Abstract
In the field of lipid research, bioorthogonal chemistry has made the study of lipid uptake and processing in living systems possible, whilst minimising biological properties arising from detectable pendant groups. To allow the study of unsaturated free fatty acids in live cells, we here report the use of sterculic acid, a 1,2‐cyclopropene‐containing oleic acid analogue, as a bioorthogonal probe. We show that this lipid can be readily taken up by dendritic cells without toxic side effects, and that it can subsequently be visualised using an inverse electron‐demand Diels–Alder reaction with quenched tetrazine‐fluorophore conjugates. In addition, the lipid can be used to identify changes in protein oleoylation after immune cell activation. Finally, this reaction can be integrated into a multiplexed bioorthogonal reaction workflow by combining it with two sequential copper‐catalysed Huisgen ligation reactions. This allows for the study of multiple biomolecules in the cell simultaneously by multimodal confocal imaging.
Keywords: Click Chemistry, Cyclopropene, Inverse Electron-Demand Diels–Alder Reaction, Lipids, Sterculic Acid
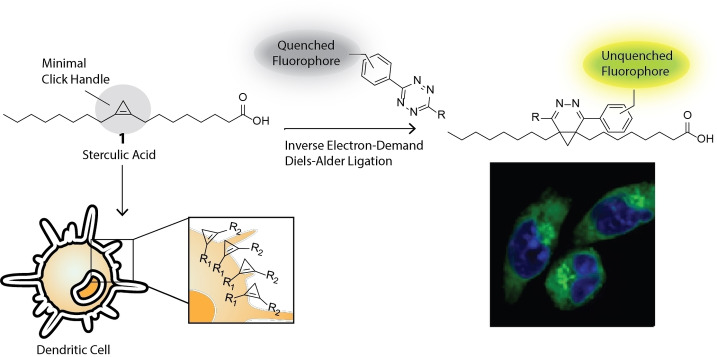
This paper describes the use of a cyclopropene‐containing oleic acid analogue for studying live cell lipid uptake by dendritic cells. The minimal oleic acid analogue is incorporated into membranes and lipoproteins, which can be studied by live cell microscopy and lipoproteomics.
Introduction
Lipids serve a myriad of roles in biology; as catabolic carbon source, [1] components of cellular and organellar membranes, [2] post‐translational protein modifications, [3] and signalling molecules. [4] The study of their contributions in biology is complicated by their lack of direct genetic encoding, their inherent lipophilicity, and the fact that chemical modifications, e.g. with pendant fluorophores, can severely alter their structure and biochemical properties. [5]
As a result, major efforts have gone into applying bioorthogonal chemistry to the study of lipid biochemistry. By introducing small terminal alkynes and azides in fatty acid tails,[ 6 , 7 , 8 , 9 , 10 , 11 ] phospholipids,[ 12 , 13 ] sphingolipids, [14] and cholesterol,[ 15 , 16 , 17 ] it has been possible to study lipids with only minimal modifications compared to the endogenous molecules. This strategy reduces the chances of the modifications affecting the native function of the lipid, and has successfully been used to study lipid localisation,[ 10 , 12 , 13 , 17 ] metabolism,[ 11 , 16 ] and trafficking,[ 18 , 19 , 20 ] as well as post‐translational lipidation of proteins (reviewed by Distefano and co‐workers [21] ). However, the downside of labelling with alkynes/azides has been the lack of live‐cell compatible chemistries that can be used with low background reactivity and fast rates.[ 22 , 23 ] Another complicating factor when performing bioorthogonal ligation on lipids is imposed by the hydrophobic environment in which the labels reside. Optimising the fluorescent reaction partners, particularly for live‐cell imaging, is thus necessary.
In 2008, the groups of Fox and Weissleder reported the inverse electron demand Diels–Alder (IEDDA) reaction—the reaction between an electron‐poor diene, such as a tetrazine, and a strained or electron‐rich dienophile—as a new bioorthogonal reaction.[ 24 , 25 ] This reaction was considered highly favourable for live‐cell use due to its high rates and the fluorescence quenching properties of the tetrazine (reviewed by Bernardes and co‐workers [26] ). The most used dienophile, trans‐cyclooctene (TCO), is a relatively large modification compared to alkynes or azides, [27] but has been used successfully to label, among others, the Golgi‐membrane with a TCO‐modified ceramide probe.[ 28 , 29 ] However, for lipids where chemical modifications to the structure can largely affect their function, smaller dienophiles were needed.[ 5 , 19 ] An important advance in the application of the IEDDA reaction to live‐cell studies of lipid function, was the development of sterically minimal dienophiles. To this end allyl‐thiols were reported by Bernardes and co‐workers. [30] However, these proved to have rather slow reaction kinetics (≤0.002 M−1 s−1). Devaraj and co‐workers, [31] and Prescher and co‐workers [32] simultaneously reported cyclopropenes, so‐called MiniTags, as minimal reactive dienophiles.[ 33 , 34 ] These cyclopropenes were incorporated into the head groups of phospholipids. So whilst capable of reacting with rate constants of up to 13 M−1 s−1 in an IEDDA reaction, [31] and only being slightly larger than alkynes and azides, [27] they could not be used to study fatty acid modifications of proteins. Still, the above properties make them an attractive alternative to TCO. Cyclopropenes are also reported to have been incorporated into glycans,[ 32 , 35 ] lignin polymers, [36] and nucleotides, [37] combined with live‐cell IEDDA reaction and imaging, emphasising their applicability as a bioorthogonal probe.
As discussed by Row and Prescher, it is often useful to look to nature for new bioorthogonal probes, as even rare motifs present in natural products indicate stability and compatibility in living systems. [27] In light of this, Nunn's 1952 discovery [38] of the plant metabolite sterculic acid (StA, 1), a carbocyclic fatty acid [39] found in the kernels of Sterculia foetida, piqued our interest. This 18‐carbon cis‐unsaturated lipid contains a naturally occurring 1,2‐substituted cyclopropene‐ring at C9‐C10. In plants, it is synthesised by addition of a methylene unit to the double bond of oleic acid (18 : 1, cis‐9), followed by enzymatic dehydrogenation to yield the cyclopropene ring. [40] We were curious whether this oleic acid analogue could be used in live‐cell compatible labelling chemistry, as it is known to be biologically stable and represents a minimal structural modification of one methylene compared to the parent oleic acid structure. Furthermore, all previously described bioorthogonal IEDDA applications have been explored for cyclopropenes with 1,3‐, 3,3‐ or 1,2,3‐substitution patterns,[ 31 , 32 , 41 ] whereas there has been no report of a 1,2‐substituted cyclopropene as a bioorthogonal probe.
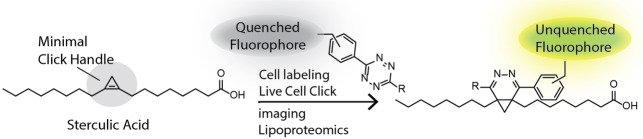
Here, we explore the use of 1 as a bioorthogonal reagent, assessing both its ligation kinetics, short‐term toxicity and live‐ and fixed‐cell imaging capacity. We show the fatty acid is taken up readily by dendritic cell lines, and can be used for live‐cell microscopy (Figure 1), as well as be used to study protein lipidation by chemical proteomics. We furthermore explore the reaction in a multiplexed reaction setup. [42] Multiple groups have previously reported the combination of up to three bioorthogonal ligation reactions in a triple mutually orthogonal system. This allows the tracking of multiple bioorthogonally labelled biomolecules in a single sample.[ 43 , 44 , 45 ] We have previously focussed our efforts in this area in the multiplexing of two copper‐catalysed Huisgen ligations (CCHL). [46] We here explore whether the reagent 1 also proved compatible with the CCHL reactions, allowing us to perform a triple‐click reaction. These experiments all indicate that 1 is a valuable reagent to study fatty acid uptake in cells.
Figure 1.
Schematic of the approach to label dendritic cells with sterculic acid (1), followed by an IEDDA reaction with tetrazine‐fluorophore conjugates to allow for live‐cell confocal imaging, due to fluorophore unquenching upon reaction.
Results and Discussion
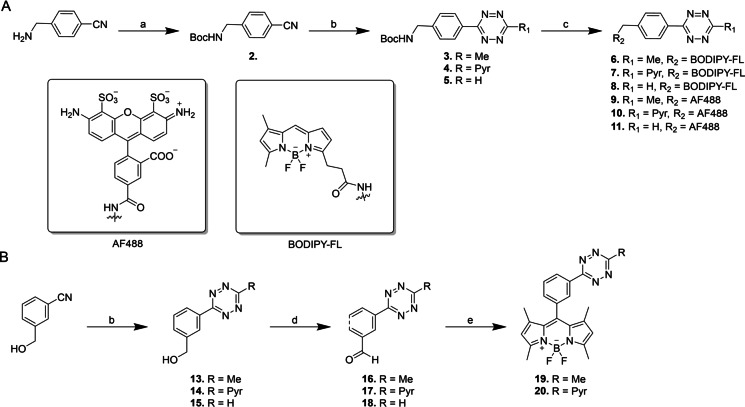
To assess whether 1 could be used in a live‐cell IEDDA reaction, we designed and synthesised a small library of tetrazine‐containing turn‐on fluorophores for optimisation of the reaction (Scheme 1). Tetrazines have a broad absorption spectrum, which peaks around 515 nm, meaning it can efficiently quench fluorescent dyes of wavelengths ≤550 nm via fluorescence resonance energy transfer (FRET).[ 47 , 48 ] Therefore, tetrazine‐fluorophore conjugates are so‐called turn‐on fluorophores, where their fluorescent intensity increases upon ligation, and their reaction with 1 can be readily quantified. We selected two different green fluorophores, BODIPY and Alexa Fluor 488 (AF488), and ligated these to three differently substituted tetrazines (H‐, methyl‐ or pyridyl‐substituted), because of the spectrum of reactivity and stability they covered. [49] Additionally, we attached the tetrazines to the BODIPY core at two different distances, as previous research has shown that decreasing the distance between the fluorophore and the tetrazine can improve the quenching effect.[ 50 , 51 ] It has been suggested that the improved quenching effect occurs via an alternate mechanism, through‐bond energy transfer (TBET), and not FRET. [51]
Scheme 1.
The synthesis of the tetrazine‐fluorophore conjugates 6–11 and 19–20. A) Synthesis of BODIPY‐FL and AF488‐tetrazines through late stage fluorophore introduction: a) Boc2O, NaOH, H2O. b) (1) NH2NH2, Zn(OTf)2, acetonitrile/2‐cyanopyridine or formamidine acetate. (2) NaNO2 in DCM/AcOH (1 : 1, v:v). c) (1) 4 m HCl, dioxane/DCM (1 : 1, v/v) (2) AF488‐NHS or BODIPY‐FL NHS, DIPEA, DCM. B) Synthesis of BODIPY‐Tetrazines 19–20, through fluorophore formation on a tetrazine scaffold: d) DMP, DCM e) (1) 2,4‐Dimethylpyrrole, TFA. (2) DDQ. (3) TEA, BF3⋅OEt2.
Based on previously published BODIPY‐FL methyl‐tetrazine conjugates, [50] we started from 4‐(aminomethyl) bezonitrile to facilitate a divergent synthesis. The free amine of 4‐(aminomethyl) benzonitrile was protected using di‐tert‐butyl dicarbonate and DIPEA to yield precursor 2. This compound was converted into N‐Boc‐protected aminoalkyl‐tetrazines 3–5 by a Lewis acid catalysed condensation of nitriles with hydrazine, followed by oxidation with sodium nitrite under acidic conditions, according to a literature procedure. [52] The protected amine of tetrazine 3–5 was deprotected using HCl/dioxane and immediately coupled to the commercially available AF488 and BODIPY‐FL via an NHS coupling, resulting in tetrazine‐fluorophore conjugates 6–11.
For BODIPY‐tetrazine conjugates 19 and 20, we first followed the synthesis previously published by Weissleder and co‐workers. [51] However, we found that the conditions needed to form a tetrazine on precursor 12 (structure can be found in Supporting Information) were too harsh for the BODIPY core, and only a trace amount of product was formed. To circumvent this issue, we followed a synthetic route published by Linden et al. [53] To this end, we synthesised tetrazines 13–15 starting from commercially available 3‐cyanobenzyl alcohol. Each respective tetrazine alcohol was then oxidised into an aldehyde using Dess–Martin periodinane, resulting in tetrazines 16–18. To form BODIPY 19 and 20, tetrazines 16 and 17 were reacted with 2,4‐dimethylpyrrole to form the dipyrromethene precursors, which were subsequently oxidised using DDQ and reacted in a chelation reaction with boron trifluoride. Unfortunately, tetrazine 18 proved too unstable to withstand the BODIPY formation. Photophysical characterisations of the tetrazine‐fluorophore conjugates can be found in Figure S1 and Table S1.
To investigate the fluorescence turn‐on of our synthesised fluorophores upon reaction with 1, the fluorophores were incubated with 1 in either PBS, DMSO/H2O (1 : 1, v/v) or complete RPMI 1640 medium augmented with 10 % fetal calf serum, and the fluorescent signal was measured over time (Figures 2 and S2). In line with previous reports, the two BODIPY‐tetrazine conjugates 19 and 20 showed the highest turn‐on ratio in PBS, followed by BODIPY FL‐tetrazine conjugates 6–7.[ 50 , 51 ] It was also found that H‐tetrazine 8, and pyridyl‐tetrazines 7 and 20 reacted faster than methyl‐tetrazines 6 and 19, as reported by the Hilderbrand group. [54] The second‐order rate constant for these reactions were calculated for tetrazines 6–7 and 19–20 as they displayed turn‐on effects in PBS (Table S1). Surprisingly the H‐tetrazine 8 showed little turn‐on in PBS. It has previously been suggested that H‐tetrazines can be too unstable to be used in biological applications, [55] and upon stability assessment of our tetrazine library in PBS (Figure S3A) it is clear that 8 is somewhat prone to degradation over time. Thereby the reaction between 1 and 8 cannot go to completion because of competition with hydrolytic degradation and unquenching of the H‐tetrazine. This is further supported by the observation that the relative fluorescent intensity (RFU) of tetrazines 8–11, all showing little turn‐on in PBS and that the starting point is high compared to RFUmax measured (Figure S3B). Tetrazines 9 and 10 also showed decreased stability over time, which could partially explain their low turn‐on ratios. However, all AF488‐tetrazine conjugates 9–11 showed very little quenching by the pendant tetrazines (Figure 2). This is difficult to explain, as non‐sulfonated analogues such as difluorinated‐fluorescein do show good turn on. [50] A possible explanation for this could either be poor resonance energy transfer, [47] photo‐induced electron transfer [56] or “energy transfer to a dark state” that have been reported to be important quenching mechanisms for longer bond‐length tetrazines. [57]
Figure 2.
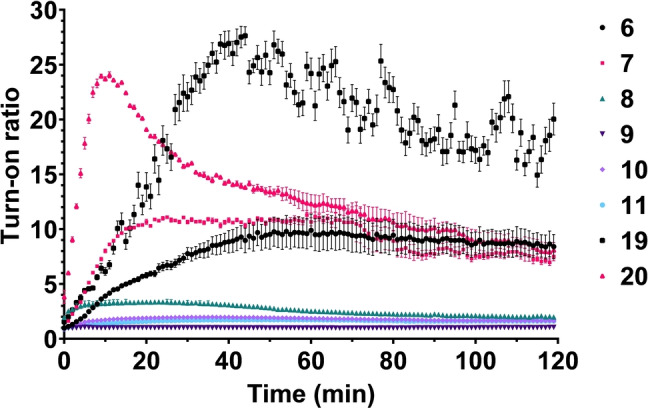
Average turn‐on ratio of tetrazine‐fluorophore conjugates 6–11 and 19–20 upon reaction with 1 in PBS at 25 °C. All conditions were measured in triplicate, and standard deviations are indicated.
Some loss of fluorescence, likely due to photobleaching, was also observed for 19 and 20. In DMSO/H2O or complete medium little‐to‐no turn‐on was observed for all the fluorophores (Figure S1). For turn‐on measurements in DMSO/H2O, this could be explained by the high intrinsic fluorescence for almost all the fluorophores (except 20) in the solvent, as shown in Figure S3A.
Due to the capricious turn‐on behaviour of the tetrazine‐fluorophore conjugates, we opted to assess them all for the live‐cell imaging of 1 (Figure S4). In contrast to cell‐free medium results, 6–8 and 19–20 exhibited successful ligation after uptake of 1 by DC2.4 dendritic cells. [58] This cell line was chosen due to its excellent imaging properties and our previous experience in using it as an in vitro model cell line for optimising bioorthogonal chemistry to study dendritic cell biology. [59] The Alexa Fluor‐based dyes 9–11 were unable to react with 1 in live cells. This is likely due to the hydrophilic nature of these fluorophores, caused by their sulfonate groups, not allowing them to diffuse over the hydrophobic plasma membrane, as previously reported.[ 60 , 61 ] Fluorophore 19 showed the brightest labelling with the lowest background fluorescence, allowing for imaging at lower laser intensities. As this leads to less bleaching of the sample, 19 was deemed to be the best fluorophore for live‐cell imaging (Figure 3A). For the brightness of the live‐cell labelling, it the substituent of the tetrazine core did not make a difference. Either H‐, methyl‐ or pyridyl‐tetrazines worked equally well. As well as being dependent on fluorophore structure, the labelling of 1 in live cells was also dependent on fluorophore concentration in a dose‐dependent manner, and on fluorophore incubation time (Figures S5 and S6).
Figure 3.
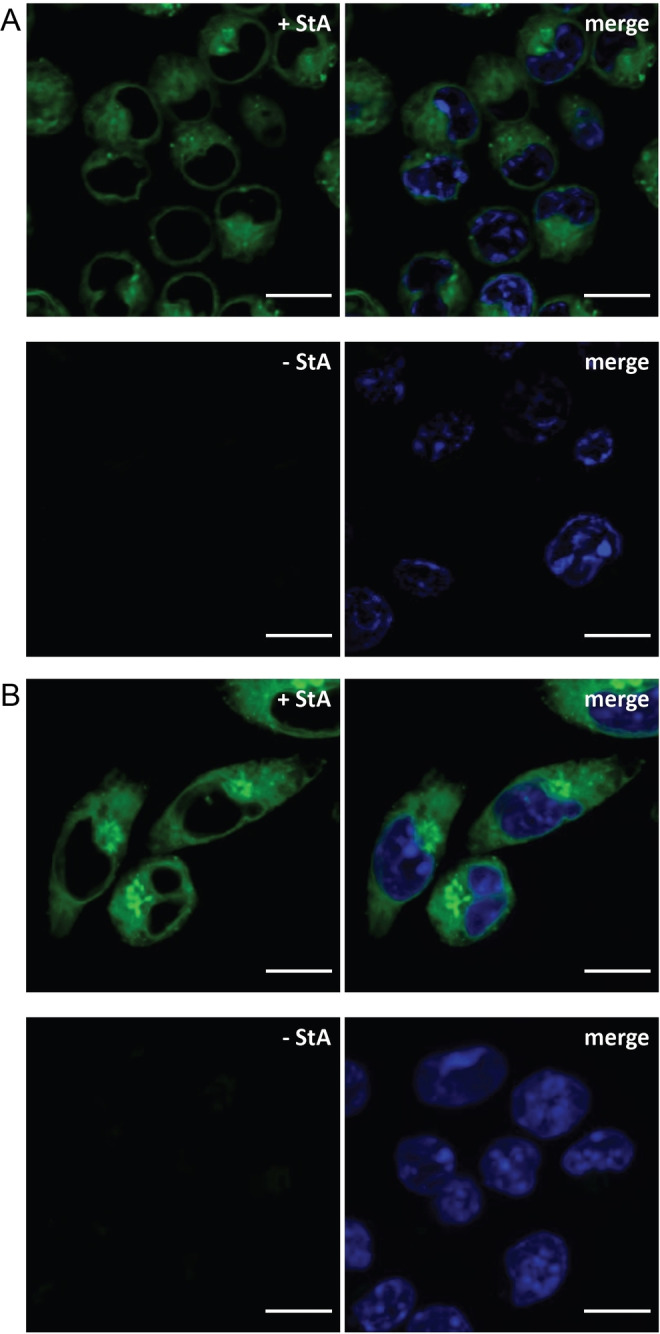
Confocal imaging of DC2.4 cells incubated with 1 (+StA, 50 μM) or without the probe (‐StA). A) Live‐cell imaging of labelled cells visualised with 19 (5 μM). B) Fixed‐cell imaging of labelled cells visualised with 8 (5 μM). The samples were washed after metabolic incorporation of 1 and after ligation with the fluorophore‐tetrazines, and were imaged at >4 distinct locations in the same well. DNA was counterstained with Hoechst 33342 (blue) for reference. Scale bars represent 10 μm.
To evaluate whether 1 was also able to react with the tetrazine‐fluorophore conjugates in fixed cells, we tested the fluorophore library on fixed and permeabilized cells (Figure S7). Consistent with the results found in live cells, fluorophores 6–8 were able to visualise the localisation of 1. As opposed to the live‐cell imaging, 9–11 showed labelling in fixed and permeabilized cells. However, the intensity of the signal of 9 and 10 was too low to be detected at the same laser settings as 6–8. Upon increased laser intensity, these fluorophores also showed signal over background (Figure S8). The signals of 19 and 20 were the brightest of the library, but showed oversaturated spots in fixed cells, which could originate from precipitation of the fluorophores under the reaction conditions. A general trend seen from our tetrazine library in fixed cells is that the H‐tetrazines give the brightest signal, followed by the pyridyl‐tetrazines. Fluorophore 8 was deemed to be the best alternative for fixed cells, due to its bright and consistent labelling (Figure 3B), despite showing low turn‐on in PBS (Figure 2). Direct comparison between the turn‐on study and cellular imaging is difficult, due to different conditions in the two situations. In the turn‐on study, there is an excess of 1 reacting with the tetrazines, whereas the concentration of 1 after metabolic incorporation for cellular imaging is the limiting factor, leading to an excess of the tetrazine instead. In addition, unreacted and potentially degraded fluorophore‐tetrazine conjugate was washed away before imaging, reducing the background signal, while this was not possible for the turn‐on study.
Even though unreacted tetrazine was washed away from both live‐ and fixed‐cell samples prior to confocal imaging to minimise background signal, sample preparation and preservation, especially for live‐cell imaging, would benefit from the reduction of washing steps. Therefore we tested whether the turn‐on effect of fluorophores 6, 7 and 19, 20 upon ligation with 1 (as shown in Figure 2) was sufficient for wash‐free live‐cell imaging. This was indeed the case, showing little detectable background signal (Figure S9).
In both live and fixed cells (Figure 3), the localisation of 1 can be observed throughout the cells, except within the nucleus. As exogenous free fatty acids can be readily incorporated into phopsholipids and other lipids via an acyl coenzyme A intermediate, [62] this would suggest that labelling of organellar membranes also occurs. This is in keeping with 1 serving as a mimic for oleic acid, which is known to be found ubiquitously in membrane lipids. [63] In fixed cells (Figure 3B), which are displayed as maximum intensity projections, it is also apparent that the fluorescent signal appears to be stronger in the endoplasmic reticulum (ER). This can be explained by the incorporation of the free fatty acids into phospholipids through Lands’ cycle and the Kennedy pathway in this organelle. [64]
In addition to our fluorophore‐ tetrazine library, 1 can also react with the commercially available (sulfo‐)Cy5 tetrazine in a similar manner, and can be used for fixed‐cell confocal imaging (Figure S10). Use of the Cy5 fluorophore shows a similar labelling pattern as described for the other tetrazine‐fluorophore conjugates, even though it has no turn‐on effect.
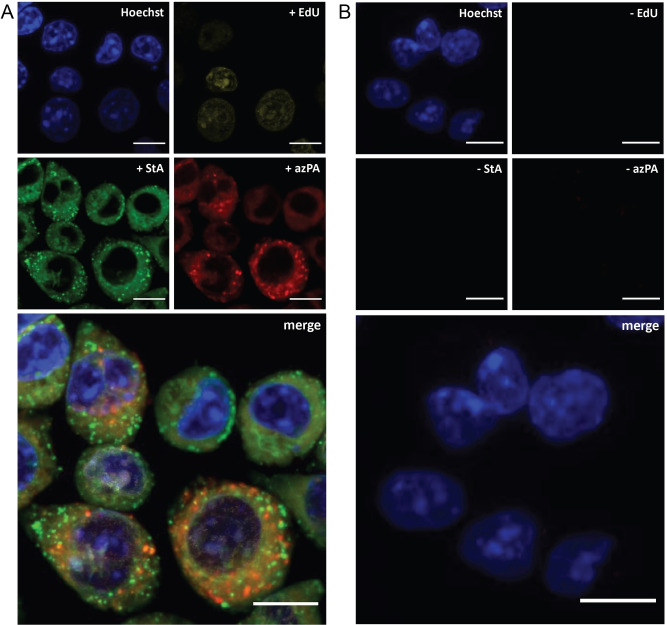
Multiplexing of bioorthogonal reactions is growing in popularity, meaning more complex systems can be studied simultaneously. [65] We have previously shown that two CCHL reactions can be combined in the same sample. [46] Here, we wanted to explore whether the sterculic acid ligation could be included in this workflow, allowing three biomolecules to be simultaneously visualised by multimodal fluorescent imaging. DC2.4 cells were metabolically labelled with the alkyne‐containing thymidine analogue 5‐ethynyl‐2′‐deoxyuridine (EdU), [66] the azide‐containing palmitic acid analogue 15‐azidopentadecanoic acid (azido palmitic acid, azPA) and 1, after which the cells were fixed before performing the corresponding bioorthogonal reactions with AZDye™ 555‐azide, AZDye™ 647‐alkyne and compound 7, respectively (Figure 4).
Figure 4.
Confocal imaging of triple‐bioorthogonally labelled DC2.4 cells incubated with A) 5‐ethynyl‐2′deoxyuridine (EdU, 10 μM, yellow) for 20 h, followed by 1 (StA, 50 μM, green) and azido palmitic acid (azPA, 100 μM, red) simultaneously for 1 h. The probes were visualised with AZDye™ 555‐azide, compound 7, and AZDye™ 647‐alkyne (all 5 μM), respectively. B) The cells were incubated without probes and treated in the same triple‐click manner as described above to show the background signal. CCHL reactions were performed using ascorbate as a reducing agent and THPTA as a ligand. The samples were washed between each metabolic incorporation and between each respective bioorthogonal reaction, and were imaged at 3 distinct locations in the same well. DNA was counterstained with Hoechst 33342 (blue) for reference. All scale bars represent 10 μm.
Figure 4 showed strong overlap between the sterculic acid and the azPA‐signal. As protein palmitoylation has previously been reported in dendritic cells, [67] and protein oleoylation in macrophages using fixed cell click chemistry, [7] we chose to investigate whether 1 could be used to study protein oleoylation in dendritic cells. Indeed, SDS‐PAGE analysis of DCs modified with 1 (Figure S12A–B) showed that we could successfully label these proteins in lysates with 11 (Figure S12A) and in live cells with 19 (Figure S13B). We were particularly interested in whether the levels of lipidated proteins changed upon stimulation of the DCs by LPS, as this could potentially offer mechanistic insights into how DCs can employ changes in fatty acid uptake to alter their immunological function.[ 68 , 69 ]
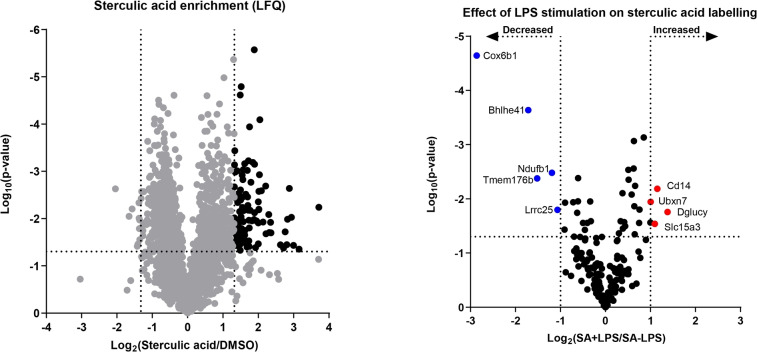
We therefore performed chemical proteomics (Figure 5) and SDS‐PAGE (Figure S12) analysis of “resting” and LPS‐activated DCs after labelling with 1. For the chemical proteomics, the labelled cells were treated with biotin‐PEG4‐tetrazine and subjected to streptavidin pull‐down, on‐bead digestion with trypsin and identification by LC‐MS/MS. From the MS data, in total 178 proteins were identified as significantly sterculic acid enriched (Figure 5). Gene enrichment analysis indicated that of the identified enriched proteins, 85 are known S‐acylated proteins, as annotated by KW‐0007. [70] This indicates that 1 is incorporated as a lipid PTM and usable to study protein modification by oleic acid. Upon activation of the DCs with LPS, SDS‐PAGE analysis showed a strong increase in the signal of lipidated proteins (Figure S13). Chemical proteomics of the LPS‐treated DCs and comparison of the recovered proteins, led to the significantly improved retrieval of four proteins (Figure 5). Of these, two (CD14, [71] SLC15A3 [72] ) are known to be upregulated upon LPS stimulation, and one in anti‐fungal immune responses (UBXN7 [73] ). The analysis also showed the reduced retrieval of an additional 5 species upon LPS activation, suggesting a complex control over protein lipidation.
Figure 5.
Proteomic analysis of proteins modified with 1 in DC2.4 cells by chemical proteomics. DC2.4 cells were stimulated with or without LPS (100 ng mL−1) for 24 h and treated with 1 (10 μM) for 20 h. Left: Volcano plot of proteins identified in pulldown experiment with 1 (10 μM) and biotin‐PEG4‐tetrazine (200 μM). Proteins with a ratio >2.5 and p‐value <0.05 are considered specifically enriched and are highlighted in black. Right: Difference in sterculic acid‐labeling of proteins between LPS‐ or vehicle‐treated DC2.4 cells. LFQ‐values of specifically sterculic acid‐enriched proteins were compared and proteins with significantly higher LFQ abundance between the two conditions are marked in red and blue for LPS‐ and vehicle‐treated conditions, respectively.
The facile inclusion of 1 in both chemical proteomics and multi‐click workflows—with good signal‐to‐noise ratios for all three click reactions, indicates that 1 does not have detectable cross‐reactivity with the CCHL components. This means that 1 can be used in combination with other click chemistries, allowing for the simultaneous study of multiple biomolecules. In addition, the chemical synthesis of 1 from its precursor stearolic acid via a Simmons–Smith reaction has previously been reported, [74] and stearolic acid can in turn be synthesised from oleic acid via bromination‐dehydrobromination. [75] This opens the door for the synthesis of cyclopropene analogues of other monounsaturated fatty acids, and their subsequent study by the IEDDA bioorthogonal reaction.
However, 1 has been reported to be an inhibitor of the enzyme stearoyl‐CoA desaturase 1 (SCD1) which catalyses the transformation of saturated fatty acids such as stearic or palmitic acid to their monounsaturated counterparts oleic or palmitoleic acid, respectively. [76] Lowered activity of SCD1 has been linked to various cellular responses such as ER stress, autophagy, and apoptosis, [76] which in turn poses the question if the addition of 1 would be toxic to the cells by its inhibiting effect on SCD1. To ensure that this was not the case, we incubated DC2.4 cells with a concentration range of 1 and measured cell viability after 24 h by 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) assay (Figure S13). No significant decrease of DC2.4 cell viability was observed for concentrations of 1 up to 50 μM and at higher concentrations there was no difference with vehicle‐induced toxicity. Nevertheless, inhibition of SCD1 should be taken into account in future use of cyclopropene‐modified lipids.
Conclusion
We have reported the first use of sterculic acid, a non‐toxic 1,2‐substituted cyclopropene‐containing fatty acid, for studying lipids in both live and fixed cells. The cyclopropene moiety readily reacts with our library of tetrazine‐fluorophore conjugates, allowing the study of fatty acid localisation by confocal microscopy. This reaction can occur in a mutually orthogonal manner with two subsequent CCHL reactions, allowing the study of multiple biomolecules simultaneously. We also established that proteins are modified with sterculic acid as a PTM, rendering it a good probe for investigating protein oleoylation by both SDS‐PAGE and chemical proteomics analysis, whilst only changing the structure from a cis‐alkene to a cyclopropene, an effective single methylene addition to the structure. The use of sterculic acid, combined with a reported generally applicable synthesis of cyclopropene‐containing fatty acids, means that unsaturated lipids can now be visualised using live‐cell microscopy and proteomics analysis; a sorely needed addition to the available tools for these elusive biomolecules.
Experimental Section
Full experimental details can be found in the Supporting Information.
Conflict of interest
The authors declare no conflict of interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
We would like to acknowledge Ken Rock for his kind gift of the DC2.4 cell line. We would also kindly like to thank the Institute for Chemical Immunology (NWO‐Zwaartekracht Grant) and the European Research Council (ERC‐CoG 865175) for funding the work described here.
K. Bertheussen, M. van de Plassche, T. Bakkum, B. Gagestein, I. Ttofi, A. J. C. Sarris, H. S. Overkleeft, M. van der Stelt, S. I. van Kasteren, Angew. Chem. Int. Ed. 2022, 61, e202207640; Angew. Chem. 2022, 134, e202207640.
A previous version of this manuscript has been deposited on a preprint server (https://doi.org/10.26434/chemrxiv‐2022‐4rr3b).
Contributor Information
Prof. Dr. Mario van der Stelt, Email: m.van.der.stelt@chem.leidenuniv.nl.
Dr. Sander I. van Kasteren, Email: s.i.van.kasteren@chem.leidenuniv.nl.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Houten S. M., Wanders R. J. A., J. Inherited Metab. Dis. 2010, 33, 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Meer G., Voelker D. R., Feigenson G. W., Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Resh M. D., Prog. Lipid Res. 2016, 63, 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wymann M. P., Schneiter R., Nat. Rev. Mol. Cell Biol. 2008, 9, 162–176. [DOI] [PubMed] [Google Scholar]
- 5. Bumpus T. W., Baskin J. M., Trends Biochem. Sci. 2018, 43, 970–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ticho A. L., Malhotra P., Manzella C. R., Dudeja P. K., Saksena S., Gill R. K., Alrefai W. A., J. Biol. Chem. 2020, 295, 4488–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thinon E., Percher A., Hang H. C., ChemBioChem 2016, 17, 1800–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pérez A. J., Bode H. B., J. Lipid Res. 2014, 55, 1897–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kostiuk M. A., Corvi M. M., Keller B. O., Plummer G., Prescher J. A., Hangauer M. J., Bertozzi C. R., Rajaiah G., Falck J. R., Berthiaume L. G., FASEB J. 2008, 22, 721–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Charron G., Zhang M. M., Yount J. S., Wilson J., Raghavan A. S., Shamir E., Hang H. C., J. Am. Chem. Soc. 2009, 131, 4967–4975. [DOI] [PubMed] [Google Scholar]
- 11. Thiele C., Papan C., Hoelper D., Kusserow K., Gaebler A., Schoene M., Piotrowitz K., Lohmann D., Spandl J., Stevanovic A., Shevchenko A., Kuerschner L., ACS Chem. Biol. 2012, 7, 2004–2011. [DOI] [PubMed] [Google Scholar]
- 12. Jao C. Y., Roth M., Welti R., Salic A., Proc. Natl. Acad. Sci. USA 2009, 106, 15332–15337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jao C. Y., Roth M., Welti R., Salic A., ChemBioChem 2015, 16, 472–476. [DOI] [PubMed] [Google Scholar]
- 14. Garrido M., Abad J. L., Fabriàs G., Casas J., Delgado A., ChemBioChem 2015, 16, 641–650. [DOI] [PubMed] [Google Scholar]
- 15. Jao C. Y., Nedelcu D., Lopez L. V., Samarakoon T. N., Welti R., Salic A., ChemBioChem 2015, 16, 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hofmann K., Thiele C., Schött H. F., Gaebler A., Schoene M., Kiver Y., Friedrichs S., Lütjohann D., Kuerschner L., J. Lipid Res. 2014, 55, 583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rakers L., Grill D., Matos A. L. L., Wulff S., Wang D., Börgel J., Körsgen M., Arlinghaus H. F., Galla H.-J., Gerke V., Glorius F., Cell Chem. Biol. 2018, 25, 952–961. [DOI] [PubMed] [Google Scholar]
- 18. Hang H. C., Wilson J. P., Charron G., Acc. Chem. Res. 2011, 44, 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liang D., Wu K., Tei R., Bumpus T. W., Ye J., Baskin J. M., Proc. Natl. Acad. Sci. USA 2019, 116, 15453–15462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pérez A. J., Bode H. B., ChemBioChem 2015, 16, 1588–1591. [DOI] [PubMed] [Google Scholar]
- 21. Suazo K. F., Park K.-Y., Distefano M. D., Chem. Rev. 2021, 121, 7178–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rigolot V., Biot C., Lion C., Angew. Chem. Int. Ed. 2021, 60, 23084–23105; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 23268–23289. [Google Scholar]
- 23. van Geel R., Pruijn G. J. M., van Delft F. L., Boelens W. C., Bioconjugate Chem. 2012, 23, 392–398. [DOI] [PubMed] [Google Scholar]
- 24. Blackman M. L., Royzen M., Fox J. M., J. Am. Chem. Soc. 2008, 130, 13518–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Devaraj N. K., Weissleder R., Hilderbrand S. A., Bioconjugate Chem. 2008, 19, 2297–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oliveira B. L., Guo Z., Bernardes G. J. L., Chem. Soc. Rev. 2017, 46, 4895–4950. [DOI] [PubMed] [Google Scholar]
- 27. Row R. D., Prescher J. A., Acc. Chem. Res. 2018, 51, 1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Erdmann R. S., Takakura H., Thompson A. D., Rivera-Molina F., Allgeyer E. S., Bewersdorf J., Toomre D., Schepartz A., Angew. Chem. Int. Ed. 2014, 53, 10242–10246; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 10407–10412. [Google Scholar]
- 29. Erdmann R. S., Toomre D., Schepartz A., Methods Mol. Biol. 2017, 1663, 65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oliveira B. L., Guo Z., Boutureira O., Guerreiro A., Jiménez-Osés G., Bernardes G. J. L., Angew. Chem. Int. Ed. 2016, 55, 14683–14687; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 14903–14907. [Google Scholar]
- 31. Yang J., Šečkutė J., Cole C. M., Devaraj N. K., Angew. Chem. Int. Ed. 2012, 51, 7476–7479; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2012, 124, 7594–7597. [Google Scholar]
- 32. Patterson D. M., Nazarova L. A., Xie B., Kamber D. N., Prescher J. A., J. Am. Chem. Soc. 2012, 134, 18638–18643. [DOI] [PubMed] [Google Scholar]
- 33. Sauer J., Heinrichs G., Tetrahedron Lett. 1966, 7, 4979–4984. [Google Scholar]
- 34. Dowd P., Gold A., Tetrahedron Lett. 1969, 10, 85–86. [Google Scholar]
- 35. Cole C. M., Yang J., Šečkutė J., Devaraj N. K., ChemBioChem 2013, 14, 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morel O., Lion C., Neutelings G., Stefanov J., Baldacci-Cresp F., Simon C., Biot C., Hawkins S., Spriet C., Plant Physiol. 2022, 188, 816–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Šečkutė J., Yang J., Devaraj N. K., Nucleic Acids Res. 2013, 41, e148–e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nunn J. R., J. Chem. Soc. 1952, 313–318. [Google Scholar]
- 39.M. U. Ahmad, S. M. Ali, A. Ahmad, S. Sheikh, I. Ahmad, AOCS Press, 2017, pp. 147–185.
- 40. Greenberg A., Harris J., J. Chem. Educ. 1982, 59, 539. [Google Scholar]
- 41. Kumar P., Jiang T., Zainul O., Preston A. N., Li S., Farr J. D., Suri P., Laughlin S. T., Tetrahedron Lett. 2018, 59, 3435–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hu Y., Schomaker J. M., ChemBioChem 2021, 22, 3254–3262. [DOI] [PubMed] [Google Scholar]
- 43. Willems L. I., Li N., Florea B. I., Ruben M., van der Marel G. A., Overkleeft H. S., Angew. Chem. Int. Ed. 2012, 51, 4431–4434; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2012, 124, 4507–4510. [Google Scholar]
- 44. Simon C., Lion C., Spriet C., Baldacci-Cresp F., Hawkins S., Biot C., Angew. Chem. Int. Ed. 2018, 57, 16665–16671; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 16907–16913. [Google Scholar]
- 45. Chio T. I., Gu H., Mukherjee K., Tumey L. N., Bane S. L., Bioconjugate Chem. 2019, 30, 1554–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bakkum T., Heemskerk M. T., Bos E., Groenewold M., Oikonomeas-Koppasis N., Walburg K. V., van Veen S., van der Lienden M. J. C., van Leeuwen T., Haks M. C., Ottenhoff T. H. M., Koster A. J., van Kasteren S. I., ACS Cent. Sci. 2020, 6, 1997–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beliu G., Kurz A. J., Kuhlemann A. C., Behringer-Pliess L., Meub M., Wolf N., Seibel J., Shi Z.-D., Schnermann M., Grimm J. B., Lavis L. D., Doose S., Sauer M., Commun. Biol. 2019, 2, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wieczorek A., Werther P., Euchner J., Wombacher R., Chem. Sci. 2017, 8, 1506–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang J., Liang Y., Šečkutė J., Houk K. N., Devaraj N. K., Chem. Eur. J. 2014, 20, 3365–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Devaraj N. K., Hilderbrand S., Upadhyay R., Mazitschek R., Weissleder R., Angew. Chem. Int. Ed. 2010, 49, 2869–2872; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2010, 122, 2931–2934. [Google Scholar]
- 51. Carlson J. C. T., Meimetis L. G., Hilderbrand S. A., Weissleder R., Angew. Chem. Int. Ed. 2013, 52, 6917–6920; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 7055–7058. [Google Scholar]
- 52. Sarris A. J. C., Hansen T., de Geus M. A. R., Maurits E., Doelman W., Overkleeft H. S., Codée J. D. C., Filippov D. V., van Kasteren S. I., Chem. Eur. J. 2018, 24, 18075–18081. [DOI] [PubMed] [Google Scholar]
- 53. Linden G., Zhang L., Pieck F., Linne U., Kosenkov D., Tonner R., Vázquez O., Angew. Chem. Int. Ed. 2019, 58, 12868–12873; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 13000–13005. [Google Scholar]
- 54. Karver M. R., Weissleder R., Hilderbrand S. A., Bioconjugate Chem. 2011, 22, 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Simon C., Lion C., Ahouari H., Vezin H., Hawkins S., Biot C., Chem. Commun. 2021, 57, 387–390. [DOI] [PubMed] [Google Scholar]
- 56. Knall A.-C., Hollauf M., Slugovc C., Tetrahedron Lett. 2014, 55, 4763–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chi W., Huang L., Wang C., Tan D., Xu Z., Liu X., Mater. Chem. Front. 2021, 5, 7012–7021. [Google Scholar]
- 58. Shen Z., Reznikoff G., Dranoff G., Rock K. L., J. Immunol. 1997, 158, 2723–2730. [PubMed] [Google Scholar]
- 59. van Leeuwen T., Araman C., Pieper Pournara L., Kampstra A. S. B., Bakkum T., Marqvorsen M. H. S., Nascimento C. R., Groenewold G. J. M., van der Wulp W., Camps M. G. M., Janssen G. M. C., van Veelen P. A., van Westen G. J. P., Janssen A. P. A., Florea B. I., Overkleeft H. S., Ossendorp F. A., Toes R. E. M., van Kasteren S. I., RSC Chem. Biol. 2021, 2, 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Grimm J. B., Lavis L. D., Nat. Methods 2022, 19, 149–158. [DOI] [PubMed] [Google Scholar]
- 61. Hapuarachchige S., Montaño G., Ramesh C., Rodriguez D., Henson L. H., Williams C. C., Kadavakkollu S., Johnson D. L., Shuster C. B., Arterburn J. B., J. Am. Chem. Soc. 2011, 133, 6780–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grevengoed T. J., Klett E. L., Coleman R. A., Annu. Rev. Nutr. 2014, 34, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Casares D., Escribá P. V., Rosselló C. A., Int. J. Mol. Sci. 2019, 20, 2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Moessinger C., Klizaite K., Steinhagen A., Philippou-Massier J., Shevchenko A., Hoch M., Ejsing C. S., Thiele C., BMC Cell Biol. 2014, 15, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smeenk M. L. W. J., Agramunt J., Bonger K. M., Curr. Opin. Chem. Biol. 2021, 60, 79–88. [DOI] [PubMed] [Google Scholar]
- 66. Chehrehasa F., Meedeniya A. C. B., Dwyer P., Abrahamsen G., Mackay-Sim A., J. Neurosci. Methods 2009, 177, 122–130. [DOI] [PubMed] [Google Scholar]
- 67. Chesarino N. M., Hach J. C., Chen J. L., Zaro B. W., Rajaram M. V. S., Turner J., Schlesinger L. S., Pratt M. R., Hang H. C., Yount J. S., BMC Biol. 2014, 12, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Almeida L., Everts B., Eur. J. Immunol. 2021, 51, 1628–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wculek S. K., Khouili S. C., Priego E., Heras-Murillo I., Sancho D., Front. Immunol. 2019, 10, 775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Huang D. W., Sherman B. T., Lempicki R. A., Nat. Protoc. 2009, 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 71. Landmann R., Knopf H. P., Link S., Sansano S., Schumann R., Zimmerli W., Infect. Immun. 1996, 64, 1762–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Song F., Yi Y., Li C., Hu Y., Wang J., Smith D. E., Jiang H., Cell Death Dis. 2018, 9, 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Srivastava M., Bencurova E., Gupta S. K., Weiss E., Löffler J., Dandekar T., Front. Cell. Infect. Microbiol. 2019, 9, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Castellucci N. T., Griffin C. E., J. Am. Chem. Soc. 1960, 82, 4107. [Google Scholar]
- 75. Silbert L. S., J. Am. Oil Chem. Soc. 1984, 61, 1090–1092. [Google Scholar]
- 76. Peláez R., Pariente A., Pérez-Sala Á., Larráyoz I. M., Cells 2020, 9, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.