Abstract
Alcoholic liver disease (ALD) is caused by alcohol abuse and can progress to hepatitis, cirrhosis, and even hepatocellular carcinoma. Previous studies suggested that Lactobacillus reuteri (L. reuteri) ameliorates ALD, but the exact mechanisms are not fully known. This study created an ALD model in mice, and the results showed L. reuteri significantly alleviating lipid accumulation in the mice. Transcriptome sequencing showed the L. reuteri treatment group had the most enriched metabolic pathway genes. We then studied the farnesoid X receptor (FXR) metabolic pathway in the mice liver tissue. Western blot analysis showed that FXR and carbohydrate response element binding protein (ChREBP) were upregulated and sterol regulatory element binding transcription factor 1 (Srebf1) and Cluster of differentiation (CD36) were downregulated in the L. reuteri-treated group. Subsequently, we administered FXR inhibitor glycine-β-muricholic acid (Gly-β-MCA) to mice, and the results show that Gly-β-MCA could reduce the therapeutic effect of L. ruteri. In conclusion, our study shows L. reuteri improved liver lipid accumulation in mice via the FXR signaling regulatory axis and may be a viable treatment option for ALD.
Keywords: Lactobacillus reuteri, alcohol, liver injury, lipid metabolism, transcriptome
Introduction
Alcoholic liver disease (ALD) reportedly affects 25% of the population, making ALD is the second most common cause of liver disease after viral hepatitis.1 Alcohol abuse is one of the main causes of liver disease and is a serious social problem worldwide; the incidence of ALD has increased in China with the increased frequency of drinking.2 ALD is a progressive disease that starts with simple steatosis and can progress to alcoholic steatohepatitis, fibrosis, cirrhosis, and even hepatocellular carcinoma.3 ALD treatment and management are crucial because its progression can be effectively prevented with appropriate treatment for steatosis.4
The main treatment and management of ALD is abstinence from alcohol, but at present, it is difficult to completely implement the method of abstinence. Supplementation with β-carotene and vitamins A, C, and E also did not show much benefit, while some drug therapies were limited by the hepatotoxicity of the drugs.5 In recent decades, dietary probiotics or prebiotic supplements have become a major strategy for the treatment of ALD.6,7 In general, supplementation with dietary probiotics normalizes the disturbed gut microbiome, competitively excluding enteric pathogens and bacteriotin production.8 Numerous studies have shown that probiotics are involved in developing ALD, and this process is complex. In 2008, Kirpich et al. first tested probiotics in humans, and patients who received probiotics had significantly lower AST and ALT activities at the end of treatment than those who received standard therapy alone.9 In addition, Lactobacillus rhamnosus has been reported to play a role in preventing hepatic steatosis and injury caused by chronic alcohol exposure by regulating hepatic AMPK activation and Bax/Bcl-2 mediated apoptosis,10 and oral A. muciniphila supplementation reduces liver injury, steatosis, and neutrophil infiltration.11 Although more probiotic strains and related products are identified as potential treatments for ALD, the exact role of probiotics in regulating intestinal flora, intestinal barrier function, the gut–brain axis, and the pathogenesis of ALD needs further study.12,13L. reuteri is a probiotic with beneficial effects for preventing and improving a wide range of diseases. The benefits of colonization of the intestine include colonization of the intestine include reduction of infections, enhanced absorption of nutrients, and maintenance of mucosal integrity.14 Our preliminary animal data showed the therapeutic effect of L. reuteri in the treatment of ALD, although the specific mechanism needs further study. FXR is highly expressed in the liver under physiological conditions and is a key regulator in fatty liver development. Existing studies have focused on the role of FXR in nonalcoholic fatty liver disease (NAFLD), revealing that FXR may moderately slow the progression of NAFLD by inhibiting steatosis.15
With the development of transcriptome sequencing technology in recent years, transcriptome sequencing has been applied to many medical fields, such as clinical diagnosis, marker screening, prognosis evaluation, and pathogenesis.16 This study assessed the effects of L. reuteri on alcohol-induced liver disease in mice using a transcriptomic analysis approach. First, we established an animal model of ALD and performed transcriptome analysis of mouse liver tissues to identify differently expressed genes. Then a molecular biology approach validated the relevant signaling pathways further. Our experimental data indicate the FXR signaling pathway also plays an important role in ALD.
Experimental Section
Materials
L. reuteri DSM 17938 provided by BioGaia AB (Stockholm, Sweden) was used in the study. A bicinchoninic acid assay (BCA) protein assay kit, radio-immunoprecipitation assay lysate, and phosphate buffer saline were purchased from Beyotime (Shanghai, China). Gly-β-MCA, a bile acid, is a potent, sable, intestine-selective, and oral bioactive FXR inhibitor that may be a candidate for the treatment of metabolic disorders (MedChemExpress, MCE, China).
Experimental Animals
Male C57BL/6 mice were purchased from Chongqing Tengxinbir Experimental Animal Sales Ltd. (Chongqing, China). A murine ALD model was established and treated with L. reuteri according to the method described in a previous study (Gao-binge model).17 Briefly, all mice were housed in groups with a controlled relative humidity (45–65%) and temperature (22 ± 2 °C) environment for a 12-h light/dark cycle. All mice were randomly divided into 5 groups (n = 8, age of all mice was 8 weeks, and initial body weight was 21.31 ± 1.39 g): (A) control group (given Lieber-DeCarli liquid control diet for 5 days, followed by Lieber-DeCarli liquid control diet for 10 days or 18 days); (B) alcohol diet group (given Lieber-DeCarli liquid control diet for 5 days to adapt to the liquid diet, followed by Lieber-DeCarli alcohol diet containing 5% alcohol for 10 days or 18 days); (C) L. reuteri intervention group (given Lieber-DeCarli liquid control diet for 5 days to adapt to the liquid diet, followed by Lieber-DeCarli alcohol diet containing 5% alcohol for 10 days or 18 days. L reuteri 2 × 107 CFUs once daily by gavage); (D) L. reuteri + Gly-β-MCA intervention group (given Lieber-DeCarli liquid control diet for 5 days to adapt to the liquid diet, followed by Lieber-DeCarli alcohol diet containing 5% alcohol for 18 days. L reuteri 2 × 107 CFUs once daily and 10 mg/kg Gly-β-MCA once every 2 days by gavage); (E) Gly-β-MCA intervention group (given Lieber-DeCarli liquid control diet for 5 days to adapt to the liquid diet, followed by Lieber-DeCarli alcohol diet containing 5% alcohol for 18 days. L reuteri 2 × 107 CFUs once daily and 10 mg/kg Gly-β-MCA once every 2 days by gavage).
Gavage Experiment and Sample Collection in Mice
In the L. reuteri intervention group, 2 × 107 CFUs of L. reuteri were gavaged daily at the same time as the mice were started on a liquid diet, and equal amounts of L. reuteri solvent were given to the control and alcoholic diet groups. Each group of mice was euthanized in 2 batches. The first batch was euthanized on the 10th day of alcoholic feeding and the rest on the 18th day of feeding. The mice in the alcohol-fed and L. reuteri intervention groups were gavaged once with a 31.5% (v/v) ethanol solution (5 g/kg body weight) 9 h before specimen collection, while the control group was gavaged with isocaloric dextrin 9 h before euthanization.
Determination of Blood Biochemical Indexes and Pathological Changes
The levels of serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were detected using an automatic biochemical analyzer in our hospital (Erba XL-640, Germany).
The liver tissue was flash-frozen in liquid nitrogen and embedded in a frozen tissue embedding agent. It was sectioned by microtome, stained with oil red and hematoxylin, sealed with neutral gum, and examined microscopically for lipid accumulation in the liver tissues of the different treatment groups.
The formalin-fixed intestinal and liver tissues were dehydrated with different concentrations of alcohol gradient, xylene transparent, embedded in paraffin, sectioned by microtome, underwent hematoxylin-eosin (H&E) and immunohistochemical (IHC) staining, neutral gum sealing, and microscopic examination to observe the pathological changes in the intestinal and liver tissues of the different treatment groups.
Serum Tumor Necrosis Factor α (TNFα) Level
The serum TNFα level was detected by the corresponding enzyme linked immunosorbent assay (ELISA) kits. ELISA kits (TNFα) were purchased from Elabscience Biotechnology Co., Ltd.
Protein Expression Assay (Western Blotting)
Western blot analysis was performed to detect the expression of FXR, CD36, Srebf1, sirtuin 1 (Sirt1), interleukin 6 (IL6), TNFα, and ChREBP in liver and the expression of FXR in intestinal. Proteins were extracted from intestinal and liver samples of the different treatment groups using Radio Immunoprecipitation Assay buffer (Beyotime, Shanghai, China). Total proteins in samples were measured with BCA. Protein were subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride membranes (Chuangrong, Guangzhou, China). Primary antibodies: Rabbit anti-FXR (1:1000, Absin, Shanghai, China), Rabbit anti-ChREBP (1:1000, Absin, Shanghai, China), Rabbit anti-Srebf1 (1:1000, Absin, Shanghai, China), Rabbit anti-CD36 (1:1000, Absin, Shanghai, China), Rabbit anti-GAPDH (1:5000, Proteintech, Wuhan, China), Rabbit anti-Sirt1 (1:3000, Proteintech, Wuhan, China), Mouse anti-IL6 (1:3000, Proteintech, Wuhan, China), Mouse anti-TNFα (1:4000, Proteintech, Wuhan, China), Goat antirabbit (1:10000, Absin, Shanghai, China), Goat antirabbit (1:10000, Absin, Shanghai, China). The chemiluminescence signal was measured with a chemilumiescent and fluorescent imaging system (ChampChemi professional, Beijing, China).
Transcriptome Analysis
RNA Extraction
Complete RNA was extracted from the liver tissue samples of each group using TRIzol (Invitrogen), and genomic DNA was removed using the DNaseI (TaKara) method. A 2100 Bioanalyser (Agilent) and ND-2000 (Nano Drop Technologies) were used to test the quality of RNA samples and ensure that qualified samples (OD260/280 = 1.8–2.2, OD 260/230 ≥ 2.0, RIN ≥ 6.5, 28S:18S ≥ 1.0, >1 μg) were used for transcriptome sequencing.
Library Creation and Illumina HiSeq X Ten/NovaSeq6000 Sequencing
RNA libraries were created using the TruSeq RNA sample preparation kit (Illumina, San Diego, CA). Enrichment of mRNA with poly-A tails was performed from 1 μg of total RNA using Oligo(dT) beads, and then fragmentation buffer was added to break the mRNA into small fragments of 300 bp randomly. Next, the SuperScript double-stranded cDNA synthesis kit (Invitrogen, CA) was used, and six-base random primers (Illumina) were added to synthesize one-stranded cDNA in reverse with mRNA as the template, followed by two-stranded synthesis to form a stable double-stranded structure. Polymerase chain reaction (PCR) then enriches the double-stranded cDNA (sample preparation kit (Illumina, San Diego, CA)), and beads (DNA clean beads) were added to the end of the cDNA. After PCR enrichment (sample preparation kit, Illumina, San Diego, CA), the cDNA was screened for 200–300 bp bands with beads (DNA clean beads). After quantification by TBS380 (Picogreen), the library was sequenced using the Illumina HiSeq X Ten/NovaSeq6000 sequencing platform for high-throughput sequencing with a read length of PE150.
Screening and Comparison of Differential Genes
The raw data were subjected to splicing and quality control to obtain high-quality sequencing data for bioinformatics analysis, including target gene clustering analysis, target gene Venn analysis, gene set enrichment analysis (GSEA) analysis, and visualization analysis.18−23
Statistic Analysis
Statistical analysis was performed using SPSS 17.0. The OPLS-DA model Student t test was used to ensure reliable results. One-way ANOVA was used to compare the differences in liver/body weight ratio, serum biochemical marker levels, and TNFα levels between the groups. Data were expressed as mean ± SD, with P < 0.05 considered a statistically significant difference.
Results
L. reuteri Can Alleviate Alcohol-Induced Liver Injury
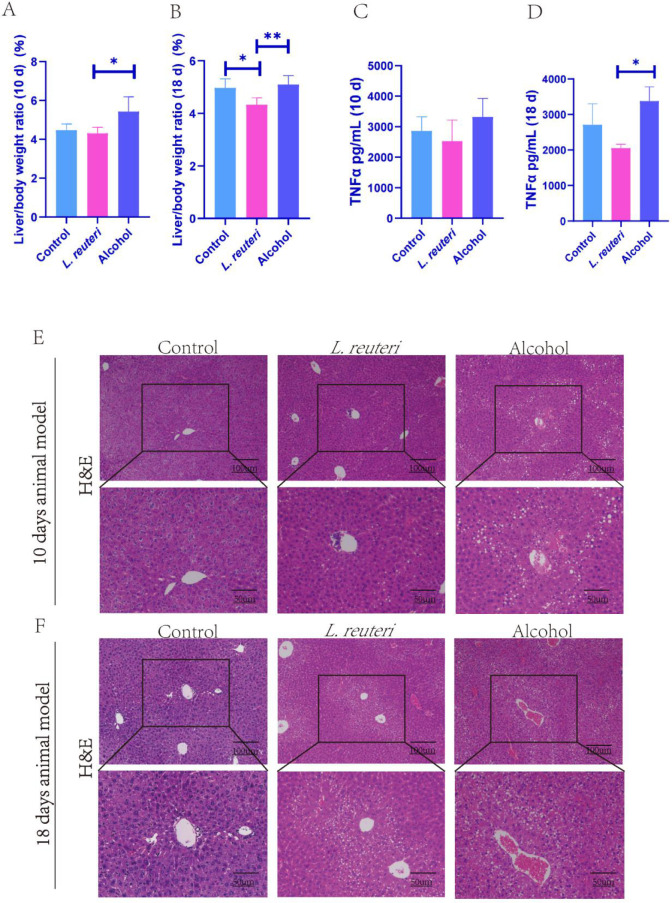
Probiotics are “living microorganisms that, when administered in sufficient amounts, provide health benefits to the host”.24 We used the NIAAA method to establish a model of ALD in C57BL/6 mice, and L. reuteri (DSM 17938) was administered to the mice by gavage. We found L. reuteri could slightly reduce the weight of the liver in the ALD model mice (Figure 1A, B). In addition, there was a decrease in the serum TNFα level after administration of L. reuteri (Figure 1C, D). In the H&E-stained sections of the liver, morphological changes showed reduced hepatic vacuole-like changes in the L. reuteri group (Figure 1E, F). In conclusion, L. reuteri improved alcohol-induced liver injury in mice. In addition, we found that L. reuteri treatment was more effective based on a time-gradient study with 18 days of alcohol induction (Figure 1B, D, F) compared with 10 days of alcohol induction (Figure 1A, C, E).
Figure 1.
L. reuteri has a protective effect from liver injury induced by alcohol in mice. (A, B) Liver weight to body weight ratio of mice in each group. (C, D) Serum TNFα levels of mice in each group (ELISA). (E, F) H&E staining of the liver tissue in all groups of mice (100 × and 200 × magnification). Ten d: 10 days animal model; 18 d: 18 days animal model. *P < 0.05, **P < 0.01.
L. reuteri Attenuates Liver Injury in ALD Mice
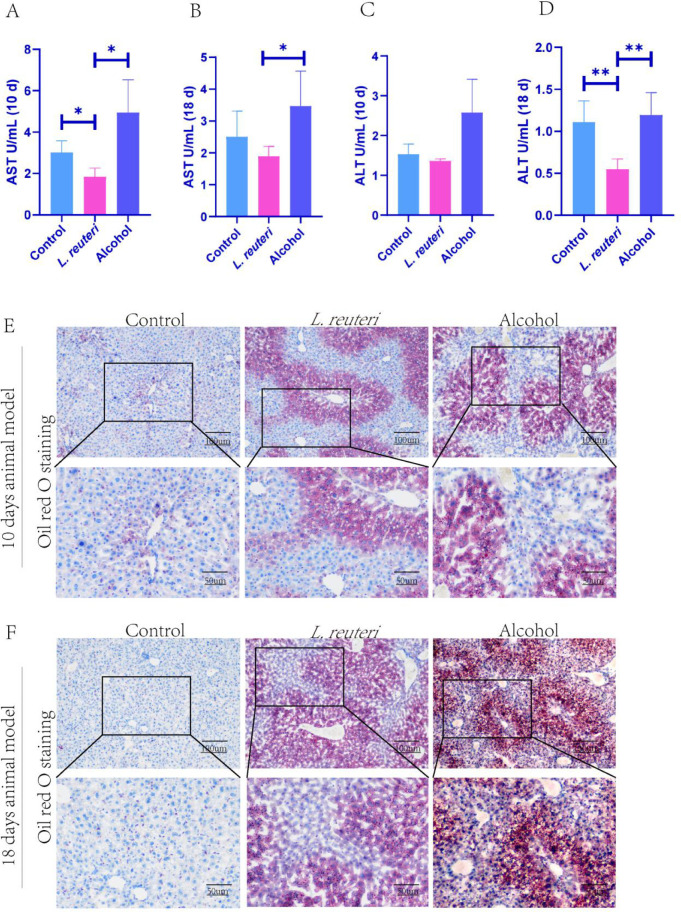
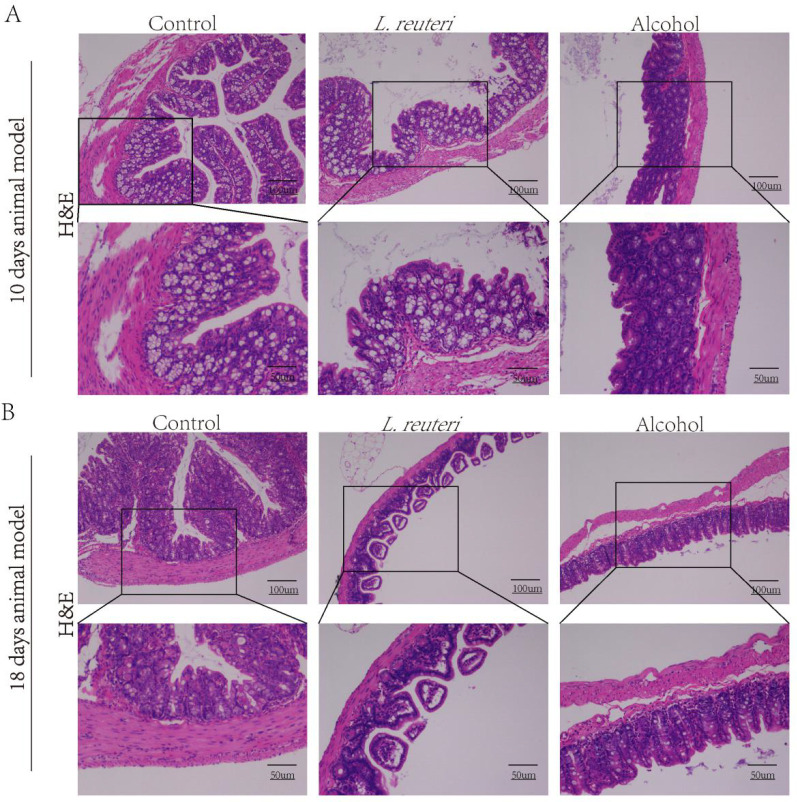
To further investigate the degree of the therapeutic effects of L. reuteri in the ALD mice livers, we measured serum AST levels (Figure 2A, B) and serum ALT levels (Figure 2C, D) to assess liver function, indicating an improvement in both and a more pronounced effect based on a time gradient. Oil red staining of the mouse liver showed less lipid accumulation in the L. reuteri-treated group than in the model group (Figure 2E, F). The mouse intestinal tissues showed no significant differences under H&E staining (Figure 3A, B). In conclusion, L. reuteri may reduce liver lipid accumulation in mice to improve liver injury.
Figure 2.
L. reuteri can alleviate liver injury and lipid accumulation in mice. (A, B) Serum aspartate aminotransferase (AST) levels of the mice in each group. (C, D) Serum alanine aminotransferase (ALT) levels of mice in each group. (E, F) Oil red O staining of liver tissue in all groups of mice (100 × and 200 × magnification).10 d: 10 days animal model; 18 d: 18 days animal model. *P < 0.05, **P < 0.01.
Figure 3.
L. reuteri did not significantly alter the histomorphological manifestation of the intestine in mice. (A, B) H&E staining of intestinal tissue in all groups of mice (100 × and 200 × magnification).
Transcriptomic Analysis of the Liver Tissue Showed Significant Differences in Gene Expression between the Probiotic and Model Groups
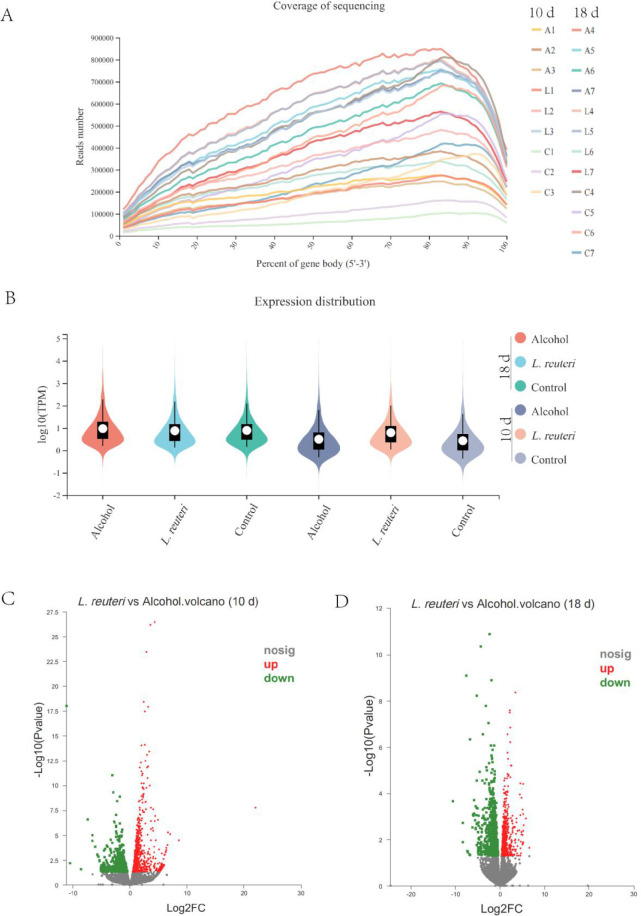
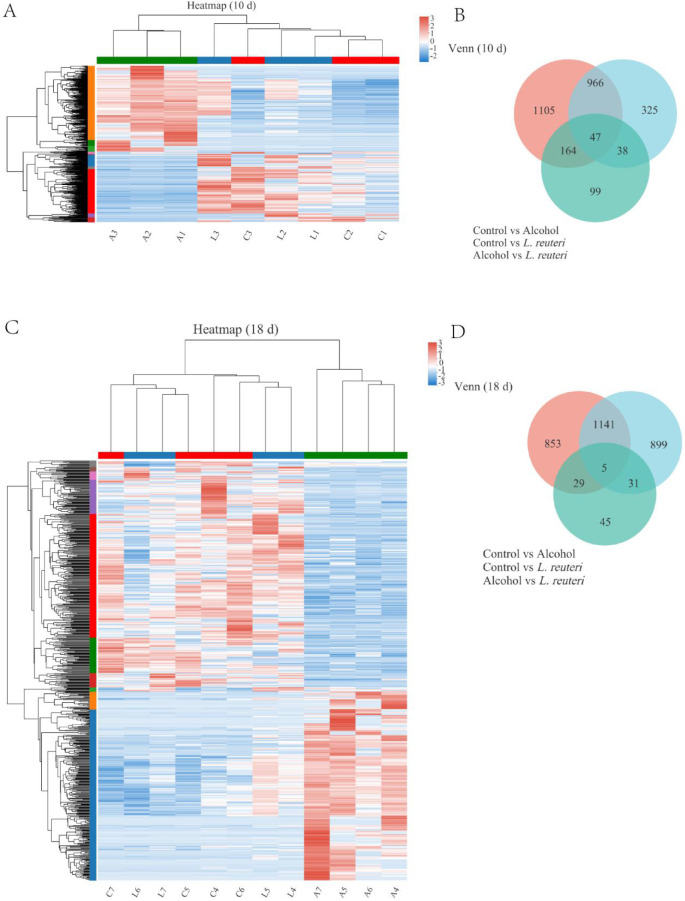
We performed transcriptomic sequencing analysis with randomly selected liver tissue samples from mice from each group. A total of 21 liver tissue samples (10 days: control group, n = 3; L. reuteri, n = 3; alcohol group, n = 3. 18 days: control group, n = 4; L. reuteri, n = 4; alcohol group, n = 4) were sequenced for preliminary quality assessment analysis and showed that the sequences were evenly distributed across genes with no significant biased fronts, indicating that the sequencing was unbiased (Figure 4A). In addition, the overall expression distribution in each group did not show significant differences (plotted according to the mean value of each subgroup) (Figure 4B). We further analyzed the differences in gene expression between the L. reuteri treatment and model groups. First, we counted all sequencing results for expression differences. The volcano plot demonstrated the differences between upregulated and downregulated gene expression at 10 days (Figure 4C) and 18 days (Figure 4D) of alcohol induction. We then created the different genes as target gene sets for further analysis, and the target gene Venn diagram shows the overall gene expression for each group (Figure 5B, D). The target gene clustering heat map analysis revealed significant differences in gene expression in the L. reuteri treatment group compared to the alcohol group. Gene expression was closer than the alcohol group, and differences in gene expression between the two groups were more pronounced as the duration of alcohol intake increased (Figure 5A, B for 10 days of alcohol induction; Figure 5C, D for 18 days of alcohol induction). ALD model mice treated with L. reuteri showed significant differences in gene expression.
Figure 4.
Differences in gene expression between the L. reuteri group and the alcohol group. (A) Sequence coverage on the 5′ to 3′ region of all genes in the liver tissue samples of each group of mice. Alcohol-induced for the 10 days control group (C1–3), L. reuteri group (L1–3), and alcohol group (A1–3). Alcohol-induced for the 18 days control group (C4–7), L. reuteri group (L4–7), and alcohol group (A4–7). (B) Distribution of expression in each group of mice. (C, D) Expression difference volcano map. Ten d: 10 days animal model; 18 d: 18 days animal model.
Figure 5.
L. reuteri reversed transcriptomic differences in the liver induced by alcohol. (A, C) Heat map of differential gene clustering analysis. Alcohol-induced for the 10 days control group (C1–3), L. reuteri group (L1–3), and alcohol group (A1–3). Alcohol-induced for the 18 days control group (C4–7), L. reuteri group (L4–7), and alcohol group (A4–7). (B, D) Venn analysis of differential genes. Ten d: 10 days animal model; 18 d: 18 days animal model.
Increased Expression of Lipid Metabolizing Genes in the Pathway Were Analyzed by Enrichment Analysis
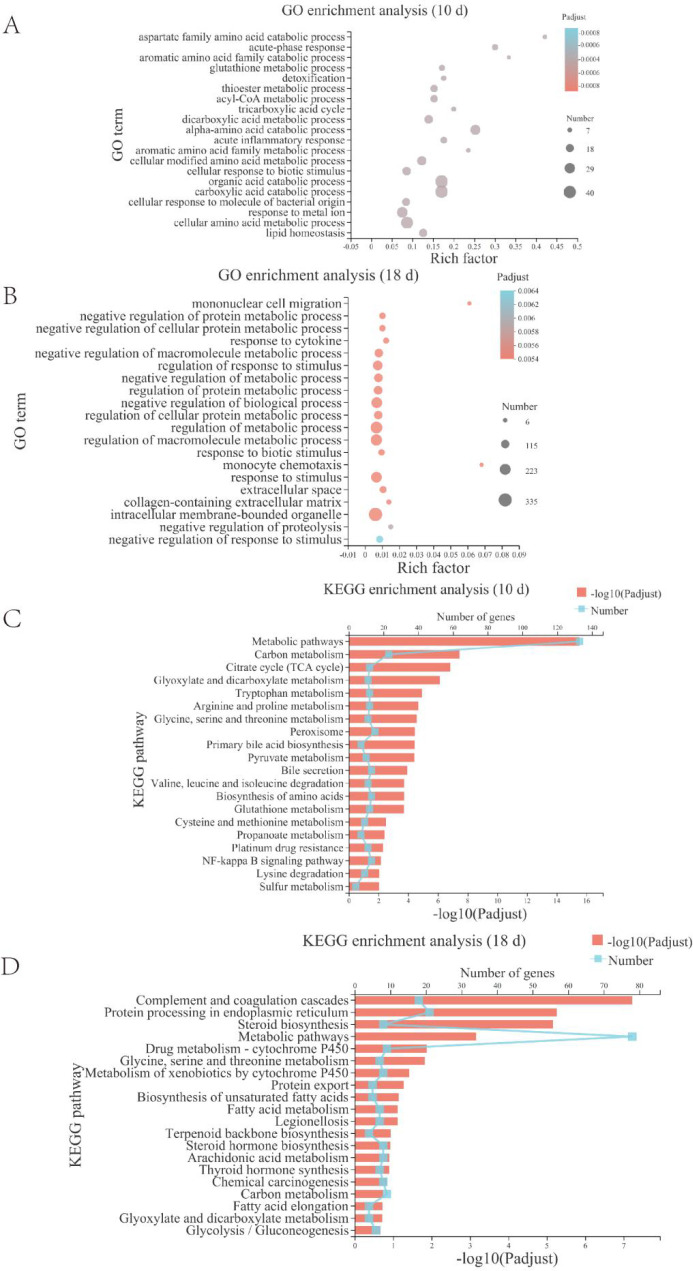
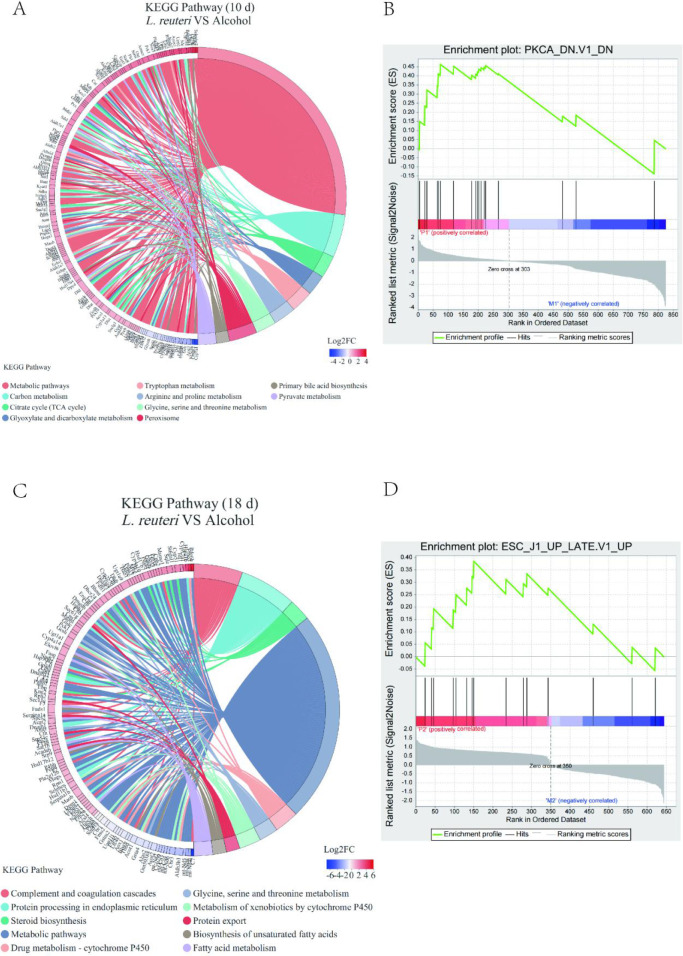
To verify that L. reuteri may improve liver injury by attenuating hepatic lipid accumulation in mice, we subjected the transcriptome sequencing results to a differential gene enrichment analysis using GO and KEGG enrichment analysis sequentially. The differential genes were enriched in various metabolic pathways such as lipid and cholesterol metabolism (Figure 6). The enrichment chord diagram showed that the genes with significant differences at 10 days of alcohol induction were primarily those in metabolic pathways, including the citric acid cycle, tryptophan and serine metabolism, and primary bile acid synthesis (Figure 7A). At 18 days of alcohol induction, the enrichment chord diagram showed that different genes were primarily enriched in the complement and coagulation cascades, steroid biosynthesis, and drug metabolism-cytochrome P450 processes (Figure 7 C). The top 10 enriched pathway different genes are shown in Table 1, and the biggest difference was in the metabolic pathways. We then proceeded to enrich the Kyoto Encyclopedia of Genes and Genomes (KEGG) different gene set using GSEA and compared it to the different gene set of L. reuteri-treated mice and alcohol group mice with significantly upregulated genes (Figure 7B, D).
Figure 6.
Functional enrichment analysis revealed the FXR signaling pathway may be involved in the mechanism by which L. reuteri alleviates ALD. (A, B) GO enrichment analysis bubble diagram. (C, D) KEGG enrichment analysis histogram. Ten d: 10 days animal model; 18 d: 18 days animal model.
Figure 7.
Enrichment chor diagram and GSEA analysis revealed that the FXR signaling pathway may be involved in how L. reuteri alleviates ALD. (A, C) KEGG enriched string diagram. (B, D) GSEA analysis. Ten d: 10 days animal model; 18 d: 18 days animal model.
Table 1. Top 10 Differential Gene Enrichment Pathways between the L. reuteri Group and the Alcohol Groupa.
| Pathway id | Number | Description | P value | P adjust |
|---|---|---|---|---|
| mmu01100 | 354 | Metabolic pathways | <0.0001 | <0.0001 |
| mmu04610 | 44 | Complement and coagulation cascades | <0.0001 | <0.0001 |
| mmu04141 | 53 | Protein processing in endoplasmic reticulum | <0.0001 | <0.0001 |
| Mmu00100 | 13 | Steroid biosynthesis | <0.0001 | <0.0001 |
| mmu05204 | 32 | Chemical carcinogenesis | <0.0001 | 0.0004 |
| rnmu00830 | 31 | Retinol metabolism | <0.0001 | 0.0004 |
| mmu00140 | 30 | Steroid hoimone biosynthesis | <0.0001 | 0.0005 |
| mmu03010 | 40 | Ribosome | <0.0001 | 0.0010 |
| mmu00260 | 18 | Glycine, serine and threonine metabolism | <0.0001 | 0.0012 |
| mmu01200 | 36 | Carbon metabolism | <0.0001 | 0.0012 |
Pathway id: pathway number; Number: number of genes enriched to the pathway; Description: specific description of the KEGG pathway; P value: uncorrected P value, P value represents whether the enriched result is statistically significant, the smaller the P value, the more statistically significant it is; P adjust: using the BH method, P value after correction
KEGG Enrichment Analysis Screens for Hepatic Lipid Metabolism Pathway Genes
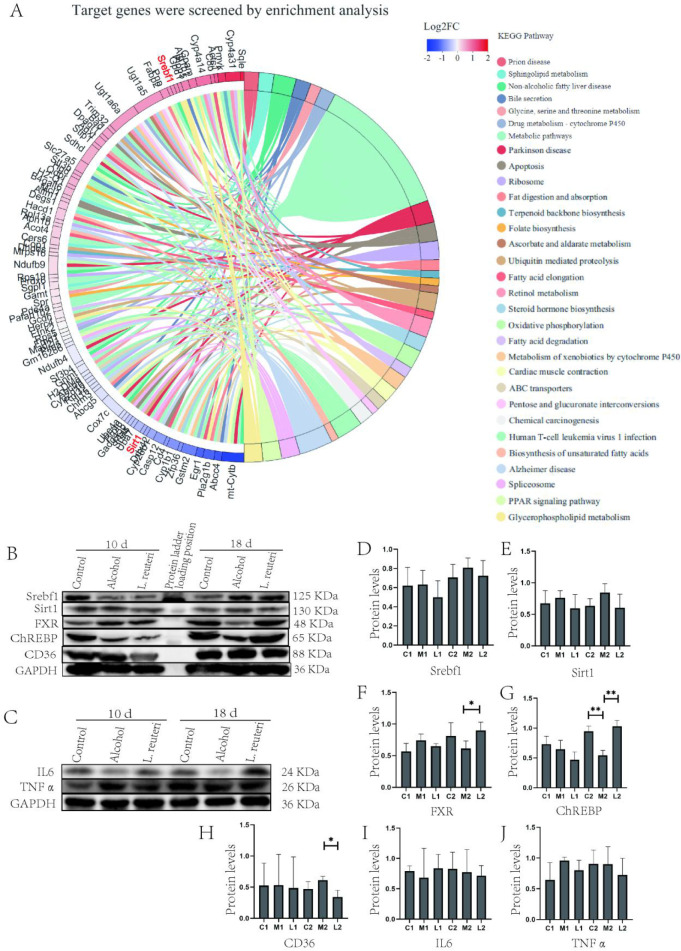
We analyzed genes related to lipid metabolism in the mouse liver samples and found that Srebf1 was upregulated in expression, while Sirt1, a gene of the Sirtuins family, was downregulated in the L. reuteri treatment group (Figure 8A). Srebf1 primarily enhances the expression of genes required for fatty acid synthesis.25 Sirt1 plays an important role in the hepatic control of lipid and cholesterol homeostasis, and Sirt1 deacetylates and activates FXR.26 Therefore, we hypothesize that L. reuteri alleviates hepatic lipid accumulation by acting on the FXR signaling pathway.
Figure 8.
FXR signaling pathway is involved in the mechanism by which L. reuteri alleviates ALD. (A) Alcohol-induced for 18 days, target pathway gene expression. (B, C) Expression of FXR signaling pathway-related proteins in mouse liver tissue. Ten d: 10 days animal model; 18 d: 18 days animal model. (D–J) Quantitative analysis of FXR signaling pathway protein levels in liver tissue. Ten days animal model: Control (C1), Model (M1), L. reuteri (L1). 18 days animal model: Control (C2), Model (M2), L. reuteri (L2). *P < 0.05, **P < 0.01.
Expression of Proteins Related to Lipid Metabolism in Mice Liver and Intestinal Tissues
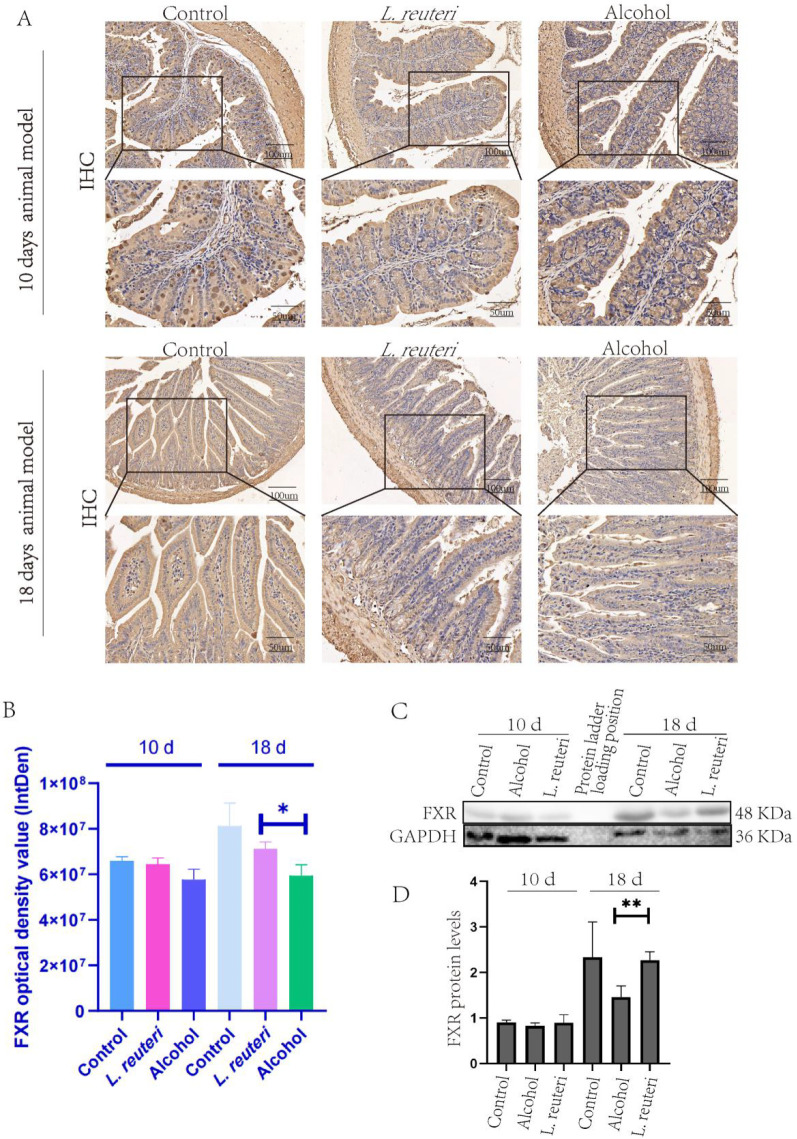
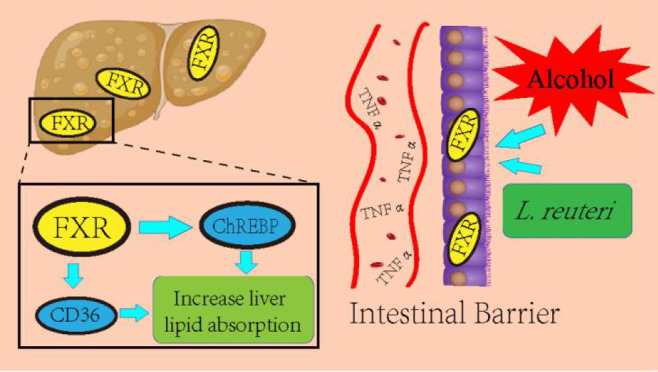
To test our hypothesis, we conducted experiments using Western blot and academic immunohistochemical methods. First, we tested the expression of Sirt1, IL 6, and TNF α (Figure 8B, C, E, I, J) in liver tissues, and the results did not show significant differences, so we concluded that the mice did not have significant inflammatory changes during alcohol induction. We then proceeded to test the expression of FXR, Srebf1, CD36, and ChREBP, and the results showed that the L. reuteri-treated group showed an upregulation of FXR, Srebf1, and ChREBP expression relative to the model group, while CD36 expression was downregulated (Figure 8B, D, F–H). The results showed upregulated expression of FXR, Srebf1, and ChREBP in the L. reuteri-treated group compared to the model group, while the expression of CD36 was downregulated (Figure 8B). We performed Western blot tests on the mouse intestinal tissues and found FXR expression was consistent with that of the liver tissues (Figure 9C, D), while the results of immunohistochemical staining also showed FXR expression was upregulated in the intestinal tissues of L. reuteri-treated mice and showed significant differences compared to the model group (Figure 9A, B). In conclusion, we suggest that L. reuteri can improve alcohol-induced hepatic lipid accumulation in mice via the FXR signaling regulatory axis. The animal experiment results were analyzed along with the transcriptomic results. We found that the L. reuteri treatment group showed significantly improved lipid accumulation compared with the model group, and this effect was primarily based on the FXR enterohepatic regulatory axis. L. reuteri or L. reuteri metabolites activated and enhanced the FXR signaling axis in the liver after passing through the intestine. Subsequently, upregulated Srebf1 and ChREBP, while inhibiting CD36 expression, increased lipid uptake in liver tissues, reducing lipid accumulation.
Figure 9.
L. reuteri enhanced the expression of FXR protein in ALD mice intestinal tissue. (A) IHC staining of FXR protein in mouse intestinal tissue. (B) Optical density of FXR immunohistochemical staining in mouse intestinal tissue. (C) FXR protein expression in the intestinal tissue of mice. (D) Quantitative analysis of the FXR protein level in intestinal tissue (Western Blotting). Ten d: 10 days animal model; 18 d: 18 days animal model. *P < 0.05, **P < 0.01.
FXR Inhibitor Gly-β-MCA Attenuates the Therapeutic Effect of L. reuteri on ALD
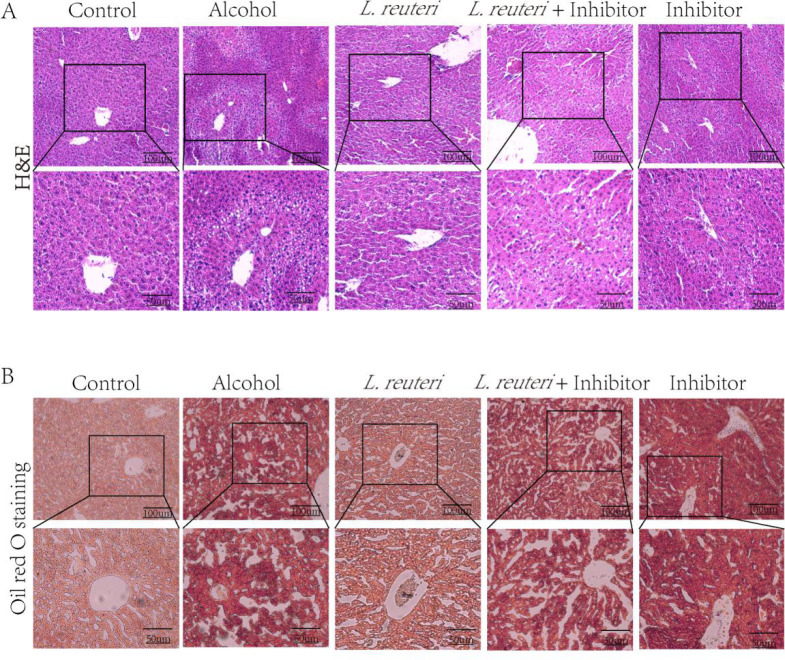
Similarly, mice treated with the FXR inhibitor Gly-β-MCA showed changes in H&E and oil red staining in liver tissue, and Gly-β-MCA reduced the therapeutic effect of L. reuteri (Figure 10).
Figure 10.
FXR inhibitor downregulates the therapeutic effect of L. reuteri on ALD. (A) H&E staining of the liver tissue in all groups of mice (100 × and 200 × magnification). (B) Oil red O staining of liver tissue in all groups of mice (100 × and 200 × magnification).
Discussion
In this study, we studied the effect of L. reuteri on ALD using a mouse model of alcoholic liver disease undergoing treatment with L. reuteri (DSM 17983). We first clarified the therapeutic effect of L. reuteri on ALD through an animal model, and L. reuteri significantly ameliorated liver injury in mice. Subsequently, based on transcriptomic analysis of differently expressed genes in response to alcohol exposure in mice, enrichment pathway analysis identified genes related to lipid metabolism in mice, and finally, validation determined that L. reuteri ameliorated ALD via the FXR signaling regulatory axis.
The therapeutic potential of probiotics in ALD has been demonstrated in clinical studies and animal experiments27 by transient colonization of the gastrointestinal tract, correction of homeostatic dysregulation, and inhibition of toxin production.28 And in our mouse ALD model, alcohol-induced mice treated with concomitant administration of L. reuteri also significantly improved the levels of AST, ALT, and blood biochemical indexes of liver injury (Figure 2A–D). We targeted differential genes such as Sirt1 and Srebf1 with transcriptomic analysis based on the morphological staining results. These factors play an important role in regulating liver metabolism.29 We then validated the upstream and downstream factors of Srebf1, including FXR, CD36, and ChREBP, by Western blot. In conclusion, these data confirm the link between gavage administration of L. reuteri and hepatic lipid metabolism.
It was previously reported30 that alcohol consumption causes quantitative and qualitative dysregulation of the gut microbiota in rodents and humans. Alcohol-fed mice developed ALD, which was associated with bacterial overgrowth in the small intestine and ecologically dysregulated cecum (the origin of the large intestine).31 In particular, the GI microbiota of alcohol-treated mice showed a decrease in firmicutes and an increase in the relative abundance of bacteria such as Bacteroidetes and verrucobacteria. Chronic alcohol consumption in humans can overgrow small intestinal bacteria from both aerobic and anaerobic bacteria in jejunum32 and alter the composition of mucosa-associated microbiota in human sigmoid biopsies. Microbiota dysregulation in alcoholics is also associated with high levels of endotoxin in the blood, suggesting that dysregulation may lead to excessive intestinal permeability and/or increased translocation of Gram-negative microbial bacterial products from the intestinal lumen into the systemic circulation.33 Previous studies on metabolomics verified that L. reuteri treatment reversed the dysregulation of linoleic acid, arachidonic acid, and triglyceride metabolism in ALD.34 There are more studies on L. reuteri for ALD, and L. reuteri can alleviate hepatic lipid metabolism by regulating intestinal and hepatic signaling factor expression, enhancing intestinal barrier function, and modulating the mucosal immune system.35 This result is similar to our findings. In addition, alcohol induces impaired intestinal barrier function, allowing toxins produced by intestinal bacteria and other microorganisms to translocate to the liver, inducing and promoting inflammation and promoting hepatocyte death and fibrotic responses.36 Hsieh et al. verified that L. reuteri upregulates glutathione and glutathione peroxidase activities, reducing oxidative stress and the inflammatory response, thereby reducing liver injury.37 Meanwhile, the results of Gu et al. suggest that Lactobacillus rhamnosus-GG may improve barrier function by releasing exosome-like nanoparticles enriched with bacterial AhR ligands, which increase the expression of Reg3 and Nrf2.38 In conclusion, our results suggest that L. reuteri may play a role in improving hepatic lipid metabolism in ALD mice through the FXR signaling pathway. This mechanism may be closely related to the impaired intestinal barrier function.
Notably, in the animal model, we verified the expression of Sirt1, IL6, and TNFα by Western blot, which suggested that none of the mice reached a significant inflammatory response. There was no statistical difference in the results of repeated trials. Ramirez et al. showed manifestations of inflammation at 10 days of alcohol induction and demonstrated more pronounced inflammation in aging mice (>12 mouths).39 In our study, the mice in the ALD model were all young mice (8 weeks), which may explain the lack of significant inflammation in our model. In this study, the indices of liver damage of the 10 day alcohol-induced mice suggested that L. reuteri treatment was effective. With the alcohol induction time extension to 18 days, we found that the L. reuteri treatment showed a significant effect. We speculate that the improvement of ALD with L. reuteri treatment becomes more pronounced as the time is extended.
In analyzing the transcriptomic results, we found those differential genes were primarily enriched in metabolic pathways (Table 2). The SREBP family, perilipin 2, perilipin 5, Delta-5 desaturase, ChREBP, and SGLT1 were involved.40 SREBP are a family of transcription factors that control all aspects of lipid metabolism by regulating the expression of a set of genes containing sterol regulatory elements in their respective promoter regions.41 The CYP1A2/PTEN/AKT/Srebf1 pathway contributes to improved hepatocyte lipid metabolism, as illustrated by Zhu et al. Alcohol induces upregulation of Srebf1 in mice.42 Steatosis occurs in the liver of Sirt1-specific knockout mice,43 which is consistent with our transcriptomic results. Sirt1 also regulates LXRα by upregulating the oxysterol receptor target gene ABCA1 to help reverse cholesterol transport in peripheral tissues.44 Sirt1 also deacetylates and activates the FXR,45 and FXR is involved in maintaining and preserving the integrity and function of the intestinal barrier and preventing bacterial translocation from the gut.45,46 Alcohol-induced bacterial translocation increased intestinal permeability, thereby promoting the development of endotoxemia and ALD.47 Kong et al. reported that FXR in extrahepatic tissues plays a minor role in ALD development.48 Our study found that the FXR signaling regulatory axis in liver tissue plays an important role in ALD lipid accumulation. Since no significant inflammation was observed in the ALD model mice in this study, we could not verify that L. reuteri and its metabolites regulate liver inflammation via the FXR signaling regulatory axis, and further studies are needed.
Table 2. Results of RNA Sample Quality Tests for Transcriptome Sequencinga.
| Sample number | Sample type | RIN | Test results | Interpretation of results |
|---|---|---|---|---|
| C1 | Liver | 7.90 | B | 6.5 ≤ RIN < 8 |
| C2 | Liver | 7.80 | B | 6.5 ≤ RIN < 8 |
| C3 | Liver | 6.60 | B | 6.5 ≤ RIN < 8 |
| C4 | Liver | 7.80 | B | 6.5 ≤ RIN < 8 |
| C5 | Liver | 8.10 | A | RIN ≥ 8 |
| C6 | Liver | 8.30 | A | RIN ≥ 8 |
| C7 | Liver | 8.20 | A | RIN ≥ 8 |
| L1 | Liver | 6. 80 | B | 6.5 ≤ RIN < 8 |
| L2 | Liver | 8.90 | A | RIN ≥ 8 |
| L3 | Liver | 6. 80 | B | 6.5 ≤ RIN < 8 |
| L4 | Liver | 7.20 | B | 6.5 ≤ RIN < 8 |
| L5 | Liver | 6.70 | B | 6.5 ≤ RIN < 8 |
| L6 | Liver | 9.10 | A | RIN ≥ 8 |
| L7 | Liver | 8.10 | A | RIN ≥ 8 |
| M1 | Liver | 6.60 | B | 6.5 ≤ RIN < 8 |
| M2 | Liver | 6. 80 | B | 6.5 ≤ RIN < 8 |
| M3 | Liver | 7.30 | B | 6.5 ≤ RIN < 8 |
| M4 | Liver | 9.00 | A | RIN ≥ 8 |
| M5 | Liver | 6.60 | B | 6.5 ≤ RIN < 8 |
| M6 | Liver | 8.60 | A | RIN ≥ 8 |
| M7 | Liver | 8.50 | A | RIN ≥ 8 |
RIN: RNA integrity number. Description of quality inspection results: A, RNA band is clear, without pigment, protein, sugar or other impurities contamination; B, RNA band is slightly degraded, without pigment, protein, sugar, or other impurities contamination; C, RNA has no pigment contamination, no obvious protein, sugar, or other impurities contamination; D, RNA band is seriously dispersed, or cloudy, or contaminated with pigment, or seriously contaminated with impurities such as protein, sugar, polysaccharide, or DNA.
We demonstrated for the first time that L. reuteri alleviates the lipid accumulation of ALD, specifically through the FXR signaling regulatory axis. The aim is to provide a richer theoretical basis for the treatment of ALD by L. reuteri and to develop better management of ALD in the future.
Glossary
Abbreviations Used
- ALD
alcoholic liver disease
- L. reuteri
Lactobacillus reuteri
- FXR
farnesoid X receptor
- ChREBP
carbohydrate response element binding protein
- Srebf1
sterol regulatory element binding transcription factor 1
- CD36
Cluster of differentiation 36
- Gly-β-MCA
glycine-β-muricholic acid
- NAFLD
non-alcoholic fatty liver disease
- BCA
bicinchoninic acid assay
- AST
aspartate aminotransferase
- ALT
alanine aminotransferase
- H&E
hematoxylin-eosin
- IHC
immunohistochemistry
- TNFα
tumor necrosis factor α
- ELISA
enzyme linked immunosorbent assay
- Sirt1
sirtuin 1
- IL6
interleukin 6
- PCR
polymerase chain reaction
- IgG
immunoglobulin G
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GSEA
gene set enrichment analysis
- SREBP
sterol-regulatory element binding protein
- Reg3
regenerating islet-derived protein 3
- Nrf2
nuclear factor erythroid-2-related factor 2
- SGLT1
recombinant sodium/glucose cotransporter 1
- CYP1A2
cytochrome P450
- PTEN
phosphatase and tensin homologue deleted on chromosome ten
- AKT
protein kinase B
- LXRα
liver X receptor α
- ABCA1
ATP-binding cassette transporter A1
Author Contributions
† Yonglang Cheng, Xin Xiang, and Chen Liu contributed equally to this work.
This research was funded by the National Natural Science Foundation of China (No. 82170587), Natural Science Foundation of Sichuan Province (No. 2022NSFSC1321), Luzhou Municipal People’s Government-Southwest Medical University Science and Technology Strategic Cooperation Project (No. 2021LZXNYDZ01, No. 2021LZXNYDJ01), Neijiang First People’s Hospital - Southwest Medical University Strategic cooperation project (No. 2021NJXNYD03), and Southwest Medical University New Academic Project (No. 2021ZKMS026).
The authors declare no competing financial interest.
Notes
The raw data supporting the conclusions of this article will be made available by the authors without undue reservation. The data have been uploaded to the National Center for Biotechnology Information (NCBI) (BioProject ID: PRJNA835972). The animal study was reviewed and approved by the Animal Care and Use Committee of Southwest Medical University (NO. 20211119-047).
References
- Song X.; Liu Z.; Zhang J.; Zhang C.; Dong Y.; Ren Z.; Gao Z.; Liu M.; Zhao H.; Jia L. Antioxidative and hepatoprotective effects of enzymatic and acidic-hydrolysis of Pleurotus geesteranus mycelium polysaccharides on alcoholic liver diseases. Carbohydr. Polym. 2018, 201, 75–86. 10.1016/j.carbpol.2018.08.058. [DOI] [PubMed] [Google Scholar]
- Wang J.; Zhang Y.; Liu R.; Li X.; Cui Y.; Qu L. Geniposide protects against acute alcohol-induced liver injury in mice via up-regulating the expression of the main antioxidant enzymes. Can. J. Physiol. Pharmacol. 2015, 93 (4), 261–267. 10.1139/cjpp-2014-0536. [DOI] [PubMed] [Google Scholar]
- Bastian J. Electrolocation in the presence of jamming signals: behavior. J. Comp. Physiol. A 1987, 161 (6), 811–824. 10.1007/BF00610223. [DOI] [PubMed] [Google Scholar]
- Xing H.; Jia K.; He J.; Shi C.; Fang M.; Song L.; Zhang P.; Zhao Y.; Fu J.; Li S. Establishment of the tree shrew as an alcohol-induced Fatty liver model for the study of alcoholic liver diseases. PLoS One 2015, 10 (6), e0128253 10.1371/journal.pone.0128253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman A.; Mehta K. J. Betaine in ameliorating alcohol-induced hepatic steatosis. Eur. J. Nutr. 2022, 61 (3), 1167–1176. 10.1007/s00394-021-02738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslamparast T.; Eghtesad S.; Hekmatdoost A.; Poustchi H. Probiotics and Nonalcoholic Fatty liver Disease. Middle East. J. Dig. Dis. 2013, 5 (3), 129–136. [PMC free article] [PubMed] [Google Scholar]
- Koutnikova H.; Genser B.; Monteiro-Sepulveda M.; Faurie J. M.; Rizkalla S.; Schrezenmeir J.; Clement K. Impact of bacterial probiotics on obesity, diabetes and non-alcoholic fatty liver disease related variables: a systematic review and meta-analysis of randomised controlled trials. BMJ. Open. 2019, 9 (3), e017995 10.1136/bmjopen-2017-017995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.; Tian H.; Kang Y.; Tian Y.; Li L.; Kang X.; Yang H.; Wang Y.; Tian J.; Zhang F.; Tong M.; Cai H.; Fan W. Probiotics alleviate autoimmune hepatitis in mice through modulation of gut microbiota and intestinal permeability. J. Nutr. Biochem. 2021, 98, 108863. 10.1016/j.jnutbio.2021.108863. [DOI] [PubMed] [Google Scholar]
- Kirpich I. A.; Solovieva N. V.; Leikhter S. N.; Shidakova N. A.; Lebedeva O. V.; Sidorov P. I.; Bazhukova T. A.; Soloviev A. G.; Barve S. S.; McClain C. J.; Cave M. Probiotics restore bowel flora and improve liver enzymes in human alcohol-induced liver injury: a pilot study. Alcohol. 2008, 42 (8), 675–682. 10.1016/j.alcohol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.; Wang C.; Wang C.; Zhao H.; Zhao C.; Chen Y.; Wang Y.; McClain C.; Feng W. Enhanced AMPK phosphorylation contributes to the beneficial effects of Lactobacillus rhamnosus GG supernatant on chronic-alcohol-induced fatty liver disease. J. Nutr. Biochem. 2015, 26 (4), 337–344. 10.1016/j.jnutbio.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grander C.; Adolph T. E.; Wieser V.; Lowe P.; Wrzosek L.; Gyongyosi B.; Ward D. V.; Grabherr F.; Gerner R. R.; Pfister A.; Enrich B.; Ciocan D.; Macheiner S.; Mayr L.; Drach M.; Moser P.; Moschen A. R.; Perlemuter G.; Szabo G.; Cassard A. M.; Tilg H. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut. 2018, 67 (5), 891–901. 10.1136/gutjnl-2016-313432. [DOI] [PubMed] [Google Scholar]
- Leclercq S.; de Timary P.; Starkel P. Targeting the gut microbiota to treat alcoholic liver diseases: evidence and promises. Acta Gastroenterol. Belg. 2020, 83 (4), 616–621. [PubMed] [Google Scholar]
- Hong M.; Han D. H.; Hong J.; Kim D. J.; Suk K. T. Are Probiotics Effective in Targeting Alcoholic Liver Diseases?. Probiotics Antimicrob. Proteins. 2019, 11 (2), 335–347. 10.1007/s12602-018-9419-6. [DOI] [PubMed] [Google Scholar]
- Mu Q.; Tavella V. J.; Luo X. M. Role of Lactobacillus reuteri in Human Health and Diseases. Front. Microbiol. 2018, 9, 757. 10.3389/fmicb.2018.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.; Lu C.; Yao L.; Zhang F.; Shao J.; Zheng S. Dihydroartemisinin protects against alcoholic liver injury through alleviating hepatocyte steatosis in a farnesoid X receptor-dependent manner. Toxicol. Appl. Pharmacol. 2017, 315, 23–34. 10.1016/j.taap.2016.12.001. [DOI] [PubMed] [Google Scholar]
- Byron S. A.; Van Keuren-Jensen K. R.; Engelthaler D. M.; Carpten J. D.; Craig D. W. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat. Rev. Genet. 2016, 17 (5), 257–271. 10.1038/nrg.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertola A.; Mathews S.; Ki S. H.; Wang H.; Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nat. Protoc. 2013, 8 (3), 627–637. 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.; Dewey C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011, 12, 323. 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I.; Huber W.; Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15 (12), 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Feng Z.; Wang X.; Wang X.; Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010, 26 (1), 136–138. 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Robinson M. D.; McCarthy D. J.; Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010, 26 (1), 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C.; Mao X.; Huang J.; Ding Y.; Wu J.; Dong S.; Kong L.; Gao G.; Li C. Y.; Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39 (Web Server issue), W316–322. 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S.; Park J. W.; Lu Z. X.; Lin L.; Henry M. D.; Wu Y. N.; Zhou Q.; Xing Y. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U. S. A. 2014, 111 (51), E5593–5601. 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemarajata P.; Gao C.; Pflughoeft K. J.; Thomas C. M.; Saulnier D. M.; Spinler J. K.; Versalovic J. Lactobacillus reuteri-specific immunoregulatory gene rsiR modulates histamine production and immunomodulation by Lactobacillus reuteri. J. Bacteriol. 2013, 195 (24), 5567–5576. 10.1128/JB.00261-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J.; Yu J.; Xu H.; Kang C.; Shaul P. W.; Guan Y.; Zhang X.; Su W. Enhanced Liver Regeneration After Partial Hepatectomy in Sterol Regulatory Element-Binding Protein (SREBP)-1c-Null Mice is Associated with Increased Hepatocellular Cholesterol Availability. Cell. Physiol. Biochem. 2018, 47 (2), 784–799. 10.1159/000490030. [DOI] [PubMed] [Google Scholar]
- Chang H. C.; Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 2014, 25 (3), 138–145. 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H.; Kim S. W.; Hong M.; Suk K. T. Microbiota-based treatments in alcoholic liver disease. World J. Gastroenterol. 2016, 22 (29), 6673–6682. 10.3748/wjg.v22.i29.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman P. M.; Ossa J. C.; Johnson-Henry K. Unraveling mechanisms of action of probiotics. Nutr. Clin. Pract. 2009, 24 (1), 10–14. 10.1177/0884533608329231. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Dentin R.; Chen D.; Hedrick S.; Ravnskjaer K.; Schenk S.; Milne J.; Meyers D. J.; Cole P.; Yates J. III; Olefsky J.; Guarente L.; Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008, 456 (7219), 269–273. 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engen P. A.; Green S. J.; Voigt R. M.; Forsyth C. B.; Keshavarzian A. The Gastrointestinal Microbiome: Alcohol Effects on the Composition of Intestinal Microbiota. Alcohol Res. 2015, 37 (2), 223–236. [PMC free article] [PubMed] [Google Scholar]
- Yan A. W.; Fouts D. E.; Brandl J.; Starkel P.; Torralba M.; Schott E.; Tsukamoto H.; Nelson K. E.; Brenner D. A.; Schnabl B. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology 2011, 53 (1), 96–105. 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode J. C.; Bode C.; Heidelbach R.; Durr H. K.; Martini G. A. Jejunal microflora in patients with chronic alcohol abuse. Hepatogastroenterology 1984, 31 (1), 30–34. [PubMed] [Google Scholar]
- Mutlu E.; Keshavarzian A.; Engen P.; Forsyth C. B.; Sikaroodi M.; Gillevet P. Intestinal dysbiosis: a possible mechanism of alcohol-induced endotoxemia and alcoholic steatohepatitis in rats. Alcohol.: Clin. Exp. Res. 2009, 33 (10), 1836–1846. 10.1111/j.1530-0277.2009.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T. X.; Pu S. L.; Tan P.; Du Y. C.; Qian B. L.; Chen H.; Fu W. G.; Huang M. Z. Liver Metabolomics Reveals the Effect of Lactobacillus reuteri on Alcoholic Liver Disease. Front. Physiol. 2020, 11, 595382. 10.3389/fphys.2020.595382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z.; Wu Y.; Wang Y.; Sun H.; You Y.; Piao C.; Liu J.; Wang Y. Lactobacillus rhamnosus Granules Dose-Dependently Balance Intestinal Microbiome Disorders and Ameliorate Chronic Alcohol-Induced Liver Injury. J. Med. Food. 2020, 23 (2), 114–124. 10.1089/jmf.2018.4357. [DOI] [PubMed] [Google Scholar]
- Sarin S. K.; Pande A.; Schnabl B. Microbiome as a therapeutic target in alcohol-related liver disease. J. Hepatol. 2019, 70 (2), 260–272. 10.1016/j.jhep.2018.10.019. [DOI] [PubMed] [Google Scholar]
- Hsieh P. S.; Chen C. W.; Kuo Y. W.; Ho H. H. Lactobacillus spp. reduces ethanol-induced liver oxidative stress and inflammation in a mouse model of alcoholic steatohepatitis. Exp. Ther. Med. 2021, 21 (3), 188. 10.3892/etm.2021.9619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z.; Li F.; Liu Y.; Jiang M.; Zhang L.; He L.; Wilkey D. W.; Merchant M.; Zhang X.; Deng Z. B.; Chen S. Y.; Barve S.; McClain C. J.; Feng W. Exosome-Like Nanoparticles From Lactobacillus rhamnosusGG Protect Against Alcohol-Associated Liver Disease Through Intestinal Aryl Hydrocarbon Receptor in Mice. Hepatol. Commun. 2021, 5 (5), 846–864. 10.1002/hep4.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez T.; Li Y. M.; Yin S.; Xu M. J.; Feng D.; Zhou Z.; Zang M.; Mukhopadhyay P.; Varga Z. V.; Pacher P.; Gao B.; Wang H. Aging aggravates alcoholic liver injury and fibrosis in mice by downregulating sirtuin 1 expression. J. Hepatol. 2017, 66 (3), 601–609. 10.1016/j.jhep.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja Gopal Reddy M.; Pavan Kumar C.; Mahesh M.; Sravan Kumar M.; Jeyakumar S. M. Expression data on liver metabolic pathway genes and proteins. Data Brief. 2016, 6, 625–629. 10.1016/j.dib.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loregger A.; Raaben M.; Nieuwenhuis J.; Tan J. M. E.; Jae L. T.; van den Hengel L. G.; Hendrix S.; van den Berg M.; Scheij S.; Song J. Y.; Huijbers I. J.; Kroese L. J.; Ottenhoff R.; van Weeghel M.; van de Sluis B.; Brummelkamp T.; Zelcer N. Haploid genetic screens identify SPRING/C12ORF49 as a determinant of SREBP signaling and cholesterol metabolism. Nat. Commun. 2020, 11 (1), 1128. 10.1038/s41467-020-14811-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q.; Huang C.; Meng X.; Li J. CYP1A2 contributes to alcohol-induced abnormal lipid metabolism through the PTEN/AKT/SREBP-1c pathway. Biochem. Biophys. Res. Commun. 2019, 513 (2), 509–514. 10.1016/j.bbrc.2019.04.034. [DOI] [PubMed] [Google Scholar]
- Purushotham A.; Schug T. T.; Xu Q.; Surapureddi S.; Guo X.; Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009, 9 (4), 327–338. 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Zhang S.; Blander G.; Tse J. G.; Krieger M.; Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell 2007, 28 (1), 91–106. 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Kemper J. K.; Xiao Z.; Ponugoti B.; Miao J.; Fang S.; Kanamaluru D.; Tsang S.; Wu S. Y.; Chiang C. M.; Veenstra T. D. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009, 10 (5), 392–404. 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T.; Moschetta A.; Lee Y. K.; Peng L.; Zhao G.; Downes M.; Yu R. T.; Shelton J. M.; Richardson J. A.; Repa J. J.; Mangelsdorf D. J.; Kliewer S. A. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. U. S. A. 2006, 103 (10), 3920–3925. 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie G.; Zhong W.; Li H.; Li Q.; Qiu Y.; Zheng X.; Chen H.; Zhao X.; Zhang S.; Zhou Z.; Zeisel S. H.; Jia W. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. FASEB J. 2013, 27 (9), 3583–3593. 10.1096/fj.13-231860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong B.; Zhang M.; Huang M.; Rizzolo D.; Armstrong L. E.; Schumacher J. D.; Chow M. D.; Lee Y. H.; Guo G. L. FXR deficiency alters bile acid pool composition and exacerbates chronic alcohol induced liver injury. Dig. Liver Dis. 2019, 51 (4), 570–576. 10.1016/j.dld.2018.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]