Abstract
Antisense inhibition of microRNAs is an emerging preclinical approach to pharmacoresistant epilepsy. A leading candidate is an "antimiR" targeting microRNA‐134 (ant‐134), but testing to date has used rodent models. Here, we develop an antimiR testing platform in human brain tissue sections. Brain specimens were obtained from patients undergoing resective surgery to treat pharmacoresistant epilepsy. Neocortical specimens were submerged in modified artificial cerebrospinal fluid (ACSF) and dissected for clinical neuropathological examination, and unused material was transferred for sectioning. Individual sections were incubated in oxygenated ACSF, containing either ant‐134 or a nontargeting control antimiR, for 24 h at room temperature. RNA integrity was assessed using BioAnalyzer processing, and individual miRNA levels were measured using quantitative reverse transcriptase polymerase chain reaction. Specimens transported in ACSF could be used for neuropathological diagnosis and had good RNA integrity. Ant‐134 mediated a dose‐dependent knockdown of miR‐134, with approximately 75% reduction of miR‐134 at 1 μmol L−1 and 90% reduction at 3 μmol L−1. These doses did not have off‐target effects on expression of a selection of three other miRNAs. This is the first demonstration of ant‐134 effects in live human brain tissues. The findings lend further support to the preclinical development of a therapy that targets miR‐134 and offer a flexible platform for the preclinical testing of antimiRs, and other antisense oligonucleotide therapeutics, in human brain.
Keywords: antimiR, ASO, epilepsy, human tissue, microRNA
1. INTRODUCTION
A promising preclinical approach to pharmacoresistant epilepsy is to target microRNA (miRNA). 1 , 2 miRNAs are endogenous ~22‐nt noncoding RNAs that repress the translation of targeted mRNAs via complementary binding to target regions in the 3′ untranslated region (UTR). 3 Individual miRNAs can regulate large numbers of mRNAs; therefore, their targeting could potentially reshape the dysregulated gene expression landscape in human temporal lobe epilepsy. Most notably, antisense oligonucleotide (ASO) inhibition of miR‐134 (using ant‐134) produces potent antiseizure and disease‐modifying effects in rodent models of seizures and epilepsy. 4 , 5 , 6 , 7 However, the binding sites for miR‐134 on 3’UTRs and hence the mechanism(s) of action of ant‐134 may not be conserved between rodents and humans. 8 Furthermore, it is unclear how efficiently ant‐134 will penetrate into human brain cells. ASO uptake mechanisms can vary between cell types 9 and may not be the same in rodent and human brain.
A platform to test these mechanisms is offered by resective brain surgeries to alleviate pharmacoresistant epilepsy. 10 The resected brain tissue can be collected and used for research purposes, offering an additional translational model to complement findings in rodent brain. 11 , 12 , 13 Here, we set out an adapted methodology to obtain human brain tissue for molecular assessment, without impacting clinical processes. Specimens processed with this method were viable and had sufficient RNA integrity for molecular studies. We used sections of these specimens to inhibit miR‐134 in human brain using ant‐134 as a proof of concept. This provides new tools for translational epilepsy research.
2. MATERIALS AND METHODS
2.1. Ethical approval
All studies were approved by the local ethical committee at Beaumont Hospital Dublin (Reference 13/75, 20/58). Patients had the capacity to give fully informed consent and were provided with a patient information leaflet. Participants were free to withdraw their consent at any time without providing a reason. Tissues used were removed as part of the normal clinical procedure, and no extra tissue was resected for research use.
2.2. Resective neurosurgery and tissue specimens obtained
Thirteen human neocortical brain specimens were obtained (nine male and four female patients, 22–62 years old; Table 1). Neocortical tissues were resected en bloc and were obtained by the researcher directly in the surgical theater (overview in Figure 1A). We immediately submerged these specimens into ice cold oxygenated sucrose artificial cerebrospinal fluid (ACSF; in mmol L−1: 205 sucrose, 10 glucose, 2.5 KCl, 5 MgCl2, .1 CaCl2, NaHCO3, NaH2PO4·H2O) and transported them to the neuropathology laboratory. This transport process took approximately 5 min, during which time the ACSF was not oxygenated further.
TABLE 1.
Details of study participants and tissue specimens used
| Participant research ID | Sex | Age at time of surgery, years | Age at onset of epilepsy, years | Nature of initial specimen | Antiseizure medications |
|---|---|---|---|---|---|
| 1 | M | 37 | 28 | LTL | ESLI, LEV, ZNS |
| 2 | F | 61 | 7 | LTL | ESLI, LTG |
| 3 | M | 59 | 44 | LTL | CLOB, LAC, LEV, PERM, VAP |
| 4 | M | 62 | 32 | Tumor | ESLI, LEV |
| 5 | M | 22 | 12 | LTL | ESLI, LTG, VAP |
| 6 | M | 43 | 33 | LTL | LTG, LEV |
| 7 | F | 39 | 7 | FL | CLOB, LTG, LEV |
| 8 | F | 22 | 7 | LTL | CLOB, ESLI, LTG |
| 9 | M | 54 | 42 | LTL | CBZ, LTG, LEV |
| 10 | F | 47 | <6 months | LTL | BRIV, ESLI, LAC |
| 11 | M | 39 | 27 | LTL | BRIV, CBZ, VAP, ZNS |
| 12 | M | 59 | UNK | LTL | ESLI, LTG, PERM |
| 13 | M | 29 | UNK | LTL | ESLI, LTG, VAP |
Abbreviations: BRIV, brivaracetam; CBZ, carbamazepine; CLOB, clobazam; ESLI, eslicarbazepine; F, female; FL, frontal lobe; LAC, lacosamide; LEV, levetiracetam; LTG, lamotrigine; LTL, lateral temporal lobe; M, male; PERM, perampanel; UNK, unknown; VAP, valproate; ZNS, zonisamide.
FIGURE 1.
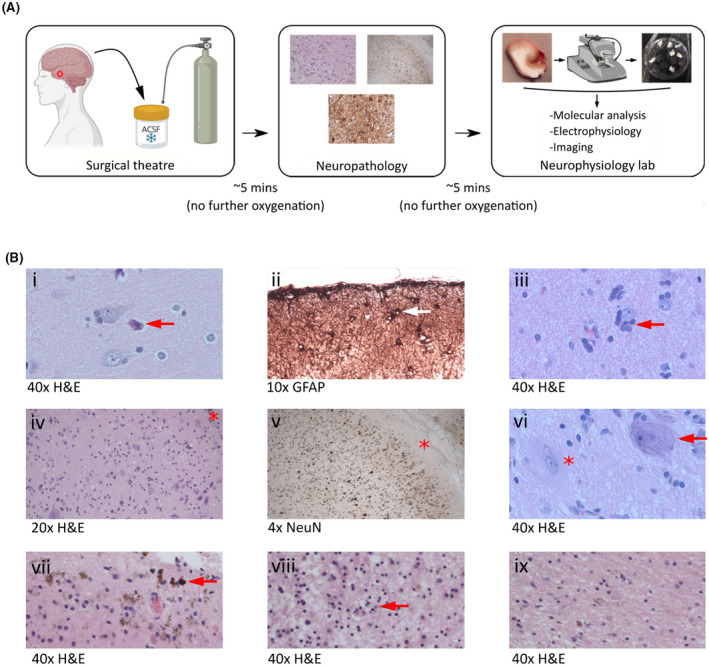
Integration of brain tissue collection for neurophysiology into the clinical workflow. (A) Resected human brain tissue specimens were obtained directly from the neurosurgical theater and immediately submerged into oxygenated ice cold artificial cerebrospinal fluid (ACSF). Specimens were transported in ACSF to the neuropathology laboratory as part of the clinical pathway. A proportion of the specimen was then returned to ACSF and taken to the research laboratory for study. (B) Examples of neuropathological observations made in specimens transported in ACSF illustrate the compatibility of our approach with the clinical pathway: (i) normal cortex and neuronal morphology with perioperative "dark cell change" (arrow); (ii) subpial gliosis (arrow); (iii) oligodendroglial hyperplasia; (iv–vi) focal cortical dysplasia type 2B characterized by disorganized lamination and dysmorphic neurons (arrow in vi; asterisk at Layer I in iv and v) and balloon cells (asterisk in vi); (vii, viii) remote hemorrhage and gliosis; (ix) a directly comparable section exhibiting remote hemorrhage and gliosis in a specimen handled in a standard way without direct ACSF exposure, indicating no qualitative impact of ACSF transport. GFAP, glial fibrillary acidic protein; H&E, hematoxylin and eosin; NeuN, neuron‐specific nuclear protein.
2.3. Neuropathological assessment
Neocortical specimens were removed from the ACSF and assessed for diagnosis. After macroscopic examination and serial sectioning, a piece of the specimen (~1 × 1 × .5 cm) was immediately resubmerged in ACSF, without additional oxygenation, and transported on ice to the neurophysiology laboratory. A distinction was made between peripheral sections in immediate contact with ACSF, and deeper sections, largely remote from ACSF. The remaining specimen was fixed overnight in 10% formalin, after which it was processed and embedded in paraffin. Four‐micrometer sections were stained with hematoxylin and eosin (H&E) ± glial fibrillary acidic protein (GFAP), microtubule‐associated protein 2, neuron‐specific nuclear protein immunohistochemistry for systematic neuropathological assessment (Figure 1B). We compared the robustness of peripheral sections that were in direct contact with ACSF against the deeper sections, otherwise processed in the same way.
2.4. AntimiR treatment of human brain specimens
In the neurophysiology laboratory, the specimen was transferred to a dish filled with fresh ice cold and oxygenated sucrose ACSF, and any remaining pia mater was removed. We divided the specimen into cubes (~1 mm3) using a scalpel. We created a low‐volume incubation chamber using a standard 12‐well plate. Specimens were placed into mesh inserts, and submerged into 4 ml normal ACSF (mmol L−1: 125 NaCl, 10 glucose, 3 KCl, 2 CaCl2, 1 MgCl2, 26 NaHCO3, 1.25 NaH2PO4·H2O) within the wells. The low volume facilitated the application of active antimiR concentrations. Specimens were incubated for 24 h at room temperature in the presence of either ant‐134 (Qiagen; catalogue number 339132: TGGTCAACCAGTCACA), or a scrambled nontargeting control (Negative Control A; Qiagen; catalogue number 339136: TAACACGTCTATACGCCCA), at .1, 1, and 3 μmol L−1. After 24 h, specimens were removed from ACSF and flash frozen at −80°C.
2.5. Rodent brain specimens
C57BL/6 mice (age range = 21–48 days) underwent unilateral stereotaxic injection of phosphate‐buffered saline (PBS) into the amygdala. Twenty‐four hours later, mice were euthanized with 0.2 ml intraperitoneal sodium pentobarbital (200 mg/ml). Mice were perfused with 20 ml PBS via cardiac puncture, and brain tissue was frozen at −80°C .
2.6. RNA processing
RNA was extracted using TRIzol, as described previously. 14 RNA integrity was assessed using the Agilent 2100 Bioanalyzer instrument and RNA 6000 Nano kit (5067–1511). The microfluidic RNA Nano chip was prepared followed by loading of 1 μl of sample. Total RNA quality was assessed by using the ratio of ribosomal peaks (18S/28S) to derive the RNA integrity number (RIN) value, an objective readout of RNA quality between 0 (completely degraded) and 10 (highly intact). miRNA expression was assessed using previously described methods, 14 with specific primers for hsa‐miR‐134, hsa‐miR‐10, hsa‐miR‐129, and hsa‐miR‐132 (Applied Biosystems, miRNA assays IDs 001186, 000387, 000590, 000457).
2.7. Statistics
All datasets contained at least one nonnormally distributed dataset. Comparisons used Kruskal–Wallis test with Dunn multiple comparisons (GraphPad Prism v9.3.1). Averages are shown as median ± interquartile range.
3. RESULTS
3.1. Neuropathological assessment
We verified that our method did not impact key neuropathological assessments and diagnoses. Qualitative analysis of sections transported in sucrose ACSF showed conserved cellular morphology, as observed with both H&E and immunohistochemical staining protocols, enabling the identification of pathologies including perioperative dark cell change, subpial gliosis with upregulation of GFAP, oligodendroglial hyperplasia, focal cortical dysplasia type 2B with abnormal cortical neuronal lamination, dysmorphic neurons and balloon cells, and remote cortical hemorrhage and gliosis with white matter cavitation (Figure 1B). Figure 1B(ix) shows a comparable section with remote hemorrhage and gliosis obtained from a specimen transported without direct ACSF contact, showing no qualitative impact of our method.
3.2. miRNA‐134 inhibition in human brain tissue
We used our method as a platform to test ant‐134 in human brain tissue. We applied ant‐134 for 24 h (Figure 2A,B) at a range of concentrations (based on previous work in induced pluripotent stem cells 15 ). To verify the viability of our method, we assessed RNA integrity in a subset of these samples, and compared them with equivalent neocortical samples that were transported without ACSF. Bioanalyzer results revealed significantly higher RIN values in samples processed with, compared to without, ACSF (Figure 2C). This provides an objective quality control measure validating the use of this approach as a platform to study RNA‐targeting oligonucleotide therapeutics. Notably, the RNA yield from human brain specimens was lower than comparable rodent samples (Figure 2C).
FIGURE 2.
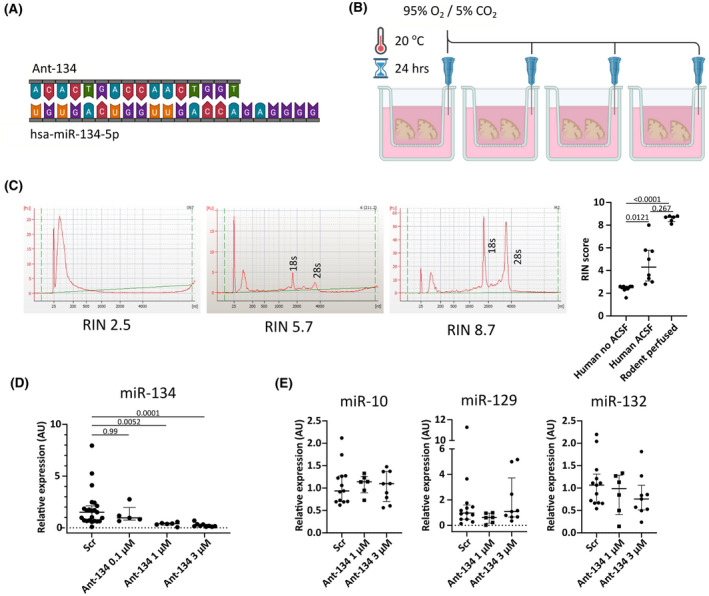
Ant‐134 mediates a dose‐dependent knockdown of microRNA‐134 in human neocortex. (A) Sequences of hsa‐miR‐134‐5p (22‐mer) and ant‐134 (16‐mer) indicates perfect complementarity between the two. (B) Experimental setup to treat acutely sectioned human brain specimens with antimiR. Sections were placed into small inserts with a permeable mesh at the bottom. Inserts were placed into individual wells of a standard 12‐well plate and submerged into 4 ml normal artificial cerebrospinal fluid (ACSF) containing ant‐134 or scrambled (Scr) nontargeting control at varying concentrations. The ACSF is each well was oxygenated using a syringe needle connected to a carbogen gas supply. This preparation was left for 24 h at room temperature. (C) BioAnalyzer traces from human brain tissue transported without ACSF (RNA integrity number [RIN] = 2.5) and with ACSF (RIN = 5.7). A trace from a mouse brain perfused for molecular biology is included as a gold standard comparison (RIN = 8.7) [FU ‐ fluorescence units]. The graph shows that human samples processed using our method had significantly higher RIN values than those processed using standard methods (Kruskal–Wallis test with Dunn multiple comparisons test). (D) Quantitative reverse transcriptase polymerase chain reaction shows robust dose‐dependent knockdown of miR‐134 after 24 h (Kruskal–Wallis test with Dunn test for multiple comparisons). (E) For the viable doses of ant‐134, we did not observe off‐target inhibition of miR‐10, miR‐129, or miR‐132 (all Kruskal–Wallis test with Dunn multiple comparisons tests).
Using quantitative reverse transcriptase polymerase chain reaction, we observed a dose‐dependent inhibition of miR‐134 by ant‐134, with significant knockdown mediated by 1 μmol L−1 (~75% reduction in expression) and 3 μmol L−1 (~90% reduction), compared with a nontargeting control antimiR (Figure 2D). Ant‐134 inhibition was specific for miR‐134, and we did not observe changes to expression of a selection of other brain‐enriched miRNAs: miR‐10, miR‐129, and miR‐132 (Figure 2E). Together, this indicates a platform for the testing of antimiR uptake and efficacy in human brain tissue, offering an important tool in preclinical research to aid in the translation of oligonucleotide therapeutics to the clinic.
4. DISCUSSION
The approach outlined here provides a methodology to interrogate the molecular effects of miRNA manipulation in the human brain. This provides a much‐needed tool to aid translation of miRNA‐targeting therapies to the clinic. 1 , 11 Studies on human‐derived tissues are key to test molecular mechanisms that may not be captured by rodent models because of species‐specific variation in the 3′UTRs of mRNAs. Notably, the binding site of miR‐134 on the 3′UTR of human LIMK1 differs from that of rodents. 8 Because derepressing Limk1 contributes to neuroprotective and antiseizure effects of ant‐134 in rodents, 4 , 7 testing in human models is critical. There is the possibility that miRNA manipulation in humans could give rise to unanticipated adverse effects not observed in rodents. Testing antimiRs in human brain tissues may therefore become an important step in the preclinical development or regulatory pathway. These specimens can also be used to acutely induce epileptiform activity for ex vivo small molecule drug testing. 11 , 12 Finally, there are ethical advantages to our approach, which represents a replacement for animal research within the concept of replacement, reduction, and refinement. 16
Although we focused here on antimiRs, the current approach could be applied to other ASO strategies. Due to their relative ease of delivery and ability to readily target RNAs in widespread brain structures, ASOs are emerging as a key approach to neurological diseases, particularly those that affect the whole brain. For example, ASO gapmers have been used in a mouse model of a genetic channelopathy that leads to epilepsy. 17 Current challenges for ASO translation from animal models to human applications include on‐ and off‐target toxicity effects, 18 which may be unique to the human brain and not possible to observe in mice. ASO mechanisms of action can include steric blocking of mRNA translation, degradation of target mRNA, or regulation of RNA splice events. 19 Different ASOs may also have different lengths and chemistries, which can affect their uptake, bioavailability, and cellular effects. 1 Our methodology provides a platform that could be used to test all of these ASO mechanisms and properties in real human brain tissue. Furthermore, this approach can be applied to other neurological diseases beyond epilepsy, through the use of nonepileptic temporal neocortex, removed to access the deeper epileptogenic zone.
Our approach fitted seamlessly into the clinical pathway and should be adoptable in comparable clinical settings. Researchers were trained to safely enter the surgical theater to collect tissue, meaning that the approach did not place any additional demands on the surgical team. This had the added advantage that resected specimens were immediately transferred to fresh sucrose ACSF and could be moved by the researcher to the neuropathology and neurophysiology laboratories quickly, maximizing tissue viability. We also worked closely as a multidisciplinary team to verify that sucrose ACSF transportation had no impact on neuropathological assessments. Together, the training of our researchers to work in the clinical environment, alongside close collaboration with our neurosurgeons and neuropathologists, was critical to our approach.
Our approach also has limitations. Notably, the resected tissues are also required for neuropathologic diagnosis. Although we present a methodology to fully integrate our approach into clinical pathways, it remains suboptimal from a research viewpoint that tissues cannot be taken directly to the neurophysiology laboratory. This may impact on tissue viability for research, although our assessments suggest that human tissues, when handled carefully, have comparable quality to freshly prepared rodent samples. Another challenge is the relatively limited availability and the heterogeneity of the specimens. The approach relies upon a research laboratory in close physical proximity to a specialist neurosurgery center. The frequency of surgeries dictates tissue availability and may not always be consistent due to the nature of the clinical pathway. Coupled with this, specimens are highly heterogeneous and are obtained from patients with different sex, age, and medication histories. This likely adds more variability to datasets and necessitates larger sample sizes, although this is mitigated in part because it is usually possible to obtain many acute sections from one individual resected specimen. Taken together, the relatively low throughput and need for more samples means human tissue‐based studies take much longer than those in rodents, and this must be considered by researchers before beginning such studies.
In conclusion, we present a method for the use of resected human brain tissues for research on RNA‐based therapies, within pre‐existing clinical pathways. These tissues are amenable to studies including molecular and electrophysiological assessment, and provide a critical tool in translational epilepsy research. Furthermore, we demonstrated for the first time the use of ant‐134, a leading preclinical candidate for pharmacoresistant epilepsy, in human brain. This paves the way for further study into the mechanisms of ant‐134, and other miRNA‐based approaches, in human brain.
AUTHOR CONTRIBUTIONS
Gareth Morris, Norman Delanty, Mark O. Cunningham, and David C. Henshall conceived the study. Gareth Morris, Elena Langa, Karen Conboy, Kelvin Lau E‐How, Amaya Sanz‐Rodriguez, Austin Lacey, and Jane B. Cryan produced and analyzed data. Conor Fearon, Donncha F. O'Brien, Kieron Sweeney, Norman Delanty, Alan Beausang, Francesca M. Brett, and Jane B. Cryan provided clinical specimens and associated metadata. Gareth Morris and David C. Henshall wrote the manuscript. All authors edited and approved the final version of the manuscript.
CONFLICT OF INTEREST
The Royal College of Surgeons in Ireland (D.C.H.) holds a patent for the inhibition of miRNA‐134 for the treatment of seizure‐related disorders and other neurologic injuries (US 9803200 B2). None of the other authors has any conflict of interest to disclose. We confirm that we have read the Journal's position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
ACKNOWLEDGMENTS
Foremost, we thank the individuals who consented to participate in this research. This publication has emanated from research supported in part by a research grant from Science Foundation Ireland under grant number 16/RC/3948 and cofunded under the European Regional Development Fund and by FutureNeuro industry partners. G.M. is supported by a Marie Skłodowska‐Curie Actions Individual Fellowship (EpimiRTherapy, H2020‐MSCA‐IF‐2018 840262) and an Emerging Leader Fellowship Award from Epilepsy Research UK (grant reference F2102 Morris).
Morris G, Langa E, Fearon C, Conboy K, Lau E‐How K, Sanz‐Rodriguez A, MicroRNA inhibition using antimiRs in acute human brain tissue sections. Epilepsia. 2022;63:e92–e99. 10.1111/epi.17317
Elena Langa and Conor Fearon joint‐second authors.
Contributor Information
Gareth Morris, Email: gareth.morris@ucl.ac.uk.
David C. Henshall, Email: dhenshall@rcsi.ie.
REFERENCES
- 1. Morris G, O'Brien D, Henshall DC. Opportunities and challenges for microRNA‐targeting therapeutics for epilepsy. Trends Pharmacol Sci. 2021;42(7):605–16. 10.1016/j.tips.2021.04.007 [DOI] [PubMed] [Google Scholar]
- 2. Brennan GP, Henshall DC. MicroRNAs as regulators of brain function and targets for treatment of epilepsy. Nat Rev Neurol. 2020;16(9):506–19. 10.1038/s41582-020-0369-8 [DOI] [PubMed] [Google Scholar]
- 3. Bartel DP. Metazoan microRNAs. Cell. 2018;173(1):20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jimenez‐Mateos EM, Engel T, Merino‐Serrais P, McKiernan RC, Tanaka K, Mouri G, et al. Silencing microRNA‐134 produces neuroprotective and prolonged seizure‐suppressive effects. Nat Med. 2012;18(7):1087–94. 10.1038/nm.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morris G, Reschke CR, Henshall DC. Targeting microRNA‐134 for seizure control and disease modification in epilepsy. EBioMedicine. 2019;45:1–9. 10.1016/j.ebiom.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reschke CR, Silva LFA, Norwood BA, Senthilkumar K, Morris G, Sanz‐Rodriguez A, et al. Potent anti‐seizure effects of locked nucleic acid antagomirs targeting mir‐134 in multiple mouse and rat models of epilepsy. Mol Ther Nucleic Acids. 2017;6:45–56. 10.1016/j.omtn.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reschke CR, Silva LFA, Vangoor VR, Rosso M, David B, Cavanagh BL, et al. Systemic delivery of antagomirs during blood‐brain barrier disruption is disease‐modifying in experimental epilepsy. Mol Ther. 2021;29(6):1–12. 10.1016/j.ymthe.2021.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, et al. A brain‐specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–9. [DOI] [PubMed] [Google Scholar]
- 9. Crooke ST, Wang S, Vickers TA, Shen W, Liang XH. Cellular uptake and trafficking of antisense oligonucleotides. Nat Biotechnol. 2017;35(3):230–7. [DOI] [PubMed] [Google Scholar]
- 10. Rosenow F, Lüders H. Presurgical evaluation of epilepsy. Brain. 2001;124:1683–700. [DOI] [PubMed] [Google Scholar]
- 11. Morris G, Rowell R, Cunningham MO. Limitations of animal epilepsy research models: can epileptic human tissue provide translational benefit? ALTEX. 2021;38:451–62. 10.14573/altex.2007082 [DOI] [PubMed] [Google Scholar]
- 12. Wickham J, Brödjegård NG, Vighagen R, Pinborg LH, Bengzon J, Woldbye DPD, et al. Prolonged life of human acute hippocampal slices from temporal lobe epilepsy surgery. Sci Rep. 2018;8(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jones RSG, da Silva AB, Whittaker RG, Woodhall GL, Cunningham MO. Human brain slices for epilepsy research: pitfalls, solutions and future challenges. J Neurosci Methods. 2016;260:221–32. 10.1016/j.jneumeth.2015.09.021 [DOI] [PubMed] [Google Scholar]
- 14. Morris G, Brennan GP, Reschke CR, Henshall DC, Schorge S. Spared CA1 pyramidal neuron function and hippocampal performance following antisense knockdown of microRNA‐134. Epilepsia. 2018;59:1518–26. 10.1111/epi.14475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Campbell A, Morris G, Sanfeliu A, Augusto J, Langa E, Kesavan JC, et al. AntimiR targeting of microRNA‐134 reduces seizures in a mouse model of Angelman syndrome. Mol Ther Nucleic Acids. 2022;28:514–29. 10.1016/j.omtn.2022.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flecknell P. Replacement, reduction and refinement. ALTEX. 2002;19(2):73–8. http://www.ncbi.nlm.nih.gov/pubmed/12098013 [PubMed] [Google Scholar]
- 17. Li M, Jancovski N, Jafar‐Nejad P, Burbano LE, Rollo B, Richards K, et al. Antisense oligonucleotide therapy reduces seizures and extends life span in an SCN2A gain‐of‐function epilepsy model. J Clin Invest. 2021;131(23):e152079. 10.1172/JCI152079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schoch KM, Miller TM. Antisense oligonucleotides: translation from mouse models to human neurodegenerative diseases. Neuron. 2017;94:1056–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Papasaikas P, Valcárcel J. The spliceosome: the ultimate RNA chaperone and sculptor. Trends Biochem Sci. 2016;41:33–45. [DOI] [PubMed] [Google Scholar]


