Abstract
The lead resistance operon, pbr, of Ralstonia metallidurans (formerly Alcaligenes eutrophus) strain CH34 is unique, as it combines functions involved in uptake, efflux, and accumulation of Pb(II). The pbr lead resistance locus contains the following structural resistance genes: (i) pbrT, which encodes a Pb(II) uptake protein; (ii) pbrA, which encodes a P-type Pb(II) efflux ATPase; (iii) pbrB, which encodes a predicted integral membrane protein of unknown function; and (iv) pbrC, which encodes a predicted prolipoprotein signal peptidase. Downstream of pbrC, the pbrD gene, encoding a Pb(II)-binding protein, was identified in a region of DNA, which was essential for functional lead sequestration. Pb(II)-dependent inducible transcription of pbrABCD from the PpbrA promoter is regulated by PbrR, which belongs to the MerR family of metal ion-sensing regulatory proteins. This is the first report of a mechanism for specific lead resistance in any bacterial genus.
The presence of toxic heavy metals in the environment has resulted in the development or acquisition by bacteria of genetic systems that counteract their effects. Many bacterial heavy metal resistance systems are based on efflux, and two groups of efflux systems have been identified. These can be either P-type ATPases, e.g., the Cu(II), Cd(II), and Zn(II) ATPases of gram-negative bacteria (31), or chemiosmotic pumps, e.g., the three-component divalent-cation efflux systems cnr, ncc, and czc of Ralstonia metallidurans (formerly Alcaligenes eutrophus) CH34 (35).
Lead resistance has been reported in both gram-negative and gram-positive bacteria isolated from lead-contaminated soils, with Pseudomonas marginalis showing extracellular lead exclusion and Bacillus megaterium demonstrating intracellular cytoplasmic lead accumulation (28). Pb(II)-resistant strains of Staphylococcus aureus and Citrobacter freundii that accumulated the metal as an intracellular lead-phosphate have also been isolated (15, 16), though the molecular mechanism of detoxification remains to be elucidated. Efflux of Pb(II) has also been reported for the CadA ATPase of S. aureus and the ZntA ATPase of Escherichia coli (27).
R. metallidurans strain CH34 contains at least seven determinants encoding resistances to toxic heavy metals, located on one of the two endogenous megaplasmids, pMOL28 and pMOL30 (for a review, see reference 35). One of these determinants, located on pMOL30 (20), mediates resistance to Pb(II). In this paper we describe the isolation and characterization of the Pb(II) resistance determinant, pbr, from pMOL30.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
R. metallidurans and E. coli were grown in 869 medium (20) at 30 and 37°C, respectively. Antibiotic resistance was selected on media supplemented with 20 μg of tetracycline or 100 μg of ampicillin per ml, as appropriate. To test for Pb(II) resistance, cells were grown on RM medium, a modified 284 gluconate minimal medium (20) in which Tris-HCl is replaced by 20 mM morpholinepropanesulfonic acid (MOPS)–NaOH (pH 7) and Na is replaced by 0.95 mM beta-glycerol-phosphate. These two components were filter sterilized and added separately after the RM medium was autoclaved. Pb(II) was added as Pb(NO3)2 at the required concentrations.
Molecular cloning techniques, sequencing, and electroporation.
Standard molecular cloning techniques and plasmid DNA isolation from E. coli were performed as described by Sambrook et al. (29). Electroporation was used to introduce plasmid DNA into R. metallidurans and E. coli (36).
Subcloning, primer walking, and cycle sequencing strategies were used for DNA sequencing. The sequences were analyzed using GCG software (Genetics Computer Group, Inc., Madison, Wis.).
Cloning of the pbr lead resistance determinant.
The pbr Pb(II) resistance determinant was subcloned from an R. metallidurans CH34 cosmid library constructed in pLAFR3 (8). The CH34 cosmid bank was introduced by electroporation into the plasmid-free, heavy metal-sensitive derivative of strain CH34, R. metallidurans AE104 (20). Transformants were selected for resistance to tetracycline and Pb (0.4 mM) on RM medium. The cosmids from 10 independent, Pb(II)-resistant transformants were isolated and digested with EcoRI. Subclones in vector pRK415 (14) were similarly introduced into R. metallidurans AE104 and tested for the lead resistance phenotype conferred.
Pb(II) uptake experiments.
The uptake of Pb(II) was based on a previously described protocol for Al(III) uptake (11). All cultures were grown at 30°C in 200 ml of RM medium, in which no precipitation of Pb(II) occurs. When the optical density at 660 nm reached a value of approximately 0.8, the cells were concentrated fivefold by centrifugation and resuspended in fresh RM medium containing 0.2 mM Pb(NO3)2. As a negative control, the experiment was also performed in RM medium without Pb(NO3)2. After 2 h the cells were collected by centrifugation (2,000 × g, 10 min, Sorvall centrifuge, SS34 rotor) and subsequently washed with 50 ml of 0.2 M EDTA and 50 ml of sterile distilled water to remove adventitiously bound Pb(II). The cells were freeze-dried and weighed, and the amount of Pb(II) was determined using inductively coupled plasma analysis. The amount of accumulated Pb(II) per milligram of biomass was calculated.
Transcript analysis.
The start point of transcription was determined by primer extension analysis. Total RNA was extracted, using the hot-acid phenol method (1) from exponential phase R. metallidurans AE2473 grown in RM medium for 2 h with or without 0.2 mM Pb(NO3)2. The RNA preparation was treated with 10 U of RQ1 RNase-free DNase I for 30 min at 37°C (Promega, Southampton, United Kingdom). The primer PbrApe, 5′-GCGCCAACCGTGCTCGGTTCTGGG-3′, was labeled with 25μCi of [γ-32P]ATP (NEN Life Sciences, London, United Kingdom) by incubation with 10 U of T4 polynucleotide kinase (Life Technologies, Paisley, Scotland) for 30 min at 37°C and used with 10 μg of total RNA for primer extension analysis. Labeled primer and RNA were heated to 85°C for 5 min and then at 45°C overnight prior to ethanol precipitation. Primer-RNA hybrids were resuspended in the reaction mixture for Superscript II reverse transcriptase (Life Technologies) and incubated at 42°C for 60 min and then at 72°C for 10 min after the addition of 400 U of Superscript II. The labeled cDNA was run on a 6% (wt/vol) denaturing polyacrylamide gel alongside a sequencing reaction mixture generated using the PbrApe primer.
The reverse transcription (RT)-PCR method described by Gupta et al. (9) for transcript analysis of the sil operon was adapted for this study of pbr transcription. RNA was prepared as described above from AE2473 and AE104 grown for 2 h with or without 0.2 mM Pb(NO3)2. RNA was annealed in the presence of deoxynucleoside triphosphates to the primer pbrD-RT, 5′-CTACAGGCGTAGGCACCGTCG-3′ (positions 7231 to 7211; see Results for numbering), for 5 min at 65°C and then cooled on ice. First-strand buffer, dithiothreitol, and RNasin (Promega) were added to the annealed primer-RNA hybrids, and the reaction mixture was incubated at 42°C for 60 min and then at 72°C for 10 min after the addition of Superscript II reverse transcriptase. cDNA (2 μl) from the RT reaction was added to a PCR with the primers pbrA-N term, 5′-ATGAGCGAATGTGGCTCGAAG-3′ (positions 2730 to 2750), and pbrA-C term, 5′-TCATCGACGCAACAGCCTCAA-3′ (positions 5126 to 5106). PCR conditions were 96°C for 3 min (step 1), followed by 96°C for 60 s (step 2), 58°C for 60 s (step 3), and 72°C for 3 min (step 4). Steps 2 to 4 were repeated 39 times followed by a further incubation at 72°C for 5 min. Appropriate controls were run (see Results). RT-PCR products were run on a 0.7% (wt/vol) agarose Tris-borate-EDTA gel.
RESULTS
Restriction map of pMOL1027 and identification of the minimal Pb(II) resistance region.
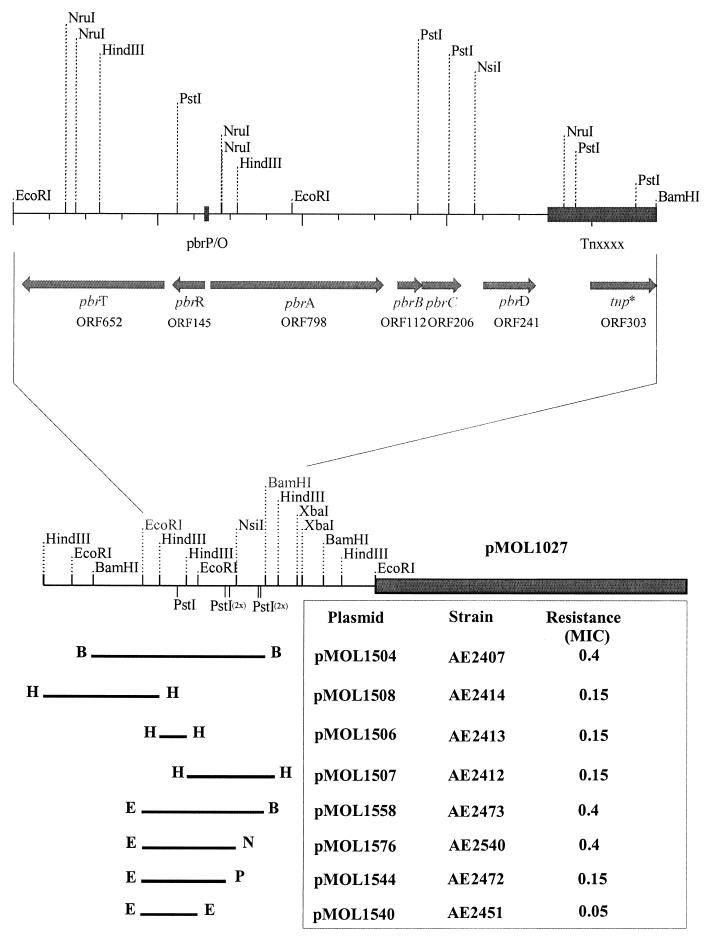
A cosmid library of R. metallidurans total DNA (8) was screened for Pb(II) resistance clones in the heavy metal-sensitive strain R. metallidurans AE104 (20). Ten identical Pb(II)-resistant clones were identified, and the restriction map of one of the cosmid clones, pMOL1027, is shown in Fig. 1. No resistance to other divalent metal salts, including Zn(II), Cd(II), Co(II), Cu(II), Ni(II), and Hg(II), was observed, even after induction by Pb(II). The minimal region of DNA required for Pb(II) resistance was determined by subcloning from pMOL1027 into the vector pRK415 (14) and testing R. metallidurans AE104 transformants for Pb(II) resistance. The Pb(II) resistance locus was localized initially to a 12-kb BamHI fragment in clone pMOL1504. Several subclones of pMOL1504 were made (Fig. 1). The 8.9-kb EcoRI-EcoRI-BamHI fragment of pMOL1558 conferring Pb(II) resistance was completely sequenced (EMBL accession no. AJ278984). pMOL1576 contains the 6.4-kb EcoRI-EcoRI-NsiI fragment (positions 1 to 6383) and also conferred Pb(II) resistance. Strains carrying plasmids pMOL1544, containing the 5.6-kb EcoRI-EcoRI-PstI fragment (positions 1 to 5598), and pMOL1540, containing the 3.8-kb EcoRI-EcoRI fragment (positions 1 to 3852), were Pb(II) sensitive.
FIG. 1.
Restriction map of pMOL1027 and identification of the minimal region required for Pb(II) resistance. The bars indicate the positions of the subcloned fragments and are flanked by the restriction sites used, as follows: BamHI (B), EcoRI (E), HindIII (H), NsiI (N), and PstI (P). On the right side of each bar, the plasmid and strain number for the corresponding subclone are indicated as well as the lead resistance phenotype, defined as MIC in millimolar Pb(NO3)2 compared with the MIC of 0.15 mM for the plasmid-free strain AE104. The physical map of the 8,890-bp EcoRI-EcoRI-BamHI fragment required for Pb(II) resistance is shown in more detail. The positions of the pbrT, pbrR, pbrA, pbrB, pbrC, and pbrD genes plus the sizes of their corresponding ORFs (in numbers of amino acids), the incomplete transposon (Tnxxxx, similar to Tn5271 [22]) flanking the pbr operon and its presumed transposase gene (tnp*), and the pbr-promoter-operator region (pbr P/O) as well as the important restriction sites used for subcloning are indicated.
Analysis of the Pb(II) resistance region.
Within the 8.9-kb DNA sequence, seven predicted open reading frames (ORFs) were identified in an operon-like structure, encoding proteins with the predicted sizes of 652 (C), 145 (C), 798, 112, 206, 241, and 303 amino acids (aa), where C represents the sequence encoded by the complementary strand. ORF652, ORF145, ORF798, ORF112, and ORF206 were located on the 6.4-kb EcoRI-EcoRI-NsiI fragment of pMOL1576, which was the minimal Pb(II) resistance clone. Both database search analysis and functional analysis were used to determine the roles of the different ORFs of the pbr operon.
ORF652 (positions 127 to 2085, complement) encodes a predicted membrane protein that contains 8 to 10 potential transmembrane regions. The N-terminal region of the ORF652 protein (aa 106 to 218) shows sequence similarity with the C-terminal cytochrome c6 domain of the diheme C-type cytochrome FixP from Azorhizobium caulinodans (30% identity over the last 113 aa) (18), while the C-terminal region (aa 223 to 619) shows sequence similarity with a large family of integral membrane proteins, including the FTR1 plasma membrane iron permease of Saccharomyces cerevisiae (30% identity) (32). Expression of ORF652 in the absence of the other structural genes of the pbr operon resulted in Pb(II) hypersensitivity in R. metallidurans AE2451 (Fig. 1). For this strain, the MIC of Pb(NO3)2 was 0.05 mM compared to 0.15 mM determined for the plasmid-free strain AE104. ORF652 is therefore suggested to encode a Pb(II) uptake protein, located in the inner membrane. As it is suggested to be functionally analogous to merT, encoding the Hg uptake protein of the mer operon (21), ORF652 was named pbrT.
The predicted ORF145 gene product (positions 2206 to 2643, complement) showed strong similarity to members of the MerR family (for reviews, see references 10 and 34). MerR is the regulator of the Hg resistance mer operon. The ORF145 protein was named PbrR.
The predicted protein encoded by ORF798 (positions 2730 to 5126) showed strong similarity to members of the P-type ATPase family involved in heavy metal efflux, such as CadA of S. aureus (23), and was named PbrA. Although plasmid pMOL1544 (Fig. 1) did not encode full Pb(II) resistance, a Pb(II) hypersensitivity phenotype like that found for pMOL1540 was not observed. This suggests that the PbrA protein is a functional Pb(II) efflux ATPase, able to counteract Pb(II) uptake by PbrT.
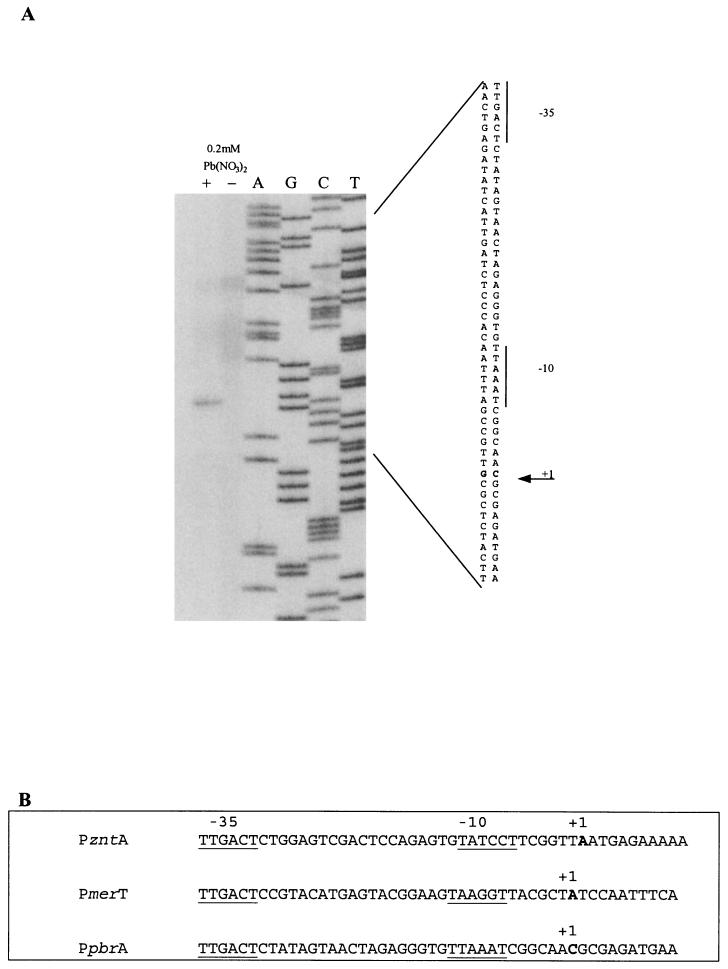
The DNA fragment that contained the pbrR-pbrA intergenic region (positions 2644 to 2729) showed striking similarity to the merO/P and zntO/P regions (5, 24) (Fig. 2B). The transcription start site of the PpbrA promoter was determined by primer extension on RNA isolated from strain AE2473 grown with and without 0.2 mM Pb(NO3)2. Transcription started at C2701 and was located at the same distance (7 bp) from the −10 sequences as the transcription start sites in the PmerT and PzntA promoters (Fig. 2A and B).
FIG. 2.
Comparison of the PzntA, PmerTPAD, and PpbrA promoters. (A) Primer extension analysis of the PpbrA promoter region. Autoradiograph of a 6% acrylamide sequencing gel showing the PpbrA promoter region DNA sequence aligned with the primer extension product generated as described in Materials and Methods. Total RNA was isolated from log-phase R. metallidurans AE2540 cells grown for 2 h with (+) and without (−) 0.2 mM Pb(NO3)2. (B) Mapping of the transcriptional start sites of the PzntA (5), PmerTPAD (17), and PpbrA (shown in panel A) promoters. The positions of the −35 and −10 regions of the promoters are underlined. The +1 nucleotide is indicated in bold.
Analysis of the predicted ORF112 protein reveals characteristics of a membrane lipoprotein. These include the presence of an N-terminal leader sequence with the processing site Ile-Ala-Ser-Cys, which fits well with the consensus sequence for recognition by prolipoprotein signal peptidases (4), and two hydrophobic regions which are potential transmembrane sequences. Inactivation of ORF112, as in plasmid pMOL1544, results in a Pb(II)-sensitive phenotype, indicating its essential role in Pb resistance. The gene was designated pbrB. PbrB shows amino acid sequence similarity to a family of membrane-bound components of bacterial transport systems of which BcrC, which is required for bacitracin resistance in Bacillus licheniformis (25), is the best characterized. The ORF206 protein showed strong similarity (30 and 46% identity) with prolipoprotein signal peptidases from Pseudomonas fluorescens (12) and E. coli (37). The ORF206 protein was named PbrC.
Downstream from the minimal region required for Pb(II) resistance, two further ORFs, ORF241 and part of ORF303, were identified. ORF241 (positions 6645 to 7232) encodes a predicted protein containing a hypothetical metal binding site with the sequence Cys-7X-Cys-Cys-7X-Cys-7X-His-14X-Cys, with a high proportion of Pro and Ser residues between the cysteines. RT-PCR (see below) showed that Pb-dependent transcription of ORF241 is coupled to that of pbrABC.
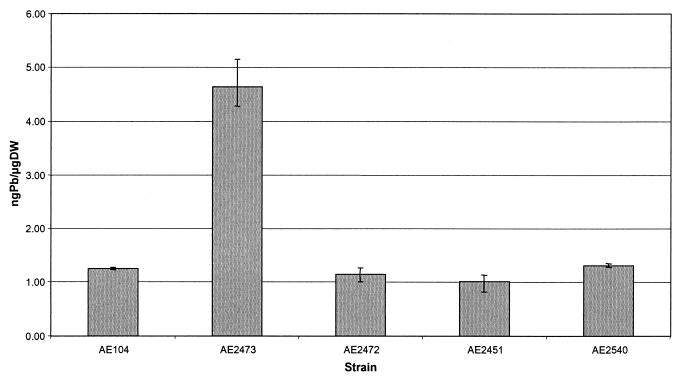
Since the predicted ORF241 protein contains a region with a potential heavy metal binding site, its role, if any, in Pb(II) management was investigated, even though it did not seem to be absolutely required for Pb(II) resistance. Pb(II) accumulation levels in the strains AE104 (plasmid free), AE2451, AE2472, AE2473, and AE2540 were compared after 2 h of incubation in the presence of 0.2 mM PbNO3. The results are presented in Fig. 3. For strains AE2451, AE2472, and AE2540, which contained plasmids with the pbrTRA′, pbrTRAB′, and pbrTRABC regions, respectively, no increase in Pb(II) accumulation was observed compared to the Pb(II)-sensitive control strain AE104. However, with strain AE2473 (pbrTRABCD), a significant increase in Pb(II) accumulation (4.6 instead of 1.2 pg/μg [dry weight]) was observed. This suggests that the region downstream of pbrTRABC, which encodes ORF241, is involved in Pb(II) accumulation, and ORF241 was renamed pbrD.
FIG. 3.
Pb(II) accumulation in the strains AE104 (plasmid free), AE2473 (pbrTRABCD), AE2472 (pbrTRAB′), AE2451 (pbrTRA′), and AE2540 (pbrTRABC) after 2 h of incubation in the presence of 0.2 mM PbNO3. Lead accumulation is expressed in nanograms of Pb/microgram (dry weight ([DW]).
The pbrABCD genes form one transcriptional unit.
RT-PCR was used to test whether the transcription of pbrB, pbrC, and pbrD was Pb dependent and coupled to that of pbrA. Total RNA was extracted from strain AE2473 grown in the presence or absence of 0.2 mM Pb(NO3)2, and cDNA synthesis was initiated from primer pbrD-RT (position 7211 at the 3′ end of pbrD). Subsequently, primers directed against the pbrA gene were used to obtain RT-PCR products. For cells grown in the absence of lead no RT-PCR product was obtained. However, when cells were induced with 0.2 mM Pb(NO3)2, an RT-PCR product with the expected size of approximately 2,400 bp was found. This experiment shows that pbrA and pbrD, and consequently pbrB and pbrC, are transcribed from one large mRNA, the synthesis of which is Pb(II) dependent.
DISCUSSION
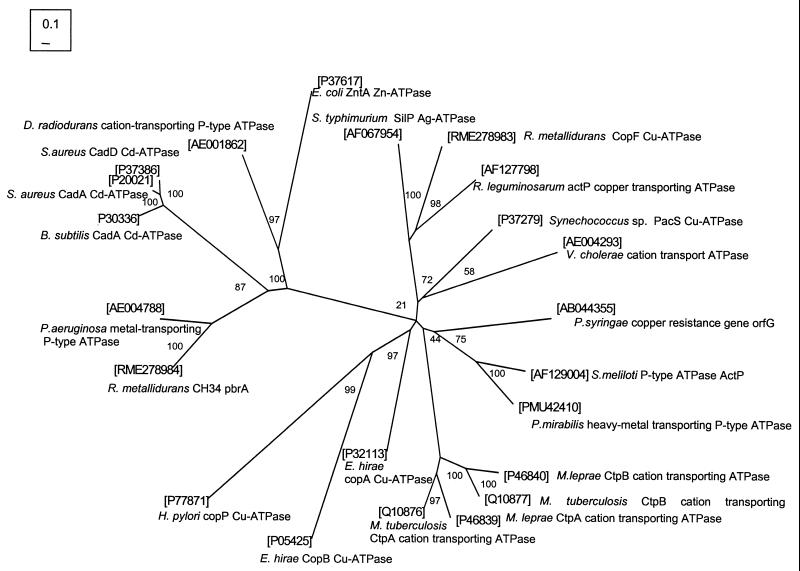
This paper describes the first bacterial resistance determinant that is specific for lead. It has recently been shown that heavy metal resistance systems, such as the cad Cd(II) resistance of S. aureus and the znt Zn efflux system of E. coli, can also confer Pb(II) resistance in E. coli (26, 27). In these cases resistance to several metals, including Pb(II), involves a metal-translocating P-type ATPase. These constitute a highly conserved and widely distributed family of efflux ATPases, which contain heavy metal-associated metal binding domains (6) that are also found in other proteins interacting with heavy metals, including the prokaryotic MerA and MerP (Hg) proteins and the eukaryotic Menkes and Wilsons (Cu) proteins (30, 31). The PbrA Pb(II) ATPase is phylogenetically grouped with the CadA-type Cd ATPases and the ZntA Zn/Cd ATPase (Fig. 4), and they form a group distinct from the Cu/Ag-type ATPases. However, unlike CadA and ZntA, PbrA possesses two heavy metal-associated motifs with the amino acid sequence Cys-Pro-Thr-Glu-Glu instead of the consensus sequence Cys-X-X-Cys (Table 1). This difference, where one Cys is replaced by two Glu, might reflect the preferential coordination of Pb(II) to oxygen rather than to sulfur (2, 13).
FIG. 4.
Phylogenetic tree of P-type heavy metal transport ATPases. Scale designation and bootstrap values are indicated for the branches. The sequence ID numbers are shown between brackets. The sequence of the R. metallidurans CopF Cu-ATPase was recently determined (Borremans and van der Lelie, EMBL accession no. AJ278983). Bar represents 0.1 nucleotide substitution per 10 nucleotides.
TABLE 1.
Comparison of heavy metal-associated motifsa of P-type heavy metal efflux ATPases
| ATPase | Resistance to: | Heavy metal binding motifb |
|---|---|---|
| PbrA of R. metallidurans CH34 | Pb(II) | IENMDCPTEEALIRDKLAKLPGV |
| IHQMDCPTEEALIRSKLAGIPGV | ||
| CadA of S. aureus | Cd, Pb(II) | VQGFTCANCAGKFEKNVKKIPGV |
| ZntA of E. coli | Zn, Cd, Pb(II) | VSGMDCAACARKVENAVRQLAGV |
A MerR-like regulator, PbrR, controls transcription of the pbr structural genes, with the first gene downstream of the pbr operator-promoter encoding a P-type ATPase, like the zntA operon (5) and copA (33). However, zntR and cueR are physically distant on the chromosome from their cognate promoters, whereas pbrR is divergently transcribed from pbrA with an operator-promoter (O/P) structure similar to that found in the gram-negative mer operons, Tn501 and Tn21 (10).
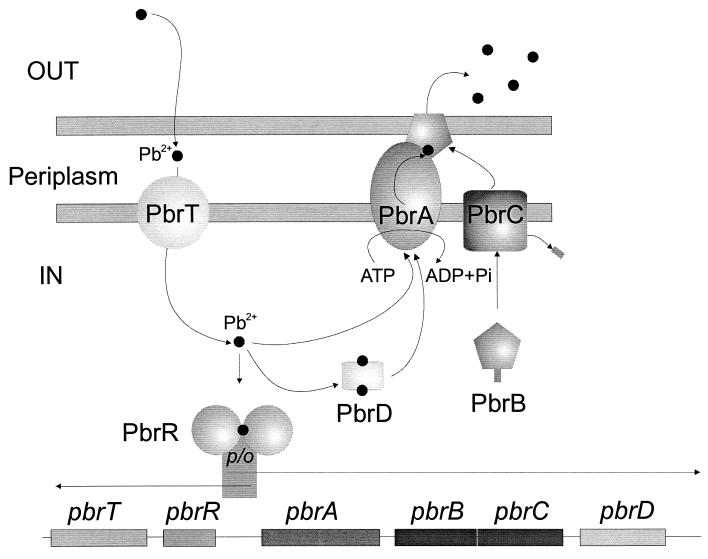
In contrast to the cad and znt operons, which both appear to comprise a regulatory gene plus an efflux-ATPase only, additional proteins are required for maximal Pb(II) resistance in R. metallidurans CH34. These are PbrT, PbrB, PbrC, and PbrD. Our model for pbr-encoded Pb(II) resistance is presented in Fig. 5 and involves a Pb(II) uptake system encoded by PbrT. Expression of PbrT in the absence of PbrABCD results in Pb(II) hypersensitivity, probably due to increased Pb(II) uptake into the cytoplasm. The result of this Pb(II) uptake would be to reduce the interaction of free Pb(II) with side chains of membrane and periplasmic proteins, which would cause extensive cellular damage. Once Pb(II) has entered the cytoplasm, it can be exported by the PbrA Pb(II) efflux ATPase or be bound by the PbrD protein, which may function as a chaperone for Pb(II). PbrD is not absolutely required for Pb(II) resistance, but cells lacking PbrD show a decreased accumulation of Pb(II) compared to that observed for wild-type cells, and this accumulation may protect against free exported Pb(II) and the futile cycle of Pb(II) uptake and export.
FIG. 5.
Model for the pbr Pb(II) resistance operon-encoded lead resistance of R. metallidurans CH34. The model involves the following proteins: PbrT, which transports Pb(II) into the cytoplasm; PbrA, the Pb(II) efflux ATPase; PbrB and PbrC, a Pb(II) transport-facilitating lipoprotein and its prolipoprotein signal peptidase, respectively; and PbrD, a protein involved in Pb(II) sequestration. The PbrR protein, which mediates Pb(II)-inducible transcription from its divergent promoter, regulates the pbr operon.
The PbrA Pb(II) efflux ATPase has been shown to be functional and able to compensate for the Pb(II) uptake driven by PbrT. However, for full Pb(II) resistance PbrB and PbrC are required. PbrB and related proteins may be part of a new family of transporter-assisting resistance proteins. Comparison of PbrB with the EMBL database resulted in 16 hits. However, with the exception of the BcrC protein of B. licheniformis, all others were hypothetical membrane (lipo)proteins. The BcrC gene of B. licheniformis encodes a hydrophobic membrane protein; this protein and the BcrB membrane protein function as membrane components of the bacitracin resistance ABC transporter (25). Inactivation of BcrC results in bacitracin sensitivity, and inactivation of PbrB results in Pb(II) sensitivity. The PbrB lipoprotein may promote transfer of Pb(II) from the periplasm to the outer membrane. This would result in a decreased Pb(II) uptake by PbrT.
At increased concentrations of Pb(II), metal removal from the solution was observed during the late log phase and the stationary phase, during which a progressive increase of the pH (up to 9) was observed (results not shown). At this increased pH the formation of lead complexes with hydroxide and carbonates will be strongly favored and should play an important role in avoiding reentry of Pb(II). A similar phenomenon has been described for the czc cadmium-zinc-cobalt resistance system of R. metallidurans CH34 (7).
The presence of pbrC, encoding a predicted prolipoprotein signal peptidase, in the Pb(II) resistance determinant of pMOL30 is the first identification of a prolipoprotein signal peptidase gene as part of a heavy metal resistance operon. Since pbrB and pbrC are part of a single transcriptional unit, we hypothesize that the PbrC prolipoprotein signal peptidase is required for the processing of the PbrB prolipoprotein. So, either PbrC is specifically required for the processing of PbrB or, alternatively, the availability of chromosomally encoded prolipoprotein signal peptidases is insufficient for the processing of PbrB, as has been suggested for the ORF1 proteins encoded by pTA1015 and pTA1040 (19), which were found in an operon-like organization with signal peptidase genes. The organization of pbrABCD in one transcriptional unit would result in fine-tuning of both the transcription and posttranslational processing of these proteins.
ACKNOWLEDGMENTS
We are grateful to Tatiana Vallaeys for having provided the phylogenetic analysis of the ATPases. Philippe Corbisier, Safieh Taghavi, Max Mergeay, and Ludo Diels are acknowledged for fruitful and cooperative discussion.
This collaboration between VITO and The University of Birmingham was supported by grant ENV4-CT95-0141 from the European Commission and by grant B10333 to N.L.B. from the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;146:1905–1910. [PubMed] [Google Scholar]
- 2.Battistuzzi G, Borsari M, Menabue L, Saladini M, Sola M. Amide group coordination of the Pb2+ ion. Inorg Chem. 1996;35:4239–4247. doi: 10.1021/ic950599h. [DOI] [PubMed] [Google Scholar]
- 3.Beard S J, Hashim R, Membrillo-Hernandez J, Hughes M N, Poole R K. Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol Microbiol. 1997;25:883–891. doi: 10.1111/j.1365-2958.1997.mmi518.x. [DOI] [PubMed] [Google Scholar]
- 4.Braun V, Wu H C. Lipoproteins, structure, function, biosynthesis and model for protein export. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B. V.; 1994. pp. 319–341. [Google Scholar]
- 5.Brocklehurst K R, Hobman J L, Lawley B, Blank L, Marshall S J, Brown N L, Morby A P. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol Microbiol. 1999;31:893–902. doi: 10.1046/j.1365-2958.1999.01229.x. [DOI] [PubMed] [Google Scholar]
- 6.Bull P C, Cox D W. Wilson disease and Menkes disease—new handles on heavy-metal transport. Trends Genet. 1994;10:246–252. doi: 10.1016/0168-9525(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 7.Diels L, Dong Q, van der Lelie D, Baeyens W, Mergeay M. The czc operon of Alcaligenes eutrophus CH34: from resistance mechanism to the removal of heavy metals. J Ind Microbiol. 1995;14:142–153. doi: 10.1007/BF01569896. [DOI] [PubMed] [Google Scholar]
- 8.Gilis A, Khan M A, Cornelis P, Meyer J M, Mergeay M, van der Lelie D. Siderophore-mediated iron uptake in Alcaligenes eutrophus CH34 and identification of aleB encoding the ferric iron-alcaligen E receptor. J Bacteriol. 1996;178:5499–5507. doi: 10.1128/jb.178.18.5499-5507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta A, Matsui A, Lo J-F, Silver S. Molecular basis for resistance to silver cations in Salmonella. Nat Med. 1999;5:183–188. doi: 10.1038/5545. [DOI] [PubMed] [Google Scholar]
- 10.Hobman J L, Brown N L. Bacterial mercury-resistance genes. In: Sigel A, Sigel H, editors. Metal ions in biological systems. Vol. 34. New York, N.Y: Marcel Dekker, Inc; 1997. pp. 527–568. [PubMed] [Google Scholar]
- 11.Hu X, Boyer G L. Siderophore-mediated aluminum uptake by Bacillus megaterium ATCC19213. Appl Environ Microbiol. 1996;62:4044–4048. doi: 10.1128/aem.62.11.4044-4048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isaki L, Beers R, Wu H C. Nucleotide sequence of the Pseudomonas fluorescens signal peptidase II gene (lsp) and flanking genes. J Bacteriol. 1990;172:6512–6517. doi: 10.1128/jb.172.11.6512-6517.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanemaguire L A P, Riley P J. High-field MNR-study of the binding of lead (II) to cysteine and glutathione. J Coordination Chem. 1993;28:105–120. [Google Scholar]
- 14.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 15.Levinson H, Mahler I, Blackwelder P, Hood T. Lead resistance and sensitivity in Staphylococcus aureus. FEMS Microbiol Lett. 1986;145:421–425. doi: 10.1111/j.1574-6968.1996.tb08610.x. [DOI] [PubMed] [Google Scholar]
- 16.Levinson H, Mahler I. Phosphatase activity and lead resistance in Citrobacter freundii and Staphylococcus aureus. FEMS Microbiol Lett. 1998;161:135–138. doi: 10.1111/j.1574-6968.1998.tb12939.x. [DOI] [PubMed] [Google Scholar]
- 17.Lund P A, Ford S J, Brown N L. Transcriptional regulation of the mercury-resistance genes of transposon Tn501. J Gen Microbiol. 1986;132:465–480. doi: 10.1099/00221287-132-2-465. [DOI] [PubMed] [Google Scholar]
- 18.Mandon K, Kaminski P A, Elmerich C. Functional analysis of the fixNOQP region of Azorhizobium caulinodans. J Bacteriol. 1994;176:2560–2568. doi: 10.1128/jb.176.9.2560-2568.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meijer W J, de Jong A, Bea G, Wisman A, Tjalsma H, Venema G, Bron S, van Dijl J M. The endogenous Bacillus subtilis (natto) plasmids pTA1015 and pTA1040 contain signal peptidase-encoding genes: identification of a new structural module on cryptic plasmids. Mol Microbiol. 1995;17:621–631. doi: 10.1111/j.1365-2958.1995.mmi_17040621.x. [DOI] [PubMed] [Google Scholar]
- 20.Mergeay M, Nies D H, Schlegel H G, Gerits J, Charles P, Van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985;162:328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misra T K, Brown N L, Fritzinger D, Pridmore R, Barnes W, Silver S. Mercuric ion reductase operons of plasmid R100 and transposon Tn501: The beginning of the operon including the regulatory region and the first two structural genes. Proc Natl Acad Sci USA. 1984;84:5975–5979. doi: 10.1073/pnas.81.19.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakatsu C, Ng J, Singh R, Straus N, Wyndham C. Chlorobenzoate catabolic transposon Tn5271 is a composite class I element with flanking class II insertion sequences. Proc Natl Acad Sci USA. 1991;88:8312–8316. doi: 10.1073/pnas.88.19.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nucifora G, Chu L, Misra T K, Silver S. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc Natl Acad Sci USA. 1989;86:3544–3548. doi: 10.1073/pnas.86.10.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Halloran T V, Walsh C T. Metalloregulatory DNA-binding protein encoded by the merR gene, isolation and characterization. Science. 1987;235:211–214. doi: 10.1126/science.3798107. [DOI] [PubMed] [Google Scholar]
- 25.Podlesek Z, Comino A, Herzog-Velikonja B, Zgur-Bertok D, Komel R, Grabnar M. Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol Microbiol. 1995;16:969–976. doi: 10.1111/j.1365-2958.1995.tb02322.x. [DOI] [PubMed] [Google Scholar]
- 26.Rensing C, Mitra B, Rosen B P. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci USA. 1997;94:14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rensing C, Sun Y, Mitra B, Rosen B P. Pb(II)-translocating P-type ATPases. J Biol Chem. 1998;273:32614–32617. doi: 10.1074/jbc.273.49.32614. [DOI] [PubMed] [Google Scholar]
- 28.Roane T M. Lead resistance in two isolates from heavy metal-contaminated soils. Microb Ecol. 1999;37:218–224. doi: 10.1007/s002489900145. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Silver S. Bacterial resistances to toxic metal ions—a review. Gene. 1996;179:9–19. doi: 10.1016/s0378-1119(96)00323-x. [DOI] [PubMed] [Google Scholar]
- 31.Silver S, Nucifora G, Phung L T. Human Menkes X-chromosome disease and the staphylococcal cadmium-resistance ATPase: a remarkable similarity in protein sequences. Mol Microbiol. 1993;10:7–12. doi: 10.1111/j.1365-2958.1993.tb00898.x. [DOI] [PubMed] [Google Scholar]
- 32.Stearman R, Yuan D S, Yamaguchi-Iwai Y, Klausner R, Dancis A. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science. 1996;271:1552–1557. doi: 10.1126/science.271.5255.1552. [DOI] [PubMed] [Google Scholar]
- 33.Stoyanov J V, Hobman J L, Brown N L. CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol Microbiol. 2001;39:502–511. doi: 10.1046/j.1365-2958.2001.02264.x. [DOI] [PubMed] [Google Scholar]
- 34.Summers A O. Untwist and shout: a heavy metal-responsive transcriptional regulator. J Bacteriol. 1992;174:3097–3101. doi: 10.1128/jb.174.10.3097-3101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taghavi S, Mergeay M, Nies D, van der Lelie D. Alcaligenes eutrophus as a model system for bacterial interactions with heavy metals in the environment. Res Microbiol. 1997;148:536–551. doi: 10.1016/S0923-2508(97)88361-1. [DOI] [PubMed] [Google Scholar]
- 36.Taghavi S, van der Lelie D, Mergeay M. Electroporation of Alcaligenes eutrophus with (mega)plasmids and genomic DNA fragments. Appl Environ Microbiol. 1994;60:3585–3591. doi: 10.1128/aem.60.10.3585-3591.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu F, Yamada H, Daishima K, Mizushima S. Nucleotide sequence of the lspA gene, the structural gene for lipoprotein signal peptidase of Escherichia coli. FEBS Lett. 1984;173:264–268. doi: 10.1016/0014-5793(84)81060-1. [DOI] [PubMed] [Google Scholar]