Abstract
Mitochondrial DNA remains a cornerstone for molecular ecology, especially for study species from which high‐quality tissue samples cannot be easily obtained. Methods using mitochondrial markers are usually reliant on reference databases, but these are often incomplete. Furthermore, available mitochondrial genomes often lack crucial metadata, such as sampling location, limiting their utility for many analyses. Here, we assembled 205 new mitochondrial genomes for platyrrhine primates, most from the Amazon and with known sampling locations. We present a dated mitogenomic phylogeny based on these samples along with additional published platyrrhine mitogenomes, and use this to assess support for the long‐standing riverine barrier hypothesis (RBH), which proposes that river formation was a major driver of speciation in Amazonian primates. Along the Amazon, Negro, and Madeira rivers, we found mixed support for the RBH. While we identified divergences that coincide with a river barrier, only some occur synchronously and also overlap with the proposed dates of river formation. The most compelling evidence is for the Amazon river potentially driving speciation within bearded saki monkeys (Chiropotes spp.) and within the smallest extant platyrrhines, the marmosets and tamarins. However, we also found that even large rivers do not appear to be barriers for some primates, including howler monkeys (Alouatta spp.), uakaris (Cacajao spp.), sakis (Pithecia spp.), and robust capuchins (Sapajus spp.). Our results support a more nuanced, clade‐specific effect of riverine barriers and suggest that other evolutionary mechanisms, besides the RBH and allopatric speciation, may have played an important role in the diversification of platyrrhines.
Keywords: mitochondrial DNA, molecular phylogenetics, platyrrhines, riverine barrier hypothesis, South American primates
1. INTRODUCTION
Although the number of whole genomes available for nonmodel organisms has grown dramatically, mitochondrial DNA (mtDNA) remains a cornerstone for many areas of research, including species diversification dynamics, phylogenetics, and conservation genetics (Cardeñosa et al., 2021; Flores‐Manzanero et al., 2022; Reese et al., 2020; Schmidt et al., 2018; Serrao et al., 2018), especially for study species from which high‐quality tissue samples cannot be easily obtained. Difficulties with invasive sampling for high‐quality tissues or blood include practical issues with trapping large‐bodied, arboreal, or marine animals, as well as ethical considerations, such as risks to the animal and to researchers (Aristizabal Duque et al., 2018). These difficulties apply to collecting invasive samples from most primates, and many genetic and genomic studies in primatology continue to rely on materials that can be collected noninvasively (Arandjelovic & Vigilant, 2018; Aylward et al., 2018; Orkin et al., 2016), or on historic samples, such as from museum skins (Burrell et al., 2015). Although there have been methodological advances that allow for the retrieval of nuclear DNA and even whole genomes from these materials (Burrell et al., 2015; Chiou & Bergey, 2018; Fontsere et al., 2021; Orkin et al., 2021), mtDNA continues to be the most accessible and cost‐effective source of genetic data.
Mitochondrial markers and genomes are especially useful for species identification and delimitation (Reese et al., 2020), for assessing population structure (Flores‐Manzanero et al., 2022; Gagneux et al., 1999; Phukuntsi et al., 2021; Serrao et al., 2018; Skovrind et al., 2021), for assessing introgression and admixture (Makhov et al., 2021; Malukiewicz et al., 2021), for monitoring of species assemblages using environmental DNA (Barnes & Turner, 2016; Thomsen & Willerslev, 2015), and for identifying the origin of animals found in wild meat markets and the illegal pet trade (Cardeñosa et al., 2021; Maligana et al., 2020; Russello et al., 2008). However, many of these methods are reliant on databases from which sequences can be integrated and against which results can be compared, and which are often incomplete (Curry et al., 2018). For example, for platyrrhine primates (a group including all monkeys found in Central and South America) only 32 mitochondrial genome assemblies are available in RefSeq, even though over 200 species have been described, and complete platyrrhine mitogenomes are only available for 76 individuals in GenBank overall. Additionally, the majority of these mitogenomes contain little or no metadata, such as sampling locality, limiting their utility for many analyses, including population genetic studies that rely on spatial data (Deichmann et al., 2017; Strohm et al., 2016; Tahsin et al., 2016).
Hypotheses about how landscape features have shaped the distribution and richness of species can be investigated with molecular data that include sampling localities, and mitochondrial DNA is a fast‐evolving marker (Brown et al., 1979), which can shed light on evolutionary relationships within young radiations more quickly than nuclear DNA. As such, mitogenomic data sets may be especially useful for assessing biogeographic and phylogeographic questions. Primates found within the Amazon are disproportionately speciose for the geographic area they occupy (Fordham et al., 2020), and Alfred Russel Wallace noted that the distributions of many Amazonian primates appear to be limited by boundaries formed by the Amazon, Madeira, and Negro rivers (Wallace, 1852). Now known as the riverine barrier hypothesis (RBH), it is a long‐standing paradigm used to explain the extraordinary species richness of not just primates (Ayres & Clutton‐Brock, 1992; Boubli et al., 2015), but also other mammals (Patton et al., 1994), birds (Cracraft, 1985; Hayes & Sewlal, 2004; Pomara et al., 2014), amphibians and reptiles (de Fraga & de Carvalho, 2021; Godinho & da Silva, 2018; Ortiz et al., 2018), and butterflies (Hall & Harvey, 2002). The RBH proposes that the rivers of the Amazon river basin acted as drivers of speciation when their formation divided existing species' ranges and formed barriers to continued gene flow, leading to allopatric speciation. As an extension of the RBH, the Amazon has been divided into proposed areas of endemism: interfluvial regions which are suggested to harbour unique species assemblages, and which have been used as units for conservation planning (da Silva et al., 2005). However, the RBH and proposed areas of endemism are not without controversy. Criticisms include limits of interspecific phylogenetic comparative methods, and that many studies are based on very few taxa or single gene markers (Losos & Glor, 2003; Santorelli et al., 2018). In addition, some large‐scale studies have found little or only species‐specific support for the RBH (Dambros et al., 2020; Gascon et al., 2000; Kopuchian et al., 2020; Naka & Brumfield, 2018; Santorelli et al., 2018; Smith et al., 2014).
Here, we assemble more than 200 new mitochondrial genomes for Amazonian primates, with locality information (Figure 1), combine these with other Amazonian primate mitogenomes currently available, and use this data set to produce a dated phylogeny (“timetree”), which we use to assess support for the RBH. Specifically, we explore support for rivers as engines of speciation by first identifying divergences in the mitochondrial phylogeny where members of the neighbouring clades are found only on opposite sides of the major river boundaries proposed by Wallace (Amazon, Negro, and Madeira rivers; (1852)), followed by assessing synchrony of congruent divergences occurring for the same river and comparing these dates to current geological evidence for the timing of river formation. We consider divergences to be congruent with the RBH if divergences meet both conditions, namely that (1) sister taxa are found only on opposite sides of a river and that (2) the timing of the divergence does not postdate the geological estimate of river formation.
FIGURE 1.
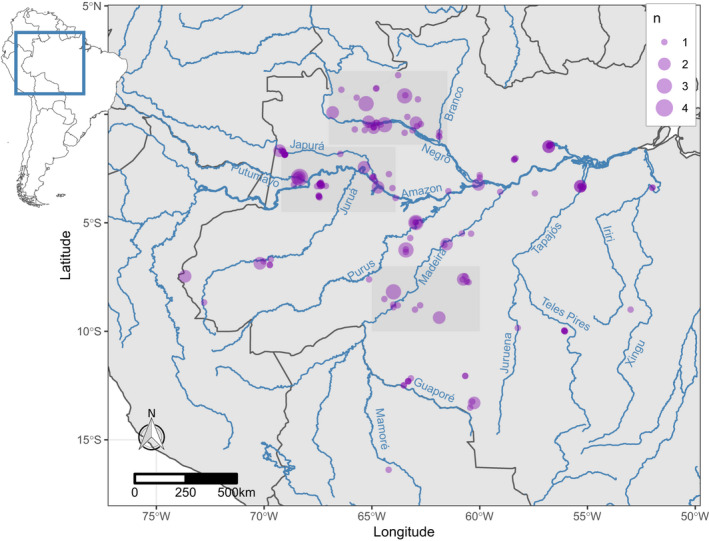
Sample locations of Amazonian primates included in this study. Point size reflects number of samples available from the same location. Major rivers that are relevant for this study are labelled. Shaded regions identify areas explored in detail in Figure 5
2. MATERIALS AND METHODS
2.1. Sample acquisition
We obtained wild‐caught primate tissue samples stored in the following Brazilian zoological collections: Instituto Nacional de Pesquisas da Amazônia (INPA), Universidade Federal do Amazonas (UFAM), Instituto de Desenvolvimento Sustentável Mamirauá (IDSM), Museu Nacional do Rio de Janeiro (MN), Museu Paraense Emílio Goeldi (MPEG), Universidade Federal de Rondônia (UFRO), Universidade Federal do Mato Grosso (UFMT) (Table S1). The majority of these samples were collected during multiple large field surveys aimed at surveying Amazonian biodiversity which were commissioned by the Brazilian government (e.g., PROBIO, SISBIOTA) from 2000 to 2017, while others were obtained from animals hunted by local communities as part of monitoring programmes. Samples consisted of muscle tissue preserved in 70%–90% ethanol. The acquisition of samples for Alouatta caraya and Alouatta guariba clamitans from Argentina has been previously described (Torosin, Argibay, et al., 2020; Torosin, Webster, et al., 2020). Samples from Alouatta palliata, Ateles geoffroyi, Saimiri oerstedii, and Saguinus geoffroyi were biobanked at Kids Saving the Rainforest (KSTR), a wildlife rehabilitation facility in Quepos, Costa Rica in 2016–2017. The A. geoffroyi, A. palliata and S. oerstedii individuals were wild‐born individuals that were brought to KSTR due to injuries or recovered from the pet trade. The Saguinus geoffroyi individual was surrendered to KSTR from a private collection of unknown origin (this species is not native to Costa Rica).
In Brazil, collection permits were obtained from the Biodiversity Authorization and Information System (SISBIO; permit nos. 55777, 42111, 32095–1, 7795–1) and exported under CITES permits (19BR033597/DF and15BR019039/DF). Costa Rican samples were collected and exported under permits from the Comisión Nacional para la Gestión de la Biodiversidad (R‐002‐2020‐OT‐CONAGEBIO) and CITES (2016‐CR2392/SJ [no. S 2477]; 2020‐CR‐4889/SJ [no. S 6825]).
2.2. Sample extraction, sequencing, and mitochondrial genome assembly
Sample extraction and sequencing for samples AC_t1 and AGC_m1 (see Table S1) were previously described (Torosin, Argibay, et al., 2020a; Torosin, Webster, et al., 2020b). Details on genomic sequence generation for the remaining samples are provided in Kuderna et al. (2022). Briefly, genomic DNA was extracted and libraries prepared using standard Illumina protocols and libraries were sequenced to ~30× coverage on an Illumina NovaSeq6000 (150 bp paired‐end reads). Reads were trimmed to remove any sequencing adapters or primers with cutadapt version 2.10 (Martin, 2011) and then subsampled to 3.5 million read pairs with reformat.sh from the bbtools suite v38.86 (Bushnell, 2014). We used mitofinder version 1.4 (Allio et al., 2020) to assemble and annotate mitochondrial genomes from the trimmed and subsampled Illumina short reads, using metaspades (Nurk et al., 2017) for the assembly step and mitfi (Jühling et al., 2012) for the tRNA annotation step. If multiple mitochondrial contigs were identified, we ran mitofinder a second time, setting the minimum contig size to 10,000 and the maximum contigs to 1, in order to force selection and annotation of only the single best contig. For each sample, we used the complete mitochondrial genome from a closely related species available in NCBI's RefSeq database as the reference genome in mitofinder (Supporting Information). All mitochondrial genomes were compared to the ncbi reference database via blast searches to confirm correct taxon identity and to check for completeness.
2.3. Alignment, trimming, and partitioning
We aligned mitochondrial genomes newly assembled with mitofinder (n = 205), as well as complete mitochondrial genomes from 32 additional platyrrhines and six primate outgroups (Table S1) available in ncbi's RefSeq database from previous phylogenomic studies (Arnason et al., 2000, 2002; Babb et al., 2011; Chan et al., 2010; Chiou et al., 2011; de Freitas et al., 2018; Finstermeier et al., 2013; Hao & Yi, 2019; Hodgson et al., 2009; Horai et al.,1995; Malukiewicz et al., 2017, 2021; Matsui et al., 2009; Menezes et al., 2013; Raaum et al., 2005; Wang et al., 2016; Zhang et al., 2016). To facilitate the alignment of circular genomes, we first shifted the genome start for all sequences to begin with the gene cytochrome B, using the fasta_shift tool (https://github.com/b‐brankovics/fasta_tools). The shifted sequences were aligned with mafft v7.309 (Katoh & Standley, 2013). We trimmed the resulting alignment with trimal v1.2 (Capella‐Gutiérrez et al., 2009) using the gappyout setting. Following (Hassanin et al., 2021), we retained only the 12 protein‐coding genes on the forward (“heavy”) strand and the 12S and 16S rRNAs for the downstream analyses, and manually removed the other regions, while visually ensuring the integrity of the alignment.
2.4. Phylogenetic analysis
We used beast 2.6.3 (Bouckaert et al., 2019) for simultaneous phylogeny estimation and divergence dating. As input, we used the trimmed alignment of the 12 forward (“heavy”) strand protein‐coding genes and rRNAs described above, partitioned by codon position for the protein‐coding genes and stems and loops for the rRNAs. We linked clock and tree models for all partitions, setting the clock model to relaxed log normal. Instead of setting an a priori substitution model for each partition, we used the bModelTest module (Bouckaert & Drummond, 2017) within beast2 to select the best model during the beast mcmc run. We set the tree prior to the Coalescent Bayesian Skyline model and added MRCA priors on the ages of 10 nodes based on well‐justified fossil calibrations (de Vries & Beck, 2021). Fossil calibration ages and distributions were based on de Vries and Beck (2021); specifically, we used a uniform distribution to constrain the timing of the divergences between Alouattinae and Atelinae (13.363–34.5 Ma), Callicebinae and Pitheciinae (13.032–34.5 Ma), Callitrichidae and Cebidae (13.183–34.5 mya), Cebinae and Saimiri (13.032–34.5 Ma), Platyrrhini and Catarrhini (33.4–56.035 Ma), Cercopithecoidea and Hominoidea (25.193–33.4 Ma) and tarsiers and anthropoids (41–66.095 Ma). Divergences between Cercopithecini and Colobini (12.47–25.2 Ma), Hominoidea and Hylobatidae (13.4–25.2 Ma), and Haplorhini and Strepsirrhini (55.935–66.1 Ma) were constrained with exponential distributions, where the minimum age was used as the offset and the mean was set to place the maximum age at the 95% quantile. The beast2 input file is available as a Supporting Information file. We ran two mcmc chains for 100 million generations each, sampling every 10,000, for a total of 20,000 trees. We assessed convergence, mixing of the chains, and ESS in tracer version 1.7.1 (Rambaut et al., 2018). We combined the chains after removing the first 25%–32% of each as burnin and constructed a maximum clade credibility tree with treeannotator version 2.6.3.
In addition to the dated tree, we constructed a maximum likelihood tree with raxml‐ng version 1.0.2 (Kozlov et al., 2019) required for use in the species delimitation program mPTP (see below). We used the same alignment and five partitions as for the beast2 analysis, assigning the GTR + G model to all partitions (Kozlov & Stamatakis, 2019), while allowing independent model parameters, and used 25 random and 25 parsimony‐based starting trees.
2.5. Lineage delimitation and assessment of riverine barriers
In order to determine whether speciation in Amazonian primates has been facilitated by riverine barriers, we first used multi‐rate Poisson Tree Processes (mPTP; Kapli et al., 2017) to identify major evolutionary lineages in our sample, rather than relying on existing species identifications or the identification of clades by eye. We did this because species limits within the platyrrhines are not always well‐resolved and/or are controversial (Fordham et al., 2020; Quintela et al., 2020; Zachos et al., 2013), and, in some cases, are based on the presence of river boundaries, even if it has not always been established definitively whether the river forms a species barrier. To avoid issues of circularity based on potential river‐guided species boundaries, we thus sought to delimit lineages in a way that is agnostic to the species assignment of our samples (see Everson et al., 2020 for a similar approach). Within mPTP, we implemented both the multi‐lambda and single‐lambda approaches, which provided a more and less conservative approach to lineage delimitation, respectively (Kapli et al., 2017). We used the maximum likelihood tree generated with raxml, removed outgroups with ‐‐outgroup_crop and determined minimum branch lengths with ‐‐minbr prior to the run.
For samples that had locality data available, phylogenetic relationships and results of delimitation with mPTP were projected onto sample localities with the phytools package (Revell, 2012) in r v4.1.0 (R Core Team, 2019). For any divergences between major lineages (as identified by mPTP) that are congruent with having occurred across a river boundary, we extracted all age estimates for the divergence of the relevant node from the posterior beast2 trees, to determine whether divergences across the same river occurred synchronously and coincided with published geological estimates for the timing of river formation.
3. RESULTS
3.1. Mitochondrial genome assembly
We successfully assembled complete mitochondrial genomes from Illumina short reads for all of our samples except for one (PD_0084), which was missing a small portion of the cytochrome b gene and most of the D‐loop. For the majority of our samples (135/207), only a single mitochondrial contig was assembled (Figure 2a); the final contigs across all samples had a mean length of 16,604 bp (Figure 2b), a mean coverage of 435.93x, and 15 genes were annotated for all final assemblies. In cases where multiple mitochondrial contigs were assembled (72/207), the additional contigs were always either substantially shorter (mean length of additional contigs = 2425 bp; Figure 2c) and/or had much lower coverage (mean coverage of additional contigs = 4.222×) than the first contig (mean coverage = 389.83×; Figure 2d). Only in a single case (PD_0429) was the second contig above 10 kb and resembled an almost complete mitochondrial genome. However, coverage was substantially lower for the second, shorter contig (15.32×) than for the first contig (2716.77×). While the higher‐coverage contig matched the taxon identification of the sample (Alouatta), the second, lower‐coverage contig matched Saimiri, so this probably reflects a low level of contamination. Results of the BLAST searches of the final assembled mitogenomes confirmed the taxonomic identification of the sample in all but three cases. For two samples (PD_0080, PD_0345) this is probably due to mislabelling, rather than contamination, so we retained these for the phylogenetic analysis (Figure 3), but did not consider them in downstream analyses. The third sample (PD_0305, Ateles) was probably contaminated, as its mitochondrial assembly was identical to another sample in a different genus (Alouatta), so we removed it from all analyses. Finally, two samples were found to have been collected from the same animal (PD_0306 & PD_0435), so only one sequence (PD_0435) was retained. Overall, we assembled mitochondrial genomes for samples from 17 genera (based on current nomenclature), representing all five families of platyrrhines. All final mitochondrial genomes that were newly generated and analysed in this project (n = 205) have been deposited in GenBank (accession numbers OM328861‐OM329065).
FIGURE 2.
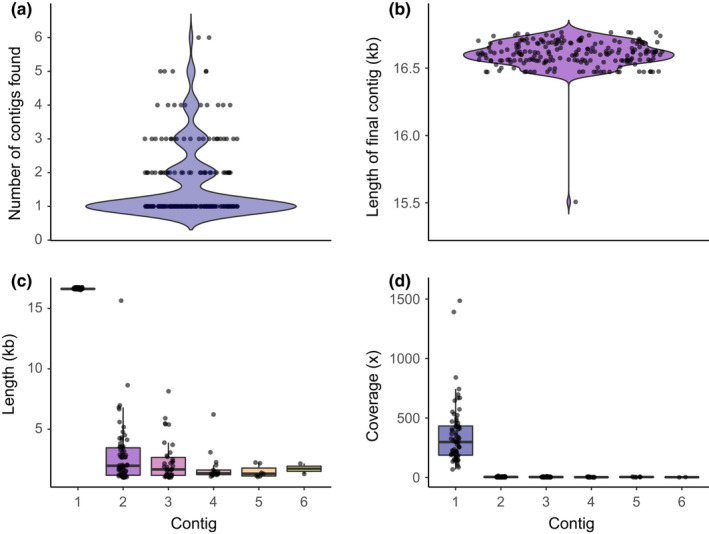
Mitochondrial genome assembly with MitoFinder. Violin plots summarize the distribution of (a) number of mitochondrial contigs found for each sample (overlaid as points) and distribution of (b) lengths of the final mitochondrial contig for all samples (overlaid as points). Boxplots describe (c) length and (d) coverage of contigs for samples in which MitoFinder identified more than one mitochondrial contig
FIGURE 3.
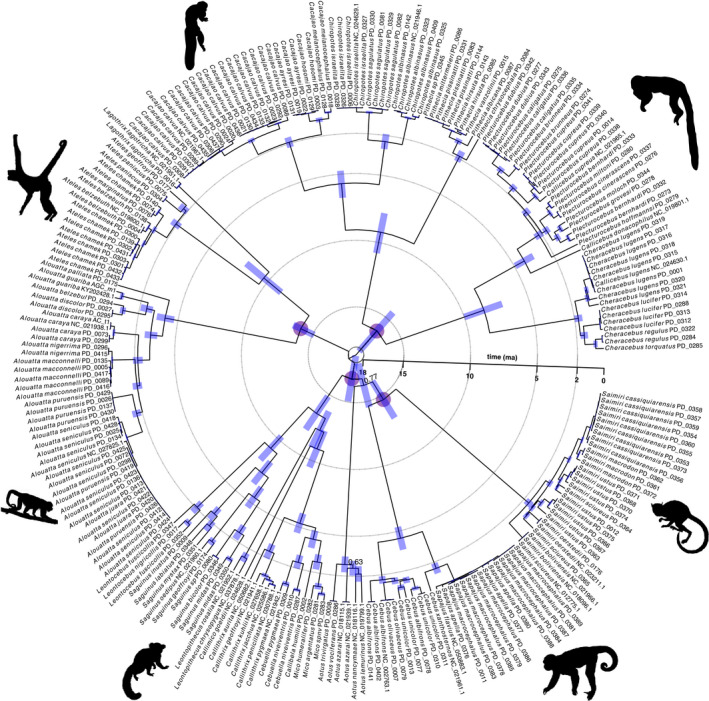
Dated mitogenomic phylogeny of platyrrhines. Blue error bars indicate 95% HPD for node ages, node numbers show posterior probability for internal nodes <0.95. Purple circles denote nodes that were calibrated with fossils. Images via PhyloPic (Saimiri, Alouatta, Ateles, and Callithrix in public domain; Cebus ‐ ©S. Werning), adapted from a. Cotta, cc‐by‐2.0 (Pitheciinae), adapted from B. Gratwicke, cc‐by‐2.0 (Callicebinae)
3.2. Phylogenetic analysis
Relationships within platyrrhines identified by the beast2 analysis support a basal split between Pitheciidae (sakis, uakaris, and titi monkeys) and a clade comprising the remaining families: Atelidae (howler, spider, and woolly monkeys), Cebidae (capuchin and squirrel monkeys), Callitrichidae (marmosets and tamarins), and Aotidae (owl or night monkeys). Aotidae form a clade with Cebidae in our analysis, albeit with somewhat low support (posterior = 0.77). The other phylogenetic relationships identified by the beast2 analysis are well‐supported (posterior > 0.95) for all divergences above genus level (Figure 3). The dated phylogenetic tree generated here is available in NEXUS format in the online repository containing all supporting data sets for this study (Janiak et al., 2022).
3.3. Lineage delimitation and assessment of riverine barriers
Lineage delimitation with mPTP identified 101 distinct lineages when using a single rate of lambda (Figure 4), and 52 lineages when using the multi‐rate setting (Figure S1). We identified 13 out of a total of 64 divergences within Amazonian platyrrhines that are congruent with having occurred across a riverine barrier, meaning that members of the respective sister clades/taxa were identified as distinct lineages by mPTP and are only found on opposite sides of a river (marked with node symbols in Figure 4, Figure S1). When using the single‐rate setting, the majority of divergences that were congruent with the RBH were found for the Amazon river, including within Saimiri, Cebus, Cheracebus, Ateles, Chiropotes, and Callitrichidae (Figure 4b–d and h–j). Using the same setting, divergences that are congruent with the RBH having occurred across the Rio Negro include clades within Cebus, Cheracebus, and Cacajao (Figure 4c,d,f); however, in the latter two cases, only a single sample is available for the area south of the Rio Negro. For the Madeira river, the divergence between two Plecturocebus lineages is congruent with the RBH (Figure 4e). When using the more conservative multi‐rate setting, six of these 13 divergences are maintained out of a total 27 divergences considered. This includes the divergences within Cebus, Cheracebus and Cacajao across the Rio Negro (Figure S1C,D,F), within Cheracebus and Chiropotes across the Amazon (Figure S1D,I), and within Plecturocebus across the Madeira river (Figure S1E). However, the divergences across the Amazon within three callitrichid genera (Saguinus, Cebuella, Leontocebus), Cebus, Saimiri, and Ateles that are congruent with the RBH when using the single‐rate setting are not identified with the multi‐rate setting, as the segregating lineages are considered a single lineage in this case.
FIGURE 4.
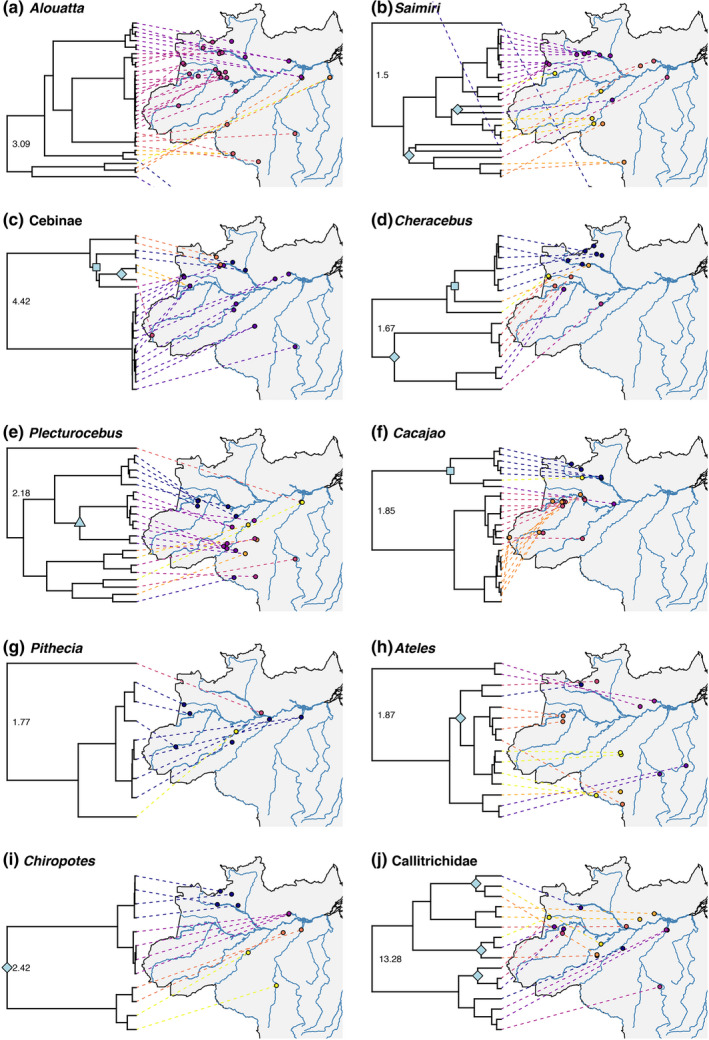
Phylogenetic relationships of platyrrhine subclades mapped onto Brazilian sampling locations. Colours indicate mPTP lineage delimitation based on the single‐rate method (see Figure S1 for multirate). Node symbols denote clades whose lineage distributions are congruent with separation by a riverine barrier, including the Amazon (diamonds), Rio Negro (squares), and Rio Madeira (triangles). Root ages for each subclade as estimated by beast2 are shown
The age estimates for divergences occurring across the Amazon river were partly synchronous, with 95% HPDs overlapping for the divergences identified as congruent with the RBH within Saguinus (2.07–3.14 Ma), Leontocebus (1.57–2.52 Ma), Cebuella (1.93–2.96 Ma), and Chiropotes (1.86–2.99 Ma). Divergences within Cheracebus (1.12–1.67 Ma), Saimiri (0.4–0.62 and 0.9–1.24 Ma), Cebus (0.36–0.65 Ma), and Ateles (0.48–0.7 Ma) were younger, with estimates within Cebus, Saimiri, and Ateles overlapping (Figure 5a). The 95% HPDs for divergences across the Rio Negro within Cheracebus (0.46–0.76 Ma) and Cacajao (0.56–0.9 Ma) overlapped, but the timing of the divergence within Cebus (1.13–1.61 Ma) was earlier (Figure 5b). Only a single divergence across the Madeira river was identified here, in Plecturocebus (0.75–1.17 Ma), so synchrony could not be assessed (Figure 5c).
FIGURE 5.
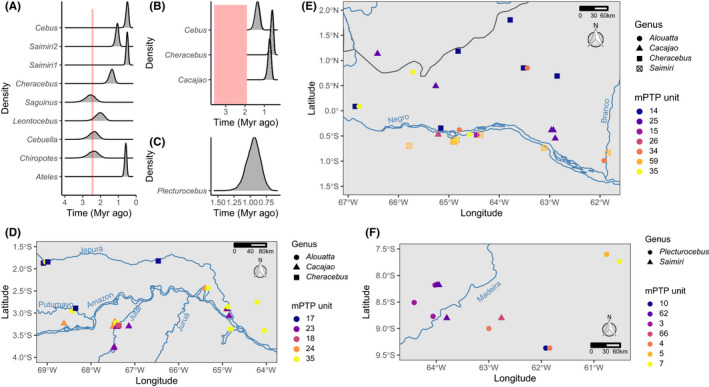
Evidence supporting the riverine barrier hypothesis is mixed for platyrrhines. Density distributions of divergence times for nodes split across the (a) Amazon, (b) Negro, and (c) Madeira rivers are only partly synchronous and many divergences postdate proposed river formation times based on geological evidence (red shading). The (d) Amazon, (e) Negro, and (f) Madeira rivers may be barriers for lineages of some genera (Cheracebus, Plecturocebus), but not for others (Alouatta, Saimiri)
For some lineages, members of the same evolutionary unit (as determined by mPTP) were found on both sides of major rivers. This included Alouatta across both the Amazon and the Rio Negro (Figures 5d,e), Saimiri across both the Rio Negro and the Madeira (Figures 5d,e), Cacajao across the Amazon (Figure 5d), Sapajus across the Amazon and Madeira (Figure 4c) and Pithecia across the Amazon (Figure 4g).
4. DISCUSSION
We assembled 205 new mitochondrial genomes for platyrrhine primates, most sampled from the Amazon region, and used them to assess support for the long‐standing riverine barrier hypothesis (RBH), which proposes that river formation was a major driver of speciation in Amazonian primates. Along the Amazon, Negro, and Madeira rivers, we found mixed evidence for the RBH, which we discuss in detail below. With the mitochondrial assemblies presented here, we have tripled the number of available mitogenomes for platyrrhines in GenBank and quadrupled the number of platyrrhine mitogenomes in RefSeq, and we provide an updated dated mitogenomic phylogeny of South American primates.
We utilized the novel mitogenomes presented here to assess support for the RBH, as originally proposed by Wallace (1852) over 150 years ago, along the Amazon, Madeira, and Negro rivers and found mixed evidence. While we identified divergences that coincide with a river barrier, only some of them occur synchronously and also overlap with the proposed dates of river formation based on geological evidence. The most compelling evidence for the RBH found here is for the Amazon river itself, within the genus Chiropotes (bearded saki monkeys), and also within Callitrichidae (marmosets and tamarins), which are the smallest extant platyrrhines. The Amazon river is thought to have taken its current form around 2.4–2.5 Ma (Campbell, 2010; Figueiredo et al., 2009), although it may have existed long before (11.8–11.3 Ma (Figueiredo et al., 2009); 10.0–4.5 Ma (Albert et al., 2018); 10.0–7.0 Ma (Hoorn et al., 2010)). Accepting 2.4 Ma as the minimum age of the Amazon (Campbell, 2010; Figueiredo et al., 2009), we identified four divergences in the platyrrhine tree that are congruent with the RBH and also align temporarily with this date, within the genera Saguinus, Leontocebus, Cebuella, and Chiropotes. We cautiously interpret divergences across the Amazon within these genera as being congruent with the RBH. However, we note that, despite this being the largest mitogenomic survey of the platyrrhines to date by far, some of the sample sizes are small, especially for callitrichids, and that many samples were collected some distance (~150–200 km) from the banks of the Amazon river, making it difficult to reject an alternative explanation of isolation by distance (Dambros et al., 2017). Furthermore, the relevant divergences within Callitrichidae are only supported when using the single‐rate mPTP model, not the more conservative multi‐rate model (Figure S1). That said, additional support for the Amazon as a species barrier for pygmy marmosets (Cebuella spp.) has recently been provided (Boubli et al., 2021; Porter et al., 2021). Notably, many divergences across the Amazon did not occur synchronously and/or postdate the known minimum time of river formation. For example, divergences within spider (Ateles spp.), squirrel (Saimiri spp.), capuchin (Cebus spp.) and titi (Cheracebus sp.) monkeys are congruent with the RBH based on sample localities, but these splits postdate even the youngest proposed date of the formation of the Amazon in its current form (2.4 Ma: Figueiredo et al., 2009) by about 1–2 million years. Additionally, we also found evidence that, despite its formidable width (~3.5 km: (Fordham et al., 2020), the Amazon does not appear to be a barrier at all for some genera, including howler monkeys (Alouatta spp.), bald uakaris (Cacajao spp.), saki monkeys (Pithecia spp.), and robust capuchin monkeys (Sapajus spp.), as members of the same evolutionary lineages occur on both sides of the river.
We identified three divergences that are congruent with the Rio Negro being a barrier, within Cebus, Cheracebus, and Cacajao, as suggested previously (Boubli et al., 2015). However, for both Cacajao and Cheracebus these divergences are based on a single sample present on the opposite river bank, and so need to be tested further with additional sampling. In the case of Cebus, the samples that form the clade south of the Rio Negro are located quite far (>500 km) from the riverbank, making it difficult to rule out alternative explanations, such as isolation by distance. Notably, we find that the Rio Negro does not appear to be a barrier for Alouatta or Saimiri, as members of the same lineage are found on both river sides. Sedimentological evidence suggests a formation date of ~3.6–1.9 Ma for the Rio Negro (Soares et al., 2017). If we accept 1.9 Ma as the minimum age for the Rio Negro, none of the divergences identified here would be temporally congruent with the RBH for this river, as the divergences within Cebus, Cacajao and Cheracebus are all more recent. Additionally, as noted above, the very wide (~3.5 km: Fordham et al., 2020) Amazon river does not appear to be a barrier for bald uakaris (Cacajao spp.) in our results, suggesting that an alternative process may explain the distribution of black uakaris along the narrower (~0.7 km: Fordham et al., 2020) Rio Negro.
Our results suggest that the Madeira river may form a barrier for titi monkeys (Plecturocebus spp.), as has been suggested previously (Byrne et al., 2016; Hoyos et al., 2016; Santorelli et al., 2018). However, the same river does not appear to present a barrier to squirrel monkeys (Saimiri spp.). Because only a single divergence congruent with the RBH was identified for the Madeira, we cannot assess synchrony here. However, the age of the Madeira river may date to the Miocene (Ruokolainen et al., 2019; Tagliacollo et al., 2015), in which case the divergence within Plecturocebus postdates river formation by several million years, and thus the river is unlikely to have acted as a vicariant agent. It has also been suggested that the bed of the Madeira has moved (Ruokolainen et al., 2019; Tagliacollo et al., 2015), complicating the ability to detect evidence for or against the RBH.
It is important to note that rivers can coincide with species barriers without having been vicariant agents (Naka & Pil, 2020), and that inferring evidence for or against vicariance from present‐day species ranges is based on the assumption that these ranges have not changed (Losos & Glor, 2003), which may not be the case (Graham et al., 1996). That said, we find evidence of evolutionarily distinct lineages in close geographic proximity within the same interfluve. This, along with our finding that even large Amazonian rivers do not appear to be barriers for several platyrrhine lineages, underscores the importance of other evolutionary mechanisms, beyond the RBH and allopatric speciation, for diversification within platyrrhines. While comparatively less attention has been paid to the role that mechanisms of sympatric speciation have played in shaping Amazonian primate diversity, speciation via sexual selection, ecological factors, and biotic interactions (Boughman, 2001; Dieckmann & Doebeli, 1999; Doebeli & Dieckmann, 2000; Gutiérrez et al., 2014; Maan & Seehausen, 2011; Rice & Salt, 1990) are important directions for future research. Our results are in line with several recent publications that find little or mixed evidence for the hypothesis that Amazonian rivers have been drivers of speciation, with many supporting a more nuanced, species‐specific effect of riverine barriers, rather than a global rule of rivers as vicariant agents (Kopuchian et al., 2020; Naka & Pil, 2020; Oliveira et al., 2017; Voss et al., 2019). Interestingly, at least some platyrrhines have been observed to be competent swimmers (Barnett et al., 2012; Benchimol & Venticinque, 2014; Gonzalez‐Socoloske & Snarr, 2010; Lynch Alfaro et al., 2015; Nunes, 2014), but floating islands and meandering rivers may offer another means for monkeys to cross large rivers (Ali et al., 2021; Ayres & Clutton‐Brock, 1992; Gascon et al., 2000). The distribution of Amazonian primates may be shaped by features beyond rivers, including moisture gradients (Silva et al., 2019), geological formations and soil properties (Ruokolainen et al., 2019), and vegetation patterns (Higgins et al., 2011), offering many avenues for future research directions.
Taxonomic revisions are outside of the scope of this study, and should not be based on mitochondrial (or even nuclear) data alone (Zachos et al., 2013); however, our mitogenomic phylogeny suggests that some species boundaries may need to be reassessed, as a handful of species were found to be paraphyletic, in particular within Alouatta. While some of these patterns may be due to incomplete lineage sorting, introgression, or hybridization, taxonomic errors are another common cause of such patterns (McKay & Zink, 2010). Mitochondrial phylogenies, like the one presented here, may be an important tool for uncovering such inconsistencies, especially in taxa for which species limits are not completely resolved, or for which ranges have been assumed to correspond to interfluvial regions or areas of endemism, which assumes a priori that rivers form dispersal barriers.
The newly assembled mitogenomes, along with their metadata, will be a valuable resource for conservation genetics and genomics, facilitating more accurate identification of sample identities and/or provenance. Novel methods to extract nuclear data and even whole genomes from low‐quality or noninvasively collected samples are available (Burrell et al., 2015; Chiou & Bergey, 2018; Fontsere et al., 2021; Orkin et al., 2021), however, the costs associated with these methods, as well as their downstream computational requirements, remain prohibitive for many researchers, especially in primate host countries. While local capacity building should be a focus for genomicists working in the Global South (de Vries et al., 2015; Hetu et al., 2019; Rodríguez et al., 2005; Şekercioğlu, 2012), these efforts will take time, and until high‐throughput methods become more accessible, mitogenomics will continue to be a pillar of conservation genomics (Pomerantz et al., 2018; Watsa et al., 2020). Importantly, the novel mitogenomes assembled here have been made publicly available on GenBank along with important metadata, including sampling locations and voucher specimens, improving their utility and value for future analyses.
5. CONCLUSIONS
Mitochondrial genomics remains a pillar of phylogenetics and conservation research. The 205 newly assembled mitogenomes for Amazonian primates presented here dramatically increase the number of available platyrrhine mitogenomes, and because they include known sampling locations are of additional value to future research. Using these novel mitogenomes, we find mixed support for the long‐standing riverine barrier hypothesis (RBH), supporting a more nuanced, clade‐specific effect of riverine barriers. This suggests that other evolutionary mechanisms, beyond the RBH and allopatric speciation, may also play key roles for explaining the extraordinary species‐diversity found in Amazonian primates.
AUTHOR CONTRIBUTIONS
Mareike C. Janiak, Robin M. D. Beck, Dorien de Vries, Ian B. Goodhead, and Jean P. Boubli conceived of the study and designed the research. Mareike C. Janiak analysed the data with input from Robin M. D. Beck, Dorien de Vries, Ian B. Goodhead, and Jean P. Boubli. Lukas F. K. Kuderna, Tomàs Marquès‐Bonet, and Nicole S. Torosin generated and provided sequencing data. Nicole S. Torosin, Amanda D. Melin, Mariluce Messias, Maria N.F. da Silva, Iracilda Sampaio, Izeni P. Farias, Rogerio Rossi, Fabiano R. de Melo, João Valsecchi, Tomas Hrbek, Felipe E. Silva, and Jean P. Boubli collected and contributed samples. Mareike C. Janiak wrote the manuscript with input from all authors.
CONFLICT OF INTEREST
Lukas F. K. Kuderna is currently an employee of Illumina Inc.
OPEN RESEARCH BADGES
This article has earned an Open Data Badge for making publicly available the digitally‐shareable data necessary to reproduce the reported results. The data are available on figshare at https://doi.org/10.6084/m9.figshare.19606063 and https://doi.org/10.6084/m9.figshare.19610187.
Supporting information
Figure S1
Table S1
ACKNOWLEDGEMENTS
We thank Rob Voss at the American Museum for Natural History for helpful discussions. This work used JASMIN, the UK collaborative data analysis facility. Further computational resources were provided by WestGrid (www.westgrid.ca) and Compute Canada Calcul Canada (www.computecanada.ca).
Janiak, M. C. , Silva, F. E. , Beck, R. M. D. , de Vries, D. , Kuderna, L. F. K. , Torosin, N. S. , Melin, A. D. , Marquès‐Bonet, T. , Goodhead, I. B. , Messias, M. , da Silva, M. N. F. , Sampaio, I. , Farias, I. P. , Rossi, R. , de Melo, F. R. , Valsecchi, J. , Hrbek, T. , & Boubli, J. P. (2022). Two hundred and five newly assembled mitogenomes provide mixed evidence for rivers as drivers of speciation for Amazonian primates. Molecular Ecology, 31, 3888–3902. 10.1111/mec.16554
Funding information
This work was supported by the Natural Environment Research Council (NE/T000341/1); Brazilian National Council for Scientific and Technological Development (Conselho Nacional de Desenvolvimento Científico e Tecnológico, CNPq) (Process nos.: 563348/2010–0, 303286/2014–8, 303579/2014–5, 200502/2015–8, 302140/2020–4, 300365/2021–7, 301407/2021–5, 301925/2021–6); Higher Education Personnel Improvement Coordination (CAPES) (Process no.: 3261/2013); International Primatological Society conservation grant; The Rufford Foundation (14861–1, 23117–2), the Margot Marsh Biodiversity Foundation (SMA‐CCO‐G0000000023, SMA‐CCO‐G0000000037), Primate Conservation Inc. (no. 1713 and no. 1689). ADM is supported by the Natural Sciences and Engineering Research Council (NSERC RGPIN‐2017‐03782) and the Canada Research Chairs Program Primate dietary ecology and genomics (950‐231257). TMB is supported by funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement No. 864203), BFU2017‐86471‐P (MINECO/FEDER, UE), "Unidad de Excelencia María de Maeztu", funded by the AEI (CEX2018‐000792‐M), Howard Hughes International Early Career, NIH 1R01HG010898‐01A1 and Secretaria d'Universitats i Recerca and CERCA Programme del Departament d'Economia i Coneixement de la Generalitat de Catalunya (GRC 2017 SGR 880). FES is supported by the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska‐Curie (grant agreement No. 801505).
Handling Editor: David Coltman
Contributor Information
Mareike C. Janiak, Email: mareike.janiak@gmail.com.
Jean P. Boubli, Email: j.p.boubli@salford.ac.uk.
DATA AVAILABILITY STATEMENT
Data used or generated for this article have been made available in the European Nucleotide Archive's SRA (BioProject PRJEB49549) and NCBI's GenBank (accession numbers OM328861‐OM329065; Table S1). Files containing metadata for all samples, sequence alignments, analysis input files, and the dated phylogenetic tree have been archived at: 10.6084/m9.figshare.19606063. Scripts used to run analyses are available on GitHub (https://github.com/MareikeJaniak/Platyrrhine‐mtDNA) and have been archived at: 10.6084/m9.figshare.19610187.
REFERENCES
- Albert, J. S. , Val, P. , & Hoorn, C. (2018). The changing course of the Amazon River in the Neogene: Center stage for neotropical diversification. Neotropical Ichthyology, 16(3), e180033. 10.1590/1982-0224-20180033 [DOI] [Google Scholar]
- Ali, J. , Fritz, U. , & Vargas‐Ramirez, M. (2021). Monkeys on a free‐floating Island in a Colombian river: Further support for over‐water colonization. Biogeographia – The Journal of Integrative Biogeography, 36, a005. 10.21426/B636051761 [DOI] [Google Scholar]
- Allio, R. , Schomaker‐Bastos, A. , Romiguier, J. , Prosdocimi, F. , Nabholz, B. , & Delsuc, F. (2020). MitoFinder: Efficient automated large‐scale extraction of mitogenomic data in target enrichment phylogenomics. Molecular Ecology Resources, 20(4), 892–905. 10.1111/1755-0998.13160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arandjelovic, M. , & Vigilant, L. (2018). Non‐invasive genetic censusing and monitoring of primate populations. American Journal of Primatology, 80(3), e22743. 10.1002/ajp.22743 [DOI] [PubMed] [Google Scholar]
- Aristizabal Duque, S. L. , Orozco‐Jimenez, L. Y. , Zapata‐Escobar, C. , & Palacio‐Baena, J. A. (2018). Conservation genetics of otters: Review about the use of non‐invasive samples. Therya, 9(1), 85–93. 10.12933/therya-18-515 [DOI] [Google Scholar]
- Arnason, U. , Adegoke, J. A. , Bodin, K. , Born, E. W. , Esa, Y. B. , Gullberg, A. , Nilsson, M. , Short, R. V. , Xu, X. , & Janke, A. (2002). Mammalian mitogenomic relationships and the root of the eutherian tree. Proceedings of the National Academy of Sciences of the United States of America, 99(12), 8151–8156. 10.1073/pnas.102164299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnason, U. , Gullberg, A. , Burguete, A. S. , & Janke, A. (2000). Molecular estimates of primate divergences and new hypotheses for primate dispersal and the origin of modern humans. Hereditas, 133(3), 217–228. 10.1111/j.1601-5223.2000.00217.x [DOI] [PubMed] [Google Scholar]
- Aylward, M. L. , Sullivan, A. P. , Perry, G. H. , Johnson, S. E. , & Louis, E. E., Jr. (2018). An environmental DNA sampling method for aye‐ayes from their feeding traces. Ecology and Evolution, 8(18), 9229–9240. 10.1002/ece3.4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres, J. M. , & Clutton‐Brock, T. H. (1992). River boundaries and species range size in Amazonian primates. The American Naturalist, 140(3), 531–537. 10.1086/285427 [DOI] [PubMed] [Google Scholar]
- Babb, P. L. , Fernandez‐Duque, E. , Baiduc, C. A. , Gagneux, P. , Evans, S. , & Schurr, T. G. (2011). mtDNA diversity in Azara's owl monkeys (Aotus azarai azarai) of the Argentinean Chaco. American Journal of Physical Anthropology, 146(2), 209–224. 10.1002/ajpa.21567 [DOI] [PubMed] [Google Scholar]
- Barnes, M. A. , & Turner, C. R. (2016). The ecology of environmental DNA and implications for conservation genetics. Conservation Genetics, 17(1), 1–17. [Google Scholar]
- Barnett, A. A. , Shaw, P. , Spironello, W. R. , MacLarnon, A. , & Ross, C. (2012). Sleeping site selection by golden‐backed uacaris, Cacajao melanocephalus ouakary (Pitheciidae), in Amazonian flooded forests. Primates, 53(3), 273–285. 10.1007/s10329-012-0296-4 [DOI] [PubMed] [Google Scholar]
- Benchimol, M. , & Venticinque, E. M. (2014). Responses of primates to landscape change in Amazonian land‐bridge islands – A multi‐scale analysis. Biotropica, 46(4), 470–478. 10.1111/btp.12122 [DOI] [Google Scholar]
- Boubli, J. P. , Janiak, M. C. , Porter, L. M. , de la Torre, S. , Cortés‐Ortiz, L. , da Silva, M. N. F. , Rylands, A. B. , Nash, S. , Bertuol, F. , Byrne, H. , Silva, F. E. , Rohe, F. , de Vries, D. , Beck, R. M. D. , Ruiz‐Gartzia, I. , Kuderna, L. F. K. , Marquès‐Bonet, T. , Hrbek, T. , Farias, I. P. , … Roos, C. (2021). Ancient DNA of the pygmy marmoset type specimen Cebuella pygmaea (Spix, 1823) resolves a taxonomic conundrum. Zoological Research, 42(6), 761–771. 10.24272/j.issn.2095-8137.2021.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubli, J. P. , Ribas, C. , Lynch Alfaro, J. W. , Alfaro, M. E. , da Silva, M. N. F. , Pinho, G. M. , & Farias, I. P. (2015). Spatial and temporal patterns of diversification on the Amazon: A test of the riverine hypothesis for all diurnal primates of Rio Negro and Rio Branco in Brazil. Molecular Phylogenetics and Evolution, 82(Pt. B), 400–412. 10.1016/j.ympev.2014.09.005 [DOI] [PubMed] [Google Scholar]
- Bouckaert, R. R. , & Drummond, A. J. (2017). bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evolutionary Biology, 17(1), 42–52. 10.1186/s12862-017-0890-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert, R. R. , Vaughan, T. G. , Barido‐Sottani, J. , Duchêne, S. , Fourment, M. , Gavryushkina, A. , Heled, J. , Jones, G. , Kühnert, D. , De Maio, N. , Matschiner, M. , Mendes, F. K. , Müller, N. F. , Ogilvie, H. A. , du Plessis, L. , Popinga, A. , Rambaut, A. , Rasmussen, D. , Siveroni, I. , … Stadler, T. (2019). BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Computational Biology, 15(4), e1006650. 10.1371/journal.pcbi.1006650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughman, J. W. (2001). Divergent sexual selection enhances reproductive isolation in sticklebacks. Nature, 411(6840), 944–948. 10.1038/35082064 [DOI] [PubMed] [Google Scholar]
- Brown, W. M. , George, M., Jr. , & Wilson, A. C. (1979). Rapid evolution of animal mitochondrial DNA. Proceedings of the National Academy of Sciences of the United States of America, 76(4), 1967–1971. 10.1073/pnas.76.4.1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell, A. S. , Disotell, T. R. , & Bergey, C. M. (2015). The use of museum specimens with high‐throughput DNA sequencers. Journal of Human Evolution, 79, 35–44. 10.1016/j.jhevol.2014.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell, B. (2014). BBTools software package. Retrieved from: http://sourceforge.Net/projects/bbmap
- Byrne, H. , Rylands, A. B. , Carneiro, J. C. , Alfaro, J. W. L. , Bertuol, F. , da Silva, M. N. F. , Messias, M. , Groves, C. P. , Mittermeier, R. A. , Farias, I. , Hrbek, T. , Schneider, H. , Sampaio, I. , & Boubli, J. P. (2016). Phylogenetic relationships of the New World titi monkeys (Callicebus): first appraisal of taxonomy based on molecular evidence. Frontiers in Zoology, 13(1), 10. 10.1186/s12983-016-0142-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, K. E. (2010). Late Miocene onset of the Amazon River and the Amazon deep‐sea fan: Evidence from the Foz do Amazonas Basin: COMMENT. Geology 38(7), e212. doi: 10.1130/G30633C.1 [Google Scholar]
- Capella‐Gutiérrez, S. , Silla‐Martínez, J. M. , & Gabaldón, T. (2009). trimAl: A tool for automated alignment trimming in large‐scale phylogenetic analyses. Bioinformatics, 25(15), 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardeñosa, D. , Fields, A. T. , Shea, S. K. H. , Feldheim, K. A. , & Chapman, D. D. (2021). Relative contribution to the shark fin trade of Indo‐Pacific and eastern Pacific pelagic thresher sharks. Animal Conservation, 24(3), 367–372. 10.1111/acv.12644 [DOI] [Google Scholar]
- Chan, Y.‐C. , Roos, C. , Inoue‐Murayama, M. , Inoue, E. , Shih, C.‐C. , Pei, K. J.‐C. , & Vigilant, L. (2010). Mitochondrial genome sequences effectively reveal the phylogeny of Hylobates gibbons. PLoS One, 5(12), e14419. 10.1371/journal.pone.0014419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou, K. L. , & Bergey, C. M. (2018). Methylation‐based enrichment facilitates low‐cost, noninvasive genomic scale sequencing of populations from feces. Scientific Reports, 8(1), 1975. 10.1038/s41598-018-20427-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou, K. L. , Pozzi, L. , Lynch Alfaro, J. W. , & Di Fiore, A. (2011). Pleistocene diversification of living squirrel monkeys (Saimiri spp.) inferred from complete mitochondrial genome sequences. Molecular Phylogenetics and Evolution, 59(3), 736–745. 10.1016/j.ympev.2011.03.025 [DOI] [PubMed] [Google Scholar]
- Cracraft, J. (1985). Historical biogeography and patterns of differentiation within the south American avifauna: Areas of endemism. Ornithological Monographs, 36, 49–84. 10.2307/40168278 [DOI] [Google Scholar]
- Curry, C. J. , Gibson, J. F. , Shokralla, S. , Hajibabaei, M. , & Baird, D. J. (2018). Identifying north American freshwater invertebrates using DNA barcodes: Are existing COI sequence libraries fit for purpose? Freshwater Science, 37(1), 178–189. 10.1086/696613 [DOI] [Google Scholar]
- Dambros, C. S. , Morais, J. W. , Azevedo, R. A. , & Gotelli, N. J. (2017). Isolation by distance, not rivers, control the distribution of termite species in the Amazonian rain forest. Ecography, 40(10), 1242–1250. 10.1111/ecog.02663 [DOI] [Google Scholar]
- Dambros, C. S. , Zuquim, G. , Moulatlet, G. M. , Costa, F. R. C. , Tuomisto, H. , Ribas, C. C. , Azevedo, R. , Baccaro, F. , Bobrowiec, P. E. D. , Dias, M. S. , Emilio, T. , Espirito‐Santo, H. M. V. , Figueiredo, F. O. G. , Franklin, E. , Freitas, C. , Graça, M. B. , d′Horta, F. , Leitão, R. P. , Maximiano, M. , … Magnusson, W. E. (2020). The role of environmental filtering, geographic distance and dispersal barriers in shaping the turnover of plant and animal species in Amazonia. Biodiversity and Conservation, 29(13), 3609–3634. 10.1007/s10531-020-02040-3 [DOI] [Google Scholar]
- da Silva, J. M. C. , Rylands, A. B. , & da Fonseca, G. A. B. (2005). The fate of the Amazonian areas of endemism. Conservation Biology, 19(3), 689–694. 10.1111/j.1523-1739.2005.00705.x [DOI] [Google Scholar]
- de Fraga, R. , & de Carvalho, V. T. (2021). Testing the Wallace's riverine barrier hypothesis based on frog and Squamata reptile assemblages from a tributary of the lower Amazon River. Studies on Neotropical Fauna and Environment. 10.1080/01650521.2020.1870838 [DOI] [Google Scholar]
- de Freitas, P. D. , Mendez, F. L. , Chávez‐Congrains, K. , Galetti, P. M., Jr. , Coutinho, L. L. , Pissinatti, A. , & Bustamante, C. D. (2018). Next‐generation sequencing of the complete mitochondrial genome of the endangered species black lion tamarin Leontopithecus chrysopygus (Primates) and mitogenomic phylogeny focusing on the Callitrichidae family. G3, 8(6), 1985–1991. 10.1534/g3.118.200153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deichmann, J. L. , Mulcahy, D. G. , Vanthomme, H. , Tobi, E. , Wynn, A. H. , Zimkus, B. M. , & McDiarmid, R. W. (2017). How many species and under what names? Using DNA barcoding and GenBank data for west central African amphibian conservation. PLoS One, 12(11), e0187283. 10.1371/journal.pone.0187283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, D. , & Beck, R. M. D. (2021). Total evidence tip‐dating phylogeny of platyrrhine primates and 27 well‐justified fossil calibrations for primate divergences. bioRxiv. 10.1101/2021.10.21.465342 [DOI] [Google Scholar]
- de Vries, J. , Tindana, P. , Littler, K. , Ramsay, M. , Rotimi, C. , Abayomi, A. , Mulder, N. , & Mayosi, B. M. (2015). The H3Africa policy framework: Negotiating fairness in genomics. Trends in Genetics, 31(3), 117–119. 10.1016/j.tig.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann, U. , & Doebeli, M. (1999). On the origin of species by sympatric speciation. Nature, 400(6742), 354–357. 10.1038/22521 [DOI] [PubMed] [Google Scholar]
- Doebeli, M. , & Dieckmann, U. (2000). Evolutionary branching and sympatric speciation caused by different types of ecological interactions. The American Naturalist, 156(S4), S77–S101. 10.1086/303417 [DOI] [PubMed] [Google Scholar]
- Everson, K. M. , Jansa, S. A. , Goodman, S. M. , & Olson, L. E. (2020). Montane regions shape patterns of diversification in small mammals and reptiles from Madagascar's moist evergreen forest. Journal of Biogeography, 47(10), 2059–2072. 10.1111/jbi.13945 [DOI] [Google Scholar]
- Figueiredo, J. , Hoorn, C. , van der Ven, P. , & Soares, E. (2009). Late Miocene onset of the Amazon River and the Amazon deep‐sea fan: Evidence from the Foz do Amazonas Basin. Geology, 37(7), 619–622. 10.1130/G25567A.1 [DOI] [Google Scholar]
- Finstermeier, K. , Zinner, D. , Brameier, M. , Meyer, M. , Kreuz, E. , Hofreiter, M. , & Roos, C. (2013). A mitogenomic phylogeny of living primates. PLoS One, 8(7), e69504. 10.1371/journal.pone.0069504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores‐Manzanero, A. , Valenzuela‐Galván, D. , Cuarón, A. D. , & Vázquez‐Domínguez, E. (2022). Conservation genetics of two critically endangered Island dwarf carnivores. Conservation Genetics, 23(1), 35–49. 10.1007/s10592-021-01401-x [DOI] [Google Scholar]
- Fontsere, C. , Alvarez‐Estape, M. , Lester, J. , Arandjelovic, M. , Kuhlwilm, M. , Dieguez, P. , Agbor, A. , Angedakin, S. , Ayimisin, E. A. , Bessone, M. , Brazzola, G. , Deschner, T. , Eno‐Nku, M. , Granjon, A.‐C. , Head, J. , Kadam, P. , Kalan, A. K. , Kambi, M. , Langergraber, K. , … Lizano, E. (2021). Maximizing the acquisition of unique reads in noninvasive capture sequencing experiments. Molecular Ecology Resources, 21(3), 745–761. 10.1111/1755-0998.13300 [DOI] [PubMed] [Google Scholar]
- Fordham, G. , Shanee, S. , & Peck, M. (2020). Effect of river size on Amazonian primate community structure: A biogeographic analysis using updated taxonomic assessments. American Journal of Primatology, 82(7), e23136. 10.1002/ajp.23136 [DOI] [PubMed] [Google Scholar]
- Gagneux, P. , Wills, C. , Gerloff, U. , Tautz, D. , Morin, P. A. , Boesch, C. , Fruth, B. , Hohmann, G. , Ryder, O. A. , & Woodruff, D. S. (1999). Mitochondrial sequences show diverse evolutionary histories of African hominoids. Proceedings of the National Academy of Sciences of the United States of America, 96(9), 5077–5082. 10.1073/pnas.96.9.5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascon, C. , Malcolm, J. R. , Patton, J. L. , d a Silva, M. N. , Bogart, J. P. , Lougheed, S. C. , Peres, C. A. , Neckel, S , & Boag, P. T. (2000). Riverine barriers and the geographic distribution of Amazonian species. Proceedings of the National Academy of Sciences of the United States of America, 97(25), 13672–13677. doi: 10.1073/pnas.230136397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho, M. B. C. , & da Silva, F. R. (2018). The influence of riverine barriers, climate, and topography on the biogeographic regionalization of Amazonian anurans. Scientific Reports, 8(1), 3427. 10.1038/s41598-018-21879-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Socoloske, D. , & Snarr, K. A. (2010). An incident of swimming in a large river by a mantled howling monkey (Alouatta palliata) on the north coast of Honduras. Neotropical Primates, 17(1), 28–31. 10.1896/044.017.0102 [DOI] [Google Scholar]
- Graham, R. W. , Lundelius, E. L., Jr. , Graham, M. A. , Schroeder, E. K. , Toomey, R. S., III , Anderson, E. , Barnosky, A. D. , Burns, J. A. , Churcher, C. S. , Grayson, D. K. , Guthrie, R. D. , Harington, C. R. , Jefferson, G. T. , Martin, L. D. , McDonald, H. G. , Morlan, R. E. , Semken, H. A. , Webb, S. D. , Werdelin, L. , & Wilson, M. C. (1996). Spatial response of mammals to late Quaternary environmental fluctuations. Science, 272(5268), 1601–1606. 10.1126/science.272.5268.1601 [DOI] [PubMed] [Google Scholar]
- Gutiérrez, E. E. , Boria, R. A. , & Anderson, R. P. (2014). Can biotic interactions cause allopatry? Niche models, competition, and distributions of south American mouse opossums. Ecography, 37(8), 741–753. 10.1111/ecog.00620 [DOI] [Google Scholar]
- Hall, J. P. W. , & Harvey, D. J. (2002). The phylogeography of Amazonia revisited: New evidence from riodinid butterflies. Evolution, 56(7), 1489–1497. 10.1111/j.0014-3820.2002.tb01460.x [DOI] [PubMed] [Google Scholar]
- Hao, Z. , & Yi, C. (2019). The complete mitochondrial genome of Sapajus flavius (Blonde Capuchin). Mitochondrial DNA. Part B, Resources, 4(2), 2970–2971. 10.1080/23802359.2019.1662748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanin, A. , Veron, G. , Ropiquet, A. , Jansen van Vuuren, B. , Lécu, A. , Goodman, S. M. , Haider, J. , & Nguyen, T. T. (2021). Evolutionary history of Carnivora (Mammalia, Laurasiatheria) inferred from mitochondrial genomes. PLoS One, 16(2), e0240770. 10.1371/journal.pone.0240770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, F. E. , & Sewlal, J.‐A. N. (2004). The Amazon River as a dispersal barrier to passerine birds: Effects of river width, habitat and taxonomy. Journal of Biogeography, 31(11), 1809–1818. 10.1111/j.1365-2699.2004.01139.x [DOI] [Google Scholar]
- Hetu, M. , Koutouki, K. , & Joly, Y. (2019). Genomics for all: International open science genomics projects and capacity building in the developing world. Frontiers in Genetics, 10, 95. 10.3389/fgene.2019.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, M. A. , Ruokolainen, K. , Tuomisto, H. , Llerena, N. , Cardenas, G. , Phillips, O. L. , Vásquez, R. , & Räsänen, M. (2011). Geological control of floristic composition in Amazonian forests. Journal of Biogeography, 38(11), 2136–2149. 10.1111/j.1365-2699.2011.02585.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson, J. A. , Sterner, K. N. , Matthews, L. J. , Burrell, A. S. , Jani, R. A. , Raaum, R. L. , Stewart, C. B. , & Disotell, T. R. (2009). Successive radiations, not stasis, in the south American primate fauna. Proceedings of the National Academy of Sciences of the United States of America, 106(14), 5534–5539. 10.1073/pnas.0810346106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoorn, C. , Wesselingh, F. P. , ter Steege, H. , Bermudez, M. A. , Mora, A. , Sevink, J. , Sanmartín, I. , Sanchez‐Meseguer, A. , Anderson, C. L. , Figueiredo, J. P. , Jaramillo, C. , Riff, D. , Negri, F. R. , Hooghiemstra, H. , Lundberg, J. , Stadler, T. , Särkinen, T. , & Antonelli, A. (2010). Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science, 330(6006), 927–931. 10.1126/science.1194585 [DOI] [PubMed] [Google Scholar]
- Horai, S. , Hayasaka, K. , Kondo, R. , Tsugane, K. , & Takahata, N. (1995). Recent African origin of modern humans revealed by complete sequences of hominoid mitochondrial DNAs. Proceedings of the National Academy of Sciences of the United States of America, 92(2), 532–536. 10.1073/pnas.92.2.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyos, M. , Bloor, P. , Defler, T. , Vermeer, J. , Röhe, F. , & Farias, I. (2016). Phylogenetic relationships within the Callicebus cupreus species group (Pitheciidae: Primates): Biogeographic and taxonomic implications. Molecular Phylogenetics and Evolution, 102, 208–219. 10.1016/j.ympev.2016.05.031 [DOI] [PubMed] [Google Scholar]
- [dataset] Janiak, M. C. , Beck, R. M. D. , de Vries, D. , Goodhead, I. B. , & Boubli, J. P. (2022). Janiak et al. 2022 Molecular Ecology ‐ mtDNA of Amazonian primates . figshare. doi: 10.6084/m9.figshare.19606063 [DOI]
- Jühling, F. , Pütz, J. , Bernt, M. , Donath, A. , Middendorf, M. , Florentz, C. , & Stadler, P. F. (2012). Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Research, 40(7), 2833–2845. 10.1093/nar/gkr1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapli, P. , Lutteropp, S. , Zhang, J. , Kobert, K. , Pavlidis, P. , Stamatakis, A. , & Flouri, T. (2017). Multi‐rate Poisson tree processes for single‐locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics, 33(11), 1630–1638. 10.1093/bioinformatics/btx025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. , & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopuchian, C. , Campagna, L. , Lijtmaer, D. A. , Cabanne, G. S. , García, N. C. , Lavinia, P. D. , Tubaro, P. L. , Lovette, I. , & di Giacomo, A. S. (2020). A test of the riverine barrier hypothesis in the largest subtropical river basin in the Neotropics. Molecular Ecology, 29(12), 2137–2149. 10.1111/mec.15384 [DOI] [PubMed] [Google Scholar]
- Kozlov, A. M. , Darriba, D. , Flouri, T. , Morel, B. , & Stamatakis, A. (2019). RAxML‐NG: A fast, scalable and user‐friendly tool for maximum likelihood phylogenetic inference. Bioinformatics, 35(21), 4453–4455. 10.1093/bioinformatics/btz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov, A. M. , & Stamatakis, A. (2019). Using RAxML‐NG in practice . doi: 10.20944/preprints201905.0056.v1 [DOI]
- Kuderna, L. F. K. , Gao, H. , Janiak, M. C. , Kuhlwilm, M. , Orkin, J. D. , Manu, S. , Valenzuela, A. , Bergman, J. , Rouselle, M. , Silva, F. E. , Agueda, L. , Blanc, J. , Gut, M. , de Vries, D. , Goodhead, I. B. , Harris, R. A. , Raveendran, M. , Jensen, A. , Chuma, I. D. , … Marquès‐Bonet, T. (2022). A global catalog of whole‐genome diversity from 233 primate species . In revision. [DOI] [PMC free article] [PubMed]
- Losos, J. B. , & Glor, R. E. (2003). Phylogenetic comparative methods and the geography of speciation. Trends in Ecology & Evolution, 18(5), 220–227. 10.1016/S0169-5347(03)00037-5 [DOI] [Google Scholar]
- Lynch Alfaro, J. W. , Boubli, J. P. , Paim, F. P. , Ribas, C. C. , Silva, M. N. F. d. , Messias, M. R. , Röhe, F. , Mercês, M. P. , Silva Júnior, J. S. , Silva, C. R. , Pinho, G. M. , Koshkarian, G. , Nguyen, M. T. T. , Harada, M. L. , Rabelo, R. M. , Queiroz, H. L. , Alfaro, M. E. , & Farias, I. P. (2015). Biogeography of squirrel monkeys (genus Saimiri): South‐central Amazon origin and rapid pan‐Amazonian diversification of a lowland primate. Molecular Phylogenetics and Evolution, 82(PB), 436–454. 10.1016/j.ympev.2014.09.004 [DOI] [PubMed] [Google Scholar]
- Maan, M. E. , & Seehausen, O. (2011). Ecology, sexual selection and speciation. Ecology Letters, 14(6), 591–602. 10.1111/j.1461-0248.2011.01606.x [DOI] [PubMed] [Google Scholar]
- Makhov, I. A. , Gorodilova, Y. Y. U. , & Lukhtanov, V. A. (2021). Sympatric occurrence of deeply diverged mitochondrial DNA lineages in Siberian geometrid moths (Lepidoptera: Geometridae): Cryptic speciation, mitochondrial introgression, secondary admixture or effect of Wolbachia? Biological Journal of the Linnean Society. Linnean Society of London, 134(2), 342–365. 10.1093/biolinnean/blab089 [DOI] [Google Scholar]
- Maligana, N. , Julius, R. S. , Shivambu, T. C. , & Chimimba, C. T. (2020). Genetic identification of freely traded synanthropic invasive murid rodents in pet shops in Gauteng Province, South Africa. African Zoology, 55(2), 149–154. 10.1080/15627020.2019.1704632 [DOI] [Google Scholar]
- Malukiewicz, J. , Cartwright, R. A. , Curi, N. H. A. , Dergam, J. A. , Igayara, C. S. , Moreira, S. B. , Molina, C. V. , Nicola, P. A. , Noll, A. , Passamani, M. , Pereira, L. C. M. , Pissinatti, A. , Ruiz‐Miranda, C. R. , Silva, D. L. , Stone, A. C. , Zinner, D. , & Roos, C. (2021). Mitogenomic phylogeny of Callithrix with special focus on human transferred taxa. BMC Genomics, 22(1), 239. 10.1186/s12864-021-07533-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malukiewicz, J. , Hepp, C. M. , Guschanski, K. , & Stone, A. C. (2017). Phylogeny of the jacchus group of Callithrix marmosets based on complete mitochondrial genomes. American Journal of Physical Anthropology, 162(1), 157–169. 10.1002/ajpa.23105 [DOI] [PubMed] [Google Scholar]
- Martin, M. (2011). Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet.journal, 17(1), 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Matsui, A. , Rakotondraparany, F. , Munechika, I. , Hasegawa, M. , & Horai, S. (2009). Molecular phylogeny and evolution of prosimians based on complete sequences of mitochondrial DNAs. Gene, 441(1‐2), 53–66. 10.1016/j.gene.2008.08.024 [DOI] [PubMed] [Google Scholar]
- McKay, B. D. , & Zink, R. M. (2010). The causes of mitochondrial DNA gene tree paraphyly in birds. Molecular Phylogenetics and Evolution, 54(2), 647–650. 10.1016/j.ympev.2009.08.024 [DOI] [PubMed] [Google Scholar]
- Menezes, A. N. , Viana, M. C. , Furtado, C. , Schrago, C. G. , & Seuánez, H. N. (2013). Positive selection along the evolution of primate mitogenomes. Mitochondrion, 13(6), 846–851. 10.1016/j.mito.2013.06.001 [DOI] [PubMed] [Google Scholar]
- Naka, L. N. , & Brumfield, R. T. (2018). The dual role of Amazonian rivers in the generation and maintenance of avian diversity. Science Advances, 4(8), eaar8575. 10.1126/sciadv.aar8575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka, L. N. , & Pil, M. W. (2020). Moving beyond the riverine barrier vicariant paradigm. Molecular Ecology, 29(12), 2129–2132. 10.1111/mec.15465 [DOI] [PubMed] [Google Scholar]
- Nunes, A. V. (2014). Report of a black spider monkey (Ateles chamek) swimming in a large river in central‐western Brazil. Neotropical Primates, 21(2), 204–206. 10.1896/044.021.0210 [DOI] [Google Scholar]
- Nurk, S. , Meleshko, D. , Korobeynikov, A. , & Pevzner, P. A. (2017). metaSPAdes: A new versatile metagenomic assembler. Genome Research, 27(5), 824–834. 10.1101/gr.213959.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, U. , Vasconcelos, M. F. , & Santos, A. J. (2017). Biogeography of Amazon birds: Rivers limit species composition, but not areas of endemism. Scientific Reports, 7(1), 2992. 10.1038/s41598-017-03098-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin, J. D. , Montague, M. J. , Tejada‐Martinez, D. , de Manuel, M. , del Campo, J. , Cheves Hernandez, S. , di Fiore, A. , Fontsere, C. , Hodgson, J. A. , Janiak, M. C. , Kuderna, L. F. K. , Lizano, E. , Martin, M. P. , Niimura, Y. , Perry, G. H. , Valverde, C. S. , Tang, J. , Warren, W. C. , de Magalhães, J. P. , … Melin, A. D. (2021). The genomics of ecological flexibility, large brains, and long lives in capuchin monkeys revealed with fecalFACS. Proceedings of the National Academy of Sciences of the United States of America, 118(7), e2010632118. 10.1073/pnas.2010632118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin, J. D. , Yang, Y. , Yang, C. , Yu, D. W. , & Jiang, X. (2016). Cost‐effective scat‐detection dogs: Unleashing a powerful new tool for international mammalian conservation biology. Scientific Reports, 6, 34758. 10.1038/srep34758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, D. A. , Lima, A. P. , & Werneck, F. P. (2018). Environmental transition zone and rivers shape intraspecific population structure and genetic diversity of an Amazonian rain forest tree frog. Evolutionary Ecology, 32(4), 359–378. 10.1007/s10682-018-9939-2 [DOI] [Google Scholar]
- Patton, J. L. , da Silva, M. N. F. , & Malcolm, J. R. (1994). Gene genealogy and differentiation among arboreal spiny rats (Rodentia: Echimyidae) of the Amazon basin: A test of the riverine barrier hypothesis. Evolution, 48(4), 1314–1323. 10.1111/j.1558-5646.1994.tb05315.x [DOI] [PubMed] [Google Scholar]
- Phukuntsi, M. A. , Du Plessis, M. , Dalton, D. L. , Jansen, R. , Sauther, M. L. , Cuozzo, F. P. , & Kotze, A. (2021). Population and genetic structure of a male‐dispersing strepsirrhine, Galago moholi (Primates, Galagidae), from northern South Africa, inferred from mitochondrial DNA. Primates, 62(4), 667–675. 10.1007/s10329-021-00912-y [DOI] [PubMed] [Google Scholar]
- Pomara, L. Y. , Ruokolainen, K. , & Young, K. R. (2014). Avian species composition across the Amazon River: The roles of dispersal limitation and environmental heterogeneity. Journal of Biogeography, 41(4), 784–796. 10.1111/jbi.12247 [DOI] [Google Scholar]
- Pomerantz, A. , Peñafiel, N. , Arteaga, A. , Bustamante, L. , Pichardo, F. , Coloma, L. A. , Barrio‐Amorós, C. L. , Salazar‐Valenzuela, D. , & Prost, S. (2018). Real‐time DNA barcoding in a rainforest using nanopore sequencing: Opportunities for rapid biodiversity assessments and local capacity building. GigaScience, 7(4), 1–14. 10.1093/gigascience/giy033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, L. M. , de la Torre, S. , Pérez‐Peña, P. , & Cortés‐Ortiz, L. (2021). Taxonomic diversity of Cebuella in the western Amazon: Molecular, morphological and pelage diversity of museum and free‐ranging specimens. American Journal of Physical Anthropology, 175(1), 251–267. 10.1002/ajpa.24266 [DOI] [PubMed] [Google Scholar]
- Quintela, F. M. , da Rosa, C. A. , & Feijó, A. (2020). Updated and annotated checklist of recent mammals from Brazil. Anais Da Academia Brasileira de Ciencias, 92(Suppl. 2), e20191004. 10.1590/0001-3765202020191004 [DOI] [PubMed] [Google Scholar]
- Raaum, R. L. , Sterner, K. N. , Noviello, C. M. , Stewart, C.‐B. , & Disotell, T. R. (2005). Catarrhine primate divergence dates estimated from complete mitochondrial genomes: Concordance with fossil and nuclear DNA evidence. Journal of Human Evolution, 48(3), 237–257. 10.1016/j.jhevol.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Rambaut, A. , Drummond, A. J. , Xie, D. , Baele, G. , & Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology, 67(5), 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2019). R: A language and environment for statistical computing (Version 3.6.0) . Retrieved from: https://www.R‐project.org/
- Reese, E. M. , Winters, M. , Booth, R. K. , & Wasser, S. K. (2020). Development of a mitochondrial DNA marker that distinguishes domestic dogs from Washington state gray wolves. Conservation Genetics Resources, 12(3), 497–501. 10.1007/s12686-020-01130-2 [DOI] [Google Scholar]
- Revell, L. J. (2012). Phytools: An R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution / British Ecological Society, 3, 217–223. 10.1111/j.2041-210X.2011.00169.x [DOI] [Google Scholar]
- Rice, W. R. , & Salt, G. W. (1990). The evolution of reproductive isolation as a correlated character under sympatric conditions: Experimental evidence. Evolution; International Journal of Organic Evolution, 44(5), 1140–1152. 10.1111/j.1558-5646.1990.tb05221.x [DOI] [PubMed] [Google Scholar]
- Rodríguez, J. P. , Simonetti, J. A. , Premoli, A. , & Marini, M. Â. (2005). Conservation in austral and neotropical America: Building scientific capacity equal to the challenges. Conservation Biology, 19(3), 969–972. [Google Scholar]
- Ruokolainen, K. , Moulatlet, G. M. , Zuquim, G. , Hoorn, C. , & Tuomisto, H. (2019). Geologically recent rearrangements in central Amazonian river network and their importance for the riverine barrier hypothesis. Frontiers of Biogeography, 11(3), e45046. 10.21425/F5FBG45046 [DOI] [Google Scholar]
- Russello, M. A. , Avery, M. L. , & Wright, T. F. (2008). Genetic evidence links invasive monk parakeet populations in the United States to the international pet trade. BMC Evolutionary Biology, 8, 217. 10.1186/1471-2148-8-217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santorelli, S., Jr. , Magnusson, W. E. , & Deus, C. P. (2018). Most species are not limited by an Amazonian river postulated to be a border between endemism areas. Scientific Reports, 8(1), 2294. 10.1038/s41598-018-20596-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, D. J. , Espinoza, T. , Connell, M. , & Hughes, J. M. (2018). Conservation genetics of the Mary River turtle (Elusor macrurus) in natural and captive populations. Aquatic Conservation: Marine and Freshwater Ecosystems, 28(1), 115–123. 10.1002/aqc.2851 [DOI] [Google Scholar]
- Şekercioğlu, Ç. H. (2012). Promoting community‐based bird monitoring in the tropics: Conservation, research, environmental education, capacity‐building, and local incomes. Biological Conservation, 151(1), 69–73. 10.1016/j.biocon.2011.10.024 [DOI] [Google Scholar]
- Serrao, N. R. , Reid, S. M. , & Wilson, C. C. (2018). Conservation genetics of redside dace (Clinostomus elongatus): Phylogeography and contemporary spatial structure. Conservation Genetics, 19(2), 409–424. 10.1007/s10592-017-1012-0 [DOI] [Google Scholar]
- Silva, S. M. , Peterson, A. T. , Carneiro, L. , Burlamaqui, T. C. T. , Ribas, C. C. , Sousa‐Neves, T. , Miranda, L. S. , Fernandes, A. M. , d'Horta, F. M. , Araújo‐Silva, L. E. , Batista, R. , Bandeira, C. H. M. M. , Dantas, S. M. , Ferreira, M. , Martins, D. M. , Oliveira, J. , Rocha, T. C. , Sardelli, C. H. , Thom, G. , … Aleixo, A. (2019). A dynamic continental moisture gradient drove Amazonian bird diversification. Science Advances, 5(7), eaat5752. 10.1126/sciadv.aat5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovrind, M. , Louis, M. , Westbury, M. V. , Garilao, C. , Kaschner, K. , Castruita, J. A. S. , Gopalakrishnan, S. , Knudsen, S. W. , Haile, J. S. , Dalén, L. , Meshchersky, I. G. , Shpak, O. V. , Glazov, D. M. , Rozhnov, V. V. , Litovka, D. I. , Krasnova, V. V. , Chernetsky, A. D. , Bel'kovich, V. , Lydersen, C. , … Lorenzen, E. D. (2021). Circumpolar phylogeography and demographic history of beluga whales reflect past climatic fluctuations. Molecular Ecology, 30(11), 2543–2559. 10.1111/mec.15915 [DOI] [PubMed] [Google Scholar]
- Smith, B. T. , McCormack, J. E. , Cuervo, A. M. , Hickerson, M. J. , Aleixo, A. , Cadena, C. D. , Pérez‐Emán, J. , Burney, C. W. , Xie, X. , Harvey, M. G. , Faircloth, B. C. , Glenn, T. C. , Derryberry, E. P. , Prejean, J. , Fields, S. , & Brumfield, R. T. (2014). The drivers of tropical speciation. Nature, 515(7527), 406–409. 10.1038/nature13687 [DOI] [PubMed] [Google Scholar]
- Soares, E. A. A. , D'Apolito, C. , Jaramillo, C. , Harrington, G. , Caputo, M. V. , Barbosa, R. O. , dos Santos, E. B. , Dino, R. , & Gonçalves, A. D. (2017). Sedimentology and palynostratigraphy of a Pliocene‐Pleistocene (Piacenzian to Gelasian) deposit in the lower Negro River: Implications for the establishment of large rivers in Central Amazonia. Journal of South American Earth Sciences, 79, 215–229. 10.1016/j.jsames.2017.08.008 [DOI] [Google Scholar]
- Strohm, J. H. T. , Gwiazdowski, R. A. , & Hanner, R. (2016). Mitogenome metadata: Current trends and proposed standards. Mitochondrial DNA. Part A, DNA Mapping, Sequencing, and Analysis, 27(5), 3263–3269. 10.3109/19401736.2015.1015003 [DOI] [PubMed] [Google Scholar]
- Tagliacollo, V. A. , Roxo, F. F. , Duke‐Sylvester, S. M. , Oliveira, C. , & Albert, J. S. (2015). Biogeographical signature of river capture events in Amazonian lowlands. Journal of Biogeography, 42(12), 2349–2362. 10.1111/jbi.12594 [DOI] [Google Scholar]
- Tahsin, T. , Weissenbacher, D. , Rivera, R. , Beard, R. , Firago, M. , Wallstrom, G. , Scotch, M. , & Gonzalez, G. (2016). A high‐precision rule‐based extraction system for expanding geospatial metadata in GenBank records. Journal of the American Medical Informatics Association: JAMIA, 23(5), 934–941. 10.1093/jamia/ocv172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen, P. F. , & Willerslev, E. (2015). Environmental DNA – An emerging tool in conservation for monitoring past and present biodiversity. Biological Conservation, 183, 4–18. 10.1016/j.biocon.2014.11.019 [DOI] [Google Scholar]
- Torosin, N. S. , Argibay, H. , Webster, T. H. , Corneli, P. S. , & Knapp, L. A. (2020a). Comparing the selective landscape of TLR7 and TLR8 across primates reveals unique sites under positive selection in Alouatta . Molecular Phylogenetics and Evolution, 152, 106920. 10.1016/j.ympev.2020.106920 [DOI] [PubMed] [Google Scholar]
- Torosin, N. S. , Webster, T. H. , Argibay, H. , Sanchez Fernandez, C. , Ferreyra, H. , Uhart, M. , Agostini, I. , & Knapp, L. A. (2020b). Positively selected variants in functionally important regions of TLR7 in Alouatta guariba clamitans with yellow fever virus exposure in Northern Argentina. American Journal of Physical Anthropology, 173(1), 50–60. 10.1002/ajpa.24086 [DOI] [PubMed] [Google Scholar]
- Voss, R. S. , Fleck, D. W. , & Jansa, S. A. (2019). Mammalian diversity and Matses ethnomammalogy in Amazonian Peru. Part 3, marsupials (Didelphimorphia). Bulletin of the American Museum of Natural History, 432, 1–87. 10.1206/0003-0090.432.1.1 [DOI] [Google Scholar]
- Wallace, A. R. (1852). On the monkeys of the Amazon. Proceedings of the Zoological Society of London, 20, 107–110. 10.1080/037454809494374 [DOI] [Google Scholar]
- Wang, W. , Liu, J.‐Y. , Wang, H.‐F. , Yang, M.‐Y. , Liu, Q.‐Y. , & Ding, M.‐X. (2016). The complete mitochondrial genome of white‐tufted‐ear marmoset, Callithrix jacchus (Primates: Callitrichinae). Mitochondrial DNA. Part A, DNA Mapping, Sequencing, and Analysis, 27(3), 1920–1921. 10.3109/19401736.2014.971287 [DOI] [PubMed] [Google Scholar]
- Watsa, M. , Erkenswick, G. A. , Pomerantz, A. , & Prost, S. (2020). Portable sequencing as a teaching tool in conservation and biodiversity research. PLoS Biology, 18(4), e3000667. 10.1371/journal.pbio.3000667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos, F. E. , Apollonio, M. , Bärmann, E. V. , Festa‐Bianchet, M. , Göhlich, U. , Habel, J. C. , Haring, E. , Kruckenhauser, L. , Lovari, S. , McDevitt, A. D. , Pertoldi, C. , Rössner, G. E. , Sánchez‐Villagra, M. R. , Scandura, M. , & Suchentrunk, F. (2013). Species inflation and taxonomic artefacts—A critical comment on recent trends in mammalian classification. Mammalian Biology = Zeitschrift Für Säugetierkunde, 78(1), 1–6. 10.1016/j.mambio.2012.07.083 [DOI] [Google Scholar]
- Zhang, X. , Pan, F. , & Wu, Z.‐W. (2016). Complete mitochondrial genome of Callithrix kuhlii (Primates: Callitrichinae) with phylogenetic consideration. Mitochondrial DNA. Part A, DNA Mapping, Sequencing, and Analysis, 27(4), 2943–2944. 10.3109/19401736.2015.1060452 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Table S1
Data Availability Statement
Data used or generated for this article have been made available in the European Nucleotide Archive's SRA (BioProject PRJEB49549) and NCBI's GenBank (accession numbers OM328861‐OM329065; Table S1). Files containing metadata for all samples, sequence alignments, analysis input files, and the dated phylogenetic tree have been archived at: 10.6084/m9.figshare.19606063. Scripts used to run analyses are available on GitHub (https://github.com/MareikeJaniak/Platyrrhine‐mtDNA) and have been archived at: 10.6084/m9.figshare.19610187.


