and define the motifs) and a C‐terminal SMR domain. Fig. 1 shows the alignment of the conserved regions of the GUN1 sequences used to generate the HMM profile, and the structural motifs that correspond, as predicted by AlphaFold (Jumper et al., 2021; Varadi et al., 2022) or hmmsearch (Eddy, 2011). AlphaFold predicts 16 helix–turn–helix motifs (Fig. S4) of which 12 are recognized as PPR motifs by hmmsearch with a P‐type PPR HMM (Cheng et al., 2016). The first helix–turn–helix motif predicted by AlphaFold is poorly conserved between species, but the final three are highly conserved within GUN1 sequences although quite divergent in sequence and length from typical PPR motifs.
The binding specificity of PPR proteins can be predicted based on the 5th and last amino acid of each PPR motif (Barkan et al., 2012). The amino acids at these positions of PPR motifs 2–12 are highly conserved between all land plant and streptophyte algae GUN1 protein sequences (Figs 1, S2). When these amino acid combinations are used to predict the nucleotide most likely to be bound by each motif, this conservation is even more striking, with the only predicted variation being at motifs 6 and 9 where different combinations are predicted to bind either C or U. This may not actually reflect functional divergence, as any of the combinations are likely to have high affinity for both C and U (Yan et al., 2019). The structure predicted by AlphaFold forms a contiguous solenoid similar to that of other PPR proteins that are known to bind RNA, albeit with some idiosyncracies. The conservation of the PPR motifs, and in particular the residues that in other PPR proteins define RNA binding specificity, is consistent with the hypothesis that GUN1 codes for an RNA binding protein with a highly conserved target.
GUN1 SMR domains are conserved
The GUN1 protein contains a C‐terminal SMR domain that is highly conserved between all the full‐length GUN1 sequences in our dataset. Four SMR‐domain‐containing proteins have been reported to have endonuclease activity in other organisms (Liu et al., 2013). Subfamily 2 SMR proteins that have endonuclease cleavage activity have two conserved motifs: the LDXH motif in the N‐terminus of the SMR domain and a central TGXG motif (Watanabe et al., 2003; Bhandari et al., 2011). The central TGXG motif is perfectly conserved as TGWG in all full‐length GUN1 proteins in our dataset. The N‐terminal LDXH motif is also almost perfectly conserved as LDLH, with a scattering of exceptions that have VDLH in this position. These observations show that the SMR domains of land plant GUN1 proteins are conserved and probably contribute to GUN1 function.
GUN1 genes from the streptophyte alga Coleochate orbicularis and the liverwort M. polymorpha restore chloroplast retrograde signalling in the Atgun1 mutant
As the predicted binding specificity of GUN1 proteins is conserved among land plants and in the streptophyte algae, we hypothesized that GUN1 proteins are also functionally conserved. To test this hypothesis, we assessed whether GUN1 genes from the streptophyte alga C. orbicularis and the liverwort M. polymorpha can complement the Arabidopsis gun1 knockout mutant. Coleochate orbicularis and M. polymorpha each contain a single‐copy GUN1 gene. The C. orbicularis GUN1 protein shares 42.5% amino acid identity (57.2% match using the BLOSUM62 substitution matrix) with AtGUN1, while the M. polymorpha GUN1 protein is 48.4% identical (66.4% match using BLOSUM62) with AtGUN1. Sequences corresponding to chloroplast transit peptides in CoGUN1 and MpGUN1 were estimated based on lack of sequence conservation (Fig. S1) and replaced with the Arabidopsis GUN1 transit peptide. These coding sequences were then expressed from the AtGUN1 promoter in the Atgun1 (SAIL_290_D09) mutant background. As a control the Atgun1 mutant was complemented with an AtGUN1 genomic fragment.
In wild‐type Arabidopsis, inhibition of plastid translation during chloroplast development activates chloroplast retrograde signalling that results in strong transcriptional repression of PhANGs. In the Atgun1 mutant this transcriptional repression is defective, although not completely abolished (Fig. 2; Koussevitzky et al., 2007). We hypothesized that CoGUN1 and MpGUN1 can restore the GUN1‐dependent repression of PhANGs in the Atgun1 mutant when plastid translation is inhibited. To test this hypothesis, we germinated the Atgun1 seeds expressing CoGUN1, MpGUN1 or AtGUN1 on media supplemented with 200 μg ml−1 lincomycin, an inhibitor of plastid translation. As expected, the inhibition of plastid translation by lincomycin resulted in strong reduction in steady‐state transcript levels of four PhANGs, namely AtLHCB1.2, AtLHCB2.2, AtCA1 and AtACP12 in wild‐type Arabidopsis, but less so in the Atgun1 mutant (Fig. 2). Expression of CoGUN1 and MpGUN1 in the Atgun1 mutant background restored the repression of these four PhANG transcripts to wild‐type levels (Fig. 2). These results indicate that CoGUN1 and MpGUN1 can activate the AtGUN1‐dependent retrograde signalling cascade that leads to transcriptional repression of PhANGs in Arabidopsis.
Fig. 2.
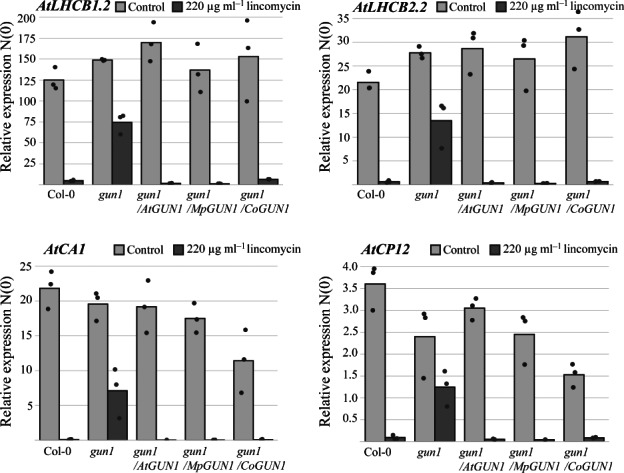
CoGUN1 or MpGUN1 expression in the Atgun1 mutant restores the repression of photosynthesis‐associated nuclear genes (PhANGs) in response to inhibition of plastid translation by lincomycin treatment. qPCR quantification of steady‐state transcript levels of four PhANGs, AtLHCB1.2, AtLHCB2.2, AtCA1 and AtACP12, in wild‐type Arabidopsis, the Atgun1 mutant, and Atgun1 mutant complemented with AtGUN1 (control), MpGUN1 or CoGUN1. The transcript levels were normalized against AtPP2AA3 and AtUBQ10. Each dot represents an independent biological replicate, and bars illustrate the average of the three biological replicates.
C. orbicularis and M. polymorpha GUN1 restore most aspects of the Atgun1 mutant phenotype
As CoGUN1 and MpGUN1 can complement the AtGUN1‐mediated repression of PhANGs when expressed in the Atgun1 mutant, we investigated whether other aspects that involve GUN1‐mediated signalling are also complemented.
Wild‐type Arabidopsis seedlings germinated on media supplemented with lincomycin in the presence of 2% sucrose develop purple cotyledons due to accumulation of anthocyanins. This anthocyanin accumulation is defective in the Atgun1 mutant (Fig. 3; Cottage et al., 2010). Atgun1 seedlings complemented with CoGUN1 or MpGUN1 develop similar purple coloration to wild‐type seedlings when germinated on lincomycin‐containing media (Fig. 3a). Treatment with lincomycin also inhibits cotyledon expansion in wild‐type Arabidopsis seedlings more than in Atgun1 mutant seedlings. CoGUN1 and MpGUN1 restore the inhibition of cotyledon expansion in response to inhibition of plastid translation by lincomycin (Fig. 3b). These findings are consistent with the hypothesis that CoGUN1 and MpGUN1 can activate AtGUN1‐dependent downstream signalling that leads to upregulation of anthocyanin biosynthesis and inhibition of cotyledon expansion when plastid translation is inhibited.
Fig. 3.
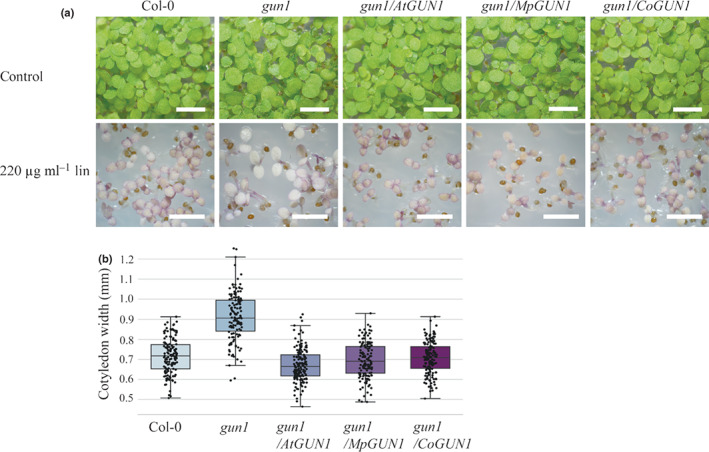
CoGUN1 and MpGUN1 expression in the Atgun1 mutant restores anthocyanin accumulation and repression of cotyledon expansion in response to lincomycin treatment. (a) Five‐day‐old seedlings of wild‐type Arabidopsis, Atgun1 mutant, and Atgun1 mutant complemented with AtGUN1 (control), MpGUN1 or CoGUN1 grown under constant light on sterile plates for 7 d (top row, Bar, 4 mm) or on media supplemented with 220 μg ml−1 lincomycin for 5 d (bottom row, Bar, 3 mm). (b) Quantification of cotyledon width of seedlings grown on media supplemented with 200 μg ml−1 lincomycin for 5 d. Centre line indicates the mean, box limits indicate the upper and lower quartiles, and whiskers indicate the data range. Dots represent individual measurements. All other lines were significantly different from gun1 (Tukey's HSD: P < 0.001).
AtGUN1 is also required for cold tolerance during germination; germinating Atgun1 mutant seedlings are hypersensitive to cold (Marino et al., 2019). This cold‐sensitive phenotype is barely noticeable in the cotyledons but becomes striking when the first true leaves appear after 6–7 wk of growth at 4°C (Fig. 4). True leaves of wild‐type plants germinated at 4°C are green and expanded, while the true leaves of the Atgun1 mutant are small, narrow and bleached. To check if CoGUN1 and MpGUN1 can restore the cold‐sensitive phenotype of the Atgun1 mutant, we germinated the complemented seeds on agar plates at 4°C. Both CoGUN1 and MpGUN1 improved the cold tolerance of the Atgun1 mutant (Fig. 4). However, CoGUN1 only partially restored cold tolerance: the first true leaves that emerged expanded and greened more than those of the gun1 mutant but were variegated. These findings indicate that MpGUN1 and CoGUN1 can at least partially complement the cold‐sensitive phenotype of the Arabidopsis gun1 mutant.
Fig. 4.

MpGUN1 and CoGUN1 complement the cold‐sensitive phenotype of the Atgun1 mutant. Phenotype of 7‐wk‐old seedlings germinated at 4°C under long‐day light regime. Bar, 4 mm.
Arabidopsis seedlings germinated in the absence of light (such as under soil) develop an etiolated phenotype characterized by an elongated hypocotyl, closed cotyledons and differentiation of proplastids into nongreen etioplasts. Upon illumination (or emerging from the soil), etiolated seedlings undergo deetiolation: hypocotyl elongation becomes inhibited, the cotyledons expand and etioplasts differentiate into photosynthetic chloroplasts. AtGUN1‐dependent signalling can be detected when plastid translation is inhibited during germination under low‐light conditions. Wild‐type Arabidopsis seedlings germinated on lincomycin‐containing media under low‐light conditions resemble dark‐grown seedlings: their hypocotyls become elongated and cotyledons remain closed (Fig. 5a–c). The Atgun1 mutant is defective in this response, indicating that inhibition of deetiolation in response to inhibition of plastid translation is dependent on functional GUN1 protein (Martín et al., 2016). CoGUN1 and MpGUN1 expressed in the Atgun1 mutant background restore the inhibition of deetiolation in response to inhibition of plastid translation by lincomycin (Fig. 5). This finding suggests that CoGUN1 and MpGUN1 can replace AtGUN1 during deetiolation.
Fig. 5.
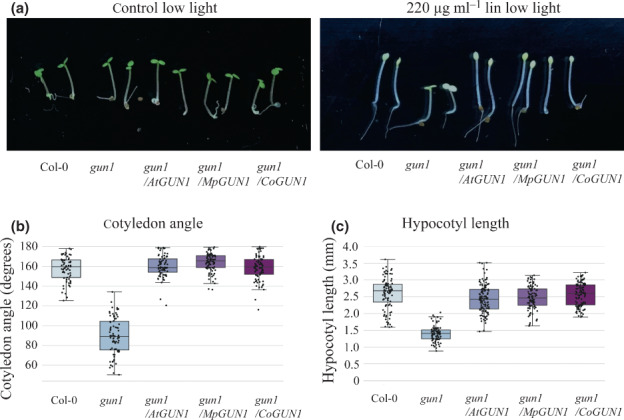
CoGUN1 and MpGUN1 expression in the Atgun1 mutant restores the repression of deetiolation when plastid translation is inhibited by lincomycin during germination. (a) Wild‐type Arabidopsis, Atgun1, and Atgun1 mutant complemented with AtGUN1 (control), MpGUN1 or CoGUN1 germinated for 3 d under constant low light (1 μmol m−2 s−1) on sterile plates without (control) or with 220 μg ml−1 lincomycin. (b) Quantification of cotyledon angle in seedlings grown on lincomycin as in (a). (c) Quantification of hypocotyl length in seedlings grown on lincomycin as in (a). In (b) and (c) the centre line indicates the mean, box limits indicate the upper and lower quartiles, and whiskers indicate the data range. Dots represent individual measurements. All other lines were significantly different from gun1 (Tukey's HSD: P < 0.001).
Tgether, our results indicate that GUN1 proteins from streptophyte algae, such as C. orbicularis, and distantly related land plants, such as M. polymorpha, can functionally replace the Arabidopsis GUN1 protein.
Mpgun1 mutants are indistinguishable from wild‐type under nonstressful conditions
To investigate if GUN1 genes are functionally conserved among land plants, we generated two different CRISPR deletion lines of the single‐copy GUN1 gene in the liverwort M. polymorpha (Fig. 6a). The resulting Mpgun1‐1 and Mpgun1‐2 deletion mutant plants were PCR‐genotyped, and PCR products of selected plants were sequenced to confirm the deletion introduced a premature stop codon (Fig. S5). Male and female plants for each line were then crossed together to obtain nonsegregating populations of Mpgun1‐1 and Mpgun1‐2 knockout mutant spores.
Fig. 6.
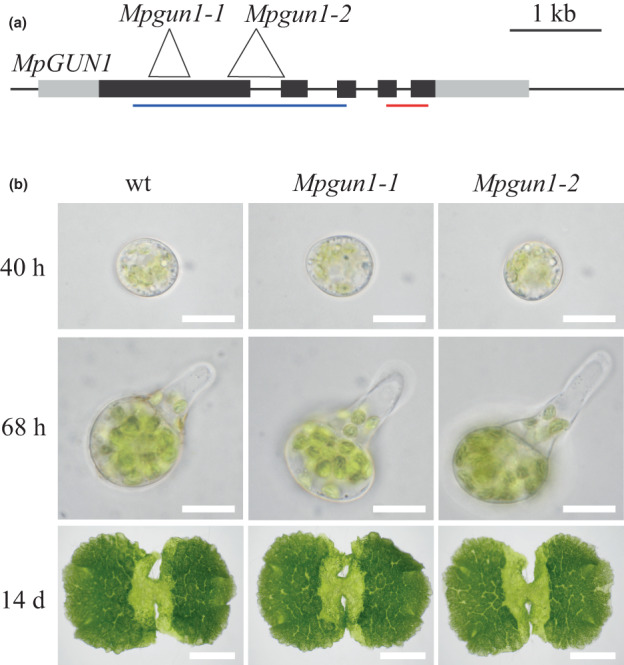
The Mpgun1 mutant phenotype is indistinguishable from the wild‐type phenotype. (a) Gene model showing the structure of the Marchantia polymorpha GUN1 gene. Exons are indicated as black boxes, and noncoding regions are indicated as grey boxes. The locations of the CRISPR‐induced deletions are shown as white triangles. Blue and red lines indicate the region containing the PPR motifs and the SMR domain, respectively. (b) 40‐h‐old (top row, Bar, 20 μm) or 68‐h‐old (middle row, Bar, 20 μm) germinating spores or 14‐d‐old gemmae (bottom row, Bar, 3 mm) of M. polymorpha wild‐type and Mpgun1 mutants.
To check if GUN1 is involved in chloroplast development in M. polymorpha, we observed germination of wild‐type and Mpgun1 mutant spores. Wild‐type M. polymorpha spores begin germination by cell expansion. After the first 24 h, Chl autofluorescence can be observed from the chloroplasts (Bowman et al., 2017). By 48 h the spores have become fully expanded and chloroplasts appear fully developed. After 48 h the spores undergo the first asymmetric cell division, followed by the emergence of the first rhizoid from the smaller daughter cell. Sometimes the emergence of the first rhizoid precedes the first cell division. The Mpgun1‐1 and Mpgun1‐2 knockout mutant spores are phenotypically indistinguishable from wild‐type spores during spore germination (Fig. 6b). This suggests that MpGUN1 is not required for chloroplast development under nonstressful conditions.
Interestingly, dark‐grown M. polymorpha spores develop green chloroplasts (Fig. S6). This indicates that unlike in Arabidopsis, proplastid‐to‐chloroplast differentiation in M. polymorpha spores is independent of light signals. Consequently, proplastid‐to‐etioplast and etioplast‐to‐chloroplast transitions do not take place in M. polymorpha spores and therefore we conclude that MpGUN1 is not involved in these processes.
MpGUN1 is not involved in global repression of PhANGs in response to inhibition of plastid translation
The Arabidopsis GUN1 protein is required for chloroplast retrograde signalling and the M. polymorpha GUN1 protein can restore retrograde signalling in the Arabidopsis gun1 mutant. Therefore, we hypothesized that GUN1 might also be involved in chloroplast retrograde signalling in M. polymorpha. To test this hypothesis, we tested if germinating Mpgun1 mutant spores are defective in chloroplast retrograde signalling when plastid translation is inhibited. Marchantia polymorpha spores are almost fully resistant to lincomycin. Therefore, we used another inhibitor of plastid translation, spectinomycin, which effectively blocks M. polymorpha spore development (Fig. S6). Spectinomycin is commonly used as a selectable marker in plastid‐transformation protocols, including in M. polymorpha (Chiyoda et al., 2006; Boehm et al., 2016), and its mechanism of function is well‐characterised (Ellis, 1970; Parker et al., 2014). Marchantia polymorpha spores that were germinated on media containing 500 μg ml−1 spectinomycin expanded and developed chloroplasts, but never grew a rhizoid or underwent the first cell division (Fig. S6). This is consistent with the hypothesis that spectinomycin effectively inhibits plastid translation in M. polymorpha. We then checked if inhibition of plastid translation by spectinomycin during chloroplast development in M. polymorpha leads to reduced transcript levels of PhANGs or other transcripts that are regulated by plastid retrograde signalling in seed plants. To this end, we prepared transcriptomes of wild‐type and Mpgun1 knockout spores germinated in the absence or presence of spectinomycin. The spores were grown under long‐day conditions and harvested 48 h after plating. Read coverage across the MpGUN1 transcript was checked to verify the lack of intact MpGUN1 sequence in Mpgun1 mutant transcriptomes (Fig. S7). Wild‐type spores germinated on spectinomycin had reduced PhANG transcript levels compared to the control (Fig. 7; Table S3). However, the reduction in PhANG transcript levels was much less profound than that observed in wild‐type Arabidopsis under similar conditions, and Mpgun1 mutant spores were not defective in this response (Fig. 7; Table S3). GO terms associated with photosynthesis were over‐represented among the genes downregulated in response to spectinomycin in both wild‐type and Mpgun1‐1 mutant spores (Table S4). Furthermore, the set of genes that was differentially expressed in the Mpgun1‐1 mutant compared to wild‐type on spectinomycin was not enriched in photosynthesis‐associated GO terms, except for RbcS‐encoding transcripts, which were more abundant in Mpgun1‐1 compared to the wild‐type (Table S4). Under control conditions, the levels of PhANG transcripts did not differ markedly in wild‐type and Mpgun1 mutant spores (Fig. 7). These findings indicate that inhibition of plastid translation does not induce MpGUN1‐dependent global repression of nuclear encoded photosynthesis‐associated genes in M. polymorpha.
Fig. 7.
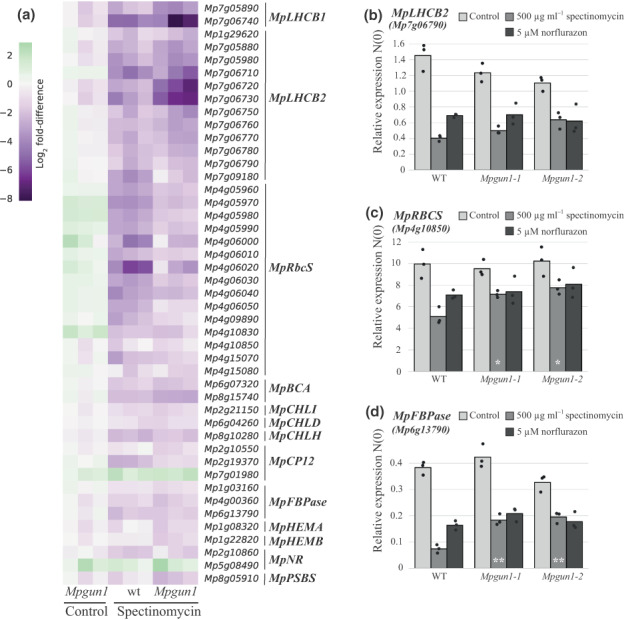
In Marchantia polymorpha inhibition of chloroplast translation by spectinomycin treatment or chloroplast stress imposed by norflurazon treatment does not result in GUN1‐mediated downregulation of the same set of genes as in seed plants. (a) Transcript levels of M. polymorpha orthologues of seed plant plastid retrograde signalling‐regulated genes LHCB1, LHCB2, RbcS, Beta‐CA (BCA), CHLI, CHLD, CHLH, CP12, FBPase, HEMA, HEMB, NR and PSBS in wild‐type and Mpgun1 mutant M. polymorpha spores germinated on control media or media supplemented with 500 μg ml−1 spectinomycin. Transcript abundances are shown as log2 fold‐difference compared to wild‐type M. polymorpha spores grown under control conditions. Each column represents an independent biological replicate transcriptome. (b–d) qPCR quantification of PhANG transcript levels in wild‐type and Mpgun1 mutant spores germinated on spectinomycin or norflurazon‐containing media. The transcript levels were normalized against MpEF1α and MpACT. Each dot represents an independent biological replicate, and bars illustrate the average of the three biological replicates. White asterisks indicate statistically significant differences between Mpgun1 and wild‐type for the same treatment (Tukey's HSD: *, P < 0.05; **, P < 0.01). The code and source data for reproducing (a) are obtainable from Dryad (10.5061/dryad.x0k6djhmk).
To independently verify that GUN1 is not required for inhibition of PhANG expression in response to inhibition of plastid translation in M. polymorpha, we carried out qPCR quantification of PhANG transcript levels in wild‐type and Mpgun1 mutant spores germinated on spectinomycin‐ or norflurazon‐containing media. Norflurazon is an inhibitor of carotenoid biosynthesis. Inhibition of carotenoid biosynthesis results in oxidative stress to the chloroplasts and leads to repression of PhANG expression in Arabidopsis. Marchantia polymorpha spores germinated on 5 μM norflurazon became bleached and their development arrested at a single‐cell stage (Fig. S6), suggesting that norflurazon probably also results in oxidative stress to the chloroplasts in M. polymorpha. We quantified the transcript levels of three PhANGs, LHCB2, RBCS and FBPase, in spectinomycin‐ and norflurazon‐treated wild‐type, Mpgun1‐1 and Mpgun1‐2 mutant spores. These PhANGs are present in M. polymorpha as multicopy genes. Therefore, primers were designed to amplify the most abundantly transcribed isoform in each gene group. Transcript levels of these PhANGs were reduced in spores germinated on spectinomycin or norflurazon, but Mpgun1‐1 or Mpgun1‐2 mutant spores were not clearly defective in this response (Fig. 7b–d). RBCS and FBPase transcripts were more abundant in gun1‐1 and gun1‐2 mutant spores germinated on spectinomycin compared to wild‐type spores (Fig. 7b–d). These differences were statistically significant, but we did not observe them in our transcriptome experiment, suggesting they may not be biologically significant. These findings indicate that GUN1 is not involved in strong transcriptional repression of PhANGs in response to inhibition of plastid translation in the liverwort M. polymorpha.
To elucidate the effects of the Mpgun1 mutation on gene expression during chloroplast development, we carried out further transcriptome analysis of germinating M. polymorpha wild‐type and gun1 mutant spores. In Arabidopsis the GUN1 protein accumulates in tissues in which proplastids differentiate into chloroplasts, including young cotyledons and shoot apical meristem 36–72 h after germination (Wu et al., 2018). As proplastid to chloroplast differentiation in germinating M. polymorpha spores takes place during the first 48 h, we harvested spores 40 h after germination when the effects of the Mpgun1 mutation could be expected to be detectable.
Transcript levels of 575 genes were significantly different between Mpgun1 mutant and wild‐type spores (log2 fold‐change > 1 or < −1, adjusted P‐value < 0.01, Table S5). Most notably, 11 out of 16 transcripts encoding putative chloroplast Chl a/b binding proteins homologous to Arabidopsis early light‐induced proteins (ELIPs) were less abundant in Mpgun1 spores compared to wild‐type spores. ELIPs are thylakoid‐associated stress‐induced proteins that appear earlier than other photosynthesis‐associated proteins during the early stages of deetiolation, and disappear before chloroplast development is completed (Grimm et al., 1989). By contrast, six out of 10 transcripts encoding putative chloroplast alpha‐carbonic anhydrases were more abundant in Mpgun1 compared to the wild‐type. The homologues of neither of the gene families are significantly differentially expressed in comparisons of deetiolation of Arabidopsis wild‐type and Atgun1 plants (Hernández‐Verdeja et al., 2022). Conversely, many genes are differentially expressed in the Arabidopsis deetiolation experiment that are not differentially expressed in M. polymorpha (Hernández‐Verdeja et al., 2022). Thus, while significant transcriptome‐level differences can be observed between Mpgun1 and wild‐type spores, these are not similar to those observed in Arabidopsis.
Discussion
GUN1 has become the most studied PPR protein since the discovery of its connection to plastid retrograde signalling 15 yr ago (Koussevitzky et al., 2007). Despite numerous attempts to pin down its function in this process, the molecular action of GUN1 remains unclear. We attempted to take a different approach by using evolutionary conservation of sequence and function to guide our interpretation of how GUN1 might act. We have shown that GUN1 is probably present in every land plant with a chloroplast genome (as well as in the streptophyte algae most closely related to land plants) and equally highly conserved in terms of its structure and sequence. Therefore, it clearly has adaptive significance to almost all plants under natural conditions. This includes many nonphotosynthetic parasitic or mycoheterotrophic plants.
The question that remains is what is this essential function? Under laboratory conditions, GUN1 is entirely dispensable in Arabidopsis and Marchantia. The only conditions under which the gun1 phenotype is truly dramatic are nonphysiological treatments with inhibitors of chloroplast development such as norflurazon, spectinomycin or lincomycin, leaving its true physiological role unclear (Pogson et al., 2008). Nevertheless, the assumption has often been made that GUN1's essential, conserved role is connected to retrograde signalling. For example, the first identification of GUN1 genes in streptophyte algae led to the suggestion that the GUN1‐mediated retrograde signalling pathway evolved before the colonization of land by plants (de Vries et al., 2018; Nishiyama et al., 2018). We found that inhibition of chloroplast translation induced much less profound repression of PhANGs in the liverwort M. polymorpha than in Arabidopsis. Furthermore, Mpgun1 mutant spores were not clearly defective in this response. This suggests that, in Marchantia, GUN1 is not involved in a strong global transcriptional repression of PhANGs in response to inhibition of chloroplast translation. Marchantia polymorpha spores develop green chloroplasts when germinated in complete darkness. Similarly, in many gymnosperms and ferns, the differentiation of proplastids into chloroplasts is not light‐dependent (Raghavan, 1993; Ranade et al., 2019). The ability to synthesize Chl in the dark correlates with the presence of chloroplast chl genes encoding subunits of a light‐independent protochlorophyllide reductase (Fujita, 1996). Knockout of chlB in M. polymorpha prevents protochlorophyllide reduction in the dark and reduces Chl accumulation under short‐day conditions (Ueda et al., 2014). Flowering plants lack chl genes, and consequently accumulate protochlorophyllide in etioplasts, making deetiolation a critical process due to potential oxidative damage from photoreactive Chl precursors. GUN1 plays a key role in Arabidopsis during this etioplast–chloroplast transition (Hernández‐Verdeja et al., 2022). We speculate that GUN1‐associated retrograde signalling may have only evolved in flowering plants under this novel selection pressure. Our findings highlight the importance of functional characterization of proteins in divergent model systems when inferring evolutionary conservation of signalling pathways.
It thus seems likely that the conservation of GUN1 from streptophyte algae through bryophytes, lycophytes, ferns and gymnosperms is due to its involvement in some other essential physiological process and the later involvement in retrograde signalling is a secondary role. The distribution of GUN1 across land plants and algae rules out some processes as candidates. It was recently proposed that GUN1 regulates RNA editing via directly interacting with MORF2 protein (Zhao et al., 2019). RNA editing has not been observed in streptophyte algae and probably only evolved in the lineage leading to land plants (Schallenberg‐Rüdinger & Knoop, 2016). Furthermore, complex thalloid (Marchantiid) liverworts, such as M. polymorpha, have entirely lost RNA editing (Groth‐Malonek et al., 2007). Moreover, MORF2 homologues are only present in seed plants (Schallenberg‐Rüdinger & Knoop, 2016; Gutmann et al., 2020). Therefore, the primordial role of GUN1 cannot be in regulating RNA editing. Similarly, GUN1 has been proposed to be involved in feedback regulation of protein import into plastids (Wu et al., 2019; Tadini et al., 2020), but the loss of GUN1 in Rafflesiaceae (which still import proteins into plastids but cannot synthesize any plastid proteins) rather implies a role in chloroplast gene expression. Thus, while these recent propositions may be relevant to understanding GUN1's secondary role in retrograde signalling, they do not appear to help identify its primary function.
Despite the fact that GUN1 is not involved in a retrograde signalling pathway in Marchantia, MpGUN1 fully complements an Arabidopsis gun1 mutant, including its retrograde signalling phenotype. This surprising result implies to us that it is not the GUN1 protein itself that directly acts in the signalling pathway, but rather the result of GUN1's (conserved) action.
What is the primary (primordial) function of GUN1? The extreme conservation of GUN1's PPR motifs (exceeding even that of the PPR splicing factors EMB2654 and PPR4, which have a similarly long evolutionary history, Lee et al., 2019) and of its SMR domain are consistent with it functioning as a typical PPR protein, that is as a sequence‐specific RNA binding protein. Based on the proposed relationships between PPR motif sequences and aligned nucleotides, we predicted an 11‐nucleotide binding site sequence putatively recognized by GUN1. This sequence is not found at conserved positions in chloroplast genomes (and is missing entirely from many), so we do not believe this sequence to accurately represent the GUN1 binding site(s), but it may resemble it. We can infer some probable features of the RNA that GUN1 might bind from evolutionary patterns. The presence of GUN1 in all land plants except those lacking a plastid genome indicates that if its target is an RNA, it is one present in all plastid genomes. This rules out almost all protein‐coding transcripts and tRNAs, leaving the rrn16 and rrn23 transcripts as the most likely targets. The extremely high conservation of the specificity‐determining residues would suggest that the GUN1 binding site is also highly conserved, even in species with highly divergent plastid genomes, again consistent with a highly conserved target transcript such as an rRNA. We predict that the essential conserved role of GUN1 will turn out to be in some way involved with the regulation of plastid ribosome biogenesis.
Author contributions
SH and IS designed the research, analysed the data and wrote the manuscript. SH performed the experiments and collected the data.
Supporting information
Fig. S1 Identification of GUN1 chloroplast transit peptide‐containing region.
Fig. S2 Multiple sequence alignment of 76 GUN1 protein sequences from diverse streptophyte algae and land plants.
Fig. S3 Identification of GUN1 sequences by hmmsearch scores.
Fig. S4 AtGUN1 protein structure prediction by AlphaFold.
Fig. S5 Location of DNA deletions in Mpgun1‐1 and Mpgun1‐2 CRISPR lines.
Fig. S6 Phenotype of Marchantia polymorpha wild‐type and Mpgun1 spores germinated under long day conditions in the absence (control) or presence of chemical inhibitors of plastid function (spec = spectinomycin 500 μg ml−1 or nor = norflurazon 5 μM) or in complete darkness.
Fig. S7 Read coverage across the MpGUN1 transcript in Marchantia polymorpha spore transcriptome data.
Methods S1 Agrobacterium‐mediated transformation of the liverwort Marchantia polymorpha.
Methods S2 Generation of transgenic Mpgun1 CRISPR/Cas9 knockout lines.
Notes S1 Sequence maps of plasmids used for complementation of the Arabidopsis gun1 mutant (GenBank format).
Table S1 List of primers used in this study.
Table S2 Identification of GUN1 sequences by hmmsearch scores.
Table S3 Differentially expressed transcripts in wild‐type and Mpgun1 mutant spores grown in the presence or absence of spectinomycin.
Table S4 Gene ontology term enrichment analysis of wild‐type and Mpgun1 spores grown in the presence or absence of spectinomycin.
Table S5 Differentially expressed transcripts in 40‐h‐old Mpgun1 mutant spores compared to wild‐type spores.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This work was supported by The Australian Research Council (FL140100179 and CE140100008 to IS) and CSIRO Synthetic Biology Future Science Platform (fellowship to SH). Vector pHB453 was kindly provided by Dr Holger Breuninger, University of Tübingen. Open access publishing facilitated by The University of Western Australia, as part of the Wiley – The University of Western Australia agreement via the Council of Australian University Librarians.
Data availability
Sequencing read data have been deposited at the Short Read Archive database at the National Center for Biotechnology Information under project nos. PRJNA800059 and PRJNA838206. The GUN1 HMM profile and all 893 identified GUN1 sequences aligned in Fasta format are available from Dryad (10.5061/dryad.x0k6djhmk). This Dryad repository also contains the code and source data for reproducing Figs 1, 7, S2 and S3; Tables S2 and S3.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Aryamanesh N, Ruwe H, Sanglard LVP, Eshraghi L, Bussell JD, Howell KA, Small I, des Francs‐Small CC. 2017. The pentatricopeptide repeat protein EMB2654 is essential for trans‐splicing of a chloroplast small ribosomal subunit transcript. Plant Physiology 173: 1164–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I. 2012. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genetics 8: e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Small I. 2014. Pentatricopeptide repeat proteins in plants. Annual Review of Plant Biology 65: 415–442. [DOI] [PubMed] [Google Scholar]
- Bartels PG, Watson CW. 1978. Inhibition of carotenoid synthesis by fluridone and norflurazon. Weed Science 26: 198–203. [Google Scholar]
- Bellot S, Renner SS. 2016. The plastomes of two species in the endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biology and Evolution 8: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari D, Guha K, Bhaduri N, Saha P. 2011. Ubiquitination of mRNA cycling sequence binding protein from Leishmania donovani (LdCSBP) modulates the RNA endonuclease activity of its Smr domain. FEBS Letters 585: 809–813. [DOI] [PubMed] [Google Scholar]
- Boehm CR, Ueda M, Nishimura Y, Shikanai T, Haseloff J. 2016. A cyan fluorescent reporter expressed from the chloroplast genome of Marchantia polymorpha . Plant & Cell Physiology 57: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F et al. 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171: 287–299.e15. [DOI] [PubMed] [Google Scholar]
- Breitenbach J, Zhu C, Sandmann G. 2001. Bleaching herbicide norflurazon inhibits phytoene desaturase by competition with the cofactors. Journal of Agricultural and Food Chemistry 49: 5270–5272. [DOI] [PubMed] [Google Scholar]
- Cai L, Arnold BJ, Xi Z, Khost DE, Patel N, Hartmann CB, Manickam S, Sasirat S, Nikolov LA, Mathews S et al. 2021. Deeply altered genome architecture in the endoparasitic flowering plant Sapria himalayana Griff. (Rafflesiaceae). Current Biology: CB 31: 1002–1011. [DOI] [PubMed] [Google Scholar]
- Carpenter EJ, Matasci N, Ayyampalayam S, Wu S, Sun J, Yu J, Jimenez Vieira FR, Bowler C, Dorrell RG, Gitzendanner MA et al. 2019. Access to RNA‐sequencing data from 1,173 plant species: the 1000 plant transcriptomes initiative (1KP). GigaScience 8: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ. 2016. Learning the languages of the chloroplast: retrograde signaling and beyond. Annual Review of Plant Biology 67: 25–53. [DOI] [PubMed] [Google Scholar]
- Charuvi D, Kiss V, Nevo R, Shimoni E, Adam Z, Reich Z. 2012. Gain and loss of photosynthetic membranes during plastid differentiation in the shoot apex of Arabidopsis . Plant Cell 24: 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Gutmann B, Zhong X, Ye Y, Fisher MF, Bai F, Castleden I, Song Y, Song B, Huang J et al. 2016. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. The Plant Journal 85: 532–547. [DOI] [PubMed] [Google Scholar]
- Chiyoda S, Linley PJ, Yamato KT, Fukuzawa H, Yokota A, Kohchi T. 2006. Simple and efficient plastid transformation system for the liverwort Marchantia polymorpha L. suspension‐culture cells. Transgenic Research 16: 41–49. [DOI] [PubMed] [Google Scholar]
- Cottage A, Mott EK, Kempster JA, Gray JC. 2010. The Arabidopsis plastid‐signalling mutant gun1 (genomes uncoupled1) shows altered sensitivity to sucrose and abscisic acid and alterations in early seedling development. Journal of Experimental Botany 61: 3773–3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Computational Biology 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ. 1970. Further similarities between chloroplast and bacterial ribosomes. Planta 91: 329–335. [DOI] [PubMed] [Google Scholar]
- Fujita Y. 1996. Protochlorophyllide reduction: a key step in the greening of plants. Plant & Cell Physiology 37: 411–421. [DOI] [PubMed] [Google Scholar]
- Fukui K, Kosaka H, Kuramitsu S, Masui R. 2007. Nuclease activity of the MutS homologue MutS2 from Thermus thermophilus is confined to the Smr domain. Nucleic Acids Research 35: 850–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm B, Kruse E, Kloppstech K. 1989. Transiently expressed early light‐inducible thylakoid proteins share transmembrane domains with light‐harvesting chlorophyll binding proteins. Plant Molecular Biology 13: 583–593. [DOI] [PubMed] [Google Scholar]
- Groth‐Malonek M, Wahrmund U, Polsakiewicz M, Knoop V. 2007. Evolution of a pseudogene: exclusive survival of a functional mitochondrial nad7 gene supports Haplomitrium as the earliest liverwort lineage and proposes a secondary loss of RNA editing in Marchantiidae . Molecular Biology and Evolution 24: 1068–1074. [DOI] [PubMed] [Google Scholar]
- Gutmann B, Royan S, Schallenberg‐Rüdinger M, Lenz H, Castleden IR, McDowell R, Vacher MA, Tonti‐Filippini J, Bond CS, Knoop V et al. 2020. The expansion and diversification of pentatricopeptide repeat RNA‐editing factors in plants. Molecular Plant 13: 215–230. [DOI] [PubMed] [Google Scholar]
- Harpster MH, Mayfield SP, Taylor WC. 1984. Effects of pigment‐deficient mutants on the accumulation of photosynthetic proteins in maize. Plant Molecular Biology 3: 59–71. [DOI] [PubMed] [Google Scholar]
- Hernández‐Verdeja T, Strand Å. 2018. Retrograde signals navigate the path to chloroplast development. Plant Physiology 176: 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández‐Verdeja T, Vuorijoki L, Jin X, Vergara A, Dubreuil C, Strand Å. 2022. GENOMES UNCOUPLED1 plays a key role during the de‐etiolation process in Arabidopsis. New Phytologist 235: 188–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Nishihama R, Yamato KT, Kohchi T. 2015. Molecular genetic tools and techniques for Marchantia polymorpha research. Plant & Cell Physiology 57: 262–270. [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A et al. 2021. Highly accurate protein structure prediction with AlphaFold . Nature 596: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. Mafft multiple sequence alignment software v.7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones‐Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto‐Martins G, Surpin M, Lim J, Mittler R, Chory J. 2007. Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719. [PubMed] [Google Scholar]
- Lee K, Park SJ, Colas des Francs‐Small C, Whitby M, Small I, Kang H. 2019. The coordinated action of PPR4 and EMB2654 on each intron half mediates trans‐splicing of rps12 transcripts in plant chloroplasts. The Plant Journal 100: 1193–1207. [DOI] [PubMed] [Google Scholar]
- Lee X‐W, Mat‐Isa M‐N, Mohd‐Elias N‐A, Aizat‐Juhari MA, Goh H‐H, Dear PH, Chow K‐S, Haji Adam J, Mohamed R, Firdaus‐Raih M et al. 2016. Perigone lobe transcriptome analysis provides insights into Rafflesia cantleyi flower development. PLoS ONE 11: e0167958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leebens‐Mack JH, Barker MS, Carpenter EJ, Deyholos MK, Gitzendanner MA, Graham SW, Grosse I, Li Z, Melkonian M, Mirarab S et al. 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Melonek J, Boykin LM, Small I, Howell KA. 2013. PPR‐SMRs. RNA Biology 10: 1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yu F, Rodermel S. 2010. An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH‐mediated chloroplast biogenesis. Plant Physiology 154: 1588–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B et al. 2004. Genome‐wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Naranjo B, Wang J, Penzler JF, Kleine T, Leister D. 2019. Relationship of GUN1 to FUG1 in chloroplast protein homeostasis. The Plant Journal 99: 521–535. [DOI] [PubMed] [Google Scholar]
- Martín G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E. 2016. Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light‐induced transcriptional network. Nature Communications 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield SP, Taylor WC. 1984. Carotenoid‐deficient maize seedlings fail to accumulate light‐harvesting chlorophyll a/b binding protein (LHCP) mRNA. European Journal of Biochemistry/FEBS 144: 79–84. [DOI] [PubMed] [Google Scholar]
- Mergner J, Frejno M, List M, Papacek M, Chen X, Chaudhary A, Samaras P, Richter S, Shikata H, Messerer M et al. 2020. Mass‐spectrometry‐based draft of the Arabidopsis proteome. Nature 579: 409–414. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. 2001. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg‐chelatase H subunit in plastid‐to‐nucleus signal transduction. Proceedings of the National Academy of Sciences, USA 98: 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Philippe H. 1999. Smr: a bacterial and eukaryotic homologue of the C‐terminal region of the MutS2 family. Trends in Biochemical Sciences 24: 298–300. [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Sakayama H, de Vries J, Buschmann H, Saint‐Marcoux D, Ullrich KK, Haas FB, Vanderstraeten L, Becker D, Lang D et al. 2018. The Chara genome: secondary complexity and implications for plant terrestrialization. Cell 174: 448–464. [DOI] [PubMed] [Google Scholar]
- Parker N, Wang Y, Meinke D. 2014. Natural variation in sensitivity to a loss of chloroplast translation in Arabidopsis. Plant Physiology 166: 2013–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias‐aware quantification of transcript expression. Nature Methods 14: 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Bayraktar OA, Prikryl J, Barkan A. 2009. Site‐specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO Journal 28: 2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz K‐J, Oelmüller R. 2006. pTAC2, ‐6, and ‐12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Förster B, Small ID. 2008. Plastid signalling to the nucleus and beyond. Trends in Plant Science 13: 602–609. [DOI] [PubMed] [Google Scholar]
- Prikryl J, Rojas M, Schuster G, Barkan A. 2011. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proceedings of the National Academy of Sciences, USA 108: 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan V. 1993. Chloroplast activities of dark‐imbibed and photoinduced spores of the fern Onoclea sensibilis . Protoplasma 175: 75–84. [Google Scholar]
- Ranade SS, Delhomme N, García‐Gil MR. 2019. Global gene expression analysis in etiolated and de‐etiolated seedlings in conifers. PLoS ONE 14: e0219272‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M, Ruwe H, Miranda RG, Zoschke R, Hase N, Schmitz‐Linneweber C, Barkan A. 2018. Unexpected functional versatility of the pentatricopeptide repeat proteins PGR3, PPR5 and PPR10. Nucleic Acids Research 155: 1520–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland N, Curien G, Finazzi G, Kuntz M, Maréchal E, Matringe M, Ravanel S, Seigneurin‐Berny D. 2012. The biosynthetic capacities of the plastids and integration between cytoplasmic and chloroplast processes. Annual Review of Genetics 46: 233–264. [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Research 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallenberg‐Rüdinger M, Knoop V. 2016. Coevolution of organelle RNA editing and nuclear specificity factors in early land plants. In: Rensing SA, ed. Advances in Botanical Research, vol. 78: genomes and evolution of charophytes, bryophytes, lycophytes and ferns. Amsterdam, the Netherlands: Elsevier, 37–93. [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al. 2012. Fiji: an open‐source platform for biological‐image analysis. Nature Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C et al. 2002. A high‐throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Kacprzak SM, Mochizuki N, Nagatani A, Watanabe S, Shimada T, Tanaka K, Hayashi Y, Arai M, Leister D et al. 2019. The retrograde signaling protein GUN1 regulates tetrapyrrole biosynthesis. Proceedings of the National Academy of Sciences, USA 116: 24900–24906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H‐J, Barkman TJ, Hao W, Jones SS, Naumann J, Skippington E, Wafula EK, Hu J‐M, Palmer JD, dePamphilis CW. 2019. Novel genetic code and record‐setting AT‐richness in the highly reduced plastid genome of the holoparasitic plant Balanophora . Proceedings of the National Academy of Sciences, USA 116: 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. 1993. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799. [DOI] [PubMed] [Google Scholar]
- Tadini L, Peracchio C, Trotta A, Colombo M, Mancini I, Jeran N, Costa A, Faoro F, Marsoni M, Vannini C et al. 2020. GUN1 influences the accumulation of NEP‐dependent transcripts and chloroplast protein import in Arabidopsis cotyledons upon perturbation of chloroplast protein homeostasis. The Plant Journal 101: 1198–1220. [DOI] [PubMed] [Google Scholar]
- Tadini L, Pesaresi P, Kleine T, Rossi F, Guljamow A, Sommer F, Mühlhaus T, Schroda M, Masiero S, Pribil M et al. 2016. GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiology 170: 1817–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nature Reviews. Genetics 5: 123–135. [DOI] [PubMed] [Google Scholar]
- Ueda M, Tanaka A, Sugimoto K, Shikanai T, Nishimura Y. 2014. chlB requirement for chlorophyll biosynthesis under short photoperiod in Marchantia polymorpha L. Genome Biology and Evolution 6: 620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, Van de Peer Y, Coppens F, Vandepoele K. 2018. Plaza 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Research 46: D1190–D1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A et al. 2022. AlphaFold protein structure database: massively expanding the structural coverage of protein‐sequence space with high‐accuracy models. Nucleic Acids Research 50: D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Curtis BA, Gould SB, Archibald JM. 2018. Embryophyte stress signaling evolved in the algal progenitors of land plants. Proceedings of the National Academy of Sciences, USA 115: E3471–E3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Wachi S, Fujita T. 2003. Identification and characterization of BCL‐3‐binding protein: implications for transcription and DNA repair or recombination. Journal of Biological Chemistry 278: 26102–26110. [DOI] [PubMed] [Google Scholar]
- Wheeler TJ, Clements J, Finn RD. 2014. Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinformatics 15: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Müller KF, de Pamphilis CW, Quandt D, Wickett NJ, Zhang Y, Renner SS, Schneeweiss GM. 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25: 3711–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA et al. 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proceedings of the National Academy of Sciences, USA 111: E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez‐Ruiz JM, Chory J. 2011. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Current Biology: CB 21: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G‐Z, Bock R. 2021. GUN control in retrograde signaling: how GENOMES UNCOUPLED proteins adjust nuclear gene expression to plastid biogenesis. Plant Cell 33: 457–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G‐Z, Chalvin C, Hoelscher MP, Meyer EH, Wu XN, Bock R. 2018. Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED 1. Plant Physiology 176: 2472–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G‐Z, Meyer EH, Richter AS, Schuster M, Ling Q, Schöttler MA, Walther D, Zoschke R, Grimm B, Jarvis RP et al. 2019. Control of retrograde signalling by protein import and cytosolic folding stress. Nature Plants 5: 525–538. [DOI] [PubMed] [Google Scholar]
- Wu W, Liu S, Ruwe H, Zhang D, Melonek J, Zhu Y, Hu X, Gusewski S, Yin P, Small ID et al. 2016. SOT1, a pentatricopeptide repeat protein with a small MutS‐related domain, is required for correct processing of plastid 23S‐4.5S rRNA precursors in Arabidopsis thaliana . The Plant Journal 85: 607–621. [DOI] [PubMed] [Google Scholar]
- Yan J, Yao Y, Hong S, Yang Y, Shen C, Zhang Q, Zhang D, Zou T, Yin P. 2019. Delineation of pentatricopeptide repeat codes for target RNA prediction. Nucleic Acids Research 16: 2089–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin S‐S, Niu Q‐W, Chua N‐H. 2006. Agrobacterium‐mediated transformation of Arabidopsis thaliana using the floral dip method. Nature Protocols 1: 641–646. [DOI] [PubMed] [Google Scholar]
- Zhao X, Huang J, Chory J. 2019. GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proceedings of the National Academy of Sciences, USA 116: 10162–10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Lu Q, Li Q, Wang L, Ding S, Zhang A, Wen X, Zhang L, Lu C. 2017. PPR‐SMR protein SOT1 has RNA endonuclease activity. Proceedings of the National Academy of Sciences, USA 114: E1554–E1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Qu Y, Zubo YO, Börner T, Schmitz‐Linneweber C. 2013. Mutation of the pentatricopeptide repeat‐SMR protein SVR7 impairs accumulation and translation of chloroplast ATP synthase subunits in Arabidopsis thaliana . Journal of Plant Research 126: 403–414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Identification of GUN1 chloroplast transit peptide‐containing region.
Fig. S2 Multiple sequence alignment of 76 GUN1 protein sequences from diverse streptophyte algae and land plants.
Fig. S3 Identification of GUN1 sequences by hmmsearch scores.
Fig. S4 AtGUN1 protein structure prediction by AlphaFold.
Fig. S5 Location of DNA deletions in Mpgun1‐1 and Mpgun1‐2 CRISPR lines.
Fig. S6 Phenotype of Marchantia polymorpha wild‐type and Mpgun1 spores germinated under long day conditions in the absence (control) or presence of chemical inhibitors of plastid function (spec = spectinomycin 500 μg ml−1 or nor = norflurazon 5 μM) or in complete darkness.
Fig. S7 Read coverage across the MpGUN1 transcript in Marchantia polymorpha spore transcriptome data.
Methods S1 Agrobacterium‐mediated transformation of the liverwort Marchantia polymorpha.
Methods S2 Generation of transgenic Mpgun1 CRISPR/Cas9 knockout lines.
Notes S1 Sequence maps of plasmids used for complementation of the Arabidopsis gun1 mutant (GenBank format).
Table S1 List of primers used in this study.
Table S2 Identification of GUN1 sequences by hmmsearch scores.
Table S3 Differentially expressed transcripts in wild‐type and Mpgun1 mutant spores grown in the presence or absence of spectinomycin.
Table S4 Gene ontology term enrichment analysis of wild‐type and Mpgun1 spores grown in the presence or absence of spectinomycin.
Table S5 Differentially expressed transcripts in 40‐h‐old Mpgun1 mutant spores compared to wild‐type spores.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
Sequencing read data have been deposited at the Short Read Archive database at the National Center for Biotechnology Information under project nos. PRJNA800059 and PRJNA838206. The GUN1 HMM profile and all 893 identified GUN1 sequences aligned in Fasta format are available from Dryad (10.5061/dryad.x0k6djhmk). This Dryad repository also contains the code and source data for reproducing Figs 1, 7, S2 and S3; Tables S2 and S3.


