Abstract
Background
During development, complex organ patterns emerge through the precise temporal and spatial specification of different cell types. On an evolutionary timescale, these patterns can change, resulting in morphological diversification. It is generally believed that homologous anatomical structures are built—largely—by homologous cell types. However, whether a common evolutionary origin of such cell types is always reflected in the conservation of their intrinsic transcriptional specification programs is less clear.
Results
Here, we developed a user‐friendly bioinformatics workflow to detect gene co‐expression modules and test for their conservation across developmental stages and species boundaries. Using a paradigm of morphological diversification, the tetrapod limb, and single‐cell RNA‐sequencing data from two distantly related species, chicken and mouse, we assessed the transcriptional dynamics of homologous cell types during embryonic patterning. With mouse limb data as reference, we identified 19 gene co‐expression modules with varying tissue or cell type‐restricted activities. Testing for co‐expression conservation revealed modules with high evolutionary turnover, while others seemed maintained—to different degrees, in module make‐up, density or connectivity—over developmental and evolutionary timescales.
Conclusions
We present an approach to identify evolutionary and developmental dynamics in gene co‐expression modules during patterning‐relevant stages of homologous cell type specification using single‐cell RNA‐sequencing data.
Keywords: cell‐intrinsic transcriptional programs, cell‐extrinsic signaling environments, evolution of gene expression, EvoDevo, gene co‐expression modules, limb development, WGCNA
Key Findings
We present an approach to identify evolutionary and developmental dynamics in gene co‐expression modules during patterning‐relevant stages of homologous cell type specification using single‐cell RNA‐sequencing data.
1. INTRODUCTION
Recent advances in single‐cell technologies now enable researchers to study the molecular dynamics of pattern formation and evolution at the level of the basic biological unit of life, the individual cell. During development, starting from a single fertilized cell, various progenitor cell populations need to proliferate, differentiate, and—for some of their progeny—undergo controlled cell elimination. These processes require tight coordination, across time and space, to result in proper pattern formation of complex organs. From a cell's perspective, this progression is linked to the integration of various extra‐cellular signals, as determined by its relative position inside the forming tissue, and the cell‐intrinsic interpretation of these cues, shaped by the lineage‐specific molecular state of the cell. Accordingly, evolutionary modifications in a given patterning process can occur through changes in either cell‐extrinsic or ‐intrinsic components—or a combination of both—, to result in morphological diversification.
This is exemplified in the vertebrate limb, a paradigm of morphological evolution, where developmental patterning has experienced important modifications across the tetrapod clade. Molecular genetics studies and experimental embryology have yielded important insights into how, for example, early limb bud outgrowth is initiated and advanced, or what modifications in these molecular programs can drive diversification of limb form and function between different species. 1 , 2 Yet only since recently, thanks to the development of single‐cell genomics technology, can we study the molecular dynamics that occur cell‐intrinsically, at cellular resolution, and in an evolutionary comparative manner.
Cell types, like anatomical structures, can be considered homologous across different taxa, with their evolutionary origins tracing back to a common ancestor. 3 , 4 Moreover, it its generally believed that homologous anatomical structures are built—to a large extent—by homologous cell types, and that changes in organ patterning often simply reflect temporal, spatial, and quantitative differences in the specification of these cells during development. 5 The overall molecular state of a homologous cell type, however, may vary substantially between species, even within a similar developmental context. This holds especially true at the transcriptional level, where selection can be weak and result in genetic drift and concerted transcriptome evolution. 6 , 7 , 8 , 9 , 10 Accordingly, identification of so‐called “species signals,” rather than functionally relevant gene expression changes, can dominate differential expression analyses, particularly when applied to similar cell types over long evolutionary distances. 6 , 11 Moreover, any given cell type might occur in a variety of so‐called “cell states,”, related to, for example, cell cycle or metabolic status, thereby further complicating these comparisons. 12 Hence, to understand developmental pattern evolution comprehensively, we require both a detailed understanding of the cell‐extrinsic changes occurring in the signaling environment, as well as appropriate methods to detect species‐specific differences in the intrinsic molecular make‐up of the recipient cell types.
Here, we present a bioinformatics workflow in R to identify gene co‐expression modules from single‐cell RNA‐sequencing (scRNA‐seq) data, and test for their conservation across developmental stages and species boundaries. Using mouse limb E15.5 data as our reference, we identify tissue and cell type‐specific co‐expression modules and demonstrate the ability to follow their compositional changes, module architecture and expression dynamics along developmental time courses in two distantly related tetrapods. Differences in module conservation—between modules, across species—indicate that patterning processes involving certain cell populations are more likely to occur through changes in extracellular environment, while others undergo high evolutionary turnover in their cell‐intrinsic molecular make‐up. Moreover, we demonstrate the power of gene co‐expression module detection to identify distinct cell states, shared across developmental, and evolutionary timescales.
2. RESULTS
2.1. Primary data acquisition, 3′ UTR annotation and data processing
We used publicly available scRNA‐seq data from mouse and chicken spanning six embryonic stages, from early limb bud initiation and outgrowth, to late stages of pattern refinement and tissue maturation. For mouse, we had access to stages E9.5, E10.5, E11.5, E13.5, E15.5, and E18.5, and used a total of 17 857 cells 13 , 14 (Figure 1A). For chicken, we used our own previously published data (HH25, HH29, and HH31), 15 and complemented the time series with newly generated data points spanning stages HH21, HH24, and HH27, to cover days 3–7 of development with a total of 32461 cells (Figure 1B). For both chicken and mouse, we have fore‐ and hindlimb data, which we analyzed interchangeably (Figure 1A, B). Although this might skew the relative amount of certain cell types, due to heterochronies between fore‐ and hindlimb development, it should not affect the actual types of cells per se (Figure 1C‐F).
FIGURE 1.
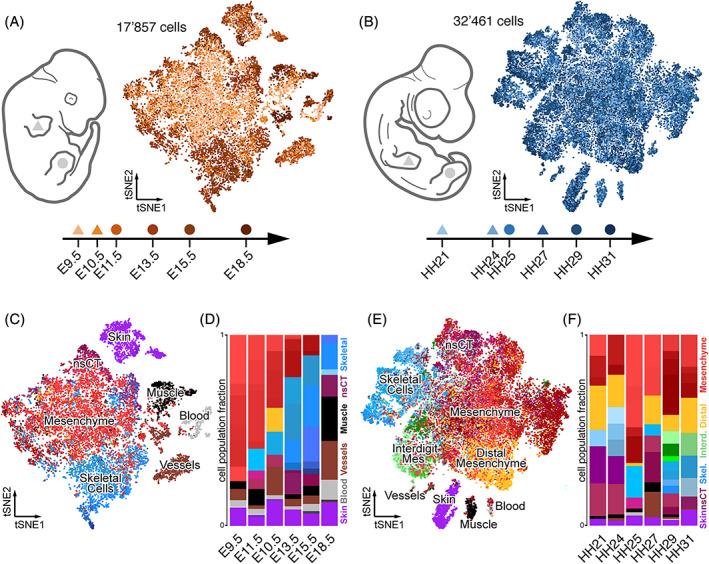
Comparative single‐cell transcriptomic atlases of the developing mouse and chicken limb. A and B, Sampling schemes and tSNE projections of 17 857 mouse (A) and 32 461 chicken (B) forelimb (triangle) and hindlimb (circle) single cell transcriptomes. C–F, Mouse (C) and chicken (E) cluster identities are highlighted by color codes in the tSNE projections, on a cell‐by‐cell basis, with cells belonging to similar tissue types sharing color codes across all samples. Relative sample composition is visualized by barplots (D, F). A total of 86 and 74 clusters were identified in mouse and chicken, respectively, distributed over six sampling time points
A preliminary inspection of the two data sets revealed that—on average—mouse samples displayed a higher percentage of skeletal cell types. We reasoned that either during the preparation of single cell suspensions the mouse tissue had been dissociated more thoroughly, thereby releasing a higher percentage of extracellular matrix‐encapsulated skeletal cells, or that our expression analyses of chick cells failed to accurately capture the expression status of genes important for skeletogenesis. Indeed, when visually examining the genomic location of our mapped chicken reads, many seemed to fall outside the annotated 3′ untranslated regions (UTR) and hence were not included in our unique molecular identifier (UMI) count tables (Figure S1A,B). Such annotation issues have been reported before, also for other species, 16 and they are particularly problematic when using sequencing technologies with high 3′ UTR‐biases like the 10x Genomics Chromium 3′ Kit. Therefore, we decided to improve the 3′ UTR annotation of the chicken genome, using publicly available bulk RNA‐seq data (see Section 4), 17 and re‐quantified all our chicken data using our new transcript models. While this did not completely alleviate the bias in skeletal cells between the species—that is differences in dissociation protocols likely also contributed to this effect –, we now managed to identify small skeletal sub‐populations, like, for example, synovial joints, more reliably in the chicken (data not shown). Moreover, we believe that this improved 3′ UTR annotation will also prove helpful for future single cell genomics studies in the chicken model system.
With these improved UMI count tables for the chicken, we continued our comparisons to the mouse limb samples. As our data sets were produced in different laboratories, we implemented a standardized filtering step of all single cells, based on quality measurements like library size, proportion of mitochondrial reads and number of genes detected. Moreover, due to the overall size of the E9.5 and E10.5 data sets, we randomly subsampled 25% of the single cell transcriptomes, to have data sets of comparable sizes. We then normalized the expression data, performed cell cycle correction and adjusted multi‐batch samples. For each species, we then integrated all cells into a single tSNE dimensionality reduction embedding (Figure 1A,B). In order to identify the different cell types in our data, we first analyzed all stages individually. Using the same parameters of unsupervised graph‐based clustering throughout, we found 13 clusters in the mouse E9.5 sample, 16 at E10.5, 12 at E11.5, and 15 at E13.5, E15.5, and E18.5 each. For our chicken samples, we found 9 clusters in the HH21 data set, 11 at HH24, 15 at HH25, 9 at HH27, 19 at HH29, and 11 at HH31. By comparing the results of our differential gene expression analyses to known marker genes, we were able to identify most of these clusters as distinct cell or tissue types. In all samples, we found one major cell population, consisting of lateral plate mesoderm‐derived limb mesenchymal cells at various stages of differentiation, as well as several smaller clusters of cells with different developmental origins (Figure 1C,E). Of those, skin cells (purple) were present in all samples, while muscle cells (black), blood (light gray), and the endothelial and lymphatic capillary cells of the vascular system (brown) were detected only in a subset of the samples. A small cluster of likely melanocytes (dark gray) was found only at mouse stages E15.5 and E18.5.
Within the lateral plate mesoderm‐derived limb mesenchymal cells, we identified undifferentiated limb mesenchyme (light red), proliferating or cycling mesenchyme (dark red), non‐skeletal connective tissue (nsCT; maroon), and skeletogenic cells like, for example, chondrocytes (blue). Mesenchymal cells with a likely distal location in the autopodial paddle (yellow) were detected only at stage E11.5 in the mouse, but in all chicken samples, while the interdigit mesenchyme cells were only found in the later chicken stages HH29 and HH31 (green). Differences in dissection strategies and dissociation protocols, as well as embryonic stage, likely account for these disparities in cell types detected in a given sample, as well as for changes in their relative abundance. For example, while in mouse samples coming from whole limbs a steady increase in the proportion of skeletal cells is observed, a more targeted sampling of certain limb sub‐domains in the older chicken samples likely obscured this effect (see Reference 15 for details; Figure 1D,F).
Overall, in 17 857 mouse cells and 32 461 chicken cells, we identified a total of 86 and 74 clusters, many of which correspond to distinct cell types at various stages of maturation across the six developmental stages. More importantly, the large majority of these cell types can be considered homologous between the two species, and hence their single‐cell transcriptomes can now be used in a comparative context, to assess cell type‐specific transcriptional dynamics across developmental and evolutionary timescales.
2.2. A bioinformatics workflow to detect cell type‐specific gene co‐expression modules from scRNA‐seq data
To detect cell type‐specific gene expression signatures, and circumvent some of the issues inherent to cross‐species differential expression analyses, we adapted weighted gene correlation network analysis (WGCNA) 18 and tested for the occurrence of transcriptome‐wide gene co‐expression patterns in single cells. WGCNA, originally developed for the detection of gene co‐expression modules in bulk RNA‐seq data, has seen a recent surge in popularity, given the high number of replicate samples, that is, single cells, available when working with scRNA‐seq data. We reasoned that a standardized, user‐friendly bioinformatics workflow would proof beneficial to first‐time users of WGCNA, as well as make the results more comparable between different types of studies and data sets. Accordingly, we developed an R package (“scWGCNA”) for gene co‐expression module detection and comparisons. In a first part, our analysis starts with a Seurat object 19 —one of the most commonly used output formats of scRNA‐seq data analyses these days—and then performs, (a) pseudocell construction, to increase overall robustness; (b) identification of highly variable genes (if not already provided by the user); (c) WGCNA module detection; (d) gene ontology (GO)‐term enrichment analyses and putative cell type identification; and, lastly, produces (e) a standardized output file in HTML format.
For pseudocell construction, 20% of the cells from each cell cluster in the sample are chosen at random (see Figure 2A), to which their 10 nearest neighboring cells in the PCA space are then aggregated. 20 The average expression of every gene is calculated for each of these cell aggregates and normalized, to result in a gene‐by‐pseudocell expression data matrix. Users of our package can, nonetheless, set the fraction of cells used as pseudocell seeds, the number of nearest neighbors (NN) calculated, as well as different dimensionality reductions and number of dimensions to use for the WGCNA detection of co‐expression modules. Additionally, we consider several metadata bins contained in the original Seurat object, including—if already calculated—a set of highly variable genes as determined from the single‐cell data. This gene set is critical for subsequent analyses, as it directly affects the modules that potentially can be detected. 18 , 21 Accordingly, “highly variable gene detection” is optional in our pipeline (see above), allowing users to opt for their method of choice. Thereafter, using the variable genes expression matrix as input, a range of powers are tested to find a suitable soft‐thresholding power that transforms the correlation network to resemble a scale‐free topology, that is, where the underlying structure and characteristics are independent of changes in network size, which is assumed—although not universally—to be the case for many biological networks. 18 , 22 , 23 , 24 This step is inherent to WGCNA and aims to reduce the noise of correlations in the adjacency matrices used. Moreover, it also serves as an important control point: if a scale‐free topology index is not reached, the genes—or cells—used should be reconsidered by the user. Next, the main WGCNA analysis follows. In short: based on the expression matrix, expression correlation, adjacency, and topological overlap matrices are calculated first. Then, based on topological overlap, genes are assigned to discrete modules of co‐expression. The membership of the genes to their modules is tested, based on the correlation of their expression to the overall expression of the module: genes without significant membership are discarded, and the process is repeated until all genes pass the membership test. Lastly, the mean expression of the different modules is calculated in single‐cell space, and plotted onto a dimensionality reduction of choice. Graph representations of all modules are generated, and—optionally—corresponding GO‐term enrichment analyses are performed. All results are then summarized in a single report in HTML format.
FIGURE 2.
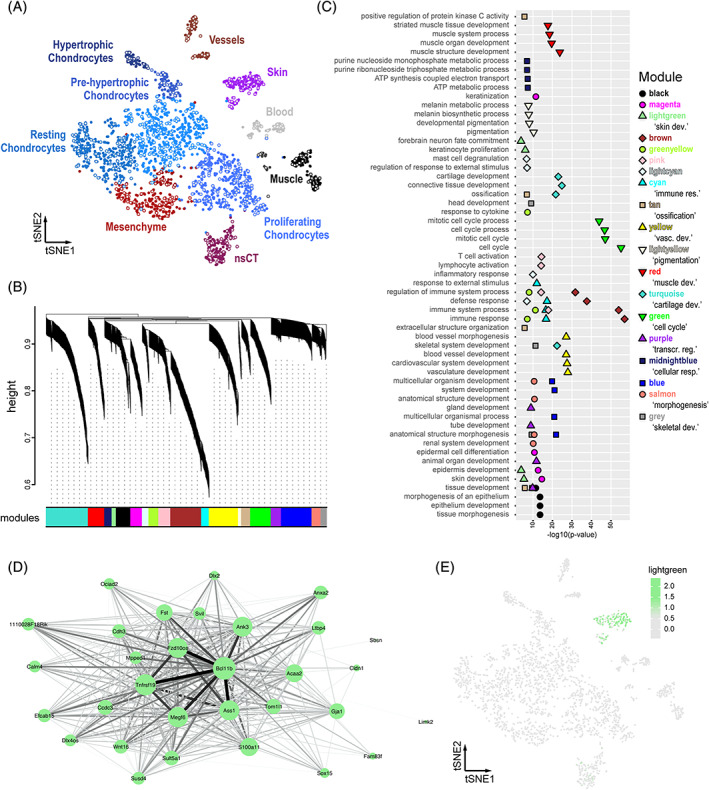
A bioinformatics workflow for iterative gene co‐expression module detection and identification. A, tSNE projection of the mouse E15.5 sample used as reference for iterative WGCNA gene co‐expression module detection. Identified clusters are color‐coded and labeled, with seed cells used for pseudocell construction highlighted in grey. B–E, Sample outputs of our “scWGCNA” package. B, Final WGCNA gene hierarchical clustering dendrogram, showing 19 modules of co‐expression. C, Cumulative results of GO‐term enrichment analyses, showing the top 4 (by P‐value) enriched GO‐terms for each module. D, Cytoscape visualization of co‐expression module “lightgreen.” Node size for each gene is proportional to its module membership, edge thickness and intensity represent topological overlap. E, E15.5 tSNE showing the averaged cellular expression of module “lightgreen.” GO, gene ontology; WGCNA, weighted gene correlation network analysis
We decided to test our workflow with only one limb data set, in order to be able to compare composition and expression status of the identified modules in different embryonic stages, as well as across species boundaries. We used the mouse E15.5 sample as our reference data, as it showed a high variety in skeletal and connective tissue cell populations, at various stages of differentiation (Figure 2A). It is important to note here that the cellular complexity and quality of the reference data set, as well as the set of variable genes to be used, will influence the overall outcome of the ensuing analyses. The user of our pipeline is, therefore, advised to make an informed decision regarding these input parameters and interpret the results accordingly. Here, a total of 513 pseudocells were constructed from the 2594 mouse E15.5 single‐cell transcriptomes. Using the 2967 top variable genes as input, we obtained 1248 genes showing significant co‐expression dynamics, distributed over 19 modules of co‐expression of varying sizes and similarities (Figure 2B). Genes within the detected modules showed signs of enrichment for functional GO‐terms that we expected to be important for limb development, distributed over the different tissue types found in the forming appendage (Figure 2C). Likewise, the averaged expression of a module, calculated over all single cells as the averaged expression of all the genes contained within it, often showed patterns of tissue or cell type specificity. Certain modules exhibited highly restricted expression, confined to a single cell cluster, while others spanned across multiple populations. Accordingly, the different modules identified are hereafter referred to by their likely affiliation to a particular biological process, based on our GO‐term enrichment analysis or by their tissue‐restricted activity. Module colors, as resultant from the primary WGCNA analysis, are also provided for easier visual inspection of the figures. For example, we labeled module “lightgreen” as a “skin development” module. This module was one of the smallest detected, with 31 genes centered around Bcl11b, an averaged module activity confined to our previously identified skin cluster, and enrichments for corresponding GO‐terms (Figure 2D,E). Other modules with tissue specificity were, for example, “muscle” (“red”), “vascular system” (“yellow”) or “cartilage” (“turquoise”); while modules “cell cycle” (“green”) and “cellular respiration” (“midnightblue”) displayed broader patterns of activity.
Hence, our workflow is able to detect gene co‐expression modules in scRNA‐seq data that reflect the transcriptional identity of a certain “cell type” or “cell state.” The analysis of a mouse E15.5 limb scRNA‐seq data set revealed the existence of several gene co‐expression modules with varying patterns of tissue or cell type specificity, as well as containing distinct proliferative or metabolic signatures.
2.3. Testing for gene co‐expression module conservation across developmental and evolutionary timescales
We then conducted a comparison of these modules of gene co‐expression across all samples, testing for the conservation of different properties in each module: gene composition, “density” and “connectivity.” First, to assess module composition and overall activity between the two species, we checked if genes within the modules were present as orthologous, and whether they are expressed in any of the chicken samples. In terms of gene content, we only considered 1‐to‐1 orthologous, based on Ensembl criteria with a confidence cutoff of 1. 25 , 26 Presence/absence of genes in the chicken genome varied greatly between the different modules, with the highest percentage of 1‐to‐1 orthologous missing in modules related to “immune function” (Figure 3A, “brown,” “greenyellow,” “pink,” “lightcyan” and “cyan”). Conversely, modules enriched for GO‐terms related to “transcriptional regulation” and “morphogenesis” all had more than 75% of their genes represented in the chick genome (Figure 3A, “purple”; and “blue,” “salmon”). In terms of expression, the highest fraction of non‐expressed genes was found in two modules related to “skin development” (“magenta”) and “pigmentation” (“lightyellow”), with ~15% and ~25% of their 1‐1 orthologous not being detected in our chicken samples. Overall, our analysis showed that the degree of gene conservation—on a module‐by‐module basis—can vary substantially between the two species, with 8 of the 19 modules having less than 60% of their genes present in our chicken samples as 1‐to‐1 expressed orthologous. However, these discrepancies did not seem to occur randomly, but agreed with what we know about the molecular evolution of the biological system the corresponding modules were affiliated to. For example, seven of these eight modules were enriched for GO‐terms related to immune function or skin development (Figure 3A), both of which are known to have diverged considerably between the two species. For the remainder of our analyses, we only considered 1‐to‐1 orthologous expressed in samples of both species.
FIGURE 3.
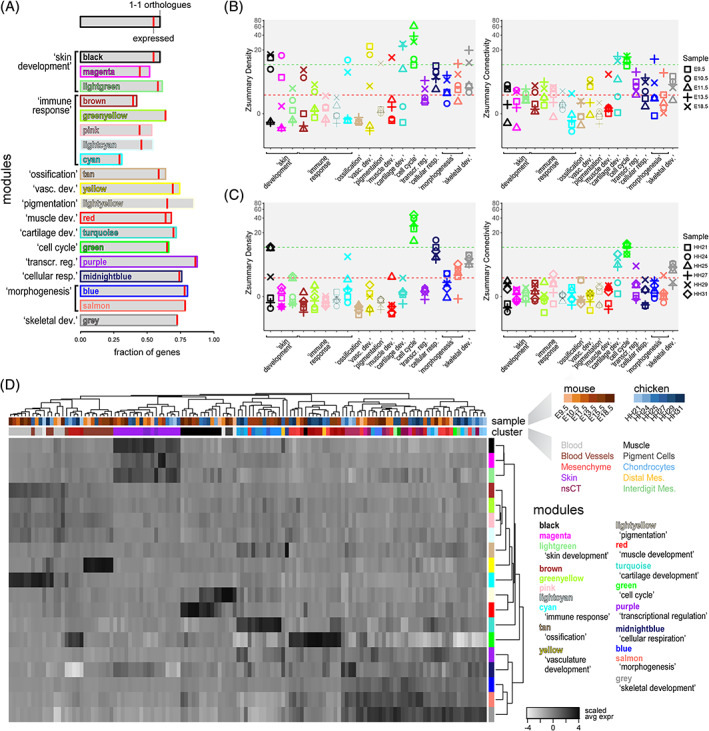
Evolutionary and developmental dynamics in gene co‐expression modules. A, Conservation of module gene composition and expression status. Barplots showing, module by module, the fraction of module genes present as 1‐1 orthologous in the chicken genome, and whether or not they are expressed in our samples (red bar). B and C, Zsummary statistics for “density” and “connectivity,” module by module, for each test sample in mice (B) and chicken (C). The different developmental stages for each sample are coded for by symbols. Green and red dotted lines indicate Zsummary values of 10 and 2, respectively, to indicate “strong evidence” of module “density” or “connectivity” conservation (above 10), potential conservation (between) or “no evidence” of conservation (below 2). D, Hierarchically clustered heatmap of scaled averaged expression for each gene co‐expression module, across mouse and chicken samples on a cluster‐by‐cluster pseudobulk basis. Top row of cluster identifiers is color‐coded for species and developmental stage, bottom row for tissue type to which the respective cluster was attributed to, based on marker gene expression. Representative gene ontology (GO)‐terms are provided for each module
We next wanted to assess to what extent the module co‐expression relationships between genes are conserved across developmental and evolutionary timescales. A co‐expression network can be visualized as a group of genes (nodes), connected with different strengths as defined by their co‐expression relationships (edges; see also Figure 2D). We tested for conservation of “density” (i.e., the average strength of all connections between all genes) and “connectivity” (i.e., the patterns of strength of connections) in all of our modules. 27 For this, we developed a second part of our workflow, again implemented in our R package. As input, a list of 1‐to‐1 orthologous genes, expression matrices of the test data sets, and the reference WGCNA analysis (see above) is required. In a first step, modules are filtered to only contain expressed 1‐to‐1 orthologous. In a second step, a “preservation test” is performed, which aggregates and summarizes four different “density preservation statistics”, to test if modules remain highly interconnected, as well as three “connectivity preservation statistics,” evaluating whether the connectivity pattern of the modules is mantained. 27 Hence, we obtain metrics for the conservation of module gene composition, as well as indices for the preservation of “density” and “connectivity” for each module, across the different test samples (Figure 3A–C).
These “preservation tests” showed that the co‐expression dynamics within our modules have different levels of conservation across our samples. To quantify these differences, we used the summarized Z statistic of conservation. This statistic can be interpreted with two thresholds, with a Z statistic greater than 10 implying strong evidence of module preservation, and lower than 2 suggesting no evidence of preservation. 27 In general, we observed that—as expected—the co‐expression relationships of the modules detected in the E15.5 sample are more conserved in the other mouse samples, than in chicken. We also found that, overall, density is more conserved than connectivity, with only a few exceptions (Figure 3B,C). By using a median rank index, we observed that the most conserved modules are related to “cell cycle” (“green”) and “skeletal development” (“grey”). In the mouse samples, we noticed that only module “transcriptional regulation” (“purple”) shows an overall higher conservation of connectivity than density, implying that the co‐expression relationships between specific genes are better conserved than the overall correlation in the module. Moreover, in chicken samples, modules “transcriptional regulation” and “cartilage development” (“purple” and “turquoise”) also showed overall higher conservation of connectivity than density. On average, however, modules related to “cartilage” and “skeletal development” showed higher conservation in density and/or connectivity in chicken samples, as compared to “transcriptional regulation”, even though the latter contained a higher fraction of expressed 1‐to‐1 orthologous. The structure of these two “cell type”‐related modules, “cartilage” and “skeletal development,” therefore, appear particularly preserved between the two species, especially at later stages of differentiation (Figure 3A‐C).
Despite this seemingly low level of overall conservation, our gene co‐expression modules still seemed to carry a substantial amount of information concerning “cell type” and “cell state,” both across developmental stages as well as for comparing samples between the two species. To try and infer cell type and cell state equivalencies, we first calculated the expression of each module for every cell. We then averaged cellular module expression across all previously identified cell populations in our samples, that is, across developmental stages and species, to define so‐called cell population‐specific “pseudobulk” representations of module activities. Importantly, not all of these pseudobulks might be equally well represented by the activities of modules calculated from the mouse E15.5 data set, due to presence/absence of certain cell types in the different samples of our study (see also comment above, Section 2.2). To account for this potential shortcoming, we defined a threshold as the median expression of all modules in a given pseudobulk, plus two times the median absolute deviation (MAD). Only pseudobulks expressing any module at a higher level than this threshold were considered for further comparisons. Out of a total of 160 pseudobulks, 128 showed high enough expression of at least one of the co‐expression modules to pass our threshold. We scaled the expression data module‐wise and calculated Pearson's correlation coefficients, Euclidian distances, and hierarchical clustering of all pseudobulks and modules. For the most part, these pseudobulks did not group by species in the hierarchical clustering, but rather by cell type in general (Figure 3D). Pseudobulks not derived from lateral plate mesoderm cells showed a particularly clear cell type‐based clustering, regardless of the species of origin. We found chicken and mouse blood cells, vessels, muscle, and skin pseudobulks grouped together, due to their elevated expression of modules enriched for GO‐terms reflecting the respective cellular functions. On the other hand, lateral plate mesoderm‐derivatives were divided into three major sub‐clusters. For the first two on the left, this sub‐division was driven mainly by the high expression of the “cell type”‐related module “cartilage development” (“turquoise”), and a proliferative “cell state”‐signature characterized by module “cell cycle” (“green”). Interestingly, the third sub‐cluster was further structured by more “cell state”‐like module signatures, for example, “cellular respiration” or “morphogenesis”. Several of our previously attributed “cell type”‐based classifications of pseudobulks intermingled here, suggesting that “cell state”‐like module activities might indeed contribute an important layer to cell cluster classifications, compared to using differential gene expression analyses alone (Figure 3D).
Collectively, using our R package to test for conservation of gene co‐expression at multiple embryonic time points, and between distantly related species, we uncovered considerable disparities between different modules. Often, these differences were in line with the likely cellular functions attributed to the respective modules, and the known evolutionary dynamics of the associated biological systems. Regardless of the degree and type of conservation, however, most of the identified modules still seemed to contain important information concerning the cell type and state from which a given single‐cell transcriptome originated from, both across different developmental stages and taxa. Importantly, using our gene co‐expression module approach, certain “cell types” could be further sub‐divided into distinct classes, based on shared “cell state” signatures.
2.4. Cross‐species developmental dynamics and ontogenetic trajectories of gene co‐expression modules
Finally, we analyzed the expression of our identified modules across embryonic time, in both species, taking advantage of the different developmental stages that were used for tissue sampling. We focused only on cells derived from the early limb bud mesenchyme, as they have a common developmental origin in the lateral plate mesoderm, play a central role in establishing the eventual limb morphology, and displayed a higher degree of heterogeneity in module activities amongst themselves (see Figure 3D). We selected modules showing high‐scaled averaged expression in these cells, and all lateral plate mesoderm‐derived pseudobulks as input. In order to appreciate developmental changes in module gene expression, we re‐grouped the corresponding cell population pseudobulks, species by species, by computing pair‐wise Pearson's correlation coefficients, Euclidian distances and hierarchical clustering. Based on tree height, this identified four major clusters for the mouse and five for the chicken. For both species, we additionally identified clusters consisting of only two or less pseudobulks, which we chose not to analyze further (Figure 4A,B). These module‐defined clusters roughly equated to “mesenchyme,” “proliferative mesenchyme,” “nsCT” and “chondrocytes,” when comparing them to the original assignments of their respective cell population pseudobulks (Figure 4A,B).
FIGURE 4.
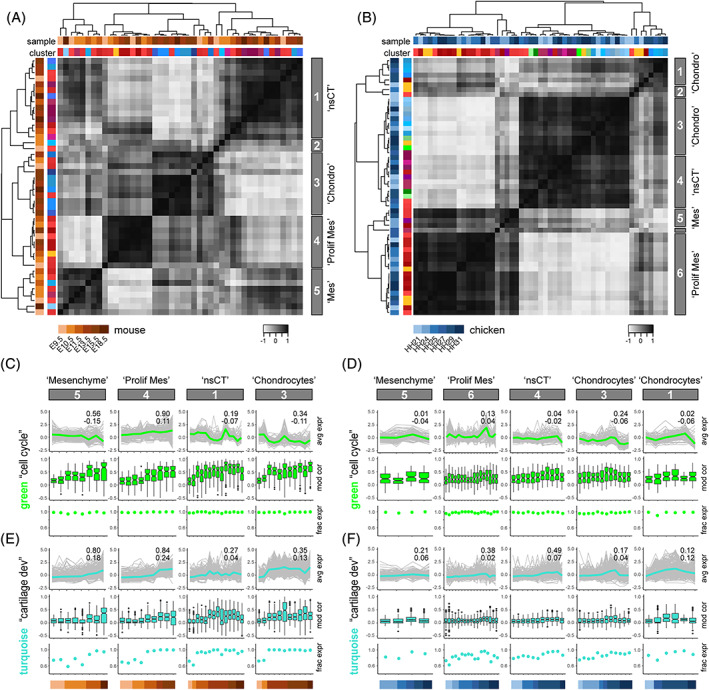
Comparative developmental transcriptome trajectories in homologous cell populations. A and B, Heat maps of Pearson's correlation coefficients and hierarchical clustering based on pairwise Euclidian distances, calculated on averaged expression of selected modules, for mouse (A) and chicken (B) lateral plate mesoderm‐derived cell population pseudobulks. Top row of cluster identifiers is color‐coded for species and developmental stage, bottom row for tissue type to which the respective cluster was attributed to, based on marker gene expression. The five (A, mouse) and six (B, chicken) major clusters emerging, based on tree height, are highlighted on the right with grey boxes. C–F, Developmental trajectories of scaled averaged module expression in ontogenetically ordered pseudobulks, separated by “tissue‐like” clusters identified in A and B. Embryonic stage of each pseudobulk is color‐coded at the bottom, for mouse (C, E) and chicken (D, F). Scaled averaged modules expression trajectories shown in the respective module color, with individual module gene traces underlaid in grey. Superimposed on each trajectory, a coefficient of determination (R 2) of the individual traces (top), as well as the slope of a linear regression (bottom) is provided. Boxplots below indicate the pseudobulk‐wise distribution of the gene‐by‐gene Pearson's correlations to the average module expression across the respective pseudocells. Additionally, the fraction of genes expressed, as defined by a variance >0 in each pseudobulk, is represented by dot‐plots
We decided to focus our analyses on the comparative developmental dynamics of two modules, one related to “cell state” (“cell cycle”, “green”), the other one to “cell type” (“cartilage development”, “turquoise”). For both mouse and chicken, within each module‐defined cluster, we ordered the pseudobulks according to their embryonic stage of collection and plotted the scaled averaged expression of modules “cell cycle” and “cartilage development“ along these ontogenetic trajectories. Additionally, we included the individual gene expression traces contained within the respective module activities (Figure 4C,D). We observed that the transcriptional activity of module “cell cycle” increased along development in the “proliferative mesenchyme,” while it decreased in “mesenchyme,” “nsCT” and “chondrocytes.” These tissue‐specific trends were conserved between the two species, even though all cell populations had an almost uniformly high fraction of the module “cell cycle” expressed (Figure 4C,D). Moreover, on a gene‐by‐gene basis, overall correlation of each gene's expression to the averaged module activity improved with progressively later sampling time points (Figure 4C). This could potentially be attributed to the increasing developmental proximity toward our reference data set, that is, mouse E15.5. However, we did not always find the highest correlations in pseudobulks from that sample and it appeared that gene‐by‐gene expression levels gradually aligned with the overall module activity, with similar developmental dynamics in both species (Figure 4C,D). For the module “cartilage development”, averaged expression was generally higher in pseudobulks of later stages, regardless of their tissue affiliations or species origins (Figure 4E,F). Likewise, gene expression correlations to the averaged module activity increased with developmental time. In contrast to the module “cell cycle,” however, the expressed fraction of genes in module “cartilage development” showed tissue‐specific temporal dynamics. While starting out with an overall lower percentage of genes being expressed (~60–80%), this quickly approached saturation in mouse “nsCT” and “Chondrocyte” pseudobulks for all but the earliest time points. Mesenchymal populations, and chicken pseudobulks in general, displayed less pronounced increments in the fraction of “cartilage development” genes being expressed (Figure 4E,F).
Overall, by contrasting the tissue‐specific developmental activities of two modules, the “cell state”‐related module “cell cycle” and the “cell type”‐related module “cartilage development”, we uncovered distinct “expression level,” “gene‐to‐module correlation” and “gene activity” dynamics.
3. DISCUSSION
To understand the molecular basis of morphological evolution, as driven by changes in embryonic and post‐embryonic development, both cell‐extrinsic and ‐intrinsic alterations need to be considered. 28 Here, we present an integrative approach to perform comparative gene co‐expression analyses at the single‐cell level. We demonstrate its functionality by testing single‐cell transcriptomic data from the developing mouse limb for the occurrence of cell type‐specific gene co‐expression modules and assess their conservation and developmental dynamics in the corresponding cell populations of the chicken.
3.1. Assessing gene co‐expression modules in scRNA‐seq data from distantly related species
Deciphering species‐specific molecular states of homologous cell types is essential, to correctly interpret their response to alterations in extracellular signaling environments. With the advent of single‐cell genomics, we now have the technological means to perform such analyses at the appropriate cellular resolution, across different species. 29 However, comparing gene expression between distantly related taxa has its challenges, especially when working with sparse data like scRNA‐seq. 6 , 16 , 30 , 31 To circumvent some of these inherent issues, we decided to test for the dynamics and conservation of gene co‐expression modules in pseudocells, across developmental stages and in two distantly related tetrapod species. 32 , 33 , 34 , 35
In a first step, we follow the logic of an iterative approach, to perform and optimize WGCNA gene co‐expression modules calculations within a reference scRNA‐seq data set of choice. We use WGCNA statistics to measure significance of gene membership to their assigned modules, and re‐group them accordingly for successive rounds of clustering and testing. 15 , 36 , 37 It is important to note here that WGCNA does not reveal de facto regulatory networks or functional relationships between genes, but rather simply reflects modules of gene co‐expression. 18 For example, while the co‐expression of transcription factors and their putative target genes might indeed reflect regulatory interactions, relying on gene expression data alone to infer this process is prone to result in a high proportion of false positives. 38 Alternative approaches, making use of properly annotated cis‐regulatory sequence information, may seem more appropriate for such purposes. 39 , 40 However, the application of such algorithms is mostly restricted to a very limited set of model species, as they rely on the availability of extensive and high‐quality transcription factor binding motif data sets.
Accordingly, in the second step of our workflow, we opted to perform comparative analyses using modules of gene co‐expression, to make it applicable to the largest number of species possible. Within these modules, we specifically tested for the preservation of the overall strength of connections, that is, “density,” as well as for the patterns of those connections between genes, that is, “connectivity”. 27 The validity of such comparisons obviously depends on the presence of corresponding cell populations between the samples, as well as the number of orthologous genes found in each species to be compared. Naturally, detection of true 1‐to‐1 orthologous is bound to decrease with increasing evolutionary distance. 41 , 42 On a module‐by‐module basis, however, differences in this overall trend may be informative in itself, to interpret the underlying evolutionary dynamics (see Figure 3A, and discussion, below). As for homologous cell types, the restricted presence—for example, hypertrophic chondrocytes in the mouse E15.5 sample—or absence—for example, distal mesenchyme—of certain cell populations in the reference data set also has implications for our comparative analyses (Figure 2A,B). For example, imagine a gene with strong topological overlap to a cell type‐specific module in the reference sample. If that gene in the test sample is co‐expressed with different genes in an additional cell population—that is, absent from the reference sample—, then this might skew connectivity of the tested module. Moreover, developmental heterochronies—for example, in samples originating from serially homologous structures like fore‐ and hindlimbs, or from different species—are further to be considered when interpreting the results. While they should not affect the activity of “cell type”‐related modules in truly homologous cell types, they can impact “cell state” signatures, or the relative numbers of a given cell type in a developing tissue (see Figures 1D,E and 4). We, therefore, advise for an informed and balanced selection of the cell populations and sets of variable genes considered, in order to obtain the most meaningful results. These decisions should be guided by the quality and complexity of the reference data set, as well as the particular question a user wishes to address.
The entirety of the workflow presented above is wrapped in an R package with functions that can be run independently, are customizable, and produce standardized output files to serve as input for further in‐depth analyses. All necessary code and documentation are publicly available. Importantly, while we applied our workflow here to one particular patterning system—that is, the developing tetrapod limb—we would like to highlight that this pipeline could be equally used for many other biological systems for which comparative scRNA‐seq data sets are available.
3.2. Conservation of gene co‐expression modules in the developing tetrapod limb
Working with mouse limb E15.5 data as our reference, we identified a total of 19 gene co‐expression modules and tested for their conservation in mouse and chicken samples, at multiple developmental time points. Already at the compositional level, important qualitative and quantitative differences emerged between the modules. For example, among modules enriched for immune functions, some showed as few as 30% of their genes to be present as 1‐to‐1 orthologous in the chicken genome (Figure 3A). Such high genomic turnover is considered a hallmark of the immune system, compared to other functional groups of genes, as it constantly adapts in an evolutionary arms race to an ever‐changing pathogen and parasite regime. 43 , 44 Likewise, modules enriched for skin‐related functions showed low levels of compositional conservation. The function of the skin, and its associated ectodermal appendages (i.e., hair follicles, glands, or feathers), have diverged considerably between mammals and sauropsids. 45 , 46 , 47 Moreover, selection for a variety of integumentary traits in domesticated chickens might have accentuated this trend further. 48
In terms of “density” and “connectivity,” module “cell cycle” (“green”) showed the overall highest degree of conservation, both for mouse and chicken samples (Figure 3B,C). This is somewhat expected, as a “cell state”‐related co‐expression module reflecting the cell cycle process likely should be conserved even between distantly related taxa. For modules predominantly active in lateral plate mesoderm derivatives, certain tendencies emerged when comparing them across developmental time. Overall, “density” and “connectivity” of these modules seemed better conserved in samples at later stages of development (Figure 3B,C). Likewise, we observed that early pseudobulks of less differentiated cell populations were under‐represented in our analysis of module expression levels (Figure 3D). Nine out of the 14 excluded pseudobulks in mouse, and six out of 18 in chicken, stem from mesenchyme populations of our earliest two time points. Moreover, at the finer scale of our hierarchical clustering, pseudobulks from earlier stages tend to cluster by species (Figure 3D). The fact that we calculated our reference modules at a rather late stage of development might potentially explain this tendency, that is, the expression of certain modules might simply not be adequately represented in these early cells. However, by recreating the same analysis using E11.5 modules as reference, we observed a similar trend (data not shown). Therefore, we suggest that advanced differentiation of cell types effectively makes them—at least module‐wise—transcriptionally more similar to their counterparts in other species, than to their less differentiated relatives in the same organism. 4 , 49
Of all the modules identified for a distinct cell or tissue type, “cartilage development” (“turquoise”) was the overall largest and showed the highest degree of conservation (Figures 2A and 3A‐C). This was particularly evident at later stages of development, and for Zsummary “connectivity,” implicating that differentiating chondrocytes indeed follows similar molecular programs in the two species. Specifically, this evolutionary conserved “connectivity” indicates that genes of module “cartilage development” share conserved co‐expression dynamics, or that they are controlled by the same up‐stream factor(s) across taxa. The cells producing the cartilage template of the limb skeleton thus seem equipped with a similar molecular make‐up, hence making patterning changes between species likely to occur predominately through alterations in extracellular signaling. However, not all signal‐receiving cell populations of patterning relevance show equal conservation in their gene co‐expression dynamics. Modules related to skin development show, as outlined above, high compositional variance and low conservation of “connectivity” (Figure 3A‐C), and integumental patterns can vary greatly, even amongst closely related species. 50 , 51 , 52 Our comparative gene co‐expression analyses in single cells can, therefore, provide important clues whether a certain patterning process is likely to be dominated by changes in the extracellular environment, or if cell‐intrinsic factors are also important to consider for its amenability to evolutionary change.
3.3. Cell types and cell states, in development and evolution
Lastly, looking at our module‐based clustering of pseudobulks, we often observed discrepancies in cluster composition, compared to our original cell type assignments. This holds especially true for pseudobulks of lateral plate mesoderm origin (Figure 3D and 4A,B). There, many of our prior assignments—based on differential expression analysis and marker gene identification—no longer seem to concur with the transcriptional clustering of our gene co‐expression modules. As a result, pseudobulks of different assigned cellular identities, for example, chondrocytes, mesenchyme or interdigit, start to intermingle. Upon closer inspection, this trend seems to be driven—to a large extent—by the differential activities of modules “cell cycle” and “cellular respiration”, respectively (Figure 3D). Both of these modules clearly seem more indicative of cell state, than cell type. 12 Therefore, using co‐expression module detection on single‐cell data appears to reveal commonalities in the expression dynamics of groups of genes that otherwise might go unnoticed. For example, if relying on differential expression analyses alone, that is, by contrasting each of the populations against the rest of the cells, groups of genes with broad expression patterns will most likely not be detected as markers of a given cell population. 53 Moreover, concerted transcriptome evolution can result in a strong “species signal,” thereby interfering with the differential expression analyses‐based identification of functionally relevant transcriptome signatures. 6 , 11 These issues seem particularly relevant for genes that relate to cell state, rather than cell type, as module‐based “cell state” signatures of gene expression can be shared by a variety of different cell types (Figure 3D and 4A,B). By specifically recognizing the impact of such shared “cell state”‐related modules on the overall transcriptome, one may thus shift the focus towards true “cell type”‐identifying signatures.
Accordingly, we advocate for a multi‐layered approach when assigning cellular identifiers to scRNA‐seq data, where a combination of differential expression analyses, cluster‐independent gene co‐expression module detection, and prior knowledge of the biological system at hand is taken into consideration. At a broader scale, even in samples from embryonic stages, the data will generally have the tendency to sort according to developmental lineage, cell type, and only then cell state. The last two categories especially, however, can be difficult to disentangle during development. Many cell types can often be present in multiple stages of differentiation, with rare trajectional intermediates—or transitional stages—interspersed in between. 54 , 55 , 56 , 57 Whether those themselves should be considered distinct cell types, or rather cell states, can be a matter of debate. 4 , 12 , 58 Clearly, though, accounting for more general, lineage‐independent cell states should result in a more comprehensive appreciation of the respective cell type behaviors, with, for example, “cell cycle” expected to be a dominant signature in any growing tissue. This will only become more relevant, as scRNA‐seq studies continue to expand into investigating the impacts of different genetic backgrounds, or environmental variables. 59 , 60 , 61
Overall, we observe that our comparative gene co‐expression module approach represents a valuable addition to discriminate distinct cell states, some of which can be shared amongst different cell types or even distinct developmental lineages. Especially among early, undifferentiated tissues, these module signatures can contain important temporal information across samples, but also—for more mature cell types—signals relevant for comparisons between distantly related species, mutant backgrounds, and environmental parameters.
4. EXPERIMENTAL PROCEDURES
4.1. Sampling and data sources
We sampled complete forelimbs at stages HH21, HH24 and HH27. Tissue dissociation and 10x Genomics Chromium 3′ Kit library preparation was performed as reported previously. 15 We obtained for HH21/HH24/HH27 a total of 2990/5352/2189 cells, with median UMI counts of 2365/1735/1315 and median number of genes detected of 978/776/637 per cell. Raw sequencing data and UMI count matrices are available under GEO accession GSE174565. Publicly available data sets used in this study were mouse E9.5 and E10.5 (GEO accession: GSE149368) 14 ; mouse E11.5, E13.5, E15.5 and E18.5 (GEO accession: GSE142425,) 13 ; and chicken HH25, HH27 and HH29 (GEO accession: GSE130439). 15
4.2. 3′ UTR elongation and improved chick genome annotation
To elongate 3′ UTR annotations, we used stage HH11, HH14, HH21/22, HH25/26, HH32, and HH36 whole embryo bulk RNA‐seq data sets. 17 RNA‐seq reads were processed and mapped individually for each stage, filtered and down‐sampled to 40 million pairs of mapped reads per sample. Resulting BAM files were merged and used to generate transcript models with Cufflinks. 62 The newly calculated transcript models were then processed for 3′ UTR elongation. Elongation of existing GRCg6a 3′ UTR annotations was conducted with the following logic: We only considered transcript models with expression >1 FPKM, which overlapped only one original gene annotation track, and where the original 3′ UTR annotation was shorter than the novel model. 3′ UTR elongation was capped at a maximum of 5000 bp, and was shortened accordingly, if it resulted in any overlap with a neighboring gene. A total of 3132 3′UTRs were elongated in such way. This resulted in a slight overall increase of the average 3′UTR length, yet with many of the extensions not exceeding 100 bp (Figure S1C,D). However, even such modest extensions in 3′ untranslated region (UTR) lengths resulted in a substantial increase of unique molecular identifier (UMI) counts detected for many genes, including some well‐known regulators of tetrapod limb development (Figure S1E). Overall, genes with increased UMI counts showed a slight enrichment for GO‐terms related to a variety of different developmental processes (Figure S1F). Additionally, we realized that with the migration form Gallus_gallus‐5.0, 225 genes stable IDs associated with a gene name were now absent from GRCg6a. Using a combination of BLAST 63 and the GenomicRanges and IRanges packages 64 in R, we managed to recover 62 of these genes and appended them to our modified GRCg6a annotation.
4.3. Single‐cell data pre‐processing
All chicken samples were processed with CellRanger (10x Genomics), using our improved GRCg6a genome annotation. Chicken and mouse UMI count matrices were processed, with cells filtered for quality based on total and relative UMI counts (i.e., >4*mean and <0.2*median of the sample) and percentage of mitochondrial UMIs (i.e., >median + 3*MAD & >0.1, except if UMI count >median), and relation of UMI count/genes detected (i.e., <0.15 & UMI count <2/3). UMI matrices for E9.5 and E10.5 samples are already filtered for total UMI counts and mitochondrial counts. Due to the overall size of these two data sets, we randomly subsampled 25% of the single cell transcriptomes, to have data sets of comparable sizes. Moreover, we excluded 4412 cells from the first replicate of the E9.5 sample showing abnormal hemoglobin genes expression.
4.4. Data normalization and correction
UMI count data was normalized cell‐wise using Seurat v3.1.4 19 with a scale factor of 10 000 and then log‐transformed using the function “NormalizeData” with the rest of the default parameters. Total UMI count, proportion of mitochondrial UMIs and cell cycle stage scores 15 , 65 , 66 were then used as variables to regress using the function “SCTransform” from Seurat with default parameters. Moreover, for samples E9.5 and HH29, sequencing batch effects were also regressed. It is important to note that cell cycle correction is only applied to calculate PCs, tSNEs, and clusters, but not for differential expression analyses and all other analyses.
4.5. Dimensionality reduction, cell clustering, and cluster annotation
We performed principal component analysis (PCA) using Seurat's “RunPCA” with default options. Significant PCs were determined for each sample as those falling outside of a Marchenko‐Pastur distribution 67 —namely, for the mouse samples: 17, 23, 14, 22, 23, and 23 PCs; and for the chicken samples: 18, 19, 20, 17, 22, and 21 PCs. tSNEs were produced to retain and represent the global structure of the data. 68 , 69 To infer cell clusters, we identified the NN of each cell, using the first significant PCs and the function “FindNeighbors” followed by the function “FindClusters” with a resolution of 0.8, a random seed of 42, and the rest of the default parameters. We calculated a hierarchical tree of clusters using “BuildClusterTree” based on significant PCs and identified “sister tips” and performed differential expression tests on each of them. If two clusters showed less than five genes differentially expressed, they were merged, and the process repeated with a new tree of clusters. Differential expression analyses were performed with the MAST 70 implementation in Seurat. Using “FindVariableFeatures,” we selected highly variable genes with a standardized variance larger than the sample median. For making comparisons across clusters we used normalized but “uncorrected” data, using the δ(S‐G2M) as a latent variable. We only tested highly variable genes expressed in at least 25% of the cells in either cell population. Only genes with an adjusted P‐value <.05 and log2 fold change >.5 were considered as differentially expressed. Differentially expressed genes were then used as “marker genes” for cell cluster annotation, in combination with spatial gene expression data repositories like Geisha (Chicken Embryo Gene Expression Database) 71 and MGI (Mouse Gene Expression Database), 72 as well as GO‐term enrichment analyses. 73 Data integration into a single tSNE per species was conducted using transformed data and “IntegrateData” with its related functions. We used as anchors all the shared expressed genes for the mouse and 3000 highly variable genes for the chicken, with 20 dimensions, a k.filter of 100 and the rest of the default options. PCA and tSNE were calculated as above.
4.6. R package “scWGCNA”
The main analytical workflow presented in this article is contained within a newly developed R package, “scWGCNA,” and is available on GitHub with accompanying documentation and sample HTML output files at https://github.com/CFeregrino/scWGCNA. The three different functions in the package can be customized by changing different parameters (see help in the package itself). The functions are outlined below with the parameters used in this study.
4.6.1. “Pseudocell” function
To increase robustness, we define so‐called “pseudocells.” The 10 NN of each cell were calculated in the PCA space using “FindNeighbors.” From each of the previously calculated cell clusters, 20% of the cells were chosen randomly as seed cells. In order to maximize the number of cells aggregated into pseudocells, we perform a sampling of 50 sets of randomly chosen seed cells and choose the set with the largest NN count. Moreover, seed cells typically share some of their NN with other seed cells, for which we do an iterative cell distribution step. First, to avoid “greedy” seed cells, starting with the seed cell with the lowest amount of remaining NN, one of its NN is chosen at random. The chosen cell is removed from the universe of cells and its assigned seed cell is recorded. Once all cells have been distributed to a seed cell, we use the function “AverageExpression” to aggregate the scaled expression of each resulting pseudocell. We recommend using between 10 and 15 NN for 20% of the cells as seeds, as a saturation of aggregated cells is achieved in this range according to our simulations (data not shown).
4.6.2. “Iterative WGCNA” function
We calculate highly variable genes from normalized single cell data using “FindVariableFeatures” and the “mvp” method with cutoffs of minimal 0.25 dispersion and minimal 0 expression. Then, with pseudocell expression data, a soft thresholding power is selected to calculate an adjacency matrix using “pickSoftThreshold” in WGCNA with the bidweight midcorrelation method and a signed network type. 18 The WGCNA analyses itself occurs in a recursive manner. A topological overlap matrix is produced from pseudocell expression data with the function “TOMsimilarityFromExpr,” with previously calculated soft thresholding power and bidweight midcorrelation. A hierarchical clustering tree is then computed using the topological overlap distances. A series of cut heights are set in steps of 0.0001 around (+‐ 0.0005) of a height of 99% of the range between the fifth percentile and the maximum heights on the clustering tree. The size of the detected modules for each cut height is recorded, and the height producing the smallest—or no—gray module (i.e., unassigned genes), and the same number of modules as the previous iteration (or 20, in the first run) is selected. Once a height is selected, modules are detected, and module membership of each gene is calculated using “geneModuleMembership.” Genes not assigned, or without significant module membership, are removed, and the remaining ones are used to start the process again. Once all remaining genes have significant module membership, eigengenes and average expression of each module are calculated in single‐cell space, and GO‐term enrichment analyses for each module us performed using Limma. 15 , 73 All output is contained within a single HTML file, with averaged module expression plotted on tSNEs. Networks are visualized using R packages “network” 74 and “GGally,” 75 with edge thicknesses and intensities scaled module‐wise to represent topological overlap. Additionally, an RDS object is generated, which contains all the data calculated during this step, including the expression matrix used for the final module detection, module assignment per gene, and other module properties. This object can then be used to further analyze the modules and create different plots.
4.6.3. “Comparative WGCNA” function
We use pseudocell data of both reference and test data sets. We subset the modules to contain only genes present as high confidence 1‐to‐1 orthologous, using orthologous genes list from ENSEMBL BioMart. 76 Using the “goodGenes” function from WGCNA, we filter genes based on expression and variance in all test samples. Conservation test is performed by “modulePreservation,” with filtered module assignments, bidweight midcorrelation, a maximal gold modules size of 300, and 20 permutations. 27 The overall conservation Zsummary and median rank, as well as the density and connectivity conservation Zsummary are summarized in a single HTML file. An RDS object is also created during this step, which can be used to recreate the plots presented in the HTML file, as well as to explore other statistical results of the preservation test.
AUTHOR CONTRIBUTIONS
Christian Feregrino: Conceptualization; data curation; writing code; formal analysis; investigation; methodology; validation; visualization; writing‐original draft; writing‐review & editing. Patrick Tschopp: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; supervision; validation; visualization; writing‐original draft; writing‐review & editing.
Supporting information
Figure S1. Elongating 3′ UTR annotations in the chicken genome. (A, B) Genome browser visualizations of mapped reads, stemming from either bulk (top) or 10x Genomics single‐cell (bottom) RNA‐seq experiments, at the SOX9 (A) and GDF5 (B) locus. The respective gene models are displayed from the original GRCg6a annotation (galGal6), the annotation of Orgeur and colleagues77 and the annotation produced in this work. (C) Boxplots of 3′ UTR lengths of the original GRCg6a and our annotation. (D) Histogram showing the distribution of elongation lengths for all elongated 3′ UTRs. (E) Resulting changes in UMI counts, when mapping our HH31 sample to either the original GRCg6a or our modified annotation. For the large majority of genes, UMI counts either stay unchanged or show an increase with our annotation, with tissue‐specific limb marker genes highlighted (for color code, please refer to Figure 1E). We visually inspected the few genes that showed a reduced UMI count and realized that these UMIs had been attributed to another gene or transcript model in our second CellRanger run, that is, they were not lost due to misguided 3′ UTR elongations. (F) Overrepresented GO‐terms, for genes experiencing a 3′ UTR annotation‐based increase in UMI counts in our HH31 sample.
ACKNOWLEDGMENTS
The authors wish to thank Henrik Kaessmann and his lab for hosting CF for his EMBO STF, Virginie Ricci and Fabio Sacher for beta‐testing the “scWGCNA” R package, Lila Allou and Stefan Mundlos for making mouse scRNA‐seq data available prior to publication, Christian Beisel and the “Genomics Facility Basel” for help with single‐cell sequencing, and all members of our group for useful discussions. All calculations were performed at sciCORE (http://scicore.unibas.ch/), scientific computing center at the University of Basel. This work was supported by an EMBO short‐term fellowship to CF (Fellowship Number: 8593), and research funds from the Swiss 3R Competence Centre (3RCC grant OC‐2018‐005), the Swiss National Science Foundation (SNSF project grant 310030_189242), and the University of Basel to PT. Open Access Funding provided by Universitat Basel.
Feregrino C, Tschopp P. Assessing evolutionary and developmental transcriptome dynamics in homologous cell types. Developmental Dynamics. 2022;251(9):1472–1489. 10.1002/dvdy.384
Funding information EMBO Short Term Fellowship, Grant/Award Number: 8593; Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung, Grant/Award Number: 310030_189242; Swiss 3R Competence Centre (3RCC), Grant/Award Number: OC‐2018‐005
Contributor Information
Christian Feregrino, Email: christian.feregrino@gmx.net.
Patrick Tschopp, Email: patrick.tschopp@unibas.ch.
REFERENCES
- 1. Zuniga A. Next generation limb development and evolution: old questions, new perspectives. Development. 2015;142(22):3810‐3820. 10.1242/dev.125757. [DOI] [PubMed] [Google Scholar]
- 2. Petit F, Sears KE, Ahituv N. Limb development: a paradigm of gene regulation. Nat Rev Genet. 2017;18(4):245‐258. 10.1038/nrg.2016.167. [DOI] [PubMed] [Google Scholar]
- 3. Arendt D. The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet. 2008;9(11):868‐882. 10.1038/nrg2416. [DOI] [PubMed] [Google Scholar]
- 4. Arendt D, Musser JM, Baker CVH, et al. The origin and evolution of cell types. Nat Rev Genet. 2016;17(12):744‐757. 10.1038/nrg.2016.127. [DOI] [PubMed] [Google Scholar]
- 5. Wagner GP. Homology, Genes, and Evolutionary Innovation; Princeton, New Jersey, USA: Princeton University Press; 2014. 10.5860/choice.52-0829. [DOI] [Google Scholar]
- 6. Musser JM, Wagner GP. Character trees from transcriptome data: origin and individuation of morphological characters and the so‐called “species signal”. J Exp Zool B Mol Dev Evol. 2015;324(7):588‐604. 10.1002/jez.b.22636. [DOI] [PubMed] [Google Scholar]
- 7. Cannavò E, Koelling N, Harnett D, et al. Genetic variants regulating expression levels and isoform diversity during embryogenesis. Nature. 2017;541(7637):402‐406. 10.1038/nature20802. [DOI] [PubMed] [Google Scholar]
- 8. Liang C, Musser JM, Cloutier A, Prum RO, Wagner GP. Pervasive correlated evolution in gene expression shapes cell and tissue type transcriptomes. Genome Biol Evol. 2018;10(2):538‐552. 10.1093/gbe/evy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Groen SC, Ćalić I, Joly‐Lopez Z, et al. The strength and pattern of natural selection on gene expression in rice. Nature. 2020;578(7796):572‐576. 10.1038/s41586-020-1997-2. [DOI] [PubMed] [Google Scholar]
- 10. Wang Z‐Y, Leushkin E, Liechti A, et al. Transcriptome and translatome co‐evolution in mammals. Nature. 2020;588(7839):642‐647. 10.1038/s41586-020-2899-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tschopp P, Tabin CJ. Deep homology in the age of next‐generation sequencing. Philos Trans R Soc Lond B Biol Sci. 2017;372(1713):1‐8. 10.1098/rstb.2015.0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xia B, Yanai I. A periodic table of cell types. Development. 2019;146(12):1‐9. 10.1242/dev.169854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kelly NH, Huynh NPT, Guilak F. Single cell RNA‐sequencing reveals cellular heterogeneity and trajectories of lineage specification during murine embryonic limb development. Matrix Biol. 2020;89:1‐10. 10.1016/j.matbio.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allou L, Balzano S, Magg A, et al. Non‐coding deletions identify Maenli lncRNA as a limb‐specific En1 regulator. Nature. 2021;592(7852):93‐98. 10.1038/s41586-021-03208-9. [DOI] [PubMed] [Google Scholar]
- 15. Feregrino C, Sacher F, Parnas O, Tschopp P. A single‐cell transcriptomic atlas of the developing chicken limb. BMC Genomics. 2019;20(1):401. 10.1186/s12864-019-5802-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tschopp P, Sherratt E, Sanger TJ, et al. A relative shift in cloacal location repositions external genitalia in amniote evolution. Nature. 2014;516(7531):391‐394. 10.1038/nature13819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmid M, Smith J, Burt DW, et al. Third report on chicken genes and chromosomes 2015. Cytogenet Genome Res. 2015;145(2):78‐179. 10.1159/000430927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9(559):559. 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single‐cell data. Cell. 2019;177(7):1888‐1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kanton S, Boyle MJ, He Z, et al. Organoid single‐cell genomic atlas uncovers human‐specific features of brain development. Nature. 2019;574(7778):418‐422. 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- 21. Langfelder P, Horvath S. Tutorial for the WGCNA package for R : 1. Data input and cleaning. Tutorials for the WGCNA package; Published 2014. https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/Tutorials/FemaleLiver-01-dataInput.pdf. Accessed October 9, 2019.
- 22. van Noort V, Snel B, Huynen MA. The yeast coexpression network has a small‐world, scale‐free architecture and can be explained by a simple model. EMBO Rep. 2004;5(3):280‐284. 10.1038/sj.embor.7400090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Payne JL, Eppstein MJ. Evolutionary Dynamics on Scale‐Free Interaction Networks. IEEE Trans Evol Comput. 2009;13(4):895‐912. 10.1109/TEVC.2009.2019825. [DOI] [Google Scholar]
- 24. Broido AD, Clauset A. Scale‐free networks are rare. Nat Commun. 2019;10(1):1017. 10.1038/s41467-019-08746-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herrero J, Muffato M, Beal K, et al. Ensembl comparative genomics resources. Database (Oxford). 2016;2016:1‐17. 10.1093/database/bav096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yates AD, Achuthan P, Akanni W, et al. Ensembl 2020. Nucleic Acids Res. 2020;48(D1):D682‐D688. 10.1093/nar/gkz966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7(1):e1001057. 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grall E, Tschopp P. A sense of place, many times over ‐ pattern formation and evolution of repetitive morphological structures. Dev Dyn. 2020;249(3):313‐327. 10.1002/dvdy.131. [DOI] [PubMed] [Google Scholar]
- 29. Marioni JC, Arendt D. How single‐cell genomics is changing evolutionary and developmental biology. Annu Rev Cell Dev Biol. 2017;33(1):537‐553. 10.1146/annurev-cellbio-100616-060818. [DOI] [PubMed] [Google Scholar]
- 30. Shafer MER. Cross‐species analysis of single‐cell transcriptomic data. Front Cell Dev Biol. 2019;7:175. 10.3389/fcell.2019.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stuart T, Satija R. Integrative single‐cell analysis. Nat Rev Genet. 2019;20(5):257‐272. 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]
- 32. Wu YE, Pan L, Zuo Y, Li X, Hong W. Detecting activated cell populations using single‐cell RNA‐seq. Neuron. 2017;96(2):313‐329.e6. 10.1016/j.neuron.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 33. Tosches MA, Yamawaki TM, Naumann RK, Jacobi AA, Tushev G, Laurent G. Evolution of pallium, hippocampus, and cortical cell types revealed by single‐cell transcriptomics in reptiles. Science (80‐ ). 2018;360(6391):881‐888. 10.1126/science.aar4237. [DOI] [PubMed] [Google Scholar]
- 34. Korrapati S, Taukulis I, Olszewski R, et al. Single Cell and Single Nucleus RNA‐Seq Reveal Cellular Heterogeneity and Homeostatic Regulatory Networks in Adult Mouse Stria Vascularis. Front Mol Neurosci. 2019;12:1‐25. 10.3389/fnmol.2019.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Niu J, Huang Y, Liu X, et al. Single‐cell RNA‐seq reveals different subsets of non‐specific cytotoxic cells in teleost. Genomics. 2020;112(6):5170‐5179. 10.1016/j.ygeno.2020.09.031. [DOI] [PubMed] [Google Scholar]
- 36. Greenfest‐Allen E, Cartailler J‐P, Magnuson M, Stoeckert C. iterativeWGCNA: iterative refinement to improve module detection from WGCNA co‐expression networks. bioRxiv . 2017. doi: 10.1101/234062 [DOI]
- 37. Kee N, Volakakis N, Kirkeby A, et al. Single‐cell analysis reveals a close relationship between differentiating dopamine and subthalamic nucleus neuronal lineages. Cell Stem Cell. 2017;20(1):29‐40. 10.1016/j.stem.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 38. Pratapa A, Jalihal AP, Law JN, Bharadwaj A, Murali TM. Benchmarking algorithms for gene regulatory network inference from single‐cell transcriptomic data. Nat Methods. 2020;17(2):147‐154. 10.1038/s41592-019-0690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Balwierz PJ, Pachkov M, Arnold P, Gruber AJ, Zavolan M, van Nimwegen E. ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res. 2014;24(5):869‐884. 10.1101/gr.169508.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aibar S, González‐Blas CB, Moerman T, et al. SCENIC: single‐cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083‐1086. 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thornton JW, DeSalle R. Gene family evolution and homology: genomics meets phylogenetics. Annu Rev Genomics Hum Genet. 2000;1:41‐73. 10.1146/annurev.genom.1.1.41. [DOI] [PubMed] [Google Scholar]
- 42. Wolf YI, Koonin EV. A tight link between orthologs and bidirectional best hits in bacterial and archaeal genomes. Genome Biol Evol. 2012;4(12):1286‐1294. 10.1093/gbe/evs100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lazzaro BP, Clark AG. Rapid evolution of innate immune response genes. Rapidly Evolving Genes and Genetic Systems. Oxford University Press; 2012:203‐210. 10.1093/acprof:oso/9780199642274.003.0020. [DOI] [Google Scholar]
- 44. Ebert D, Fields PD. Host‐parasite co‐evolution and its genomic signature. Nat Rev Genet. 2020;21(12):754‐768. 10.1038/s41576-020-0269-1. [DOI] [PubMed] [Google Scholar]
- 45. Wu P, Hou L, Plikus M, et al. Evo‐Devo of amniote integuments and appendages. Int J Dev Biol. 2004;48(2‐3):249‐270. 10.1387/ijdb.041825pw. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vandebergh W, Bossuyt F. Radiation and functional diversification of alpha keratins during early vertebrate evolution. Mol Biol Evol. 2012;29(3):995‐1004. 10.1093/molbev/msr269. [DOI] [PubMed] [Google Scholar]
- 47. Eckhart L, Ehrlich F. Evolution of Trichocyte Keratins. Adv Exp Med Biol. 2018;1054:33‐45. 10.1007/978-981-10-8195-8_4. [DOI] [PubMed] [Google Scholar]
- 48. Núñez‐León D, Aguirre‐Fernández G, Steiner A, et al. Morphological diversity of integumentary traits in fowl domestication: Insights from disparity analysis and embryonic development. Dev Dyn. 2019;248(11):1044‐1058. 10.1002/dvdy.105. [DOI] [PubMed] [Google Scholar]
- 49. Briggs JA, Weinreb C, Wagner DE, et al. The dynamics of gene expression in vertebrate embryogenesis at single‐cell resolution. Science. 2018;360(6392):967‐968. 10.1126/science.aar5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mallarino R, Henegar C, Mirasierra M, et al. Developmental mechanisms of stripe patterns in rodents. Nature. 2016;539(7630):518‐523. 10.1038/nature20109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Haupaix N, Curantz C, Bailleul R, Beck S, Robic A, Manceau M. The periodic coloration in birds forms through a prepattern of somite origin. Science. 2018;361(6408):1202‐1203. 10.1126/science.aar4777. [DOI] [PubMed] [Google Scholar]
- 52. Busby L, Aceituno C, McQueen C, Rich CA, Ros MA, Towers M. Sonic hedgehog specifies flight feather positional information in avian wings. Development. 2020;147(9):1‐11. 10.1242/dev.188821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kiselev VY, Andrews TS, Hemberg M. Challenges in unsupervised clustering of single‐cell RNA‐seq data. Nat Rev Genet. 2019;20(5):273‐282. 10.1038/s41576-018-0088-9. [DOI] [PubMed] [Google Scholar]
- 54. Athanasiadis EI, Botthof JG, Andres H, Ferreira L, Lio P, Cvejic A. Single‐cell RNA‐sequencing uncovers transcriptional states and fate decisions in haematopoiesis. Nat Commun. 2017;8:1‐11. 10.1038/s41467-017-02305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Goldring MB, Tsuchimochi K, Ijiri K. The control of chondrogenesis. J Cell Biochem. 2006;97(1):33‐44. 10.1002/jcb.20652. [DOI] [PubMed] [Google Scholar]
- 56. Gómez‐Picos P, Eames BF. On the evolutionary relationship between chondrocytes and osteoblasts. Front Genet. 2015;6:297. 10.3389/fgene.2015.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kozhemyakina E, Lassar AB, Zelzer E. A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development. 2015;142(5):817‐831. 10.1242/dev.105536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morris SA. The evolving concept of cell identity in the single cell era. Development. 2019;146(12):1‐5. 10.1242/dev.169748. [DOI] [PubMed] [Google Scholar]
- 59. Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835‐849.e21. 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Grosswendt S, Kretzmer H, Smith ZD, et al. Epigenetic regulator function through mouse gastrulation. Nature. 2020;584(7819):102‐108. 10.1038/s41586-020-2552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wisdom AJ, Mowery YM, Hong CS, et al. Single cell analysis reveals distinct immune landscapes in transplant and primary sarcomas that determine response or resistance to immunotherapy. Nat Commun. 2020;11(1):6410. 10.1038/s41467-020-19917-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511‐515. 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7(1–2):203‐214. 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 64. Lawrence M, Huber W, Pagès H, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9(8):e1003118. 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lun ATL, McCarthy DJ, Marioni JC. A step‐by‐step workflow for low‐level analysis of single‐cell RNA‐seq data with Bioconductor. F1000Research. 2016;5:2122. 10.12688/f1000research.9501.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scialdone A, Natarajan KN, Saraiva LR, et al. Computational assignment of cell‐cycle stage from single‐cell transcriptome data. Methods. 2015;85:54‐61. 10.1016/j.ymeth.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 67. Shekhar K, Lapan SW, Whitney IE, et al. Comprehensive classification of retinal bipolar neurons by single‐cell transcriptomics. Cell. 2016;166(5):1308‐1323.e30. 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kobak D, Berens P. The art of using t‐SNE for single‐cell transcriptomics. Nat Commun. 2019;10(1):5416. 10.1038/s41467-019-13056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Linderman GC, Rachh M, Hoskins JG, Steinerberger S, Kluger Y. Fast interpolation‐based t‐SNE for improved visualization of single‐cell RNA‐seq data. Nat Methods. 2019;16(3):243‐245. 10.1038/s41592-018-0308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Finak G, McDavid A, Yajima M, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single‐cell RNA sequencing data. Genome Biol. 2015;16(1):278. 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Darnell DK, Kaur S, Stanislaw S, et al. GEISHA: an in situ hybridization gene expression resource for the chicken embryo. Cytogenet Genome Res. 2007;117(1‐4):30‐35. 10.1159/000103162. [DOI] [PubMed] [Google Scholar]
- 72. Smith CM, Hayamizu TF, Finger JH, et al. The mouse Gene Expression Database (GXD): 2019 update. Nucleic Acids Res. 2019;774‐779. 10.1093/nar/gky922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Butts CT. network : A package for managing relational data in R. J Stat Softw. 2008;24(2):1‐36. 10.18637/jss.v024.i02.18612375 [DOI] [Google Scholar]
- 75. Schloerke B, Briatte F, bigbeardesktop , et al. ggobi/ggally: v1.5.0. Published online 2020. doi: 10.5281/zenodo.3727162. [DOI]
- 76. Kinsella RJ, Kähäri A, Haider S, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford). 2011;2011:bar030. 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Orgeur M, Martens M, Börno ST, Timmermann B, Duprez D, Stricker S. A dual transcript‐discovery approach to improve the delimitation of gene features from RNA‐seq data in the chicken model. Biol Open. 2018;7(1):bio028498. 10.1242/bio.028498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Elongating 3′ UTR annotations in the chicken genome. (A, B) Genome browser visualizations of mapped reads, stemming from either bulk (top) or 10x Genomics single‐cell (bottom) RNA‐seq experiments, at the SOX9 (A) and GDF5 (B) locus. The respective gene models are displayed from the original GRCg6a annotation (galGal6), the annotation of Orgeur and colleagues77 and the annotation produced in this work. (C) Boxplots of 3′ UTR lengths of the original GRCg6a and our annotation. (D) Histogram showing the distribution of elongation lengths for all elongated 3′ UTRs. (E) Resulting changes in UMI counts, when mapping our HH31 sample to either the original GRCg6a or our modified annotation. For the large majority of genes, UMI counts either stay unchanged or show an increase with our annotation, with tissue‐specific limb marker genes highlighted (for color code, please refer to Figure 1E). We visually inspected the few genes that showed a reduced UMI count and realized that these UMIs had been attributed to another gene or transcript model in our second CellRanger run, that is, they were not lost due to misguided 3′ UTR elongations. (F) Overrepresented GO‐terms, for genes experiencing a 3′ UTR annotation‐based increase in UMI counts in our HH31 sample.


