Abstract
Specific labeling of a single row of cellulose-synthesizing complexes (terminal complexes, TC subunits, TCs, or TC arrays) in Acetobacter xylinum by antibodies raised against a 93-kDa protein (the cyclic dignanylic acid-binding protein) has been demonstrated by using the sodium dodecyl sulfate (SDS)–freeze-fracture labeling (FRL) technique. The antibodies to the 93-kDa protein specifically recognized the TC subunits on the protoplasmic fracture (PF) face of the outer membrane in A. xylinum; however, nonlabeled TCs were also observed. Two types of TC subunits (particles or pits) are observed on the PF face of the outer membrane: (i) immunogold-labeled TCs showing a line of depressions (pits) with an indistinct particle array and (ii) nonlabeled TC subunits with a distinct single row of particle arrays. The evidence indicates that the labeling patterns differ with respect to the presence or absence of certain TC subunits remaining attached to the replica after SDS treatment. This suggests the presence of at least two TC components, one in the outer membrane and the other in the cytoplasmic membrane. If the TC component in the outer membrane is preferentially fractured and remains attached to the ectoplasmic fracture face (or outer leaflet) of the outer membrane, subsequent replica formation reveals a pit or depression with positive antibody labeling on the PF face of the outer membrane. If the TC component in the outer membrane remains with the PF face (or inner leaflet) of the outer membrane, the innermost TC component is removed during SDS treatment and labeling does not occur. SDS-FRL of TCs in A. xylinum has enabled us to provide the first topological molecular analysis of component proteins in a cellulose-synthesizing TC structure in a prokaryotic organism.
Cellulose is the most abundant biological polymer on earth and is the major component of cell walls in higher plants, as well as in some algae. Cellulose-synthesizing organisms are widely distributed in all biological kingdoms, including not only higher plants and algae but also bacteria, protists, fungi, and tunicates (28). It is generally recognized that native cellulose is synthesized and crystallized by a multimeric enzyme complex located in the cytoplasmic membrane (CM) (6). Terminal complexes (TCs) have been found in most cellulose-synthesizing organisms (4) and are classified into two types: linear TCs and rosette TCs. Linear TCs have been observed among various algae (6, 7, 20), Dictyostelium sp. (a social amoeba) (5, 17), ascidians (primitive chordate animals) (21), and Acetobacter xylinum (a bacterium) (8). TCs of A. xylinum consist of a single row of particles on the outer membrane (OM), and a flat cellulose microfibril is produced from at least three of these TC subunits (8). Each subunit of the TCs is a transmembrane protein complex that spans both the OM and the CM. Rosette TCs have been found in land plants (26) and freshwater algae (15), including solitary rosette TCs virtually identical to those in land plants (18, 19), as well as primitive land plants (13).
Investigations of A. xylinum as a model organism for cellulose biosynthesis (for reviews, see references 28, 29, and 35) have led to the following: (i) in vitro cellulose biosynthesis (1, 2, 9, 16, 34), (ii) discovery of cyclic diguanylic acid (c-di-GMP) as a specific activator of cellulose biosynthesis in A. xylinum in vitro (30), (iii) purification and identification of cellulose synthase (23, 24, 25), and (iv) the first isolation of the cellulose synthase gene (31, 36).
In spite of numerous efforts using biochemical and molecular biological approaches, there is no direct evidence for the participation of a TC structure in cellulose biosynthesis in A. xylinum. Because freeze-fracture replication is the only method known for visualization of a TC, it has been impossible to obtain information relating this structure directly to the underlying chemistry and/or identity of a component of cellulose biosynthesis.
Sodium dodecyl sulfate (SDS)-solubilized, freeze-fracture replica labeling, which was initially developed for animal cells by Fujimoto (14), has allowed immunocytochemical labeling of freeze-fracture replicas by antibodies. The application of this novel technique has revealed a direct correlation between the structure (rosette TC) and its biochemical component (cellulose synthase) in a vascular plant cell (22).
Purification of cellulose synthase from A. xylinum by the product entrapment technique allowed the identification of two polypeptides with molecular masses of 83 and 93 kDa (23, 24, 25). Specific antibodies were raised against these proteins (10, 25), and several immunochemical studies have been done with these antibodies (10, 11, 12). More specifically, we have used the anti-93-kDa antibody to study the thermal stability of the cellulose synthase complex from A. xylinum.
The present study examined immunogold labeling of the linear TC in A. xylinum by using an antibody prepared against the 93-kDa protein. The 93-kDa protein is part of the cellulose synthase and is proposed to bind c-di-GMP, which is known to be a specific activator of cellulose biosynthesis in A. xylinum (25, 30). Based on this evidence, we anticipated that the 93-kDa proteins should be localized near the cellulose synthase of A. xylinum in the TC.
MATERIALS AND METHODS
Culture and isolation of cells.
A. xylinum NQ5 (ATCC 53582) was grown statically in SH medium (33) at 27°C. The flasks used, which contained an active cellulose pellicle, were shaken vigorously by hand in order to separate the cellulose-synthesizing cells. The medium and squeezed medium from pellicles were filtered using a 50-μm nylon mesh and centrifuged at 2,000 × g for 5 min. The bacterial pellet was resuspended in half of the volume of fresh medium and incubated at 27°C for 1 h.
Antibody production.
Polyclonal antibody against the 93-kDa polypeptide from A. xylinum NQ5 (ATCC 53582) was prepared as described by Chen and Brown (10).
Freeze-fracture and immunogold labeling.
The cell suspensions described above were placed onto gold specimen carriers and immediately quick-frozen by liquid propane in a Reichert KF80 quick-freezing unit (Leica). The frozen samples were fractured in a Balzers BAF400D freeze-etch unit (Baltec, Liechtenstein) at −110°C, replicated by evaporation of platinum-carbon from an electron gun positioned at a 45° angle, and carbon coated. The replicated sample with the specimen carrier was transferred to a solution containing lysozyme (Sigma) at 1 mg/ml, 10 mM EDTA, and 25 mM Tris-HCl (pH 8.0). Digestion of the peptidoglycan was performed for 2 h at room temperature with continuous shaking on a rotary shaker at 100 to 200 rpm.
After peptidoglycan digestion, the replica pieces were transferred to 2.5% SDS containing 10 mM Tris-HCl (pH 8.3). SDS solubilization was conducted for 2 h at room temperature with continuous shaking on a rotary shaker at 100 to 200 rpm. After treatment with SDS, replicas were washed four or more times with phosphate-buffered saline (PBS) and placed on drops of 1% bovine serum albumin in PBS for 30 min at room temperature. The replicas were then labeled with anti-93-kDa protein antibody (diluted 1:100 in PBS) for 2 h at room temperature or overnight at 4°C. After labeling, the replicas were washed three times with PBS containing 0.05% Tween 20 and incubated for 2 h at room temperature with anti rabbit immunoglobulin G antibody conjugated to 10-nm colloidal gold (Zymed Laboratories, San Francisco, Calif.) diluted 1:50 in PBS. After immunogold labeling, the replicas were washed three times with PBS–0.05% Tween 20, fixed with 0.5% glutaraldehyde in PBS for 10 min at room temperature, washed twice with distilled water, and placed onto Formvar-coated grids. The replicas were observed with a transmission electron microscope (model 2000EXII; JEOL, Akishima, Japan).
RESULTS
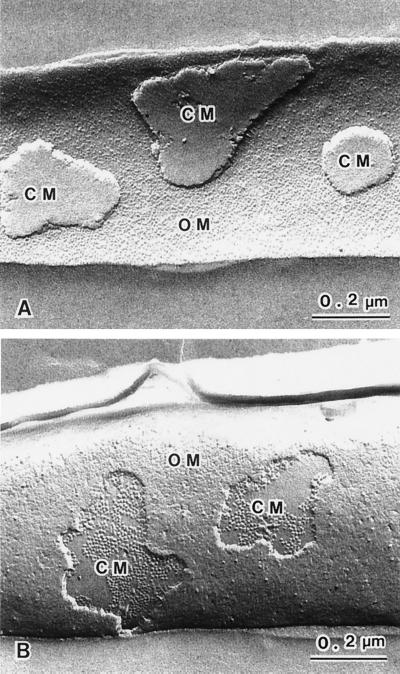
In addition to a CM, gram-negative bacteria, including A. xylinum, have an OM. Sandwiched between the OM and the CM is a concentrated, gel-like matrix (the periplasm) found in the periplasmic space (3). The peptidoglycan in the periplasm forms an envelope around the CM. Since the freeze-fracture plane follows regions of weakly bonded components within a bacterial cell, membranes are most frequently cleaved through their hydrophobic domains and intrinsic membrane proteins are exposed. Fortuitous freeze-fractures through gram-negative bacteria can reveal the inner and outer faces of membranes by exposing concave and convex surfaces. Figures 1A and B show freeze-fractured images of two A. xylinum cells in which the fracture planes have passed through the cell envelope. The cell in Fig. 1A shows a concave fracture plane through the OM and pieces of the CM, whereas the cell in Fig. 1B shows a convex fracture plane through the OM and part of the CM. Due to their characteristic features, four fracture planes can be recognized in A. xylinum, (i) the outer leaflet of the OM (the exoplasmic fracture [EF] face of the OM), (ii) the EF face of the CM, (iii) the inner leaflet of the OM (the protoplasmic fracture [PF] face of the OM), and (iv) the PF face of the CM. The EF face of the OM appears concave and shows the presence of a large number of membrane particles. The EF face of the CM appears concave, with a relatively smooth structure. The PF face of the OM appears to have a rough surface and is convex, with membrane particles and depressions or pits that are randomly arrayed with low density. The PF face of the CM appears convex, with membrane particles of uniform size present at a high density. In the PF faces of both the OM and the CM, the difference between individual membranes is clearly identifiable (Fig. 1B). The identification of the type of membrane is important for discussing the topology of cellulose-synthesizing complexes, as well as the localization with antibody labeling visualized by freeze-fracture electron microscopy.
FIG. 1.
Freeze-fracture images showing various fractured membrane faces. Panel A shows the EF faces of both the OM and the CM. A large number of particles can be seen on the concave plane of the OM. The EF face of the CM has a smooth appearance. Panel B shows the PF faces of both the OM and the CM. The PF face of the OM has a rough appearance, with a low density of variable-sized particles, while the PF face of the CM shows uniform-sized particles, with a high density.
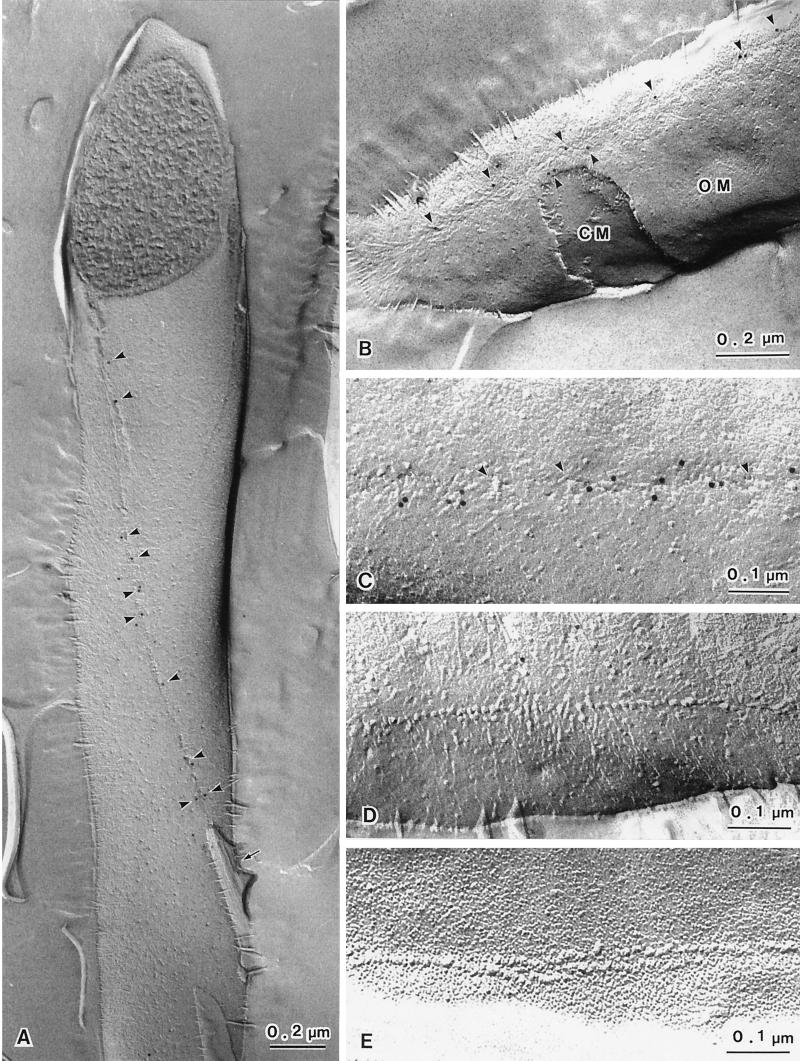
The digestion of the peptidoglycan by lysozyme prior to SDS solubilization was found to be a prerequisite for observing freeze-fracture labeling of membrane proteins in A. xylinum. The lysozyme digests the peptidoglycan and therefore allows the cell debris to be removed, which in conventional freeze-fracture techniques is done by harsh acid treatments. The replicas obtained by SDS–freeze-fracture replica labeling (Fig. 2A) appear similar to those obtained by conventional freeze-fracture techniques. The linear TCs of A. xylinum exhibit ordered particle arrays with a single row or double rows. The bacterial cell in Fig. 2A shows the PF face of the OM and a typical single row of TC subunits with a cellulose ribbon attached at its terminus (arrow). Upon closer examination, the gold particles were observed to be attached along a single row of TCs (arrowheads). In the case of A. xylinum, almost all of the fracture planes occurred through the OM. In other words, the fractured CM was rarely observed in A. xylinum although it is more commonly observed in other gram-negative bacteria (3). The frequency of CM fractures was less than 5% based on the observation of more than 100 cells. Even in the case in which we successfully visualized the fractured plane of the CM, only part of this membrane was exposed (Fig. 1B). Furthermore, TC structures were never observed on the CM of A. xylinum because of the very low frequency of CM fractures. Figure 2B shows a fracture plane occurring mostly in the OM and occasionally in the CM. Note the antibody-labeled TC components (arrowheads) in the vicinity of the pits.
FIG. 2.
Fracture-labeled images (PF face, panels A to D; EF face, panel E) showing reaction with the c-di-GMP-binding protein (93-kDa protein) antibody. Panel A shows a typical fracture-labeled image of A. xylinum. The TC appears to be a single row along the longitudinal axis of the bacterial cell. A ribbon of cellulose microfibrils is attached to the end of the TC row (A, arrow). The 93-kDa protein antibody is distributed along the TC row on the PF face of the OM (A, arrowheads). Panel B also shows labeled TCs on the PF face of the OM but not on the PF face of the CM, which is seen as a window through the OM. The labeled TCs are visible as a row of slight depressions with indistinct particles and small holes that may be due to particle displacement (C, arrowheads). The TCs showing a distinct particle row on the PF face of the OM are not labeled with antibodies (D). Panel E shows TCs with double rows on the EF face of the OM. The TCs on the EF face of the OM are never labeled by the antibodies.
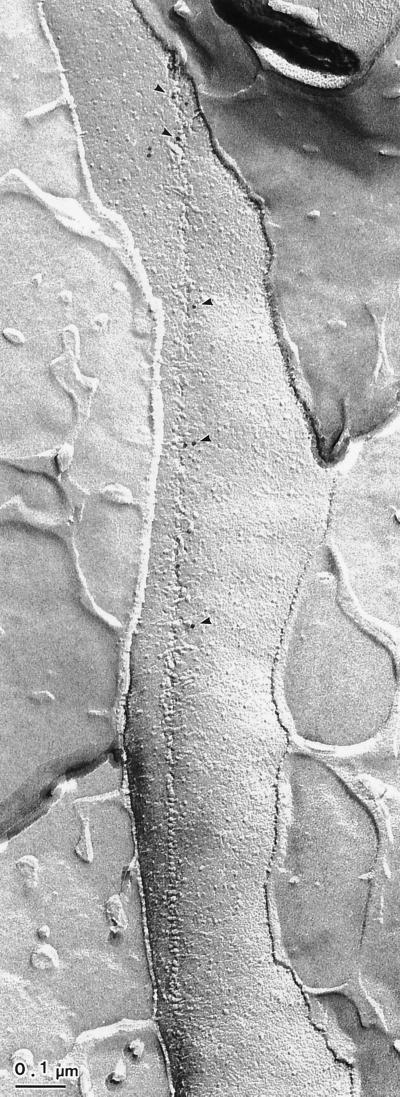
Antibody labeling of TCs on the PF face of the OM is shown in Fig. 2A to C. The labeling is not strictly coincident with the TCs arranged in a linear row. Two different features associated with the linear rows on the PF face of the OM are notable: (i) a pit or depressed region with indistinct particles (Fig. 2B and C) and (ii) a distinct single row of particles (Fig. 2D). The gold particles were localized only in the former and not in the latter. In addition, these same TC particles can be found associated with the outer leaflet of the OM in all cases; however, TCs on the EF face are never labeled (Fig. 2E). These membrane particles often appear to be complementary to the pit-like formations or depressions in the PF face of the OM (not shown). A very rare case of freeze-fracture showed a distinct row of TC particles and pits in a single line on the same PF face of the OM (Fig. 3). The TCs in the region of the pits were exclusively labeled by antibodies.
FIG. 3.
Fracture-labeled image showing two types of TC structures revealed on the PF face of the OM. The distinct TC particles (upper half) and depressions with particles (lower half) are visible as a single row on the same PF face of the OM. Moreover, the labeling of gold particles can be seen only in the region with depressions.
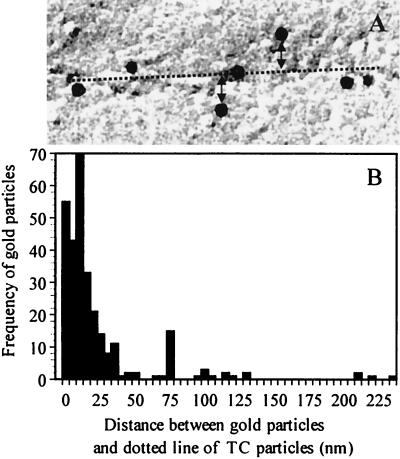
The distribution of gold particles associated with TCs is shown in Fig. 4. The distance between the TCs and gold particles was calculated by measuring the vertical distance between the edge of gold particles and a linear row of TC particles (Fig. 4A, double arrowheads). For frequency analysis, 277 gold particles were randomly sampled from 30 different cells that have a single row of TCs. Measurement of the distance is difficult where the bacterium has double rows of TCs. In this case, 75% of the 277 gold particles were found within 20 nm of the linear row (shown as a dashed line in Fig. 4A. Most gold particles were found within 10 to 14 nm, with a median distance of 9.3 nm from the linear row (Fig. 4B).
FIG. 4.
Frequency distribution of 93-kDa protein antibody-labeled particles. (A) Schematic diagram for measurement of the distance between gold particles and linear TCs. The distance (double arrowheads) between the edge of gold particles and the linear TCs is indicated by the dotted line. (B) Frequency distribution of the number of gold particles associated with the 93-kDa protein antibody shown as a function of the measured distance (nanometers) to the linear TC. The total number of gold particles measured, taken from 30 different cells, was 277.
DISCUSSION
Using freeze-fracture replicas of A. xylinum, we have successfully labeled linear TCs with the antibody against a 93-kDa protein. This protein is a c-di-GMP-binding protein, and c-di-GMP is known to be the activator of cellulose synthase in A. xylinum (25). Our experiments provide the first direct evidence that the linear TCs contain the c-di-GMP-binding protein. The c-di-GMP-binding protein is associated with the cytoplasmic membrane, as suggested by biochemical and sequencing data (9, 32).
Specific labeling of the TCs with antibodies against the 93-kDa protein is found on the PF face of the OM in A. xylinum. Two different parts of the TCs occur on the same PF face of the OM. Almost all of the colloidal gold is associated with an arrangement of depressions that we refer to as pits (Fig. 2A to C). Labeling was never observed where a single row of TC particles is clearly visible on the PF face of the OM (Fig. 2D). Very few fractures were observed across the CM.
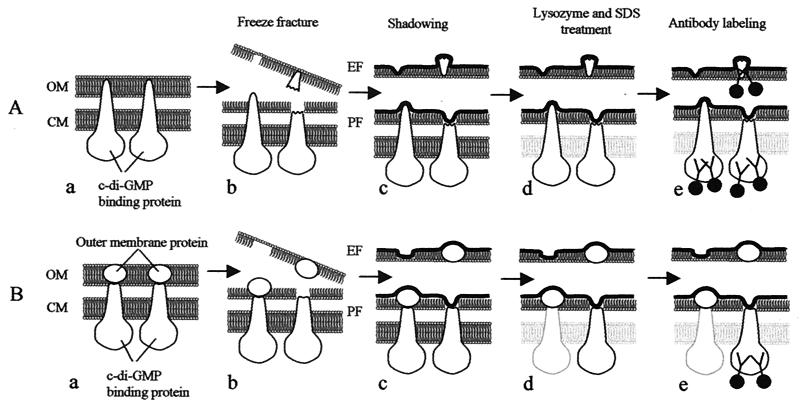
These observations are based on interpretations of a large amount of freeze-fracture data; however, it is difficult to visualize the complete assembly of TCs in relation to the sites of antibody binding. Our interpretation is diagrammed in Fig. 5, which shows two possible arrangements of TC subcomponents and the predicted membrane localization of these components based on freeze-fracture and antibody labeling. The first arrangement is schematically illustrated in Fig. 5A. In this model, the c-di-GMP-binding proteins are proposed to span not only the CM but also the OM (Fig. 5A-a). This model also implies that a single protein spans both membranes, including the periplasm. This could be highly unlikely, but even so, assuming that this is possible during fracture and labeling, we should observe antibody labeling in both the pits and the particles, as shown in Fig. 5A-e.
FIG. 5.
Schematic illustration showing membrane proteins revealed in freeze-fractured images. A hypothetical illustration is shown in panel A-a, where the c-di-GMP-binding proteins span both the CM and the OM of bacterial cells. After freeze-fracturing, three different views of TCs could be expected (A-b). The first is a distinct TC particle row on the PF face of the OM (A-b, left [note that the view is of the flipped replica as a complementary half of the lower replica]), the second is a depression remaining from the tip fracture of the TC on the PF face of the OM (A-b, lower right), and the third is a distinct TC depression area on the EF face of the OM (A-b, upper left). Pieces of c-di-GMP-binding proteins are fixed after shadowing (A-c, upper right) and remain attached to the replica after treatment with lysozyme and SDS (A-d, upper right). Only proteins remaining on the PF and EF faces of the OM are expected to be labeled by the antibodies (A-e). No hypothetical case showing the situation described above was found in the present investigation. The second hypothetical illustration is shown in Panel B-a, where TC particles are composed of two types of subunits, a c-di-GMP-binding protein and an OM protein. After freeze-fracture, OM proteins remain attached to c-di-GMP-binding proteins (B-b, left) or separate from the latter (B-b, right). After shadowing (B-c), OM proteins are fixed on the PF face (B-c, left) or the EF face (B-c, upper right) of the OM. In the latter case, c-di-GMP-binding proteins are fixed on the PF face of the OM (B-c, lower right). After lysozyme and SDS treatment, the c-di-GMP-binding proteins will be removed, as shown in the left half of panel B-d, or they will not be removed, as shown in the lower right of panel B-d. Therefore, only the depression in the TC structure in the latter case will remain on the PF face of the OM and be labeled by the antibodies (B-e, lower right).
By assuming that the TC is a multicomponent complex, the interpretation of labeling shown in Fig. 5B is more likely. In Fig. 5B, only two of the many proteins in the TC are depicted. One is shown to be an OM protein which is linked to the CM component of the TC. This interaction can be either direct or indirect. This is not unlike the multicomponent flux pumps of gram-negative bacteria (27). The model proposed in Fig. 5B is useful in understanding not only that at least two separate components are part of the TC but also where these components reside and more specifically in which membranes.
After freeze-fracturing, the OM proteins remain either on the PF face of the OM (Fig. 5B-b, left) or on the EF face of the OM (Fig. 5B-b, right). After shadowing, the OM proteins are fixed on the replica on both the PF and the EF of the OM (Fig. 5B-c). When the OM proteins are fixed on the replica and remain on the PF face of the OM, the c-di-GMP-binding proteins are never covered by shadowing materials. Therefore, the c-di-GMP-binding proteins will be washed out after treatment with both lysozyme and SDS (Fig. 5B-d, left). When the OM proteins are fixed and remain on the EF face of the OM (Fig. 5B-b, right), the c-di-GMP-binding proteins are fixed and remain on the replica on the PF face of the OM even after treatment with lysozyme and SDS (Fig. 5B-d, right). The antibody recognizes the CM protein stuck to the pit area after replication (Fig. 5B-e, right). The distinct TC particles on both the PF and EF faces of the OM are never labeled by the antibody because those particles are composed of OM proteins (Fig. 5B-e, left and upper right). In summary, these observations help to formulate the topology and composition of the TC with respect to the distribution of proteins in the two membranes.
What is the nature of the proteins in the cellulose-synthesizing complex? It has been suggested that this complex in A. xylinum is made up of three or four different proteins based on the organization of genes in the cellulose-synthesizing operon (32, 36). According to our model, the cellulose synthase catalytic subunit and the c-di-GMP-binding protein (AcsAB) are present in the CM. These proteins possibly interact with AcsC and AcsD in forming the transenvelope complex (6). The identities of the AcsC and AcsD proteins have not been ascertained; however, mutational analysis indicates that these proteins are required for normal cellulose ribbon production (32). It is possible that the AcsC protein is present in the OM and contacts the cellulose synthase catalytic subunit and c-di-GMP-binding proteins that are present in the CM (32). Together, these proteins form a pore complex through which nascent glucan chains emerge to crystallize into the metastable cellulose I allomorph.
A novel combination of freeze-fracture and SDS treatment coupled with a specific antibody label has shown that these methods can be used for topological analysis of large membrane complexes. The cellulose-synthesizing complex in A. xylinum can be useful in understanding the diverse structures and functions of membrane complexes that traverse both the OM and the inner membrane, including the multidrug efflux pumps of gram-negative bacteria (27).
ACKNOWLEDGMENTS
We thank Goro Kikuchi for preparing freeze-fractured replicas.
This work was supported by grants-in-aid for the Research for the Future program from the Japan Society for the Promotion of Science (JSPS-RFTF 96L00605), a grant-in-aid from the Ministry of Education, Science, Sports and Culture of Japan (12306009) to T. Itoh, and grants from the Welch Foundation (F1217) and the Department of Energy (DE-FG03-94ER20145) to R. M. Brown, Jr.
REFERENCES
- 1.Aloni Y, Delmer D P, Benziman M. Achievement of high rates of in vitro synthesis of 1,4-β-glucan: activation by cooperative interaction of the Acetobacter xylinum enzyme system with GTP, polyethylene glycol, and a protein factor. Proc Natl Acad Sci USA. 1982;79:6448–6452. doi: 10.1073/pnas.79.21.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aloni Y, Cohen R, Benziman M, Delmer D. Solubilization of the UDP-glucose: 1,4-β-d-glucan 4-β-d-glucosyltransferase (cellulose synthase) from Acetobacter xylinum. J Biol Chem. 1983;258:4419–4423. [PubMed] [Google Scholar]
- 3.Beveridge T J. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown R M., Jr Cellulose microfibril assembly and orientation: recent developments. J Cell Sci Suppl. 1985;2:13–32. doi: 10.1242/jcs.1985.supplement_2.2. [DOI] [PubMed] [Google Scholar]
- 5.Brown R M., Jr . Algae as tools in studying the biosynthesis of cellulose, nature's most abundant macromolecule. In: Wiessner W, Robinson D G, Starr R C, editors. Experimental phycology I: cell walls and surfaces, reproduction, photosynthesis. Berlin, Germany: Springer-Verlag; 1990. pp. 20–39. [Google Scholar]
- 6.Brown R M., Jr The biosynthesis of cellulose. Pure Appl Chem. 1996;10:1345–1373. [Google Scholar]
- 7.Brown R M, Jr, Montezinos D. Cellulose microfibril: visualization of biosynthetic and orienting complexes in association with the plasma membrane. Proc Natl Acad Sci USA. 1976;73:143–147. doi: 10.1073/pnas.73.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown R M, Jr, Willison J H M, Richardson C L. Cellulose biosynthesis in Acetobacter xylinum: visualization of the site of synthesis and direct measurement of the in vivo process. Proc Natl Acad Sci USA. 1976;73:4565–4569. doi: 10.1073/pnas.73.12.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bureau T E, Brown R M., Jr In vitro synthesis of cellulose II from a cytoplasmic membrane fraction of Acetobacter xylinum. Proc Natl Acad Sci USA. 1987;84:6985–6989. doi: 10.1073/pnas.84.20.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H P, Brown R M., Jr Immunochemical studies of the cellulose synthase complex in Acetobacter xylinum. Cellulose. 1996;3:63–75. [Google Scholar]
- 11.Chen H P, Brown R M., Jr Occurrence of polypeptides in other organisms cross-reacting with antibodies against A. xylinum cellulose synthase. Cellulose. 1998;5:263–279. [Google Scholar]
- 12.Chen H P, Brown R M., Jr Thermal stability of the cellulose synthase complex of Acetobacter xylinum. Cellulose. 1999;6:137–152. [Google Scholar]
- 13.Emons A M C. Role of particle rosettes and terminal globules in cellulose synthesis. In: Haigler C H, Weimer P J, editors. Biosynthesis and biodegradation of cellulose. New York, N.Y: Marcel Dekker, Inc; 1991. pp. 71–98. [Google Scholar]
- 14.Fujimoto K. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. J Cell Sci. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- 15.Giddings T H, Brower D L, Staehelin L A. Visualization of particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls. J Cell Biol. 1980;84:327–339. doi: 10.1083/jcb.84.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaser L. The synthesis of cellulose in cell-free extracts of Acetobacter xylinum. J Biol Chem. 1958;232:627–636. [PubMed] [Google Scholar]
- 17.Grimson M J, Haigler C H, Blanton R L. Cellulose microfibrils, cell motility, and plasma membrane protein organization change in parallel during culmination in Dictyostelium discoideum. J Cell Sci. 1996;109:3079–3087. doi: 10.1242/jcs.109.13.3079. [DOI] [PubMed] [Google Scholar]
- 18.Hotchkiss A T, Jr, Brown R M., Jr . Evolution of the cellulosic cell wall in the Charophyceae. In: Schuerch C, editor. Cellulose and wood—chemistry and technology. New York, N.Y: John Wiley & Sons, Inc.; 1988. pp. 591–609. [Google Scholar]
- 19.Hotchkiss A T, Jr, Brown R M., Jr The association of rosette and globule terminal complexes with cellulose microfibril assembly in Nitella translucens var. axillaris (Charophyceae) J Phycol. 1987;23:229–237. [Google Scholar]
- 20.Itoh T. Cellulose-synthesizing complexes in some giant marine algae. J Cell Sci. 1990;95:309–319. [Google Scholar]
- 21.Kimura S, Itoh T. New cellulose-synthesizing complexes (= terminal complexes) involved in animal cellulose biosynthesis in the tunicate, Metandrocarpa uedai. Protoplasma. 1996;194:151–163. [Google Scholar]
- 22.Kimura S, Laosinchai W, Itoh T, Cui X, Linder C R, Brown R M., Jr Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell. 1999;11:2075–2085. doi: 10.1105/tpc.11.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin F C, Brown R M, Jr, Drake R R, Jr, Haley B E. Identification of the uridine-5′-diphosphoglucose (UDP-glc) binding subunit of cellulose synthase in Acetobacter xylinum using the photoaffinity probe 5-azido-UDP-glc. J Biol Chem. 1990;265:4782–4784. [PubMed] [Google Scholar]
- 24.Lin F C, Brown R M., Jr . Purification of cellulose synthase from Acetobacter xylinum. In: Scheurch C, editor. Cellulose and wood—chemistry and technology. New York, N.Y: John Wiley & Sons, Inc.; 1989. pp. 473–492. [Google Scholar]
- 25.Mayer R, Ross P, Weinhouse H, Amikam D, Volman G, Ohana P, Calhoon R D, Wong H G, Emerick A W, Benziman M. Polypeptide composition of bacterial cyclic diguanylic acid-dependent cellulose synthase and the occurrence of immunologically crossreacting proteins in higher plants. Proc Natl Acad Sci USA. 1991;88:5472–5476. doi: 10.1073/pnas.88.12.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller S C, Brown R M., Jr Evidence for an intramembranous component associated with a cellulose microfibril synthesizing complex in higher plants. J Cell Biol. 1980;84:315–326. doi: 10.1083/jcb.84.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richmond P A. Occurrence and functions of native cellulose. In: Haigler C H, Weimer P J, editors. Biosynthesis and biodegradation of cellulose. New York, N.Y: Marcel Dekker, Inc; 1991. pp. 5–23. [Google Scholar]
- 29.Ross P, Mayer R, Benziman M. Cellulose biosynthesis and function in bacteria. Microbiol Rev. 1991;55:35–58. doi: 10.1128/mr.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross P, Weinhouse H, Aloni Y, Michaeli D, Ohana P, Mayer R, Braun S, de Vroom E, vander Marel G A, van Boom J H, Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 31.Saxena I M, Lin F C, Brown R M., Jr Cloning and sequencing of the cellulose synthase catalytic subunit gene of Acetobacter xylinum. Plant Mol Biol. 1990;15:673–683. doi: 10.1007/BF00016118. [DOI] [PubMed] [Google Scholar]
- 32.Saxena I M, Kudlicka K, Okuda K, Brown R M., Jr Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J Bacteriol. 1994;176:5735–5752. doi: 10.1128/jb.176.18.5735-5752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schramm M, Hestrin S. Factors affecting production of cellulose at the air/liquid interface of a culture of Acetobacter xylinum. J Gen Microbiol. 1954;11:123–129. doi: 10.1099/00221287-11-1-123. [DOI] [PubMed] [Google Scholar]
- 34.Swissa M, Aloni Y, Weinhouse H, Benziman M. Intermediary steps in Acetobacter xylinum cellulose synthesis: studies with whole cells and cell-free preparations of the wild type and a celluloseless mutant. J Bacteriol. 1980;143:1142–1150. doi: 10.1128/jb.143.3.1142-1150.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volman G, Ohana P, Benziman M. Biochemistry and molecular biology of cellulose biosynthesis. Carbohydr Eur. 1995;12:20–27. [Google Scholar]
- 36.Wong H C, Fear A L, Calhoon R D, Eichinger G H, Mayer R, Amikam D, Benziman M, Gelfand D H, Meade J H, Emerick A W, Bruner R, Ben-Bassat A, Tal R. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc Natl Acad Sci USA. 1990;87:8130–8134. doi: 10.1073/pnas.87.20.8130. [DOI] [PMC free article] [PubMed] [Google Scholar]