Abstract
Drimane‐type sesquiterpenes are a class of compounds produced by a wide range of organisms, initially isolated and characterized in plants. Meanwhile, in the past 20–30 years, a large number of novel structures from many divergent fungi have been elucidated. Recently, the biosynthesis of drimane‐type sesquiterpenes and their esters has been explained in two filamentous fungi, namely Aspergillus oryzae and Aspergillus calidoustus, disclosing the basic biosynthetic principles needed to identify similar pathways in the fungal kingdom.
Keywords: Bioactivity, biosynthesis, chemical diversity, drimane, fungal natural products
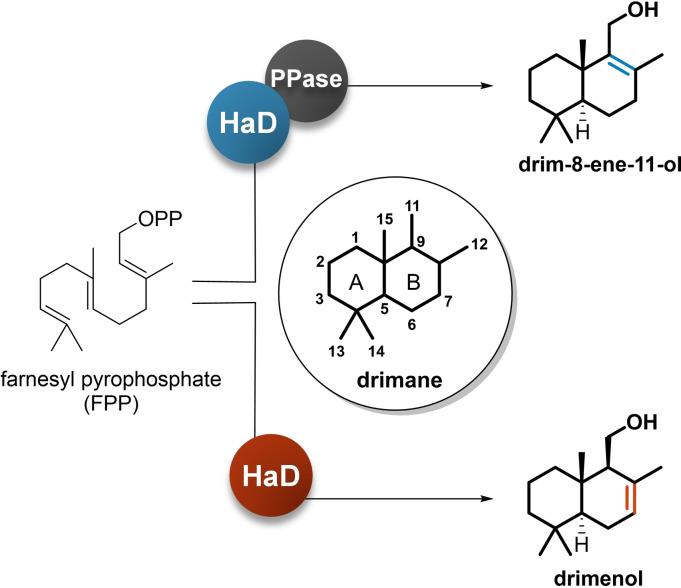
Fungal drimane‐type sesquiterpenes (DTSs) are emerging as a very interesting class of chemicals with many notable activities. Elucidation of DTS biosynthesis in fungi revealed that the two main precursors, drimenol and drim‐8‐ene‐11‐ol, are the preferred backbones. Hence, hundreds of different fungal DTS derivatives have been isolated and identified, suggesting that they are relevant secondary metabolites in the fungal kingdom.
1. Introduction
Fungi are considered an endless source of natural active compounds. [1] These molecules are generally synthetized by the secondary metabolism; thus, named as well secondary metabolites (SMs). The ecological importance of SMs resides in their use in increasing fungal fitness. For this purpose, fungi produce different classes of chemicals that may specifically inhibit the growth of competing organisms.
SMs can be classified as polyketides, nonribosomal peptides (NPRs), terpenes, ribosomally synthesized and post‐translationally modified peptides (RiPPs), alkaloids and phenylpropanoids. The formation of SMs scaffold is synthesized by a central enzyme from a set of simple building blocks, such as acetyl‐, propionyl‐ and malonyl‐CoA for polyketides, isoprene units for terpenes, amino acids for NRPs. Nonetheless, some other compounds may directly use single amino acids as the core structure, such as the tryptophan‐derived alkaloid psilocybin. [2]
Genes involved in the biosynthesis of SMs in fungi tend to be neighboured in the genome. That's why we commonly name the group of genes involved in a specific biosynthesis as a biosynthetic gene cluster (BGCs). Each cluster usually consists of a gene encoding a core enzyme, and adjacent genes encoding modifying enzymes. This unique genetic trait makes the computational analysis and identification of putative BGCs relatively easy. However, we generally use the amino acid sequences of distinct enzymes as baits to identify potential BGCs and, if scaffold‐producing enzymes are unknown, we may miss out on identifying correct hits. This is exactly the issue that we face with the identification of putative fungal BGCs responsible for the production of drimane‐type sesquiterpenes.
Drimane‐type sesquiterpenes (DTSs) constitute an important group of natural products with a unique C15 bicyclic skeleton (Figure 1). They have been firstly identified in plants [3] and quickly attracted interest because of their cytotoxic activity and potential use as anticancer drugs. [4] Later on, numerous additional DTSs have been discovered, many of which with reported insecticidal activity. [5]
Figure 1.
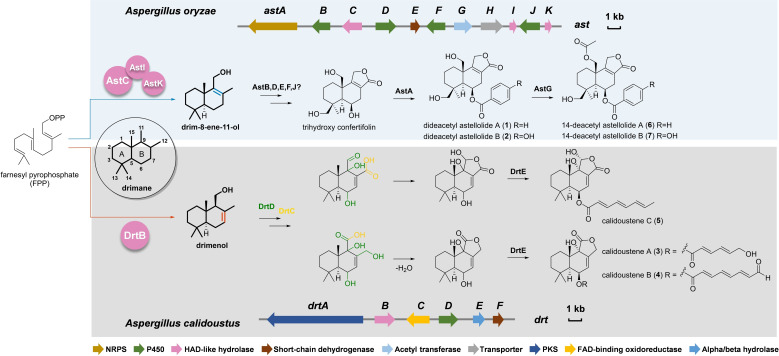
Reported biosynthetic pathways of DTS‐esters in Aspergillus oryzae and Aspergillus calidoustus. NRPS: nonribosomal peptide synthetase; HAD: haloacid dehalogenase; PKS: polyketide synthase; FAD: flavin adenine dinucleotide; ast: astelloide biosynthetic gene cluster; drt: drimane‐type sesquiterpene biosynthetic gene cluster.
The interest in DTSs has been growing with the time, and a large number of synthetic derivatives manufactured and tested. [6] However, besides plants, DTSs have been also isolated in fungi. Early examples are the antibiotic siccanin isolated from Helminthosporium siccans, which displayed antifungal activity, [7] and the cryptoporic acid series, DTS‐ethers from Cryptoporus volvatus exhibiting anti‐tumorigenic effects. [8] Hence, hundreds of fungal DTS have been identified and some of them tested, revealing a wide range of activities including antibacterial, antifungal, antiviral, anti‐inflammatory, and antiproliferative activity. Unfortunately, comparative assays on the many isolated compounds, using standardized methods, are still missing; consequently, it is difficult to estimate structure‐activity relationships between the different derivatives.
Biosynthesis of DTSs in plants starts with the formation of drimenol from farnesyl pyrophosphate (FPP). [9] The biosynthesis involves terpene (or terpenoid) cyclases, enzymes that promote the cyclization of FPP with consequential release of diphosphate group. [10] Considering the structural similarities between the DTSs isolated from plants and fungi, we expected terpene cyclase‐like enzymes to be involved in their biosynthesis; however, this was not the case. The recent advances in elucidating DTSs biosynthesis in fungi revealed that the two main drimane precursors, drimenol and drim‐8‐ene‐11‐ol, are synthesized by enzymes classified as haloacid dehalogenase (HAD)‐like proteins. [11] The HAD‐like superfamily includes a very large and broad compendium of enzymes in which most members primarily manage phosphoryl‐transfer activities. [12] Indeed, the two HAD‐like proteins elucidated in the biosynthesis of DTSs in fungi are quite exceptional in their functions but not in their amino acid sequences; therefore, identifying novel BGCs responsible for DTS biosynthesis using computational analysis still remains challenging.
In this review, we focus specifically on fungal DTSs, from simpler structures to more complex compounds. Among the very large number of DTSs so far isolated in fungi, we focus here on those structures that we consider relevant for their chemistry and proved activity.
2. Biosynthesis of Fungal Drimane‐Type Sesquiterpene Backbones
2.1. Two reported precursors of fungal drimanes
Considering the abundance of DTSs in nature, the first question to address was to assign their biosynthetic origin. Feeding experiments conducted with C‐labelled mevalonate in plants, fungi and marine molluscs could clearly attribute the use of FPP as a precursor. [13] As with other terpenes, the formation of the bicyclic sesquiterpene in drimane biosynthesis may lead to diverse skeletons, differentiated by the presence of a double bond in one of the rings. According to the reported literatures, we know that in fungi there are at least two types of HAD‐like drimane synthases: one forming drim‐8‐ene‐11‐ol, hosting a Δ8,9 double bond, and one forming drimenol, having a double bond at Δ7,8 (Figure 1).
The first fungal drimane synthase was identified during a study aimed at elucidating the biosynthesis of astellolides, antiproliferative compounds produced in Aspergillus oryzae. [11] The ast BGC consists of 11 open reading frames, including three HAD‐like encoding genes. However, one of these, astC, has been validated in vitro and its activity has been confirmed. Nonetheless, this enzyme is not able to hydrolyse FPP by itself and its activity must be supported by the presence of two phosphatases (AstI and AstK) (Figure 1). The obtained drim‐8‐ene‐11‐ol is then further modified ending with the synthesis of dideacetyl astellolide A (1) and B (2). [11]
We assumed that DTSs biosynthesis in fungi would be similar to that reported in A. oryzae after the AstC characterization, but the elucidation of a second BGC changed our perspectives. The drt BGC in Aspergillus calidoustus is responsible for the synthesis of DTSs and their esters. [14] The in vitro activity of the isolated HAD‐like enzyme, DrtB, demonstrated the formation of drimenol. This implies that, similar to plant terpene cyclases involved in polygodial biosynthesis, [15] DrtB is self‐sufficient and produces drimenol by catalysing both dephosphorylation and cyclization of FPP (Figure 1). This explained the biosynthesis of the known DTSs and novel structures named calidoustenes A (3), B (4), and C (5), identified from A. calidoustus.
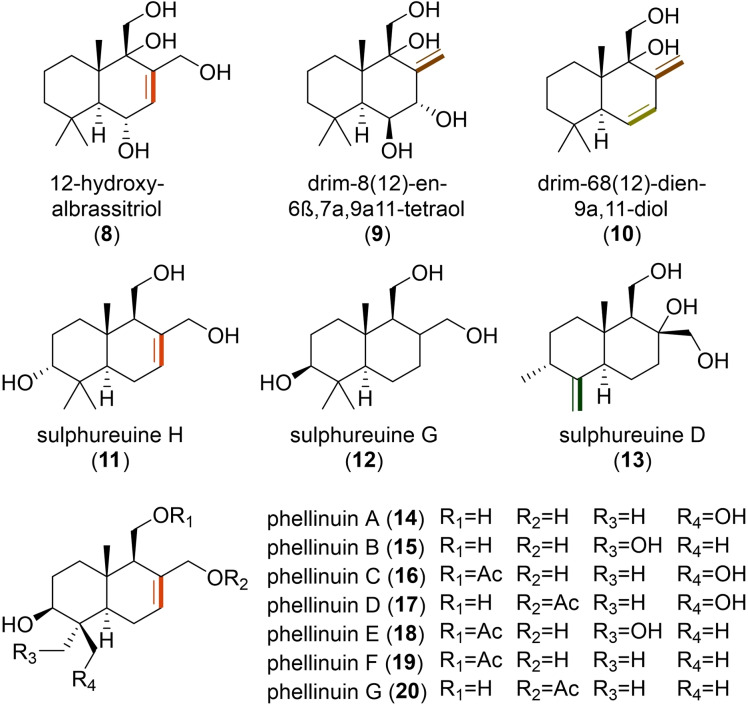
The two main differences highlighted in these biosyntheses seem to be predominant in the fungal DTSs identified so far and likely relevant to drive further chemical modifications. However, we could already observe variations of the bicyclic rings from compounds isolated from the same culture (Figure 2). As an example, in Penicillium sp., the isolation of 12‐hydroxyalbrassitriol (8) containing a Δ7,8 was accompanied with derivatives presenting either a Δ8,12 (9), or two double bonds (Δ6,7 and Δ8,12) (10). [16] This was observed as well for sulphureuine H (11), sulphureuine G (12) and sulphureuine D (13), isolated from Laetiporus sulphureus, which possess a Δ7,8, no double bond, or a Δ4,13 on the rearranged backbone, respectively. [17] Oppositely, there were also drimanes with similar structures and chemical modifications that have been isolated from different fungi, such as phellinuins (14–20). [18]
Figure 2.
Structures of fungal DTSs with bicyclic variations. Some of these structures have been isolated from the same culture, namely Penicillium sp. (8–10) and Laetiporus sulphureus (11–13), while phellinuins (14–20) were identified in different fungi of the division Basidiomycota.
2.2. Tri‐ and tetracyclic DTSs
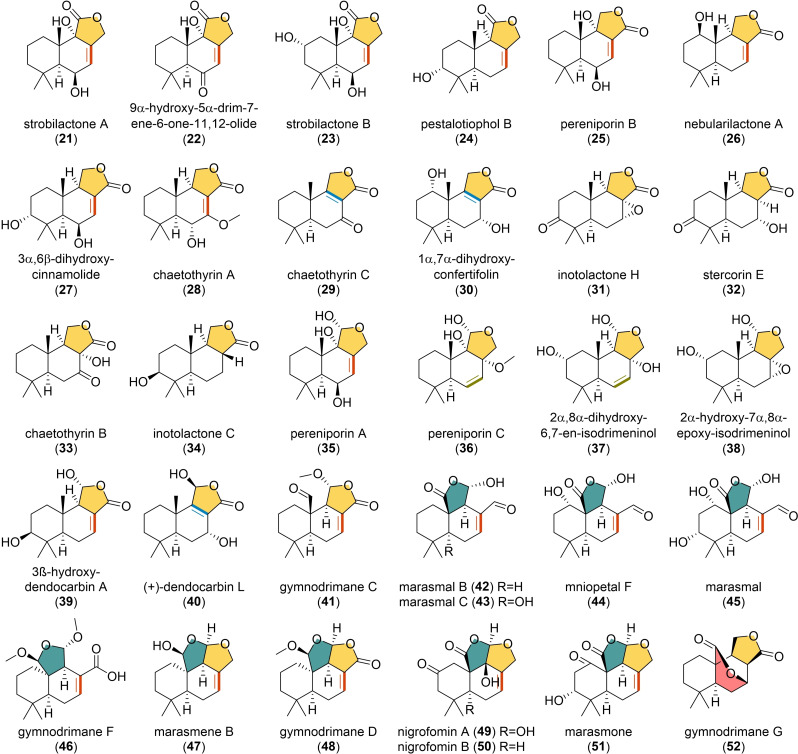
Among the simple DTSs isolated in fungi, the great majority of them are characterized by the presence of a lactone ring (Figure 3). The formation of this structure is generally favoured by the close proximity of an alcohol and an aldehyde or an alcohol and a carboxylic group, leading to the formation of a α‐hydroxytetrahydrofuran or a γ‐butyrolactone, respectively. The position of the lactone carbonyl group diverges depending on whether C‐11 or C‐12 formed a carboxylic acid and condensed with the hydroxy group at C‐12 or C‐11, which would give γ‐butyrolactone ring in 21–24, [19] and 25–34, [20] respectively. These compounds display various activities, for example, strobilactone A (21) and B (23), both isolated from the mushroom Strobilurus ohshimae, have showed antibacterial and antifungal activity,[ 19a , 21 ] the 3α,6β,dihydroxycinnamolide (27) from the fruiting bodies of Inonotus rickii presented a moderate activity on human colon cancer cells [20c] and neurotrophic [22] activity. Differently, pereniporin B (25), from Perenniporia medullaepanis, displayed plant growth inhibitory and cytotoxic activities. [23]
Figure 3.
Structures of representative fungal tri‐ and tetracyclic DTSs.
Slightly different structures could be obtained by the condensation of C‐11 aldehyde with C‐12 hydroxy group or carboxylic acid, which would result in different ring variations, as observed in 35–38[ 20a , 24 ] and 39–41. [25] Among them, pereniporin A (35) was isolated together with 25 and also showed plant growth inhibitor activity, [23] while 6‐epi‐pereniporin A from Perenniporia maackiae exhibited anticancer property. [24a] Concerning their potential biosynthesis, in A. calidoustus, a multistep cytochrome P450 (DrtD) and a FAD‐binding oxidoreductase (DrtC) were solely responsible for subsequent hydroxylations and oxidations that led to the lactone ring formation. [14] In contrast, in A. oryzae, the presence of more genes coding for cytochrome P450 enzymes (AstB, AstD, AstF and AstJ) accompanied by a dehydrogenase (AstE) in the cluster, led to the hypothesis of more specialized single‐step reactions (Figure 1). [11] From an evolutionary perspective, this would explain the occurrence of hydroxylated drimenol derivatives in fungi (Figure 2), suggesting the absence of ketone‐forming enzymes in their corresponding BGCs. Furthermore, in presence of promiscuous cytochrome P450 enzymes, as reported in A. calidoustus, we may find γ‐butyrolactones on diverse orientations from the same organism. [14]
Among these tricyclic DTSs, drimenol derivatives (21–28, 35, 39 and 41) and drim‐8‐ene‐11‐ol derivatives (29, 30 and 40) might have a similar biosynthetic mechanism to that reported in A. calidoustus and A. oryzae, respectively. However, there are numerous reported DTSs with diverging changes on their B ring (31–34 and 36–37). For these molecules, specific enzymes catalysing epoxidations, hydroxylations and reductions can possibly cause double bonds to disappear or shift, as it may be suggested from the isolation of the above mentioned 8–10 from Penicillium sp., [16] sulphureuine 11–13 from Laetiporus sulphureus, and the lactone‐containing derivatives 24, 37 and 38 from Pestalotiopsis sp., [24b] as well as chaetothyrins (28, 29, and 33) from Chaetothyriales sp. [20d] Nonetheless, the biosynthesis of alternative bicyclic sesquiterpenes must be considered; in particular, some structures containing a Δ6,7‐double bond may come from still uncharacterized synthases.
Another type of tricyclic DTSs (Figure 3) have been isolated from a few Agaricomycetes (Basidiomycota). As shown in gymnodrimane C (41), the C‐15 methyl group can also be oxidized to an aldehyde, which can be further oxidized to a carboxylic acid and condensed with the C‐11 aldehyde thereby forming the lactone ring in 42–46.[ 25c , 26 ] However, this chemical characteristic does not inhibit the formation of a supplementary tetrahydrofuran ring, resulting in the tetracyclic DTSs possessing a rare dioxabicyclooctane moiety (47–51).[ 25c , 26a , 27 ] Among them, gymnodrimanes (41, 46 and 48) were isolated from cultures of Gymnopilus sp., together with gymnodrimane G (52), which present a peculiar δ‐lactone ring [25c] that may occur upon the condensation between C‐7 hydroxyl group and C‐15 carboxylic acid. Moreover, marasmals (42 and 43) and marasmene B (47) were isolated from Marasmius sp. and tested for their antifungal activity and found to interfere with conidial germination, [26a] while mniopetal F (44) showed inhibitory effects on reverse transcriptases and exhibited antimicrobial and cytotoxic properties.[ 26b , 28 ] Also, nigrofomins (49 and 50), from Nigrofomes melanoporus, were found to inhibit the growth of acute leukaemia T‐cells. [27a]
Based on the above structures, we can assume that the C‐2 and C‐3 hydroxylations on the A ring are very common in the isolated DTSs. The C‐3 hydroxy group of cryptoporic acid was supposed to be formed catalytically by a cytochrome P450 rather than by the cyclization of epoxyfarnesyl pyrophosphate in Ganoderma neo‐japonicum, but the involved enzyme has not been identified yet. [13b] Additionally, in A. calidoustus such variations led to the synthesis of shunt products that could not be further modified. [14] The heterologous expression of the entire drt BGC in Aspergillus fumigatus, where those shunt products were absent, suggested that such modifications may occur either due to non‐cluster‐associated hydroxylases or through the interaction of other biosynthetic pathways. The latter hypothesis would explain the many analogues present in nature, but so far it has not been proven.
3. Drimane‐Type Sesquiterpene Esters and Ethers
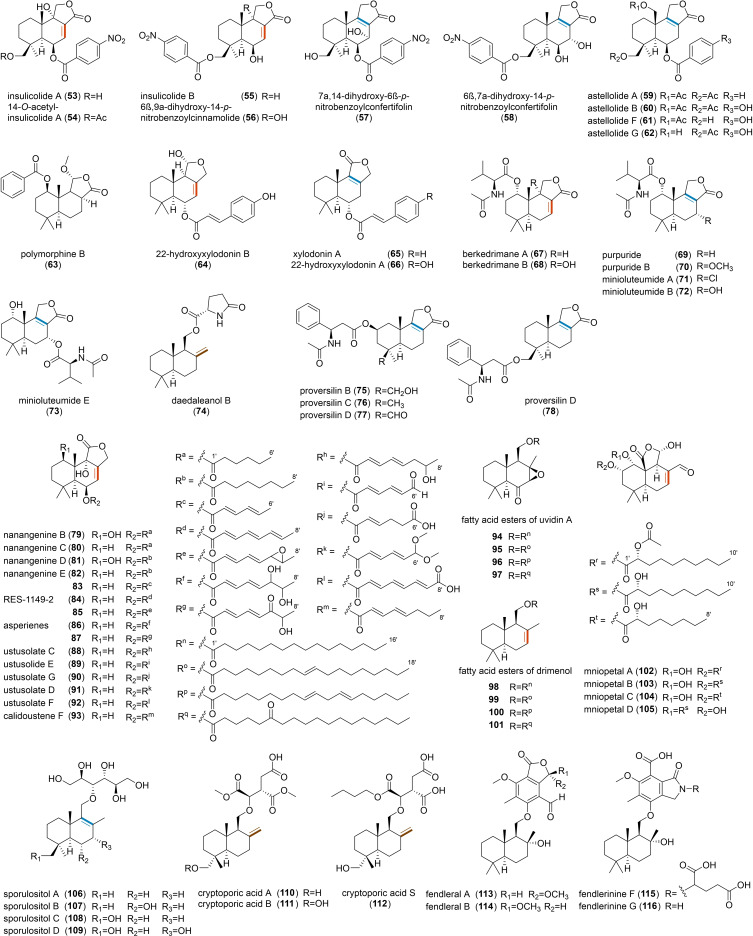
Esterification of DTS backbone has been observed in various fungi, but mainly in the division Ascomycota (Figure 4). As an example, nitrobenzoyl sesquiterpenoids (53–58), having an unusual p‐nitrobenzoic acid moiety at C‐6 and C‐14, were identified in Aspergillus insulicola, [29] Aspergillus ochraceus [30] and Aspergillus versicolor. [31] These compounds have been tested for their antiproliferative activity and found to exert a good cytotoxicity against various cancer cell lines as well as antiviral activity.[ 31 , 32 ] Similarly, benzoyl or hydroxybenzoyl DTSs, the astelloide series (59–62), were isolated from Aspergillus variecolor [33] and A. oryzae. Benzoic acid can also form esters at the C‐1 position, as in polymorphine B (63), which was isolated from Xylaria polymorpha and exhibited anti‐acetylcholinesterase and α‐glucosidase inhibitory activities. [34] Additional DTS‐esters, such as xylodonin derivatives (64–66), presenting similar structures but possessing a p‐coumaroyl or cinnamoyl moiety, have been found to inhibit the osteoclastogenesis and thus are promising therapeutics for treating osteoclast‐related diseases such as osteoporosis. [35] From a biosynthetic perspective, the mechanisms of ester bond formation and acetylation on the DTS backbone have been proposed in A. oryzae, [11] where a nonribosomal peptide synthetase (NRPS), AstA, should be responsible for the AMP‐esterification and transferring of benzoic acid to form 1 and 2, and the acetyl transferase AstG promotes O‐acetylation at position C‐15 to form 6 and 7 (Figure 1), a similar mechanism for compounds 53–66 was hypothesized.
Figure 4.
Structures of representative fungal DTS‐esters and ‐ethers.
DTSs can also form esters with amino acids and their derivatives, but such structures are rare. To date, only about twelve N‐acetyl‐L‐valine‐conjugated DTS‐esters have been identified and found to be present only in Penicillium species. Purpuride (69) was the first example of this type of natural products and was originally isolated from Penicillium purpurogenum. [36] Other examples like berkedrimanes A (67) and B (68), [37] purpuride B (70) [38] as well as minioluteumides A (71) and B (72) [39] are all conjugated with N‐acetyl‐L‐valine at C‐1, while minioluteumide E (73) [39] contains the N‐acetyl‐L‐valine at C‐7. These compounds have showed antimicrobial activities, [40] with 67 and 68 found to have anti‐inflammatory activity. [37] Moreover, L‐pyroglutamate has been also associated to DTS‐ester, as in daedaleanol B (74). [41] Interestingly, A. versicolor produced proversilins (75–78), the first four examples of natural products with an N‐acetyl‐β‐phenylalanine moiety, of which 76 and 77 showed moderate cytotoxic activity. However, the putative BGCs for these amino‐acid‐conjugated DTS‐esters are still unknown.
The involvement of PKS in DTS‐esters biosynthesis have been reported in different Aspergilli (79–93),[ 14 , 42 ] and esterification usually occurs at C‐6 (Figure 4). All those molecules are characterized by the presence of reduced or partially reduced polyketides. Biosynthetic studies in A. calidoustus have pointed out the involvement of an acyl‐transferase (DrtE) able to transfer the carboxylic acid from the ACP domain of the polyketide synthase to the DTS backbone. Interestingly, DrtE could also transfer various CoA‐activated substrates, suggesting that alternative routes for the synthesis of DTS‐esters with benzoic or cinnamic acid moieties may exist in nature. Moreover, computational analysis on available genomes revealed that the DTS‐ester pathway from A. calidoustus is much conserved in other Aspergilli and that the polyketide derivatives are associated with modifications of the enyolreductase (ER) domain of the cluster‐associated PKS. [14] The esterified forms of uvidin A (94–97) and even simple drimenol (98–101), with C16 and C18 acyl chains added at C‐11, have been isolated from the mushroom Lactarius uvidus [43] (Figure 4). These compounds suggest that the structures in Figure 3 represent potential precursors for further modifications. This is somehow confirmed by mniopetal F (44), isolated from the basidiomycetes Marasmius oreades, which can be further esterified with C8 and C10 acyl chains at C‐1 or C‐2 (102–105). [26b] These compounds have been tested for various activities and found to be effective in inhibiting the mammalian RNA‐directed DNA‐polymerases. [44] The biosynthesis of these esters with long acyl chains is still unknown. However, we may hypothesize that the acyl chains are either originated from PKSs or fatty acid synthases; however, in both cases, an acyl‐transferase should be involved in the biosynthesis.
Different from the large chemical variety of DTS‐esters, ethers are usually formed at C‐11 (Figure 4). Reported examples include sporulositols (106–109), isolated from Paraconiothyrium sporulosum, containing a D‐mannitol group. [45] Other relevant ethers are cryptoporic acids (110–112), with an isocitric acid moiety, initially isolated from C. volvatus. [46] Noteworthy, cryptoporic acid derivatives have been identified in both ascomycetes [47] and basidiomycetes, [48] and presented various activities including superoxide release inhibitory,[ 8b , 8c ] anti‐tumour, [8c] antioxidant, [46b] antiplasmodial, [49] antimyco‐bacterial, [49] and general cytotoxic activities. [49] Furthermore, DTS‐ethers with more complex moieties have been isolated from Hypoxylon fendleri, with fendlerals A (113) and B (114) containing a phthalide group and showing a strong antibacterial activity against Bacillus cereus; however, fendlerinines F (115) and G (116), which possess an isoindolinone moiety, have shown no significant activity. Among the identified ethers, we could observe saturated drimane backbones, like in fendlerals and fendlerinines (113–116); this may be due to the specific hydratase catalysing the hydration reaction, thus causing their double bonds to disappear and produce a C‐8 hydroxyl group.
4. Di‐ and Trimeric Structures
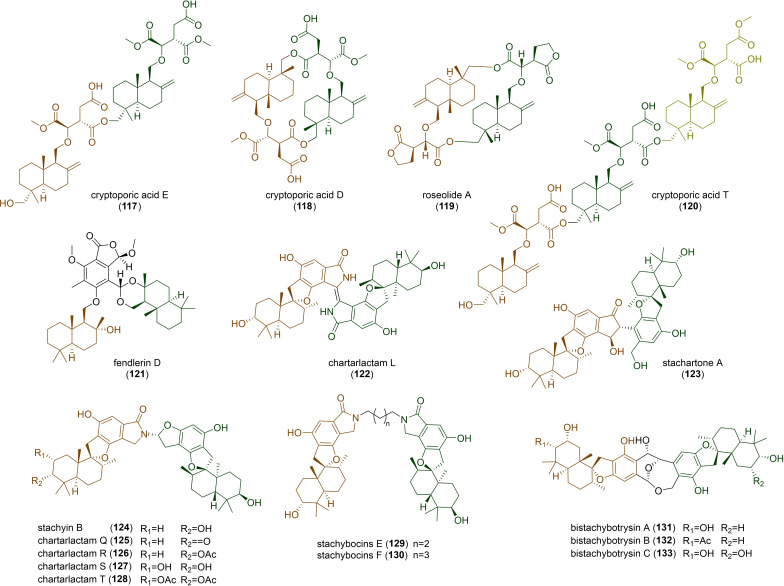
DTS‐ether cryptoporic acids and their derivatives often form linear (117), [8b] cyclic dimers (118 and 119),[ 8b , 50 ] or even trimers (120) [51] (Figure 5). Unlike two ether molecules that are polymerized together, phthalide‐containing DTS‐ester can combine with another unit of DTS to form fendlerin D (121) [52] (Figure 5). Drimane‐related merosesquiterpenoids should be biosynthetically distinct from the pathways of the DTSs we have mentioned, as suggested in macrophorins, where the cyclization was catalysed by a membrane‐bound type II terpene cyclase, MacJ, through direct olefinic bond protonation of the terminal olefinic bond in acyclic yanuthones. [53] It's of interest that these compounds can also form dimers, usually phenylspirodrimane dimers, as observed in the genus Stachybotrys. [54] For this type of dimers, they can be polymerized by two units of identical phenylspirodrimanes (122), [55] or two units of different phenylspirodrimanes (123). [56] Unfortunately, the biosynthesis of these compounds has not been clarified so far. In other cases, the dimerization of two units of phenylspirodrimanes can also be linked by a C−N bond to form stachyin B (124) and chartarlactams (125–128). Notably, some phenylspirodrimanes exhibited antibacterial activities against the methicillin‐resistant Staphylococcus aureus. [57] Lastly, stachybocins (129 and 130), which are presumably formed by the reaction between phenylspirodrimane and lysine with subsequent decarboxylation, [58] and bistachybotrysins (131–133), which harbour an unusual [6,6,7,7]‐tetracyclic skeleton and exhibited a potent cytotoxicity, [59] have also been isolated from the genus Stachybotrys.
Figure 5.
Structures of representative fungal polymeric drimanes.
5. Perspectives
The discovery of DTSs has broadened our knowledge of chemical diversity in fungi. Like other classic terpenes, the DTSs share a cyclized hydrocarbon backbone, which is then modified by a series of tailoring enzymes to produce hundreds of distinct metabolites from a single backbone. This is different from other fungal secondary metabolites, where the backbones originating from PKSs, NRPSs and RiPPs are, in their respective groups, already quite divergent. [1] Admittedly, the diversity of terpenes is also influenced by the way linear polyenes are isomerized and cyclized, [60] as well as meroterpenes, derived from hybrid isoprenic precursors, whose skeletons are affected by the non‐terpene portion. [61]
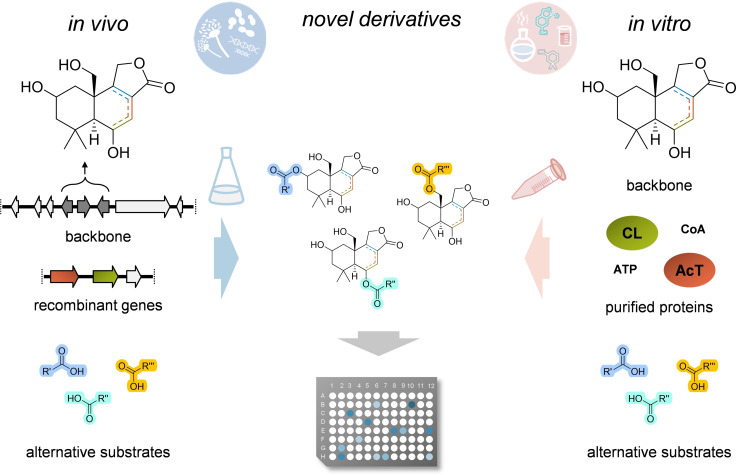
According to the many DTS structures identified so far, we may conclude that their chemical specializations strongly rely on modifications, occurring in nature to create DTS‐derivatives with very diverse activities, many of them with pharmacological potency. In this perspective, fungal DTSs appear to be extremely suitable for combinatorial biosynthesis through genome engineering or cell‐free multistep catalyses (Figure 6). The only limiting factor still remains the poor knowledge of naturally occurring DTS‐modifying enzymes. We have already learned that in A. calidoustus only three enzymes are responsible for the formation of the drimane‐type backbone and the associated γ‐butyrolactone, and that the involved acyltransferase can transfer various acyl‐CoA to the main scaffold. [14] However, this activity appears to be limited to the modification of the hydroxyl group at the C‐6 position, but, as we have observed in other fungal DTSs, hydroxyl groups at other positions can also be used for the esterification. In the future, we may well use this class of molecules to establish a synthetic biology platform to rationally modify active DTSs. First, we can easily create fungal strains producing the required backbones, with specific ring‐saturations and targeted hydroxylations, and then in the same modified organism we may express any suitable CoA‐ligases (CL) and acyltransferases (AcT). Complex substrates can be added by precursor‐directed feeding in the growing medium, a strategy that has been successfully used for other derivatizations, as reported for plant derived flavonoids and bacterial indole alkaloids and lasso peptides. [62] This will permit to obtain DTS chemical libraries that can be directly used for the screening of activities (Figure 6). In this perspective, it would be also of interest to investigate if DTS‐ethers can be added and modified as well.
Figure 6.
Combinatorial biosynthesis of novel bioactive DTSs through genome engineering or cell‐free multistep catalyses. CoA, coenzyme A; CL, acyl‐CoA ligase; AcT, acyl‐transferase.
In recent years, the opportunity to produce active compounds in vitro is emerging. Cell‐free approaches are more favourable than classical fermentation, because they give the possibility to reduce the size of biofermenters, optimize the production buffers, and reduce the purification costs of the final bioproduct. [63]
Furthermore, since in vitro enzymes tend to use only the available precursors, cell‐free multi‐step catalyses are very efficient regarding the use of unnatural substrates for chemical derivatizations. This would be a more advisable way to produce DTS by‐products. The backbone structure can be either produced in vivo or chemically synthetized, incubated with the purified enzymes and their suitable substrates and cofactors, and then the products directly analysed for their activity (Figure 6). However, once again, this method can result in a high number of different molecules only when more modifying enzymes will be identified and their substrate promiscuity assayed.
In conclusion, fungal DTSs are emerging as a very interesting class of chemicals with many notable activities. The high number of DTSs identified so far suggests that they are important secondary metabolites in the fungal kingdom. Their chemical structure is quite simple and can potentially be modified through rewiring of existing pathways and / or heterologous expression. Therefore, fungal DTSs are ideal candidates to build chemical libraries for the screening of new activities.
Conflict of interest
The authors declare no conflict of interest.
Biographical Information
Ying Huang obtained her MSc in medicinal chemistry from Kunming Institute of Botany, Chinese Academy of Sciences. Her Master's thesis was focused on the secondary metabolites of higher fungi and their bioactivities. She is currently a PhD candidate in Biochemistry at the Leibniz Institute for Natural Product Research and Infection Biology (HKI) under the guidance of Dr. Vito Valiante. Her PhD research focuses on the biosynthesis of natural products.

Biographical Information
Vito Valiante obtained his MSc in Plant Biotechnology and his PhD in Biotechnology at the University of Naples Federico II. He was a postdoctoral researcher in the Department of Molecular and Applied Microbiology (2006–2015), at the Leibniz Institute for Natural Products and Infection Biology, in Jena and has been an independent junior research group leader since 2015, heading the group Biobricks of Microbial Natural Product Syntheses. His scientific interests are chemical biology, biochemistry, synthetic biology, and microbial genetics.

Acknowledgements
We thank Katarina Jojić for proofreading the manuscript and the International Leibniz Research School (ILRS) for their support. Open Access funding enabled and organized by Projekt DEAL.
Y. Huang, V. Valiante, ChemBioChem 2022, 23, e202200173.
References
- 1. Keller N. P., Nat. Rev. Microbiol. 2019, 17, 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fricke J., Blei F., Hoffmeister D., Angew. Chem. Int. Ed. 2017, 56, 12352–12355; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 12524–12527. [Google Scholar]
- 3. Appel H., Scientia 1948, 15, 31–32. [Google Scholar]
- 4. Mahmoud I. I., Kinghorn A. D., Cordell G. A., Farnsworth N. R., J. Nat. Prod. 1980, 43, 365. [DOI] [PubMed] [Google Scholar]
- 5.
- 5a. Kubo I., Ganjian I., Experientia 1981, 37, 1063–1064; [DOI] [PubMed] [Google Scholar]
- 5b. Kubo I., Lee Y. W., Pettei M., Pilkiewicz F., Nakanishi K., J. Chem. Soc. Chem. Commun. 1976, 1013–1014. [Google Scholar]
- 6. Jansen B., De Groot A., Nat. Prod. Rep. 2004, 21, 449–477. [DOI] [PubMed] [Google Scholar]
- 7. Hirai K., Nozoe S., Tsuda K., Iitaka Y., Ishibashi K., Shirasaka M., Tetrahedron Lett. 1967, 8, 2177–2179. [DOI] [PubMed] [Google Scholar]
- 8.
- 8a. Narisawa T., Fukaura Y., Kotanagi H., Asakawa Y., Jpn. J. Cancer Res. 1992, 83, 830; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8b. Hashimoto T., Tori M., Mizuno Y., Asakawa Y., Fukazawa Y., J. Chem. Soc. Chem. Commun. 1989, 258; [Google Scholar]
- 8c. Asakawa Y., Hashimoto T., Mizuno Y., Tori M., Fukazawa Y., Phytochemistry 1992, 31, 579. [Google Scholar]
- 9.
- 9a. Kwon M., Cochrane S. A., Vederas J. C., Ro D.-K., FEBS Lett. 2014, 588, 4597–4603; [DOI] [PubMed] [Google Scholar]
- 9b. Henquet M. G., Prota N., van der Hooft J. J., Varbanova-Herde M., Hulzink R. J., de Vos M., Prins M., de Both M. T., Franssen M. C., Bouwmeester H., Plant J. 2017, 90, 1052–1063. [DOI] [PubMed] [Google Scholar]
- 10. Christianson D. W., Chem. Rev. 2017, 117, 11570–11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shinohara Y., Takahashi S., Osada H., Koyama Y., Sci. Rep. 2016, 6, 32865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chan P. W. Y., Chakrabarti N., Ing C., Halgas O., To T. K. W., Wälti M., Petit A. P., Tran C., Savchenko A., Yakunin A. F., Edwards E. A., Pomès R., Pai E. F., ChemBioChem 2022, 23, e202100414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.
- 13a. Fontana A., Villani G., Cimino G., Tetrahedron Lett. 2000, 41, 2429–2433; [Google Scholar]
- 13b. Hirotani M., Ino C., Furuya T., Phytochemistry 1993, 32, 891; [Google Scholar]
- 13c. Opiyo S. A., Trends Phytochem. Res. 2019, 3, 147–180. [Google Scholar]
- 14. Huang Y., Hoefgen S., Valiante V., Angew. Chem. Int. Ed. 2021, 60, 23763–23770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Henquet M. G. L., Prota N., van der Hooft J. J. J., Varbanova-Herde M., Hulzink R. J. M., de Vos M., Prins M., de Both M. T. J., Franssen M. C. R., Bouwmeester H., Jongsma M., Plant J. 2017, 90, 1052–1063. [DOI] [PubMed] [Google Scholar]
- 16. Ding J. H., Ding Z. G., Chunyu W. X., Zhao J. Y., Wang H. B., Liu S. W., Wang F., J. Asian Nat. Prod. Res. 2017, 19, 780–785. [DOI] [PubMed] [Google Scholar]
- 17. He J. B., Tao J., Miao X. S., Bu W., Zhang S., Dong Z. J., Li Z. H., Feng T., Liu J. K., Fitoterapia 2015, 102, 1–6. [DOI] [PubMed] [Google Scholar]
- 18.
- 18a. He J. B., Feng T., Zhang S., Dong Z. J., Li Z. H., Zhu H. J., Liu J. K., Nat. Prod. Bioprospect. 2014, 4, 21–25; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18b. Zhao J. Y., Ding J. H., Li Z. H., Dong Z. J., Feng T., Zhang H. B., Liu J. K., J. Asian Nat. Prod. Res. 2013, 15, 305–309. [DOI] [PubMed] [Google Scholar]
- 19.
- 19a. Shiono Y., Hiramatsu F., Murayama T., Koseki T., Funakoshi T., Ueda K., Yasuda H., Z. Naturforsch. B 2007, 62, 1585–1589; [Google Scholar]
- 19b. Zhuravleva O. I., Afiyatullov S., Denisenko V. A., Ermakova S. P., Slinkina N. N., Dmitrenok P. S., Kim N. Y., Phytochemistry 2012, 80, 123–131; [DOI] [PubMed] [Google Scholar]
- 19c. Xiao J., Lin L., Hu J., Jiao F., Duan D., Zhang Q., Tang H., Gao J., Wang L., Wang X., RSC Adv. 2017, 7, 29071–29079. [Google Scholar]
- 20.
- 20a. Kida T., Shibai H., Seto H., J. Antibiot. 1986, 39, 613–615; [DOI] [PubMed] [Google Scholar]
- 20b. Wangun H. V. K., Doerfelt H., Hertweck C., Eur. J. Org. Chem. 2006, 2006, Issue 7, 1643–1646; [Google Scholar]
- 20c. Chen H. P., Dong W. B., Feng T., Yin X., Li Z. H., Dong Z. J., Li Y., Liu J. K., J. Asian Nat. Prod. Res. 2014, 16, 581–586; [DOI] [PubMed] [Google Scholar]
- 20d. Zhou Y. H., Li X. B., Zhang J. Z., Li L., Zhang M., Chang W. Q., Wang X. N., Lou H. X., J. Asian Nat. Prod. Res. 2016, 18, 409–414; [DOI] [PubMed] [Google Scholar]
- 20e. Chang D. D., Zuo W. J., Mei W. L., Dai H. F., J. Asian Nat. Prod. Res. 2012, 14, 577–580; [DOI] [PubMed] [Google Scholar]
- 20f. Zou C. X., Wang X. B., Lv T. M., Hou Z. L., Lin B., Huang X. X., Song S. J., Bioorg. Chem. 2020, 96, 103588; [DOI] [PubMed] [Google Scholar]
- 20g. Yin X., Qi J., Li Y., a Bao Z., Du P., Kou R., Wang W., Gao J. M., Nat. Prod. Res. 2021, 35, 4524–4533; [DOI] [PubMed] [Google Scholar]
- 20h. Ying Y. M., Zhang L. Y., Zhang X., Bai H. B., Liang D. E., Ma L. F., Shan W. G., Zhan Z. J., Phytochemistry 2014, 108, 171–176. [DOI] [PubMed] [Google Scholar]
- 21. Cao F., Zhao D., Chen X. Y., Liang X. D., Li W., Zhu H. J., Chem. Nat. Compd. 2017, 53, 1189–1191. [Google Scholar]
- 22. Kou R. W., Du S. T., Li Y. X., Yan X. T., Zhang Q., Cao C. Y., Yin X., Gao J. M., J. Antibiot. 2019, 72, 15–21. [DOI] [PubMed] [Google Scholar]
- 23. Kida T., Shibai H., Seto H., J. Antibiot. 1986, 39, 613–615. [DOI] [PubMed] [Google Scholar]
- 24.
- 24a. Kwon J., Lee H., Seo Y. H., Yun J., Lee J., Kwon H. C., Guo Y., Kang J. S., Kim J. J., Lee D., J. Nat. Prod. 2018, 81, 1444–1450; [DOI] [PubMed] [Google Scholar]
- 24b. Kuang C., Jing S. X., Liu Y., Luo S. H., Li S. H., Nat. Prod. Bioprospect. 2016, 6, 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.
- 25a. Chen Z., Dong Z., Wen J., Feng T., Liu J., Rec. Nat. Prod. 2016, 10, 307–310; [Google Scholar]
- 25b. Riga R., Happyana N., Hakim E. H., Nat. Prod. Res. 2020, 34, 2229–2231; [DOI] [PubMed] [Google Scholar]
- 25c. Isaka M., Sappan M., Suvannakad R., Boonpratuang T., Thummarukcharoen T., Phytochem. Lett. 2020, 35, 141–146. [Google Scholar]
- 26.
- 26a. Liermann J. C., Thines E., Opatz T., Anke H., J. Nat. Prod. 2012, 75, 1983–1986; [DOI] [PubMed] [Google Scholar]
- 26b. Velten R., Klostermeyer D., Steffan B., Steglich W., Kuschel A., Anke T., J. Antibiot. 1994, 47, 1017. [DOI] [PubMed] [Google Scholar]
- 27.
- 27a. Chan H. H., Juang S. H., Thang T. D., Chen M. Y., Kuo P. C., Yang M. L., Nguyen T. N., Nguyen N. L., Wu T. S., Planta Med. 2012, 78, 737–739; [DOI] [PubMed] [Google Scholar]
- 27b. Ayer W. A., Craw P. A., Stout T. J., Clardy J., Can. J. Chem. 1989, 67, 773. [Google Scholar]
- 28. Kuschel A., Anke T., Velten R., Klostermeyer D., Steglich W., König B., J. Antibiot. 1994, 47, 733–739. [DOI] [PubMed] [Google Scholar]
- 29. Rahbæk L., Christophersen C., Frisvad J., Bengaard H. S., Larsen S., Rassing B. R., J. Nat. Prod. 1997, 60, 811–813. [Google Scholar]
- 30.
- 30a. Tan Y., Yang B., Lin X., Luo X., Pang X., Tang L., Liu Y., Li X., Zhou X., J. Nat. Prod. 2018, 81, 92–97; [DOI] [PubMed] [Google Scholar]
- 30b. Fang W., Lin X., Zhou X., Wan J., Lu X., Yang B., Ai W., Lin J., Zhang T., Tu Z., MedChemComm 2014, 5, 701–705. [Google Scholar]
- 31. Belofsky G. N., Jensen P. R., Renner M. K., Fenical W., Tetrahedron 1998, 54, 1715–1724. [Google Scholar]
- 32.
- 32a. Fang W., Lin X., Zhou X., Wan J., Lu X., Yang B., Ai W., Lin J., Zhang T., Tu Z., Liu Y., MedChemComm 2014, 5, 701–705; [Google Scholar]
- 32b. Tan Y., Yang B., Lin X., Luo X., Pang X., Tang L., Liu Y., Li X., Zhou X., J. Nat. Prod. 2018, 81, 92–97. [DOI] [PubMed] [Google Scholar]
- 33. Gould R. O., Simpson T. J., Walkinshaw M. D., Tetrahedron Lett. 1981, 22, 1047–1050. [Google Scholar]
- 34. Yang N., Kong F., Ma Q., Huang S., Luo D., Zhou L., Dai H., Yu Z., Zhao Y., Youji Huaxue 2017, 37, 1033–1039. [Google Scholar]
- 35. Kwon J., Lee H., Ryu S. M., Jang Y., Kwon H. C., Guo Y., Kang J. S., Kim J. J., Lee D., J. Nat. Prod. 2019, 82, 2835–2841. [DOI] [PubMed] [Google Scholar]
- 36. King T., Roberts J. C., Thompson D., J. Chem. Soc. Perkin Trans. 1 1973, 78–80. [DOI] [PubMed] [Google Scholar]
- 37. Stierle D. B., Stierle A. A., Girtsman T., McIntyre K., Nichols J., J. Nat. Prod. 2012, 75, 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang H., Wang Y., Liu P., Wang W., Fan Y., Zhu W., Chem. Biodiversity 2013, 10, 1185–1192. [DOI] [PubMed] [Google Scholar]
- 39. Ngokpol S., Suwakulsiri W., Sureram S., Lirdprapamongkol K., Aree T., Wiyakrutta S., Mahidol C., Ruchirawat S., Kittakoop P., Mar. Drugs 2015, 13, 3567–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.
- 40a. Kaleem S., Ge H., Yi W., Zhang Z., Wu B., Nat. Prod. Res. 2021, 35, 2498–2506; [DOI] [PubMed] [Google Scholar]
- 40b. Ma M., Ge H., Yi W., Wu B., Zhang Z., Tetrahedron Lett. 2020, 61, 151504. [Google Scholar]
- 41. Huang Y., Zhang S. B., Chen H. P., Zhao Z. Z., Li Z. H., Feng T., Liu J. K., Nat. Prod. Res. 2019, 33, 74–79. [DOI] [PubMed] [Google Scholar]
- 42.
- 42a. Felix S., Sandjo L. P., Opatz T., Erkel G., Beilstein J. Org. Chem. 2013, 9, 2866; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42b. Felix S., Sandjo L. P., Opatz T., Erkel G., Bioorg. Med. Chem. 2014, 22, 2912–2918; [DOI] [PubMed] [Google Scholar]
- 42c. Lacey H. J., Gilchrist C. L., Crombie A., Kalaitzis J. A., Vuong D., Rutledge P. J., Turner P., Pitt J. I., Lacey E., Chooi Y. H., Beilstein J. Org. Chem. 2019, 15, 2631–2643; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42d. Hayes M. A., Wrigley S. K., Chetland I., Reynolds E. E., Ainsworth A. M., Renno D. V., Latif M. A., Cheng X. M., Hupe D. J., J. Antibiot. 1996, 49, 505–512; [DOI] [PubMed] [Google Scholar]
- 42e. Liu H., Edrada-Ebel R., Ebel R., Wang Y., Schulz B., Draeger S., Müller W. E., Wray V., Lin W., Proksch P., J. Nat. Prod. 2009, 72, 1585–1588; [DOI] [PubMed] [Google Scholar]
- 42f. Lu Z., Wang Y., Miao C., Liu P., Hong K., Zhu W., J. Nat. Prod. 2009, 72, 1761–1767; [DOI] [PubMed] [Google Scholar]
- 42g. Liu X. H., Miao F. P., Qiao M. F., Cichewicz R. H., Ji N. Y., RSC Adv. 2013, 3, 588–595; [Google Scholar]
- 42h. Liu Y. F., Yue Y. F., Feng L. X., Zhu H. J., Cao F., Mar. Drugs 2019, 17, 550; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42i. Zhou H., Zhu T., Cai S., Gu Q., Li D., Chem. Pharm. Bull. 2011, 59, 762–766. [DOI] [PubMed] [Google Scholar]
- 43. Garlaschelli L., Mellerio G., Vidari G., Vita-Finzi P., J. Nat. Prod. 1994, 57, 905. [DOI] [PubMed] [Google Scholar]
- 44. Kuschel A., Anke T., Velten R., Klostermeyer D., Steglich W., König B., J. Antibiot. 1994, 47, 733–739. [DOI] [PubMed] [Google Scholar]
- 45. Zhang L. H., Chen G., Sun Y., Wang H. F., Bai J., Hua H. M., Pei Y. H., Molecules 2019, 24, 1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.
- 46a. Hashimoto T., Tori M., Mizuno Y., Asakawa Y., Tetrahedron Lett. 1987, 28, 6303; [Google Scholar]
- 46b. Wang J. C., Li G. Z., Lv N., Shen L. G., Shi L. L., Si J. Y., J. Asian Nat. Prod. Res. 2017, 19, 719–724. [DOI] [PubMed] [Google Scholar]
- 47. Takahashi H., Toyota M., Asakawa Y., Phytochemistry 1993, 33, 1055. [Google Scholar]
- 48.
- 48a. Morita Y., Hayashi Y., Sumi Y., Kodaira A., Shibata H., Biosci. Biotechnol. Biochem. 1995, 59, 2008; [Google Scholar]
- 48b. Meng J., Li Y. Y., Ou Y. X., Song L. F., Lu C. H., Shen Y. M., Mycology 2011, 2, 30–36. [Google Scholar]
- 49. Isaka M., Chinthanom P., Danwisetkanjana K., Choeyklin R., Phytochem. Lett. 2014, 7, 97–100. [Google Scholar]
- 50. Nozoe S., Agatsuma T., Takahashi A., Ohishi H., In Y., Kusano G., Tetrahedron Lett. 1993, 34, 2497. [Google Scholar]
- 51. Pham H. T., Lee K. H., Jeong E., Woo S., Yu J., Kim W. Y., Lim Y. W., Kim K. H., Kang K. B., J. Nat. Prod. 2021, 84, 298–309. [DOI] [PubMed] [Google Scholar]
- 52. Intaraudom C., Punyain W., Bunbamrung N., Dramae A., Boonruangprapa T., Pittayakhajonwut P., Fitoterapia 2019, 138, 104353. [DOI] [PubMed] [Google Scholar]
- 53. Tang M. C., Cui X., He X., Ding Z., Zhu T., Tang Y., Li D., Org. Lett. 2017, 19, 5376–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Speck K., Magauer T., Beilstein J. Org. Chem. 2013, 9, 2048–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li Y., Wu C., Liu D., Proksch P., Guo P., Lin W., J. Nat. Prod. 2014, 77, 138–147. [DOI] [PubMed] [Google Scholar]
- 56. Ding Z. G., Zhao J. Y., Ding J. H., Chunyu W. X., Li M. G., Gu S. J., Wang F., Wen M. L., Nat. Prod. Res. 2018, 32, 2370–2374. [DOI] [PubMed] [Google Scholar]
- 57.
- 57a. Liu D., Li Y., Guo X., Ji W., Lin W., Chem. Biodiversity 2020, 17, e2000170; [DOI] [PubMed] [Google Scholar]
- 57b. Wu B., Oesker V., Wiese J., Malien S., Schmaljohann R., Imhoff J. F., Mar. Drugs 2014, 12, 1924–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ma X., Li L., Zhu T., Ba M., Li G., Gu Q., Guo Y., Li D., J. Nat. Prod. 2013, 76, 2298–2306. [DOI] [PubMed] [Google Scholar]
- 59. Zhao J., Feng J., Tan Z., Liu J., Zhang M., Chen R., Xie K., Chen D., Li Y., Chen X., Dai J., Bioorg. Med. Chem. Lett. 2018, 28, 355–359. [DOI] [PubMed] [Google Scholar]
- 60. Quin M. B., Flynn C. M., Schmidt-Dannert C., Nat. Prod. Rep. 2014, 31, 1449–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jiang M., Wu Z., Liu L., Chen S., Org. Biomol. Chem. 2021, 19, 1644–1704. [DOI] [PubMed] [Google Scholar]
- 62.
- 62a. Kufs J. E., Hoefgen S., Rautschek J., Bissell A. U., Graf C., Fiedler J., Braga D., Regestein L., Rosenbaum M. A., Thiele J., Valiante V., ACS Synth. Biol. 2020, 9, 1823–1832; [DOI] [PubMed] [Google Scholar]
- 62b. Si Y., Kretsch A. M., Daigh L. M., Burk M. J., Mitchell D. A., J. Am. Chem. Soc. 2021, 143, 5917–5927; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62c. Khatri Y., Hohlman R. M., Mendoza J., Li S., Lowell A. N., Asahara H., Sherman D. H., ACS Synth. Biol. 2020, 9, 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ji X., Liu W. Q., Li J., Curr. Opin. Microbiol. 2022, 67, 102142. [DOI] [PubMed] [Google Scholar]