Significance
The faithful replication of eukaryotic genomes requires balancing the replication capacities of different genomic regions, such as repetitive versus single-copy genetic elements, which may compete for limiting replication resources, possibly leading to replication stress and genome instability. We examined the function of histone deacetylases Rpd3 and Sir2 in balancing replication between unique genome sequences and the multicopy ribosomal DNA genes. Our findings support prior conclusions that Sir2 directly suppresses early firing of rDNA origins, thereby enabling balanced replication of the genome. We further show that Rpd3’s function in delaying firing of later-firing, single-copy origins is independent of Sir2 and rDNA load. Instead, Rpd3 appears to oppose the Fkh1/2 origin activation pathway by regulating binding of the origin-stimulator Fkh1.
Keywords: Replication origin, Chromatin domains, Genome instability, Repetitive DNA, Histone deacetylase
Abstract
Eukaryotic chromosomes are organized into structural and functional domains with characteristic replication timings, which are thought to contribute to epigenetic programming and genome stability. Differential replication timing results from epigenetic mechanisms that positively and negatively regulate the competition for limiting replication initiation factors. Histone deacetylase Sir2 negatively regulates initiation of the multicopy (∼150) rDNA origins, while Rpd3 histone deacetylase negatively regulates firing of single-copy origins. However, Rpd3’s effect on single-copy origins might derive indirectly from a positive function for Rpd3 in rDNA origin firing shifting the competitive balance. Our quantitative experiments support the idea that origins compete for limiting factors; however, our results show that Rpd3’s effect on single-copy origin is independent of rDNA copy-number and of Sir2’s effects on rDNA origin firing. Whereas RPD3 deletion and SIR2 deletion alter the early S phase dynamics of single-copy and rDNA origin firings in opposite fashion, unexpectedly only RPD3 deletion suppresses overall rDNA origin efficiency across S phase. Increased origin activation in rpd3Δ requires Fkh1/2, suggesting that Rpd3 opposes Fkh1/2-origin stimulation, which involves recruitment of Dbf4-dependent kinase (DDK). Indeed, Fkh1 binding increases at Rpd3-regulated origins in rpd3Δ cells in G1, supporting a mechanism whereby Rpd3 influences initiation timing of single-copy origins directly through modulation of Fkh1-origin binding. Genetic suppression of a DBF4 hypomorphic mutation by RPD3 deletion further supports the conclusion that Rpd3 impedes DDK recruitment by Fkh1, revealing a mechanism of Rpd3 in origin regulation.
The replication of functionally distinct chromosomal regions presents challenges to genome stability. For example, repeated and highly expressed DNA sequences, such as the ribosomal DNA genes, appear to be destabilized by transcription–replication collisions and unequal sister chromatid exchange or intrachromosomal recombination (reviewed in 1). In Saccharomyces cerevisiae, the ribosomal DNA (rDNA) gene cluster consists of about 150 direct repeats of the ∼9.1-kbp rDNA sequence, each containing one potential origin (reviewed in 2), which represents a significantly higher origin density than the genome average among unique sequences (reviewed in 3). Thus, rDNA origins have the potential to impose a significant load on the replication system and impede the replication of other chromosomal regions, as several replication-initiation factors are present in limiting quantities (reviewed in 4). However, only ∼20% of the rDNA origins fire in each S phase in wild-type (WT) cells (5, 6), and this has been attributed to an intrinsically weak autonomously replicating sequence (ARS) (7, 8), and to suppression of origin firing by the histone deacetylase Sir2 (9). Deletion of SIR2 results in a higher proportion of the rDNA origins initiating replication, which has been suggested to contribute to instability of the rDNA repeats. Histone deacetylase Rpd3, in contrast, has been implicated in the delayed initiation of late-firing, single-copy origins dispersed throughout the genome. RPD3 deletion advances the firing of many late origins while increasing the levels of histone acetylation at these origins (10–12). Targeting of Gcn5 histone acetylase to a late replication origin advanced its timing, supporting the idea that histone acetylation regulates initiation rate (12). However, specific DNA-binding, protein-mediated recruitment of Rpd3 to origins has not been demonstrated (nor ruled out), so Rpd3 has been suggested to act directly at single-copy origins through an untargeted (or “global”) interaction mechanism (10, 12).
More recently, however, this model of Rpd3 function has been challenged by a study claiming that Rpd3 acts directly to stimulate rDNA origins and that effects of RPD3 deletion on single-copy origins are an indirect consequence of reduced competition from rDNA origins for a limited pool of replication-initiation factors (13). Indeed, recent studies have identified several initiation factors as rate limiting for origin firing, leading to models in which unequal competition for limiting factors among all licensed origins underlies their differential activities (14–16). In the Yoshida study, deletion of SIR2 increased early firing of rDNA origins while decreasing early firing of single-copy origins, consistent with the competition model. Additional deletion of RPD3 suppressed the effects of SIR2 deletion, consistent with these factors acting in opposition at the rDNA. Thus, it was concluded that advanced firing of single-copy origins in rpd3Δ cells is a consequence of reduced Rpd3-stimulated rDNA origin firing competing for limiting factors, rather than a direct repressive effect of Rpd3 on single-copy origins. A previous study implicating Rpd3 in transcriptional silencing of the rDNA was also cited as supporting evidence for the idea that Rpd3 acts directly on rDNA origins (17).
The main conclusions of the earlier study relied on quantitative comparisons of independently generated bromodeoxyuridine-immunoprecipitation–sequencing (BrdU-IP-seq) datasets with WT, sir2Δ, rpd3Δ, and sir2Δ rpd3Δ strains. Such comparisons require signal normalization between the independently produced experimental datasets, which can be problematic, typically requiring arbitrary, if not dubious, assumptions about the data. More recently, we developed quantitative BrdU-seq (qBrdU-seq) (or “QBU”) to overcome this barrier to an unambiguous, quantitative comparison of independent BrdU-IP-seq samples (18). Our method involves sample-specific barcoding of BrdU-labeled genomic DNA, followed by pooling for IP, amplification, and high-throughput sequencing. Additionally, prior to IP, a small aliquot of the pooled “input” sample is retained for amplification and sequencing in parallel, providing a direct reference for normalization of IP sequencing read counts based on actual input. QBU eliminates and/or corrects for most sources of technical or experimental error that may hinder (or at least cast doubt upon) quantitative comparison between individually processed samples.
In this study, we have re-examined the impacts of Rpd3 and Sir2 on genome replication, with particular focus on the origin competition model and Rpd3 function at single-copy versus rDNA origins. Our findings support the idea of competition between rDNA and single-copy origins, but not the notion that Rpd3 acts positively through rDNA origins to squelch single-copy origins. We have also probed more deeply into the mechanism of Rpd3 action at origins and present evidence that Rpd3 antagonizes single-copy origin firing by regulating Fkh1 binding and, hence, recruitment of Dbf4-dependent kinase (DDK) to origins. In accord, deletion of RPD3 rescues defective DDK function.
Results
Rpd3 Has Differential Effects on Single-Copy Origins.
A rigorous test of the competition model between rDNA and single-copy origins and of the roles of Rpd3 and Sir2 in regulating these origins requires an unambiguous, quantitative comparison of origin firing levels. Therefore, we performed QBU analysis of WT, rpd3Δ, sir2Δ, and sir2Δ rpd3Δ strains. Cells were synchronized in G1 phase with α-factor and released into S phase in the presence of hydroxyurea (HU) to block replication and distinguish between early origins, which fire efficiently in HU, and late origins, which do not due to intra-S checkpoint inhibition (19). DNA content analysis of cells similarly synchronized and released into S phase without HU shows similar overall timing of S phase execution for the four strains, with rpd3Δ cells completing S phase slightly more rapidly than WT, as observed previously (SI Appendix, Fig. S1A) (12).
Distribution of the QBU values comparing the individual replicates of each strain shows high correlations, demonstrating the high experimental reproducibility of the QBU analysis (SI Appendix, Fig. S1B). Chromosomal plots of the QBU data show qualitatively similar results as observed previously in comparison of WT and rpd3Δ cells, with increased BrdU incorporation at several single-copy, later-firing origins in rpd3Δ cells (Fig. 1A). We determined the origin loci with significant BrdU incorporation and identified 235 in WT cells and 301 in rpd3Δ cells; a Venn diagram shows that almost all the origins identified in WT cells were also identified in rpd3Δ cells (Fig. 1B). Origins were assigned to timing quartiles based on their previously determined replication timings (TRep) among 625 confirmed or likely origin in the oriDB (20) and displayed as stack graphs of origin counts and pie charts of origin proportions (Fig. 1C). These results show that almost all of the additional origins detected to fire in rpd3Δ cells fall into later (Q2, Q3, and Q4) timing quartiles (in WT cells), consistent with Rpd3 acting to delay origin firing.
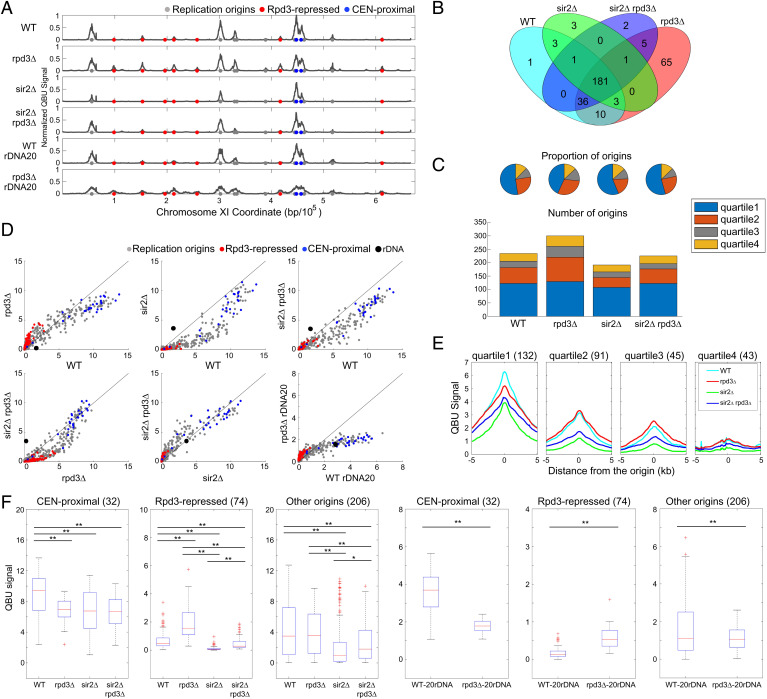
Fig. 1.
Rpd3 and Sir2 have different effects on single-copy origins. Strains CVy43 (WT), CVy44 (rpd3Δ), YHy3 (sir2Δ), and YHy6 (sir2Δ rpd3Δ) were synchronized in G1 phase and released into S phase without HU for DNA content analysis (SI Appendix, Fig. S1A) or with HU for 60 min for QBU analysis. Strains with reduced rDNA copies PP1758 (WT rDNA20) and JPy115 (rpd3Δ rDNA20) were synchronized in G1 phase and released into S phase for 120 min in the presence of HU for QBU analysis. (A) QBU values averaged for three replicates and scale normalized across strains are shown for representative chromosome XI; origins and origin subgroups are indicated with color-coded circles below the x axis. (B) Venn diagrams depicting overlaps between origins identified as active in each strain; the union of 311 origins detected was used in subsequent analyses. (C) Origin distributions according to TRep quartiles shown as pie charts with proportions of identified origins in each strain and as stack graphs showing total counts of origins identified in each TRep quartile. (D) 2D scatter plots comparing QBU signals for 500-bp regions centered on 311 individual origins plus the rDNA origins, which are represented by two (overlapping) data points; origins and subgroups are color coded as indicated. (E) Average QBU signals for 311 origins according to their TRep quartile assignments. (F) Box plot distributions of QBU counts for 500-bp regions aligned on origins of the indicated subgroups; n of origins in each group is indicated within parentheses; two-sided t tests were performed on all pairs of strains, and significant results are indicated as *P < 0.05 and **P < 0.01.
As BrdU signals at origins in HU-blocked cells inversely correlate with TRep (11), we also examined the relationship between the effect of RPD3 deletion and origin timing by plotting QBU signals for individual origins in rpd3Δ cells against their corresponding values in WT cells (Fig. 1D). The two-dimensional scatter plot reveals a relationship between WT BrdU levels and the effect of RPD3, with origins having higher QBU values in WT being decreased in rpd3Δ cells and origins having lower QBU values in WT being increased in rpd3Δ cells. To directly examine the relationship between the QBU values and origin timing, we plotted average QBU values for origins divided according to their assigned TRep quartiles (Fig. 1E). The results show that, in rpd3Δ cells, QBU values are lower for origins in the earliest TRep quartile, while QBU values are slightly higher for origins in the later quartiles, although only the difference in the first quartile is statistically significant (SI Appendix, Fig. S1C). We also examined the effects of RPD3 deletion on specific origin classes by highlighting these in the scatter plots (Fig. 1D) and by producing distribution box plots for those groups (Fig. 1F); t tests of the distributions show that centromere (CEN)-proximal origins, which are early firing, are significantly decreased in activity, while Rpd3-repressed origins, which are later firing, show significantly increased replication activity in rpd3Δ cells (Fig. 1F). The relatively minor but significant reduction in early origin firing in rpd3Δ cells has not been previously reported, likely because normalization methods obscured the difference. The opposite effect on earlier versus later origins is consistent with the idea that origins compete for limiting factors and that increased competition from normally later-firing origins in rpd3Δ cells consequently reduces activation of normally earlier-firing origins.
In contrast to the results in rpd3Δ cells, the replication profiles of sir2Δ cells showed decreased BrdU incorporation at virtually all single-copy origins (Fig. 1 A, D–F), though the effect was smaller among origins with higher BrdU incorporation values (Fig. 1D). Only 192 origins were called as active in sir2Δ cells, and there was a decrease in origins called in each TRep quartile (Fig. 1C). Overall, these results suggest that Sir2 acts nonspecifically with respect to single-copy origins because almost all single-copy origins are decreased in activity.
Combined deletion of SIR2 and RPD3 resulted in a replication profile intermediate to the single-mutant strains, and similar to WT (Fig. 1 A and D), as supported by correlation analysis (SI Appendix, Fig. S1D). Two hundred twenty-six origins were detected in rpd3Δ sir2Δ cells with almost all these origins also being detected in WT cells and having a similar TRep distribution as in WT cells (Fig. 1 B and C). Overall, QBU signals at individual origins were also more like WT than either single mutant, although more origins were reduced in activity as in sir2Δ cells than increased in activity as in rpd3Δ cells (Fig. 1 D and F and SI Appendix, Fig. S1D). Comparison of QBU origin signals between rpd3Δ sir2Δ and sir2Δ cells and between rpd3Δ sir2Δ and rpd3Δ cells shows that RPD3 deletion in sir2Δ cells and SIR2 deletion in rpd3Δ cells affect origins similarly as they do in WT cells, consistent with RPD3 and SIR2 acting through independent mechanisms (Fig. 1 D and F and SI Appendix, Fig. S1D).
SIR2 Deletion Is Epistatic to RPD3 Deletion for rDNA Origin Firing.
Next, we examined the effects of RPD3 and SIR2 deletion on initiation levels of rDNA origins. In WT cells, the QBU signal for rDNA origins (QBU corrects for copy-number so the signal represents the average firing level per rDNA copy) was between that of single-copy origins in the first and second timing quartiles (Fig. 2A, compare with Fig. 1E), consistent with the average mid-S phase peak replication timing of these origins (18). In rpd3Δ cells, the rDNA QBU signal was strongly diminished (Fig. 2 A and B). In contrast, the rDNA QBU signal in sir2Δ cells was about twice the level in WT cells (Fig. 2 A and B), in rough agreement with previous reports on the effect of SIR2 deletion on rDNA origin firing levels (9, 13). Unlike the additive effect of double deletion of RPD3 and SIR2 on single-copy origins, the rDNA QBU signal in rpd3Δ sir2Δ was almost identical to the sir2Δ cells (Fig. 2 A and B). This result suggests that SIR2 deletion is epistatic to, not additive with, RPD3 deletion with respect to rDNA origin firing, differing from previous conclusion (13). Whereas the significantly reduced early firing of rDNA origins in rpd3Δ cells is consistent with the idea that Rpd3 is a direct stimulator of rDNA origin firing, the high level of rDNA origin firing in sir2Δ rpd3Δ cells conflicts with this idea. Moreover, the effect of RPD3 deletion on single-copy origins in rpd3Δ sir2Δ cells (Fig. 1D) occurred independently of any effect on rDNA origin firing (Fig. 2B), suggesting a direct effect of Rpd3 on single-copy origins.
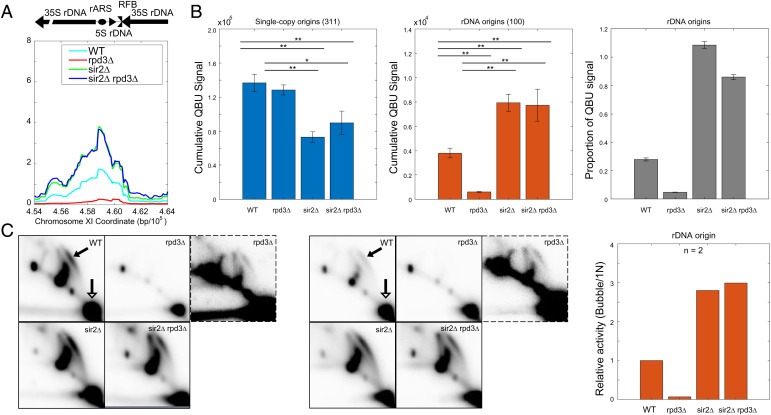
Fig. 2.
SIR2 deletion is epistatic to RPD3 deletion for rDNA origin firing. See Fig. 1 legend for strains and experimental description. (A) Average QBU signal for the rDNA depicted as a single locus. (B) Cumulative (+/−SD) QBU signal (500-bp windows) for 311 single-copy origins (Left panel) and 100 rDNA origins (Center panel) was determined and plotted; two-sided t tests were performed on all pairs of strains, and significant results are indicated as *P < 0.05 and **P < 0.01. Total QBU signal associated with rDNA origins was divided by total QBU signal associated with 311 single-copy origins plus the rDNA origins (Right panel). (C) 2D gel electrophoresis analysis probing for the rDNA was performed on the strains as described above. Two replicate sets are shown; the images framed with broken lines are darker images of the corresponding rpd3Δ image in each set. The arrows in the WT panels indicate the bubble arcs (filled arrowhead) and 1N spots (unfilled arrowhead) that are used for quantification (see SI Appendix, Fig. S2B for images with exact areas measured for quantification). The graph indicates quantification of the 2D gels by measuring intensity of “bubble arcs” in relation to the corresponding 1N spots; values are relative to WT arbitrarily set to 1.
We also quantified QBU signal from single-copy origins versus rDNA origins, after confirming that all four strains had similar rDNA copy counts, estimated at ∼100 in WT (SI Appendix, Fig. S2A). BrdU incorporation into single-copy origins was significantly lower in the sir2Δ and sir2Δ rpd3Δ strains versus WT, while rpd3Δ cells were like WT (Fig. 2B). We also calculated the absolute proportion of rDNA origin activity as a proportion of total genome origin activity for each strain, which shows the substantial shifts in balance between replication of the rDNA in relation to the genome resulting from deletion(s) of RPD3 and SIR2 (Fig. 2B, Right panel). Overall, the finding that RPD3 deletion in sir2Δ cells affects single-copy (Rpd3-repressed) origins without affecting rDNA origins argues against the idea that the effect of RPD3 deletion on single-copy origin firing derives indirectly from diminished rDNA origin firing.
To confirm that the results of QBU at the rDNA accurately reflect origin initiation levels, we used two-dimensional agarose gel (2D gel) electrophoresis to directly examine replication initiation structures at rDNA origins. As above, cells were synchronized in G1 phase with α-factor and released into S phase in the presence of HU to evaluate early replication events. Comparison of the intensity of replication “bubble arcs” representing initiation structures versus the 1N spots representing total molecules yields a relative measurement of origin firing activity (Fig. 2C and SI Appendix, Fig. S2B). The 2D gel results mirror the QBU results with rDNA origin firing decreased significantly in rpd3Δ cells, increased significantly in sir2Δ cells, and similarly increased in sir2Δ rpd3Δ cells (Fig. 2C and SI Appendix, Fig. S2B). These results reinforce the conclusion that SIR2 deletion is epistatic to, not additive with, RPD3 deletion with respect to rDNA origin activation.
RPD3 Regulates Single-Copy Origins Independently of the rDNA Array.
Yoshida et al. elegantly tested their model of competition between rDNA and single-copy origins by evaluating the effect of a reduction in rDNA copies on the suppression of single-copy origin firing resulting from SIR2 deletion, the premise being that fewer rDNA origins would create less competition. In support of the competition model and a role for Sir2 in regulating the rDNA directly and single-copy origin indirectly, they showed that, in a strain with only ∼20 copies of rDNA, the effect of SIR2 deletion on single-copy origins was largely suppressed, as predicted. We adopted the same rationale and approach to test whether Rpd3 acts on single-copy origins through effects on the rDNA by evaluating the effect of RPD3 deletion in the strain background with the reduced rDNA copies; we confirmed that the strains with reduced rDNA copies contained about 25% of the rDNA complement as the normal copy strains (SI Appendix, Fig. S2A). Chromosome plots show that similar derepression of dormant/late-firing origins occurred in rpd3Δ cells compared with WT cells in the strains with ∼20 rDNA copies (Fig. 1A). Analysis of QBU values for individual origins in the scatter plots revealed a similar relationship of RPD3 deletion on origins according to their QBU levels, with the greatest increases among those origins that normally have low QBU values, reflecting later-firing origins (Fig. 1D). Distribution box plots of QBU signals affirm the global increase in firing of Rpd3-repressed origins in rpd3Δ cells relative to WT (Fig. 1F), and correlation analysis demonstrates closer similarity of the rpd3Δ strain with reduced rDNA copies to the rpd3Δ strain with normal rDNA than the WT strain with normal rDNA (SI Appendix, Fig. S1D). Notably, rDNA origin activity level was only modestly reduced relative to WT in the rpd3Δ rDNA 20 strains. These results indicate that the effect of RPD3 deletion on single-copy origins does not rely on repression of a large array of rDNA origins. Instead, the findings support the conclusion that Rpd3 regulates origin firing directly at single-copy origins, rather than indirectly through the rDNA.
RPD3 Suppresses Pan-S Phase rDNA Origin Efficiency.
The analyses above were all conducted in the presence of HU to evaluate the levels of origin firing occurring in an early S phase interval. To determine the impacts of RPD3 and SIR2 on the overall efficiency of rDNA origin firing throughout the duration of S phase, we used 2D gels to examine and quantify replication intermediates at the rDNA origins in unperturbed cycling cells (pan-S phase); here, we compared the intensities of initiation bubble structures with replication fork structures to provide a relative measure of origin efficiency (Fig. 3A and SI Appendix, Fig. S3A). In striking contrast to the 2D gel results in early S phase (HU) described above, pan-S phase rpd3Δ cells exhibited modest but significantly increased overall efficiency of rDNA origin firing relative to WT cells (Fig. 3A). The overall increased firing of rDNA origins in rpd3Δ cells is inconsistent with the notion that RPD3 is a direct activator of rDNA origins. Interestingly, sir2Δ cells showed no difference in pan-S phase rDNA origin efficiency compared with WT (Fig. 3A), which contrasts with a previous report (9). Moreover, rpd3Δ sir2Δ cells showed similar pan-S phase rDNA origin efficiency as sir2Δ and WT cells (Fig. 3A). Thus, the striking, and opposite, effects of SIR2 deletion and RPD3 deletion on rDNA origin firing levels in early S phase are not reflected in their overall efficiencies through the duration of S phase.
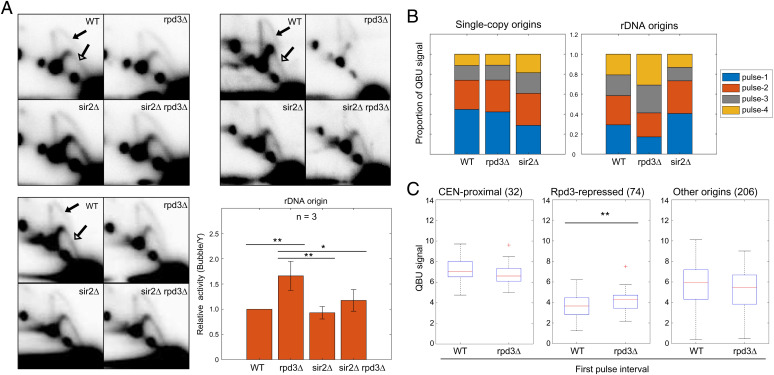
Fig. 3.
Rpd3 suppresses rDNA origin firing. (A) Strains described in Fig. 1 legend were grown logarithmically and harvested for 2D gel analysis and probing for the rDNA. Three replicate sets are shown. The arrows in the WT panels indicate the bubble arcs (filled arrowhead) and early Y-arcs (unfilled arrowhead) that are used for quantification (see SI Appendix, Fig. S3A for images with exact areas measured for quantification). The graph shows quantification of the 2D gels by measuring intensity of bubble arcs in relation to the corresponding Y-arcs (replication fork structures); values are relative to WT arbitrarily set to 1; two-sided t tests were performed on all pairs of strains, and results are indicated as *P < 0.05 and **P < 0.01. (B) WT, rpd3Δ, and sir2 strains described above were synchronized in G1 phase, released into S phase, and incubated with BrdU for the following time intervals for QBU analysis: pulse 1 = 15–30 min, pulse 2 = 25–40 min, pulse 3 = 35–50 min, and pulse 4 = 45–60 min; 5-min overlap is included to account for lag time in BrdU entry to cells and incorporation into DNA. The stack graphs show total QBU signal for 625 potential single-copy origins (Left) and total QBU signal for all rDNA origins during each time interval. (C) Box plot distributions of QBU signals (500-bp regions) in the first pulse interval of the time course for the indicated origin groups.
The differences in origin firing in the early-S versus pan-S phase analysis, particularly in rpd3Δ cells, suggests that alterations in rDNA origin usage early are subsequently compensated. For example, more early-firing of normally later-firing, single-copy, Rpd3-regulated origins should leave fewer such origins unfired in late S phase in competition with unfired rDNA origins, thus shifting the competitive balance. To more precisely examine the temporal dynamics of replication in the rDNA versus single-copy origins in rpd3Δ cells, we analyzed BrdU incorporation through a time series of pulse labeling in cells progressing synchronously through S phase. This analysis also eliminates possible perturbations due to HU. The analysis shows relative levels of BrdU incorporation at single-copy versus rDNA origins over the time course (Fig. 3B). Replication origin usage profiles are similar in the initial time pulse as in HU (SI Appendix, Fig. S3B). As expected, rDNA replication occurs throughout S phase in WT cells. In rpd3Δ cells, the relative proportions of replication at rDNA versus single-copy origins shift in favor of single-copy origins in earlier pulse periods and, conversely, in favor of rDNA origins in the later periods (Fig. 3B). The results are consistent with the idea that reduced usage of rDNA origins in early S phase preserves more rDNA origins to initiate later, likely with fewer unfired, single-copy origins in competition. Analysis of single-copy origin firing in the earliest interval shows significantly increased activity of Rpd3-repressed origins, showing that the effect of RPD3 deletion is similar in early S phase with or without HU (Fig. 3C).
Although deletion of SIR2 had no effect on overall origin efficiency in pan-S phase analysis, examination of BrdU incorporation over time shows a striking shift toward earlier replication of the rDNA in sir2Δ compared with WT cells (Fig. 3B), consistent with the early S phase results in HU. SIR2 deletion also results in notably delayed replication of the total genome, while RPD3 deletion’s effect on the genome is minor and in the same direction as its effect on the rDNA in the earliest pulse (Fig. 3B). We also note that this type of global BrdU incorporation analysis does not distinguish active versus passive replication of origin sequences.
Origin Derepression in rpd3Δ Cells Requires Fkh1/2.
FKH1 and/or FKH2 (FKH1/2) are responsible for the early activation of most non-CEN-proximal, early-firing origins throughout the genome (21). Fkh1 promotes early origin firing by recruiting limiting factor Dbf4 to stimulate execution of the DDK-dependent Sld3-Cdc45 loading step of replication initiation (22, 23). Overexpression of Fkh1 stimulates earlier firing of many later-firing origins, including Rpd3-repressed origins (SI Appendix, Fig. S4A) (18), suggesting an antagonistic relationship between Fkh1 and Rpd3. Because Fkh1 levels can modulate firing of Rpd3-repressed origins, we tested whether Fkh1 protein level was altered in rpd3Δ cells and found no difference relative to WT (SI Appendix, Fig. S4B). To gain further insight into the relationship between Rpd3 and Fkh1/2 function in replication timing, we tested for genetic interaction by determining the effect of their combined mutation. We performed QBU with WT, rpd3Δ, fkh1Δ fkh2-dsm, and rpd3Δ fkh1Δ fkh2-dsm strains; the fkh2-dsm allele was used rather than fkh2Δ because it is virtually null in replication timing control but functional in regulating gene expression and, therefore, does not, in combination with FKH1 deletion, exhibit more-severe phenotypes of fkh1Δ fkh2Δ cells, such as pseudohyphal growth, which complicates analysis and potential interpretation (24). QBU values comparing the individual replicates of each strain are well correlated (SI Appendix, Fig. S4C).
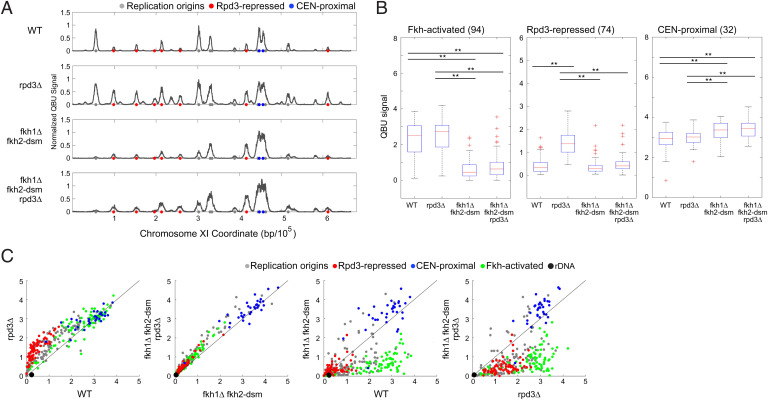
The results show that derepression of Rpd3-repressed origins in rpd3Δ cells was largely eliminated in rpd3Δ fkh1Δ fkh2-dsm cells (Fig. 4). Analysis of Rpd3-repressed origins shows no significant increase in BrdU incorporation in rpd3Δ fkh1Δ fkh2-dsm versus fkh1Δ fkh2-dsm cells (Fig. 4B). This finding suggests that Rpd3 opposes origin stimulation by Fkh1/2. As origin stimulation by Fkh1/2 involves its direct binding to DNA sequences at origins, we posited that repression by Rpd3 likely involves direct effect on the origin.
Fig. 4.
Origin derepression in rpd3Δ cells requires Fkh1/2. Strains CVy43 (WT), CVy44 (rpd3Δ), OAy1114 (fkh1Δ fkh2-dsm), and YHy13 (fkh1Δ fkh2-dsm rpd3Δ) were synchronized in G1 phase and released into S phase with HU for 90 min for QBU analysis. (A) Average QBU values are shown for representative chromosome XI; origins and origin subgroups are indicated with color-coded circles below the x axis. (B) Box plot distributions of QBU counts for 500-bp regions aligned on origins of the indicated subgroups; n of origins in each group is indicated within parentheses; two-sided t tests were performed on all pairs of strains, and results are indicated as *P < 0.05 and **P < 0.01. (C) 2D scatter plots comparing QBU signals centered on 311 individual origins plus the rDNA origins represented by two (overlapping) data points; origins and subgroups are color coded as indicated.
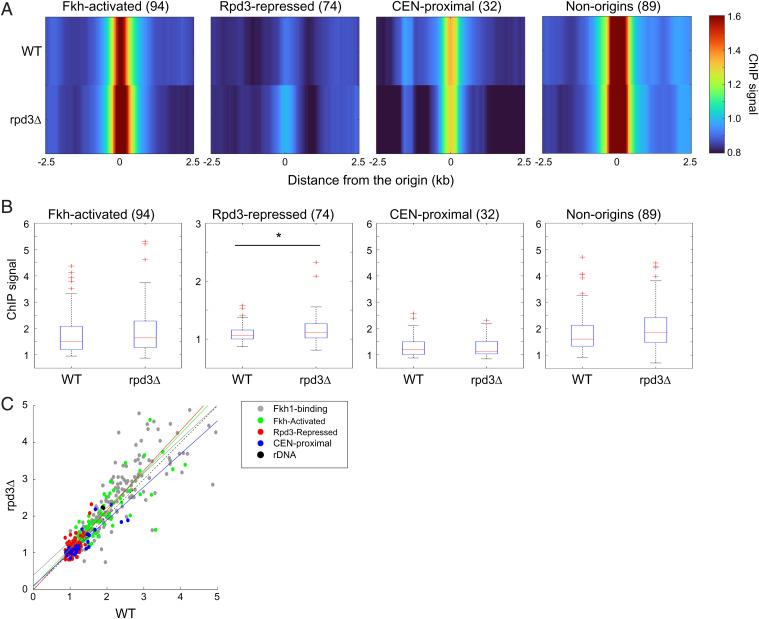
Rpd3 Modulates Fkh1 Binding to Origins.
Given the opposing activities of Fkh1/2 and Rpd3 in origin activation, we considered the possibility that Rpd3 might regulate binding of Fkh1/2 to origins, particularly Rpd3-regulated origins, including the possibility of differential regulation of Fkh1/2 binding to rDNA origins versus single-copy origins. Our previous studies have shown Fkh1/2 binding to Fkh-activated origins in G1 phase (25). In that study, we also observed more-extensive origin binding in S-phase-arrested cells, suggesting that Fkh1/2 may bind additional origins during S phase to stimulate activation of later origins. Therefore, we hypothesized that Rpd3 regulates Fkh1 binding to later-firing origins in G1 and/or early S phase and performed chromatin immunoprecipitation sequencing (ChIP-seq) of Fkh1-3xFLAG in WT and rpd3Δ cells synchronized in G1 to test this idea. Comparison of ChIP signal from experimental replicates shows high correlation, demonstrating high reproducibility of the data (SI Appendix, Fig. S5A). In WT cells in G1, heatmaps of average ChIP signal show local Fkh1 enrichment at previously identified Fkh1 binding sites, including Fkh-activated origins, as expected, as well as some enrichment at Rpd3-repressed origins, previously unnoticed (Fig. 5A). In rpd3Δ cells in G1, Fkh1 profiles are similar; however, box plot analysis and statistical testing show there is a modest but significant increase in Fkh1 enrichment at Rpd3-repressed origins, but not at other origin groups or Fkh1 binding sites more generally (Fig. 5 A and B). Regression analysis of the ChIP enrichment values for Rpd3-repressed origin correlates with the increased activation of Rpd3-repressed origins in early S phase in rpd3Δ cells (Fig. 5C). These results are consistent with the idea that the mechanism of Rpd3 in regulation of origin initiation is to oppose origin binding of Fkh1.
Fig. 5.
Rpd3 modulates Fkh1 binding to origins. Strains OAy1112 (FKH1-3xFLAG) and YHy17 (rpd3Δ, FKH1-3xFLAG) were synchronized in G1 phase and subjected to ChIP-seq. (A) Heatmaps of ChIP-seq enrichment at selected origin and Fkh1 binding loci; n of each group is indicated within parentheses. The “non-origins” group refers to Fkh1-binding sites called as peaks by MACS in the Fkh1 ChIP-seq dataset that do not overlap with a replication origin. (B) Box plots distributions of ChIP-seq enrichments for 500-bp regions aligned on origins of the indicated subgroups; n of origins or Fkh1 binding loci in each group is indicated within parentheses. Two-sided t tests were performed on all pairs of strains, and results are indicated as *P < 0.05 and **P < 0.01. (C) 2D scatter plots of ChIP-seq signals (500-bp regions) for 174 Fkh1-binding sites (called as peaks by MACS as described above but including origins) plus the indicated origin subgroups, color coded as indicated. A linear regression deriving the best-fit line is shown for each origin group.
Fkh1 binding at origins occurs in G1 and S phases and is dependent on the prereplication complex (pre-RC), defined by origin-loaded MCM complexes (25, 26). Thus, a plausible mechanism for the increased Fkh1 binding to Rpd3-repressed origins in rpd3Δ cells might derive from an effect of Rpd3 on pre-RC levels. To address this possibility, we performed ChIP-seq of Mcm4-3xHA in WT and rpd3Δ cells synchronized in G1. The results show robust enrichment for Mcm4 at origins of various classes, as expected; however, no significant difference in Mcm4 enrichment is detected at Rpd3-repressed origins or at the other groups examined (SI Appendix, Fig. S5). Taken together, these results indicate that Rpd3 modulates Fkh1 binding without altering pre-RC levels and are consistent with the idea that Rpd3 acts downstream of pre-RC assembly to impede Fkh1 binding.
RPD3 Deletion Suppresses DBF4 Deficiency Implicating the Fkh1-DDK Pathway.
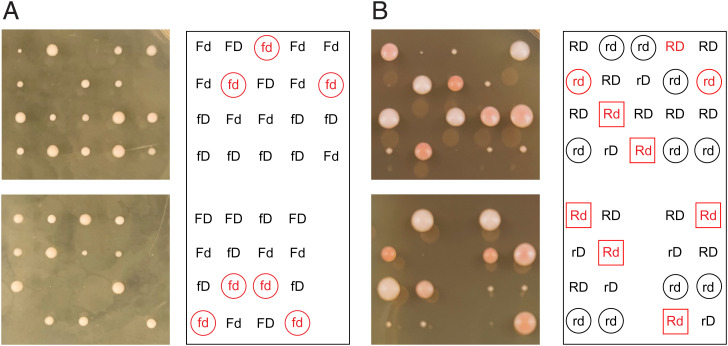
As a further test of our hypothesis that Rpd3 acts in opposition to Fkh1/2-dependent stimulation of Dbf4 recruitment, we tested whether deletion of RPD3 suppresses the temperature sensitivity of a carboxyl-terminally truncated DBF4 allele (dbf4ΔC) (27). Deletion of FKH1 (or FKH1 and FKH2) does not cause temperature sensitivity, indicating that Dbf4ΔC is defective in function beyond its proposed defect in interaction with Fkh1. In attempting to isolate a fkh1Δ dbf4ΔC strain through meiosis and sporulation of a heterozygous diploid, we determined that FKH1 deletion causes lethality in combination with dbf4ΔC (at normally permissive temperature for dbf4ΔC), strongly suggesting that Dbf4ΔC depends on Fkh1 for recruitment to origins (Fig. 6A). Thus, we created a diploid heterozygous for RPD3/rpd3Δ and DBF4/dbf4ΔC and induced meiosis and sporulation at permissive temperature. Individual spores from mature tetrads were dissected onto rich medium, incubated at 30 °C (restrictive temperature for dbf4ΔC), and imaged and genotyped. The results show that RPD3 deletion suppresses the lethality of dbf4ΔC at 30 °C, as virtually no dbf4ΔC segregants were recovered while the expected quantity of rpd3Δ dbf4ΔC segregants were recovered, although these formed much smaller colonies than WT or rpd3Δ isolates (Fig. 6B). Out of 13 tetrads dissected, 37 viable spores were genotyped, resulting in 15 WT, 9 rpd3Δ, 12 rpd3Δ dbf4ΔC, and 1 dbf4ΔC viable isolates. Fisher’s exact test indicates that the RPD3 and DBF4 genotypes being tested are not independent (P = 0.00164), strongly supporting the notion that loss of Rpd3 function restores viability of Dbf4-deficient cells, likely by facilitating Dbf4ΔC recruitment to origins by Fkh1.
Fig. 6.
RPD3 deletion suppresses DBF4 deficiency. (A) Diploid strain YHy38 (FKH1/fkh1Δ DBF4/dbf4ΔC) was induced into meiosis and sporulation. Tetrads were dissected onto rich medium and incubated at 23 °C and imaged after 3 days and genotyped. Tetrads are arranged in columns; determined haploid genotypes are indicated on the panel to the right as F/f for FKH1/fkh1Δ and D/d for DBF4/dbf4ΔC; FKH1 and DBF4 are unlinked. Genotypes in red for inviable spores were inferred based on assumption of 2:2 segregation of alleles. The “fd” hypotheticals are circled. (B) Diploid strain OAy1188 (RPD3/rpd3Δ DBF4/dbf4ΔC) was induced into meiosis and sporulation. Tetrads were dissected onto rich medium and incubated at 30 °C and imaged and genotyped after 3 days. Tetrads are arranged in columns; determined haploid genotypes are indicated on the panel to the right as R/r for RPD3/rpd3Δ and D/d for DBF4/dbf4ΔC; RPD3 and DBF4 are unlinked. Genotypes in red for inviable spores were inferred based on assumption of 2:2 segregation of alleles. The “rd” isolates and hypotheticals are circled; the “Rd” hypotheticals are boxed.
Discussion
A full understanding of genome-wide replication dynamics requires the application of methods that accurately quantify the process and that may be accurately compared across independent samples. The QBU method has now allowed us to re-examine how the Sir2 and Rpd3 histone deacetylases affect genome replication, particularly the balance between dispersed, single-copy origins and the tandemly repetitive rDNA origins, which has been identified as a key factor in genome stability (28). Overall, our results support the current concept of differential replication timing among origins as the consequence of the differential accrual of limiting replication initiation factors. Deletions of regulators, such as Sir2 and Rpd3, that affect the differential accrual of factors disrupt the existing balance, potentially creating conflicts between different genomic regions. Thus, understanding how different chromatin regulators help to balance the activity of origins intrinsically in competition with each other is an important goal.
A clear conclusion of our study is that Rpd3 acts independently of Sir2 and of the rDNA in exerting its effects on single-copy origins. By quantifying and normalizing for the absolute levels of replication associated with single-copy versus the rDNA origins, we showed that the effects of RPD3 deletion on single-copy origins are additive with, and hence independent of, SIR2 deletion, whereas SIR2 deletion’s effect on the rDNA origins is epistatic to RPD3 deletion. Specifically, RPD3 deletion in sir2Δ cells affects single-copy origins without altering rDNA origin firing, which remains elevated in the sir2Δ background. This is not consistent with Rpd3 acting as an rDNA origin activator but is consistent with Rpd3 acting as a repressor of single-copy origins. Indeed, the increased pan-S phase efficiency of rDNA origins in rpd3Δ cells is consistent with Rpd3 acting as a repressor of rDNA origins also. Moreover, we showed that the effect of RPD3 deletion on single-copy origins occurred in strains with much reduced rDNA complement, even while the effect on rDNA origins was also reduced. Thus, deregulation of single-copy origins in rpd3Δ cells does not appear to derive from primary effects on rDNA origins, though we cannot rule out the possibility of direct effect(s) of Rpd3 on rDNA origins.
Our quantitative conclusions on the effects of Rpd3 and Sir2 on rDNA origin firing from QBU were confirmed by the “gold standard” method of 2D gels for analysis of origin initiation levels. First, we showed that the differences in early firing levels (in HU) matched those determined by QBU, with significantly higher initiation of the rDNA origins in sir2Δ and sir2Δ rpd3Δ cells but lower initiation in rpd3Δ cells compared with WT. Remarkably, we found that rpd3Δ cells showed a higher level of rDNA origin firing than WT in a pan-S phase culture and conversely found that sir2Δ and sir2Δ rpd3Δ cells were unchanged from WT in this analysis. In both cases, it appears that changes in origin usage subsequent to the levels observed in the early-S-phase–arrested cells, compensate for the earlier imbalances We used a BrdU pulse-labeling approach throughout S phase that provided evidence in support of the idea that the early reduction in rDNA origin firing in rpd3Δ cells is followed by an increase in later rDNA origin firing relative to WT cells. We note that the levels of early- versus late-origin usage are intrinsically linked in that reduced usage early reserves a larger complement of origins for later usage and vice versa. It is also anticipated that, as replication progresses, the abundance of replication factors will increase relative to the remaining unfired origins, while passive replication of potential origins by active forks simultaneously plays an important role in these dynamics (29, 30). In considering passive replication of the rDNA, it is notable that most replication is unidirectional due to the presence of the replication fork barrier (31), so passive replication of rDNA origins is reduced, potentially reserving more origins for later firing. Interestingly, the reduced early replication of rDNA in rpd3Δ cells ultimately is associated with a higher overall efficiency, which we think is quite unusual and are unaware of similar examples.
The exact mechanism involved in the repression of origin firing by Rpd3 has remained elusive and is generally thought to act through deacetylation to reduce access to chromatin (32). Several potential targets have been proposed among replication-initiation factors, including those acting in both cyclin-dependent kinase (CDK)-dependent and DDK-dependent initiation pathways (10, 12, 14). For example, we showed previously that RPD3 deletion partially suppresses the late-origin initiation defect of clb5Δ cells (10), presumably facilitating access of Clb6-Cdk1, which we showed was limiting in clb5Δ cells (33). Mantiero and colleagues showed that RPD3 acts in opposition to simultaneous overexpression of multiple initiation factors on both initiation branches (14). More recently, we showed that overexpression of Fkh1/2 alone stimulates significantly more origins to detectably fire earlier (18). As origin stimulation by Fkh1/2 involves Dbf4 recruitment directly to origins (22, 23), we tested whether RPD3 affects the DDK pathway through impedance of Fkh1 access to bind origin chromatin. ChIP analyses indicate that RPD3 deletion leads to a modest but significantly increased enrichment of Fkh1 at Rpd3-repressed origins in G1 phase, anticipating the modest increases in QBU signals at these origins in early S phase. Together with the dependence of increased origin firing in rpd3Δ cells on FKH1/2, these findings support a mechanism whereby RPD3 regulates origin function by impeding Fkh1 binding, to varying degrees at different loci. The relatively subtle effects of RPD3 deletion on Fkh1 binding may reflect the limiting nature of Fkh1 being distributed across numerous potential binding loci differing in the No. and/or distribution of Fkh1/2 binding sites and/or in local recruitment of Rpd3. In fact, Fkh1/2 were originally identified as potential origin regulators due to anticorrelation of their binding with Rpd3-regulated origin loci (21). We also showed previously that Fkh1 binding is associated with decreased nucleosome occupancy (25), and Soriano et al. reported changes in nucleosome occupancy at Fkh1-binding sites in rpd3Δ cells (32).
Finally, we presented evidence that Fkh1 and Rpd3 have opposing functions in relation to Dbf4, as deletion of FKH1 causes lethality of dbf4ΔC cells, whereas elimination of RPD3 function rescues the viability of cells having the dbf4ΔC allele, which is hypomorphic for DDK function. The implication is that RPD3 deletion facilitates Fkh1 recruitment to origins, which becomes critical in Dbf4-deficient cells. This finding reinforces the model we have proposed wherein Rpd3 acts by impeding Fkh1, though the results do not exclude other factors from being similarly affected, such as implied for the CDK pathway by suppression of clb5Δ defects as discussed above. Given the pervasive role of histone deacetylases in most aspects of genome regulation, the mechanistic insights gained here in yeast are expected to serve as paradigms for derived functions in more-complex systems.
Concluding on the possible evolutionary and developmental roles of Fkh1/2 in regulation of rDNA origin activity, we note that rDNA dynamics, such as expansion and contraction of repeats, and regulation of gene expression are both potentially influenced by rDNA origin activity and replication timing. For example, higher initiation levels are expected to lead to higher levels of blocked forks and thus increased potential for DNA breaks, leading to changes in rDNA copies. Increased early firing of the rDNA may also contribute to higher rRNA expression levels by increasing template copies early in the cell cycle and confer a growth advantage. Intriguingly, deletion of both FKH1 and FKH2 leads to pseudohyphal growth, which is normally a starvation response, on rich medium, suggesting that FKH1/2 play a central role to integrate growth signals with cell-cycle progression and DNA replication, which will be interesting to study further.
Materials and Methods
Yeast Strain Construction.
Strain constructions were carried out by genetic crosses or lithium acetate transformations with linearized plasmids or PCR products generated with hybrid oligonucleotide primers having homology to target loci (34, 35); primer sequences for strain constructions are given in Table 1. Yeast strains are described in Table 2. All strains used for experiments are congenic with W303 background, and most are derived from BrdU-incorporating strains CVy43, CVy61, and CVy68, which are derived from SSy161 (36). OAy1096 was derived from a cross of OAy1069 and OAy1070 (18). OAy1100 was created by transformation of SSy161 with PCR product of primers 2xL-3xFLAG-FKH1-F and 2xL-3xFLAG-FKH1-R using plasmid p2xL-3xFLAG(TRP1) as template (gift from T. Tsukiyama); expression of Fkh1-3xFLAG was confirmed by Western blotting with anti-FLAG M2 at 1:1,000 (Sigma F1804). OAy1102 is an ADE2 derivative of OAy1100. The clean replacement of FKH2 with fkh2-dsm was constructed in two steps: FKH2 was deleted in CVy68 using primers pAG61-Fkh2Δ5′ and pAG61-Fkh2Δ3′ with URA3MX to create strain OAy1107, which was transformed with fkh2-dsm DNA amplified using primers Fkh2-up400bp and Fkh2-down375bp from p405-Fkh2-dsm (24), followed by selection of 5-fluoroorotic acid to yield strain OAy1109. OAy1114 is haploid segregant from a cross of strains OAy1109 and OAy1096. Strain YHy36 was constructed by PCR amplification of HIS3MX from pFA6a-HIS3MX6 (35) with primers HIS-F and HIS-R and transformation into JHy4. RPD3 and SIR2 deletions were constructed by PCR amplification of rpd3Δ::KanMX from the CVy44 or JHy4 genomes or rpd3Δ::HIS3MX from the YHy36 genome using primers RPD3-del-up and RPD3-del-down or sir2Δ::KanMX from the JHy3 genome using primers SIR2-del-up and SIR2-del-down, followed by transformations into PP1758, CVy61, MPy102, OAy1102, OAy1114, and YHy3 to create strains JPy115, YHy33, YHy37, YHy17, YHy13, and YHy6, respectively (see Table 2). HMLα deletions were constructed by transformation of YHy33 with PCR-amplified hmlɑΔ::URA3MX from pAG61 (37), using primers HMLalpha-del-F and HMLalpha-del-R to create YHy3. MCM4 was carboxyl-terminally tagged with 3xHA epitope by transformation with PCR product of primers 3HA-MCM4-F and 3HA-MCM4-R and template pFA6-3HA-kanMX6 to create strain MPy102; expression of Mcm4-3xHA was confirmed by Western blotting with anti-HA (16B12 or 12CA5) at 1:2,000. DBF4 carboxyl-terminal truncation (dbf4ΔC) was created using primers Dbf4ΔC-F1 and Dbf4ΔC-R1 to amplify a DNA fragment for insertion of a stop codon with HIS3MX into a diploid strain based on crossing CVy43 and CVy68. Haploid segregants were generated to yield YHy19. CVy44 was backcrossed with CVy68 to generate OAy1186, which was mated with YHy19 to create diploid OAy1188. Genomic alterations were confirmed by PCR analysis and/or DNA sequence analysis as appropriate.
Table 1.
Sequences of DNA oligonucleotides used in this study
| Oligonucleotides | Sequence |
|---|---|
| SIR2-deletion-F | ACATCTAGCACTCCTTCCAAC |
| SIR2-deletion-R | ACCTGCCCTTCTTACATTAAGC |
| RPD3-deletion-F | TCGCGGGCTGAACTGAATC |
| RPD3-deletion-R | GCTTTATCAACAGCGGTGGG |
| sir2-del-up | TTACTTGTAGCCTGCAACTCC |
| rpd3-del-up | TCAGCATAACGAATTGACGG |
| sir2-del-down | ATTCGACTTCTTTCCTTCGTTGT |
| rpd3-del-down | TGCAATTAGAAGAGAGTGAATC |
| int-sir2-down | ATCACAGGGTTCAATGTCGG |
| int-rpd3-down | TAGTGTTCAGTTGAATCACAC |
| SIR2-del-TRP-F | ATTCAAACCATTTTTCCCTCATCGGCACATTAAAGCTGGGACAAGTAACTGCAGGAATT |
| SIR2-del-TRP-R | ATATTAATTTGGCACTTTTAAATTATTAAATTGCCTTCTACACTATAGGGCGAATTGGGT |
| HIS-F | AGAATACCCTCCTTGACAGTC |
| HIS-R | TAGTATCGAATCGACAGCAG |
| HML-alpha-del-F | TATAGGGCAGTGTGTGACTTATGAATTGTTGTAGAAGGACgacatggaggcccagaatac |
| HML-alpha-del-R | ATGGCACAAGGAACACGCATTTTCCCAAGGCTTAGTATACcggcgttagtatcgaatcga |
| extend-HML-F | TTTTGGGACGATATTGTCATTATAGGGCAGTGTGTGACTT |
| extend-HML-R | TTTTATGAAGTAGCTTTCGGATGGCACAAGGAACACGCAT |
| probe-F | CTCACACTTGTACTCCATGA |
| probe-R | TGATGTGGAGAATAAGGTGC |
| 2xL-3xFLAG-FKH1-5′ | TTTCTACTACGACATCCATGGACGTAACAACAAACGCAAACGTGAACAATTCCTCTCTGAGTAGGGAACAAAAGCTGGAG |
| 2xL-3xFLAG-FKH1-3′ | TTCTTAACGGGTCTTTGTTCTTTATTGTTTAATAATACATATGGGTTCGACGACGCTGAATTCTATAGGGCGAATTGGGT |
| pAG61-Fkh1Δ5′ | AATAATAGTGTGTAAATTGTGCGTTCAATTAGCAAAGAAAgacatggaggcccagaatac |
| pAG61-Fkh1Δ3′ | TATTGTTTAATAATACATATGGGTTCGACGACGCTGAATTcggcgttagtatcgaatcga |
| Fkh2-up400bp | CATTACCGAAAATCTTCGATTTCGC |
| Fkh2-down375bp | CCGAAGCGTTGAGAAACAGC |
| Dbf4ΔC-F1 | AGCACAGACAGCACAGCCGGTGAAGAAAGAAACGGTAtgacggatccccgggttaattaa |
| Dbf4ΔC-R1 | GATTTTATCACTAAAAGCTACTGCACTTTACGTCGTGTCCcggcgttagtatcgaatcga |
| 3HA-MCM4-F | CGAGGGTGTAAGGAGATCAGTTCGCCTGAATAACCGTGTCCGGATCCCCGGGttaattaac |
| 3HA-MCM4-R | GATTTTATCACTAAAAGCTACTGCACTTTACGTCGTGTCCcggcgttagtatcgaatcga |
Oligonucleotides were obtained from International DNA Technologies.
Table 2.
Genotypes of S. cerevisiae strains used in this study
| Name | Genotype | Source |
|---|---|---|
| SSy161 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1Δ::hisG | Viggiani et al., 2006 |
| CVy43 | ura3::BrdU-inc(URA3) | “ |
| CVy61 | trp1::BrdU-inc(TRP1) | “ |
| CVy68 | MATα leu2::BrdU-inc(LEU2) | “ |
| CVy44 | rpd3Δ::KanMX ura3::BrdU-inc(URA3) | Knott et al., 2009 |
| JHy3 | his3-1 leu2-0 ura3-0 met15-0 sir2Δ::KanMX | Mark Rose |
| JHy4 | his3-1 leu2-0 ura3-0 met15-0 rpd3Δ::KanMX | “ |
| PP1758 | fob1Δ::URA3 ura3::7xTK (URA3) rDNA20 (∼20copies) | Philippe Pasero |
| JPy115 | rpd3Δ::KanMX fob1Δ::URA3 ura3::7xTK (URA3) rDNA20 (∼20copies) | This study |
| MPy55 | FKH1-3xFLAG(TRP1) ADE2::FLOPv2x2 | “ |
| MPy102 | MCM4-3HA(KanMX) TRP1::HOPv1 | “ |
| MPy188 | fkh1Δ::URA3 | “ |
| OAy1096 | fkh1Δ::KanMX fkh2Δ::HIS3MX ars305Δ::BrdU-inc(URA3) | “ |
| OAy1100 | FKH1-3xFLAG(TRP1) | “ |
| OAy1102 | FKH1-3xFLAG(TRP1) ADE2::FLOPv1 | “ |
| OAy1107 | MATα fkh2Δ::URA3MX leu2::BrdU-inc(LEU2) | “ |
| OAy1109 | MATα fkh2-dsm leu2::BrdU-inc(LEU2) | “ |
| OAy1114 | fkh1Δ::KanMX fkh2-dsm ars305Δ::BrdU-inc(URA3) | “ |
| OAy1186 | MATα rpd3ΔKanMX leu2::BrdU-inc(LEU2) | “ |
| OAy1188 | OAy1186 x YHy19 diploid | “ |
| OAy1189 | dbf4ΔC::HIS3MX leu2::BrdU-inc(LEU2) | “ |
| YHy3 | sir2Δ::KanMX hmlɑΔ::URA3MX trp1::BrdU-inc(TRP1) | “ |
| YHy6 | sir2Δ::KanMX rpd3Δ::HIS3MX hmlɑΔ::URA3MX trp1::BrdU-inc(TRP1) | “ |
| YHy13 | fkh1Δ::KanMX fkh2-dsm rpd3Δ::HIS3MX ars305Δ::BrdU-inc(URA3) | “ |
| YHy17 | rpd3Δ::HIS3MX FKH1-3xFLAG(TRP1) ADE2::FLOPv1 | “ |
| YHy19 | dbf4ΔC::HIS3MX ura3::BrdU-inc(URA3) | “ |
| YHy27 | rpd3Δ::HIS3MX FKH1-3xFLAG(TRP1) ADE2::FLOPv2x2 | “ |
| YHy33 | sir2Δ::KanMX trp1::BrdU-inc(TRP1) | “ |
| YHy36 | his3-1 leu2-0 ura3-0 met15-0 rpd3Δ::HIS3MX | “ |
| YHy37 | rpd3Δ::HIS3MX MCM4-3HA(KanMX) TRP1::HOPv1 | “ |
| YHy38 | OAy1189 x MPy188 diploid | “ |
All strains are in the W303 (RAD5) background; the parental genotype of SSy161 is shown, whereas for other strains, only differences from this genotype are given, with the exceptions of strains JHy3, JHy4, and YHy36, which are in the BY4741 background and only used as source DNA for strain constructions.
Other Methods.
Cultures were grown at 23 °C, and synchronization was performed as described previously (38). Five micrograms per milliliter α-factor was used for synchronization of BAR1 strains and 5 ng/mL for bar1Δ strains. QBU analysis was performed as described (38) using KAPA Hyper Prep Kit (KK8504). DNA content analysis by flow cytometry (FACScan) has been described previously (10). 2D gels were performed as described previously (39), except that only the rDNA fraction in the cesium chloride gradient was isolated and benzoylated naphthoylated diethylaminoethyl cellulose enrichment was omitted. One microgram rDNA was digested with HindIII; the probe for the 2D gel was generated as described previously by radioactive labeling of a DNA fragment produced by PCR with primers Probe-F and Probe-R using genomic DNA as template. Radioactive signal was captured on phosphor screens, scanned with Typhoon scanner, and imaged and quantified with ImageQuant software (Bio-Rad). ChIP-seq was performed as described (40) using KAPA Hyper Prep Kit (KK8504) using anti-FLAG M2 monoclonal antibody (Sigma F1804) at 1:100 and anti-HA monoclonal antibodies 12CA5 (Millipore-Sigma ROAHA) and 16B12 (BioLegend MMS-101P) at 1:100. High-throughput DNA sequencing was performed on one of several Illumina platforms at several different facilities.
Computation and Statistics.
All sequencing data were binned (50 bp) and median smoothed over a 1-kb window. BrdU peaks were called by MACS 1.4.2 with no-model mode (P < 0.01). The called peaks in each set were cross-referenced against origins listed in OriDB to eliminate any peaks not aligning with a “confirmed” or “likely” origin. Overlapping origins between the different datasets were determined by bedtools 2.25.0 using intersect function; from these results, the union of origins identified in WT and rpd3Δ cells was determined for use as the total origins list of 306 only for the following purpose: Rpd3-repressed origins were called by applying a two-sample, two-tailed t test (P < 0.001) comparing WT and rpd3Δ QBU signals within a 5-kb window centered on these 306 replication origins; 80 differential origins were detected, of which 74 were increased in rpd3Δ and thus denoted Rpd3 repressed (SI Appendix, Table S1). The list of origins and their TRep values and set assignments (FKH activated; n = 94, excludes rDNA origin) are from refs. (20, 21). CEN-proximal origin refers to the closest origin on each side of each centromere (SI Appendix, Table S2). FKH1-OE-activated origins were taken from ref. (18) (SI Appendix, Table S3). Matlab was used for generation of most data displays and analyses. ChIP-seq data were normalized against data untagged control strains. Fkh1 ChIP peaks were called by MACS 2.2.7.1 with no-model mode and instead an extension size of 100 bp. ChIP data files were compared with control data files (untagged strain), allowing for background to be estimated (false discovery rate adjusted P < 0.05).
Supplementary Material
Acknowledgments
We thank Takehiko Kobayashi, Philippe Pasero, and Mark Rose for providing yeast strains; Yan Gan for technical assistance; Atish Jain for assistance with data analysis; Daniel Campo of University of Southern California’s (USC’s) UPC Genome and Cytometry Core and Jason Kim of the NexGen Sequencing Core of the Norris Comprehensive Cancer Center at USC Keck School of Medicine, supported by Norris Comprehensive Cancer Center Cancer Center Support Grant (National Cancer Institute grant No. P30CA014089), for assistance with high-throughput DNA sequencing; and Phuong Pham and Myron Goodman for assistance with and access to Illumina MiniSeq. This work was supported by NIH R01-GM05494 (to O.M.A.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2212134119/-/DCSupplemental.
Data, Materials, and Software Availability
[DNA sequencing] data have been deposited in [GEO](GSE200351) (41).
References
- 1.Tsang E., Carr A. M., Replication fork arrest, recombination and the maintenance of ribosomal DNA stability. DNA Repair (Amst.) 7, 1613–1623 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Nomura M., Ribosomal RNA genes, RNA polymerases, nucleolar structures, and synthesis of rRNA in the yeast Saccharomyces cerevisiae. Cold Spring Harb. Symp. Quant. Biol. 66, 555–565 (2001). [DOI] [PubMed] [Google Scholar]
- 3.MacAlpine D. M., Bell S. P., A genomic view of eukaryotic DNA replication. Chromosome Res. 13, 309–326 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Aparicio O. M., Location, location, location: It’s all in the timing for replication origins. Genes Dev. 27, 117–128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fangman W. L., Brewer B. J., Activation of replication origins within yeast chromosomes. Annu. Rev. Cell Biol. 7, 375–402 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Linskens M. H., Huberman J. A., Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 8, 4927–4935 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwan E. X., et al. , A natural polymorphism in rDNA replication origins links origin activation with calorie restriction and lifespan. PLoS Genet. 9, e1003329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller C. A., Umek R. M., Kowalski D., The inefficient replication origin from yeast ribosomal DNA is naturally impaired in the ARS consensus sequence and in DNA unwinding. Nucleic Acids Res. 27, 3921–3930 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasero P., Bensimon A., Schwob E., Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev. 16, 2479–2484 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aparicio J. G., Viggiani C. J., Gibson D. G., Aparicio O. M., The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol. Cell. Biol. 24, 4769–4780 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knott S. R., Viggiani C. J., Tavaré S., Aparicio O. M., Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev. 23, 1077–1090 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogelauer M., Rubbi L., Lucas I., Brewer B. J., Grunstein M., Histone acetylation regulates the time of replication origin firing. Mol. Cell 10, 1223–1233 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Yoshida K., et al. , The histone deacetylases sir2 and rpd3 act on ribosomal DNA to control the replication program in budding yeast. Mol. Cell 54, 691–697 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Mantiero D., Mackenzie A., Donaldson A., Zegerman P., Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 30, 4805–4814 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel P. K., et al. , The Hsk1(Cdc7) replication kinase regulates origin efficiency. Mol. Biol. Cell 19, 5550–5558 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka S., Nakato R., Katou Y., Shirahige K., Araki H., Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr. Biol. 21, 2055–2063 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Smith J. S., Caputo E., Boeke J. D., A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19, 3184–3197 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peace J. M., Villwock S. K., Zeytounian J. L., Gan Y., Aparicio O. M., Quantitative BrdU immunoprecipitation method demonstrates that Fkh1 and Fkh2 are rate-limiting activators of replication origins that reprogram replication timing in G1 phase. Genome Res. 26, 365–375 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santocanale C., Diffley J. F., A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395, 615–618 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Nieduszynski C. A., Hiraga S., Ak P., Benham C. J., Donaldson A. D., OriDB: A DNA replication origin database. Nucleic Acids Res. 35, D40–D46 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knott S. R., et al. , Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 148, 99–111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang D., et al. , Dbf4 recruitment by forkhead transcription factors defines an upstream rate-limiting step in determining origin firing timing. Genes Dev. 31, 2405–2415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H., et al. , Dynamic relocalization of replication origins by Fkh1 requires execution of DDK function and Cdc45 loading at origins. eLife 8, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostrow A. Z., et al. , Conserved forkhead dimerization motif controls DNA replication timing and spatial organization of chromosomes in S. cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 114, E2411–E2419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostrow A. Z., et al. , Fkh1 and Fkh2 bind multiple chromosomal elements in the S. cerevisiae genome with distinct specificities and cell cycle dynamics. PLoS One 9, e87647 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinapae A., et al. , Recruitment of Fkh1 to replication origins requires precisely positioned Fkh1/2 binding sites and concurrent assembly of the pre-replicative complex. PLoS Genet. 13, e1006588 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones D. R., Prasad A. A., Chan P. K., Duncker B. P., The Dbf4 motif C zinc finger promotes DNA replication and mediates resistance to genotoxic stress. Cell Cycle 9, 2018–2026 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Lindström M. S., et al. , Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 37, 2351–2366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyrien O., Goldar A., Mathematical modelling of eukaryotic DNA replication. Chromosome Res. 18, 147–161 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Rhind N., Gilbert D. M., DNA replication timing. Cold Spring Harb. Perspect. Biol. 5, a010132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brewer B. J., Lockshon D., Fangman W. L., The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71, 267–276 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Soriano I., Morafraile E. C., Vázquez E., Antequera F., Segurado M., Different nucleosomal architectures at early and late replicating origins in Saccharomyces cerevisiae. BMC Genomics 15, 791 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibson D. G., Aparicio J. G., Hu F., Aparicio O. M., Diminished S-phase cyclin-dependent kinase function elicits vital Rad53-dependent checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 24, 10208–10222 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.H. Ito et al., Transformation of intact yeast cells treated with alkali cations. J Bacteriol, 1983. 153(1): p. 163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longtine M. S., et al. , Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Viggiani C. J., Aparicio O. M., New vectors for simplified construction of BrdU-Incorporating strains of Saccharomyces cerevisiae. Yeast 23, 1045–1051 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Goldstein A. L., Pan X., McCusker J. H., Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast 15, 507–511 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Haye-Bertolozzi J. E., Aparicio O. M., “Quantitative bromodeoxyuridine immunoprecipitation analyzed by high-throughput sequencing (qBrdU-seq or QBU)” in Genome Instability: Methods and Protocols, Methods in Molecular Biology, M. Muzi-Falconi and G.W. Brown, Editors. 2018, Springer Protocols. p. 209–225. [DOI] [PubMed] [Google Scholar]
- 39.Villwock S. K., Aparicio O. M., Two-dimensional agarose gel electrophoresis for analysis of DNA replication. Methods Mol. Biol. 1205, 329–340 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Ostrow A. Z., Viggiani C. J., Aparicio J. G., Aparicio O. M., ChIP-seq to analyze the binding of replication proteins to chromatin. Methods Mol. Biol. 1300, 155–168 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Y. He, J. M. Peace, O. M. Aparicio, Rpd3 regulates single-copy origins independently of the rDNA array by opposing origin stimulation by Fkh1. NCBI: GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE200349. Accessed 6 April 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
[DNA sequencing] data have been deposited in [GEO](GSE200351) (41).