Abstract
This study describes the characterization of the recently described Salmonella genomic island 1 (SGI1) (D. A. Boyd, G. A. Peters, L.-K. Ng, and M. R. Mulvey, FEMS Microbiol. Lett. 189:285–291, 2000), which harbors the genes associated with the ACSSuT phenotype in a Canadian isolate of Salmonella enterica serovar Typhimurium DT104. A 43-kb region has been completely sequenced and found to contain 44 predicted open reading frames (ORFs) which comprised ∼87% of the total sequence. Fifteen ORFs did not show any significant homology to known gene sequences. A number of ORFs show significant homology to plasmid-related genes, suggesting, at least in part, a plasmid origin for the SGI1, although some with homology to phage-related genes were identified. The SGI1 was identified in a number of multidrug-resistant DT120 and S. enterica serovar Agona strains with similar antibiotic-resistant phenotypes. The G+C content suggests a potential mosaic structure for the SGI1. Emergence of the SGI1 in serovar Agona strains is discussed.
Multiple-drug-resistant (MDR) Salmonella enterica serovar Typhimurim phage type DT104 (hereafter abbreviated as Typhimurium DT104) is currently the second most prevalent Salmonella serotype isolated in England and Wales (35, 36) and is increasingly prevalent in the United States (15, 18) and Canada (26). Outbreaks of MDR Typhimurium DT104 have also been reported in poultry, beef, cheese, and swine in numerous countries (9, 12, 16, 24, 39). This strain is resistant to a core group of antimicrobials, including ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline (commonly abbreviated ACSSuT); however, isolates have been identified which are also resistant to fluoroquinolones, trimethoprim, and kanamycin (25, 34).
Many isolates of Typhimurium DT104 conferring the ACSSuT phenotype have a similar genetic makeup comprised of the floR and tet(G) genes bracketed by two class 1 integrons carrying the pse-1 and aadA2 cassettes clustered on a 14-kb region of the Typhimurium DT104 genome (3, 5, 25, 28, 30). Cotransduction experiments using P22-like phages ES18 and PDT17 demonstrated the antimicrobial resistance gene clustered on a fragment of less than 46 kb (31). Recently, this region, termed Salmonella genomic island 1 (SGI1), has been cloned from the genome of a Canadian isolate and has been shown to be comprised of a 43-kb region between thdF and a novel retron sequence (4). The genomic location, the fact that the resistance cannot be transferred (37), and the demonstration that excision cannot be detected at the genetic level (4) has led to speculation that even if antimicrobial selective pressure is removed, the resistance will persist (4, 23, 37). However, it should be noted that persistence of the antibiotic resistance genes depends on the relative fitness cost in the absence of antimicrobials. MDR Typhimurium DT104 isolates from different countries that harbor the pseI and aadA2 integrons have been shown to be similar using several molecular typing techniques, and this has led investigators to suggest a clonal dissemination of this organism (4, 10, 11, 21, 30). Recently, a number of S. enterica serovar Agona (hereafter referred to as Agona) strains have been characterized as harboring the same antimicrobial resistance region, suggesting horizontal gene transfer of this region (8).
Some questions exist about the nature of possible increased virulence of drug-resistant DT104 strains. Case control studies have suggested that MDR Typhimurium DT104 is possibly a hypervirulent strain compared to susceptible strains of Typhimurium DT104 or other Salmonella serotypes (12, 33, 39). However, this virulence does not appear to be related to a hyperinvasive phenotype as shown in tissue culture assays, as resistant DT104 is no more invasive than susceptible serovar Typhimurium strains with or without exposure to antibiotics (6, 7). Thus, as suggested, the overall pathogenicity may be enhanced by invasion-independent virulence-related factors, one of which may be treatment failure due to multidrug resistance (7).
In an attempt to identify genetic factors responsible for the increased virulence and to obtain a better understanding of the origins of the SGI1, we have sequenced the entire genomic island harboring the resistance genes as well as analyzed other MDR strains of Salmonella to determine if the drug-resistant genes are associated with SGI1.
MATERIALS AND METHODS
Bacteria, bacteriophages, and media.
The Salmonella strains used in this study are listed in Table 1. Escherichia coli LE392 was used for propagating phages, E. coli XL1-Blue and plasmid pBluescript II (Stratagene) were used in cloning experiments, and λEMBL3 (Promega) was used in phage cloning experiments. All strains were grown at 37°C in brain heart infusion broth or Luria-Bertani (LB) medium. Stock cultures were stored at −70°C in Microbank vials (Pro-Lab Diagnostics, Richmond Hill, Ontario, Canada).
TABLE 1.
Strains used in this study and some associated characteristics
| Strain no. | Serovara | Resistance profileb | Isolation site | SGI1 junction PCR resultsc
|
XbaI fragments(s) hybridizing withd: | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Left | Right | qac/sulI | p1–9 | |||||
| 96-5227 | DT104 | ACSSuT | Canada | + | + | 11.7, 4.3 | 9, 4 | 24 |
| BN3791 | DT104 | ACSSuT | France | + | + | 11.7, 4.3 | 9, 4 | This work |
| 1641SA96 | DT104 | ACSSuT | Belgium | + | + | 11.7, 4.3 | 9, 4 | This work |
| 1276SA96 | DT104 | ACSSuT | Belgium | + | + | 11.7, 4.3 | 9, 4 | This work |
| 1390SA96 | DT104 | ACSSuT | Belgium | + | + | 11.7, 4.3 | 9, 4 | This work |
| 2019SA96 | DT104 | ACSSuT | Belgium | + | + | 11.7, 4.3 | 9, 4 | This work |
| S/952569 | DT104 | ACSSuT | Scotland | + | + | NDe | NDe | This work |
| S/960081 | DT104 | ASSuT | Scotland | + | + | 9 | 9, 4 | This work |
| S/960725 | DT104 | ASu | Scotland | + | + | 4.3 | 9, 4 | This work |
| S/954435 | DT104 | SSu | Scotland | + | + | 7 | 9, 4 | This work |
| S/921495 | DT104 | Sensitive | Scotland | − | − | None | None | This work |
| 424SA93 | DT120 | ACSSuT | Belgium | + | + | 11.7, 4.3 | 9, 4 | 7 |
| 1439SA96 | DT120 | ACSSuT | Belgium | + | + | 11.7, 4.3 | 9, 4 | 7 |
| 959SA97 | Agona | ACSSuT | Belgium | + | + | 11.7, 4.3 | 9, 4 | 7 |
| 251SA97 | Agona | ACSSuT | Belgium | + | + | 11.7, 8.4 | 9, 4 | 7 |
| 1169SA97 | Agona | ACSSuT | Belgium | + | + | 11.7, 8.4 | 9, 4 | This work |
| 1146SA97 | Agona | ACSSuT | Belgium | + | + | 11.7, 8.4 | 9, 4 | This work |
| 0059SA98 | Agona | Sensitive | Belgium | − | − | None | None | This work |
S. enterica serovars Typhimurium DT104, Typhimurium DT120, and Agona.
A, ampicillin; C, chloramphenicol; S, spectinomycin and streptomycin; Su, sulfonamides; T, tetracycline.
+, product obtained; −, no product obtained. Left, U7-L12 and LJ-R1 primers; right, 104-RJ and C9-L2 or 104-RJ and 104-D primers.
The qac/sulI probe was an amplicon generated with QS-1 and QS-2 primers; p1–9 is a 2-kb EcoRI fragment (see Fig. 1) cloned in pBluescript II.
ND, not done.
Antimicrobial susceptibility testing.
The strains were tested for their antibiotic susceptibility on Mueller-Hinton agar by the disk diffusion method. Resistance to the following antibiotics was tested with disks containing ampicillin (10 μg), chloramphenicol (30 μg), florfenicol (30 μg), spectinomycin (100 μg), streptomycin (10 IU), sulfonamides (200 μg), and tetracyclines (30 IU). The media and disks were from Sanofi Diagnostics Pasteur (Marnes-la-Coquette, France), except for disks with florfenicol, which were purchased from Schering-Plough Santé Animale (Segré, France).
Recombinant DNA methodology.
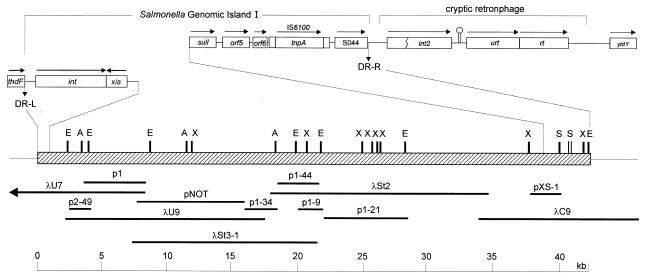
Genomic DNA was isolated as previously described (4). A genomic DNA library was constructed in λEMBL3 using 15- to 20-kb Sau3A genomic DNA fragments from serovar Typhimurium 96–5227, and clones spanning the entire SGI1 were isolated as previously described (4) (Fig. 1). Templates for sequencing were obtained by subcloning various fragments from lambda clones into pBluescript II (Stratagene) (Fig. 1) and by PCR of specific regions using primers designed on previously sequenced DNA. In addition, a ∼8.7-kb amplicon was obtained by long PCR using the primers St31-Not, 5′-TAATgcggccgcAAGCAATAGCCAGTACGCTG-3′, and p134-Not, 5′-AATAgcggccgcTCTCCGATGCTGTCGAATG-3′ (italics indicate non-Salmonella sequence; lowercase indicates a NotI site), digested with NotI and cloned into pBluescript II. The resulting plasmid, pNOT (Fig. 1), was then subjected to random mutagenesis using the EZ::TN Insertion System (Epicentre Technologies) to allow for rapid sequencing of the insert. Synthesis of oligonucleotides and DNA sequencing were carried out in the DNA Core Facility, National Microbiology Laboratory, Health Canada, Winnipeg, Canada. Standard PCRs were carried out using 2.5 U of AmpliTaq Gold DNA polymerase in PCR Buffer II (PE Applied Biosystems) containing 3 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 0.5 μM concentrations of primers, and 50 ng of DNA. PCR cycling conditions were 95°C for 10 min followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 5 min. Other annealing temperatures may have been used depending on the melting temperature (Tm) of the primers in the reaction mixture. Long PCR was carried out using the Expand Long Template PCR System (Roche Diagnostics, Laval, Quebec) as recommended by the manufacturer. Plaque and Southern blotting was carried out by standard methods (29) with probes labeled and detected by ECL kits using the manufacturer's instructions (Amersham Pharmacia Biotech).
FIG. 1.
Cloning strategy of the complete SGI1. Letters above the map indicate restriction enzyme sites. X, XbaI; E, EcoRI; S, SalI; A, AvrII. Boxes above the map depict previously characterized regions (4) and are not drawn to scale. Arrows denote the direction of transcription.
IRS-PCR. (i) Adapters and primers.
Infrequent restriction site (IRS)-PCR was performed as described previously by Mazurek et al. (22) with some modification. In brief, the HhaI adapter (AH), which consists of a 22-base oligonucleotide (AH1; 5′-AGA ACT GAC CTC GAC TCG CACG-3′) with a 7-base oligonucleotide (AH2; 5′-TGC GAG T-3′), and the XbaI adapter (AX), which consisted of a phosphorylated 18-base oligonucleotide (AX1; 5′-PO4 -CTA GTA CTG GCAGAC TCT-3′) with a 7-base oligonucleotide (AX2; 5′-GCC AGT A-3′), were designed to ligate specifically to the cohesive ends of the corresponding restricted fragments. To prepare the adapters, oligonucleotides AH1 and AH2 or AX1 and AX2 were mixed in equal molar amounts in 1× PCR buffer (Promega) and were allowed to anneal as the mixture cooled from 95°C to room temperature over 1 h. The mixture was briefly centrifuged and was stored at −20°C until use.
(ii) Preparation of template DNA.
One microgram of DNA was digested with 10 U of XbaI (QuantumAppligene) and 10 U of HhaI (QuantumAppligene) in 1× buffer II for 120 min at 37°C in a volume of 15 μl. Sterile distilled water, 1.5 U of T4 DNA ligase (Boehringer Mannheim), 1× ligase buffer, the XbaI adapter (20 pmol), and the HhaI adapter (20 pmol) were added for a total volume of 20 μl. The mixture was incubated at 16°C for 90 min and then at 65°C for 20 min to inactivate T4 DNA ligase. The sample was redigested with 5 U of XbaI and 5 U of HhaI at 37°C for 15 min to cleave any restriction sites re-formed by ligation and then submitted to amplification.
(iii) PCR amplification.
Each PCR mixture included 2.5 μl of a 1:10 dilution of template DNA, 0.5 U of Taq DNA polymerase (Promega), deoxynucleoside triphosphates (50 mM each) (Promega), and the oligonucleotide primers in 1× PCR buffer. Typically, the oligonucleotides AH1 and 6-carboxyfluorescein (Fam)-labeled PX were used together as primers. Amplification was performed in a Geneamp 9700 thermocycler (Perkin-Elmer) with an amplification profile that consisted of an initial denaturation step at 94°C for 5 min and then 30 cycles of denaturation at 94°C for 30 s, primer annealing at 60°C for 30 s, and extension at 72°C for 90 s. All experiments included negative controls, which were processed with the samples.
(iv) Separation of PCR products.
Following amplification, 1 to 1.5 μl of undiluted IRS-PCR product was mixed with 0.5 μl of internal lane standard (Genescan-Rox 500; PE Applied Biosystems), and deionized formamide was added to a final volume of 20 μl. The resulting mixture was heated at 95°C for 4 min and then quickly cooled on ice. Separation and detection of 6-carboxyfluorescein (Fam)-labeled PCR products were performed by capillary electrophoresis on an ABI 310 automated sequencer, and electrophoresis was conducted for 30 min per sample at 60°C as described by the manufacturer.
(v) Data capture and analysis.
The results were automatically collected with the Genescan collection and fragment analysis software. The internal lane standards, included in each lane, allowed an accurate sizing of individual IRS-amplified fragments. The results were viewed in the form of an electrophoregram, tabular data, or a combination of both. Interpretation of Genescan data was performed using the Genotyper software, which allowed construction of tabular binary data based on presence or absence of bands between each studied strain. The Genotyper analysis parameters were set to medium smoothing, and the baseline fluorescence was set to 150 U. The software filter used to remove PCR and background noise was as follows: “remove labels from peaks preceded by higher (at least 5%), labeled peak within 0 to 2.5 bp, and remove labels from peaks followed by higher (at least 5%), labeled peak within 0 to 2.5 bp.” IRS-PCR similarities between strain pairs were calculated using the Jaccard coefficient, and cluster analysis was performed using the unweighted pair group method with averages (UPGMA) algorithm (33).
Computer-aided analysis and annotation.
Homology searches were carried out using the BLAST suite of programs (2), and open reading frames (ORFs) were detected with ORFinder via the World Wide Web interface of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih). All ORFs larger than 180 bp (more than 60 amino acids) were used as queries in BLAST searches. Significant homology was defined as an expect value (E value) of <1e-05 (e means exponential; 1e-05 is 0.00001) for the top-scoring protein and/or >20% identity over at least 60% of the length of the protein.
Nucleotide accession number.
The complete nucleotide sequence of SGI1 has been deposited in the GenBank database under accession number AF261825.
RESULTS AND DISCUSSION
General properties of the SGI1 sequence.
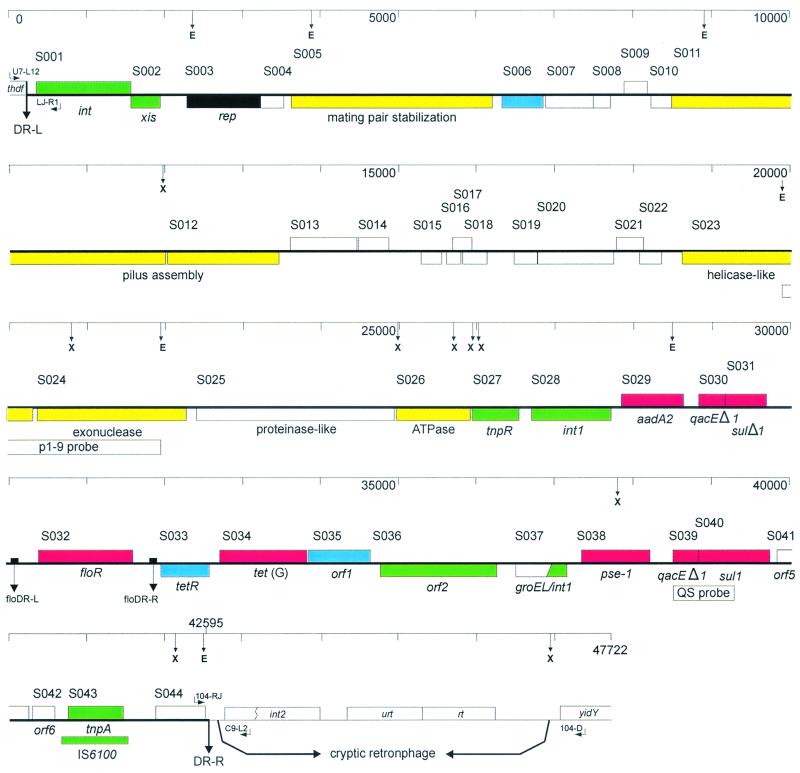
Overlapping lambda clones and plasmids used in this study are shown in Fig. 1. The complete sequence of the 42,415-bp region between the previously characterized direct repeats DR-L and DR-R, which define the boundaries of the genomic island inserted between the thdF gene and a cryptic retronphage in the Typhimurium DT104 genome (4), has been determined. A total of 44 ORFs initiated by 42 putative ATG, 1 TTG (S023), and 1 CTG (S036) start codons have been identified. Table 2 lists the ORFs whose putative products show similarity to protein sequences available in GenBank, and Table 3 lists the ORFs whose putative products show no significant similarity to available protein sequences. Figure 2 depicts a linear map with all ORFs labeled with some named based on function, and the putative functions of others are indicated. No conclusions regarding whether S017, which overlaps S016 and S018, is an expressed ORF or vice versa, though the latter two are preceded by putative Shine-Dalgarno sequences while S017 is not. Similarly, S021 and S022, which overlap by 88 bp, are likely mutually exclusive as ORFs, though S022 is preceded by a putative Shine-Dalgarno sequence while S021 is not. Nonetheless, the putative ORFs account for ∼87% of the SGI1 sequence.
TABLE 2.
The ORFs in SGI1 DNA whose putative products exhibit significant homologya to extant protein sequences
| ORFb
|
Locationc (start–stop) | Size in:
|
% G+C | E valued | % Identity/rangee | Exhibits homology to (accession no. and/or reference): | ||
|---|---|---|---|---|---|---|---|---|
| No. | Name | bp | aa | |||||
| S001 | int | 367–1584 | 1,218 | 405 | 43 | 1.0e-40 | 29/365 | Integrase from Tn4555; Bacteroides fragilis (U75371) |
| S002 | xis | 1949–1581 | 369 | 122 | 46.1 | 4.0e-05 | 31/94 | Excisionase from Tn4555; B. fragilis (U75371) |
| S003 | rep | 3260–2307 | 954 | 317 | 44.4 | 7.0e-43 | 34/277 | Replication protein RepA pMG101; Rhodopseudomonas palustris (AB031076) |
| S005 | 6384–3625 | 2,760 | 919 | 46 | 2.0e-52 | 24/862 | Mating pair stabilization protein TrhN, plasmid R27; serovar Typhi (AF105019) | |
| S006 | 7014–6481 | 534 | 177 | 43.8 | 0.01 | 25/159 | Flagellar transcriptional activator Fr1B; Bordetella bronchiseptica (U17998) | |
| S011 | trhG | 11992–8588 | 3,405 | 1,134 | 45.5 | 2.0e-27 | 21/822 | Pilus assembly protein, plasmid R27; serovar Typhi (AF250878) |
| S012 | trhH | 13420–11996 | 1,425 | 474 | 43.2 | 2.0e-33 | 26/446 | Pilus assembly protein TrhH, plasmid R27; serovar Typhi (AF112468) |
| S020 | 17712–16747 | 966 | 321 | 53 | 2.0e-21 | 28/321 | Orf3 protein, putative integrase, pFNL10; Francisella tularensis (AF121418) | |
| S023 | 20328–18688 | 1,641 | 546 | 47.1 | 5.0e-20 | 24/448 | DNA helicase II UvrD; Haemophilus influenzae (U32798) | |
| S024 | 22271–20346 | 1,926 | 641 | 46.4 | 2.0e-05 | 22/468 | Overcoming lysogenization defect protein OLD, exonuclease; bacteriophage P2 (AF063097) | |
| S025 | 24892–22349 | 2,544 | 847 | 38.9 | 6.0e-15 | 24/416 | Hypothetical protein Y4bN, symbiosis plasmid; Rhizobium sp. NGR234 (AE000066) | |
| S026 | 25902–24913 | 990 | 329 | 39.2 | 1.0e-41 | 32/330 | ATPase Y4kL, symbiosis plasmid; Rhizobium sp. NGR234 (AE000081) | |
| S027 | tnpR | 26565–25981 | 585 | 194 | 46.8 | 0 | 100/171 | Resolvase (3, 5) |
| S028 | int1 | 27852–26839 | 1,014 | 337 | 61.2 | 0 | 100/193 | Integrase of class I integrons (3, 5, 29) |
| S029 | aadA2 | 27998–28789 | 792 | 263 | 51.9 | 0 | 100/263 | Spectinomycin/streptomycin resistance protein (3, 5, 29) |
| S030 | qacEΔ1 | 28953–29300 | 348 | 115 | 50 | 0 | 100/115 | Quaternary ammonium compound and disinfectant resistance partial protein (3, 5, 29) |
| S031 | sulIΔ1 | 29294–29821 | 528 | 176 | 63.1 | 0 | 100/176 | Sulfonamide resistance partial protein (3, 5, 29) |
| S032 | floR | 30482–31696 | 1,215 | 404 | 58.2 | 0 | 100/404 | Chloramphenicol and florfenicol resistance (3, 5) |
| S033 | tetR | 32529–31903 | 627 | 208 | 59.2 | 0 | 100/209 | Tetracycline resistance regulator; repressor (3, 5) |
| S034 | tet(G) | 32681–33808 | 1,128 | 375 | 57.5 | 0 | 100/375 | Tetracycline resistance (3, 5) |
| S035 | orf1 | 33829–34620 | 792 | 263 | 61.6 | 4.0e-25 | 35/194 | LysR-type transcriptional regulator; Deinococcus radiodurans (5) |
| S036 | orf2 | 36196–34712 | 1,482 | 494 | 68.9 | 3.0e-90 | 58/484 | Putative transposase OrfA, plasmid 10507-1, E. coli (5) |
| S037 | groEL/intI | 37124–36471 | 654 | 217 | 63.3 | 0 | 100/217 | GroEL-like/integrase fusion protein (5) |
| S038 | pse-1 | 37330–38196 | 867 | 288 | 41 | 0 | 100/288 | Carpencillinase (3, 5, 29) |
| S039 | qacEΔ1 | 38413–38760 | 348 | 115 | 50 | 0 | 100/115 | Quaternary ammonium compound and disinfectant resistance partial protein (3, 5, 29) |
| S040 | sulI | 38754–39593 | 840 | 279 | 61.7 | 0 | 100/279 | Sulfonamide resistance (3, 4, 5, 29) |
| S041 | orf5 | 39721–40221 | 501 | 166 | 65.1 | 0 | 100/166 | Putative acetyltransferase in Tn21 (4) |
| S042 | orf6 | 40245–40532 | 288 | 95 | 60.8 | 0 | 100/95 | Hypothetical protein in pSCH884 integron In5 (4) |
| S043 | tnpA | 40698–41492 | 795 | 264 | 61 | 0 | 100/264 | Transposase from IS6100 (4) |
Significant homology is defined as an E value of ≤1e-05 and/or >20% identity over at least 60% of the length of the protein.
ORFs of at least 180 bp (≥60 amino acids). Named ORFs are based on those previously characterized or designated here based on a putative function.
Nucleotide position in the sequence deposited under accession no. AH261825.
Expect value. Values of 0.0 were as returned by BLAST.
% Identity as returned by BLAST search and range (number of amino acids and gaps) over which identity value was calculated.
TABLE 3.
The ORFs in SGI1 DNA whose deduced products showed no significant homologya to extant protein sequences
| ORFb | Locationc (start–stop) | Size in:
|
% G+C | |
|---|---|---|---|---|
| bp | aa | |||
| S004 | 3537–3247 | 291 | 96 | 41.2 |
| S007 | 7628–7017 | 612 | 203 | 41 |
| S008 | 7849–7628 | 222 | 73 | 41 |
| S009 | 8024–8314 | 291 | 96 | 40.2 |
| S010 | 8588–8334 | 255 | 84 | 37.3 |
| S013 | 13664–14521 | 858 | 285 | 44.3 |
| S014 | 14518–14913 | 396 | 131 | 46 |
| S015 | 15597–15328 | 270 | 89 | 43.3 |
| S016 | 15860–15675 | 186 | 61 | 45.7 |
| S017 | 15720–15974 | 255 | 84 | 47.8 |
| S018 | 16188–15871 | 318 | 105 | 46.9 |
| S019 | 16743–16447 | 297 | 98 | 47.1 |
| S021 | 17805–18140 | 336 | 111 | 55.7 |
| S022 | 18327–18052 | 276 | 91 | 57.3 |
| S044 | 41840–42469 | 630 | 209 | 47.3 |
No significant homology was defined as an E value of >1e-2 for the top- scoring hit in a BLAST search.
ORFs of at least 180 bp long (≥60 amino acids).
Nucleotide position in the sequence deposited under accession no. AF261825.
FIG. 2.
Linear representation of the complete SGI1 and flanking regions. Upper rectangles indicate ORFs transcribed from right to left, and lower rectangles are transcribed left to right. GenBank entries of ORFs were assigned unique identifiers in the form SXXX. Color coding indicates ORFs with similar function as follows: green, DNA recombination; black, DNA replication; yellow, conjugal transfer; blue, regulatory; red, drug resistance; white, not known or other functions.
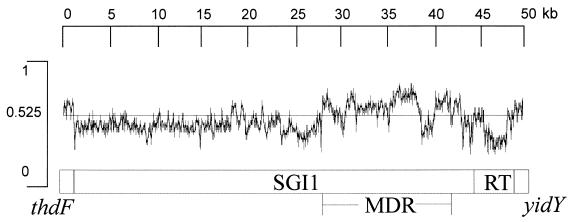
Overall the G+C content for SGI1 is 49.17%, compared with 51 to 53% for the S. enterica Typhimurium genome (Fig. 3) (27). Within SGI1, regions of different G+C content can be identified, notably the MDR region (S028 to S042), which is 58.7% G+C, although within this region the pse-1 gene is 41% G+C. The region encompassing S001 to S027 is 44% G+C, although within this region the S020 to S022 region is 53.2% G+C. Thus even outside of the MDR region a mosaic structure is indicated.
FIG. 3.
G+C content of the SGI1. MDR and RT represent the multidrug resistance and retron sequences, respectively.
The multidrug resistance region.
The MDR region of Typhimurium DT104 96–5227 (S028 to S042) has previously been characterized by PCR mapping and partial sequencing (4, 25) and has been found to be similar in content and gene order to that in other Typhimurium DT104 isolates displaying a pentaresistance phenotype (3, 5, 30, 34). Essentially, the floR gene (S032) and the tetR-tet(G) genes (S033 to S034) are bracketed by two class I integrons, one containing an aadA2 cassette (S028 to S031) and the other a pse-1 cassette (S037 to S042). A resolvase gene, tnpR, is located upstream of the aadA2 integron. Two additional genes, orf1 (S035), encoding a putative lysR-type transcriptional regulator, and orf2 (S036), carrying a transposase-like gene, are also found in this region. We have, however, detected a number of nucleotide differences between the 96–5227 sequence in this region and the sequences from other strains. In bovine Typhimurium DT104 strain BN9181 isolated in Europe, the published sequence of the tnpR gene (accession no. AF121001) contains an extra T in the position equivalent to that beside the T at position 26051 of the 96–5227 sequence (for base pair coordinates refer to accession no. AF261825) and which introduces a stop codon at this point in the BN9181 sequence (3). Thus, the predicted BN9181 TnpR protein is 172 amino acids, while that of 96–5227 is 194 amino acids. This extra T is absent in the human clinical Typhimurium DT104 strain H3380 (accession no. AF071555) isolated in the USA (5), which, however, has a G instead of a C at the position equivalent to position 26054 of the 95–5227 sequence. This results in an amino acid difference at residue 171 of the TnpR protein, with 96–5227 having an arginine here (as does BN9181) and H3380 a proline. In the aadA2 gene region, strain 96–5227 has a G at position 28211, while a C is found in strain H3380, resulting in a difference at residue 72 with a glutamic acid in 96-5227 substituted for a lysine in H3380. Upstream of the floR gene, the published sequence from BN9181 (accession no. AF118107) contains an extra A at a position equivalent to one between 30350 and 30354 of the 96–5227 sequence. The extra base, absent in the H3380 sequence, is in a noncoding region and would appear not to have any functional significance. Within the floR gene itself we have found two nucleotide differences between the 96–5227 and H3380 sequences. Position 30679 is a G in 96–5227 and an A in H3380 but results in no change in residue 66 of the FloR protein, which is a glutamine in both strains. Position 31299 is a C in the 96–5227 sequence and a T at the equivalent position in the H3380 sequence, which results in residue 273 being alanine in the 96–5227 FloR protein and valine in the H3380 protein. The BN9181 FloR protein sequence is identical to that of the 96–5227 protein sequence. Finally, strain 96–5227 has an A at position 34842, which is absent in H3380 and results in a change in reading frames here between the orf2 genes such that the predicted 96–5227 Orf2 transposase-like protein consists of 494 residues while the predicted H3380 protein consists of 531 residues.
The region between the pse-1 integron and DR-R has been previously characterized (accession no. AF261825) and contains the only insertion element found in SGI1, namely IS6100, and an ORF, S044 (previously designated SgiI1), encoding a hypothetical protein (4).
The remainder of SGI1.
A number of ORFs have been identified which show similarity to plasmid genes involved in mating pair formation and DNA transfer. Three ORFs, S005, S0011, and S012, have putative products that showed similarity to the mating pair stabilization protein TrhN and the pilus assembly proteins R0128 and TrhH, of the IncH plasmid R27 found in S. enterica serovar Typhi and other Enterobacteriaceae, respectively (32). The S023 product shows similarity to bacterial DNA helicases, and the S026 product shows similarity to an ATPase from the Rhizobium symbiosis plasmid pNGR234a (14). Interestingly, the Tra2 region of pR27 contains a helicase (TrhI) and an ATPase (TrhC), the latter of which is involved in pilus synthesis and assembly. The C-terminal region of the S025 product, which exhibits similarity to the N-terminal region of the Y4BN hypothetical protein of pNGR234a, contains a peptidase (S8) conserved domain from the subtilase family of proteinases. The S003 product exhibits similarity to the DNA replication protein RepA from the Rhodopseudomonas plasmid pMG101 (19). The S024 product is similar to the phage P2 OLD protein, which may be an exonuclease involved in overcoming defects in lysogenization (19). One other ORF product, that of S020, showed similarity to a plasmid protein of unknown function, Orf3, from the Francisella plasmid pFNL10 (AF121418). At least one regulator protein has been identified in this region, the product of S006, which shows similarity to a transcriptional activator regulating flagellum biosynthesis in Bordetella bronchiseptica (1).
The region between DR-L and S003 has been previously characterized (accession no. AF261825) (4) and found to contain two ORFs, S001 and S002, encoding a putative integrase (int) and excisionase (xis), respectively.
We could find no significant homology to proteins in GenBank for the putative products of 15 other ORFs in SGI1 (Table 3). Attempts to identify putative origins of replication (ori) were not successful.
Distribution of SGI1 amongst serovars Typhimurium DT104 and DT120 and Agona with various resistance profiles.
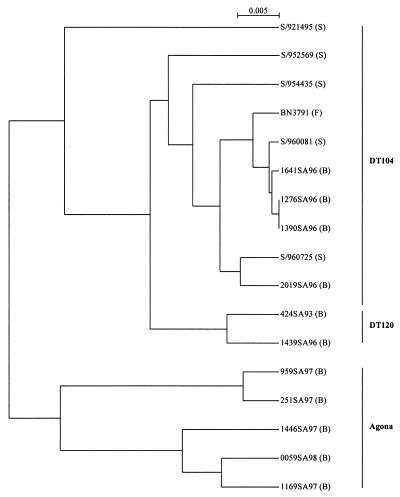
Genetically diverse strains, as determined by IRS-PCR (Fig. 4), of pentaresistant Typhimurium DT104, DT120, and Agona, as well as Typhimurium DT104 with different MDR profiles, were analyzed by PCR to determine the presence of SGI1 (Table 1). PCR to detect the left junction (thdF-S001) was carried out with the primer pair U7-L12 and LJ-R1, and PCR to detect the right junction (S044-int2 or S044-yidY) was carried out with the primer pair 104-RJ and C9-L2 or 104-RJ and 104-D (Table 4). All drug-resistant strains produced a product of the expected size for the left junction (thdF-S001), while the serovar Typhimurium strains were positive for the right junction (S044-int2) and the Agona strains were positive for the S044-yidY product. Drug-sensitive strains were negative for the above PCRs. Thus, in Agona strains SGI1 appears to be inserted at the 3′ end of the thdF gene as in serovar Typhimurium strains, but they do not contain the cryptic retronphage in the thdF-yidY intergenic region (4). Southern hybridization analysis of XbaI digests of the above strains was carried out using the 2-kb EcoRI fragment from p1–9 and the qacEdeltaI/sulI amplicon produced using primers QS-1 and QS-2 (Fig. 2). The 2-kb EcoRI fragment probe hybridized with the 9-kb and 4-kb XbaI fragments in the resistant strains as expected, while sensitive strains did not show any hybridization signals (Table 1). The qacEdeltaI/sulI probe hybridized with the 11.7-kb fragment containing most of the MDR region in all the pentaresistant serovar Typhimurium and Agona strains, but only in the serovar Typhimurium strains and Agona 959SA97 did the expected 4.3-kb fragment at the right end of SGI1 hybridize (Table 1). In three other Agona strains besides the 11.7-kb fragment, a fragment of about 8.4 kb hybridized, suggesting that an additional 4 kb of DNA was present in this region (Table 1). This larger-than-expected product was not due to deletion of the XbaI site in S044 as determined by sequence analysis (data not shown). In the serovar Typhimurium strains containing the single integron S/960725 (ASu), a 4.3-kb fragment hybridized, and in S/954435 (SSu) a 7-kb fragment hybridized with the qacEdelta1/sulI probe. Analysis of these strains is being undertaken to determine the nature of the variant MDR regions. Thus, from the results above, it appears that all drug-resistant strains in this study harbor SGI1. However, some variability exists in the MDR region, suggesting a high level of recombination can occur in this particular region of SGI1.
FIG. 4.
Clustering of Salmonella isolates by analysis of IRS-PCR patterns. The dendrogram was constructed by UPGMA on a matrix based on Jaccard's coefficient. Strains were isolated in Belgium (B), France (F), or Scotland (S).
TABLE 4.
Primers used in this study
| Name | Sequence (5′ to 3′) |
|---|---|
| U7-L12 | ACACCTTGAGCAGGGCAAAG |
| LJ-R1 | AGTTCTAAAGGTTCGTAGTCG |
| 104-RJ | TGACGAGCTGAAGCGAATTG |
| C9-L2 | AGCAAGTGTGCGTAATTTGG |
| 104-D | ACCAGGGCAAAACTACACAG |
| QS-1 | ATGAAAGGCTGGCTTTTTCTTG |
| QS-2 | TGAGTGCATAACCACCAGCC |
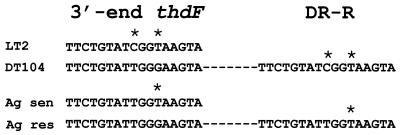
The SGI1 is flanked by an imperfect 18-bp direct repeat which appeared to be a duplication of the last 18 bp of the thdF in strain 96-5227 (Fig. 2) (4). This type of direct repeat is similar to those found in pathogenicity islands (17). Sequence analysis of the left and right junctions of Agona 1146SA97 also revealed a similar structure. However, comparison of the direct repeats from these two strains with thdF sequences from respective sensitive strains has revealed some interesting findings. The DR-R sequence is identical to the sequence from the respective thdF sequences from sensitive serovar Typhimurium or Agona strains, suggesting the origin of the DR-R is actually the end of thdF (Fig. 5). These sequences are slightly divergent between serovar Typhimurium and Agona, with a T located at position 9 of the direct repeat in serovar Typhimurium as opposed to a C at this position in Agona. The DR-L sequence is identical in both serotypes, suggesting the origin of this sequence may be from the donor DNA and not the result of a duplication event. Taken together, these results suggest that the SGI1 insertions were separate events and not a result of genetic exchange between the two serotypes.
FIG. 5.
Alignment of the direct repeats (DR) flanking the SGI1 in serovars Typhimurium and Agona. Asterisks represent nucleotide substitutions (see the text). sen, sensitive; res, resistant.
The possible independent emergence of MDR Typhimurium and Agona serotypes suggests this multidrug phenotype may emerge in other strains of Salmonella. If this region also carries genes responsible for increased virulence or transmission possibly observed with MDR Typhimurium DT104 (see below), the spread of other phagetypes or serotypes acquiring this unique region may be observed in the future. The ACSSuT phenotype has been identified on a transferable 140-kb plasmid in non-phage-typeable strains of serovar Typhimurium (38). The plasmid was shown to contain integrons other than the ones described here, suggesting SGI1 did not originate from this plasmid. An S. enterica serovar Typhi strain has been described harboring the ACSSuT phenotype on a 98- to 100-MDa transferable plasmid (20). It will be interesting to determine the genes responsible for this phenotype and if any homology exists between this plasmid and SGI1.
Infections associated with MDR Typhimurium DT104 have been associated with higher rates of admission to hospitals and mortality than other salmonellas (40). In addition, a study involving a small number of infections with MDR Typhimurium DT104 demonstrated a higher number of blood infections compared to those with sensitive strains (23). We could not identify any ORFs whose products might be directly related to pathogenesis in the SGI1 and possibly explain the increase in virulence demonstrated by the above studies. It is interesting to note the similarities of SGI1 to pathogenicity islands. Both harbor large segments of DNA flanked by small direct repeats which have different G+C contents compared to the chromosomal DNA, and both harbor cryptic and functional genes encoding mobility factors (17 and this study). Although no virulence factors were identified in SGI1, this study has revealed 15 potential ORFs with no homology to any known gene (Table 3), which may function as potential virulence factors. Both in vitro and in vivo studies are under way to try to elucidate the role, if any, of the SGI1 in virulence.
REFERENCES
- 1.Akerley B J, Cotter P A, Miller J F. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell. 1995;80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arcangioli M A, Leroy-Setrin S, Martel J L, Chaslus-Dancla E. A new chloramphenicol and florrfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol Lett. 1999;174:327–332. doi: 10.1111/j.1574-6968.1999.tb13586.x. [DOI] [PubMed] [Google Scholar]
- 4.Boyd D A, Peters G A, Ng L-K, Mulvey M R. Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhimurium DT104. FEMS Microbiol Lett. 2000;189:285–291. doi: 10.1111/j.1574-6968.2000.tb09245.x. [DOI] [PubMed] [Google Scholar]
- 5.Briggs C E, Fratamico P M. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson S A, Browning M, Ferris E, Jones B D. Identification of diminished tissue culture invasiveness among multiple antibiotic resistant Salmonella typhimurium DT104. Microb Pathog. 2000;28:37–44. doi: 10.1006/mpat.1999.0322. [DOI] [PubMed] [Google Scholar]
- 7.Carlson S A, Willson R M, Crane A J, Ferris K E. Evaluation of invasion-conferring genotypes and antibiotic-induced hyperinvasive phenotypes in multiple antibiotic resistant Salmonella typhimurium DT104. Microb Pathog. 2000;28:373–378. doi: 10.1006/mpat.2000.0355. [DOI] [PubMed] [Google Scholar]
- 8.Cloeckaert A, Boumedine K S, Flaujac G, Imberechts H, D'Hooghe I, Chaslus-Dancla E. Occurrence of a Salmonella enterica serovar Typhimurium DT104-like antibiotic resistance gene cluster including the floRR gene in S. enterica serovar Agona. Antimicrob Agents Chemother. 2000;44:1359–1361. doi: 10.1128/aac.44.5.1359-1361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cody S H, Abbott S L, Marfin A A, Schulz B, Wagner P, Robbins K, Mohle-Boetani J C, Vugia D J. Two outbreaks of multidrug-resistant Salmonella serotype Typhimurium DT104 infections linked to raw-milk cheese in Northern California. JAMA. 1999;281:1805–1810. doi: 10.1001/jama.281.19.1805. [DOI] [PubMed] [Google Scholar]
- 10.Daly M, Buckley J, Power E, O'Hare C, Cormican M, Cryan B, Wall P G, Fanning S. Molecular characterization of Irish Salmonella enterica serotype Typhimurium: detection of class I integrons and assessment of genetic relationships by DNA amplification fingerprinting. Appl Environ Microbiol. 2000;66:614–619. doi: 10.1128/aem.66.2.614-619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daly M, Fanning S. Characterization and chromosomal mapping of antimicrobial resistance genes in Salmonella enterica serotype Typhimurium. Appl Environ Microbiol. 2000;66:4842–4848. doi: 10.1128/aem.66.11.4842-4848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies A, O'Neill P, Towers L, Cooke M. An outbreak of Salmonella typhimurium DT104 food poisoning associated with eating beef. Comm Dis Rep. 1996;6:R159–R162. [PubMed] [Google Scholar]
- 13.Evans S, Davies R. Case control study of multiple-resistant Salmonella typhimurium DT104 infection of cattle in Great Britain. Vet Rec. 1996;139:557–558. [PubMed] [Google Scholar]
- 14.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 15.Glynn M K, Bopp C, Dewitt W, Dabney P, Mokhtar M, Angulo F J. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–1338. doi: 10.1056/NEJM199805073381901. [DOI] [PubMed] [Google Scholar]
- 16.Grein T, O'Flanagan D, McCarthy T, Bauer D. An outbreak of multidrug-resistant Salmonella typhimurium food poisoning at a wedding reception. Ir Med J. 1999;92:238–241. [PubMed] [Google Scholar]
- 17.Hacker J, Kaper J B. The concept of pathogenicity islands. In: Kaper J B, Hacker J, editors. Pathogenicity islands and other mobile virulence elements. Washington, D.C.: American Society for Microbiology; 1999. pp. 1–11. [Google Scholar]
- 18.Hosek G, Leschinsky D D, Irons S, Safranek T J. Multidrug-resistant Salmonella serotype Typhimurium—United States, 1996. Morb Mortal Wkly Rep. 1997;46:308–310. [PubMed] [Google Scholar]
- 19.Inui M, Roh J H, Zahn K, Yukawa H. Sequence analysis of the cryptic plasmid pMG101 from Rhodopseudomonas palustris and construction of stable cloning vectors. Appl Environ Microbiol. 2000;66:54–63. doi: 10.1128/aem.66.1.54-63.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kariuki S, Gilks C, Revathi G, Hart A. Genotypic analysis of multidrug-resistant Salmonella enterica serovar Typhi, Kenya. Emerg Infect Dis. 2000;6:649–651. doi: 10.3201/eid0606.000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markogiannakis A, Tassios P T, Lambiri M, Ward L R, Kourea-Kremastinou J, Legakis N J, Vatopoulos A C The Greek Nontyphoidal Salmonella Study Group. Multiple clones within multidrug-resistant Salmonella enterica serotype Typhimurium phage type DT104. J Clin Microbiol. 2000;38:1269–1271. doi: 10.1128/jcm.38.3.1269-1271.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazurek G H, Reddy V, Marston B J, Haas W H, Crawford J T. DNA fingerprinting by infrequent-restriction-site amplification. J Clin Microbiol. 1996;34:2386–2390. doi: 10.1128/jcm.34.10.2386-2390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller M A. Widespread emergence in the United States of multiple drug-resistant type of Salmonella typhimurium. FDA Vet. 1997;12:5–6. [Google Scholar]
- 24.Møbak K, Baggesen D L, Aarestrup F M, Ebbesen J M, Engberg J, Frydendahl K, Gerner-Smidt P, Petersen A M, Wegener H C. An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype Typhimurium DT104. N Engl J Med. 1999;341:1420–1425. doi: 10.1056/NEJM199911043411902. [DOI] [PubMed] [Google Scholar]
- 25.Ng L-K, Mulvey M R, Martin I, Peters G A, Johnson W. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob Agents Chemother. 1999;43:3018–3021. doi: 10.1128/aac.43.12.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng L-K, Khakhria R, Woodward D, Johnson W. National laboratory surveillance of enteric pathogens. Can J Infect Dis. 1997;8:133–136. [Google Scholar]
- 27.Ochman H, Lawrence J G. Phylogenetics and the amelioration of the bacterial genome. In: Neidhardt F C, Curtiss III R, Ingraham J, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2627–2637. [Google Scholar]
- 28.Ridley A, Threlfall E J. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella typhimurium DT104. Microb Drug Resist. 1998;4:113–118. doi: 10.1089/mdr.1998.4.113. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Sandvang D, Aarestrup F M, Jensen L B. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol Lett. 1998;160:37–41. doi: 10.1111/j.1574-6968.1998.tb12887.x. [DOI] [PubMed] [Google Scholar]
- 31.Schmieger H, Schicklmaier P. Transduction of multiple drug resistance of Salmonella enterica serovar Typhimurium DT104. FEMS Microbiol Lett. 1999;170:251–256. doi: 10.1111/j.1574-6968.1999.tb13381.x. [DOI] [PubMed] [Google Scholar]
- 32.Sherburne C K, Lawley T D, Gilmour M W, Blattner F R, Burland V, Grotbeck E, Rose D J, Taylor D E. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 2000;28:2177–2186. doi: 10.1093/nar/28.10.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokal R R, Sneath P H A, editors. Principles of numerical taxonomy. San Francisco, Calif: Freeman; 1963. The construction of a taxonomic system; pp. 169–210. [Google Scholar]
- 34.Threlfall E J, Ward L R, Rowe B. Multiresistant Salmonella typhimurium DT104 and salmonella bacteremia. Lancet. 1998;352:287–288. doi: 10.1016/s0140-6736(05)60261-9. [DOI] [PubMed] [Google Scholar]
- 35.Threlfall E J, Ward L R, Skinner J A, Rowe B. Increase in multiple antibiotic resistance in nontyphoidal salmonellas from humans in England and Wales: a comparison of data for 1994 and 1996. Microb Drug Resist. 1997;3:263–266. doi: 10.1089/mdr.1997.3.263. [DOI] [PubMed] [Google Scholar]
- 36.Threlfall E J, Frost J A, Ward L R, Rowe B. Increasing spectrum of resistance in multiresistant Salmonella typhimurium. Lancet. 1996;347:1053–1054. doi: 10.1016/s0140-6736(96)90199-3. [DOI] [PubMed] [Google Scholar]
- 37.Threlfall E J, Frost J A, Ward L R, Rowe B. Epidemic in cattle and humans of Salmonella typhimurium DT104 with chromosomally integrated multiple drug resistance. Vet Rec. 1994;137:577. doi: 10.1136/vr.134.22.577. [DOI] [PubMed] [Google Scholar]
- 38.Tosini F, Visca P, Luzzi I, Dionisi A M, Pezzella C, Petrucca A, Carattoli A. Class 1 integron-borne multiple-antibiotic resistance carried by IncFI and IncL/M plasmids in Salmonella enterica serotype Typhimurium. Antimicrob Agents Chemother. 1998;42:3053–3058. doi: 10.1128/aac.42.12.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villar R G, Macek M D, Simons S, Hayes P S, Goldoft M J, Lewis J H, Rowan L L, Hursh D, Patnode M, Mead P S. Investigation of multidrug-resistant Salmonella serotype Typhimurium DT104 infections linked to raw-milk cheese in Washington State. JAMA. 1999;281:1811–1816. doi: 10.1001/jama.281.19.1811. [DOI] [PubMed] [Google Scholar]
- 40.Wall P G, Morgan D, Lamden K, Ryan M, Griffin M, Threlfall E J, Ward L R, Rowe B. A case control study of infection with an epidemic strain of multi-resistant Salmonella typhimurium DT104 in England and Wales. Comm Dis Rep CDR Rev. 1994;4:R130–R135. [PubMed] [Google Scholar]