Abstract
The complete 108,845-nucleotide sequence of catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP was determined. Plasmid pADP-1 was previously shown to encode AtzA, AtzB, and AtzC, which catalyze the sequential hydrolytic removal of s-triazine ring substituents from the herbicide atrazine to yield cyanuric acid. Computational analyses indicated that pADP-1 encodes 104 putative open reading frames (ORFs), which are predicted to function in catabolism, transposition, and plasmid maintenance, transfer, and replication. Regions encoding transfer and replication functions of pADP-1 had 80 to 100% amino acid sequence identity to pR751, an IncPβ plasmid previously isolated from Enterobacter aerogenes. pADP-1 was shown to contain a functional mercury resistance operon with 99% identity to Tn5053. Complete copies of transposases with 99% amino acid sequence identity to TnpA from IS1071 and TnpA from Pseudomonas pseudoalcaligenes were identified and flank each of the atzA, atzB, and atzC genes, forming structures resembling nested catabolic transposons. Functional analyses identified three new catabolic genes, atzD, atzE, and atzF, which participate in atrazine catabolism. Crude extracts from Escherichia coli expressing AtzD hydrolyzed cyanuric acid to biuret. AtzD showed 58% amino acid sequence identity to TrzD, a cyanuric acid amidohydrolase, from Pseudomonas sp. strain NRRLB-12227. Two other genes encoding the further catabolism of cyanuric acid, atzE and atzF, reside in a contiguous cluster adjacent to a potential LysR-type transcriptional regulator. E. coli strains bearing atzE and atzF were shown to encode a biuret hydrolase and allophanate hydrolase, respectively. atzDEF are cotranscribed. AtzE and AtzF are members of a common amidase protein family. These data reveal the complete structure of a catabolic plasmid and show that the atrazine catabolic genes are dispersed on three disparate regions of the plasmid. These results begin to provide insight into how plasmids are structured, and thus evolve, to encode the catabolism of compounds recently added to the biosphere.
Many bacteria contain plasmids that carry genes functional in antibiotic resistance, virulence for animal or plant hosts, or the catabolism of diverse chemical compounds. While much has been learned since Lederberg's initial discovery of plasmids in 1952 (36), genomic approaches will further enhance our understanding of plasmid structure and evolution. Recently, the complete nucleotide sequences of approximately 90 bacterial and 10 archaeal plasmids have been obtained (5, 20, 29, 41, 46, 52, 59; http://www.ncbi.nlm.nih.gov:80/PMGifs/Genomes/eub_p.html). However, most of the sequenced plasmids are relatively small, are used as vectors in molecular biology, or are of medical importance. The latter group of plasmids consist predominantly of those known to contain antibiotic resistance genes or encode virulence determinants associated with infectious diseases (5, 29, 52). In contrast, catabolic plasmids, which have been identified in many nonpathogenic soil bacteria (49), have been shown to transfer among bacteria, thus disseminating genes encoding the metabolism of environmentally relevant compounds (11). For example, plasmids encoding the catabolism of toluene, camphor, naphthalene, and 2,4-dichlorophenoxyacetate are known (3, 12, 45, 64). Many of the genes involved in their respective catabolic pathways have been cloned and sequenced, but to date only one catabolic plasmid, pNL1, from Sphingomonas aromaticivorans strain F199 has been completely sequenced (46). This plasmid contains genes encoding enzymes for the metabolism of biphenyl, naphthalene, m-xylene, and p-cresol.
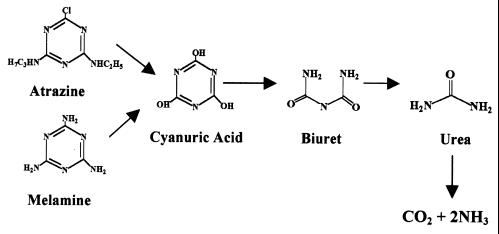
Metabolism of the herbicide atrazine has also been shown to be linked to catabolic plasmids. Mandelbaum et al. (38) isolated Pseudomonas sp. strain ADP, which metabolizes atrazine to carbon dioxide and ammonia. The first three enzymatic steps, encoded by the genes atzA, atzB, and atzC, transform atrazine to cyanuric acid (9, 15, 47). Cyanuric acid has been shown to be a common intermediate in the degradation pathways for melamine (2,4,6-triamino-s-triazine) and atrazine (21, 22, 47) in Pseudomonas sp. strain NRRLB-12227 and Pseudomonas sp. strain ADP, respectively (Fig. 1). However, while the hydrolysis of cyanuric acid in Pseudomonas sp. strain NRRLB-12227 proceeds through biuret and urea intermediates, the reactions involved in cyanuric acid degradation in Pseudomonas sp. strain ADP were not established.
FIG. 1.
Proposed pathway for the degradation of cyanuric acid by atrazine- and melamine-degrading bacteria. Cyanuric acid is hydrolyzed to biuret and is hypothesized to be subsequently hydrolyzed to urea, carbon dioxide, and ammonia.
The atzA, atzB, and atzC genes in Pseudomonas sp. strain ADP have been localized to an approximately 100 kb plasmid, pADP-1 (17), and DNA regions with homology to IS1071 have been shown to flank atzA (18). Recently, plasmid-localized genes homologous to the atzA, atzB, and atzC genes have been identified in different genera of atrazine-degrading bacteria isolated from geographically diverse locations (16; B. Martinez, M. de Souza, L. Wackett, and M. Sadowsky, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. Q352, 1999). Since the atrazine catabolic genes have >99% sequence identity among different species of bacteria isolated independently from different continents, this suggests a recent evolution and dissemination of the atzA, atzB, and atzC genes.
To further our understanding about the assembly of the atrazine catabolic pathway and the accessory functions encoded by pADP-1, we undertook the complete sequencing and annotation of this plasmid. Sequence analysis revealed that the gene regions encoding plasmid replication, transfer, and maintenance functions of pADP-1 were nearly identical to those found on pR751, an IncPβ plasmid from Enterobacter aerogenes. Plasmid pADP-1 is predicted to encode 104 proteins. A functional mercury resistance operon is also present on pADP-1. Structural and functional studies showed that the genes encoding the initial reactions of atrazine catabolism are not organized in an operon, but are dispersed and flanked by transposases. Moreover, genes for the complete catabolism of cyanuric acid to CO2 and NH3 were localized to pADP-1.
MATERIALS AND METHODS
Isolation of pADP-1 DNA.
The pADP-1 plasmid was introduced into Escherichia coli AD256 by conjugation as described previously (17). Cells were grown overnight in one-fourth-strength Luria-Bertani (LB) medium (48) containing 500 μg of atrazine per ml. Cells were harvested by centrifugation at 8,000 × g for 10 min at 4°C. Large-scale plasmid DNA isolation preparations were done as described by Hirsch et al. (27), and pADP-1 was further purified using CsCl buoyant-density ultracentrifugation (48).
Construction of pADP-1 shotgun library.
Plasmid pADP-1 DNA was nebulized using 4.4 × 104 Pa of N2 for 4.0 min and a nebulizer (IPT Medical Products Inc., Chicago, Ill.). The DNA ends were filled in, ligated into plasmid pUC18, and transferred to DH10B (Gibco-BRL, Grand Island N.Y.) as described previously (48). Clones containing inserts were picked randomly using the Genetix Q-Bot robot (Genetix Ltd., New Milton, United Kingdom) and stored in 96-well microtiter plates.
DNA sequencing.
DNA templates from randomly selected shotgun clones were prepared from 3-ml overnight cultures grown in LB medium containing 50 μg of ampicillin per ml using an AutoGene 740 DNA isolation system (Integrated Separation Systems, Framingham, Mass.). DNA from each preparation was dissolved in 80 μl of sterile H2O and stored at −20°C until used. Plasmid DNA, 500 ng, was sequenced using M13 forward and reverse primers and an ABI BigDye cycle sequencing kit (Applied Biosystems, Foster City, Calif.) and an ABI model 377 DNA sequencer (Applied Biosystems).
Sequence assembly.
The Phred/Phrap/Consed sequence analysis software package was used to cross-match vector sequences and assemble the pADP-1 sequence into contigs (25). For gap closure, custom primers were designed from the ends of each contig in the pADP-1 sequence using Primer Designer software, version 2.01 (Scientific and Educational Software, State Line, Pa.). Primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, Iowa). DNA fragments containing sequences necessary to close gaps between contigs were obtained by using the PCR high-fidelity rTth polymerase and purified plasmid pADP-1 DNA as a template. PCR was done using a Perkin-Elmer/Applied Biosystems XL Polymerase DNA amplification kit (PE/Applied Biosystems, Foster City, Calif.) and a PTC-100 thermocycler (MJ Research, Incline Village, Nev.). PCR products were purified using a Qiagen gel extraction kit (Qiagen, La Jolla, Calif.) and sequenced directly as described above. The pBluescript vector (Stratagene) was used to clone PCR products longer than 3 kb and facilitate sequencing.
Analysis of ORFs.
The analysis of the open reading frames (ORFs) present in the pADP-1 sequence was completed using the web-based versions of GeneMark (7; http://genemark.biology.gatech.edu/GeneMark/), GeneMark.hmm (37; http://genemark.biology. gatech.edu/GeneMark/hmmchoice.html), Pfam (http://pfam.wustl.edu/), and the NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) programs. ORFs that were consistently predicted by these programs were selected and met the following criteria: (i) the start codon was ATG, GTG, or TTG; (ii) the stop codon was TAA, TAG, or TGA; and (iii) the size of the ORF was between 150 and 5,000 bp. Start codon positions were assigned using manual identification of the Shine-Dalgarno sites within 15 bp of a potential start site. GeneMark.hmm (37) was also used to examine potential frameshifts leading to misidentification of ORFs. Those ORFs predicted to encode genes were analyzed further to determine their homology to proteins of known function using BLAST and BLASTP (http://seqsim.ncgr.org/newBlast.html).
Cloning of the cyanuric acid hydrolase gene.
The putative cyanuric acid hydrolase gene, atzD, was cloned from pADP-1 using a PCR approach and primers CAAHF (5′-GCGGATCCTGCGTTCATCGACAGAG-3′) and CAAHR (5′-GCGGATCCAGATGGCCTGTATCGCT-3′). The primers contained BamHI restriction sites at each end to facilitate gene cloning. PCR was performed using a high-fidelity XL polymerase DNA amplification kit (PE/Applied Biosystems). Amplification of the 1.4-kb DNA fragment was achieved using the following conditions: 94°C for 1 min, and then 28 cycles consisting of 94°C for 15 s and 60°C for 5 min. A final extension of the PCR product was done at 72°C for 10 min. The PCR product was resolved on a 0.8% agarose gel, and the band was excised from the gel and purified using the Qiagen gel extraction kit (Qiagen, La Jolla, Calif.). The PCR product was cloned into the BamHI site of pKT230 (4) using standard cloning procedures (48). Potential clones were screened for the insert by restriction enzyme digestions using BamHI, and one clone, pBMZ1, was used for subsequent functional analyses.
Cyanuric acid degradation assays.
To determine if the putative atzD gene encoded a cyanuric acid amidohydrolase, the hydrolysis of cyanuric acid by crude extracts of E. coli DH5α(pBMZ1) and E. coli DH5α was examined. Crude extracts were prepared as described previously (9). Reaction mixtures contained 10 mM potassium phosphate buffer (pH 7.2), 3 mM cyanuric acid (Sigma-Aldrich, St. Louis, Mo.), and 100 μg of protein in a final reaction volume of 1.0 ml. Replicate reactions were incubated at 30°C and at various times terminated by heating at 95°C for 2 min. Samples were centrifuged at 14,000 × g for 10 min, filtered through 0.2-μm filters, and placed in vials for analysis by high-performance liquid chromatography (HPLC). The disappearance of cyanuric acid was monitored by HPLC analysis using an analytical Absorbosphere C18 reverse-phase HPLC column (5-μm spherical packing; 250 by 4.6 mm) (Alltech Associates, Deerfield, Ill.). The isocratic mobile phase was 5 mM potassium phosphate buffer (pH 6.8) containing 5 mM dodecyltriethylammonium phosphate and Q12 ion pair cocktail (Regis Chemical Technologies, Morton Grove, Ill.) at a flow rate of 1.0 ml per min. Spectral data of the column eluent was acquired at 224 nm. Under these conditions, cyanuric acid eluted from the column at about 6.0 min and biuret at 3.3 min. The concentration of cyanuric acid in the sample analyzed was obtained by integrating peak areas at 224 nm. Biuret was resolved from reaction mixtures using an Absorbosphere C18 reverse-phase HPLC column as described above, and its identity was confirmed by direct insertion mass spectroscopy with a Kratos MS25 mass spectrometer (Ramsey, N.J.) using electron impact ionization at 70 eV.
Biuret degradation assays.
Plate-clearing, ammonia release, thin-layer chromatography (TLC), and HPLC assays were used to determine if ORF101 (atzE) encoded biuret hydrolase activity. One clone from the pADP-1 sequencing library, p11A07, containing a complete copy of ORF101 was used for these analyses. For the plate-clearing assays, 500 μl of an E. coli(p11A07) culture (optical density at 600 nm [OD600] = 1.0) was placed onto the surface of one-fourth-strength LB medium plates containing 60 mM sodium phosphate buffer and 4,000 μg of biuret per ml. Plates were incubated at 37°C for up to 1 week. The appearance of clearing zones surrounding cell growth indicated biuret degradation.
Ammonia release assays were done in 50 mM sodium phosphate buffer (pH 8.0) containing 3 mM biuret and cell extract (1 mg of protein) from E. coli(p11A07). Reaction mixtures were incubated at 30°C for 6 h. Samples were taken at several time points, and the reactions were stopped by addition of 1 N H2SO4. For urea-coupled assays, reaction mixtures were treated with 100 U of type III jack bean urease (Sigma, St. Louis, Mo.) during incubations. Analysis of released ammonia was determined using the Berthelot reactions described by Weatherburn (61) with the following modifications: 500 μl of the reaction mixture was treated with 200 μl of 0.3 M Na2WO4 and 200 μl of 1 N sulfuric acid to precipitate proteins. The samples were mixed and centrifuged for 5 min at 14,000 × g, and 500 μl of the supernatant was treated with 1.5 ml of solution A (1% phenol and 0.005% sodium nitroprusside in water) and 2.0 ml of solution B (0.5% NaOH and 0.84% sodium hypochlorite in water). Reactions were incubated at room temperature for 1 h, and the resulting indophenol was monitored at 630 nm.
The degradation of biuret was also examined by HPLC analyses. Cell extracts of E. coli DH5α(p11A07) and E. coli DH5α(pUC18) were prepared as described previously (9). Reaction mixtures containing 50 mM sodium phosphate buffer (pH 7.0), 0.6 mM biuret, and cell extract (1 mg of protein), in a final volume of 10 ml, were incubated at 30°C. Reactions were terminated by heating at 95°C for 3 min. Samples were centrifuged at 14,000 × g for 10 min, filtered through 0.2-μm filters, and analyzed by HPLC. The disappearance of biuret was followed by using a Waters IC-Pak A HC anion-exchange column (150 mm by 4.6 mm; Waters Corp., Milford, Mass.). The isocratic mobile phase consisted of 5 mM sodium phosphate buffer (pH 7.0) at a flow rate of 0.5 ml per min (35). Spectral data of the column eluent were collected at 200 nm. Under these conditions, urea eluted after 3.9 min and biuret eluted after 5.0 min. The concentration of biuret was obtained by integrating peak areas at 200 nm.
TLC analyses were performed as described by Radosevich (44) to detect the products of biuret degradation. Reaction mixtures containing 1 mM sodium phosphate buffer (pH 8.0), 3 mM biuret, and cell extract (1 mg of protein), in a final reaction volume of 10 ml, were incubated overnight at 30°C. Samples of the reaction mixture (1.0 ml) were evaporated to dryness using a SpeedVac concentrator (Savant Instruments Inc., Farmingdale, N.Y.) and resuspended in 25 μl of NH4OH. Samples were chromatographed on Cellulose 300, F-254 TLC plates (Selecto Scientific, Suwanee, Ga.) using a mobile phase consisting of 40% t-butyl alcohol, 30% methyl ethyl ketone, 10% ammonium hydroxide, and 20% water. Standards and reactions products were visualized by spraying the plate with a solution containing 1 g of 4-diethylaminobenzaldehyde in 75 ml of methanol and 25 ml of concentrated HCl.
Allophanate degradation assays.
Ammonia release, HPLC, and urea agar plate assays were done to determine if ORF102 (atzF) encoded allophanate hydrolase activity. For these studies, a clone from the pADP-1 sequencing library, p14D12, containing only the complete copy of ORF102 was used. To test for urea hydrolysis, 500 μl of E. coli(p14D12) and E. coli(pUC18) cultures (OD600 = 1.0) were spotted onto the surface of urea agar (Difco, Detroit, Mich.). The cultures were incubated for up to 1 week at 37°C, and urea hydrolysis was monitored by observing a change in the color of the medium, indicating an increase in pH due to urea hydrolysis.
Ammonia release assays were done to determine if urea or allophanate was a substrate for ORF102. These studies were done using cell extracts from E. coli(p14D12) and E. coli(pUC18). Ultrapure urea was obtained from Gibco-BRL (Grand Island, N.Y.). Potassium allophanate was synthesized from ethyl allophanate (Fisher Scientific, Pittsburgh, Pa.) using the following method. Twenty millimoles of ethyl allophanate was mixed with 25 mmol of KOH and heated at 40°C for 3 h. The mixture was dissolved in 250 ml of 100% ethanol and 50 ml of diethylether. The reaction mixture was incubated at 4°C for 8 h, filtered, and dried in a desiccator until ready to use. The ammonia release procedure described above was used with the following modifications: reactions were done in 10 mM sodium phosphate buffer (pH 8.0) containing 4 mM potassium allophanate or 3 mM urea, and each sample contained 32 μg of protein from cell extracts. Reactions were terminated by adding 200 μl of 1 N sulfuric acid.
HPLC analyses done to detect allophanate degradation were performed essentially as described above except that the isocratic mobile phase consisted of 10 mM sodium phosphate buffer (pH 8.0). Samples were assayed in 10 mM sodium phosphate buffer (pH 8.0) containing 4 mM potassium allophanate and 32 μg of protein. Reactions were terminated by adding 4 μl of 3 M NaOH. Under these conditions, allophanate eluted after 11.2 min.
RT-PCR analyses.
RNA was isolated from exponentially growing Pseudomonas sp. strain ADP cells using the diethyl polycarbonate (DEPC) of Summers (56). RNA samples were treated with 10 U of RNase-free DNase (Takara Biomedical Group, Shiga, Japan) for 1 h at 37°C, extracted twice with phenol-chloroform, and ethanol precipitated. The resulting RNA was quantified spectrophotometrically.
Reverse transcriptase PCR (RT-PCR) reactions were done using the GeneAmp RNA PCR kit (PE/Applied Biosystems, Branchburg, N.J.). Reverse transcriptase reactions were carried out with 500 ng of RNA and random hexamer primers using the following protocol: 10 min at 25°C, 15 min at 42°C, 5 min at 99°C, and 5 min at 5°C. The complete reverse transcriptase reaction mixture was used as the template for cDNA PCR. This was done using primer pairs specific for the DNA region upstream of atzD and ORF101 (atzE) [5′atzD-f (5′-CGGCGTACCTAACTCGT-3′) and ORF101-r (5′-GCGTATGGAACCGTTGG-3′] and the region between atzE and atzF [atzE-f (5′-GCCAGCGAAGTCGTCAT-3′) and atzF-r (5′-TCTGTACCGGCGGCATA-3′)]. Since a single mRNA from atzD to atzF would be approximately 5.2 kb and difficult to amplify, two primer pairs were used to amplify overlapping cDNA regions. The following PCR protocol was used: 1 min at 94°C, 20 cycles of 15 s at 94°C and 5 min at 63°C, and 18 cycles at 95°C for 15 s and 63°C for 5 min, with an extension of 15 s per cycle. The PCR mixture was subjected to a final extension of 72°C for 10 min. Reaction products were separated by horizontal gel electrophoresis on 0.85% agarose, stained, and photographed under UV light.
Mercury resistance analyses.
The functionality of the putative mercury resistance operon in pADP-1 was determined by disk (62) and mercuric reductase activity assays (23). The disk assay was used to determine if the presence of pADP-1 allowed recipient bacteria to grow in media containing different concentrations of HgCl2. E. coli strains AD256 (17), AD256(pADP-1) (17), S17-1 (53) and S17-1(pADP-1) were grown to mid-log phase in LB medium containing 100 μg of atrazine per ml. Aliquots, 100 μl, of each strain were spread-plated onto the surface of LB plates, and filter disks with 0, 50, 100, 150, 200, 250, and 300 μg of HgCl2 per ml were overlaid onto the inoculated medium. Plates were incubated overnight at 37°C, and sensitivity to Hg was determined by the size of the growth inhibition zones surrounding the disks. Mercuric reductase activity was determined by monitoring Hg-dependent oxidation of NADPH. E. coli strains AD256(pADP-1) and S17-1(pADP-1) were grown in 100 ml of LB liquid medium containing 10 μM HgCl2 (Sigma-Aldrich, St. Louis, Mo.) to mid-log phase (OD600 = 0.5), after which an additional 20 μM HgCl2 was added to the medium. Cultures were grown for an additional 1 h and centrifuged at 10,000 × g, and cell pellets were washed twice in 10 mM potassium phosphate buffer (pH 7.4). Cells were resuspended in 10 ml of potassium phosphate buffer (pH 7.4), and crude cell extracts were prepared as previously described (9). Reaction mixtures contained 150 μM NADPH, 200 μM disodium EDTA, 1.0 mM β-mercaptoethanol, 2 μM flavin adenine dinucleotide, and 0.05 mg of total cell protein in 50 mM sodium phosphate buffer (pH 7.2), and were preincubated at 4°C for 10 min. Reactions were initiated by the addition of 100 μM HgCl2. The oxidation of NADPH was followed by monitoring the decrease in absorbance at 340 nm for 10 min. Assays were done in triplicate.
Nucleotide sequence accession number.
The complete nucleotide sequence of pADP-1 from Pseudomonas sp. strain ADP has been deposited with GenBank as accession number U66917.
RESULTS
Nucleotide sequence, physical map, and genetic organization of pADP-1.
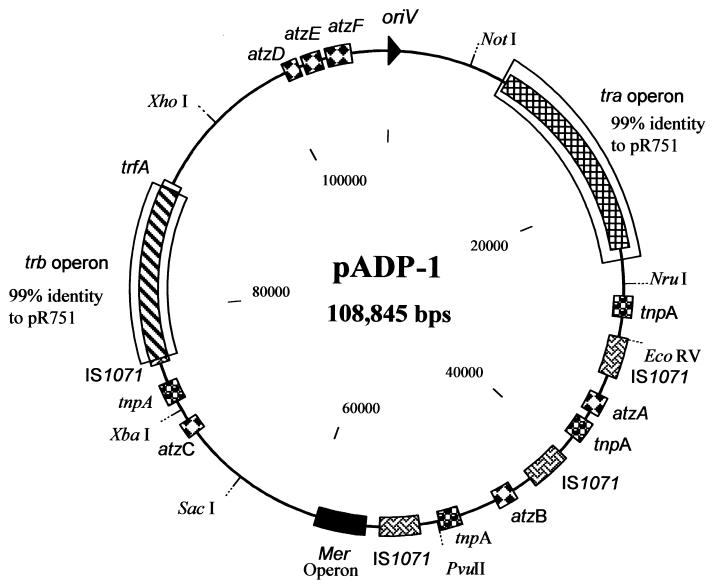
The atrazine catabolic plasmid pADP-1 is 108,845 bp, with an overall G+C content of 62.6 mol%. This value is within the range of those found in several genera of gram-negative soil bacteria, including Pseudomonas and Alcaligenes strains (28, 40). A circular physical map of pADP-1 is shown in Fig. 2. The assembly of pADP-1 was verified by using restriction enzyme analyses and PCR (data not shown). This plasmid contains a classical IncPβ backbone that consists of two regions involved in plasmid conjugation (the tra and trb operons), an origin of replication (oriV), and a region involved in plasmid control, stable inheritance, and partitioning. The nucleotide sequence of the pADP-1 backbone is 80 to 100% identical to the backbone of the archetype IncPβ plasmid R751 except for kleG. The main catabolic region of pADP-1 lies outside the trb and tra operons. This region contains the first three genes necessary for the degradation of atrazine, atzA, atzB, and atzC. In addition, the region contains a mercury resistance operon and several insertion sequence elements. Restriction enzyme analysis of pADP-1 showed that the catabolic region of this plasmid contains numerous restriction sites, while only a few enzymes cut the backbone.
FIG. 2.
Physical circular map of the catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. The map positions of selected restriction sites, genes, and operons on pADP-1 are indicated. Regions with 99% identity to the tra and trb operons of plasmid pR751 are boxed. Genes involved in atrazine catabolism are indicated. Copies of similar putative transposons have the same shading.
Analyses of ORFs.
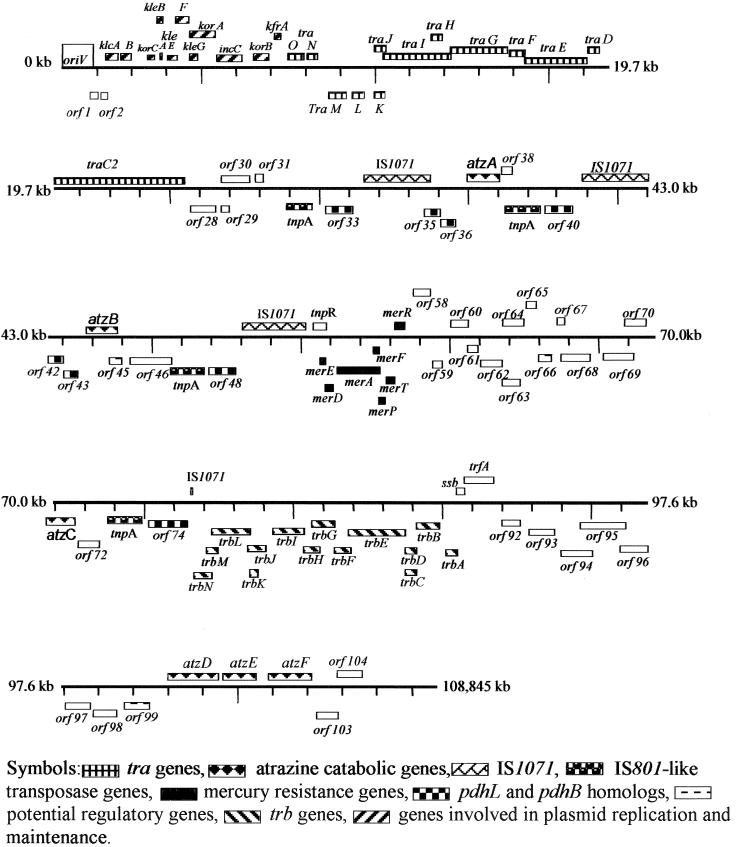
A total of 104 ORFs were identified in the pADP-1 sequence using GeneMark, GeneMark.hmm, and NCBI ORF Finder (Fig. 3). All three ORF-finding programs identified the same ORFs. Based on significant similarities to proteins of known function, putative functions were assigned to a majority of the ORFs (Table 1); 15% of the ORFs were predicted to be involved in catabolism, 15% were predicted to be involved in transposition, 5% encode putative transporters, 3% are putative transcriptional regulators, 13% are proteins involved in general metabolism, and 39% are proteins necessary for plasmid maintenance, transfer, and replication. Several ORFs (8%) had >38% amino acid identity to hypothetical proteins, and 2% had no significant homology to any known proteins in databases. The first ORF after the origin of replication, oriV, was arbitrarily assigned as ORF1 (Table 1 and Fig. 3). More detailed descriptions of many of the ORFs and genes on pADP-1 are found below.
FIG. 3.
Linear map showing graphical representation of ORFs present on the pADP-1 sequence. Boxes above the lines refer to ORFs on the top strand, while those below the line are from the complementary strand. Genes of the same type (operons, insertion sequence [IS] elements, and catabolic genes) have similar shading, and ORFs with putative functions are listed by their numbers. ORFs and genes have been numbered relative to the origin of replication (oriV). Distance between tick marks is 1.3 kb.
TABLE 1.
Localization and predicted functions of ORFs in pADP-1
| ORF no. | Positions (bp)a | Function of closest relative | Source microoganism | No. of amino acids, pADP-1/relativec | % Amino acid identityd | GenBank accession no. | E valuee |
|---|---|---|---|---|---|---|---|
| 1 | 869–1183 c | Hypothetical protein—no significant homology to others | N/Ab | N/A | N/A | N/A | N/A |
| 2 | 1180–1461 c | Hypothetical protein—no significant homology to others | N/A | N/A | N/A | N/A | N/A |
| 3 | 1618–2046 d | Protein invoved in stable inheritance (KlcA) | Enterobacter aerogenes pR751 | 142/142 | 100 (142/142) | AAC64430.1 | 1.00E−77 |
| 4 | 2098–2517 d | Protein invoved in stable inheritance (KlcB) | Enterobacter aerogenes pR751 | 139/371 | 91 (106/116) | AAC64429.1 | 3.00E−55 |
| 5 | 3427–3684 d | Transcriptional repressor (KorC) | Enterobacter aerogenes pR751 | 85/84 | 92 (78/84) | AAC64428.1 | 8.00E−41 |
| 6 | 3844–4080 d | Protein invoved in stable inheritance (KleA) | Enterobacter aerogenes pR751 | 78/78 | 100 (78/78) | AAC64427.1 | 2.00E−38 |
| 7 | 4139–4354 d | Protein invoved in stable inheritance (KleB) | Enterobacter aerogenes pR751 | 71/71 | 100 (71/71) | AAC64426.1 | 1.00E−34 |
| 8 | 4442–4828 d | Protein invoved in stable inheritance (KleE) | Enterobacter aerogenes pR751 | 128/109 | 82 (90/109) | AAC64425.1 | 7.00E−48 |
| 9 | 4830–5360 d | Protein invoved in stable inheritance (KleF) | Enterobacter aerogenes pR751 | 176/97 | 91 (88/96) | AAC64424.1 | 2.00E−47 |
| 10 | 5104–5367 d | Protein invoved in stable inheritance (KleG) | Enterobacter aerogenes pR751 | 87/89 | 43 (37/86) | AAC64423.1 | 2.00E−05 |
| 11 | 5461–6537 d | Inclusion membrane protein (IncC) | Enterobacter aerogenes pR751 | 358/358 | 100 (358/358) | AAC64421.1 | 0.00 |
| 12 | 5474–5776 d | Transcriptional repressor (KorA) | Enterobacter aerogenes pR751 | 100/100 | 100 (100/100) | AAC64422.1 | 3.00E−50 |
| 13 | 6534–7583 d | Transcriptional repressor protein (KorB) | Enterobacter aerogenes pR751 | 349/349 | 99 (347/349) | AAC64419.1 | 0.00 |
| 14 | 7763–8794 d | Transcriptional regulator (KfrA) | Enterobacter aerogenes pR751 | 343/343 | 99 (341/343) | AAC64418.1 | 1.00E−180 |
| 15 | 8965–9312 d | TraO protein | Enterobacter aerogenes pR751 | 115/115 | 99 (14/115) | AAC64417.1 | 2.00E−59 |
| 16 | 9341–9994 d | TraN protein | Enterobacter aerogenes pR751 | 217/217 | 96 (210/217) | AAC64416.1 | 1.00E−112 |
| 17 | 10197–10637 c | Essential transfer protein (TraM) | Enterobacter aerogenes pR751 | 146/146 | 100 (146/146) | AAC64481.1 | 2.00E−74 |
| 18 | 10637–11362 c | Transfer origin protein (TraL) | Enterobacter aerogenes pR751 | 241/241 | 99 (240/241) | AAC64480.1 | 1.00E−135 |
| 19 | 11362–11760 c | oriT binding protein (TraK) | Enterobacter aerogenes pR751 | 132/132 | 100 (132/132) | AAC64479.1 | 9.00E−75 |
| 20 | 12134–12508 d | oriT binding protein (TraJ) | Enterobacter aerogenes pR751 | 124/130 | 87 (113/129) | AAC64478.1 | 1.00E−54 |
| 21 | 12543–14783 d | DNA relaxase (TraI) | Enterobacter aerogenes pR751 | 747/747 | 99 (743/747) | AAC64475.1 | 0.00E 00 |
| 22 | 14089–14481 d | Relaxosome stabilization protein (TraH) | Enterobacter aerogenes pR751 | 130/130 | 100 (130/130) | AAC64476.1 | 1.00E 73 |
| 23 | 14780–16693 d | DNA transport protein (TraG) | Enterobacter aerogenes pR751 | 637/637 | 100 (636/637) | AAC64474.1 | 0.00 |
| 24 | 16690–17226 d | Maturation peptidase (TraF) | Enterobacter aerogenes pR751 | 178/178 | 100 (178/178) | AAC64473.1 | 3.00E−98 |
| 25 | 17238–19301 d | DNA topoisomerase (TraE) | Enterobacter aerogenes pR751 | 687/687 | 85 (593/687) | AAC64472.1 | 0.00 |
| 26 | 19323–19712 d | Unknown function (TraD) | Enterobacter aerogenes pR751 | 129/129 | 100 (129/129) | AAC64471.1 | 7.00E−69 |
| 27 | 19716–24062 d | DNA replication primase (TraC) | Enterobacter aerogenes pR751 | 1,448/1,448 | 99 (1,446/1,448) | AAC64468.1 | 0.00 |
| 28 | 24327–25175 c | Cointegrate formation and resolution protein (IstB) | Ralstonia eutropha NH9 | 282/264 | 69 (170/245) | BAA33970.1 | 1.00E−90 |
| 29 | 25159–25416 c | Cointegrate formation and resolution protein (IstA) | Ralstonia eutropha NH9 | 84/518 | 44 (34/76) | BAA33969.1 | 5.00E−09 |
| 30 | 25315–26307 d | Conserved hypothetical protein | Synechocystis sp. strain 6803 | 353/535 | 46 (131/283) | BAA10081.1 | 7.00E−66 |
| 31 | 26288–26764 d | Putative cytosine deaminase | A. aveneae subsp. citrulli 12227 | 158/158 | 100 (158/158) | AAK00500.1 | 8.00E−82 |
| 32 | 27514–29061 c | Transposase similar to TnpA from IS801 | P. pseudoalcaligenes JS45 | 515/515 | 99 (514/515) | AAB94124.1 | 1.00E−150 |
| 33 | 29170–30372 c | Dihydrolipoamide dehydrogenase (PdhL) | Ralstonia eutropha H16 | 400/594 | 68 (276/401) | AAA21600.1 | 1.00E−154 |
| 34 | 30521–33436 d | Transposase from IS1071 | Alcaligenes sp. strain BR60 | 971/970 | 99 (971/971) | AAA70396.1 | 0.00 |
| 35 | 33549–34253 c | Dihydrolipoamide dehydrogenase (PdhL) | Ralstonia eutropha H16 | 234/594 | 63 (145/229) | AAA21600.1 | 9.00E−75 |
| 36 | 34270–34920 c | Dihydrolipoamide acetyltransferase (PdhB) | Ralstonia eutropha H16 | 216/553 | 82 (174/212) | AAA21599.1 | 3.00E−99 |
| 37 | 34964–36388 d | Atrazine chlorohydrolase (AtzA) | Pseudomonas sp. strain ADP | 474/474 | 100 (474/474) | AAC64663.1 | 0.00 |
| 38 | 36392–36856 d | Amidase | R. rhodochrous J1 | 154/515 | 46 (70/154) | BAA03744.1 | 7.00E−28 |
| 39 | 37037–38584 c | Transposase similar to TnpA from IS801 | P. pseudoalcaligenes JS45 | 515/515 | 99 (514/515) | AAB94124.1 | 0.00 |
| 40 | 38693–39895 c | Dihydrolipoamide dehydrogenase (PdhL) | Ralstonia eutropha H16 | 400/594 | 68 (276/401) | AAA21600.1 | 1.00E−154 |
| 41 | 40043–42959 d | Transposase from IS1071 | Alcaligenes sp. strain BR60 | 971/970 | 99 (971/971) | AAA70396.1 | 0.00 |
| 42 | 43072–43776 c | Dihydrolipoamide dehydrogenase (PdhL) | Ralstonia eutropha H16 | 234/594 | 63 (145/229) | AAA21600.1 | 9.00E−75 |
| 43 | 43793–44443 c | Dihydrolipoamide acetyltransferase (PdhB) | Ralstonia eutropha H16 | 216/553 | 82 (174/212) | AAA21599.1 | 1.00E−100 |
| 44 | 44487–45932 d | Hydroxyatrazine hydrolase (AtzB) | Pseudomonas sp. strain ADP | 481/481 | 100 (481/481) | AAC45138.1 | 0.00 |
| 45 | 45919–46494 c | Transcriptional regulator Tet/Acr family | Aquifex aeolicus VF5 | 191/192 | 30 (27/90) | AAC07881.1 | 2.00E−06 |
| 46 | 46883–48223 c | Conserved hypothetical protein | D. radiodurans R1 | 446/464 | 43 (198/457) | AAF12190.1 | 2.00E−89 |
| 47 | 48776–50323 c | Transposase similar to TnpA from IS801 | P. pseudoalcaligenes JS45 | 515/515 | 100 (515/515) | AAB94124.1 | 0.00 |
| 48 | 50432–51688 c | Dihydrolipoamide dehydrogenase (PdhL) | Ralstonia eutropha H16 | 319/594 | 69 (236/341) | AAA21600.1 | 1.00E−129 |
| 49 | 51784–54699 d | Transposase from IS1071 | Alcaligenes sp. strain BR60 | 971/970 | 99 (969/971) | AAA70396.1 | 0.00 |
| 50 | 55067–55681 d | Resolvase from Tn5053 transposon | Xanthomonas sp. strain W17 | 204/204 | 99 (203/204) | AAA98330.1 | 1.00E−110 |
| 51 | 55734–55970 c | MerE protein from Tn5053 transposon | Xanthomonas sp. strain W17 | 78/78 | 100 (78/78) | AAA98328.1 | 4.00E−40 |
| 52 | 55967–56332 c | Mercury coregulator protein from Tn5053 (MerD) | Xanthomonas sp. strain W17 | 121/121 | 100 (121/121) | AAA98327.1 | 9.00E−63 |
| 53 | 56348–57994 c | Mercury reductase from Tn5053 (MerA) | Xanthomonas sp. strain W17 | 548/548 | 99 (547/548) | AAA98326.1 | 0.00 |
| 54 | 57991–58236 c | Mercury ion transport protein from Tn5053 (MerT) | Xanthomonas sp. strain W17 | 81/81 | 100 (81/81) | AAA98325.1 | 1.00E−30 |
| 55 | 58239–58514 c | Periplasmic mercury ion binding (MerP) | Xanthomonas sp. strain W17 | 91/91 | 100 (91/91) | AAA98324.1 | 8.00E−45 |
| 56 | 58530–58880 c | Mercury ion transport protein | Xanthomonas sp. strain W17 | 116/116 | 100 (116/116) | AAA98323.1 | 2.00E−63 |
| 57 | 58952–59386 d | Mercury regulatory protein (MerR) | Xanthomonas sp. strain W17 | 144/144 | 100 (144/144) | AAA98322.1 | 9.00E−78 |
| 58 | 59783–60556 d | Conserved hypothetical protein | P. aeruginosa PAO1 | 257/205 | 61 (127/205) | AAG06457.1 | 2.00E−68 |
| 59 | 60661–61119 c | Conserved hypothetical protein | P. aeruginosa PAO1 | 152/152 | 86 (98/113) | AAG03518.1 | 8.00E−53 |
| 60 | 61471–62265 d | Putative glutamine amidotransferase | P. aeruginosa PAO1 | 264/250 | 43 (107/248) | AAG03686.1 | 2.00E−54 |
| 61 | 62296–62769 c | Conserved hypothetical protein | P. aeruginosa PAO1 | 157/166 | 64 (90/128) | AAG03517.1 | 9.00E−48 |
| 62 | 62824–63771 c | Conserved hypothetical protein; glycerol sensor protein homolog | P. aeruginosa PAO1 | 315/287 | 62 (183/274) | AAG05365.1 | 2.00E−89 |
| 63 | 63790–64593 c | Conserved hypothetical protein | P. aeruginosa PAO1 | 267/278 | 61 (175/286) | AAG03555.1 | 3.00E−95 |
| 64 | 63796–64755 d | Mutator protein (MutT) | Synechocystis sp. strain 6803 | 319/136 | 32 (30/94) | BAA16660.1 | 4.08E−02 |
| 65 | 64864–65307 d | Short-chain dehydrogenase | P. aeruginosa PAO1 | 147/265 | 66 (97/147) | AAG08217.1 | 3.00E−48 |
| 66 | 65433–65999 c | Putative transcriptional regulator | P. aeruginosa PAO1 | 188/186 | 67 (126/188) | AAG08216.1 | 9.00E−64 |
| 67 | 66253–66582 d | Dihydrolipoamide dehydrogenase 3 | P. aeruginosa PAO1 | 109/467 | 78 (85/109) | AAG08214.1 | 3.00E−54 |
| 68 | 66429–67739 c | Putative transposase | A. pasteurianus 11380 | 436/461 | 45 (130/284) | BAA00934.1 | 6.00E−60 |
| 69 | 67797–68942 c | Mg/citrate secondary transporter | Bacillus subtilis 6GM | 381/433 | 21 (82/386) | AAC44564.1 | 3.00E−05 |
| 70 | 69053–70021 d | Transposase | R. solanacearum PS68 | 322/321 | 66 (211/318) | AAD49338.1 | 1.00E−117 |
| 71 | 70219–71430 c | N-Isopropylammelide isopropylamino hydrolase (AtzC) | Pseudomonas sp. strain ADP | 403/403 | 100 (403/403) | AAB96621.1 | 0.00 |
| 72 | 71488–72447 c | Carbamoyl phosphate synthetase | Pyrococcus abassy GE5 | 319/314 | 51 (162/315) | AAF23075.1 | 2.00E−86 |
| 73 | 72897–74444 c | Transposase similar to TnpA from IS801 | P. pseudoalcaligenes JS45 | 515/515 | 100 (515/515) | AAB94124.1 | 0.00 |
| 74 | 74260–75759 c | Dihydrolipoamide dehydrogenase (PdhL) | Ralstonia eutropha H16 | 499/594 | 68 (228/335) | AAA21600.1 | 1.00E−122 |
| 75 | 75908–76264 d | Transposase from IS1071 | Alcaligenes sp. strain BR60 | 118/970 | 100 (118/118) | AAA70396.1 | 6.00E−60 |
| 76 | 76280–76915 c | Mating pair formation protein (TrbN) | Enterobacter aerogenes pR751 | 211/211 | 99 (209/211) | AAC64455.1 | 1.00E−120 |
| 77 | 76929–77516 c | Mating pair formation protein (TrbM) | Enterobacter aerogenes pR751 | 195/196 | 90 (178/196) | AAC64454.1 | 1.00E−95 |
| 78 | 77534–79252 c | DNA topoisomerase (TrbL) | Enterobacter aerogenes pR751 | 572/572 | 99 (570/572) | AAC64453.1 | 0.00 |
| 79 | 79263–79490 c | Protein involved in plasmid entry/ exclusion (TrbK) | Enterobacter aerogenes pR751 | 75/75 | 97 (73/75) | AAC64452.1 | 4.00E−37 |
| 80 | 79500–80264 c | Mating pair formation protein (TrbJ) | Enterobacter aerogenes pR751 | 254/254 | 100 (254/254) | AAC64451.1 | 1.00E−139 |
| 81 | 80285–81706 c | Mating pair formation protein (TrbI) | Enterobacter aerogenes pR751 | 473/473 | 99 (471/473) | AAC64450.1 | 0.00 |
| 82 | 81711–82199 c | Mating pair formation protein (TrbH) | Enterobacter aerogenes pR751 | 162/162 | 99 (161/162) | AAC64449.1 | 3.00E−86 |
| 83 | 82202–83122 c | Mating pair formation protein (TrbG) | Enterobacter aerogenes pR751 | 306/306 | 99 (303/306) | AAC64448.1 | 1.00E−173 |
| 84 | 83119–83901 c | Mating pair formation protein (TrbF) | Enterobacter aerogenes pR751 | 260/260 | 99 (258/260) | AAC64447.1 | 1.00E−150 |
| 85 | 83898–86456 c | Mating pair formation protein (TrbE) | Enterobacter aerogenes pR751 | 852/852 | 99 (847/852) | AAC64446.1 | 0.00 |
| 86 | 86453–86764 c | Mating pair formation protein (TrbD) | Enterobacter aerogenes pR751 | 103/103 | 98 (101/103) | AAC64445.1 | 1.00E−53 |
| 87 | 86768–87232 c | Mating pair formation protein (TrbC) | Enterobacter aerogenes pR751 | 154/154 | 99 (152/154) | AAC64444.1 | 6.00E−82 |
| 88 | 87249–88211 c | ATPase autophosphorylase (TrbB) | Enterobacter aerogenes pR751 | 320/320 | 99 (319/320) | AAC64443.1 | 0.00 |
| 89 | 88521–88883 c | Regulatory protein for mating pair formation (TrbA) | Enterobacter aerogenes pR751 | 121/121 | 98 (118/120) | AAC64442.1 | 4.00E−61 |
| 90 | 88997–89338 d | Single-strand DNA-binding protein (SSB) | Enterobacter aerogenes pR751 | 113/113 | 100 (113/113) | AAC64441.1 | 2.00E−61 |
| 91 | 89488–90399 d | DNA-binding protein for plasmid replication (TrfA) | Enterobacter aerogenes pR751 | 303/407 | 96 (292/304) | AAA17040.1 | 1.00E−162 |
| 92 | 90795–92165 c | Putative transposase | Streptococcus pyogenes AP1 | 456/410 | 29 (110/378) | AAC38767.1 | 6.00E−33 |
| 93 | 92490–93821 c | Cytosine deaminase | R. leguminosarum T24 | 443/450 | 32 (141/428) | AAB17512.1 | 5.00E−46 |
| 94 | 94005–94928 c | Putative permease of ABC transporter | P. aeruginosa PAO1 | 327/308 | 55 (171/306) | AAG03528.1 | 3.00E−86 |
| 95 | 94918–96039 c | Putative permease of ABC transporter | P. aeruginosa PAO1 | 373/365 | 52 (187/357) | AAG03527.1 | 1.00E−97 |
| 96 | 96026–97648 c | Putative ATP-binding domain of ABC transporter | P. aeruginosa PAO1 | 540/523 | 58 (289/510) | AE004451.7 | 1.00E−154 |
| 97 | 97655–98737 c | Conserved hypothetical protein | P. aeruginosa PAO1 | 360/365 | 53 (184/347) | AAG03536.1 | 2.00E−99 |
| 98 | 98776–99834 c | Conserved hypothetical protein | P. aeruginosa PAO1 | 352/277 | 38 (98/256) | AAG03623.1 | 2.00E−40 |
| 99 | 99885–100853 c | Putative transcriptional regulator (LysR family) | R. leguminosarum T24 | 322/300 | 39 (116/295) | AAB17511.1 | 4.00E−44 |
| 100 | 101053–102144 d | Cyanuric acid amidohydrolase (AtzD) | Pseudomonas sp. strain 12227 | 363/370 | 58 (212/363) | AAC61577.1 | 1.00E−110 |
| 101 | 102427–103800 d | Nicotinamidase/pyrazinamidase | Mycobacterium smegmatis MC55 | 457/468 | 37 (171/454) | AAC77368.1 | 5.00E−64 |
| 102 | 104283–106100 d | Urea amidolyase | Saccharomyces cerevisiae | 605/1835 | 44 (254/572) | CAA85172.1 | 1.00E−126 |
| 103 | 106158–106941 c | Similar to OrfQ | Pseudomonas sp. strain KHP41 | 260/349 | 36 (70/190) | CAA67456.1 | 2.00E−24 |
| 104 | 107219–108286 d | Putative integrase-like protein | Pseudomonas sp. strain KHP41 | 355/351 | 65 (209/317) | CAA67462.1 | 1.00E−113 |
Letters indicate coding strand: c, complementary strand; d, direct strand.
N/A, not applicable.
Number of amino acids in pADP-1 ORF/number in the closest relative.
Values in parentheses refer to numbers of amino acids with identity per total number examined.
Similarities and differences between pADP-1 and pR751.
Sequence analyses revealed that the regions of pADP-1 containing replication, basic transfer, and maintenance functions were nearly identical to gene regions encoding these functions in the IncPβ plasmid pR751 from Enterobacter aerogenes. The tra and trb operons of pADP-1 and pR751 contain genes necessary for plasmid transfer and replication (59). In addition, the origin of replication and the IncC, KorA, KorB, and Klc proteins, which are involved in the regulation of replication and maintenance of pR751, are highly conserved in pADP-1 (Table 1). These observations suggest that pADP-1 and pR751 have a common ancestor.
Despite having similarity in backbone structure, the regions between oriV and trfA and between trb and tra of plasmids pR751 and pADP-1 are different. In pR751, two transposons, Tn4321 and Tn402/Tn5090, are located in the regions between oriV-trfA and trb-tra, respectively; these transposons are not present in pADP-1. Moreover, in pADP-1 two complete copies of a transposase similar to TnpA from IS801 flank the region between trb and tra. In pADP-1, this 52-kb region resembled a nested transposon structure and contains all of the genes necessary for the degradation of atrazine to cyanuric acid, a complete mercury resistance operon, and several copies of different transposable elements.
Some of the inverted repeats postulated to be involved in the acquisition of foreign DNA in pR751 are also present in pADP-1 (59). All the inverted repeats in the area between oriV and trfA on pR751 except for inverted repeat 4, are present and highly conserved in pADP-1. In addition, the inverted repeats in pR751 between upf54.4 (traO) and traM are present and highly conserved in pADP-1. However, the inverted repeats present in the areas between Tra1-Tn402 and Tn402-Tra2 in pR751 were not found in the pADP-1 genome.
Transposition and integration-related ORFs.
About 15% of the identified ORFs in the pADP-1 sequence had significant identity to proteins involved in DNA integration and transposition. Three ORFs (ORF34, ORF41, and ORF49) in the pADP-1 sequence had >99% amino acid sequence identity to TnpA from IS1071. Two IS1071 insertion elements flank the atrazine chlorohydrolase gene, atzA (ORF37), while the third copy is located approximately 6 kb downstream of the hydroxyatrazine ethylaminohydrolase (atzB) gene (ORF44). A truncated copy of transposase TnpA from IS1071 was localized at nucleotide positions 75908 to 76264 near the trb operon (Fig. 3). The 110-bp inverted repeats normally associated with IS1071 are present in the homologs found on pADP-1. In addition, a partial copy (69 nucleotides) of these inverted repeats was found between the istB and the traC genes. DNA fragments homologous to the pdhL and pdhB genes were found to flank most of the copies of IS1071 present on pADP-1. These genes encode different components of the pyruvate dehydrogenase enzyme complex (26). The IS1071 copies present in pADP-1 (Table 1) always interrupt pdhL at the same position, suggesting that pdhL is a target for the insertion of this transposon.
Four ORFs, ORF32 (nucleotides 27514 to 29061), ORF39 (37037 to 38584), ORF47 (48776 to 50323), and ORF73 (72897 to 74444), had >99% amino acid sequence identity to an IS801-like transposase (TnpA) previously identified in Pseudomonas pseudoalcaligenes JS45 (14). Complete copies of this transposase were identified on the opposite strand flanking the atzA and atzB genes and on the same strand upstream of the atzC gene (ORF71) (Fig. 3). No inverted repeat structures were found in the DNA regions surrounding these transposases. However, 187-bp and 108-bp conserved DNA regions were consistently found, upstream and downstream, respectively, of the IS801-like tnpA genes in pADP-1. These sequences were also found in the same positions in IS801-like genes present in Acidovorax avenae subsp. citrulli (accession number AF086815.2), Pseudomonas huttiensis (accession number AF028594), and Pseudomonas pseudoalcaligenes (14). These conserved regions have 40% identity to each other, do not contain direct or inverted repeats, and have no homology to other well-characterized transposons.
We also identified additional ORFs with homology to transposases from organisms of diverse genera. For example, ORF 68 (nucleotides 66429 to 67739) had 45% identity to a putative transposase from Acetobacter pasteurianus (58), ORF70 (69053 to 70021) had 66% identity to a transposase from Ralstonia solanacearum (Y. A. Lee, unpublished data; GenBank accession no. AAD49338), and ORF92 (90795 to 92165) was 29% identical to a putative transposase from Streptococcus pyogenes (6). In addition, our analysis revealed the presence of ORF104 (107219 to 108286), which had 69% sequence identity to an integrase-like protein from Tn5041 in Pseudomonas sp. strain KHP41 (33). Two ORFs, ORF28 (24327 to 25175) and ORF29 (25159 to 25416), had homology to the IstB and IstA proteins from Ralstonia eutropha, respectively (51). In plasmids carrying the insertion sequence IS21, istAB are usually transcribed as part of an operon (51). In pADP-1, the IstB homolog appears complete, while the IstA homolog (ORF29) is truncated.
The complete nucleotide sequence of pADP-1 revealed the presence of several genes involved in catabolism (Table 1 and Fig. 3). These genes and gene families are discussed below in detail.
Atrazine catabolic genes.
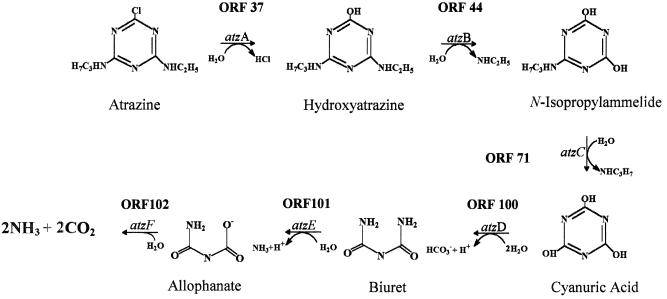
Sequence analysis showed that three previously identified genes involved in the initial steps of atrazine catabolism, atzA, atzB, and atzC (9, 15, 47), are localized to different regions of pADP-1 and are not organized in an operon-like structure. The first enzyme, AtzA, catalyzes the hydrolytic dechlorination of atrazine, yielding hydroxyatrazine. The second enzyme, AtzB, catalyzes hydroxyatrazine deamidation, yielding N-isopropylammelide, and the third, AtzC, encodes N-isopropylammelide isopropylaminohydrolase activity, stoichiometrically catabolizing N-isopropylammelide to cyanuric acid and N-isopropylamine. Consistent with our previously reported cosmid sequencing efforts, the atzA and atzB genes are located about 8 kb apart on pADP-1 (9). The atzC gene is located about 34 and 25 kb from the atzA and atzB genes, respectively. Sequence analysis confirmed that the G+C content of atzC (44%) was lower than those of atzA (58%) and atzB (61%) and most of the other genes present in the pADP-1 sequence. This result suggests that atzC was acquired from an organism with vastly different G+C content (47).
New catabolic genes involved in atrazine catabolism.
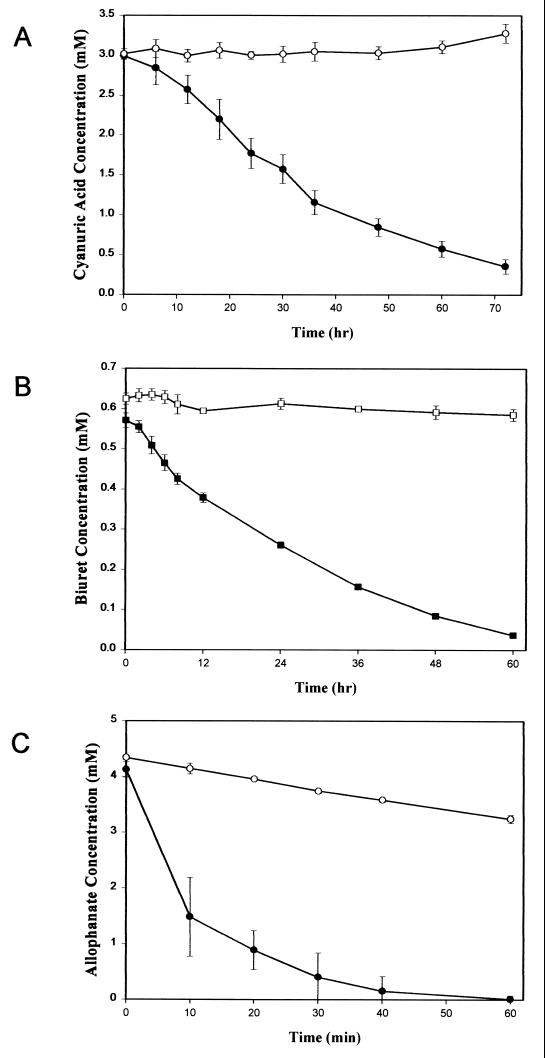
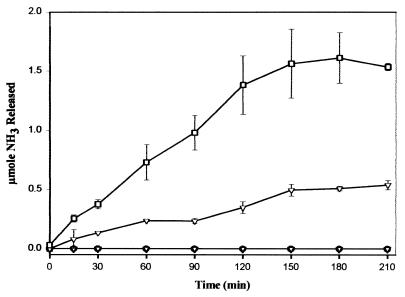
ORF100 (nucleotides 101053 to 102144) had 58% amino acid identity to TrzD, a cyanuric acid amidohydrolase from Pseudomonas sp. strain NRRLB-12227 (30). Since AtzC in Pseudomonas sp. strain ADP transforms N-isopropylammelide to cyanuric acid, we postulated that cyanuric acid was transformed by a similar enzyme in this bacterium. However, we previously were unsuccessful in using a trzD gene probe to find this gene in Pseudomonas sp. strain ADP or to demonstrate transformation of cyanuric acid in E. coli AD256(pADP-1) (M. L. de Souza and I. R. Fruchey, personal communication). To determine whether ORF100 functionally encoded cyanuric acid amidohydrolase in Pseudomonas sp. strain ADP, PCR was used to clone a 1.4-kb DNA fragment from pADP-1 into the BamHI site of pKT230, resulting in plasmid pBMZ1. Crude extracts of E. coli DH5α and E. coli DH5α(pBMZ1) were tested for their ability to hydrolyze cyanuric acid, as evidenced by HPLC analysis. Results in Fig. 4A show that crude extracts of E. coli(pBMZ1) had the ability to hydrolyze cyanuric acid, whereas the control E. coli strain did not transform this substrate. Mass spectrophotometric studies done using crude cell extracts of E. coli(pBMZ1) and cyanuric acid as the substrate indicated that biuret is the product of this reaction (I. R. Fruchey, unpublished data). These results indicate that pADP-1 encodes a functional cyanuric acid amidohydrolase that is homologous to TrzD. This enzyme, AtzD, is encoded in the region between oriV and trfA. We are currently in the process of purifying and characterizing AtzD, and these results will be presented elsewhere.
FIG. 4.
(A) Degradation of cyanuric acid by crude cell extracts from E. coli DH5α(pBMZ1). Plasmid pBMZ1 contains atzD from pADP-1 cloned into the BamHI site of pKT230. Symbols: ●, E. coli DH5α(pBMZ1); ○, E. coli DH5α. (B) Degradation of biuret by crude cell extracts from E. coli DH5α (atzE). Symbols: ▪, E. coli DH5α (atzE); □, E. coli DH5α(pUC18). (C) Degradation of allophanate by crude cell extracts of E. coli DH5α (atzF). Symbols: ●, E. coli DH5α (atzF); ○, E. coli DH5α(pUC18). Values are the means of three replicates. Bars indicate standard deviations of the mean.
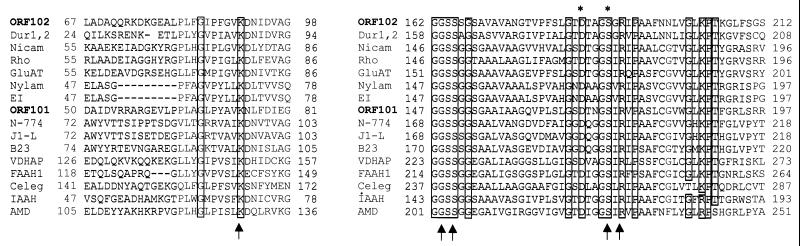
Two ORFs, ORF101 and ORF102, were located directly downstream of atzD. ORF101 (102427 to 103800) had 37% sequence identity with a nicotinamidase/pyrazinamidase from Mycobacterium smegmatis (8) (GenBank accession no. AAC77368), while ORF102 (104283 to 106100) had 44% sequence identity to urea amidolyase from Saccharomyces cerevisiae (GenBank accession no. CAA85172) (24, 57, 63). Multiple sequence alignments revealed that ORF101 and ORF102 had conserved sequence signatures diagnostic of an amidase protein family (Fig. 5) (34, 43). The residues corresponding to Asp184 and Ser188 in ORF102 and Asp169 and Ser173 in ORF101 are conserved in all the members of this amidase family. In addition, the residues corresponding to Lys90, Ser164, Ser165, Ser188, and Arg190 in ORF102 and Lys73, Ser149, Ser150, Ser173, and Arg175 in ORF101 are conserved in all the members of this same amidase family (34, 43). Mutagenesis studies have shown that these five residues are critical for the amidase activity of fatty acid amide hydrolase, a mammalian amidase that hydrolyses fatty acid amides (43). Based on our observations, we propose here that ORF101 and ORF102 are new members of this amidase family.
FIG. 5.
Sequence alignment of ORF101 and ORF102 with members of the amidase protein family. Identical amino acids are boxed. The arrows indicate residues that have been shown to be important for amidase activity in a fatty acid amide hydrolase (FAAH). Residues common to the amidases and aspartic proteases are denoted by the asterisks. The amidase sequences used for the alignment were as follows: ORF102 from the pADP-1 plasmid sequence; Dur (1, 2), urea amidolyase from Saccharomyces cerevisiae (accession number CAA85172); Nicam, nicotinamidase from Mycobacterium smegmatis (accession number AAC77368.1); Rho, amidase from Rhodococcus (accession number M74531); GluAT, Glu-tRNA amidotransferase from Bacillus subtilis (accession number gi:25899195); Nylam, 6-aminohexanoate cyclic dimer hydrolase, Flavobacterium sp. (accession number gi:148711), EI, 6-aminohexanoate-cyclic-dimer hydrolase from Flavobacterium sp. (accession number M26953); ORF101 from the pADP-1 plasmid sequence; N-774, amidase from Rhodoccocus sp. strain N-774 (accession number X54074); J1-L, amidase from Rhodococcus rhodochrous J1 (accession number D16207); B23, amidase from Pseudomonas chlororaphis (accession number D90216); VDHAP, vitamin D3 hydroxylase-associated protein, Gallus domesticus (accession number gi:1079452); FAAH1, fatty acid amide hydrolase, Rattus norvegicus (accession number gi:1680722); Celeg, predicted amidase from Caenorhabditis elegans (accession number gi:6425411); IAAH, indoleacetamide hydrolase, Pseudomonas syringae (accession number gi:77820); AMD, acetamidase, Emericella nidulans (accession number gi:101782).
Growth studies indicated that Pseudomonas putida PRS2000(pADP-1) transconjugants had the ability to use cyanuric acid as the sole nitrogen source (data not shown), while the wild-type strain did not. This result indicated that the gene(s) necessary for further catabolism of cyanuric acid was present on pADP-1. Since biuret is the product of cyanuric acid degradation in Pseudomonas sp. strains ADP and NRRLB-12227 and contains multiple amide bonds, the data suggested that ORF101 and ORF102 were involved in biuret metabolism. To experimentally determine whether these ORFs encoded a functional biuret amidohydrolase, E. coli(p11A07) and E. coli(p14D12), containing ORF101 and ORF102, respectively, were spotted onto the surface of LB agar medium containing 4,000 μg of biuret per ml. While a clear zone surrounding cell growth of E. coli DH5α(p11A07) was found after 7 days of incubation, no such zone was detected with E. coli(p14D12) or the E. coli(pUC18) negative control (data not shown). To confirm these results, cell extracts from E. coli DH5α(p11A07) were examined for biuret hydrolysis using HPLC and ammonia release assays. HPLC analyses showed that crude extracts of E. coli DH5α(p11A07) hydrolyzed biuret, while E. coli DH5α(pUC18) did not degrade this substrate (Fig. 4B). Ammonia release assays showed that ORF101 hydrolyzed biuret with a specific activity of approximately 3 nmol of ammonia per min per mg of protein (data not presented). The product(s) of biuret hydrolysis by ORF101 was examined by using TLC and HPLC analyses. Allophanate was detected by both methods, but only in the reaction mixtures containing biuret and crude-cell extracts from E. coli DH5α(p11A07) (data not shown). Taken together, these results indicate that ORF101 encodes a functional biuret hydrolase that is part of the atrazine degradation pathway present in Pseudomonas sp. strain ADP. Based on these observations, ORF101 was designated atzE.
ORF102 (nucleotides 104283 to 106100), located downstream of atzE, was found to have 44% sequence identity to the allophanate hydrolase domain of urea amidolyase from S. cerevisiae. To determine if ORF102 was also involved in the atrazine degradation pathway, we examined E. coli(p14D12), containing a complete copy of ORF102, for its ability to hydrolyze urea or allophanate. HPLC results in Fig. 4C show that cell extracts from E. coli(p14D12) had allophanate hydrolase activity. In contrast, only limited loss of allophanate was seen with extracts from E. coli(pUC18), most likely due to the long assay time and well-known instability of allophanate in aqueous solution (63). Based on this result, ORF102 was designated atzF. Ammonia release assays indicated that cell extracts of E. coli(p14D12) failed to hydrolyze urea or biuret (Fig. 6).
FIG. 6.
Ammonia released after incubation of biuret with cell extracts from E. coli DH5α (atzE) and E. coli DH5α (atzF). Symbols: □, 3 mM biuret plus crude cell extracts from E. coli DH5α (atzE) and E. coli DH5α (atzF); ▿, 3 mM biuret plus cell extract from E. coli DH5α (atzE); ▾, 3 mM urea plus cell extract from E. coli DH5α (atzF); ○, 3 mM biuret plus cell extract from E. coli DH5α (atzF); ●, 3 mM biuret plus cell extract from E. coli DH5α(pUC18). Values are the means of results from three replicates. Error bars indicate standard deviations of the means.
The results in Fig. 6 also show that a cell extract from E. coli(p11A07) (producing AtzE) incubated with biuret released three times more ammonia when combined with E. coli(p14D12) (producing AtzF) than in reactions containing only biuret hydrolase. Since AtzF does not hydrolyze biuret or urea, these results confirm that allophanate is the product of biuret hydrolysis by AtzE. Taken together, these results indicate that all genes necessary for the catabolism of atrazine to ammonia and carbon dioxide are localized on pADP-1 and that the last three genes in the atrazine degradation pathway, atzD, atzE, and atzF, are located in a contiguous cluster in the pADP-1 plasmid.
ORF99 (nucleotides 99885 to 100853) had 39% identity to members of the LysR family of transcriptional regulators (50). ORF99 is divergently transcribed from atzD and located 166 bp from the start codon of this gene. Moreover, ORF99, atzD, atzE, and atzF have similar mol% G+C contents (59 to 61%) and codon usage, suggesting that these four genes constitute an operon-like structure on pADP-1. To determine whether the atzD, atzE, and atzF genes are transcribed as a single mRNA, we used RT-PCR and primers designed to amplify contiguous cDNAs between atzD and atzE and from atzE to atzF. RT-PCR analyses showed that a single transcript of 2.3 kb was obtained using the atzD and atzE primers and a 1.1-kb transcript was obtained using the atzE and atzF primer pair (data not shown). A single mRNA from atzD to atzF would be approximately 5.2 kb and difficult to amplify. Consequently, two overlapping primer pairs were used. Since these two cDNAs were derived from a contiguous DNA region containing atzDEF, this result indicates that these three genes are cotranscribed as a single mRNA in Pseudomonas sp. strain ADP.
Other regulatory proteins.
Several other ORFs with homology to transcriptional regulators were also identified in different regions of the pADP-1 sequence. ORF45 (nucleotides 45919 to 46494) had 30% identity to the transcriptional regulators of the Tet/Acr family (2, 42). ORF66 (65433 to 65999), which is located approximately 4.2 kb downstream of the atzC gene, had 67% identity to a putative transcriptional regulator from Pseudomonas aeruginosa. Domain analysis done using Pfam showed that this ORF was related to members of the Tetr family of transcriptional regulators (2, 42).
Mercury resistance genes.
A DNA region with high homology to genes and an operon involved in mercury resistance was identified in the pADP-1 sequence (ORFs 50 to 57, nucleotides 55067 to 59386). This approximately 4.3-kb DNA region contains eight ORFs, each of which had >99% sequence identity to proteins from the mercury resistance operon identified in Xanthomonas sp. strain W17 (32) (Table 1). The functionality of the putative mercury resistance operon in pADP-1 was determined by measuring mercuric reductase activity in crude extracts of E. coli(pADP-1) strains and by determining growth in LB medium containing different concentrations of HgCl2. Mercuric reductase activity in E. coli AD256(pADP-1) and E. coli S17-1(pADP-1) was 301 ± 18 and 387 ± 33 nmol/min/mg of protein, respectively, while the specific activity of this enzyme was 10-fold lower in the control strains (21 ± 4 and 30 ± 2 nmol/min/mg of protein for E. coli S17-1 and E. coli AD256, respectively). In addition, results from the disk diffusion assays indicated that while E. coli AD256(pADP-1) and E. coli S17-1(pADP-1) were resistant to greater than 200 μg of HgCl2 per ml (inhibition zone < 1 mm), both parental control strains were inhibited for growth by 100 μg of HgCl2 per ml (inhibition zone of 2.0 mm), and at 200 μg of HgCl2 per ml, inhibition zones were 4.0 and 6.0 mm for E. coli AD256 and E. coli S17-1, respectively (data not shown). Taken together, these results showed that the mercury resistance operon present in pADP-1 is functional and confers on strains the ability to detoxify mercury.
Transporters.
A cluster of type ABC transporters was identified on pADP-1. ORF94 (nucleotides 94005 to 94928) and ORF95 (94918 to 96039) are homologous to permeases of type ABC transporters from Pseudomonas aeruginosa (55) (GenBank accession no. AAG03528 and AAG03527, respectively), while ORF96 (96026 to 97648) had 58% identity to the ATP-binding component of the same type ABC transporter (GenBank accession no. AE004451). In addition, two ORFs (ORF46 and ORF69) had homology to a hypothetical protein and a secondary magnesium/citrate transporter, respectively. These ORFs were classified as transporters based on results obtained from domain searches done using Pfam. ORF46 (46883 to 48223) showed a significant match to a xanthine/uracil permease family of proteins, having many residues conserved in the signature sequence of this family of proteins (1, 19). In addition, hydrophobicity plots of ORF46 predicted 12 transmembrane domains, a characteristic that is commonly observed in the xanthine/uracil permease family of proteins (1, 19). Similarly, hydrophobicity plots of ORF69 (67797 to 68942) showed 12 membrane-spanning domains, and BLAST and Pfam searches showed predicted homology to a variety of transporters.
Hypothetical proteins.
Approximately 8% of the predicted ORFs on pADP-1 sequence had homology to conserved hypothetical proteins identified in other genomes, especially to hypothetical proteins from Pseudomonas aeruginosa. In an attempt to identify the potential function of these predicted ORFs, sequences were analyzed to identify motifs and conserved sequence domains. ORF58 (nucleotides 59783 to 60556), ORF59 (60661 to 61119), ORF61 (62296 to 62769), ORF63 (63790 to 64593), ORF97 (97655 to 98737), and ORF98 (98776 to 99714) had no significant matches to proteins present in the Prosite, Pfam, PRINTS, and BlOCKS databases. Therefore, the function of these proteins remains unknown. However, ORF62 (62824 to 63771) and ORF30 (25315 to 26307) had a low percentage of identity to many membrane-bound proteins and transporters. The hydrophobicity plots of these ORFs predicted 6 to 10 transmembrane domains and suggested that these ORFs may be membrane-bound proteins or transport proteins. About 2% of the ORFs had no significant homology to any known proteins in the databases.
DISCUSSION
In this study we describe the complete nucleotide sequence of catabolic plasmid pADP-1 from a Pseudomonas sp. strain ADP. One of the most remarkable features of plasmid pADP-1 is its high degree of relatedness to the 53.3-kb broad-host-range IncPβ plasmid pR751, initially isolated from Enterobacter aerogenes (59). Nearly one-half of pADP-1 consists of the pR751 backbone (Fig. 2). Members of the IncP plasmid group have been shown to be highly conserved despite the fact that they have been isolated from diverse genera of bacteria in different parts of the world (11, 54, 60). Antibiotic resistance, mercury resistance, and other catabolic genes have previously been identified on diverse IncP plasmids (11, 54). Burlage et al. (11) reported that a 30-kb DNA fragment from pR751 was conserved in the catabolic plasmids pJP4, pSS50, and pSS60. Moreover, antibiotic resistance plasmids pR906 and pR772 have several loci identical to those found in pR751 (54). Similarly, Tralau and coworkers (60) mapped pTSA from Comamonas testosteroni T2, encoding p-toluenesulfonate degradation, and reported that the 72.4-kb catabolic plasmid also had a similar backbone structure.
The region of pADP-1 between the tra and trb operons (nucleotide positions 24062 to 76280) contains the atzA, atzB, and atzC genes. Since transposases and gene cassettes that resemble nested catabolic transposons flank atzA, atzB, and atzC, the atrazine catabolic genes may have been recruited at different times through independent transposition or homologous recombination events. Previously, transposase proteins from IS1071 were shown to participate in DNA insertion and recombination events in the chlorobenzoate catabolic plasmid pBR60 (39), and the operon for p-toluenesulfonate catabolism in Comamonas testosteroni has been shown to be part of a composite transposon flanked by two IS1071 elements (60).
While our previous data suggested that the gene for cyanuric degradation was not located on pADP-1 (17), in the present study we show, by sequence and functional analysis, that atzD (ORF100) hydrolyzes cyanuric acid to biuret (Fig. 4A and 7). AtzD has 58% sequence identity to TrzD from Pseudomonas sp. strain NRRLB-l2227 (30). TrzD, which is functional in the melamine catabolic pathway, also hydrolyzes cyanuric acid (21, 22). Recently, trzD was localized to a self-transmissible IncIα plasmid, pPDL12, in Klebsiella pneumoniae (31).
FIG. 7.
Complete catabolic pathway for atrazine degradation by Pseudomonas sp. strain ADP. Genes and potential ORFs involved at each catabolic step are indicated.
Results from our cloning and functional studies show that two amidases, AtzE (ORF101) and AtzF (ORF102), are involved in the further catabolism of biuret to CO2 and NH3 (Fig. 7). These proteins are members of the same amidase family. AtzE (nucleotides 102427 to 103800) had 37% sequence identity to a nicotinamidase/pyrazinamidase from Mycobacterium smegmatis and was shown by three independent methods to encode a biuret hydrolase, resulting in the formation of allophanate. Previously, biuret hydrolases have been reported in cyanuric acid-degrading bacteria (13, 21, 22).
The functional studies reported here indicate that AtzF hydrolyzes allophanate to NH3 and CO2. AtzF (nucleotides 104283 to 106100) had 44% sequence identity to the allophanate hydrolase domain of urea amidolyase from S. cerevisiae (24, 57, 63). This domain has been shown to catalyze the hydrolysis of allophanate to 2 mol each of ammonia and carbon dioxide (63). While genome sequencing efforts have identified homologs to the S. cerevisiae allophanate hydrolase gene in Bacillus subtilis and Campylobacter jejuni (GenBank accession no. AL139078 and Z99106, respectively), the amino acid identity of these proteins to AtzF was low (20 and 25%, respectively). To our knowledge, this is the first report of the functional characterization of an allophanate hydrolase from a bacterium.
Previously, Cook et al. (13) reported that urea was the product of biuret degradation in Pseudomonas sp. strain NRRLB-12228 (strain D) and that Klebsiella pneumoniae strain 99 had negligible allophanate amidohydrolase activity. Likewise, Eaton and Karns (21, 22) reported that cyanuric acid was hydrolyzed to biuret and urea in Pseudomonas sp. strain NRRLB-12227. However, these metabolites were identified by using HPLC analyses at pH 2.0, precluding detection of the acid-labile allophanate which decomposes to urea.
The atzD (ORF100), atzE (ORF101), and atzF (ORF102) genes form a gene cluster, 166 bp from the divergently transcribed ORF99. RT-PCR analyses indicated that atzDEF are cotranscribed on a single mRNA. This physical relationship suggests that ORF99 may regulate atzDEF. ORF99 is predicted to encode a LysR-type transcriptional regulator and contains a DNA-binding helix-turn-helix motif at its N-terminal domain (amino acids 26 to 168). Moreover, the intergenic region between ORF99 and atzD (166 bp upstream of atzD) contains a T-N11-A DNA motif (at positions 100875 to 100887) as part of an interrupted dyad sequence. This motif has been found upstream of most LysR-type-regulated genes (50), suggesting that ORF99 may regulate the atzDEF genes. The exact regulatory function of ORF99 awaits further study.
While our data indicate that atzDEF are organized in an operon-like structure, the atzA, atzB, and atzC genes are dispersed on plasmid pADP-1 and flanked by many transposable elements (Fig. 2 and 3). Moreover, there is no sequence evidence for regulatory elements upstream of the atzA, atzB, or atzC genes, and previous Northern hybridization studies have shown that in Pseudomonas sp. strain ADP, the atzA and atzB genes are transcribed in the absence of atrazine or hydroxyatrazine as inducers, respectively (M. L. de Souza, unpublished data). Taken together, these observations are consistent with the idea that the constitutively expressed atzA, atzB, and atzC genes were recently acquired by Pseudomonas sp. strain ADP and other soil bacteria. We previously reported that pADP-1 is self-transmissible to gram-negative bacteria (17) and that several genera of bacteria isolated from different geographical regions contain nearly identical atrazine catabolism genes (16). While direct plasmid transfer may appear to be a plausible mechanism for the dissemination of atrazine genes among these bacteria, we have reported that the atzA, atzB, and atzC genes are localized to different-sized plasmids in phylogenetically diverse microorganisms (B. Martinez, M. de Souza, L. Wackett, and M. Sadowsky, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. Q352, 1999). Moreover, variants of Pseudomonas sp. strain ADP have been identified that contain the atzB and atzC but not the atzA gene (M. L. de Souza, unpublished data), some wild-type bacteria contain only the atzA gene (10), and individual bacteria in an atrazine-metabolizing consortium each contain an incomplete set of atrazine catabolism genes (18). Taken together, these data suggest that the atrazine catabolic genes can be recruited or lost independently and that direct plasmid transfer may not be the only mechanism by which atrazine degradation genes are disseminated in the environment.
In summary, the plasmid sequencing studies reported here revealed that all the genes for the complete metabolism of atrazine are localized on pADP-1 in Pseudomonas sp. strain ADP. Moreover, we show that atrazine catabolism in Pseudomonas sp. strain ADP occurs via a novel pathway involving an allophanate intermediate. These findings provide a window onto the apparent recent evolution of catabolic pathways in nutritionally diverse soil bacteria.
ACKNOWLEDGMENTS
This work was supported, in part, by grant 98-35107-6368 from the U.S. Department of Agriculture–NRI/CGP/CSREES, by a grant from Syngenta Crop Protection, Greensboro, N.C., and by the University of Minnesota Agricultural Experiment Station.
We thank Janis McFarland from Syngenta Crop Protection for providing substrates and Becky Parales for providing Pseudomonas putida PRS2000(pADP-1). We also thank Isaac Fruchey for help with atzD and Sung-Sick Woo and Yeisoo Yu for help with sequencing and the sequence assembly software. We also thank Claire Fant and Megan Bruce-Carver for technical assistance in the sequencing laboratory, Lynda Ellis for assistance with open reading frame analyses, Gil Johnson and Jack Richman for the synthesis and NMR analysis of allophanate, and Charlotte Rosendahl for helpful discussions.
REFERENCES
- 1.Andersen P S, Frees D, Fast R, Mygind B. Uracil uptake in Escherichia coli K-12: isolation of uraAmutants and cloning of the gene. J Bacteriol. 1995;177:2008–2013. doi: 10.1128/jb.177.8.2008-2013.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aramaki H, Yagi N, Suzuki M. Residues important for the function of a multihelical DNA binding domain in the new transcription factor family of Cam and Tet repressors. Protein Eng. 1995;8:1259–1266. doi: 10.1093/protein/8.12.1259. [DOI] [PubMed] [Google Scholar]
- 3.Austen R A, Dunn N W. Isolation of mutants with altered metabolic control of the NAH plasmid-encoded catechol meta-cleavage pathway. Aust J Biol Sci. 1977;30:583–592. doi: 10.1071/bi9770583. [DOI] [PubMed] [Google Scholar]
- 4.Bagdasarian M, Lurz R, Ruckert B, Franklin F C, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 5.Berg T, Firth N, Apisiridej S, Hettiaratchi A, Leelaporn A, Skurray R A. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J Bacteriol. 1998;180:4350–4359. doi: 10.1128/jb.180.17.4350-4359.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berge A, Rasmussen M, Bjorck L. Identification of an insertion sequence located in a region encoding virulence factors of Streptococcus pyogenes. Infect Immun. 1998;66:3449–3453. doi: 10.1128/iai.66.7.3449-3453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borodovsky M, McIninch J. GeneMark: parallel gene recognition for both DNA strands. Comput Chem. 1993;17:123–133. [Google Scholar]
- 8.Boshoff H I M, Mizrahi V. Purification, gene cloning, targeted knockout, overexpression, and biochemical characterization of the major pyrazinamidase from Mycobacterium smegmatis. J Bacteriol. 1998;180:5809–5814. doi: 10.1128/jb.180.22.5809-5814.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boundy-Mills K L, de Souza M L, Mandelbaum R T, Wackett L P, Sadowsky M J. The atzB gene of Pseudomonassp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway. Appl Environ Microbiol. 1997;63:916–923. doi: 10.1128/aem.63.3.916-923.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouquard C, Quazzani J, Prome J C, Briand Y M, Plesiat P. Dechlorination of atrazine by a Rhizobiumisolate. Appl Environ Microbiol. 1997;63:862–866. doi: 10.1128/aem.63.3.862-866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burlage R S, Bemis L A, Layton A C, Sayler G S, Larimer F. Comparative genetic organization of incompatibility group P degradative plasmids. J Bacteriol. 1990;172:6818–6825. doi: 10.1128/jb.172.12.6818-6825.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhry G R, Huang G H. Isolation and characterization of a new plasmid from a Flavobacteriumsp. which carries the genes for degradation of 2,4-dichlorophenoxy-acetate. J Bacteriol. 1988;170:3897–3902. doi: 10.1128/jb.170.9.3897-3902.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook A M, Beilstein P, Grossenbacher H, Hutter R. Ring cleavage and degradative pathway of cyanuric acid in bacteria. Biochem J. 1985;231:25–30. doi: 10.1042/bj2310025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis J K, Paoli G C, He Z, Nadeau L J, Somerville C C, Spain J C. Sequence analysis and initial characterization of two isozymes of hydroxylaminobenzene mutase from Pseudomonas pseudoalcaligenesJS45. Appl Environ Microbiol. 2000;66:2965–2971. doi: 10.1128/aem.66.7.2965-2971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Souza M L, Wackett L P, Boundy-Mills K L, Mandelbaum R T, Sadowsky M J. Cloning, characterization, and expression of a gene region from Pseudomonassp. strain ADP involved in the dechlorination of atrazine. Appl Environ Microbiol. 1995;61:3373–3378. doi: 10.1128/aem.61.9.3373-3378.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Souza M L, Sadowsky M J, Seffernick J, Martinez B, Wackett L P. The atrazine catabolism genes are widespread and highly conserved. J Bacteriol. 1998;180:1951–1954. doi: 10.1128/jb.180.7.1951-1954.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Souza M L, Wackett L P, Sadowsky M J. The atzABC genes encoding atrazine catabolism are located on a self-transmissible plasmid in Pseudomonassp. strain ADP. Appl Environ Microbiol. 1998;64:2323–2326. doi: 10.1128/aem.64.6.2323-2326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Souza M L, Newcombe D, Alvey S, Crowley D E, Hay A, Sadowsky M J, Wackett L P. Molecular basis of a bacterial consortium: interspecies catabolism of atrazine. Appl Environ Microbiol. 1998;64:178–184. doi: 10.1128/aem.64.1.178-184.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diallinas G, Gorfinkiel L, Arst H N, Cecchetto G, Scazzocchio C. Genetic and molecular characterization of a gene encoding a wide specificity purine permease of Aspergillus nidulansreveals a novel family of transporters conserved in prokaryotes and eukaryotes. J Biol Chem. 1995;270:8610–8622. doi: 10.1074/jbc.270.15.8610. [DOI] [PubMed] [Google Scholar]
- 20.Dougherty B A, Hill C, Weidman J F, Richardson D R, Venter J C, Ross R P. Sequence and analysis of the 60 kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactisDPC3147. Mol Microbiol. 1998;29:1029–1038. doi: 10.1046/j.1365-2958.1998.00988.x. [DOI] [PubMed] [Google Scholar]
- 21.Eaton R W, Karns J S. Cloning and analysis of s-triazine catabolic genes from Pseudomonassp. strain NRRLB-12227. J Bacteriol. 1991;173:1215–1222. doi: 10.1128/jb.173.3.1215-1222.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eaton R W, Karns J S. Cloning and comparison of the DNA encoding ammelide aminohydrolase and cyanuric acid amidohydrolase from three s-triazine-degrading bacterial strains. J Bacteriol. 1991;173:1363–1366. doi: 10.1128/jb.173.3.1363-1366.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox B, Walsh C T. Mercury reductase: purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J Biol Chem. 1982;257:2498–2503. [PubMed] [Google Scholar]
- 24.Genbauffe F S, Cooper T G. The urea amidolyase (DUR1,2) of Saccharomyces cerevisiae. DNA Sequence. 1991;2:19–32. doi: 10.3109/10425179109008435. [DOI] [PubMed] [Google Scholar]
- 25.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 26.Hein S, Steinbuchel A. Biochemical and molecular characterization of the Alcaligenes eutrophuspyruvate dehydrogenase complex and identification of a new type of dihydrolipoamide dehydrogenase. J Bacteriol. 1994;176:4394–4408. doi: 10.1128/jb.176.14.4394-4408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsch P R, van Montagu M, Johnston A W B, Brewin N J, Schell J. Physical Identification of bacteriocinogenic, nodulation, and other plasmids in strains of Rhizobium leguminosarum. J Gen Microbiol. 1980;120:403–412. [Google Scholar]
- 28.Holben W E, Harris D. DNA-based monitoring of total bacteria community structure in environmental samples. Mol Ecol. 1995;4:627–631. doi: 10.1111/j.1365-294x.1995.tb00263.x. [DOI] [PubMed] [Google Scholar]
- 29.Hu P, Elliot J, McCready P, Skowronski E, Garnes J, Kobayashi A, Brunaker R R, Garcia E. Structural organization of virulence-associated plasmids of Yersinia pestis. J Bacteriol. 1998;180:5192–5202. doi: 10.1128/jb.180.19.5192-5202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karns J. Gene sequence and properties of a s-traizine ring-cleavage enzyme from Pseudomonassp. strain NRRLB-12227. Appl Environ Microbiol. 1999;65:3512–3517. doi: 10.1128/aem.65.8.3512-3517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karns J S, Eaton R W. Genes encoding s-triazine degradation are plasmid-borne in Klebsiella pneumoniaestrain 99. J Agric Food Chem. 1997;45:1017–1022. [Google Scholar]
- 32.Kholodii G Y, Yurieva O V, Lomovskaya O L, Gorlenko Z H M, Mindlin S Z, Nikiforov V G. Tn5053, a mercury resistance transposon with integron's ends. J Mol Biol. 1993;230:1103–1107. doi: 10.1006/jmbi.1993.1228. [DOI] [PubMed] [Google Scholar]
- 33.Kholodii G Y, Mindilin S Z, Gorlenko Z M, Bass I A, Kalyaeva E S, Nikiforov V. Molecular genetic analysis of the Tn5041transposition system. Russ J Genet. 2000;36:365–373. [PubMed] [Google Scholar]
- 34.Kobayashi M, Fujiwara Y, Goda M, Komeda H, Shimizu S. Identification of active sites in amidase: evolutionary relationship between amide bond-and peptide bond-cleaving enzymes. Proc Natl Acad Sci USA. 1997;94:11986–11991. doi: 10.1073/pnas.94.22.11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobel M, Elsener M. Determination of urea and its thermal decomposition products by high performance liquid chromatography. J Chromatogr A. 1995;689:164–169. [Google Scholar]
- 36.Lederberg J. Cell genetics and hereditary symbiosis. Physiol Rev. 1952;32:403–430. doi: 10.1152/physrev.1952.32.4.403. [DOI] [PubMed] [Google Scholar]
- 37.Lukashin A V, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandelbaum R T, Allan D L, Wackett L P. Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl Environ Microbiol. 1995;61:1451–1457. doi: 10.1128/aem.61.4.1451-1457.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakatsu C, Ng J, Singh R, Strais N, Wyndham C. Chlorobenzoate catabolic transposon Tn5271is a composite class I element with flanking class II insertion sequences. Proc Natl Acad Sci USA. 1991;88:8316–8321. doi: 10.1073/pnas.88.19.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nusslein K, Tiedje J M. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl Environ Microbiol. 1998;64:1283–1289. doi: 10.1128/aem.64.4.1283-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okinaka R T, Cloud K, Hampton O, Hoffmaster A R, Hill K K, Keim P, Koehler T M, Lamke G, Kumano S, Mahillon J, Manter D, Martinez Y, Ricke D, Svensson R, Jackson P J. Sequence and organization of pX01: the large Bacillus anthracisplasmid harboring the anthrax toxin genes. J Bacteriol. 1999;181:6509–6515. doi: 10.1128/jb.181.20.6509-6515.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pabo C O, Sauer R T. Transcriptional factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 43.Patricelli M P, Cravatt B F. Clarifying the catalytic roles of conserved residues in the amidase signature family. J Biol Chem. 2000;275:19177–19184. doi: 10.1074/jbc.M001607200. [DOI] [PubMed] [Google Scholar]
- 44.Radosevich M, Traina S M, Hao Y L, Tuovinen O H. Degradation and mineralization of atrazine by a soil bacteria isolate. Appl Environ Microbiol. 1995;61:297–302. doi: 10.1128/aem.61.1.297-302.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rheinwald J G, Chakrabarty A M, Gunsalus I C. A transmissible plasmid controlling camphor oxidation in Pseudomonas putida. Proc Natl Acad Sci USA. 1973;70:885–889. doi: 10.1073/pnas.70.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romine M F, Stillwell L C, Wong K K, Thurston S J, Sisk E C, Sensen C, Gaasterland T, Fredrickson J K, Saffer J D. Complete sequence of 184-kilobase catabolic plasmid from Sphingomonas aromaticivoransF199. J Bacteriol. 1999;181:1585–1602. doi: 10.1128/jb.181.5.1585-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadowsky M J, Tong Z, de Souza M, Wackett L P. AtzC is a new member of the amidohydrolase protein superfamily and is homologous to other atrazine-metabolizing enzymes. J Bacteriol. 1998;180:152–158. doi: 10.1128/jb.180.1.152-158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Sayler G S, Hooper S W, Layton A C, King J M. Catabolic plasmids of environmental and ecological significance. Microb Ecol. 1990;19:1–20. doi: 10.1007/BF02015050. [DOI] [PubMed] [Google Scholar]
- 50.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 51.Schmid S, Seitz T, Hass D. Cointegrase, a naturally occurring, truncated form of IS21 transposase, catalyzes replicon fusion rather than simple insertion of IS21. J Mol Biol. 1998;282:571–583. doi: 10.1006/jmbi.1998.2041. [DOI] [PubMed] [Google Scholar]
- 52.Sherburn C K, Lawley T D, Gilmour M W, Blattner F R, Burland V, Grotbeck E, Rose D J, Taylor D. The complete nucleotide sequence and analysis of R27, a large IncHI plasmid from Salmonella typhithat is temperature sensitive for transfer. Nucleic Acids Res. 2000;28:2177–2186. doi: 10.1093/nar/28.10.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivogenetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 54.Smith C M, Thomas C M. Comparison of the organisation of the genomes of phenotypically diverse plasmids of incompatibility group P: members of the IncPβ subgroup are closely related. Mol Gen Genet. 1987;206:419–427. doi: 10.1007/BF00428881. [DOI] [PubMed] [Google Scholar]
- 55.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K, Wu Z, Paulsen I T. Complete genome sequence of Pseudomonas aeruginosaPAO1: an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 56.Summers W. A simple method for extraction of RNA from E. coliutilizing diethylpyrocarbonate. Anal Biochem. 1970;33:459–463. doi: 10.1016/0003-2697(70)90316-7. [DOI] [PubMed] [Google Scholar]
- 57.Sumrada R A, Cooper T G. Urea carboxylase and allophanate hydrolase are components of multifunctional protein in yeast. J Biol Chem. 1982;257:9119–9127. [PubMed] [Google Scholar]
- 58.Takemura H, Horinouchi S, Beppu T. Novel insertion sequence IS1380 from Acetobacter pasteurianusis involved in the loss of ethanol-oxidizing ability. J Bacteriol. 1991;173:7070–7076. doi: 10.1128/jb.173.22.7070-7076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thorsted P B, Marcarteney D P, Akhtar P, Haines A S, Ali N, Davidson P, Stafford T, Pocklington M J, Pansegrau W, Wilkings B M, Lanka E, Thomas C. Complete sequence of the IncPβ plasmid R751: implications for the evolution and organisation of the IncP backbone. J Mol Biol. 1998;282:969–990. doi: 10.1006/jmbi.1998.2060. [DOI] [PubMed] [Google Scholar]
- 60.Tralau T, Cook A M, Ruff J. Map of the IncP1β plasmid pTSA encoding the widespread genes (tsa) for p-toluenesulfonate degradation in Comamonas testosteroniT2. Appl Environ Microbiol. 2001;67:1508–1516. doi: 10.1128/AEM.67.4.1508-1516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weatherburn M W. Phenol-hypochlorite reaction for the determination of ammonia. Anal Chem. 1967;39:971–974. [Google Scholar]
- 62.Weiss A A, Murphy S D, Silver S. Mercury and organomercurial resistances determined by plasmids in Staphylococcus aureus. J Bacteriol. 1977;122:197–208. doi: 10.1128/jb.132.1.197-208.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitney P A, Cooper T G. Urea carboxylase and allophanate hydrolase. Two components of adenosine triphosphate:urea amidolyase in Saccharomyces cerevisiae. J Biol Chem. 1972;247:1349–1353. [PubMed] [Google Scholar]
- 64.Williams P A, Murray K. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida(arvilla) mt-2: evidence for the existence of a TOL plasmid. J Bacteriol. 1974;1:416–423. doi: 10.1128/jb.120.1.416-423.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]