Abstract
Background
An outbreak of monkeypox virus infections in non-endemic countries was recognised on May 12, 2022. As of September 29, more than 67 000 infections have been reported globally, with more than 3400 confirmed cases in the UK by September 26. Monkeypox virus is believed to be predominantly transmitted through direct contact with lesions or infected body fluids, with possible involvement of fomites and large respiratory droplets. A case of monkeypox in a health-care worker in the UK in 2018 was suspected to be due to virus exposure while changing bedding. We aimed to measure the extent of environmental contamination in the isolation rooms of patients with symptomatic monkeypox.
Methods
We investigated environmental contamination with monkeypox virus from infected patients admitted to isolation rooms at the Royal Free Hospital (London, UK) between May 24 and June 17, 2022. Surface swabs of high-touch areas in five isolation rooms, of the personal protective equipment (PPE) of health-care workers in doffing areas in three rooms, and from air samples collected before and during bedding changes in five rooms were analysed using quantitative PCR to assess monkeypox virus contamination levels. Virus isolation was performed to confirm presence of infectious virus in selected positive samples.
Findings
We identified widespread surface contamination (56 [93%] of 60 samples were positive) in occupied patient rooms (monkeypox DNA cycle threshold [Ct] values 24·7–37·4), on health-care worker PPE after use (Ct 26·1–35·6), and in PPE doffing areas (Ct 26·3–36·8). Of 20 air samples taken, five (25%) were positive. Three (75%) of four air samples collected before and during a bedding change in one patient's room were positive (Ct 32·7–36·2). Replication-competent virus was identified in two (50%) of four samples selected for viral isolation, including from air samples collected during bedding change.
Interpretation
These data show contamination in isolation facilities and potential for suspension of monkeypox virus into the air during specific activities. PPE contamination was observed after clinical contact and changing of bedding. Contamination of hard surfaces in doffing areas supports the importance of cleaning protocols, PPE use, and doffing procedures.
Funding
None.
Introduction
An unprecedented number of cases of monkeypox have been confirmed outside of the endemic areas of west and central Africa since May 12, 2022. As of Sept 29, 67 328 infections have been reported by 99 non-endemic countries; up to Sept 26, 3485 confirmed cases were reported by the UK.1, 2
Monkeypox virus is an enveloped double-stranded DNA virus classified within the Orthopoxvirus genus of the Poxviridae family. Monkeypox infection causes a clinical illness that is typically milder than smallpox, consisting of an influenza-like prodrome followed by a distinctive vesiculopustular rash. Lymphadenopathy typically occurs in monkeypox but not in smallpox.3 Mortality in monkeypox is thought to be 1–10%, influenced by clade and patient characteristics. As of Sept 29, 2022, 12 deaths had been reported from non-endemic countries during the current outbreak, plus 15 deaths from endemic countries.4
The number of monkeypox cases increased in various endemic countries after the cessation of the smallpox vaccination programme, and there have been concerns about a potential increase in cases in travellers over the past decade.5, 6 Primary cases arise from contact with animal reservoirs, such as rope squirrels. Rodents are thought to have an important role, although further research is needed.7 Sustained human-to-human transmission had not been reported before outbreaks identified in May, 2022, and the secondary household attack rate in endemic settings has been reported as 0–10% in the majority of cases, and as high as 50% in one outbreak.8 Transmission to secondary cases is believed to be predominately via direct contact with body fluids or lesions, respiratory droplets, and fomites. Infection via inhalation of high-tire, aerosolised monkeypox virus of the central African clade has been demonstrated in non-human primates, raising the possibility of potential aerosol transmission between humans; however, existing epidemiological investigations suggest long-range aerosol transmission does not occur.9, 10
Research in context.
Evidence before this study
We searched PubMed for all relevant papers published between Jan 1, 1970 (first case of human monkeypox was in 1970) and July 30, 2022, using the keywords “monkeypox”, “contamination”, “environmental”, “surface”, and “airborne”, with no language restrictions. We selected articles describing environmental sampling to detect monkeypox virus or describing human infection with a potential link to environmental contamination. Articles describing human-to-human transmission, or the stability of monkeypox virus on surfaces or in air, were also of interest. Our search returned 128 studies, 11 of which were relevant to this topic. A 2022 paper on environmental sampling in a domestic environment described widespread contamination on various surfaces and showed that competent virus was retrievable in certain positive samples. Regarding the hospital environment, findings published in June, 2022 from the sampling of surfaces in rooms occupied by individuals with monkeypox in Germany showed detection of virus on most surfaces sampled, including on touch surfaces in an anteroom used for the removal of personal protective equipment (PPE; known as doffing). Secondary infections in household contacts have, in most reports, ranged between 0% and 10%. A hospital worker in the UK with confirmed monkeypox infection in 2018 was thought to have been exposed to the virus while changing bedding, without wearing PPE (beyond gloves and an apron) as the patient was yet to be diagnosed. The durability of monkeypox virus on surfaces is unknown. Under laboratory conditions, aerosolised monkeypox virus remained viable for 90 h. In non-human primates, there has been proven transmission via aerosol (Zaire strain), albeit using a high quantity of virus. Human-to-human transmission of monkeypox virus is believed to be via direct contact, body fluids, fomites, and large respiratory droplets. To date, there have been no previous reports of positive air samples or viable virus detected from hospital settings where patients with monkeypox have been managed.
Added value of this study
Our findings contribute further evidence of substantial environmental contamination around patients with monkeypox, and that frequency of viral DNA detection and quantity of virus detected is variable. This variation might be due to differing clinical characteristics, patient behaviour, or cleaning procedures. Contamination of PPE and of the floor of the area where PPE is removed add to the information base around this important component of infection prevention and control. To our knowledge, we report for the first time the detection of monkeypox virus DNA and viable virus in air samples, with sampling around bedding changes indicating that this procedure and others (such as cleaning) could cause re-aerosolisation of virus. Our data is of public health importance and should inform policy to protect health-care workers and reduce the risk of nosocomial transmission of monkeypox.
Implications of all the available evidence
The available evidence shows widespread contamination in environments occupied by individuals with symptomatic monkeypox. Viral DNA has been detected on the PPE of health-care workers and in areas used for the removal of PPE. Competent virus has now been retrieved from environmental sampling, including an air sample. The detection of monkeypox virus (DNA and virus by isolation) in air samples is novel and detection of monkeypox virus DNA at distances of more than 1·5 m from the patient's bed and a height taller than 2 m supports the theory that virus DNA is suspended in aerosols, skin flakes, or dust. Current evidence supports the use of PPE, including respiratory protection, regular surface cleaning, and appropriate doffing, and the disposal of articles that are likely to be contaminated. Further investigation should consider the extent of contamination in clinical spaces occupied for shorter periods of time than those investigated here (as occurs during outpatient consultations), the effectiveness of cleaning protocols in decontaminating environments, and explore further the risk of respiratory transmission.
Orthopox viruses are stable in the environment and can remain viable in aerosols for up to 90 hours.11, 12 A hospital worker in the UK who developed monkeypox in October, 2018 was thought to have been exposed to virus while changing the bedding used by a patient with monkeypox, before the diagnosis had been considered and appropriate infection control measures initiated.13 Widespread surface contamination in hospital rooms occupied by two patients with monkeypox virus infection was reported in Germany in June, 2022.14 Infections in health-care workers have been reported during the 2022 outbreak, although the route of transmission is not known for all and many appear to have had community exposure.15 In the UK, confirmed cases requiring prolonged hospital care have been managed in respiratory isolation by staff wearing recommended personal protective equipment (PPE), measures that are designed to reduce the risk of transmission to specialist health-care workers, many of whom have now been vaccinated against orthopox infections. Therefore, the absence of health-care worker infections in the current UK outbreak does not mean exposure risk is low or absent.
We aimed to investigate the extent of environmental contamination in the rooms of symptomatic patients within the airborne isolation units of National Health Service (NHS) England's High Consequence Infectious Diseases (HCID) Network, to inform practice around isolation, PPE, decontamination protocols, and public health management of community exposures. We also aimed to investigate whether aerosol transmission risks occur and whether certain activities—such as changing bedding—increase the risk of exposure.
Methods
Study setting and design
Sampling of patient rooms
Hospitalised adults with confirmed monkeypox and active skin lesions were identified, and verbal informed consent obtained to sample the air and environment within isolation rooms at the Royal Free Hospital (London, UK) between May 24 and June 17, 2022. Further information on the ward layout and room specifications are given in the appendix (pp 4–5). Air and surface sampling was performed in four positive-pressure ventilated lobby,16 single-occupancy, respiratory isolation rooms (rooms A, C, D, and E; room A was sampled twice), including the bedroom, bathroom, and anteroom of each isolation room. Air sampling was performed in the anterooms and external corridor for three isolation rooms (rooms A–C), in conjunction with swabs of used PPE and of the floor of the doffing area in the anteroom immediately after the removal of PPE. Patient rooms had at least ten air changes per hour and the median pressure differential between negative pressure areas (bedroom and bathroom) and other areas was maintained at 8 Pa or higher. Sampling of room A and the anteroom of room C was performed twice, each with different occupants. Rooms were cleaned every 12 h during occupancy using 5000 ppm available chlorine sodium hypochlorite on all hard surfaces and floors and 10 000 ppm available chlorine sodium hypochlorite for the toilet, shower, wash basins, and floors. A full room clean was done with 5000 ppm available chlorine sodium hypochlorite after patient discharge, followed by decontamination using vapourised hydrogen peroxide (Bioquell BQ-50). Further details are given in the appendix (p 2). The first three patients sampled were in rooms in which no patients with monkeypox had previously been admitted, excluding the possibility of detecting contamination from previous occupants.
Surface sampling using Copan UTM swabs (Copan, Carlsbad, CA, USA) targeted high-touch areas (patient call bell, television remote control, arm of patient chair, door handle, light switch, tap handles, shower handles, and floor), an air vent above the door leading to the ensuite bathroom, and a potential deposition area that is unlikely to have been directly touched by patients. Where practicable, all surface samples were approximately 10 cm × 10 cm; samples that were not conducive for standardised sampling (such as door and tap handles) were sampled by swabbing the entire item. Air sampling using the MD8 Airport (with gelatine filters, flow rate 50 L/min for 10 min; Sartorius, Goettingen, Germany) was performed before and during the changing of bedding for the first visit to room A, all subsequent air sampling in patient rooms lasted 5 min (flow rate 50 L/min). Two Sartorius air samplers were simultaneously placed near to the bed (height: 1 m, distance from patient bed: 1 m) and further away (height: 2 m, distance from bed: >1·5 m). Distances are approximate. These placements were chosen within the limitations of the room layout and size to provide one sample that is more likely to capture particles present in air at close range to the patient, and one that would probably only capture particles that remained suspended in air over a longer distance. Specifically, a height of 2 m and horizontal distance of at least 1·5 m meant one sampler was placed on a cupboard against the wall, above the level of activity of the health-care worker and patient. The other sampler was placed, near to the bed, on the opposite side of the bed to which the health-care worker was standing to change the bedding. Further details on the protocol for bedding changes are given in the appendix (p 3). Wearable button samplers (with gelatine filters, flow rate 4 L/min for 10 min; SKC, Blandford Forum, UK) were used during the bedding change, on the first visit to room A, by two health-care workers and a third was hung in the room. Minimal anonymised epidemiological and clinical data were provided by the treating clinical team, including date of admission, date of onset of illness, most recent virology results, and whether the patient had received tecovirimat. All patients provided written informed consent for the ISARIC Clinical Characterisation Protocol17 that includes air and environmental sampling. Before environmental sampling, informed consent was confirmed verbally. The study was undertaken as an Urgent Public Health Investigation with UK Health Security Agency Research Ethics and Governance of Public Health Practice Group approval (NR0327).
Sampling of PPE
As per protocol for infectious diseases, staff entering an isolation room donned the following disposable, single-use PPE before entering the anteroom: surgical gown, plastic apron, visor, double nitrile gloves, FFP3 respirator, hair cover, and autoclavable plastic clogs. To exit a patient room, staff entered the anteroom, which is split into toxic (close to patient room) and non-toxic (close to exit to the corridor) zones using tape on the floor. PPE doffing protocols within the anteroom are designed to minimise contamination of the non-toxic area, with potentially contaminated items entering waste streams located in the toxic area. Staff transfer into clean clogs after all PPE has been removed, and they transition to the non-toxic part of the anteroom before a final hand wash prior to exit. This process is monitored by a buddy (outside of the room) to ensure the process occurs in the correct order.
Before the removal of PPE in the anteroom, swabs were taken of the front of the gown, gloves, and visor of health-care workers who had either had clinical contact (rooms B and C) or changed the bedding (room A). Swabs were taken of the floor in the doffing area immediately after PPE removal in each case. Air samples were taken simultaneously in the anteroom and in the adjacent corridor before and during the doffing procedure, using the MD8 Airport (with gelatine filters, 50 L/min for 5 min), in rooms A–C.
Sample processing
For surface samples, 140 μL of sample in universal transport media was inactivated using Buffer AVL (Qiagen, Hilden, Germany) with nucleic acid using the Viral RNA Mini Kit (Qiagen), in accordance with the manufacturer's instructions. For air samples, gelatine filters were dissolved in 20 mL of warmed minimum essential media (Gibco, Carlsbad, CA, USA) for MD8 filters, or 5 mL for personal sampler gelatine filters, with 140 μL then used for inactivation and extraction as described. We analysed the extracted nucleic acid for the presence of monkeypox virus DNA using a published assay, with minor modifications to conform to local standardised diagnostic processes.18
Viral Isolation
Four samples—one high-touch area, one air sample, and two anteroom floor samples—were selected for viral isolation to determine whether infection-competent virus was detectable (ie, viral particles capable of infecting a cell and resulting in the production of new viral particles).
0·5 mL of universal transport media (for swab samples) or minimum essential media containing dissolved gelatine filter (for air samples) was added to a 70% confluent monolayer of Vero C1008 cells (ECACC 85020206) in a T-25 non-vented tissue culture flask (25 cm2 available growth area; Corning, New York, NY, USA) and incubated for 1 h at 37°C. After 1 h, the inoculum was removed and washed with sterile phosphate-buffered saline before 5 mL viral culture medium was added, consisting of 1 × minimum essential media plus GlutaMAX supplemented with 5% heat-inactivated fetal bovine serum, 25 mM HEPES, and 4 × antibiotic-antimycotic solution (all Gibco). A negative control flask was also prepared in parallel by the same method using 0·5 mL of minimum essential media as the inoculum. All flasks were incubated at 37°C and monitored for cytopathic effect using a phase contrast inverted light microscope. 140 μL timepoints were collected every 48–72 h to monitor for a change in detectable DNA via quantitative PCR. Previous work suggests that monkeypox virus is likely to be culturable from samples with a Ct value below 30 for this reference laboratory PCR assay.19 Over-confluent cell monolayers (5 days post-infection) that did not display viral cytopathic effect were passaged by inoculating supernatant onto fresh sub-confluent cells, providing continuous assessment for cytopathic effect for 10 days.
Results
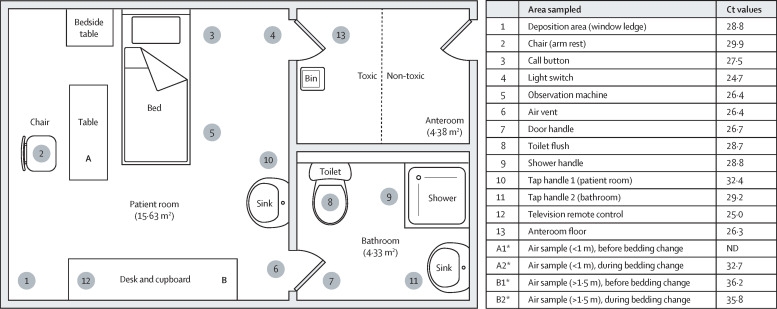
All seven patients had monkeypox lesions present on multiple areas of their bodies at the time of environmental sampling (between May 24 and June 17, 2022). Monkeypox virus DNA was detected in 56 (93%) of 60 surface swab samples obtained within patient bedrooms and bathrooms, with Ct values of 24·7–37·4 (table 1 ). These positive detections included samples from areas unlikely to have been directly touched by patients—such as the air vent above the door between the bedroom and the bathroom—suggesting non-contact contamination, possibly via respiratory droplets or suspension of particles in the air from changing bedding. Swabs of the anteroom floor were positive on the toxic side for all rooms, and on the non-toxic side of three rooms (table 1).
Table 1.
Clinical characteristics and results of surface and air environmental sampling in patients' rooms
| Room A (P1)* | Room A (P2) | Room C (P2) | Room D* | Room E* | ||
|---|---|---|---|---|---|---|
| Clinical characteristics | ||||||
| Date of sampling | May 24 | June 17 | June 16 | June 17 | June 16 | |
| Days since symptom onset | 9 | 30 | 6 | 26 | 7 | |
| Days since hospital admission | 2 | 7 | 7 | 18 | 3 | |
| Hours since room was cleaned | 6 | 3 | 3 | 5 | 5 | |
| Ct at hospital admission | ||||||
| Throat | 27 | Negative | 22 | 37 | 30 | |
| Lesion | 22 | 23 | 28 | 23 | 31 | |
| Plasma | 32 | 34 | 35 | Negative | 31 | |
| Days on tecovirimat | 2 | 4 | NR | NR | 3 | |
| Ct values of surfaces in patient room | ||||||
| Floor | NA | 26·9 | 30·9 | 34·9 | 32·5 | |
| Call button | 27·5 | 29·4 | 32·4 | Negative | 26·1 | |
| Light switch | 24·7 | 31·6 | 34·5 | 36·3 | 30·2 | |
| Television remote control | 25·0 | 28·9 | 37·4 | 32·2 | 28·2 | |
| Observation machine | 26·4 | NA | NA | NA | NA | |
| Tap handle 1 (patient room) | 32·4 | 34·2 | 35·6 | 36·7 | 27·1 | |
| Deposition area (window ledge) | 28·8 | 29·7 | Negative | 35·6 | 35·5 | |
| Chair (arm rest) | 29·9 | 33·5 | 33·8 | 31·6 | 24·9 | |
| Door handle (patient room to bathroom) | 26·7 | 33·3 | 32·6 | Negative | 28·1 | |
| Ct values of surfaces in bathroom | ||||||
| Vent or grille (room to bathroom) | 26·4 | 25·9 | 27·9 | 33·3 | 33·6 | |
| Toilet flush handle | 28·7 | 32·6 | 31·8 | 34·8 | 26·4 | |
| Shower handle | 28·8 | 33·5 | 34·0 | 33·8 | 32·7 | |
| Tap handle (bathroom) | 29·2 | 29·3 | 32·8 | Negative | 25·9 | |
| Ct values of surfaces in anteroom | ||||||
| Floor, toxic side | 26·3 | 28·7 | 33·2 | 32·9 | 30·6 | |
| Floor, non-toxic side | NA | 33·6 | Negative | 36·8 | 36·8 | |
| Ct values of surface in ward | ||||||
| Floor, corridor | NA | Negative | 37·5 | Negative | 36·7 | |
| Air sampling Ct values | ||||||
| Before bedding change | ||||||
| Near (<1m) | Negative | Negative | Negative | Negative | Negative | |
| Far (>1·5 m) | 36·2 | 36·5 | Negative | Negative | Negative | |
| During bedding change | ||||||
| Near (<1 m) | 32·7 | 36·2 | Negative | Negative | Negative | |
| Far (>1·5 m) | 35·8 | Negative | Negative | Negative | Negative | |
Details of environmental sampling performed in five patient rooms at the Royal Free Hospital (London, UK) on May 24–June 17, 2022. Air samples were collected using a Sortorius MD8 Airport at a rate of 50 L/min. Room A was sampled on two occasions, with different patients (P1 and P2) patients occupying this room on each visit. Rooms were cleaned every 12 h during occupancy using 5000 ppm available chlorine sodium hypochlorite on all hard surfaces and floors and 10 000 ppm available chlorine sodium hypochlorite for the toilet, shower, wash basins, and floors. A full room clean was done with 5000 ppm available chlorine sodium hypochlorite after patient discharge, followed by decontamination using vapourised hydrogen peroxide. Ct=quantitative PCR crossing threshold value of moneypox DNA detected. NA=not applicable (sample not taken for this room). NR=tecovirimat not received.
Denotes occupant of this room was the first patient admitted into this room with monkeypox.
Four of 12 samples collected from the PPE of health-care workers were positive, but all three samples taken from visors were negative. Monkeypox virus DNA was detected on two glove samples where swabbing included both palmar surface and fingertips, and was not detected on a sample collected by swabbing only the palmar surface of the gloves.
After the doffing of PPE by hospital staff, monkeypox virus DNA was detected on the floor of the anteroom where doffing took place (table 2 ). Monkeypox virus DNA was detected (Ct 38·2) in one air sample taken in the corridor before doffing, but not in other air samples taken before and during doffing in anterooms used for doffing. The volume of air filtered in each air sample collected in the anteroom and corridor was 250 litres (50 L/min for 5 min). Monkeypox virus DNA was not detected in samples from any of the three wearable air samplers. By contrast, five (63%) of eight air samples collected over a period of 10 min by the large volume air sampler (flow rate 50 L/min) in room A contained monkeypox virus DNA (table 1). These samples were collected in two sets about 3 weeks apart, with different patients in this room at the time each sampling was performed. These results include samples taken further away from the patient (>1·5 m). A lower Ct value was reported for the sample obtained during the bedding change than that of samples collected before the bedding change (figure ). Monkeypox virus DNA was not detected in air samples collected over a period of 5 min at the same distances before and during bedding changes in rooms C–E (table 1).
Table 2.
Clinical characteristics and results of sampling around PPE doffing procedure
| Room A (P1)* | Room B* | Room C (P1)* | ||
|---|---|---|---|---|
| Clinical characteristics | ||||
| Date of sampling | May 24 | May 25 | May 25 | |
| Days since symptom onset | 9 | 15 | 15 | |
| Days since hospital admission | 2 | 2 | 1 | |
| Ct at hospital admission | ||||
| Throat | 27 | 27 | 23 | |
| Lesion | 22 | 31 | 18 | |
| Plasma | 32 | 30 | 32 | |
| Days on tecovirimat | 2 | 2 | NA | |
| Surface sampling Ct values | ||||
| Gloves | 30·8 | 27·1 | Negative† | |
| Gown | Negative | 35·6 | 34·3 | |
| Visor | Negative | Negative | Negative | |
| Anteroom floor, post-doffing | 26·1 | 26·9 | 30·9 | |
| Air sampling Ct values | ||||
| Corridor, pre-doffing | Negative | Negative | 38·2 | |
| Corridor, during doffing | Negative | Negative | Negative | |
| Anteroom, pre-doffing | Negative | Negative | Negative | |
| Anteroom, during doffing | Negative | Negative | Negative | |
Environmental sampling performed around PPE doffing procedure at the Royal Free Hospital (London, UK) on May 24–25, 2022. Rooms A and C were both sampled on two occasions, with different patients (P1 and P2) occupying these rooms on each visit. PPE was swabbed after the health-care worker had changed bedding (room A) or after the health-care worker had conducted a clinical visit with direct patient contact (rooms B and C). Rooms were cleaned every 12 h (hard surfaces and floors) using 5000 ppm available chlorine sodium hypochlorite during occupancy. A full room clean with 5000 ppm available chlorine sodium hypochlorite was done after patient discharge, followed by decontamination using vapourised hydrogen peroxide. Ct=quantitative PCR crossing threshold value of monkeypox DNA detected. PPE=personal protective equipment.
The occupant of this room was the first patient admitted into this room with monkeypox.
By contrast to the other glove samples taken, only palmar surface and not fingertips were swabbed.
Figure.
Plan of room A representing the sites of samples and Ct values
*Air samples were collected over a period of 10 min at a rate of 50 L/min (500 L total). Ct=quantitative PCR crossing threshold value of monkeypox DNA detected.
Virus isolation was attempted for four samples: room A light switch (original Ct 24·7), room A anteroom floor after PPE doffing (original Ct 26·3), room A near-bed air sample during bedding change (original Ct 32·7), and room B anteroom floor after PPE doffing (original Ct 26·9). Although no flask showed marked cytopathic effect during the attempted viral culture in cells, viral DNA replication occurred for the room B anteroom floor sample after PPE doffing and the room A near-bed air sample during bedding change—with Ct values recorded as at least 31·0 cycles for all timepoints between day 0 and day 5, but with Ct values of less than 18·0 for day 10 indicating at least 100 000 times more detectable DNA between day 5 and day 7 (table 3 ). Only a subtle cytopathic effect was observed in the room B sample, with no obvious cytopathic effect for the room A air sample. The other two swab samples (room A anteroom floor and light switch), and the negative control, showed no increase in detectable DNA during the 10-day isolation.
Table 3.
Monkeypox virus Ct values at specified timepoints from viral isolation cultures
| No infection control | Room A (P1), bedding change air sample | Room A (P1), anteroom floor swab | Room A (P1), light switch swab | Room B, anteroom floor swab | |
|---|---|---|---|---|---|
| Environmental sample Ct | NA | 32·7 | 26·3 | 24·7 | 26·9 |
| Day 0, P0 | ND | ND | 33·5 | 32·2 | 32·2 |
| Day 3, P0 | ND | ND | 33·6 | 33·1 | ND |
| Day 5, P0 | ND | 35·5 | 36·3 | 38·1 | 31·4 |
| Day 7, P0 | ND | 27·6 | ND | 35·6 | 22·4 |
| Day 7, P1 (5+2) | ND | ND | ND | ND | 39·5 |
| Day 10, P1 (5+5) | ND | 17·5 | 36·2 | 36·5 | 14·9 |
| Cultured monkeypox virus? | No | Yes | No | No | Yes |
Ct values from viral isolation cultures of environmental samples taken at the Royal Free Hospital (London, UK) in May–June, 2022. No infection control denotes that a mock infection was used as a negative control. Ct=quantitative PCR crossing threshold value of monkeypox DNA detected. NA=not applicable. ND=not detected. P0=passage 0. P1=passage 1.
No procedures listed on the UK Health Security Agency or WHO list of medical aerosol-generating procedures were performed before sampling, for any patient. None of the patients in the rooms sampled had diarrhoea, vomiting, cough, sneezing, or respiratory distress, according to information provided by the attending clinicians and observations of the investigating team during sampling. A consistent bedding change technique was observed for all rooms where bedding was changed. No variability in cleaning practices was observed.
Discussion
We identified widespread monkeypox virus DNA contamination of the environment in respiratory isolation rooms occupied by individuals with symptomatic monkeypox. Monkeypox virus PCR Ct values obtained from these samples are within the range previously shown to be associated with recovery of infection-competent monkeypox virus.19 We found evidence of replication-competent virus from two samples: from an air sample collected during a bedding change and from a swab sample of an anteroom floor. Detection of infectious monkeypox virus in air samples collected during a bedding change highlights the importance of suitable respiratory protection equipment for health-care workers when performing activities that might suspend infectious material within contaminated environments. The variability we observed in the frequency of detection, and the Ct values observed in surface samples from different patient rooms, might be due to individual patient factors, the timepoint during patient infection at which environmental sampling was performed, staff or patient behaviour, and the recency of cleaning. No variability in cleaning practices was observed that could explain the differences in environmental sampling results seen. None of the patients' clinical characteristics explain why there were differences in air sampling results for different isolation rooms sampled, based on the clinical data available to us and the observations made by the investigating team during sampling.
Detection of virus, even replication-competent virus, in environmental samples does not mean that transmission leading to infection would necessarily occur if someone were exposed to that virus, as there are many factors that can influence successful infection of a human. These factors include routes of transmission, host susceptibility, environmental factors that could weaken the virus' ability to infect cells and replicate, and the amount of virus to which one is exposed. The infectious dose of monkeypox virus in humans is not known, and could vary according to the site of the body exposed to virus.
Contamination of PPE was also found following clinical contact with a patient (rooms B and C) or bedding (room A). The use of PPE by staff was consistent with the standardised approach followed by all NHS Airborne HCID Treatment Centres.20 Monkeypox virus DNA was detected on the glove samples where swabbing included both palmar surface and fingertips, and not detected on a sample collected only from the palmar surface. The detection of monkeypox virus DNA with relatively low Ct values (≤30 Ct) in hard-surface samples from the doffing environment reinforces the importance of surface cleaning protocols, the use of appropriate PPE, and robust doffing procedures to maintain the safety of staff and avoid potential onward transmission. Although our findings are specific to sampling in a specialist health-care environment and sampling within occupied rooms was restricted to a small number of patients (seven total), the environmental contamination findings could be relevant to public health measures for other spaces and settings where individuals with monkeypox spend prolonged periods, such as residential bedrooms and bathrooms. Further investigation is required into the contamination of areas occupied for shorter periods of time, such as outpatient clinics, and health-care spaces that do not have mechanical negative pressure ventilation. Previous investigations of surface contamination in a domestic setting and in two hospital rooms occupied by infected symptomatic individuals have demonstrated a high frequency of monkeypox viral DNA detection and isolation of virus.14, 19
Our study has limitations. The reallocation of laboratory space and personnel to clinical diagnostics was required for the public health response, which restricted our ability to obtain and process high numbers of environmental samples. In particular, only four samples were assessed for the presence of viable virus due to the time and space requirements. This study does not account for potential residual monkeypox virus DNA that could be present after room decontamination. Although three rooms were sampled during first occupation of that space, two were sampled during second occupation. All sampling techniques—especially the air sampling methodologies—involve substantial dilution of sampled material, which might result in a false-negative outcome for low-level contamination. It is not possible to relate the extent of contamination identified to absolute transmission risk, and the low number of nosocomial and household transmission events documented to date (especially in cases of delayed diagnosis), imply that the risk might not be substantial; however, the recommendations listed here to mitigate secondary transmission should be viewed as best practice where achievable.
To the best of our knowledge, this is the first time that detection of monkeypox virus (DNA and virus by isolation) in environmental air samples from health-care settings has been reported for any clade of monkeypox virus. Detection of monkeypox virus DNA in air samples collected at distances of greater than 1·5 m from the patient's bed and at a height of about 2 m supports the theory that monkeypox virus can be present in either aerosols or suspended skin particles or dust containing virus, and not only in large respiratory droplets that fall to the ground within 1–1·5 m of an infected individual. Although the low-flow-rate wearable button samplers provided negative results, they were used for less than 10 min, which might be an insufficient sampling time (<40 L of air sampled). Our study was not designed to show that environmental contamination of hospital rooms by monkeypox virus results in transmission events; however, our findings support precautionary recommendations21 for health-care workers interacting with hospitalised patients with confirmed monkeypox virus infection to use suitable PPE (including respiratory protective equipment) and other infection prevention and control measures designed to minimise exposure to pathogens that might become aerosolised in hospital inpatient settings.
Data sharing
Data will be made available on receipt of reasonable request to the corresponding author.
Declaration of interests
We declare no competing interests.
Acknowledgments
Acknowledgments
We gratefully thank the medical, nursing, and multidisciplinary staff supporting the Airborne HCID Treatment Centres at the Royal Free London NHS Foundation Trust, the Royal Liverpool University Hospital, the NHS England Airborne HCID Network, the UK Health Security Agency, and the Rare and Imported Pathogens Laboratory at UK Health Security Agency Porton Down. We also acknowledge assistance from Ian Nicholls and Wilhemina D'Costa (UK Health Security Agency) for nucleic acid extractions, and from Sian Summers (UK Health Security Agency) for providing cell culture flasks. SG, JD, and TF are supported by the National Institute for Health Research Health Protection Research Unit in Emerging and Zoonotic Infections. SG is also supported by the Research, Evidence, and Development Initiative, which is funded by UK aid from the UK government (project number 300342–104). The views expressed here do not necessarily reflect the UK government's official policies.
Acknowledgments
Contributors
JD, BA, TF, SG, and AB contributed to study design and research concept. BA, SG, JD, CT, and IM collected samples. BA, OO, AS, JF, and JG processed samples and analysed data. SG, BA, OO, AS, JF, CT, IM, AB, TF, and JD reviewed and edited the manuscript. SG, JD, BA, AB, and TF wrote the original draft of the manuscript. All authors had access to all underlying data in the study. BA, OO, AS, JF, JD, SG, AB, and TF accessed and verified the data.
Contributor Information
NHS England Airborne High Consequence Infectious Diseases Network:
Jake Dunning, Nicholas Price, Michael Beadsworth, Matthias Schmid, Marieke Emonts, Anne Tunbridge, David Porter, Jonathan Cohen, Elizabeth Whittaker, and Ruchi Sinha
Supplementary Material
References
- 1.US Centers for Disease Control and Prevention 2022 monkeypox outbreak global map. 2022. https://www.cdc.gov/poxvirus/monkeypox/response/2022/world-map.html
- 2.UK Health Security Agency Monkeypox outbreak: epidemiological overview. Sept 27, 2022. https://www.gov.uk/government/publications/monkeypox-outbreak-epidemiological-overview/monkeypox-outbreak-epidemiological-overview-27-september-2022
- 3.Cann JA, Jahrling PB, Hensley LE, Wahl-Jensen V. Comparative pathology of smallpox and monkeypox in man and macaques. J Comp Pathol. 2013;148:6–21. doi: 10.1016/j.jcpa.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO Multi-country monkeypox outbreak: situation update. June 10, 2022. https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON392
- 5.Rimoin AW, Mulembakani PM, Johnston SC, et al. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc Natl Acad Sci USA. 2010;107:16262–16267. doi: 10.1073/pnas.1005769107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reynolds MG, Doty JB, McCollum AM, Olson VA, Nakazawa Y. Monkeypox re-emergence in Africa: a call to expand the concept and practice of One Health. Expert Rev Anti Infect Ther. 2019;17:129–139. doi: 10.1080/14787210.2019.1567330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCollum AM, Damon IK, McCollum AM, Damon IK. Human monkeypox. Clin Infect Dis. 2014;58:260–267. doi: 10.1093/cid/cit703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunge EM, Hoet B, Chen L, et al. The changing epidemiology of human monkeypox—a potential threat? A systematic review. PLoS Negl Trop Dis. 2022;16 doi: 10.1371/journal.pntd.0010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnewall RE, Fisher DA, Robertson AB, Vales PA, Knostman KA, Bigger JE. Inhalational monkeypox virus infection in cynomolgus macaques. Front Cell Infect Microbiol. 2012;2:117. doi: 10.3389/fcimb.2012.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nalca A, Livingston VA, Garza NL, et al. Experimental infection of cynomolgus macaques (Macaca fascicularis) with aerosolized monkeypox virus. PLoS One. 2010;5:1–12. doi: 10.1371/journal.pone.0012880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verreault D, Killeen SZ, Redmann RK, Roy CJ. Susceptibility of monkeypox virus aerosol suspensions in a rotating chamber. J Virol Methods. 2013;187:333–337. doi: 10.1016/j.jviromet.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood JP, Choi YW, Wendling MQ, Rogers JV, Chappie DJ. Environmental persistence of vaccinia virus on materials. Lett Appl Microbiol. 2013;57:399–404. doi: 10.1111/lam.12126. [DOI] [PubMed] [Google Scholar]
- 13.Vaughan A, Aarons E, Astbury J, et al. Human-to-human transmission of monkeypox virus, United Kingdom, October 2018. Emerg Infect Dis. 2020;26:782–785. doi: 10.3201/eid2604.191164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nörz D, Pfefferle S, Brehm TT, et al. Evidence of surface contamination in hospital rooms occupied by patients infected with monkeypox, Germany, June 2022. Euro Surveill. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.26.2200477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WHO Second meeting of the International Health Regulations (2005) (IHR) Emergency Committee regarding the multi-country outbreak of monkeypox. July 23, 2022. https://www.who.int/news/item/23-07-2022-second-meeting-of-the-international-health-regulations-(2005)-(ihr)-emergency-committee-regarding-the-multi-country-outbreak-of-monkeypox
- 16.NHS England Health building note 04–01 supplement 1. May 21, 2021. https://www.england.nhs.uk/publication/adult-in-patient-facilities-planning-and-design-hbn-04-01/
- 17.International Severe Acute Respiratory and Emerging Infection Consortium Clinical characterisation protocol (CCP) 2020. https://isaric.org/research/covid-19-clinical-research-resources/clinical-characterisation-protocol-ccp/
- 18.Li Y, Zhao H, Wilkins K, Hughes C, Damon IK. Real-time PCR assays for the specific detection of monkeypox virus west African and Congo Basin strain DNA. J Virol Methods. 2010;169:223–227. doi: 10.1016/j.jviromet.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atkinson B, Burton C, Pottage T, et al. Infection-competent monkeypox virus contamination identified in domestic settings following an imported case of monkeypox into the UK. Environ Microbiol. 2022 doi: 10.1111/1462-2920.16129. published online July 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.NHS England NHS improvement. National infection prevention and control manual for England. April 14, 2022. https://www.england.nhs.uk/publication/national-infection-prevention-and-control/
- 21.UK Health Security Agency. Public Health Wales. Public Health Agency (Northern Ireland) Principles for monkeypox control in the UK: 4 nations consensus statement. May 30, 2022. https://www.gov.uk/government/publications/principles-for-monkeypox-control-in-the-uk-4-nations-consensus-statement
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on receipt of reasonable request to the corresponding author.