Abstract
Background
Psoriasis is an immune‐mediated inflammatory skin disease, in which an interplay between infiltrating immune cells and keratinocytes sustains chronic skin inflammation. Interleukin (IL)‐17A is a key inflammatory cytokine in psoriasis and its main cellular targets are keratinocytes.
Objectives
To explore the role of miR‐378a in psoriasis.
Methods
Keratinocytes obtained from psoriatic skin and healthy epidermis were separated by magnetic sorting, and the expression of miR‐378a was analysed by quantitative polymerase chain reaction. The regulation and function of miR‐378a was studied using primary human keratinocytes. The expression of miR‐378a was modulated by synthetic mimics, and nuclear factor kappa B (NF‐κB) activity and transcriptomic changes were studied. Synthetic miR‐378a was delivered to mouse skin in conjunction with induction of psoriasiform skin inflammation by imiquimod.
Results
We show that miR‐378a is induced by IL‐17A in keratinocytes through NF‐κB, C/EBP‐β and IκBζ and that it is overexpressed in psoriatic epidermis. In cultured keratinocytes, ectopic expression of miR‐378a resulted in the nuclear translocation of p65 and enhanced NF‐κB‐driven promoter activity even in the absence of inflammatory stimuli. Moreover, miR‐378a potentiated the effect of IL‐17A on NF‐κB nuclear translocation and downstream activation of the NF‐κB pathway. Finally, injection of miR‐378a into mouse skin augmented psoriasis‐like skin inflammation with increased epidermal proliferation and induction of inflammatory mediators. Mechanistically, miR‐378a acts as a suppressor of NFKBIA/IκBζ, an important negative regulator of the NF‐κB pathway in keratinocytes.
Conclusions
Collectively, our findings identify miR‐378a as an amplifier of IL‐17A‐induced NF‐κB signalling in keratinocytes and suggest that increased miR‐378a levels contribute to the amplification of IL‐17A‐driven skin inflammation in psoriasis.
What is already known about this topic?
Interleukin (IL)‐17A induces production of inflammatory mediators in keratinocytes and is directly or indirectly responsible for many of the features observed in psoriatic skin.
MicroRNAs (miRNAs) are regulators of gene expression and can regulate the duration or extent of inflammatory responses.
The level of several miRNAs is altered in psoriatic skin lesions.
What does this study add?
We show that IL‐17A induces miR‐378a in keratinocytes and that miR‐378a is overexpressed in psoriatic epidermis.
Our findings demonstrate that miR‐378a amplifies basal and IL‐17A‐induced nuclear factor kappa B signalling and cytokine production in keratinocytes.
Injection of synthetic miR‐378a into mouse skin exacerbates psoriasis‐like skin inflammation.
What is the translational message?
Fine tuning the IL‐17A‐responsiveness of keratinocytes by modulating miR‐378a and/or its targets may represent a new approach to break the vicious circle of inflammation in psoriasis.
Our findings prompt further research to explore the potential of miRNA modulation in psoriasis and other inflammatory skin diseases.
Psoriasis is a chronic immune‐mediated skin disease in which keratinocytes play an important role as targets for immune‐cell derived cytokines and amplifiers of skin inflammation. Here, we identify the miRNA named miR‐378a as an amplifier of IL‐17A‐induced NF‐κB signalling in keratinocytes and suggest that increased miR‐378a levels contribute to the amplification of IL‐17A‐driven skin inflammation in psoriasis.
Linked Comment: C. Johansen. Br J Dermatol 2022; 187:137–138.
Psoriasis is a common, chronic papulosquamous skin disease occurring worldwide, which can present at any age, and leads to a substantial burden for individuals and society. A combination of genetic susceptibility and environmental triggers such as streptococcal infection, stress, smoking, obesity and alcohol consumption lead to the disease, 1 in which chronic inflammation is driven by activation of the interleukin (IL)‐23/T helper (Th)17/IL‐17 axis, leading to hyperproliferation, altered differentiation and activation of keratinocytes. 2 Keratinocytes are more than just passive bystanders in the pathogenesis of psoriasis; in response to inflammatory mediators, stress, damage and innate triggers, they produce a wide array of inflammatory cytokines, chemokines and antimicrobial peptides, further amplifying skin inflammation by recruiting immune cells to the skin. 3 One of the key cytokines in psoriasis is IL‐17A, as evidenced by the high efficacy of monoclonal antibodies targeting IL‐17A or its receptor IL‐17RA for the treatment of patients with psoriasis. 4 IL‐17A signals through a receptor complex that includes IL‐17RA and IL‐17RC. 5 While IL‐17RA is expressed on multiple cell types, IL‐17RC is expressed mainly on epithelial cells. 6 In the skin, the main target cells for IL‐17A are the keratinocytes, in which IL‐17A signalling leads to activation of innate immune responses with the induction of antimicrobial peptides, cytokines and chemoattractants through the activation of the nuclear factor kappa B (NF‐κB), p38‐MAPK, C/EBP‐β and other pathways. 7 , 8 Little is known about the factors defining the intensity of keratinocyte responses to IL‐17A, even though this question is of interest, as identification of such factors could lead to therapies that attenuate this response and normalize psoriasis keratinocytes.
MicroRNAs (miRNAs) are small noncoding RNAs that suppress the expression of a set of target genes at the post‐transcriptional level via sequence‐specific interaction with miRNA‐recognition elements in their 3′UTR. 9 Moreover, miRNAs are potent regulators of biological functions, including inflammatory signalling pathways, in which they often act as modulators of the inflammatory response by being involved in regulatory feedback loops. 9 Our previous research, in addition to the work of others, has identified miRNAs that are deregulated in psoriasis, which can contribute to the pathogenesis by regulating cellular processes relevant to the disease, in keratinocytes and/or in immune cells. 10 , 15
Recently, we performed small RNA sequencing on keratinocytes isolated from lesional and nonlesional psoriatic skin and from healthy skin, and identified miRNAs with altered levels in psoriatic keratinocytes. 16 One of the miRNAs we identified to be overexpressed in psoriatic keratinocytes was miR‐378a‐3p (previously known as miR‐422b, and hereafter referred to as miR‐378a). In this study, we aimed to explore the regulation and function of miR‐378a in keratinocytes and its potential role in psoriasis.
Materials and methods
Patients and biopsies
For reverse‐transcription quantitative real‐time polymerase change reaction (RT‐qPCR) miRNA expression analysis, two 4‐mm full‐depth punch biopsies were taken from lesional (PP) and nonlesional (PN) skin on the back or buttock of the patients with chronic plaque psoriasis (n = 20) and from skin of age‐matched healthy volunteers (n = 19). All patients with psoriasis were examined by a dermatologist, and Psoriasis Area and Severity Index score was determined at the time of biopsy collection (Table S1; see Supporting Information). Patients with psoriasis had received no systemic immunosuppressive or ultraviolet (UV)B/psoralen plus UVA treatments for at least 4 weeks or topical therapy for at least 2 weeks before skin biopsy. All the participants included in the study were white. The study was approved by the Stockholm Regional Ethics Committee and conducted according to the Declaration of Helsinki principles. Written informed consent was obtained from all participants prior to enrolment.
Cell isolation and RNA extraction
Epidermal sheets were prepared from the biopsies and CD45− cells were obtained by magnetic cell sorting as previously described. 16 Total RNA was isolated from the CD45− cell fraction using the miRNeasy Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. RNA was quantified by Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Transcriptomic profiling and enrichment analysis
Transcriptomic profiling was performed using Affymetrix GeneChip® Human Gene 2·0 ST Array (Stockholm, Sweden). Data analysis was performed using Transcriptome Analysis Console (Thermo Fisher Scientific). Gene enrichment analyses were performed using EnrichR. 17 Further details are provided in Appendix S1 (see Supporting Information).
Cell culture, treatments and transfections
Primary human keratinocytes (Catalog# C0055C, Thermo Fisher Scientific) were cultured and treated as indicated in Appendix S1.
Reverse‐transcription quantitative real‐time polymerase change reaction
Total RNA was reverse‐transcribed using TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific) or RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific), including primers for miR‐378a and RNU48 (Thermo Fisher Scientific). Amplification of cDNA was performed using real‐time PCR QuantStudio 7 Flex Real‐Time PCR System (Thermo Fisher Scientific). The ΔΔCt method allowed the normalization of the expression of miR‐378a against RNU48. NFKBIZ, C/EBP‐β, IL‐8 and CCL20 expression were analysed using TaqMan‐based predesigned quantitative PCR assays (Integrated DNA Technologies, Coralville, IA, USA) and normalized to 18S rRNA (18S, forward primer: CGGCTACCACATCCAAGGAA; reverse primer: GCTGGAATTACCGCGGCT; probe: TGCTGGCACCAGACTTGCCCTC) using the ΔΔCt method.
Mice and the imiquimod‐induced model of psoriasis
Wildtype C57BL/6J 8‐week‐old mice were procured from Charles River (Wilmington, MA, USA). To overexpress miR‐378a, 5–7 μg of synthetic human miR‐378a mimic or scrambled control (mirVana; Life Technologies, Grand Island, NY, USA) packed in the transfection agent Max Suppressor In Vivo RNA‐LANCEr II (Bioo Scientific, Austin, TX, USA) were injected intradermally into the shaved back skin of female C57BL/6J mice on days 0 (or –1), 1 and 3. Topical treatment with imiquimod (IMQ) was performed as previously described. 14 Back skin thickness was measured daily using Vernier callipers (AgnTho’s AB, Lidingö, Sweden). The back skin was folded and measured at three different sites and the average of the three measurements was recorded. Clinical scores were assessed by three independent researchers and assigned a score ranging from 0 to 4 (0, none; 1, mild; 2, moderate; 3, severe; and 4, very severe) for erythema, scaling and thickness. All animal experiments were approved by the local ethics committee of Stockholm, Sweden (Swedish Board of Agriculture).
Statistical analysis
Results represent the mean ± SD from at least three independent experiments. Statistical analyses were conducted using the package ‘stats’ (version 3·6·2) for R software (version 3·5·1) (R Foundation for Statistical Computing, Vienna, Austria) and GraphPad Prism version 8·3·1 for Windows (GraphPad Software, La Jolla, CA, USA). P‐values <0·05 were considered statistically significant.
Results
miR‐378a is overexpressed in keratinocytes of psoriatic skin lesions and its local delivery exacerbates skin inflammation
Previously, we identified miRNAs differentially expressed in keratinocytes in psoriatic skin using small RNA sequencing. 16 miR‐378a was identified as an miRNA that was significantly overexpressed in keratinocytes from psoriatic skin lesions compared with keratinocytes from nonlesional psoriatic skin or healthy skin (Figure 1a and Table S1; see Supporting Information). Results of RT‐qPCR analysis on an extended cohort of patients confirmed the significantly increased levels of miR‐378a in the keratinocytes of psoriatic skin lesions compared with keratinocytes from nonlesional or healthy skin (Figure 1b).
Figure 1.
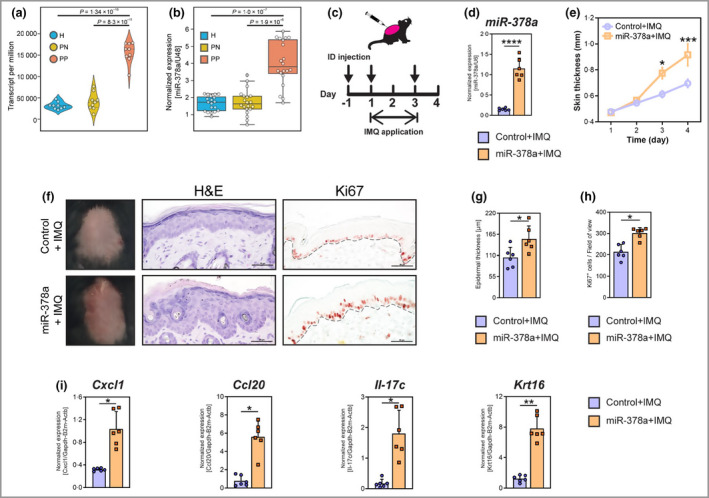
miR‐378a is upregulated in keratinocytes in human lesional psoriatic skin and increases skin thickness through promoting inflammation and keratinocyte hyperproliferation in mice. CD45− epidermal cells (mainly keratinocytes) were separated from skin biopsies obtained from lesional (PP) and nonlesional (PN) skin of patients with psoriasis (n = 9), and from healthy (H) controls (n = 9) by magnetic cell sorting. (a) Read count (transcripts per million) of miR‐378a in keratinocytes (epidermal CD45− cells) from H, PN and PP skin. Data are extracted from small RNA sequencing. (b) Reverse‐transcription quantitative real‐time polymerase change reaction (RT‐qPCR) analysis of miR‐378a in keratinocytes from PP and PN skin (n = 20), and from H skin (n = 19). Expression is normalized to U48 RNA and shown as relative expression units. (c) Timeline for topical application of imiquimod (IMQ) and intradermal (ID) injection of miR‐378a mimics or control oligos (arrows) on mouse back skin. (d) RT‐qPCR analysis of miR‐378a levels in mouse skin upon ID delivery of miR‐378a mimics. (e) Changes in skin thickness upon induction of psoriasis‐like skin inflammation by the topical application of IMQ on mice injected with miR‐378a mimic (orange) or control oligos (scramble mimic, purple). (f) Representative macroscopic imaging (left), haematoxylin and eosin (H&E) staining (centre) and immunohistochemistry for Ki67 protein (right) of back skin sections from mice treated with miR‐378a mimic or control oligo in combination with IMQ application. (g) Quantification of the epidermal thickness on H&E staining of back skin. (h) Quantification of Ki67 positive cells in the epidermis of back skin. (i) RT‐qPCR analysis of Cxcl1, Ccl20, Il‐17c and Krt16 in back skin samples. Scale bars = 50 μm. *P < 0·05, **P < 0·01, and ***P < 0·001, ****P < 0·0001; n = 6 per group. Mann–Whitney U‐test was used. [Colour figure can be viewed at wileyonlinelibrary.com]
As miRNAs with altered expression in psoriatic keratinocytes could play important roles in the pathogenesis of psoriasis, we set out to explore the potential role of miR‐378a. To this end, we investigated the effect of miR‐378a delivery in the IMQ‐induced mouse model of psoriasis. Synthetic miR‐378a was injected intradermally into the back skin of mice, in conjunction with topical application of IMQ (Aldara® cream) (Figure 1c). 18 As expected, topical application of IMQ led to erythema, increased skin thickness and scaling, with increased epidermal proliferation and induction of the inflammatory mediators Cxcl1, Ccl20, Il‐17c and Krt16, reflecting psoriasis‐like inflammation (Figure S1; see Supporting Information). Intradermal delivery of synthetic miR‐378a led to increased miR‐378a levels in mouse skin (Figure 1d) and exacerbated skin inflammation with higher severity scores and increased skin thickness (Figure 1e and Figure S2; see Supporting Information). Histological analysis revealed that miR‐378a significantly increased epidermal thickening (Figure 1f, g) and immunohistochemical staining for Ki67 demonstrated an increase in the number of proliferating keratinocytes upon miR‐378a delivery (Figure 1f, h). Furthermore, delivery of miR‐378a led to a significant increase in the levels of proinflammatory cytokines and chemokines such as Cxcl1, Ccl20, Il‐17c and the psoriasis‐associated keratin Krt16 (Figure 1i). Taken together, our results demonstrate that increased miR‐378a levels lead to more severe psoriasis‐like skin inflammation, suggesting that the increased levels of miR‐378a in psoriatic skin lesions may contribute to the pathogenesis of psoriasis.
Interleukin‐17A induces miR‐378a through nuclear factor kappa B, C/EBP‐β and IκBζ in primary human keratinocytes
Next, we aimed to identify the molecular events leading to increased miR‐378a expression in psoriatic keratinocytes. To investigate the potential effect of the inflammatory microenvironment in psoriatic skin, primary human keratinocytes were treated with the psoriasis‐associated cytokines IL‐17A, IL‐1β, IL‐36α, IL‐22, tumour necrosis factor (TNF)‐α or a combination of IL‐17A, IL‐22 and TNF‐α, 15 and the expression of miR‐378a was analysed using RT‐qPCR. Of the inflammatory mediators tested, the key psoriasis cytokine IL‐17A was identified as an inducer of miR‐378a (Figure 2a and Figure S3; see Supporting Information). IL‐17A significantly induced the level of mature miR‐378a 24 h and 48 h post‐treatment, while the levels of the primary miR‐378a transcript (pri‐miR‐378a) were already increased after 6 h, indicating direct transcriptional regulation (Figure 2a). Consistent with our results obtained with monolayer keratinocytes, miR‐378a was significantly induced by IL‐17A (Figure 2b and Figure S4; see Supporting Information), and by a combination of IL‐17A, TNF‐α and interferon‐γ (Figure 2c) in three‐dimensional epidermal equivalents.
Figure 2.
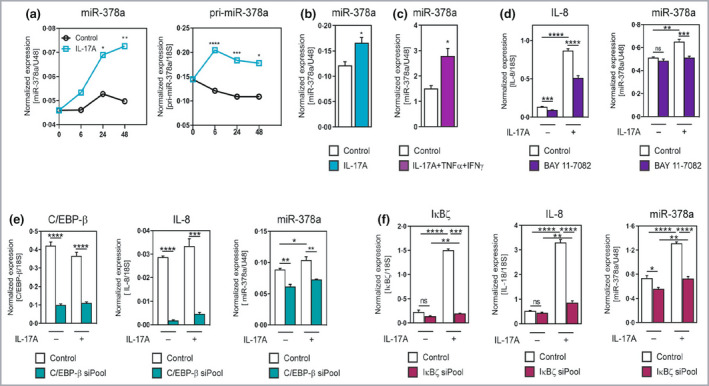
Interleukin (IL)‐17A induces miR‐378a expression in primary keratinocytes through nuclear factor kappa B (NF‐κB) and C/EBP‐β. (a) Primary human epidermal keratinocytes were treated with the cytokine IL‐17A at a concentration of 100 ng mL−1 for the indicated times; the primary transcript pri‐miR‐378a and mature miR‐378a expression was analysed by reverse‐transcription quantitative real‐time polymerase change reaction (RT‐qPCR). Expression is normalized to 18S for the primary transcript and U48 RNA for the mature miRNA and data points show the mean of four replicates. (b) RT‐qPCR analysis of miR‐378a expression in three‐dimensional (3D) epidermal equivalents treated with IL‐17A (20 ng mL−1) for 72 h. (c) RT‐qPCR analysis of miR‐378a expression in 3D epidermal equivalent treated for 72 h with a combination of cytokines associated with psoriasis (psoriasis‐like), i.e. IL‐17A (30 ng mL−1) + tumour necrosis factor (TNF)‐α (5 ng mL−1) + interferon (IFN)‐γ (20 ng mL−1). (d) RT‐qPCR analysis of miR‐378a expression in primary human keratinocytes treated with IL‐17A (100 ng mL−1) with or without pretreatment with the NF‐κB inhibitor BAY11‐7082. The expression of miR‐378a was normalized to U48 RNA. Bars show mean ± SD of four replicates. (e) RT‐qPCR analysis of miR‐378a and IL‐8 expression in keratinocytes upon inhibition of C/EBP‐β by small interfering RNA (siRNA) pool. (f) RT‐qPCR analysis of miR‐378a and IL‐8 expression in keratinocytes upon inhibition of IκBζ by an siRNA pool. *P < 0·05, **P < 0·01, ***P < 0·001, ****P < 0·0001, ns: not significant, Student’s two‐sided t‐test was used. [Colour figure can be viewed at wileyonlinelibrary.com]
To explore the mechanisms of IL‐17A‐mediated regulation of miR‐378a, we interrogated ENCODE Transcription Factor ChIP‐seq data to identify transcription factors binding to the promoter region of miR‐378a. The transcription factors NF‐κB and C/EBP‐β, both known to act downstream of IL‐17A, 19 were found to bind to the putative promoter region of miR‐378a (data not shown). Next, we investigated whether NF‐κB and/or C/EBP‐β are indeed involved in the IL‐17A‐mediated induction of miR‐378a. Induction of miR‐378a by IL‐17A was completely blocked by the NF‐κB inhibitor BAY 11‐7082, while basal levels of miR‐378a were not affected (Figure 2d, right panel). In comparison, BAY 11‐7082 inhibited both basal and IL‐17A‐induced expression of IL‐8 (Figure 2d, left panel). Inhibition of C/EBP‐β by a pool of small interfering RNAs (siRNAs) led to significant suppression of miR‐378a levels in both the presence and absence of IL‐17A (Figure 2e, right panel). The siRNA pool used significantly inhibited C/EBP‐β (Figure 2e, left panel) and both basal and IL‐17A‐induced expression of IL‐8 (Figure 2e, middle panel).
IκBζ is a transcriptional coactivator involved in downstream effects of IL‐17A by enhancing transcription of inflammatory secondary response genes, 8 , 20 and the encoding gene NFKBIZ is located in a psoriasis susceptibility locus, indicating the importance of this gene in the pathogenesis of psoriasis. 21 IκBζ has been shown to bind to promoters harbouring the NF‐κB and C/EBP‐β binding sites. 20 Therefore, we tested whether IκBζ is involved in IL‐17A‐mediated induction of miR‐378a. To this end, we used an siRNA pool targeting IκBζ, in combination with IL‐17A treatment (Figure 2f). The siRNA pool suppressed IL‐17A‐induced IκBζ and IL‐8 expression (Figure 2f, left and middle panels). Notably, IL‐17A‐induced miR‐378a was significantly suppressed by inhibition of IκBζ (Figure 2f, right panel).
Taken together, our results suggest that miR‐378a is overexpressed in psoriatic skin lesions at least partly owing to increased IL‐17A expression, which can induce miR‐378a through NF‐κB, C/EBP‐β and IκBζ.
miR‐378a enhances interleukin‐17A‐mediated inflammatory responses in keratinocytes via activation of the nuclear factor kappa B pathway
Next, we aimed to investigate the molecular consequences of miR‐378a overexpression in keratinocytes, under homeostatic and inflammatory conditions. To this end, miR‐378a was overexpressed by transient transfection with miR‐378a mimics (Figure S5; see Supporting Information), which were subsequently treated or not treated with IL‐17A, and transcriptomic analysis was performed (Figure 3a). miR‐378a significantly regulated 266 genes in unstimulated keratinocytes, 186 of which were upregulated and 80 were downregulated (Figure 3b, left panel). miR‐378a regulated 194 genes in IL‐17A‐treated keratinocytes, 127 of which were upregulated and 67 of which were downregulated (Figure 3b, right panel and Table S2; see Supporting Information). Overall, 39 genes were significantly regulated by miR‐378a under both unstimulated and IL‐17A‐stimulated conditions. Enrichment analyses revealed that biological functions enriched in miR‐378a‐regulated genes included ‘skin development’, ‘regulation of inflammatory response’, ‘regulation of epidermal cell differentiation’, ‘negative regulation of NF‐κB signalling’ and ‘response to cytokines’ (Figure 3c).
Figure 3.
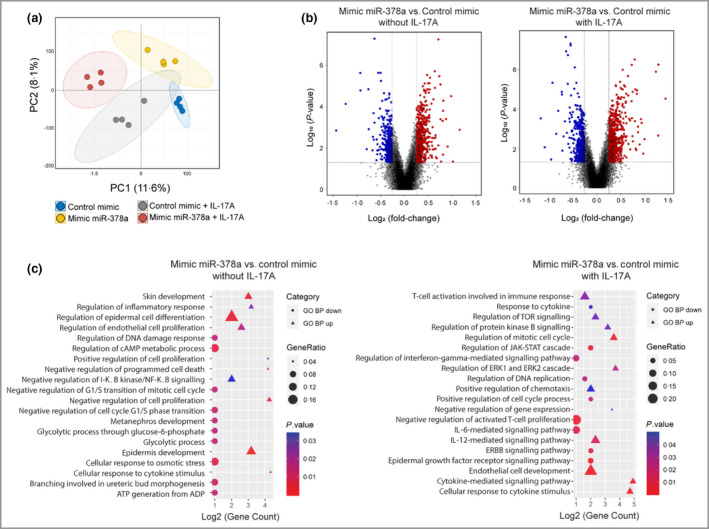
Transcriptomic changes upon overexpression of miR‐378a in keratinocytes. Keratinocytes were transfected with miR‐378a mimics (to overexpress miR‐378a) or control oligos alone or in combination with interleukin (IL)‐17A treatment, and transcriptome analysis was performed. (a) Principal component analysis of the transcriptome of cultured primary human keratinocytes transfected with control mimic alone (blue dots) or in combination with IL‐17A (grey dots), and transfected with miR‐378a mimic alone (yellow dots) or in combination with IL‐17A (red dots). (b) Volcano plots showing the log2 fold‐change (FC) (vertical grey lines) and the nominal P‐value (grey horizontal line) for all the transcripts detected by the Affymetrix platform in the miR‐378a mimic vs. control mimic group comparison, in the absence (left) and in the presence (right) of IL‐17A. Grey dots indicate transcripts not significantly changed, blue dots represent transcripts with FC < 0·67 and red dots represent transcripts with FC > 1·5. (c) Significantly enriched gene ontology biological processes (GO BP) of miR‐378‐overexpressing keratinocytes treated (right) or not treated (left) with IL‐17A. The shape of the plotted GO BP terms indicate the direction of regulation (circles for upregulated genes and triangles for downregulated genes), the size indicates the percentage of total deregulated genes in the given GO BP term (Gene Ratio) and the colour corresponds to P‐value. TOR, target of rapamycin; cAMP, cyclic adenosine monophosphate; JAK, Janus kinase; STAT, signal transducer and activator of transcription; ERK, extracellular signal‐regulated kinase. [Colour figure can be viewed at wileyonlinelibrary.com]
One of the significantly enriched pathways among miR‐378a‐regulated genes was the NF‐κB pathway, a pathway crucial for innate immune responses in the skin and activated in psoriatic skin. 22 Genes regulated in the ‘negative regulation of NF‐κB signalling’ category include PER1, TMSB4X, TNFAIP3, CCDC22 (upregulated) and TNFSF10 and IRAK4 (downregulated). To investigate whether miR‐378a can modulate basal and/or IL‐17A‐induced NF‐κB activation in keratinocytes, immunofluorescence analysis was performed to assess the nuclear translocation of p65, a major subunit of NF‐κB. As expected, p65 was localized mainly to the cytoplasm in untreated cells, while IL‐17A induced its nuclear translocation after 30 min (Figure 4a). Strikingly, overexpression of miR‐378a alone was able to induce nuclear translocation of p65 (Figure 4a). In combination with IL‐17A, miR‐378a further induced the proportion of cells with nuclear p65 localization (Figure 4a).
Figure 4.
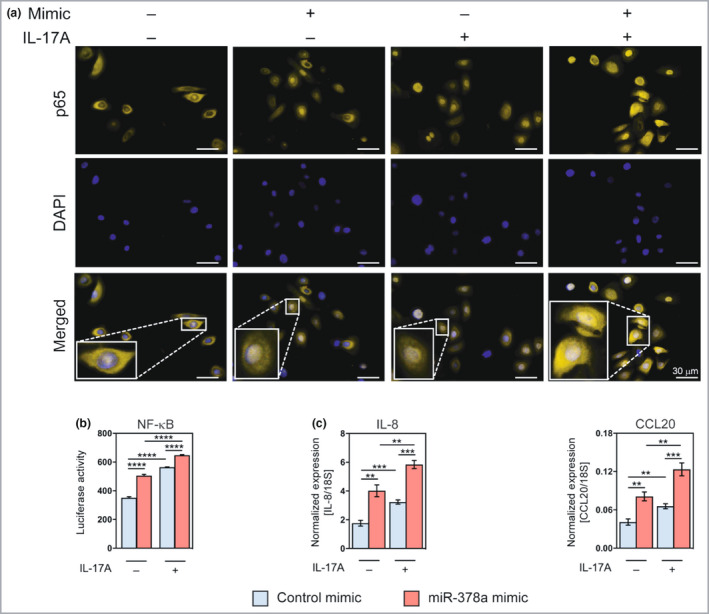
miR‐378a enhances interleukin (IL)‐17A‐mediated induction of nuclear factor kappa B (NF‐κB) activity in keratinocytes. (a) Immunofluorescence staining of p65 in keratinocytes overexpressing miR‐378a with or without treatment with IL‐17A (100 ng mL−1) for 30 min; ‘mimic’ refers to miR‐378a mimic. Control mimic (scramble oligo) was used as a control. (b) Luciferase activity was measured in keratinocytes transfected with the NF‐κB reporter and miR‐378a mimic or control mimic for 24 h, alone or treated with IL‐17A (100 ng mL−1) for 6 h. (c) The expression of CXCL8/IL‐8 and CCL20 were analysed by reverse‐transcription quantitative real‐time polymerase change reaction (RT‐qPCR) in cultured primary human keratinocytes upon transfection with miR‐378a mimic or mimic control, alone or in combination with IL‐17A (100 ng mL−1). *P < 0·05, **P < 0·01, ***P < 0·001, ****P < 0·0001 (Student’s two‐sided t‐test). DAPI, 4′,6‐diamidino‐2‐phenylindole. [Colour figure can be viewed at wileyonlinelibrary.com]
Consistent with the immunofluorescence data, NF‐κB luciferase reporter assay showed that miR‐378a alone could induce NF‐κB‐dependent promoter activity. Moreover, miR‐378a could further enhance IL‐17A‐induced activation of NF‐κB‐dependent promoter activity (Figure 4b). The activation of the NF‐κB pathway upon miR‐378a overexpression was further supported by RT‐qPCR analysis showing that miR‐378a induced both basal and IL‐17A‐induced expression of IL‐8/CXCL8 and CCL20, chemokines known to be regulated by NF‐κB activation (Figure 4c and Figure S6; see Supporting Information). 22
In line with these data, inhibition of endogenous miR‐378a suppressed the IL‐17A‐induced nuclear translocation of p65 in keratinocytes (Figure 5a) and suppressed IL‐17A‐induced NF‐κB promoter activity as shown by NF‐κB luciferase promoter assay (Figure 5b). Furthermore, inhibition of endogenous miR‐378a suppressed IL‐17A‐induced IL‐8 and CCL20 expression (Figure 5c).
Figure 5.
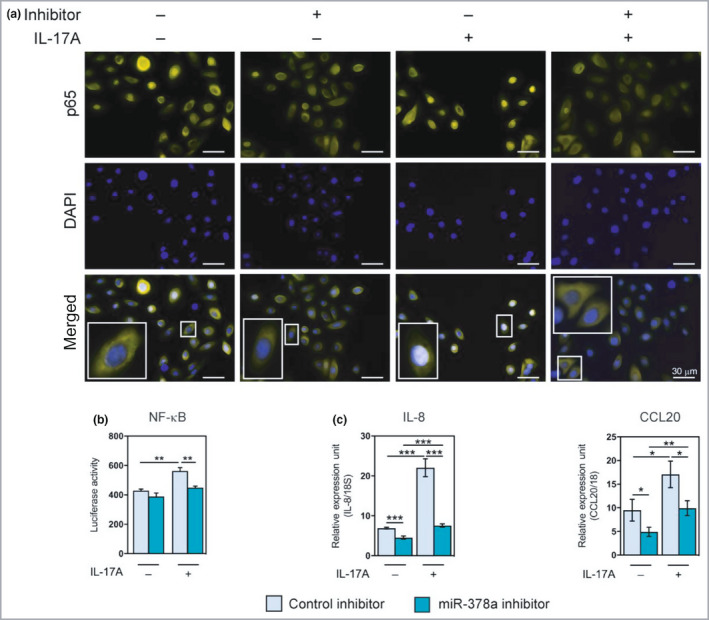
Inhibition of miR‐378a attenuates interleukin (IL)‐17A‐mediated induction of nuclear factor kappa B (NF‐κB) activity in keratinocytes. (a) Immunofluorescence staining of p65 in keratinocytes transfected with miR‐378a inhibitor, alone or in cotreatment with IL‐17A (100 ng mL−1) for 30 min; ‘inhibitor’ refers to miR‐378a inhibitor. Control inhibitor (scramble oligo) was used as a control. (b) Luciferase activity of transfection with NF‐κB reporter and miR‐378a inhibitor or control inhibitor, alone or in cotreatment with IL‐17A (100 ng mL−1). (c) RT‐qPCR analysis of the expression of CXCL8/IL‐8 and CCL20 in cultured primary human keratinocytes treated with miR‐378a inhibitor or inhibitor control, alone or in combination with IL‐17A (100 ng mL−1). *P < 0·05, **P < 0·01, ***P < 0·001 (Student’s two‐sided t‐test). DAPI, 4′,6‐diamidino‐2‐phenylindole. [Colour figure can be viewed at wileyonlinelibrary.com]
Collectively, these results suggest that miR‐378a amplifies skin inflammation in psoriasis by enhancing the activation of the NF‐κB pathway in an inflammatory milieu in the skin.
miR‐378a directly targets NFKBIA /IκBα
As miRNAs function by suppressing the expression of their target genes, we next aimed to identify direct targets of miR‐378a, which can mediate the observed effects on inflammation and NF‐κB pathway activity. One of the potential target genes predicted by TargetScan (www.targetscan.org) was NFKBIA (Figure 6a and Figure S7; see Supporting Information), a gene encoding IκBα, an inhibitor of NF‐κB signalling, which is strongly genetically associated with psoriasis. 23 , 24 Western blot analysis demonstrated that overexpression of miR‐378a suppressed IκBα, while inhibition of endogenous miR‐378a led to increased IκBα levels in human keratinocytes, supporting the link between miR‐378a and IκBα (Figure 6b). To confirm the 3′UTR sequence of NFKBIA containing the miR‐378a binding site in primary keratinocytes, RT‐PCR was performed on cDNA from cultured primary keratinocytes, using specific primers spanning a sequence of 1456 bp as predicted by TargetScan. Sequencing of the product confirmed the presence of the 3′UTR containing the predicted miR‐378a binding site (Figure S7; see Supporting Information). To verify direct targeting of the NFKBIA mRNA by miR‐378a, 3′UTR luciferase assays were performed. Overexpression of miR‐378a suppressed luciferase activity in keratinocytes transfected with a construct carrying the wildtype 3′UTR of NFKBIA, but not in cells transfected with a plasmid with the NFKBIA 3′UTR with a mutated miR‐378a binding site (Figure 6c), indicating direct targeting of NFKBIA by miR‐378a.
Figure 6.
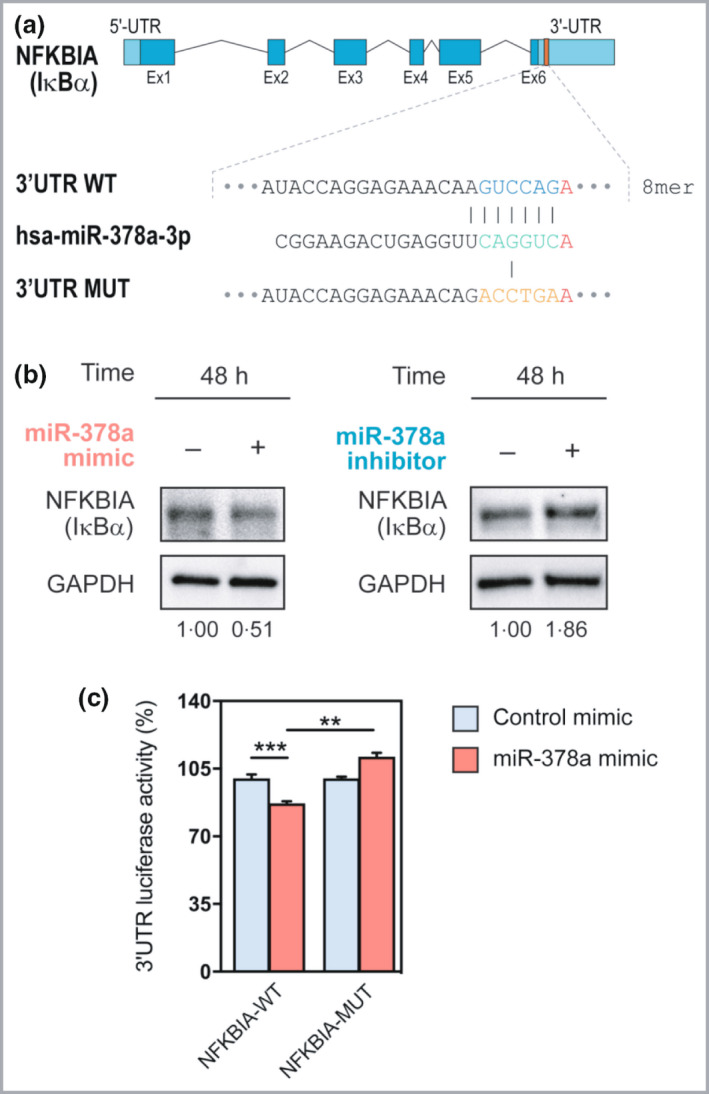
miR‐378a directly targets NFKBIA/IκBα. (a) Wildtype (WT) and mutated (MUT) sequence of the putative miR‐378a binding site in the 3′UTR of NFKBIA. (b) Western blot analysis of IκBα upon overexpression (left) and inhibition (right) of miR‐378a. (c) The effect of miR‐378a on NFKBIA 3′UTR‐dependent luciferase activity in keratinocytes, identified by three independent experiments. **P < 0·01, ***P < 0·001 (Student’s two‐sided t‐test). [Colour figure can be viewed at wileyonlinelibrary.com]
These results suggest a model in which that miR‐378a regulates both baseline and IL‐17A‐induced keratinocyte inflammatory responses via targeting NFKBIA/IκBα, a key negative regulator of the NF‐κB signal pathway (Figure 7).
Figure 7.
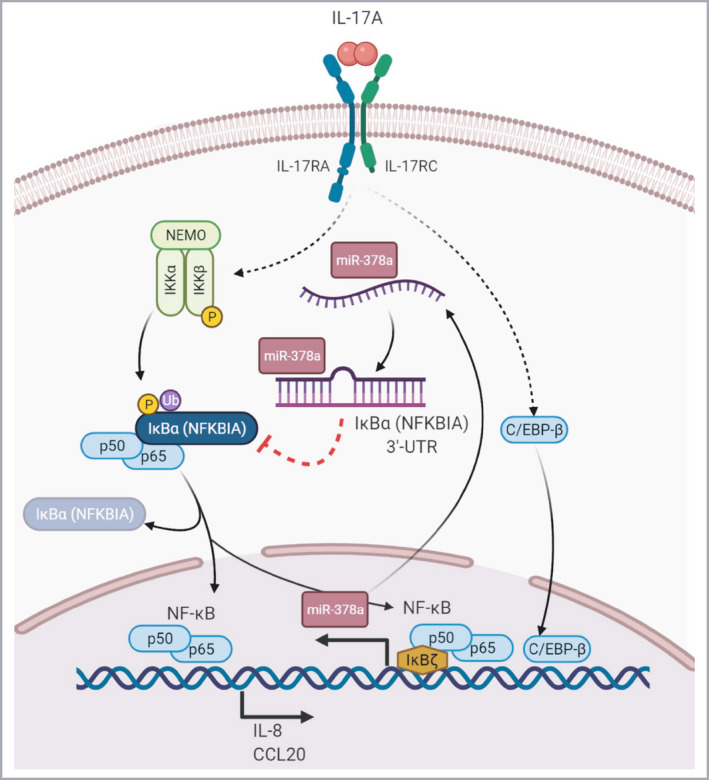
Amplification of interleukin (IL)‐17A signalling pathway by miR‐378a in keratinocytes. Our findings suggest a model in which miR‐378a is induced by IL‐17A via nuclear factor kappa B (NF‐κB), C/EBP‐β and IκBζ in keratinocytes. Induction of miR‐378a leads to the suppression of NFKBIA/IκBα, which allows for activation of the NF‐κB pathway including the nuclear translocation of p65, leading to further induction of inflammatory mediators such as IL‐8/CXCL8 and CCL20. The baseline intracellular level of miR‐378a may regulate the magnitude of the response to triggers activating the NF‐κB‐pathway. [Colour figure can be viewed at wileyonlinelibrary.com]
Discussion
In this study, we identify miR‐378a as an amplifier of IL‐17A‐induced inflammatory responses in keratinocytes. Keratinocytes had long been thought to be passive targets of T‐cell‐derived cytokines in psoriasis. However, this view has been challenged and a number of compelling studies demonstrated that keratinocytes are central players in skin immunity both as sentinels and as a source of inflammatory mediators. 25 The essential role of keratinocytes in psoriatic skin inflammation is supported by keratinocyte‐specific ablation or overexpression of psoriasis susceptibility genes modulating or inducing psoriasis‐like skin inflammation. 26 , 28 Importantly, keratinocytes are critical target cells for IL‐17A in inducing psoriasis, as evidenced by attenuation of IMQ‐induced skin inflammation in mice with deletion of IL‐17RA in keratinocytes but not with IL‐17RA deletion in other cell types. 29
Here, we show that miR‐378a is a strong inducer of the NF‐κB pathway in keratinocytes. The transcription factor NF‐κB is crucial for epidermal homeostasis in the skin and an important regulator of inflammatory responses. 30 Both its excessive activation and inhibition result in inflammation in mouse models, which highlights the narrow range of optimal activity of the NF‐κB pathway. 30 , 31
The activation of the NF‐κB pathway in keratinocytes and immune cells is essential in the pathogenesis of psoriasis. 22 This is supported by the following factors: (i) increased NF‐κB activity in psoriatic epidermis; 32 (ii) several genes encoding mediators of this pathway are genetically associated with psoriasis; 22 (iii) multiple inflammatory cytokines with key roles in psoriasis signal through the NF‐κB pathway; and (iv) inhibition of the NF‐κB pathway occurs as a result of antipsoriatic therapies. 22 , 32 Our finding that miR‐378a is an inducer of the NF‐κB pathway in keratinocytes identifies an additional layer regarding the regulation of this pathway. The observation that miR‐378a itself is induced by IL‐17A, through NF‐κB/IκBζ/C/EBP‐β suggests the existence of a self‐amplifying loop, in which IL‐17A is driving activation of NF‐κB both via its canonical signalling through TRAF6 and via induction of miR‐378a, which suppresses a negative regulator of NF‐κB signalling (Figure 7). The physiological role of this self‐amplifying loop could be to ensure a robust response to innate triggers such as fungal and/or bacterial infection, whereas in psoriasis, induction of miR‐378a could reinforce an aberrantly activated pathway and may contribute to the chronic nature of the inflammation. Interestingly, miR‐378a was not identified as being induced in atopic dermatitis skin, 33 suggesting a role for miR‐378a in IL‐17A‐induced skin inflammation rather than inflammation in general.
Surprisingly, the overexpression of miR‐378a alone resulted in nuclear translocation of p65 and increased NF‐κB‐dependent promoter activity in keratinocytes, even in the absence of IL‐17A, in unstimulated cells. This suggests that miR‐378a levels need to be kept low under homeostatic conditions to prevent skin inflammation, while upon proper trigger, such as IL‐17A, miR‐378a is induced and allows the maximal activation of the NF‐κB pathway (Figure 7). The observation that miR‐378a induces NF‐κB activation in resting cells may partly be due to the suppression of NFKBIA/IκBα, which is a major inhibitor of NF‐κB activation during homeostatic conditions. 30 Interestingly, the NFKBIA/IκBα gene is strongly associated with psoriasis susceptibility, highlighting its relevance for the disease. 22 However, because of the preiotropic action of miRNAs, it is probable that the effect of miR‐378a on NF‐κB activation is achieved through simultaneous regulation of several target genes, which will require identification to allow for the complete understanding of the mode of action of this miRNA. Interestingly, miR‐378a has been shown to induce NF‐κB activity in liver cells by regulating p65 acetylation; whether this mechanism is relevant for keratinocytes remains to be investigated. 34
It is plausible that miR‐378a may also be involved in other IL‐17A‐driven inflammatory diseases in which NF‐κB/C/EBP‐β/IκBζ activation can induce its expression. In line with our findings, miR‐378a has recently been shown to promote hepatic inflammation by enhancing NF‐κB signalling. 34 Interestingly, miR‐378a has also been shown to be a regulator of energy homeostasis, and has been linked to obesity, the metabolic syndrome and atherosclerosis, which are inflammatory conditions associated with psoriasis. 35 , 37 Moreover, in a very recent study, miR‐378a was implicated in the regulation of cell cycle in keratinocytes through targeting BMP2. 38 Future studies will be required to identify further potential inducers of miR‐378a in keratinocytes and whether the function of this miRNA is conserved across other cell types.
In summary, our results suggest a new miRNA‐mediated mechanism for the regulation of IL‐17A‐responsiveness and NF‐κB activity in keratinocytes. Elevated IL‐17A levels in psoriatic skin induce miR‐378a, which acts as a positive feedback to further amplify inflammation by suppression of NFKBIA/IκBα and subsequent increased NF‐κB activity. These results highlight miRNA‐mediated regulation of cytokine responses in inflammation and prompt further research in this field to explore the potential of miRNA modulation in inflammatory skin diseases.
Author contributions
Ping Xia: Investigation (equal); methodology (equal); writing – original draft (equal). Lorenzo Pasquali: Investigation (equal); methodology (equal); writing – original draft (equal). Chenying Gao: Investigation (equal); methodology (equal); writing – original draft (equal). Ankit Srivastava: Conceptualization (supporting); formal analysis (supporting); investigation (supporting); methodology (supporting); writing – original draft (supporting); writing – review and editing (supporting). Nupur Khera: Investigation (supporting); methodology (supporting); writing – review and editing (supporting). Jan Cedric Freisenhausen: Investigation (supporting). Longlong Luo: Investigation (supporting). Einar Rosén: Investigation (supporting); writing – review and editing (supporting). A. VanLierop: Investigation (supporting); methodology (supporting); writing – review and editing (supporting). B. Homey: Funding acquisition (supporting); methodology (supporting); resources (supporting); writing – review and editing (supporting). Andor Pivarcsi: Conceptualization (equal); formal analysis (equal); funding acquisition (supporting); resources (equal); supervision (equal); writing – original draft (equal); writing – review and editing (equal). Enikö Sonkoly: Conceptualization (lead); funding acquisition (lead); project administration (lead); resources (lead); supervision (equal); writing – original draft (lead); writing – review and editing (equal).
Supporting information
Appendix S1 Supporting materials and methods.
Figure S1 Topical application of imiquimod induces skin inflammation in mice with increased epidermal thickness and induction of inflammatory mediators.
Figure S2 Local delivery of miR‐378a mimic exacerbates imiquimod‐induced skin inflammation in mice.
Figure S3 Expression of miR‐378a in primary human keratinocytes treated with psoriasis‐associated cytokines.
Figure S4 Expression of miR‐378a in three‐dimensional epidermal equivalents treated with cytokines.
Figure S5 Expression of miR‐378a in primary human keratinocytes upon transfection with miR‐378a mimics.
Figure S6 Interleukin‐8/CXCL8 secretion is induced by miR‐378a in primary human keratinocytes.
Figure S7 Reverse‐transcription quantitative real‐time polymerase change reaction for the 3′UTR of the NFKBIA transcript containing the predicted miR‐378a binding site in human keratinocytes.
Table S1 Demographic data and Psoriasis Area and Severity Index score for the psoriasis patients and healthy donors included.
Table S2 List of top 50 deregulated genes from transcriptome analysis of primary human keratinocytes transfected with miR‐378a or control mimics with or without interleukin‐17A treatment.
Acknowledgments
We thank the patients and healthy volunteers who took part in this study, the nurses at the Swedish Psoriasis Association for assisting with patient recruitment, and Claus Johansen, Department of Dermatology, Aarhus University Hospital, Denmark, for kindly providing vehicle cream as a control for Aldara.
Funding sources This work was funded by the Stockholm County Council (ALF), the Swedish Medical Research Council (Vetenskapsrådet), the Swedish Psoriasis Association (Psoriasisfonden), the Swedish Cancer Foundation (Cancerfonden), the LEO Foundation, Welander and Finsen Foundations/Hudfonden (Skin Foundation) and the National Psoriasis Foundation of the USA.
Conflicts of interest The authors declare they have no conflicts of interest. P.X., L.P. and C.G. contributed equally.
Data availability Microarray data have been deposited at NCBI GEO (GSE164400). Any additional data are available from the corresponding author upon reasonable request.
Ethics statement The study was approved by the Stockholm Regional Ethics Committee and conducted according to the Declaration of Helsinki principles. Written informed consent was obtained from all participants prior to enrolment.
Plain language summary available online
DATA AVAILABILITY STATEMENT
Microarray data have been deposited at NCBI GEO (GSE164400). Any additional data is available from the corresponding author upon reasonable request.
References
- 1. Griffiths CEM, Armstrong AW, Gudjonsson JE, Barker J. Psoriasis. Lancet 2021; 397:1301–15. [DOI] [PubMed] [Google Scholar]
- 2. Bugaut H, Aractingi S. Major role of the IL17/23 axis in psoriasis supports the development of new targeted therapies. Front Immunol 2021; 12:621956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dainichi T, Kitoh A, Otsuka A et al. The epithelial immune microenvironment (EIME) in atopic dermatitis and psoriasis. Nat Immunol 2018; 19:1286–98. [DOI] [PubMed] [Google Scholar]
- 4. Ghoreschi K, Balato A, Enerback C, Sabat R. Therapeutics targeting the IL‐23 and IL‐17 pathway in psoriasis. Lancet 2021; 397:754–66. [DOI] [PubMed] [Google Scholar]
- 5. Tollenaere MAX, Hebsgaard J, Ewald DA et al. Signalling of multiple interleukin (IL)‐17 family cytokines via IL‐17 receptor A drives psoriasis‐related inflammatory pathways. Br J Dermatol 2021; 185:585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuestner RE, Taft DW, Haran A et al. Identification of the IL‐17 receptor related molecule IL‐17RC as the receptor for IL‐17F. J Immunol 2007; 179:5462–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chiricozzi A, Guttman‐Yassky E, Suarez‐Farinas M et al. Integrative responses to IL‐17 and TNF‐α in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J Invest Dermatol 2011; 131:677–87. [DOI] [PubMed] [Google Scholar]
- 8. Bertelsen T, Iversen L, Johansen C. The human IL‐17A/F heterodimer regulates psoriasis‐associated genes through IκBζ. Exp Dermatol 2018; 27:1048–52. [DOI] [PubMed] [Google Scholar]
- 9. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–97. [DOI] [PubMed] [Google Scholar]
- 10. Sonkoly E. The expanding microRNA world in psoriasis. Exp Dermatol 2017; 26:375–6. [DOI] [PubMed] [Google Scholar]
- 11. Sonkoly E, Wei T, Janson PC et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLOS ONE 2007; 2:e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Løvendorf MB, Mitsui H, Zibert JR et al. Laser capture microdissection followed by next‐generation sequencing identifies disease‐related microRNAs in psoriatic skin that reflect systemic microRNA changes in psoriasis. Exp Dermatol 2015; 24:187–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joyce CE, Zhou X, Xia J et al. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet 2011; 20:4025–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Srivastava A, Nikamo P, Lohcharoenkal W et al. MicroRNA‐146a suppresses IL‐17‐mediated skin inflammation and is genetically associated with psoriasis. J Allergy Clin Immunol 2017; 139:550–61. [DOI] [PubMed] [Google Scholar]
- 15. Srivastava A, Luo L, Lohcharoenkal W et al. Cross‐talk between IFN‐γ and TWEAK through miR‐149 amplifies skin inflammation in psoriasis. J Allergy Clin Immunol 2021; 147:2225–35. [DOI] [PubMed] [Google Scholar]
- 16. Srivastava A, Meisgen F, Pasquali L et al. Next‐generation sequencing identifies the keratinocyte‐specific miRNA signature of psoriasis. J Invest Dermatol 2019; 139:2547–50.e12. [DOI] [PubMed] [Google Scholar]
- 17. Kuleshov MV, Jones MR, Rouillard AD et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016; 44(W1):W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van der Fits L, Mourits S, Voerman JS et al. Imiquimod‐induced psoriasis‐like skin inflammation in mice is mediated via the IL‐23/IL‐17 axis. J Immunol 2009; 182:5836–45. [DOI] [PubMed] [Google Scholar]
- 19. Chiricozzi A, Nograles KE, Johnson‐Huang LM et al. IL‐17 induces an expanded range of downstream genes in reconstituted human epidermis model. PLOS ONE 2014; 9:e90284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johansen C. IκBζ: a key protein in the pathogenesis of psoriasis. Cytokine 2016; 78:20–1. [DOI] [PubMed] [Google Scholar]
- 21. Tsoi LC, Spain SL, Ellinghaus E et al. Enhanced meta‐analysis and replication studies identify five new psoriasis susceptibility loci. Nat Commun 2015; 6:7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goldminz AM, Au SC, Kim N et al. NF‐κB: an essential transcription factor in psoriasis. J Dermatol Sci 2013; 69:89–94. [DOI] [PubMed] [Google Scholar]
- 23. Klement JF, Rice NR, Car BD et al. IkappaBalpha deficiency results in a sustained NF‐kappaB response and severe widespread dermatitis in mice. Mol Cell Biol 1996; 16:2341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stuart PE, Nair RP, Ellinghaus E et al. Genome‐wide association analysis identifies three psoriasis susceptibility loci. Nat Genet 2010; 42:1000–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang Y, Tsoi LC, Billi AC et al. Cytokinocytes: the diverse contribution of keratinocytes to immune responses in skin. JCI Insight 2020; 5:e142067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang M, Zhang S, Zheng G et al. Gain‐of‐function mutation of card14 leads to spontaneous psoriasis‐like skin inflammation through enhanced keratinocyte response to IL‐17A. Immunity 2018; 49:66–79.e5. [DOI] [PubMed] [Google Scholar]
- 27. Devos M, Mogilenko DA, Fleury S et al. Keratinocyte expression of A20/TNFAIP3 controls skin inflammation associated with atopic dermatitis and psoriasis. J Invest Dermatol 2019; 139:135–45. [DOI] [PubMed] [Google Scholar]
- 28. Sano S, Chan KS, Carbajal S et al. Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nat Med 2005; 11:43–9. [DOI] [PubMed] [Google Scholar]
- 29. Moos S, Mohebiany AN, Waisman A, Kurschus FC. Imiquimod‐induced psoriasis in mice depends on the IL‐17 signaling of keratinocytes. J Invest Dermatol 2019; 139:1110–17. [DOI] [PubMed] [Google Scholar]
- 30. Sur I, Ulvmar M, Toftgard R. The two‐faced NF‐κB in the skin. Int Rev Immunol 2008; 27:205–23. [DOI] [PubMed] [Google Scholar]
- 31. Pasparakis M. Role of NF‐κB in epithelial biology. Immunol Rev 2012; 246:346–58. [DOI] [PubMed] [Google Scholar]
- 32. Lizzul PF, Aphale A, Malaviya R et al. Differential expression of phosphorylated NF‐κB/RelA in normal and psoriatic epidermis and downregulation of NF‐κB in response to treatment with etanercept. J Invest Dermatol 2005; 124:1275–83. [DOI] [PubMed] [Google Scholar]
- 33. Sonkoly E, Janson P, Majuri ML et al. MiR‐155 is overexpressed in patients with atopic dermatitis and modulates T‐cell proliferative responses by targeting cytotoxic T lymphocyte‐associated antigen 4. J Allergy Clin Immunol 2010; 126:581–9.e1–20. [DOI] [PubMed] [Google Scholar]
- 34. Zhang T, Hu J, Wang X et al. MicroRNA‐378 promotes hepatic inflammation and fibrosis via modulation of the NF‐κB‐TNFα pathway. J Hepatol 2019; 70:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carrer M, Liu N, Grueter CE et al. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci USA 2012; 109:15330–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krist B, Florczyk U, Pietraszek‐Gremplewicz K et al. The role of miR‐378a in metabolism, angiogenesis, and muscle biology. Int J Endocrinol 2015; 2015:281756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen W, Li X, Wang J et al. miR‐378a modulates macrophage phagocytosis and differentiation through targeting CD47‐SIRPα axis in atherosclerosis. Scand J Immunol 2019; 90:e12766. [DOI] [PubMed] [Google Scholar]
- 38. Soonthornchai W, Tangtanatakul P, Meesilpavikkai K et al. MicroRNA‐378a‐3p is overexpressed in psoriasis and modulates cell cycle arrest in keratinocytes via targeting BMP2 gene. Sci Rep 2021; 11:14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supporting materials and methods.
Figure S1 Topical application of imiquimod induces skin inflammation in mice with increased epidermal thickness and induction of inflammatory mediators.
Figure S2 Local delivery of miR‐378a mimic exacerbates imiquimod‐induced skin inflammation in mice.
Figure S3 Expression of miR‐378a in primary human keratinocytes treated with psoriasis‐associated cytokines.
Figure S4 Expression of miR‐378a in three‐dimensional epidermal equivalents treated with cytokines.
Figure S5 Expression of miR‐378a in primary human keratinocytes upon transfection with miR‐378a mimics.
Figure S6 Interleukin‐8/CXCL8 secretion is induced by miR‐378a in primary human keratinocytes.
Figure S7 Reverse‐transcription quantitative real‐time polymerase change reaction for the 3′UTR of the NFKBIA transcript containing the predicted miR‐378a binding site in human keratinocytes.
Table S1 Demographic data and Psoriasis Area and Severity Index score for the psoriasis patients and healthy donors included.
Table S2 List of top 50 deregulated genes from transcriptome analysis of primary human keratinocytes transfected with miR‐378a or control mimics with or without interleukin‐17A treatment.
Data Availability Statement
Microarray data have been deposited at NCBI GEO (GSE164400). Any additional data is available from the corresponding author upon reasonable request.


