Abstract
The expression of the maltose system in Escherichia coli is regulated at both transcriptional and translational levels by the pH of the growth medium (pHo). With glycerol as the carbon source, transcription of malT, encoding the transcriptional activator of the maltose regulon, is weaker in acidic medium than in alkaline medium. malT transcription became high, regardless of the pHo, when glycerol-3-phosphate or succinate was used as the carbon source. Conversely, malT expression was low, regardless of the pHo, when maltose was used as the carbon source. The increase in malT transcription, associated with the pHo, requires the presence of glycerol in the growth medium and the expression of the glycerol kinase (GlpK). Changes in the level of glpK transcription had a great effect on malT transcription. Indeed, a glpFKX promoter-down mutation has been isolated, and in the presence of this mutation, malT expression was increased. When glpK was expressed from a high-copy-number plasmid, the glpK-dependent reduced expression of the maltose system became effective regardless of the pHo. Analysis of this repression showed that a malTp1 malTp10 promoter, which is independent of the cyclic AMP (cAMP)-cAMP receptor protein (CRP) complex, was no longer repressed by glpFKX amplification. Thus, GlpK-dependent repression of the maltose system requires the cAMP-CRP complex. We propose that the pHo may affect a complex interplay between GlpK, the phosphotransferase-mediated uptake of glucose, and the adenylate cyclase.
In Escherichia coli, the level of expression of many genes is influenced by the pH of the growth medium (pHo) (11, 19). E. coli has a constitutive homeostatic mechanism which allows cells to maintain their internal pH between 7.4 and 7.8 over a pHo range of 5 to 8.5 (35). Within this pHo range, cells are able to sense a stimulus, triggered by the pHo, and transmit it to the target genes. Among these target genes are the maltose and the porin regulons (1, 12, 14).
ompF and ompC porin genes are controlled by the cognate sensor kinase EnvZ and the response regulator OmpR (22). A higher OmpR phosphate level at low pHo than at high pHo could be responsible for the pHo regulation of porin genes (14). In addition, the alternate phosphodonor acetyl phosphate may play a crucial role in the modulation of the OmpR-phosphate level and the subsequent pHo regulation of porin genes (14).
The maltose regulon of E. coli consists of genes encoding proteins involved in the uptake and metabolism of maltose and maltodextrins (4, 5). These genes are clustered in five transcriptional units controlled by the transcriptional activator MalT. malT expression is subjected to catabolite repression and then requires the presence of the cyclic AMP (cAMP)-cAMP receptor protein (CRP) complex. Recently, Mlc, a global negative regulator for the transcription of several genes whose products are involved in carbon utilization, has been found to be a repressor for malT (6). The expression of some operons and genes of the maltose regulon is also directly under the control of the cAMP-CRP complex. Lowering the pHo from 8 to 5 decreases malTp activity. This pHo effect relies on the cAMP-CRP binding (1). Indeed, with a cAMP-CRP-independent malT promoter, the pHo effect is not observed. malT pHo regulation triggers the pHo regulation of all the MalT-dependent promoters. In the absence of Mlc, the pHo regulation of malT is still effective (1). Recently, Eppler and Boos demonstrated that growth in tryptone broth containing glycerol reduced malT expression two- to threefold compared to tryptone broth without glycerol (10). To establish this repression, the enzyme IIAGlc, the cAMP-CRP complex, and the phosphorylation of glycerol to sn-glycerol-3-phosphate (G3P) by glycerol kinase (GlpK) are all necessary, but further metabolism to dihydroxyacetone by glycerol phosphate dehydrogenase is not (10).
The glycerol regulon is organized in multiple loci around the chromosome, and its expression is negatively controlled by GlpR (17). The glpTQ and glpACB operons located near 51 min encode the G3P permease/glycerophosphodiesterase and the subunits of the anaerobic G3P dehydrogenase, respectively (9). The glpD gene, encoding the aerobic G3P dehydrogenase, is located near 77 min and is transcribed divergently from the glpEGR operon (27). GlpE is a sulfur transferase (23). glpG encodes a protein of unknown function, and glpR encodes the repressor (28, 34). The glpFKX operon located near 88 min encodes the glycerol diffusion facilitator (GlpF), the glycerol kinase (GlpK), and a fructose 1,6-biphosphatase (GlpX) (7). This operon is subjected to multiple controls, including catabolite repression mediated by cAMP-CRP and repression by cooperative binding of GlpR to tandem operator sites which overlap the promoter. G3P, the product of the reaction catalyzed by glycerol kinase, is the inducer of the glp regulon (17). GlpK, as well as MalK, is involved in interactions with the unphosphorylated form of enzyme IIAGlc, which is an intermediate in the phosphorylation cascade of the phosphotransferase (PTS)-mediated uptake and concomitant phosphorylation of glucose. This interaction inhibits the metabolism of non-PTS-carbohydrates, such as glycerol and maltose, by preventing the induction of their respective catabolic operons (21). This process is known as inducer exclusion (15). Rohwer et al. have shown that a high level of glycerol kinase could result in IIAGlc sequestration into an inactive complex (25). The phosphorylated form of IIAGlc is considered to be involved in the activation of adenylate cyclase, and this leads to increased intracellular levels of cAMP, which binds to CRP and elaborates the cAMP-CRP complex (24).
As the involvement of the cAMP-CRP complex in the glycerol-dependent repression of malT transcription in rich medium (10) and in the pHo regulation of malT transcription during growth with glycerol as a carbon source (1) has been clearly established, we were interested in studying the connections between the glycerol effect and the pHo regulation of maltose regulon. This study shows that the phosphoenolpyruvate:carbohydrate PTS system and adenylate cyclase are involved in the complex molecular interplay between the different regulatory mechanisms that regulate gene expression according to the pHo.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are listed in Table 1. Standard media were used in this study, including Luria-Bertani (LB) broth (18), MacConkey medium with lactose (final concentration, 2% [wt/vol]) added (Difco Laboratories, Detroit, Mich.), and M63 minimal medium (18) supplemented with 3 μM thiamine hydrochloride, the appropriate amino acids, and sugars as the carbon source. Buffered minimal medium (MM) was as previously described (13). It was adjusted with 1 N NaOH to pH 4.75, 5, 5.25, 5.5, 5.75, 6.5, 7, 7.5, and 8. MM-MES is a solid MM containing 100 mM MES [2-(N-morpholino)ethane sulfonic acid; pKa, 6.1], instead of 50 mM in MM and no TAPS [tris(hydroxymethyl)methylaminopropane sulfonic acid; pKa, 8.4]. This buffered medium was adjusted to pH 5 with 1 N NaOH. MM and MM-MES were supplemented with 3 μM thiamine hydrochloride. Carbon sources used in these media were 44 mM glycerol, 12 mM G3P, 21 mM succinate, 22 mM glucose, and 58 mM maltose. When required, 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside) ml−1 or the following antibiotics were added: 30 μg (pJEL derivatives) or 100 μg of ampicillin ml−1, 50 μg of kanamycin ml−1, 15 μg of tetracycline ml−1, and 20 μg of chloramphenicol ml−1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotypea | Source or reference |
|---|---|---|
| Strains | ||
| CGSC5023 | fhuA22 relAl T2R | B. Bachmann, Yale University |
| DH5α | F−endAl (rK− mK+) supE44 thi-l hsdR17 gyrA (Nalr) recAl Δ(lacZYA-argF)U169 deoR relAl [φ80Δlac(lacZ)M15] | BRL, Inc |
| GPH1768 | MC4100/pGPH1768 | 1 |
| GPH8549 | MC4100 Δcya zie296::Tn10 crp∗ | 1 |
| GPH8804 | MC4100 lac+ | Lac+ with PI/CGSC5023 |
| GPH8805 | MC4100 φ(ompA-lacZ) (Hyb) | 1 |
| GPH8818 | MC4100 malTp1 malT p10/pGPH1768 | 1 |
| GPH8840 | MC4100 iea18 (glpFp18)/pGPH1768 | This work |
| GPH8881 | MC4100/pGPH8881 | 1 |
| GPH9244 | Cured GPH8840 | This work |
| GPH9401 | GPH9244(iea18)/pGPH8881 | This work |
| GPH9463 | MC4100/pGPH9463 | 1 |
| GPH9473 | GPH9244(iea18)/pGPH9463 | This work |
| GPH9604 | MC4100 Φ(ompF′-lacZ+)7.14 | Lac+ with Pl on MA2946 |
| GPH9605 | GPH9244(iea18) Φ(ompF′-lacZ+)7.14 | Lac+ with Pl on MA2946 |
| GPH9909 | GPH9244(iea18) Φ(ompC′-lacZ+)10.21 | Lac+ with Pl on MA2948 |
| GPH9910 | MC4100 Φ(ompC′-lacZ+)10.21 | Lac+ with Pl on MA2948 |
| GPH11296 | MC4100 anλ::(malK′-lacZ+)1 | This laboratory |
| JC2296 | HFr P4X thi metB1 relA1 spoT1 Δ(lac)U169 | This laboratory |
| MA2946 | JC2296 Φ(ompF′-lacZ+)7.14 | 12 |
| MA2948 | JC2296 Φ(ompC′-lacZ+)10.21 | 12 |
| MC4100 | F−araD139 Δ(argF-lac)205 λ−fthD5301 fruA25 relA1 rpsL150 rbsR22 deoC1 | M. Casadaban, University of Chicago |
| Plasmids | ||
| pACT3 | tac promoter expression vector, 5.3 kb, Camr (20 μg ml−1) | 8 |
| pACYC177 | Cloning vector, 3.9 kb, Ampr (50 μg ml−1) Kanr (50 μg ml−1) | 26 |
| pBR322 | Cloning vector, 4.4 kb, Ampr (50 μg ml−1) Tetr (15 μg ml−1) | 3 |
| pCJ102 | glpK constitutively expressed in pBR322 | 20 |
| pGPH1768 | Φ(malK′-lacZ+)1 in pJEL250 | 1 |
| pGPH8881 | Φ(malT′-lacZ+)1 in pJEL250 | 1 |
| pGPH9260 | Φ(malTpl malTp10′-lacZ+)1 in pJEL250 | 1 |
| pGPH9463 | Φ(malP′-lacZ+)1 in pJEL250 | 1 |
| pGPH9925 | 4,374-bp PCR product from MC4100 containing glpFKX cloned into HincII-cut pACYC177 | This work |
| pGPH9928 | 4,374-bp PCR product from GPH8840 containing glpFKXp18 cloned into HincII-cut pACYC177 | This work |
| pGPH9993 | glpF+ (2,707-bp BstEII deletion of pGPH11250) | This work |
| pGPH11247 | Φ(glpF′-lacZ+)1 in pJEL250 | This work |
| pGPH11248 | Φ(glpFp18′-lacZ+)1 in pJEL250 | This work |
| pGPH11249 | glpX+ (1,033-bp EcoRI/EcoRV deletion of pGPH9925) | This work |
| pGPH11250 | glpF+ glpK+ (805-bp AatII deletion of pGPH9925) | This work |
| pGPH11264 | glpK+ (505-bp EcoRI/RsrII deletion of pGPH11250) | This work |
| pGPH11542 | glpFKX cloned into SmaI/SacI-cut pACT3 | This work |
| pJEL250 | Temperature-sensitive copy number; lacZYA operon fusion vector, 13 kb, Ampr (30 μg ml−1) | 29 |
| pUC18 | Cloning vector, 2.7 kb, Ampr (50 μg ml−1) | 33 |
Genetic nomenclature is from the work of Berlyn (2). Kanr, Camr, Ampr, and Tetr, resistance to kanamycin, chloramphenicol, ampicillin, and tetracycline, respectively.
Growth conditions.
Overnight subcultures, grown at 30°C in LB broth supplemented with the appropriate antibiotics, were used to inoculate MM adjusted to different pHo values and supplemented with various carbon sources and antibiotics. These cultures were incubated at 30°C until the stationary phase was reached. At this point, the different buffered cultures were diluted with the same fresh medium to an optical density at 600 nm of 0.02 (path length, 1 cm; Jouan spectrophotometer). These new cultures were incubated at 30°C up to an optical density at 600 nm between 0.2 and 0.3. Up to this cell density, the pHo remains constant.
Enzyme assays.
β-Galactosidase activities associated with operon or gene fusions were measured on toluenized cells, as previously described (12). One unit of β-galactosidase activity was defined as the amount of enzyme that hydrolyzed 1 nmol of substrate min−1. Results from at least three independent experiments were averaged to obtain the values presented below.
PCR amplification.
PCRs were performed using standard conditions (16) and Pfu enzyme (Promega Corp, Madison, Wis.). Primers were phosphorylated by the T4 polynucleotide kinase prior to amplification. The amplified fragments were separated by agarose gel electrophoresis, extracted (QiaEx gel purification kit; Qiagen), and ligated into the appropriate vector. The ligated DNA was transformed into E. coli DH5α, and antibiotic-resistant clones were isolated by growth at 37°C in LB broth containing the appropriate antibiotics.
In vitro plasmid construction.
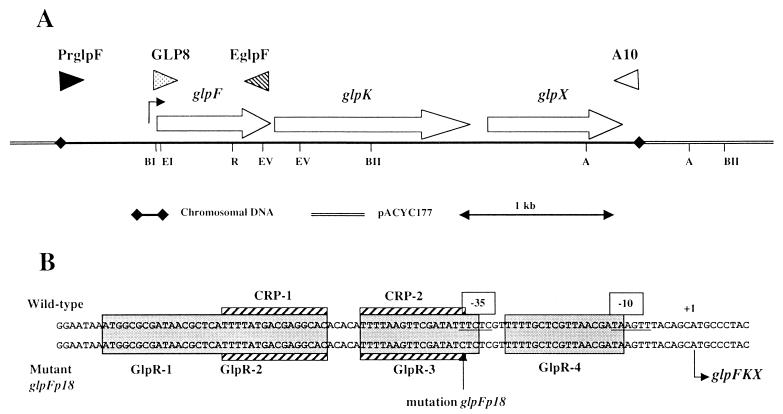
Fragments of 4,374 bp, including the glp promoter, were amplified from MC4100 and GPH8840 chromosomes by PCR (Fig. 1A). Primers used to amplify this fragment hybridized to a region 615 bases upstream of the transcriptional start point (+1) of glpFKX (PrGlpF, 5′-AGATGAAGCGTAATCAGACC-3′) and to a region 152 bases downstream of the glpX last codon (A10, 5′-TGAACGGTGAAGACTAAACAG-3′). The 4,374-bp fragments were ligated into HincII-cut and dephosphorylated pACYC177. Kanamycin-resistant clones were isolated. pGPH9925 corresponds to the PCR product from MC4100, and pGPH9928 corresponds to that from GPH8840.
FIG. 1.
Schematic representation of the glpFKX operon cloned in pACYC177 and sequence of the parental and mutant glp promoter regions. (A) Open reading frames within the glpFKX operon, with the direction of transcription, are indicated by open arrows. Restriction sites: EI, EcoRI; BI, BamHI; R, RsrII; EV, EcoRV; BII, BstEII; A, AatII. The glp transcriptional start site is indicated with an arrow upstream of glpF (32). The primers used in PCR amplification are indicated with triangles. (B) Grey boxes indicate the binding site of the GlpR repressor. Striped boxes indicate the cAMP-CRP binding site. The transcriptional start point is at position +1 (32). Putative −10 and −35 sequences are underlined.
The glp promoter region was amplified from MC4100 and GPH8840 chromosomes by PCR. The primers used were PrGlpF and EGlpF (5′-GTAGTCATATTACAGCGAAGCTT-3′) (Fig. 1A), which hybridized to a region encompassing the last codon of glpF. These primers gave 1,577-bp products which were digested by BamHI. The 718-bp promoter fragments were separated by agarose gel electrophoresis, extracted, and ligated into SmaI- and BamHI-digested pUC18. The 723-bp promoter fragments were excised from pUC18 with EcoRI and BamHI and ligated into EcoRI- and BamHI-digested pJEL250 to create pGPH11247, with the parental glpFp operon fusion, and pGPH11248, with the mutant glpFp18 operon fusion.
A 3,751-bp fragment, including the glp operon without its promoter, was amplified from the MC4100 chromosome by PCR. Primer A10 was used in combination with primer GLP8 (5′-CATCGTGGAGCTCCGTGACTTTC-3′) (Fig. 1A). This primer hybridized to a region extending from +22 to +44 relative to the transcriptional start point. At position +32, a C was introduced instead of a G in order to create a SacI site for subcloning. The 3,751-bp fragment was SacI digested, and a fragment containing a promoterless glp operon, but with an intact Shine-Dalgarno site, was obtained. The 3,741-bp fragment was purified and ligated into SmaI- and SacI-digested pACT3 to create pGPH11542.
In order to test the effect of each glpFKX gene, plasmid constructs from pGPH9925 were generated (Fig. 1A). pGPH9925 was digested with EcoRI and EcoRV, end filled with a Klenow fragment, and self-ligated to create pGPH11249, which expresses only glpX. pGPH9925 was digested with AatII and self-ligated to create pGPH11250, which keeps only glpF and glpK intact. pGPH11250 was digested with EcoRI and RsrII, end filled with a Klenow fragment, and self-ligated to create pGPH11264, which contains only glpK. pGPH11250 was digested with BstEII and self-ligated to create pGPH9993, which contains glpF and part of glpK.
Nucleotide sequencing.
Sequencing was carried out by Genome Express (Grenoble, France) using fluorescent dye terminator technology and then analyzed on a Applied Biosystems 373 automated sequencer.
In vivo genetic methods.
Exponentially growing MC4100 cells were harvested by centrifugation, washed twice, and resuspended in 0.1 M sodium citrate (pH 5.5). N-Methyl-N′-nitrosoguanidine was added to a final concentration of 40 μg ml−1. After 12 min at 37°C without shaking, cells were centrifuged and resuspended in 100 mM phosphate buffer (pH 7.0). This suspension was used to inoculate five independent expression cultures that were incubated at 37°C for at least 20 h. From each culture, electrocompetent cells were prepared and transformed with the pGPH1768 plasmid [(malK′-lacZ+)1 operon fusion]. After 1 h at 30°C, cells were centrifuged and resuspended in M63 medium. Appropriate dilutions were spread on MM-MES (pH 5.0) plates containing ampicillin, glycerol, and X-Gal. After 3 days at 30°C, blue colonies were picked and restreaked twice on the same medium.
Generalized transductions with P1vir were performed as described by Miller (18). Hfr mapping was performed using a set of Hfr Tn10 strains (31) carrying the F plasmid at various sites scattered around the chromosome.
Curing strain from pGPH1768.
An overnight GPH8840 subculture, grown in LB broth at 30°C, was used to inoculate fresh LB broth (103-fold dilution). These cultures were incubated at 30°C until the stationary phase was reached. Appropriate dilutions were spread on MacConkey medium with lactose added. Cultures were incubated at 30°C, and white colonies among the pink ones were picked and restreaked. The analysis showed that the white colonies did not harbor any plasmid. Strain GPH9244 was conserved for further study.
RESULTS
malT and malK pHo regulation without glpFKX induction.
The pHo regulation of the maltose regulon with glycerol as the carbon source was described (1). Because of the link between the mal and glp regulons, malT transcription and that of malK, according to the pHo, were compared with and without glpFKX expression. The glpFKX operon is not induced when succinate, pyruvate, lactate, ribose, or maltose is used as the carbon source, and glycerol and G3P are the carbon sources known to induce glpFKX expression.
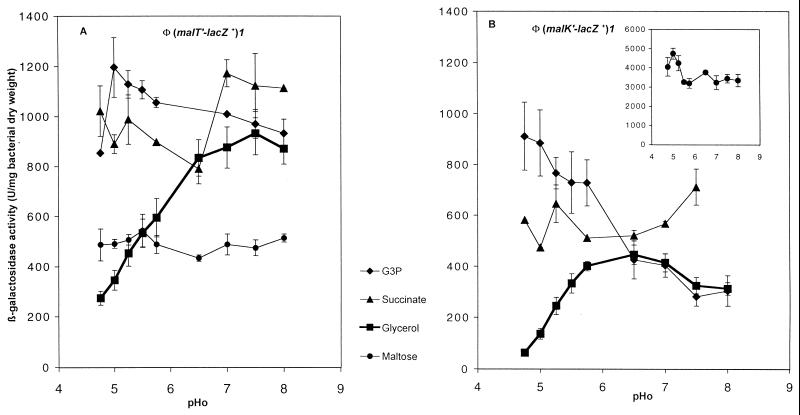
When glpFKX was induced in the presence of glycerol (Fig. 2), malT and malK expressions were repressed at low pHo and reached higher levels as the pHo was increased. With G3P as the carbon source, malT expression was high regardless of the pHo (Fig. 2A), and malK expression was quite high at low pHo and decreased to the level observed with glycerol as the carbon source as the pHo became higher than 6.5 (Fig. 2B).
FIG. 2.
pHo regulation of malT and malK during growth with different carbon sources. β-Galactosidase activity was assayed during growth at 30°C in MM adjusted to different pH values and supplemented with ampicillin and the indicated carbon source. (A) (malT′-lacZ+)1 operon fusion on pJEL250 (GPH8881); (B) (malK′-lacZ+)1 operon fusion on pJEL250 (GPH1768). (Inset) Induced malK expression with maltose as the carbon source. Error bars show standard deviations. The absence of error bars indicates that the deviation fell below the resolution limit of the graphing program.
Without glpFKX induction, in the presence of succinate (Fig. 2), pyruvate, lactate, or ribose (data not shown), malT and malK expression was not repressed during growth in acidic medium. With maltose as the carbon source, malT expression was low regardless of the pHo. Thus, with these carbon sources, the maltose regulon was not pHo regulated. We hypothesize that the synthesis of GlpF, GlpK, or GlpX may induce the repression of the maltose regulon during growth in acidic medium.
The different results obtained with G3P and glycerol demonstrated that the repression of the maltose regulon by a Glp protein may be linked to the entry of glycerol via GlpF or to glycerol phosphorylation by GlpK. Further metabolism of glycerol, beyond G3P, would not be involved in maltose regulon repression during growth at low pHo.
Isolation and characterization of a mutant altered in the glpFKX promoter.
In a mutant search aiming to identify genes whose products are involved in the putative pHo transduction pathway(s), we looked for mutants altered in the pHo regulation of the malK gene. MC4100 cells were treated with N-methyl-N′-nitrosoguanidine and then transformed with plasmid pGPH1768 carrying the (malK′-lacZ+)1 operon fusion. On buffered MM, adjusted to pH 5 and supplemented with X-Gal and glycerol as the carbon source, the basal (malK′-lacZ+)1 expression was too low for staining in blue bacterial colonies. Uninduced expression of the maltose regulon is used for expression in MM with glycerol as the carbon source. Mutants which displayed high expression of the fusion during growth at pHo 5 and in the absence of maltose were visualized as blue colonies. Seventeen independent mutants with an increased expression of malK in acidic medium (iea) were isolated. The iea18 mutation is described in this paper.
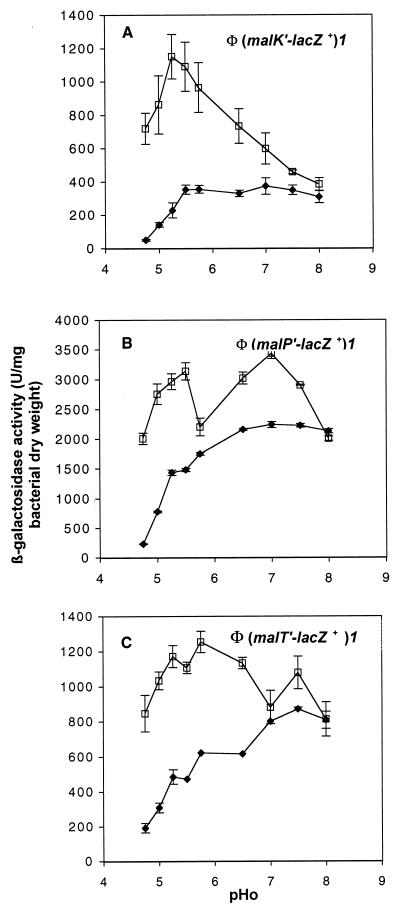
In the parental background of strain MC4100, (malK′-lacZ+)1 expression (Fig. 3A) was very low at a pHo of 4.75 and increased as the pHo reached 5.5. In the iea18 strain, uninduced (malK′-lacZ+)1 expression was high at low pHo and decreased to the level observed in the parental strain at high pHo (Fig. 3A). To study the effect of iea18 on malP and malT transcription, strain GPH8840 was cured from pGPH1768 and transformed with pGPH9463 (strain GPH9473) or pGPH8881 (strain GPH9401), harboring, respectively, the (malP′-lacZ+)1 or the (malT′-lacZ+)1 operon fusion. Data from Fig. 3B show that malP promoter activity was high whatever the pHo in the iea18 background and in the presence of glycerol plus maltose in the growth medium. The induced level of malP promoter activity was measured because its uninduced expression is too low to be detected. In the iea18 background with glycerol as the carbon source, malT expression was as high, regardless of the pHo (Fig. 3C), as those observed in the presence of G3P as the carbon source (compare Fig. 3C and 2A).
FIG. 3.
Influence of the iea18 mutation on malK, malP, and malT pHo regulation. β-Galactosidase activity was assayed during growth at 30°C in MM adjusted to different pHs and supplemented with ampicillin and glycerol (A and C) or glycerol and maltose (B). ⧫, parental strain; □, iea18 strain. (A) (malK′-lacZ+)1 operon fusion on pJEL250 (GPH1768 and GPH8840); (B) (malP′-lacZ+)1 operon fusion on pJEL250 (GPH9463 and GPH9473); (C) (malT′-lacZ+)1 operon fusion on pJEL250 (GPH8881 and GPH9401). Error bars are as in Fig. 2.
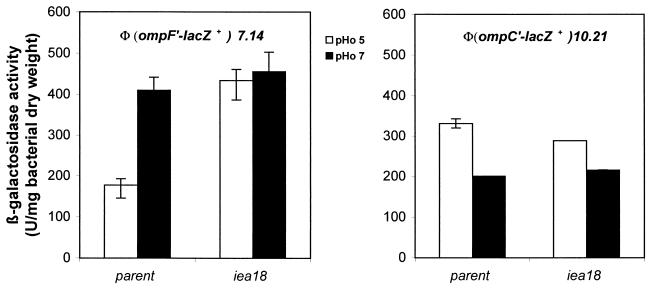
The effect of the iea18 mutation on the transcription of ompF and ompC porin genes was measured (Fig. 4). In the parental background, and with glycerol as the carbon source, ompF expression was higher in neutral than in acidic medium (Fig. 4A) whereas ompC expression was higher at low pHo than at pHo 7 (Fig. 4B). In the presence of the iea18 mutation, ompF transcription was not further decreased in acidic medium, but there was no evidence of an effect on ompC expression.
FIG. 4.
ompF and ompC expression at pHo 5 and 7 in parental and iea18 strains. Strains were grown at 30°C in MM adjusted to pH 5 or 7 and supplemented with glycerol as the carbon source. (A) GPH9604 parental strain and GPH9605 iea18 strain; (B) GPH9910 parental strain and GPH9909 iea18 strain. Error bars are as in Fig. 2.
The iea18 mutation was associated with slower growth with glycerol as the carbon source. Hfr and P1 transduction mapping (31) showed that the mutation iea18 was linked to a Tn10 marker inserted close to metF (min 89).
To determine if the iea18 mutation altered the glpFKX operon (min 88.7), chromosomal DNA from strains MC4100 and GPH8840 was isolated and used as a template in PCR amplifications with the primer pair PrGlpF-A10 (Fig. 1A) to amplify the glpFKX operon and its 5′ upstream region. Wild-type and mutant fragments of 4,374 bp were sequenced, and comparison of the sequences enabled us to identify a single mutation located 34 bp upstream of the transcription start site (Fig. 1B). The iea18 mutation was renamed glpFp18. Such a mutation, located in the −35 region of the promoter and in the GlpR-3 binding site of GlpR and close to the CRP2 binding site of cAMP-CRP (Fig. 1B), could potentially yield an altered level of glpFKX operon transcription.
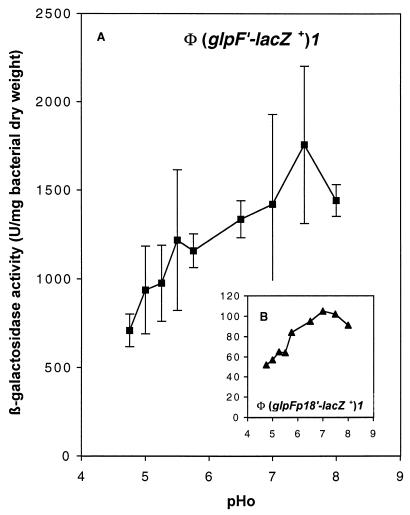
To test this hypothesis, transcriptional fusions of lacZ as the reporter gene and the wild-type glpFp or mutant glpFp18 promoter were constructed. Fragments of 718 bp, from PrGlpF to the BamHI site in glpF (Fig. 1A), were cloned in pJEL250 to yield a wild-type (glpF′-lacZ+)1 and a mutant (glpFp18′-lacZ+)1 operon fusion (pGPH11247 and pGPH11248, respectively). During growth in MM with glycerol as the carbon source, the glpFp18 mutation reduced, regardless of the pHo, the β-galactosidase activity of the corresponding fusion by a factor of 10 to 12, compared to the wild-type promoter (Fig. 5). glpFp18 is a promoter-down mutation of the glpFKX operon. Our results also indicate that growth at neutral or higher pHo, with glycerol as the carbon source, stimulated the transcription initiated at both glpFp (Fig. 5A) and glpFp18 (Fig. 5B). Thus, the glpFKX operon belongs to the pH stimulon.
FIG. 5.
Parental and mutant glp promoter activities according to the pHo. β-Galactosidase activity was assayed during growth at 30°C in MM adjusted to different pHs and supplemented with glycerol and ampicillin. (A) (glpF′-lacZ+)1 operon fusion on pJEL250 (pGPH11247 in MC4100); (B) (glpFp18′-lacZ+)1 operon fusion on pJEL250 (pGPH11248 in MC4100). Error bars are as in Fig. 2.
These results show that the glpFp18 mutation leading to a decrease in glpFKX expression is responsible for an increase in ompF, malT, and MalT-dependent gene expression during growth in acidic medium. These results confirm that at least one of the proteins encoded by the glpFKX operon and expressed at the wild-type level could contribute to the repression, directly or indirectly, of mal and ompF gene expression during growth at low pHo.
The amplification of glpK in the cell decreases mal gene expression.
To further analyze the involvement of proteins encoded by glpFKX in the repression of the maltose regulon, the glpFKX copy number was increased in the wild-type strain by introducing pGPH9925. The β-galactosidase activities of strains harboring either the (malK′-lacZ+)1, the (malT′-lacZ+)1, or the (malP′-lacZ+)1 operon fusion, in the presence of multiple copies of glpFKX, are shown in Table 2. At pHo 5 and 7, as GlpF, GlpK, and GlpX were overexpressed (pGHP9925), malK was fully repressed and malT and malP expression was reduced by factors of 3.1 and 2.1.
TABLE 2.
Repression of the maltose regulon when glp genes are amplified
| Fusion and introduced plasmida | β-Galactosidase activityb
|
Repression by glpc
|
||
|---|---|---|---|---|
| pHo 5 | pHo 7 | pHo 5 | pHo 7 | |
| Φ(malK′-lacZ+)1 (pGPH1768) | ||||
| pACYC177 | 189 ± 11 | 339 ± 51 | ||
| pGPH9925 (glpFKX) | VL | VL | Full | Full |
| pGPH11250 (glpFK) | VL | VL | Full | Full |
| pGPH9993 (glpF) | 136 ± 18 | 328 ± 29 | 1.4 | 1 |
| pGPH11264 (glpK) | VL | 90 ± 32 | Full | 3.8 |
| pGPH11249 (glpX) | 153 ± 40 | 295 ± 74 | 1.2 | 1.1 |
| Φ(malT′-lacZ+)1 (pGPH8881) | ||||
| pACYC177 | 417 ± 86 | 593 ± 41 | ||
| pGPH9925 (glpFKX) | 162 ± 35 | 194 ± 25 | 2.6 | 3.1 |
| pGPH11250 (glpFK) | 150 ± 21 | 244 ± 58 | 2.8 | 2.4 |
| pGPH9993 (glpF) | 328 ± 7 | 657 ± 53 | 1.3 | 1.1 |
| pGPH11264 (glpK) | 128 ± 37 | 232 ± 34 | 3.3 | 2.6 |
| pGPH11249 (glpX) | 395 ± 58 | 656 ± 93 | 1.1 | 0.9 |
| Φ(malP′-lacZ+)1 (pGPH9463) | ||||
| pACYC177 | 1,261 ± 21 | 2,321 ± 163 | ||
| pGPH9925 (glpFKX) | ND | 1,116 ± 26 | 2.1 | |
Fusions were in an MC4100 background in pJEL250.
β-Galactosidase activity was assayed during growth at 30°C in MM adjusted to pH 5 or 7 and supplemented with glycerol [and maltose for Φ(malP′-lacZ+)1], ampicillin, and kanamycin. ND, not done; VL, very low β-galactosidase activity (below the sensitivity threshold of the method used).
Repression values correspond to the ratio between β-galactosidase activities of the strain with pACYC177 and the strain with amplified glp genes.
In order to determine which gene of the glpFKX operon has to be amplified to exert the repressional effect on the maltose regulon, internal deletions have been carried out in the glpFKX operon with the glpFKX promoter kept intact (Table 2). Whatever the pHo, glpF (pGPH9993) or glpX (pGPH11249) amplification did not reduce maltose gene expression. The repression of maltose genes was still established in the presence of GlpF and GlpK when the glpX gene was deleted. The amplification of glpK alone triggers malK and malT repression. Thus, GlpK is the protein playing the key role in the repression observed, even if the strongest repression was observed when the full glpFKX operon was amplified (full repression compared to 3.8-fold).
To investigate whether the repression was linked to GlpK itself or to its kinase activity on glycerol, a promoterless glpFKX operon was cloned under the IPTG (isopropyl-β-d-galactopyranoside)-inducible tac promoter of pACT3, resulting in pGPH11542. This construct allows glpFKX expression without the presence of glycerol in the growth medium. It was introduced into a strain with the (malK′-lacZ+)1 operon fusion on the chromosome at attλ. Table 3 shows β-galactosidase production by the strain carrying the (malK′-lacZ+)1 operon fusion with different carbon sources in the presence of IPTG. Without IPTG added to the growth medium, malK expression was not affected by the presence of pACT3 or pGPH11542 (data not shown). In the presence of IPTG, whatever the carbon source (with the exception of glucose), repression of malK was observed when glpFKX was present (5-fold with glycerol, 2.4-fold with maltose, 5.1-fold with G3P, 18.4-fold with ribose, 5.9-fold with succinate, and 2.1-fold with mannose). These data demonstrate that the maltose regulon repression by GlpK was not specific to growth with glycerol as the carbon source. Thus, neither phosphorylation of exogenous glycerol by GlpK nor glycerol metabolism would be required for repression.
TABLE 3.
Repression of malK when the glycerol regulon is controlled by an IPTG-inducible tac promoter
| Carbon source | β-Galactosidase activitya with introduced plasmid
|
Repression by glpb | |
|---|---|---|---|
| pACT3 | pGPH11542 (glpFKX) | ||
| Glycerol | 120 ± 13 | 24 ± 7 | 5 |
| Maltose | 2,870 ± 31 | 1,177 ± 231 | 2.4 |
| G3P | 92 ± 33 | 18 ± 7 | 5.1 |
| Ribose | 661 ± 47 | 36 ± 3 | 18.4 |
| Succinate | 177 ± 54 | 30 ± 7 | 5.9 |
| Mannose | 237 ± 18 | 112 ± 6 | 2.1 |
| Glucose | 27 ± 7 | 55 ± 10 | 0.5 |
β-Galactosidase activity of a strain with Φ(malK′-lacZ+)1 in a GPH11296 background was assayed during growth at 30°C in MM adjusted to pH 7 and supplemented with the indicated carbon source (0.4%) and chloramphenicol. IPTG (5 or 10 μg/ml) was added to the medium.
Repression values correspond to the ratio between β-galactosidase activities of the strain with pACT3 and the strain with pGPH11542.
Repression of the maltose regulon required the cAMP-CRP complex.
The involvement of the cAMP-CRP complex in the repression of the maltose regulon by a high level of GlpK was characterized using strains carrying both malTp1 and malTp10 mutations in the control region of malT. These mutations render malT expression cAMP-CRP independent. Table 4 shows the effect of glpFKX amplification on a (malTp1 malTp10′-lacZ+)1 operon fusion and on a (malK′-lacZ+)1 operon fusion with the malTp1 malTp10 mutations introduced into the control region of the chromosomal malT gene. The repression became residual on malK (1.7-fold instead of full repression) and was cancelled on malT (0.6- instead of 3-fold). The repression observed on malK would be a consequence of the repression exerted on malT expression. These data demonstrate that binding of cAMP-CRP to the malT promoter was required for mal genes to be repressed by glpFKX amplification.
TABLE 4.
Genetic analysis of malT and malK repression by glpFKX amplification
| Fusiona tested and genetic background | Plasmid introduced | β-Galactosidase activityb | Repression by glpc |
|---|---|---|---|
| Φ(malK′ -lacZ+)1 | |||
| MC4100 | pACYC177 | 339 ± 51 | |
| pGPH9925 (glpFKX) | VL | Full | |
| GPH8818 (malTp1 malTp10) | pACYC177 | 377 ± 77 | |
| pGPH9925 (glpFKX) | 220 ± 57 | 1.7 | |
| GPH8549 (cya crp∗) | pACYC177 | 122 ± 10 | |
| pGPH9925 (glpFKX) | 60 ± 11 | 2 | |
| Φ(malT′-lacZ+)1 | |||
| MC4100 | pACYC177 | 593 ± 41 | |
| pGPH9925 (glpFKX) | 194 ± 25 | 3 | |
| GPH8549 (cya crp∗) | pACYC177 | 404 ± 95 | |
| pGPH9925 (glpFKX) | 345 ± 71 | 1.2 | |
| Φ(malTpl malTp10′- lacZ+)1, MC4100 | pACYC177 | 720 ± 137 | |
| pGPH9925 (glpFKX) | 1,145 ± 250 | 0.6 |
The indicated operon fusion on pJEL250.
β-Galactosidase activity was assayed during growth at 30°C in MM adjusted to pH 7 and supplemented with glycerol, ampicillin, and kanamycin. VL, very low β-galactosidase activity (below the sensitivity threshold of the method used).
Repression values correspond to the ratio between β-galactosidase activities of the indicated fusion in the presence of pGPH9925 and in the presence of pACYC177.
The use of Δcya crp* derivatives allows the expression of cAMP-CRP-dependent genes independently of the amount of cAMP present in the cell. A crp* mutation affects the coding sequence of crp and enables the Crp* protein to activate cAMP-CRP-dependent promoters, even in the absence of cAMP. In the Δcya crp* background, malK expression was 2-fold repressed instead of fully repressed and malT expression became 1.2-fold repressed by glpFKX instead of 3-fold (Table 4). The data show that the repression exerted by glpFKX amplification is cancelled when malT expression is no longer dependent on the amount of cAMP.
Is the repression exerted by glpFKX amplification restricted to mal genes?
We were interested in establishing whether the repression observed with the amplification of glpFKX operates for all pHo-regulated genes. Table 5 shows that glpFKX amplification lowered ompF transcription twofold while ompC expression was not significantly modified. These results agree with an increased ompF expression in acidic medium in the presence of the glpFp18 mutation (Fig. 4A). ompA expression, which was previously reported to be pHo independent, was not repressed by glpK amplification (Table 5). However, lactose operon expression, which is also known to be pHo independent, was repressed by a factor of 1.8 when glpFKX was amplified. Thus, the repression exerted by glpFKX amplification is neither specific to pHo-regulated genes (lac operon) nor effective on all pHo-regulated loci (ompC gene).
TABLE 5.
Effect of glpFKX amplification on ompF, ompC, ompA, and lacZ expression
| Fusion or gene (in MC4100) tested | Plasmid introduced | β-Galactosidase activitya | Repression by glpb |
|---|---|---|---|
| Φ(ompF′-lacZ+)7.14 | pACYC177 | 372 ± 82 | |
| pGPH9925 (glpFKX) | 191 ± 20 | 2 | |
| Φ(ompC′-lacZ+)10.21 | pACYC177 | 160 ± 42 | |
| pGPH9925 (glpFKX) | 209 ± 30 | 0.8 | |
| Φ(ompA′-lacZ+) (Hyb) | pACYC177 | 81 ± 4 | |
| pCJ102 (glpK) | 76 ± 18 | 1 | |
| lacZ | pACYC177 | 4,520 ± 400c | |
| pGPH9925 (glpFKX) | 2,497 ± 71c | 1.8 |
β-Galactosidase activity was assayed during growth at 30°C in MM adjusted to pH 7 and supplemented with glycerol and appropriate antibiotics.
Repression values correspond to the ratio of β-galactosidase activities produced by the indicated operon fusion without glp amplification to the activities produced with glp amplification.
1 mM IPTG was added to the medium.
DISCUSSION
The results presented in this paper demonstrate that mal and ompF gene expression is correlated with glpFKX expression levels. A mutation that lowers glpFKX promoter activity has been isolated. This mutation allows an increased expression of malT, MalT-dependent genes, and ompF during growth in acidic medium. Conversely, glpFKX amplification repressed this set of genes. Our experiments show that GlpK is the protein which is involved, directly or indirectly, in this repression.
We attempted to discover whether glycerol phosphorylation was required for establishing this repression. The cloning of an IPTG-inducible promoter in front of a promoterless glpFKX operon allowed us to overexpress this operon without glycerol or G3P in the growth medium, and this demonstrated that repression of the maltose system occurred even without exogenous glycerol. This repression was strongest with ribose as the carbon source and no longer occurred with glucose as the carbon source. Thus, GlpK in excess exerted its effect independently of glycerol phosphorylation. Conversely, our experiments show that repression of malT transcription at low pHo was abolished with G3P as the carbon source, even when glpK expression was induced. These data indicate that repression at low pHo of malT transcription would require both glycerol and GlpK.
Eppler and Boos (10) previously analyzed the glycerol-dependent repression of malT transcription in rich medium. They demonstrated that glycerol needs to be phosphorylated to G3P but no further metabolism of G3P is required to establish repression. Repression is controlled by the level of IIAGlc phosphorylation and, consequently, by the cAMP level. In this work, we show that the cAMP-CRP complex is also involved in the glpFKX amplification-dependent repression of malT transcription. The cAMP-CRP complex is linked to GlpK via the enzyme IIAGlc of the PTS. Indeed, glycerol kinase together with glycerol formed a complex with the unphosphorylated form of enzyme IIAGlc (25). As the phosphorylated form of IIAGlc is thought to stimulate adenylate cyclase (21), we propose that malT repression by glpFKX amplification could be mediated by modification of the cAMP level. Overproduction of GlpK in the cell may titrate the unphosphorylated form of IIAGlc and avoid its phosphorylation. A smaller amount of phosphorylated IIAGlc would lead to weaker adenylate cyclase activation. In addition, the amount of cAMP would become too limited to fully activate a cAMP-dependent gene, such as malT. To test this hypothesis, we are currently investigating the phosphorylation state of IIAGlc according to the amount of GlpK present and the effect of glpFKX amplification without IIAGlc in the cell (crr background).
As the level of mal gene expression was shown to be linked to the amount of GlpK, we searched for a potential link between an increased amount of GlpK in acidic medium and maltose gene pHo regulation. We have shown that glpK expression is modulated by the pHo but with a lower glpFKX promoter activity at low pHo than at high pHo. Thus, if it exists, the link between the amount of GlpK and maltose gene pHo regulation would be indirect. In acidic medium, the weaker glpFp18 promoter allowed the alleviation of malT repression that occurs in the parental background. This result indicates an interplay between malT pHo regulation and GlpK. This interplay could be at the level of cAMP synthesis. Thus, we formulated the hypothesis that malT transcription may follow the cAMP levels in the cell that would, in turn, change with the pHo. However, a decrease in malT expression driven by a smaller amount of cAMP-CRP could be demonstrated only if the amount of complex becomes too small to fully activate the promoter. As long as there is enough cAMP-CRP to bind to the low-affinity CRP-binding sites in front of malT, then malTp would be fully activated. In the wild-type strain, with glycerol in the growth medium, GlpK is induced and interacts with IIAGlc, leading to low cAMP synthesis. The effect of pHo on the amount of cAMP would be observed as pHo regulation of malT expression. With the glpFp18 mutation, small amounts of GlpK are produced and more IIAGlc would be converted to its phosphorylated form, leading to enough cAMP, whatever the pHo, to fully activate malT transcription. With maltose as the carbon source, the level of malT transcription was low regardless of the pHo. This low expression level could be explained by the same mechanism. Indeed, induced MalK would be able to interact, as does GlpK, with IIAGlc, limiting its conversion to the phosphorylated form. With maltose, no pHo regulation would be seen because the IIAGlc affinity for MalK is higher than that for GlpK (30) and the pHo would have no effect on a weakly active adenylate cyclase.
In conclusion, GlpK is not directly responsible for pHo regulation. However, it may contribute in releasing the pHo effect on adenylate cyclase through its interaction with IIAGlc, leading to a level of cAMP below that required for full malT activation. pHo could also influence the phosphorylation state of the enzyme IIAGlc or the interactions of IIAGlc with adenylate cyclase by unknown mechanisms. We are currently investigating these hypotheses.
ACKNOWLEDGMENTS
We thank M. Berlyn, M. Casadaban, C. Cosma, and D. W. Pettigrew for providing bacterial strains and plasmids. We thank Simone Rouzies for excellent technical assistance and wish her a wonderful retirement.
C.C. was supported by a grant from the Ministère en charge de l'Enseignement Supérieur et de la Recherche. S.A. was a lecturer at University Claude Bernard Lyon 1. This work was supported by a grant from the Centre National de la Recherche Scientifique (UMR 5122) and the University Claude Bernard Lyon 1.
REFERENCES
- 1.Alonzo S, Heyde M, Laloi P, Portalier R. Analysis of the effect exerted by extracellular pH on the maltose regulon in Escherichia coli K-12. Microbiology. 1998;144:3317–3325. doi: 10.1099/00221287-144-12-3317. [DOI] [PubMed] [Google Scholar]
- 2.Berlyn M K. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol Mol Biol Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heynecker H L, Boyer H W. Construction of useful cloning vectors. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 4.Boos W, Böhm A. Learning new tricks from an old dog: MalT of the Escherichia coli maltose system is part of a complex regulatory network. Trends Genet. 2000;16:404–409. doi: 10.1016/s0168-9525(00)02086-2. [DOI] [PubMed] [Google Scholar]
- 5.Boos W, Schuman H A. The maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decker K, Plumbridge J, Boos W. Negative transcriptional regulation of a positive regulator: the expression of malT, encoding the transcriptional activator of the maltose regulon of Escherichia coli, is negatively controlled by Mlc. Mol Microbiol. 1999;27:381–390. doi: 10.1046/j.1365-2958.1998.00694.x. [DOI] [PubMed] [Google Scholar]
- 7.Donahue J L, Bownas J L, Niehaus W G, Larson T J. Purification and characterization of glpX-encoded fructose 1,6-biphosphatase, a new enzyme of the glycerol-3-phosphate regulon of Escherichia coli. J Bacteriol. 2000;182:5624–5627. doi: 10.1128/jb.182.19.5624-5627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dykxhoorn D M, St Pierre R, Linn T. A set of compatible tac promoter expression vectors. Gene. 1996;177:133–136. doi: 10.1016/0378-1119(96)00289-2. [DOI] [PubMed] [Google Scholar]
- 9.Ehrmann M, Boos W, Ormseth E, Schweizer H, Larson T J. Divergent transcription of the sn-glycerol-3-phosphate active transport (glpT) and anaerobic sn-glycerol-3-phosphate dehydrogenase (glpA glpC glpB) genes of Escherichia coli K-12. J Bacteriol. 1987;169:526–532. doi: 10.1128/jb.169.2.526-532.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eppler T, Boos W. Glycerol-3-phosphate-mediated repression of malT in Escherichia coli does not require metabolism, depends on enzyme IIAGlc and is mediated by cAMP levels. Mol Microbiol. 1999;33:1221–1231. doi: 10.1046/j.1365-2958.1999.01570.x. [DOI] [PubMed] [Google Scholar]
- 11.Foster J W. When protons attack: microbial strategies of acid adaptation. Curr Opin Microbiol. 1999;2:170–174. doi: 10.1016/S1369-5274(99)80030-7. [DOI] [PubMed] [Google Scholar]
- 12.Heyde M, Portalier R. Regulation of major outer membrane porin proteins of Escherichia coli K-12 by pH. Mol Gen Genet. 1987;208:511–517. doi: 10.1007/BF00328148. [DOI] [PubMed] [Google Scholar]
- 13.Heyde M, Coll J L, Portalier R. Identification of Escherichia coli genes whose expression increases as a function of external pH. Mol Gen Genet. 1991;229:197–205. doi: 10.1007/BF00272156. [DOI] [PubMed] [Google Scholar]
- 14.Heyde M, Laloi P, Portalier R. Involvement of carbon source and acetyl phosphate in the external-pH-dependent expression of porin genes in Escherichia coli. J Bacteriol. 2000;182:198–202. doi: 10.1128/jb.182.1.198-202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogema B M, Arents J C, Bader R, Eijkemans K, Yoshida H, Takahashi H, Aiba H, Postma P W. Inducer exclusion in Escherichia coli by non-PTS substrates: the role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIAGlc. Mol Microbiol. 1998;30:487–498. doi: 10.1046/j.1365-2958.1998.01053.x. [DOI] [PubMed] [Google Scholar]
- 16.Innis M A, Gelfand D H. Optimization of PCRs, p 3–12. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. [Google Scholar]
- 17.Lin E C C. Dissimilatory pathways for sugars, polyols, and carboxylates. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 307–342. [Google Scholar]
- 18.Miller J H. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratories; 1992. [Google Scholar]
- 19.Olson E R. Influence of pH on bacterial gene expression. Mol Microbiol. 1993;8:5–14. doi: 10.1111/j.1365-2958.1993.tb01198.x. [DOI] [PubMed] [Google Scholar]
- 20.Pettigrew D W, Ma D P, Conrad C A, Johnson J R. Escherichia coli glycerol kinase. Cloning and sequencing of the glpK gene and the primary structure of the enzyme. J Biol Chem. 1988;263:135–139. [PubMed] [Google Scholar]
- 21.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1149–1174. [Google Scholar]
- 22.Pratt L A, Silhavy T J. Porin regulon of Escherichia coli. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 105–127. [Google Scholar]
- 23.Ray W K, Zeng G, Potters M B, Mansuri A M, Larson T J. Characterization of the 12-kilodalton rhodanese encoded by glpE of Escherichia coli and its interaction with thioredoxin. J Bacteriol. 2000;182:2277–2284. doi: 10.1128/jb.182.8.2277-2284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy P, Kamireddi M. Modulation of Escherichia coli adenylyl cyclase activity by catalytic-site mutants of protein IIAGlc of the phosphoenolpyruvate:sugar phosphotransferase system. J Bacteriol. 1998;180:732–736. doi: 10.1128/jb.180.3.732-736.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohwer J M, Bader R, van Westerhoff H, Postma P W. Limits to inducer exclusion: inhibition of the bacterial phosphotransferase system by glycerol kinase. Mol Microbiol. 1998;29:641–652. doi: 10.1046/j.1365-2958.1998.00963.x. [DOI] [PubMed] [Google Scholar]
- 26.Rose R E. The nucleotide sequence of pACYC177. Nucleic Acids Res. 1988;16:356. doi: 10.1093/nar/16.1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweizer H, Sweet G, Larson T J. Physical and genetic structure of the glpD-malT interval of the Escherichia coli K-12 chromosome. Identification of two new structural genes of the glp-regulon. Mol Gen Genet. 1986;202:488–492. doi: 10.1007/BF00333282. [DOI] [PubMed] [Google Scholar]
- 28.Truninger V, Boos W, Sweet G. Molecular analysis of the glpFKX regions of Escherichia coli and Shigella flexneri. J Bacteriol. 1992;174:6981–6991. doi: 10.1128/jb.174.21.6981-6991.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valentin-Hansen P, Albrechtsen B, Løve Larsen J E. DNA-protein recognition: demonstration of the three genetically separator elements that are required for repression of the Escherichia coli deoCABD promoters by the DeoR repressor. EMBO J. 1986;5:2015–2021. doi: 10.1002/j.1460-2075.1986.tb04458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Vlag J, van Dam K, Postma P W. Quantification of the regulation of glycerol and maltose metabolism by IIAGlc of the phosphoenolpyruvate-dependent glucose phosphotransferase system in Salmonella typhimurium. J Bacteriol. 1994;176:3518–3526. doi: 10.1128/jb.176.12.3518-3526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wanner B L. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J Mol Biol. 1986;191:39–58. doi: 10.1016/0022-2836(86)90421-3. [DOI] [PubMed] [Google Scholar]
- 32.Weissenborn D L, Wittekindt N, Larson T J. Structure and regulation of the glpFKX operon encoding glycerol diffusion facilitator and glycerol kinase of Escherichia coli K-12. J Biol Chem. 1992;267:6122–6131. [PubMed] [Google Scholar]
- 33.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 34.Zeng G, Ye S, Larson T J. Repressor of the sn-glycerol 3-phosphate regulon of Escherichia coli K-12: primary structure and identification of the DNA-binding domain. J Bacteriol. 1996;178:7080–7089. doi: 10.1128/jb.178.24.7080-7089.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zilberstein D, Agmon V, Schuldiner S, Padan E. Escherichia coli intracellular pH, membrane potential and cell growth. J Bacteriol. 1984;158:246–252. doi: 10.1128/jb.158.1.246-252.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]