Abstract
Infective endocarditis (IE) is a serious infection of the inner surface of heart, resulting from minor lesions in the endocardium. The damage induces a healing reaction, which leads to recruitment of fibrin and immune cells. This sterile healing vegetation can be colonized during temporary bacteremia, inducing IE. We have previously established a novel in vitro IE model using a simulated IE vegetation (IEV) model produced from whole venous blood, on which we achieved stable bacterial colonization after 24 h. The bacteria were organized in biofilm aggregates and displayed increased tolerance toward antibiotics. In this current study, we aimed at further characterizing the time course of biofilm formation and the impact on antibiotic tolerance development. We found that a Staphylococcus aureus reference strain, as well as three clinical IE isolates formed biofilms on the IEV after 6 h. When treatment was initiated immediately after infection, the antibiotic effect was significantly higher than when treatment was started after the biofilm was allowed to mature. We could follow the biofilm development microscopically by visualizing growing bacterial aggregates on the IEV. The findings indicate that mature, antibiotic‐tolerant biofilms can be formed in our model already after 6 h, accelerating the screening for optimal treatment strategies for IE.
Keywords: Biofilm, host response, infective endocarditis, histopathology, antibiotic tolerance, organoid
Infective endocarditis (IE) is a relatively rare, but life‐threatening infection of the endocardium, the inner layer of the heart, often involving native or prosthetic heart valves or implanted devices, such as pacemakers [1]. Left‐sided IE is increasing in incidence over the last two decades [2, 3, 4] and has substantial fatality rates even under modern treatment [5]. The most frequently identified microbial etiology in IE is Staphylococcus aureus [1, 6, 7, 8]. The pathophysiological basis for development of IE on native tissue is a lesion or minor defect in the endothelium of the endocardium, inducing a sterile healing vegetation consisting of fibrin, platelets, and leukocytes. This vegetation on the exposed heart surface, most often the valve, is susceptible to bacterial colonization during temporary bacteremia [9]. The bacteria attach to the healing defect and form a vegetation of bacteria, fibrin, and recruited immune cells, resulting in a biofilm mode of growth [10, 11]. The biofilm grows with further recruitment of immune cells and with bacteria producing the extracellular matrix, both leading to further damage of the heart valves. Biofilms are considered a treatment challenge in IE [12, 13] as they are characterized by slow growth and increased tolerance toward a wide range of antimicrobial agents and the host immune response [11].
The biofilm mode of growth is considered a significant contributor to the antibiotic tolerance of bacteria in IE, making surgery (in approximately one quarter of the cases) and high‐dose long‐term antibiotic treatment necessary for clearance of the infection [14]. The choice of antibiotic regimen for the individual patients is based on the minimal inhibitory concentration (MIC) of the infecting organism, as well as on clinical experience; however, this is often not optimal. MICs of a bacterium are determined based on planktonic, exponentially growing bacteria in a laboratory setting and do not take into account that metabolically more inactive, biofilm growing bacteria exhibit an increased tolerance toward a wide range of antibiotics [15, 16].
In order to improve the treatment and to identify novel treatment options, it is important to find experimental setups that can more realistically mimic the antibiotic tolerance development in slow growing biofilms. We have previously shown that we can establish a biofilm on biological simulated IE vegetations (IEVs), consisting of condensed blood, thereby containing fibrin, viable thrombocytes, and leukocytes [17], which are the essential components seen in the sterile healing vegetation in IE. We have provided evidence that the most relevant IE species, S. aureus, Enterococcus faecalis, and Streptococcus mitis grow in biofilm aggregates on our IEVs and exhibit an increased tolerance toward tobramycin, ciprofloxacin, and penicillin [17]. Previously, for proof‐of‐concept, we allowed the bacteria to form biofilms on the vegetation for 24 h, without following the dynamics of biofilm formation by means of temporal development of aggregates and antimicrobial tolerance. The purpose of this current project was to further characterize the key features of biofilm mode of growth in this novel IE simulation model by visualizing the formation of bacterial aggregates and the time‐dependent development of antibiotic tolerance of the bacterial biofilms.
MATERIALS AND METHODS
Preparation of the IE vegetation simulation model
The simulated IE vegetations (IEV) were prepared as we previously described [17]. In short, the IEVs were prepared from whole blood through a specific centrifugation process, resulting in a patch which consists of three distinct layers of fibrin, thrombocytes, and leukocytes. One full patch was 2.5 cm in diameter and 1–2 mm thick. Each patch was divided into seven 6 mm IEVs using punch biopsies (Kai industries, Solingen, Germany) and randomized to the different treatment groups.
Preparation of assay medium
All bacteria and antibiotics were diluted in a medium containing equal parts of Krebs–Ringer buffer (SSC Panum, Copenhagen, Denmark; containing 0.2% (5.2 mM) D‐Glucose (Merck, Darmstadt, GER)), and lysogeny broth (LB).
Bacterial preparation
Clinical IE isolates of Staphylococcus aureus were derived from the sample collection of the Partial Oral antibiotic Endocarditis Treatment (POET) trial [14, 18]. The bacterial strains used in this project are listed in Table 1.
Table 1.
Staphylococcus aureus strains used in this work
| Strain | Specifics | MIC (μg/mL) | ||
|---|---|---|---|---|
| Penicillin G | Tobramycin | Ciprofloxacin | ||
| Staphylococcus aureus | NCTC 8325–4 | 0.03 | 0.125 | 0.25 |
| AH 2547 (GFP‐tagged NCTC 8325–4) | 0.03 | 0.125 | 0.25 | |
| Clinical IE isolate | R | 0.5 | 0.64 | |
| Clinical IE isolate | R | 0.25 | 0.19–0.25 | |
| Clinical IE isolate | R | 0.75 | 0.64–0.94 | |
Abbreviations:MIC, minimal inhibitory concentration; GFP, Green‐fluorescent protein; IE, Infective endocarditis.
The minimal inhibitory concentrations (MICs) for penicillin, tobramycin, and ciprofloxacin were determined by E‐tests (Biomérieux, Ballerup, Denmark). The bacteria were incubated overnight (shaking, 38°C) in LB. After preparation of the patches, the bacterial culture was diluted in the assay medium.
Preparation of antibiotics
Penicillin (Penicillin G, 1.2 g, Panpharma, Luitré, France), Tobramycin (40 mg/mL, Eurocept international, Ankeveen, Netherlands), and Ciprofloxacin (2 mg/mL, Fresenius Kabi, Uppsala, Sweden) were diluted in the assay medium to final well concentrations of 10× MIC of the respective bacterial strain.
Antibiotic exposure assay
The workflow of the antibiotic exposure assay is depicted in Fig. 1.
Day 0—Preparation of an overnight culture of the respective bacteria in LB.
Day 1—Inoculation and Biofilm formation: The IEVs were randomized into the wells of a 96‐well plate (fibrin‐side facing up). Fifty μL of a 1:1000 diluted bacterial overnight culture (corresponding to approx. 106 CFU/mL; in assay medium) was added to each IEV. Initially (0 h), or after 3 or 6 h, 150 μL of either assay‐medium or the respective antibiotics (10x MIC) were added, and the plates were incubated for 24 h at 37°C under gentle tilting.
Day 2—After a treatment period of 24 h, the IEVs were transferred into 2 mL tubes (Nerbe plus, Winsen/Luher, Germany), containing 200 μL 0.9% saline (SSC Panum, Copenhagen, Denmark) and 1 glass bead (VWR International, Søborg, Denmark). The samples were homogenized in a TissueLyser II (Qiagen, Copenhagen, Denmark) (30 min; 30/s). Serial dilutions were prepared in 0.9% saline containing 2.5% Tween80 (Merck, Darmstadt, Germany). Samples were plated on blood agar (SSI, Hillerød, Denmark) and incubated at 38 °C overnight. Colony forming units (CFU) were counted the next day.
Fig. 1.
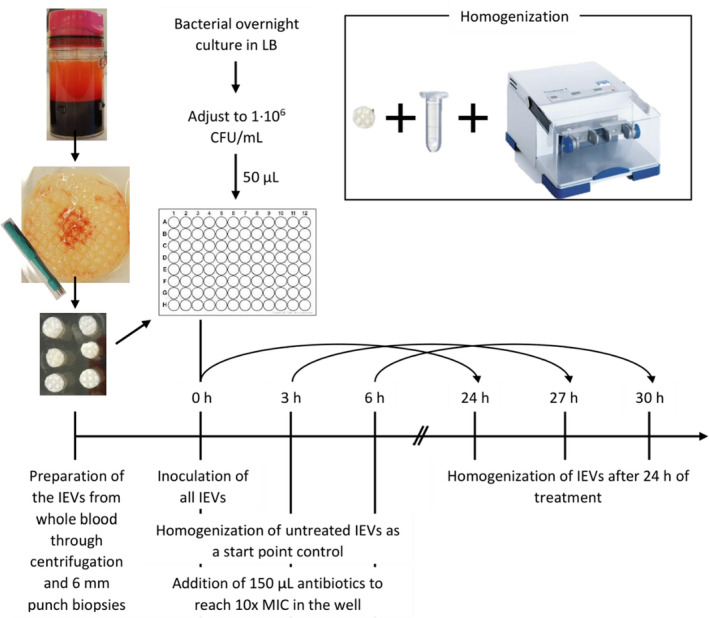
Experimental workflow for antibiotic exposure assay. Fibrin‐rich patches were freshly produced from whole blood of healthy, voluntary donors and divided into 6 mm punch biopsies. At the start of the experiment, each biopsy was inoculated with 5 × 104 CFU of a Staphylococcus aureus suspension to create the IEVs. At the start and after 3 or 6 h, some IEVs were homogenized (2 mL tubes containing one glass bead and 200 μL saline, TissueLyserII, 30/s, 10 min) and plated for CFU count as a start control. Other biopsies were treated with 10× MIC concentrations of tobramycin, ciprofloxacin, penicillin, or their combinations. After 24 h of treatment, the IEVs were homogenized and CFU were evaluated the next day.
Antibiotic tolerance was defined as a ≤ 2 log reduction of CFU/g.
Microscopy
IEVs inoculated with the GFP‐tagged S. aureus strain were transferred into Eppendorf tubes containing 10% buffered formalin either initially or 3, 6, or 9 h post‐bacterial inoculation. IEV were stored in formalin at 4°C in the dark until use.
Tissue preparation was performed at the University of Copenhagen (Histolab Panum Institute, Copenhagen, Denmark). From each IEV, slides were made by cutting vertical sections in the middle of the IEV for either hematoxylin/eosin (H&E) staining (Histolab Panum Institute, Copenhagen, Denmark) or keeping them unstained for detection of GFP‐tagged bacteria.
Unstained slides for visualization of GFP‐tagged bacteria were deparaffinized by sequential washing: Xylene (two times, 3 min), 50:50 mixture of Xylene and 100% Ethanol (3 min), 100% ethanol (two times, 3 min), then 3 min each in 95%, 70%, and 50% ethanol. The slides were rinsed and stored in tap water until further use.
Slides were evaluated for the formation of bacterial aggregates on the IEVs by light microscopy (Olympus BX53, Ballerup, Denmark) or fluorescent microscopy (100× magnification: Olympus BX53 with fluorescence cube, Ballerup, Denmark; 630× magnification: see below under CLSM) (Imaging software: cellSense, Olympus, Ballerup, Denmark).
Confocal laser scanning microscopy
For confocal laser scanning microscopy (CLSM), IEVs incubated with GFP‐tagged S. aureus for 24 h were washed, loaded onto microscopy slides (fibrin side facing up), and covered with a cover slip.
CLSM (Axio Imager.Z2, LSM710 CLSM; Zeiss, Jena, Germany/3D reconstruction software Zen Black 2010 (version 6.0; Zeiss)/image analysis software Imaris (version 8, Bitplane, Zuerich, Switzerland)) was carried out at the Costerton Biofilm Center, University of Copenhagen (Denmark). CLSM images were recorded at an excitation wavelength of 488 nm and an emission wavelength of 492–589 nm with the corresponding beam‐splitter MBS 594 (Zeiss) using a 63×/1.4 plan‐apochromat oil objective (Zeiss). Images with a resolution of 1024 × 1024 pixels (scanned twice) and a color depth of 16 bits were subsequently deconvoluted into Tiff‐files.
Ethical considerations
This project exclusively utilizes blood from anonymous, voluntary healthy donors, which cannot be linked to specific persons and is therefore exempted from approval by the Committee on Health Research Ethics, according to Danish legislation (Komitéloven §14, Stk.3). In addition, all biological material was discarded after ending the experiments.
Statistics
Statistical analyses were carried out in Prism 9 (GraphPad, San Diego, USA; version 9.0.0). Quantitative non‐parametric data were analyzed by Mann–Whitney test or Kruskal–Wallis for multiple comparisons. A p‐value ≤ 0.05 was regarded statistically significant.
RESULTS
Formation of microscopically visible bacterial aggregates
Light microscopy of the IEVs revealed visible biofilm‐like bacterial aggregates as early as 6 h after inoculation, while no bacteria could be seen after 0 and 3 h. Biofilm was exclusively found on the fibrin layer, not on the leukocyte layer (Fig. 2, left panels). Using fluorescence microscopy, we could confirm that the bacteria seen in light microscopy are S. aureus due to the strong GFP‐signal (Fig. 2, right panels).
Fig. 2.
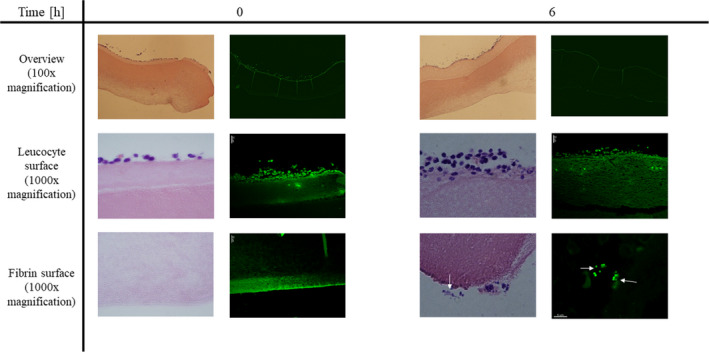
Biofilm development is microscopically visible after 6 h. IEVs were inoculated with 5 × 104 CFU GFP‐tagged Staphylococcus aureus and 2 IEVs were transferred into 4% formalin either right after infection (0 h), or after 3, 6, and 9 h. Fixated IEVs were stored in formalin in the fridge until usage. IEVs were then embedded in paraffin, and vertical slices were prepared for microscopy by H&E staining (light microscopy, left panels) or deparaffination for detection of GFP‐tagged bacteria (fluorescence microscopy, right panels) to ensure that the observed aggregates are indeed S. aureus. Displayed are representative areas of 2 biopsies at 0 and 6 h after inoculation at 100‐ and 1000‐fold (H&E) or 630‐fold (GFP) magnification. Biofilm‐like bacterial aggregates (white arrows) were observed after 6 and 9 (data not shown) hours and located exclusively on the fibrin‐surface of the IEV.
Confocal laser scanning microscopy of the IEVs incubated with GFP‐tagged S. aureus for 24 h, confirmed the dispersed aggregate formation on the fibrin surface in high resolution (Fig. 3).
Fig. 3.
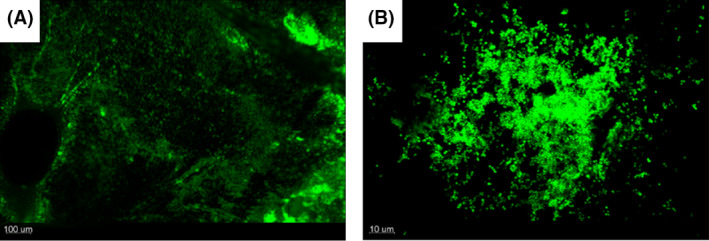
Confocal laser scanning microscopy reveals uneven distribution of biofilms on the fibrin surface of the IEV. IEVs were inoculated with 5 × 104 CFU GFP‐tagged Staphylococcus aureus and incubated for 24 h. IEVs were washed twice in 200 μL saline and loaded onto microscopy slides with the fibrin surface facing up. Bacterial biofilm formation was observed with confocal laser scanning microscopy in 100x magnification (A) and 630× magnification (B). Displayed is the top‐down view of a representative area of one IEV (A) and one of the aggregates in close up (B).
Development of antibiotic tolerance after 6 h
When initiating a 24‐h antibiotic treatment period at the same timepoint as inoculation or 3 or 6 h after, an overall increase in antibiotic tolerance was seen (Fig. 4). Figure 4A displays the results for the laboratory S. aureus strain MH 8325, while Fig. 4B shows the combined results for all four S. aureus strains tested. Penicillin alone and in combination with tobramycin and ciprofloxacin was only tested for the laboratory strain, as the clinical isolates were resistant toward this antibiotic. The log10 reductions of viable bacteria compared with the bacterial number at treatment right before initiation and compared with the untreated controls at the endpoint, are displayed in Table 2.
Fig. 4.
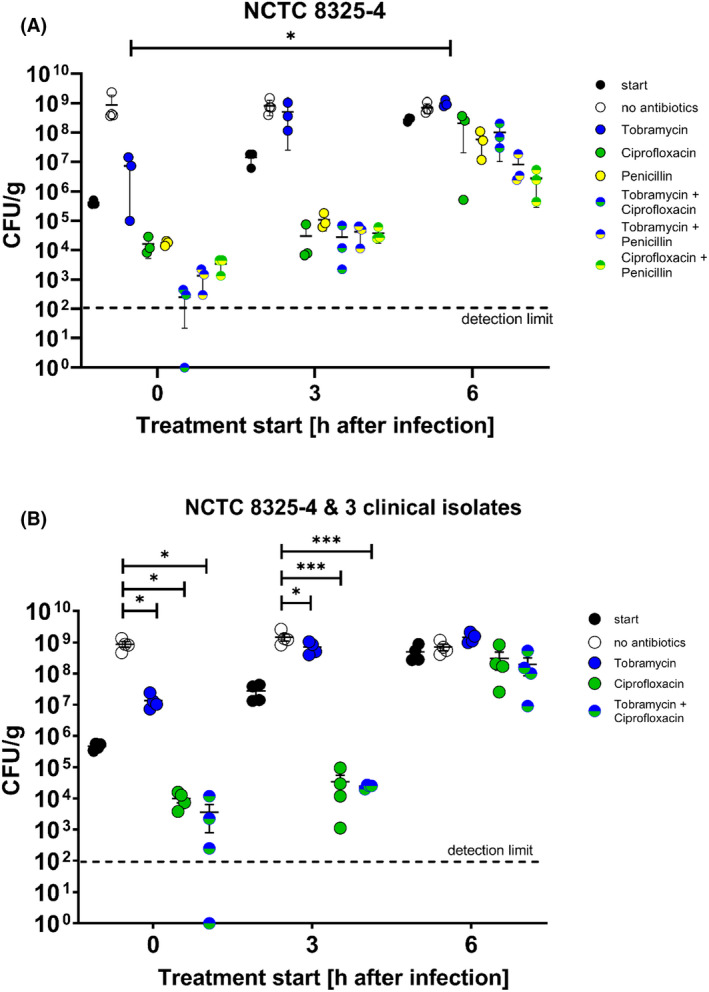
Development of antimicrobial tolerance during Staphylococcus aureus biofilm formation. IEVs were inoculated with 5 × 105 CFU S. aureus NCTC 8325–4 (A) or NCTC 8325–4 and 3 different clinical IE isolates (B). Treatment with penicillin, tobramycin, ciprofloxacin, or their combinations (all at 10× MIC concentrations) was initiated either right at the start, or after 3 or 6 h and applied for 24 h. “Start” control‐IEVs were homogenized before treatment‐initiation. Untreated (“no antibiotics”) controls were supplied with a corresponding volume of fresh media. All IEVs were homogenized 24 h after treatment start and CFU were counted the day after. The experiment was performed with IEVs produced from blood of three (NCTC 8325–4) or two (clinical isolates) different donors. One dot represents the mean of duplicates or triplicates from one donor. Displayed are the means ± SD (A) or SEM (B). Relevant statistically significant differences are indicated with: *: p ≤ 0.05; **: p ≤ 0.01, and ***: p ≤ 0.001.
Table 2.
Log10 reductions of Fig. 4
| Antibiotic | Treatment start (h after inoculation) | Log10 reduction compared to | |
|---|---|---|---|
| Treatment start | Untreated endpoint | ||
| Tobramycin | 0 | −/− | 2.6/1.85 |
| 3 | −/− | 0.37/0.36 | |
| 6 | −/− | −/− | |
| Ciprofloxacin | 0 | 1.48/1.72 | 4.8/5 |
| 3 | 2.95/3.29 | 4.72/5.03 | |
| 6 | 0.89/0.48 | 1.29/0.63 | |
| Penicillin | 0 | 1.39 | 4.71 |
| 3 | 2.17 | 3.93 | |
| 6 | 0.84 | 1.24 | |
| Tobramycin + Ciprofloxacin | 0 | 3.91/3.46 | 7.23/6.74 |
| 3 | 3.07/3.05 | 4.83/4.79 | |
| 6 | 0.57/0.73 | 0.97/0.88 | |
| Tobramycin + Penicillin | 0 | 2.61 | 5.94 |
| 3 | 2.63 | 4.4 | |
| 6 | 1.72 | 2.11 | |
| Ciprofloxacin + Penicillin | 0 | 2.14 | 5.47 |
| 3 | 2.62 | 4.38 | |
| 6 | 2.19 | 2.59 | |
Note:The means of the values displayed in Fig. 4A and B were calculated. Log10 CFU reductions of the treated samples were set in comparison with colony counts before treatment initiation and to untreated controls at the endpoint. Values seperated by a slash indicate the results for Fig. 4A/B. The “−” indicates no reduction.
Figure 4A + B and Table 2 show that tobramycin alone could only achieve a reduction of biofilm growth, but no killing. While treatment initiated right after inoculation achieved a 2.6 (Fig. 4A)/1.85 (Fig. 4B) log10 CFU growth inhibition compared with untreated controls, no bacterial reduction was detected after 6 h.
Ciprofloxacin and penicillin (only Fig. 4A), as well as the combination treatments, were able to reduce the viable bacteria in the biofilm, compared with the start point before treatment initiation. However, the efficiency of the antibiotics decreased when treatment was started 6 h after inoculation, compared with right after inoculation (Table 2). Notably, only the combination treatment of ciprofloxacin and penicillin on the laboratory strain achieved a comparable 2 log CFU reduction compared with treatment start for all three timepoints.
DISCUSSION
We have previously shown that our model is suitable to establish an antibiotic‐tolerant biofilm on the surface of the IEVs after 24 h, with bacteriologies comparable with observations in valve vegetations seen in IE patients [17]. Here, we have further evaluated the dynamics by means of correlating the time course of biofilm establishment to the development of antibiotic tolerance. In vitro biofilm development is considered to be a three‐step process, consisting of bacterial attachment, biofilm maturation, and biofilm detachment [19]. For the laboratory S. aureus strain 8325–4, that was used here, multiple important virulence factors to enable the initial colonization, are well known (e.g. [20, 21, 22]). We could show that S. aureus attached to the IEVs within a few seconds (Fig. 4: “before”; data not shown). This was expected, as the bacterial adhesion to different surfaces is rapidly mediated by charge forces and has previously been shown to occur on artificial surfaces within seconds [23].
Correspondingly, in Fig. 4, it can be seen that even bacteria that were treated immediately after being added to the IEV, were not eradicated by a single exposure to above‐MIC concentrations of the antibiotics tobramycin, ciprofloxacin (all strains, Fig. 4A,B), and penicillin (only laboratory strain, Fig. 4A). They were, however, more susceptible to antibiotics than bacteria that were allowed to form biofilms for 3 or 6 h prior to infection (Fig. 4A,B). This was demonstrated by decreasing log10 reductions of the CFU of antibiotic‐treated IEVs compared with the start and to untreated controls at the same endpoint. This correlates with the generally known phenomenon that the minimal biofilm inhibitory concentration (MBIC) and the minimal biofilm eradication concentration (MBEC) are often 2‐1000‐fold higher compared with the MIC of the bacteria [24, 25, 26].
With this current work, we wanted to evaluate if biofilm development in our IEV model requires full 24 h, which is the duration of several other studies to observe the mature biofilms [27, 28]. We could, however, demonstrate that biofilms could already develop after 6 h under the chosen conditions, which is in line with similar observations made in P. aeruginosa biofilm development [29]. This is influenced by the initial inoculum of 106 CFU/mL, resulting in 5 × 104 CFU/IEV, corresponding to approx. 2 × 106 CFU/g IEV. It is uncertain which bacterial burden in the blood stream is sufficient to establish adhesion to the endocardium, resulting in initiation of IE [30]. In one study, rats were intravenously injected with 103 and 104 CFU S. aureus [31]. In an average rat (300 g) with a total blood volume of around 20 mL, this inoculum approximately corresponds to 50 to 5 × 102 CFU/mL blood. The study demonstrated that even low inoculation concentrations were able to establish endocarditis in approximately half of the rats. However, we have previously shown that an inoculum above 104 CFU/g IEV was required to establish stable infections in our model, characterized by bacterial loads comparable with animal models and other in vitro models [17]. Bacterial inocula between the used concentration of 2 × 106 CFU/g, but above the threshold concentration of 104 CFU/g resulted in an up to 3 h delay of biofilm formation (data not shown), while inocula below the threshold only infrequently and unstably resulted in biofilm formation. Having mature biofilms already after 6 h, compared with the previously used 24 h, allows us to expose the biofilms to antibiotics one day earlier, speeding up the screening of anti‐biofilm activity. Our in vitro data suggest that bacteria are able to change into a metabolically slow growing biofilm state within a relatively short time; however, a possible clinical relevance of this observation remains to be evaluated further in future experiments.
In the present study, we tested our simulation IEV model using three clinical isolates from IE patients from the POET study [14, 18]. All produced penicillinase and were therefore resistant to penicillin. As around one third of clinical S. aureus isolates remain penicillin‐susceptible [32, 33], we chose to also include our penicillin‐susceptible laboratory reference strain in this project. As could be expected, all four employed S. aureus strains resulted in stable biofilm formation and reacted similarly to the tested antibiotics. This was furthermore independent of the blood donor, indicating that the model can be a reliable way to screen anti‐biofilm activities and antibiotic efficacies against a wide range of bacterial isolates from IE patients.
Regarding the response of the IE vegetation simulation model to the chosen antibiotics, different patterns were observed. Our data suggest that aminoglycosides, such as tobramycin, have very limited effect on biofilm‐resident S. aureus (Fig. 4). This is in line with our observations using a rat model of S. aureus aorta IE where tobramycin was ineffective in killing bacteria on the heart valves and in the myocardium [34, 35], supporting that aminoglycosides are not an eligible antibiotic class for monotherapy of S. aureus IE. Both the fluoroquinolone and penicillin revealed better antibacterial effects (Fig. 4) until tolerance was obtained. After clarifying the essential temporal dynamics between bacterial aggregate formation and development of antibiotic tolerance to tobramycin, ciprofloxacin, and penicillin, we can proceed to use our IEV model to test optimal antibiotic combinations, concentrations, and treatment timing and to study additional important IE pathogens as mentioned above.
CONCLUSION
In this project, we investigated the temporal dynamics of our novel IEV model. We were able to evaluate the time‐course of biofilm development and correlate it directly to the development of antibiotic tolerance. We showed that under the chosen conditions, mature biofilms were established after 6 h, which was characterized by antibiotic tolerance and microscopically visible biofilm‐like bacterial aggregates on the fibrin surface of the patch. Based on these results, we can proceed to use the IEV model to investigate optimal antibiotic and anti‐biofilm treatment strategies against other clinical IE bacteria, such as E. faecalis or Streptococcus spp.
FUNDING
This work was supported by the Novo Nordisk Fonden (NNF17OC0025074) to MD, PhD Claus Moser and the A.P. Møller Fonden (19‐L‐0283) to MD, PhD Christian J. Lerche, the Swedish Society for Medical Research (PD20‐0031), the Tornspiran Foundation, the Foundation for the memory of Sigurd and Elsa Golje (LA2021‐0027), the Lund University foundations (RMh2021‐0001 and RMv2021‐0002), the Längmanska Foundation (BA21‐0430), the Royal Physiographic Society of Lund (42001), and the Thelma Zoégas Fund for Medical Research (TZ2021‐0008) to MD, PhD Torgny Sunnerhagen and the Rigshospitalet introduktionsstipendium (E‐22416‐07) to MSc, Franziska A. Schwartz.
CONFLICTS OF INTEREST
All authors declare that they have no conflict of interest.
ACKNOWLEDGEMENTS
We thank Heidi M. Poulsen from the Panum Histolab for the sample preparations for the microscopy.
Schwartz FA, Nielsen L, Andersen JS, Bock M, Christophersen L, Sunnerhagen T, Lerche CJ, Bay L, Bundgaard H, Lerche CJ, Høiby N, Moser C. Dynamics of a Staphylococcus aureus infective endocarditis simulation model. APMIS. 2022; 130: 515–523.
DATA AVAILABILITY STATEMENT
The data are available on request to the authors.
References
- 1. Holland T, Baddour L, Bayer A, Hoen B, Miro J, Fowler VG. Infective endocarditis. Nat Rev Dis Primers. 2017;2:1–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jensen AD, Bundgaard H, Butt JH, Bruun NE, Voldstedlund M, Torp‐Pedersen C, et al. Temporal changes in the incidence of infective endocarditis in Denmark 1997–2017: a nationwide study. Int J Cardiol. 2021;326:145–52. [DOI] [PubMed] [Google Scholar]
- 3. Olmos C, Vilacosta I, Fernández‐Pérez C, Bernal JL, Ferrera C, García‐Arribas D, et al. The evolving nature of infective endocarditis in Spain: a population‐based study (2003 to 2014). J Am Coll Cardiol. 2017;70(22):2795–804. [DOI] [PubMed] [Google Scholar]
- 4. Erichsen P, Gislason GH, Bruun NE. The increasing incidence of infective endocarditis in Denmark, 1994–2011. Eur J Intern Med. 2016;35(2016):95–9. [DOI] [PubMed] [Google Scholar]
- 5. Lamas C. Infective endocarditis: still a deadly disease. Arq Bras Cardiol. 2020;114(1):10–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cecchi E, Chirillo F, Castiglione A, Faggiano P, Cecconi M, Moreo A, et al. Clinical epidemiology in Italian registry of infective endocarditis (RIEI): focus on age, intravascular devices and enterococci. Int J Cardiol. 2015;190(1):151–6. [DOI] [PubMed] [Google Scholar]
- 7. Bor DH, Woolhandler S, Nardin R, Brusch J, Himmelstein DU. Infective endocarditis in the U.S., 1998‐2009: a Nationwide study. PLoS One. 2013;8(3):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Toyoda N, Chikwe J, Itagaki S, Gelijns A, Adams DH, Egorova NN. Trends in infective endocarditis in California and New York state, 1998–2013. JAMA. 2017;16(317):1652–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Werdan K, Dietz S, Löffler B, Niemann S, Bushnaq H, Silber R‐E, et al. Mechanisms of infective endocarditis: pathogen‐host interaction and risk states. Nat Rev Cardiol. 2014;11(1):35–50. [DOI] [PubMed] [Google Scholar]
- 10. Bjarnsholt T. The role of biofilms in chronic infections. Oxford: Blackwell Publ Ltd; 2013. [DOI] [PubMed] [Google Scholar]
- 11. Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35(4):322–32. [DOI] [PubMed] [Google Scholar]
- 12. Elgharably H, Hussain ST, Shrestha NK, Blackstone EH, Pettersson GB. Current hypotheses in cardiac surgery: biofilm in infective endocarditis. Semin Thorac Cardiovasc Surg. 2016;28(1):56–9. [DOI] [PubMed] [Google Scholar]
- 13. Elgharably H, Hussain ST, Shrestha NK, Pettersson GB. Biofilm in infective endocarditis and clinical implications. Biofilm, Pilonidal Cysts and Sinuses. Berlin: Springer. 2018. p. 109–20. [Google Scholar]
- 14. Bundgaard H, Ihlemann N, Gill SU, Bruun NE, Elming H, Madsen T, et al. Long‐term outcomes of partial Oral treatment of endocarditis. N Engl J Med. 2019;380(14):1373–4. [DOI] [PubMed] [Google Scholar]
- 15. Høiby N, Bjarnsholt T, Moser C, Bassi GLL, Coenye T, Donelli G, et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect. 2015;21:S1–25. [DOI] [PubMed] [Google Scholar]
- 16. Ciofu O, Moser C, Østrup Jensen P, Høiby N. Tolerance and resistance of microbial biofilms. Nat Rev Microbiol. 2022. doi: 10.1038/s41579-022-00682-4. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 17. Schwartz FA, Christophersen L, Laulund AS, Lundquist R, Lerche C, Rude Nielsen P, et al. Novel human in vitro vegetation simulation model for infective endocarditis. APMIS. 2021;129(11):653–62. [DOI] [PubMed] [Google Scholar]
- 18. Iversen K, Ihlemann N, Gill SU, Madsen T, Elming H, Jensen KT, et al. Partial Oral versus intravenous antibiotic treatment of endocarditis. N Engl J Med. 2019;380(5):415–24. [DOI] [PubMed] [Google Scholar]
- 19. O'Toole GO, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. [DOI] [PubMed] [Google Scholar]
- 20. Caiazza NC, O'Toole GA. Alpha‐toxin is required for biofilm formation by staphylococcus aureus. J Bacteriol. 2003;185(10):3214–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cue D, Lei MG, Luong TT, Kuechenmeister L, Dunman PM, O'Donnell S, et al. Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J Bacteriol. 2009;191(20):6363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gross M, Cramton SE, Götz F, Peschel A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun. 2001;69(5):3423–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boks NP, Busscher HJ, Van Der MHC, Norde W. Bond‐strengthening in staphylococcal adhesion to hydrophilic and hydrophobic surfaces using atomic force microscopy. Langmuir. 2008;24(16):12990–4. [DOI] [PubMed] [Google Scholar]
- 24. Ciofu O, Rojo‐Molinero E, Macià MD, Oliver A. Antibiotic treatment of biofilm infections. APMIS. 2017;125(4):304–19. [DOI] [PubMed] [Google Scholar]
- 25. Ceri H, Olson ME, Stremick C, Read RR, Morck D. The Calgary biofilm device. J Clin Microbiol. 1999;37(6):1771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hengzhuang W, Wu H, Ciofu O, Song Z, Høiby N. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2011;55(9):4469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oliveira M, Nunes SF, Carneiro C, Bexiga R, Bernardo F, Vilela CL. Time course of biofilm formation by Staphylococcus aureus and Staphylococcus epidermidis mastitis isolates. Vet Microbiol. 2007;124(1–2):187–91. [DOI] [PubMed] [Google Scholar]
- 28. Chen X, Xu Y, Winkler H, Thomsen TR. Influence of biofilm growth age, media and antibiotics exposure time on Staphylococcus aureus and Pseudomonas aeruginosa biofilm removal in vitro. BMC Microbiol. 2020;20(264):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Høiby N, Henneberg K‐Å, Wang H, Stavnsbjerg C, Bjarnsholt T, Ciofu O, et al. Formation of Pseudomonas aeruginosa inhibition zone during tobramycin disk diffusion is due to a transition from planktonic to biofilm mode of growth. Int J Antimicrob Agents. 2019;53:564–73. [DOI] [PubMed] [Google Scholar]
- 30. Yallowitz AW, Decker LC. Infectious endocarditis. Treasure Island (FL): StatPearls Publishing; 2022. [PubMed] [Google Scholar]
- 31. Veloso TR, Amiguet M, Rousson V, Giddey M, Vouillamoz J, Moreillon P, et al. Induction of experimental endocarditis by continuous low‐grade bacteremia mimicking spontaneous bacteremia in humans. Infect Immun. 2011;79(5):2006–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iversen K, Ihlemann N, Gill SU, Madsen T, Elming H, Jensen KT, et al. Supplementary appendix partial Oral versus intravenous antibiotic treatment of endocarditis. N Engl J Med. 2019;380(5):415–24. [DOI] [PubMed] [Google Scholar]
- 33. Resman F, Thegerström J, Månsson F, Ahl J, Tham J, Riesbeck K. The prevalence, population structure and screening test specificity of penicillin‐susceptible Staphylococcus aureus bacteremia isolates in Malmö. Sweden J Infect. 2016;73(2):129–35. [DOI] [PubMed] [Google Scholar]
- 34. Lerche CJ, Christophersen LJ, Kolpen M, Nielsen PR, Trøstrup H, Thomsen K, et al. Hyperbaric oxygen therapy augments tobramycin efficacy in experimental Staphylococcus aureus endocarditis. Int J Antimicrob Agents. 2017;50(3):406–12. [DOI] [PubMed] [Google Scholar]
- 35. Lerche CJ, Christophersen LJ, Trøstrup H, Thomsen K, Jensen P, Hougen HP, et al. Low efficacy of tobramycin in experimental Staphylococcus aureus endocarditis. Eur J Clin Microbiol Infect Dis. 2015;34(12):2349–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data are available on request to the authors.


