Significance
Induction of transplantation tolerance with costimulation blockade therapy requires the presence of regulatory T cells (Tregs) and is associated with reduced function of donor-reactive CD4+ conventional T cells (Tconvs). Here, we show that alloreactive Tconvs from tolerant mice also exhibit an enhanced susceptibility to Treg suppression when compared with memory Tconvs from rejector mice, via Tconvs’ downregulation of the transcription factor Special AT-rich sequence-binding protein 1. Our findings uncover a cell-intrinsic modulator of Tconv responsiveness to Treg suppression and a novel characteristic of transplantation tolerance.
Keywords: transplantation, T cell dysfunction, Treg suppression, Satb1
Abstract
Limiting CD4+ T cell responses is important to prevent solid organ transplant rejection. In a mouse model of costimulation blockade-dependent cardiac allograft tolerance, we previously reported that alloreactive CD4+ conventional T cells (Tconvs) develop dysfunction, losing proliferative capacity. In parallel, induction of transplantation tolerance is dependent on the presence of regulatory T cells (Tregs). Whether susceptibility of CD4+ Tconvs to Treg suppression is modulated during tolerance induction is unknown. We found that alloreactive Tconvs from transplant tolerant mice had augmented sensitivity to Treg suppression when compared with memory T cells from rejector mice and expressed a transcriptional profile distinct from these memory T cells, including down-regulated expression of the transcription factor Special AT-rich sequence-binding protein 1 (Satb1). Mechanistically, Satb1 deficiency in CD4+ T cells limited their expression of CD25 and IL-2, and addition of Tregs, which express higher levels of CD25 than Satb1-deficient Tconvs and successfully competed for IL-2, resulted in greater suppression of Satb1-deficient than wild-type Tconvs in vitro. In vivo, Satb1-deficient Tconvs were more susceptible to Treg suppression, resulting in significantly prolonged skin allograft survival. Overall, our study reveals that transplantation tolerance is associated with Tconvs’ susceptibility to Treg suppression, via modulated expression of Tconv-intrinsic Satb1. Targeting Satb1 in the context of Treg-sparing immunosuppressive therapies might be exploited to improve transplant outcomes.
CD4+ T cells are an important driver of transplant rejection. In preclinical models of cardiac transplantation, CD4+ T cells are necessary and sufficient for rejection (1, 2). Upon antigen encounter, alloreactive CD4+ Tconvs differentiate into distinct effector subsets, including T helper (Th)1, Th2 and Th17, that can each cause allograft damage, albeit by different mechanisms associated with the presence of macrophages, eosinophils, or neutrophils, respectively (3–5). Moreover, CD4+ T cells provide help to CD8+ T cells (1, 6), and may engage in direct allograft cytotoxicity (7), while T follicular helper (Tfh) cells help B-cells produce alloantibodies (8). Therefore, better strategies to target CD4+ Tconvs should improve clinical transplant outcomes.
Transplantation tolerance is a state of permanent allograft acceptance following cessation of immunosuppression. Its induction remains a challenge in humans, although proof-of-principle exists that it can be achieved (9–13). In experimental models, transplantation tolerance can be attained by therapeutic approaches (14–16) and genetic manipulations (17, 18). One such mouse model of tolerance relies on coadministration of blocking anti-CD154 monoclonal antibody (αCD154) and alloantigen in the form of donor splenocyte transfusion (DST), which leads to indefinite survival of first cardiac allografts and enables spontaneous acceptance of subsequent secondary donor-matched cardiac allografts in the absence of immunosuppression (19). In this model, Tregs are essential for the induction and maintenance of transplantation tolerance, as well as the re-establishment of tolerance after acute graft rejection precipitated by a systemic infection (19, 20). The surprising observation that Treg numbers increased comparably during the induction of transplantation tolerance and during acute rejection (19), and that absence of the CD154-CD40 pathway did not enhance Treg suppressive activity (21), prompted us to investigate the functional consequences of anti-CD154/DST-mediated tolerance induction on alloreactive Tconvs. To this end, we used this model in combination with a tracer population of alloreactive CD4+ TCR75 Tconvs, which recognize the donor Kd54-68 peptide presented by host I-Ab, that are seeded into hosts just before transplantation (22). We recently reported that these TCR75 T cells acquired a state of dysfunction when exposed to the tolerogenic regimen, compared to TCR75 T cells from transplanted untreated mice that rejected their graft and developed memory to donor alloantigens (23). Acquisition of dysfunction by TCR75 T cells during transplant tolerance induction required both alloantigen persistence for >2 to 3 wk in the primary host and the presence of the anti-CD154 blocking antibody at the time of alloantigen encounter, thus combining parameters of CD4+ T cell exhaustion described in settings of chronic viral infection (24), and of CD4+ T cell anergy defined as activation in the absence of costimulation (25). In transplantation, CD4+ Tconv dysfunction has also been observed in models where allograft tolerance was established using anti-CD3 F(ab′)2 administration (26), or by deleting Fut7 or IRF4 selectively in T cells (17, 27).
In cardiac allograft recipient mice treated with anti-CD154/DST, dysfunction of alloreactive T cells appeared Tconv-intrinsic, as TCR75 cells from tolerant mice displayed reduced expansion, even when removed from their tolerogenic environment and rechallenged with alloantigen in secondary naïve hosts (23). However, the secondary hosts contained Tregs, such that it remained to be formally demonstrated whether TCR75 cells from tolerant mice were intrinsically dysfunctional, and/or had acquired enhanced susceptibility to Treg suppression. Moreover, the transcriptional program leading to alloreactive Tconv dysfunction remained to be delineated. To address these questions, we subjected TCR75 cells from tolerant and rejector mice to RNA-seq. Here we report that transplant-specific CD4+ Tconvs in tolerant mice have a unique transcriptional profile, including lower expression of the genome organizer Special AT-rich sequence-binding protein-1 (Satb1) compared to Tconvs from rejector mice. Satb1-deficiency in T cells did not result in cell-autonomous dysfunction of CD4+ Tconvs but, instead, enhanced Tconvs’ sensitivity to Treg suppression in vitro and in vivo. Notably, TCR75 cells recovered from tolerant mice displayed better susceptibility to Treg suppression when compared to memory TCR75 cells recovered from rejector mice. This demonstrates the existence of a transcriptional tuning rheostat for the susceptibility of Tconvs to Treg suppression. Thus, targeting T cell-Satb1 may complement Treg therapies to improve allograft survival.
Results
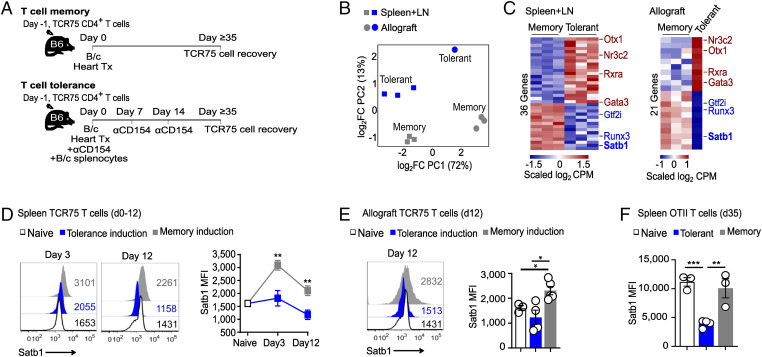
To explore the functional consequences of the anti-CD154/DST tolerogenic regimen on alloreactive CD4+ Tconvs, we used a tracer population of alloreactive TCR75 CD4+ Tconvs expressing the congenic marker CD45.1 and seeded into C57BL/6 (B6, CD45,2, H-2b) mice just before transplantation with a BALB/c (B/c, H-2d) cardiac allograft. Hosts were then treated or not with the tolerogenic regimen anti-CD154/DST. We have recently shown that TCR75 cells isolated from tolerant mice >1 mo posttransplantation displayed reduced expansion when transferred into secondary naïve B6 mice and restimulated with donor splenocytes, when compared with memory TCR75 cells isolated from untreated rejector mice (23). Exhausted CD8+ T cells in models of chronic viral infection or tumor have been associated with increased expression of the transcription factor TOX (28–31) while deficiency in the transcription factor IRF4 has been reported to promote a state of CD4+ hyporesponsiveness in a mouse model of heart transplantation (27). To identify the transcriptional program associated with CD4+ alloreactive Tconv dysfunction in anti-CD154/DST-induced donor-specific tolerance, TCR75 cells were sorted at d35 posttransplantation via their congenic marker CD45.1+ from the periphery (spleen + lymph nodes) and from the allografts of tolerant and rejector mice (gating strategy is shown in SI Appendix, Fig. 1A, and percentage of TCR75 cells within each location in SI Appendix, Fig. 1B) and subjected to RNA-seq (Fig. 1A). Principal component analysis of gene expression data revealed distinct transcriptional profiles in TCR75 T cells, with the X axis distinguishing cells based on tissue of origin and the y axis based on tolerant versus rejector status of the host (Fig. 1B). A total of 466 and 324 genes were differentially expressed between tolerant and memory TCR75 cells from the periphery and the graft, respectively. Tolerant TCR75 T cells resembled published exhausted phenotypes of CD4+ T cells following LCMV infection (24) and were enriched in inhibitory molecules and phosphatases (Btla, Cd200, Ctla4, Pilra, Pdcd1Ig2, Sirpa, Dusp1, Dusp14, and Ptpn11). Conversely, expression of effector function genes, such as chemokine/cytokine receptors and TCR signaling molecules (Cxcr5, Ccr2, Ccr5, Cd127, Il18r1, Il12rb, Ifngr1, Akt, Camk4, Grap2, Trib2, and Txk) were substantially lower in tolerant than memory TCR75 T cells (SI Appendix, Fig. 2 A and C and Table 1). Ingenuity pathway analysis (IPA) pointed to an enrichment in the T cell exhaustion pathway and reduced Th1 activation and CD28 signaling pathways in tolerant cells (SI Appendix, Fig. 2 B and D). We further identified 7 genes encoding transcription factors that were differentially expressed in tolerant versus memory TCR75 T cells in a similar fashion in the periphery and allograft. Expression of genes encoding Otx1, Nr3c2, Rxra, and Gata3 was up-regulated, while that of Gtf2i, Runx3 and Satb1 was down-regulated in tolerant TCR75 T cells (Fig. 1C and SI Appendix, Fig. 3 and Table 2). In contrast, tolerant and memory TCR75 cells expressed similar levels of Tox and Irf4 (SI Appendix, Fig. 3). Thus, the transcriptional profile from tolerant TCR75 cells was distinct from that of dysfunctional CD8+ and CD4+ T cells identified in other settings.
Fig. 1.
Differential expression of Satb1 in TCR75 T cells from tolerant and rejecting mice. (A) Experimental design. Congenic TCR75 CD4+ T cells seeded 1 d before transplantation were sorted on day 35 posttransplantation from spleen and lymph nodes (LNs) or allografts of tolerant or rejecting mice and RNA-seq was performed. (B) Principal component analysis. (C) Heatmap of differentially expressed transcription factors (fold change ≥ 1.5; adjusted P ≤ 0.05). Color key is normalized log2 transformed counts per million (CPM). n = 3 (memory: TCR75 T cells from 1 mouse/replicate; tolerant: pooled from 5 mice/replicate, except for tolerant allograft n = 1 with TCR75 T cells pooled from 15 allografts). (D and E) TCR75 T cells were seeded into host B6 mice 1 d before transplantation with B/c heart ±donor splenocytes and anti-CD154. Mice were euthanized at induction of memory and tolerance (day 3 ± 1 and day 12 ± 2) and Satb1 protein expression (MFI) was measured by flow cytometry of TCR75-gated T cells in spleen (D) and allograft (E). Naive group in (D and E) is the same and shows Satb1 expression of splenic TCR75 T cells before transfer. (F) Congenic OTII T cells were seeded 1 d before transplantation of 2W-OVA-expressing skin grafts, and hosts were treated or not with anti-CD154 and 2W-OVA-expressing splenocytes. OTII cells were sorted from spleen on d35 posttransplantation and analyzed by flow cytometry for expression of Satb1. All results are displayed as mean +/− SEM. n = 3 to 9/group (D), n = 3 to 5/group (E) and n = 3 to 4/group (F). *P < 0.05; **P < 0.01 (ANOVA, two-way for D, one-way for E and F, with pairwise comparisons).
It has been reported that tumor-reactive CD8+ T cells deficient in Satb1 lost effector function more rapidly than Satb1-sufficient T cells, thus linking reduced expression of Satb1 to loss of functionality (32) and drawing our attention to differences in Satb1 expression in CD4+ TCR75 cells from tolerant versus rejector mice. Indeed, Satb1 was down-regulated in TCR75 cells from tolerant hosts at d35 whether isolated from the periphery or the graft (Fig. 1C and SI Appendix, Fig. 3), suggesting this feature may be important for tolerance. Using TCR75 cells, we found that Satb1 protein was expressed constitutively (SI Appendix, Fig. 4 A and B) and was transiently up-regulated by day 3 postheart transplantation in splenic TCR75 cells from rejector mice but not mice treated with the tolerogenic regimen (Fig. 1D). Satb1 protein levels were also rapidly lower in TCR75 T cells infiltrating tolerant versus rejecting allografts (Fig. 1E). Lower expression of Satb1 in TCR75 cells from tolerant than rejector mice was not a consequence of antigen ignorance as the majority of TCR75 T cells from both sets of mice were CD44hi (SI Appendix, Fig. 5 A and B), demonstrating antigen experience. In vitro, Satb1 up-regulation in TCR75 T cells stimulated with allogeneic splenocytes was dependent on signal strength, which integrates TCR and costimulatory signals. Indeed, activation-mediated induction of Satb1 was curbed by addition of the calcineurin inhibitor tacrolimus, or of anti-CD154 (SI Appendix, Fig. 6 A and B), showing both NFAT and costimulation dependence for its induction. Overall, dysfunction in TCR75 cells from tolerant mice correlated with lower levels of Satb1 than in TCR75 cells from rejector mice, both at the mRNA and protein levels.
Lower expression of Satb1 in T cells from tolerant versus rejector mice was not unique to cells expressing the TCR75 transgene, as splenic endogenous polyclonal CD4+ T cells reactive to the same peptide from donor Kd presented by host I-Ab as TCR75 cells also displayed reduced Satb1 expression 30 d post-B/c heart transplantation in hosts treated with anti-CD154/DST when compared to memory counterparts (SI Appendix, Fig. 7 A and B). To determine if CD4+ T cells from other specificities and other transplant models regulated Satb1 in the same manner, we tracked ovalbumin (OVA)-reactive CD4+ OTII T cells. CD45.1+ OTII cells were seeded into B6 mice one day before transplantation of 2W-OVA-expressing B6 skin grafts in hosts treated or not with anti-CD154/DST (SI Appendix, Fig. 7C). In this minor mismatched model, skin grafts were accepted long term in anti-CD154/DST-treated hosts (SI Appendix, Fig. 7D). At day 35 posttransplantation, expression of Satb1 was lower in splenic OTII cells from tolerant versus rejector mice (Fig. 1F). Moreover, splenic endogenous CD4+ T cells specific for donor 2W peptide presented by host I-Ab (gating strategy is shown in SI Appendix, Fig. 7E) also displayed reduced Satb1 when compared to their memory counterparts, despite overnight restimulation (SI Appendix, Fig. 7F), supporting the resilience of this differential expression. Together, these data suggested that down-regulation of Satb1 was reproducible over several indirect specificities of graft-reactive CD4+ T cells and over two distinct models of heart and skin transplantation tolerance.
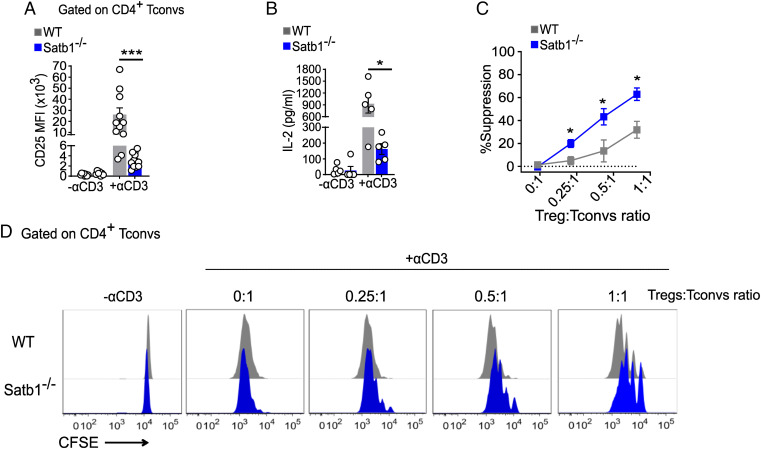
Satb1 is a chromatin organizer, and in nontumoral cells, its expression is primarily restricted to lymphocytes (33). Although Satb1-deficiency dysregulates thymic T cell development (34) and blunts anti-tumor CD8 T cell immunity (32), whether reduced Satb1 leads to decreased functionality of CD4+ Tconvs or affects their susceptibility to extrinsic suppression by Tregs is not known. Satb1 has been shown to directly bind the CD25 locus (34, 35). Hypothetically, reduced Satb1 expression in tolerant Tconvs may result in their lower expression of CD25, thereby reducing their cell-intrinsic proliferation upon activation, and/or facilitating IL-2 scavenging by Tregs that express high levels of CD25, thus promoting cell-extrinsic Treg-dependent inhibition of Tconv proliferation. To address these hypotheses, we obtained CD4CreSatb1fl/fl with T cell-restricted deficiency in Satb1. CD4CreSatb1fl/fl mice had similar distribution of Vβ families in both CD4+ and CD8+ splenic T cells as control littermates (SI Appendix, Fig. 8 A and B), suggesting an analogous polyclonal TCR repertoire and opportunity for alloreactivity. Consistent with the reported role for Satb1 in thymic T cell development (34), CD4CreSatb1fl/fl mice displayed a slightly increased percentage of peripheral CD4+CD8+ T cells, and modestly reduced frequency of CD8+ T cells (SI Appendix, Fig. 9 A–E). Thus, to analyze the function of comparable populations of Satb1-competent and -deficient CD4+ Tconvs, we sorted naïve (CD127hi CD44lo CD25− Nrp1−) CD4+ Tconvs from CD4CreSatb1fl/fl or Cre-negative control littermates. These cells were then stimulated for 72 h with soluble anti-CD3 in the presence of syngeneic splenocytes from Rag−/− mice, prior to assessment of CD25 expression and IL-2 production. Compared to control CD4+ Tconvs, Satb1−/− CD4+ Tconvs displayed significantly reduced levels of CD25 and IL-2 (Fig. 2 A and B). Consistent with Satb1 repressing PD-1 expression (32), activated Satb1−/− Tconvs expressed higher levels of PD-1 than control Tconvs (SI Appendix, Fig. 10A), but blockade of PDL1 only partially restored CD25 and IL-2 expression in activated Satb1−/− Tconvs (SI Appendix, Fig. 10B). Thus, other pathways than PD-1/PDL1 also contribute to the reduced expression of CD25 by Satb1−/− Tconvs.
Fig. 2.
Satb1 deficiency in CD4+ Tconvs promotes their sensitivity to Treg suppression in vitro. Sorted CFSE-labeled naïve WT or Satb1−/− CD4+ Tconvs were stimulated with soluble anti-CD3 in the presence of B6 Rag−/− splenocytes. (A) CD25 expression by flow cytometry on day 3. Number in histograms represents MFI. (B) IL-2 levels as measured by ELISA in culture supernatants on day 1. (C and D) Naive WT or Satb1−/− CD4+ Tconvs were stimulated as in A at indicated ratios of Tregs:Tconvs. Data (mean ± SEM) are pools of multiple independent experiments (n = 5 to 12 in A and B; n = 4 in C). *P < 0.05, **P < 0.005, ***P = 0.001 (two-way ANOVA with pairwise comparisons).
To distinguish between the hypotheses that Satb1−/− CD4+ Tconvs had a cell-intrinsic defect in proliferation or exhibited differential sensitivity to Treg suppression than Satb1-sufficient T cells, carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled sorted naïve (CD127hi CD44lo CD25− Nrp1−) CD4+ Satb1−/− and Cre-negative control littermate Tconvs were stimulated with anti-CD3 and syngeneic Rag−/− splenocytes in the presence or absence of sorted GFP+ Tregs from Foxp3GFP-transgenic mice (Satb1-sufficient). Whereas proliferation in the absence of Tregs was similar in Tconvs from Satb1-competent and -deficient littermates, addition of Foxp3GFP+ Tregs was more effective at limiting expansion of Satb1−/− than of control CD4+ Tconvs (Fig. 2 C and D). Thus, absence of Satb1 did not impart cell-intrinsic hyporesponsiveness to CD4+ T cells, but rather modulated their susceptibility to Treg suppression in vitro. Consistent with the hypothesis that CD25hi Tregs outcompeted CD25lo Satb1−/− Tconvs for IL-2, whereas a very low dose of exogenous IL-2 maximized Treg suppression, increasing doses of IL-2 up-regulated CD25 on Tconvs (SI Appendix, Fig. 11A) and eliminated differences in susceptibility to Treg suppression between Satb1−/− and control Tconvs, with Treg suppression being completely abrogated at the highest dose of IL-2 (SI Appendix, Fig. 11 A and B).
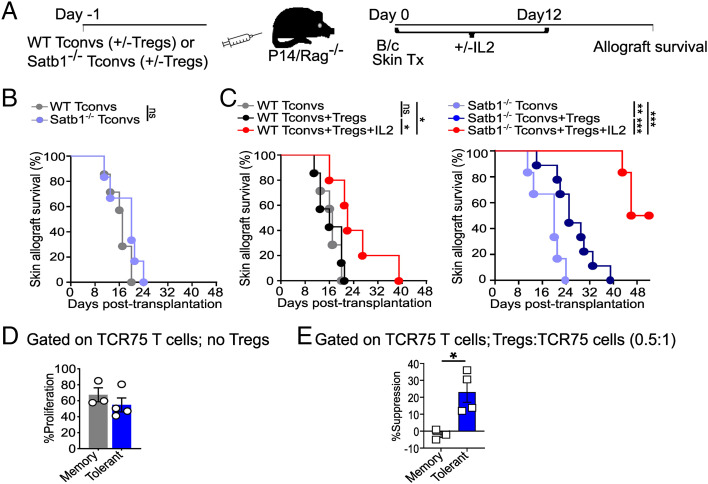
To investigate the impact of Satb1 deficiency in CD4+ Tconvs in vivo, we adoptively transferred sorted naïve (CD127hi CD44lo CD25− Nrp1−) Satb1−/− or littermate control CD4+ Tconvs ± sorted Foxp3GFP+ Tregs into P14Rag−/− recipients that subsequently received fully mismatched B/c skin allografts (Fig. 3A), a highly immunogenic transplant model. P14Rag−/− mice harbor irrelevant endogenous LCMV-reactive CD8+ P14 cells and were used as recipients to ensure that graft rejection would be mediated by the transferred CD4+ Tconvs, as well as to prevent homeostatic expansion of the transferred T cells, and because they lack endogenous Tregs. Consistent with our in vitro observations that Satb1−/− and control Tconvs proliferate similarly in the absence of Tregs, Satb1−/− and control CD4+ Tconvs rejected the allografts with similar kinetics in the absence of Treg transfer (Fig. 3B). In contrast, cotransfer of a limiting number of Tregs significantly prolonged allograft survival in recipients of Satb1−/− but not control Tconvs (Fig. 3C). Remarkably, cotransfer of Tregs along with a low dose of IL-2, a regimen shown to increase Treg fitness (36, 37) and that maximized Treg suppression in vitro (SI Appendix, Fig. 11B), promoted allograft survival much more substantially in recipients of Satb1−/− than of control Tconvs, with 50% of allografts surviving beyond day 50 posttransplantation, in the absence of immunosuppressive drugs (Fig. 3C). Overall, lack of Satb1 increased CD4+ Tconv sensitivity to Treg suppression in vivo, resulting in prolonged skin allograft survival.
Fig. 3.
Satb1-deficient CD4+ Tconvs and TCR75 cells from tolerant mice exhibit enhanced susceptibility to Treg suppression in vivo. (A) Experimental strategy. (B and C) Survival of B/c skin allografts transplanted into P14Rag−/− recipients that received 4.5 × 105 naive control or Satb1−/− CD4+ Tconvs alone (in B) or without/with 2.25 × 105 Foxp3GFP+ Tregs +/− IL-2 (in C). n = 5 to 9/group; *P < 0.05, **P < 0.005, ***P ≤ 0.0005 (log-rank test). Curves from B are reproduced in C for comparison. (D and E) Experimental setting as in Fig. 1A. At day ≥ 35 postheart transplantation, TCR75 T cells isolated from spleen and lymph nodes of tolerant or rejector mice were CFSE-labeled and stimulated in vitro with soluble anti-CD3+B6 Rag−/− splenocytes +/− Tregs for 3 d. CFSE dilution of TCR75 T cells was determined using flow cytometry. (D) Intrinsic proliferative capacity of TCR75 T cells in the absence of Tregs. (E) Percent suppression of TCR75 T cells in the presence of Tregs. n = 3 for memory and 4 for tolerant; *P < 0.05 (unpaired Student's t test).
To determine whether TCR75 T cells from cardiac allograft tolerant mice, which displayed reduced expression of Satb1 when compared with their memory counterparts from rejected mice (Fig. 1 and SI Appendix, Fig. 3), had increased susceptibility to Treg suppression, TCR75 cells were sorted through their congenic marker from the spleen and lymph nodes of tolerant or rejector mice on day ≥35 posttransplantation, as in Fig. 1A. Sorted TCR75 cells were labeled with CFSE, and subjected to Treg suppression in vitro as in Fig. 2 C and D and SI Appendix, Fig. 11B. To overcome the proliferative defect observed in TCR75 cells from tolerant mice when compared to untreated animals that had rejected their grafts (23), we restimulated TCR75 cells with a higher dose of anti-CD3 (1 μg/mL), which resulted in similar proliferation of TCR75 cells from tolerant and rejected mice when in the absence of Tregs (Fig. 3D). Notably, addition of Tregs at a 0.5:1 Treg:Tconv ratio significantly suppressed TCR75 cells from tolerant mice but not memory TCR75 cells from rejector mice (Fig. 3E). This result supports the conclusion that TCR75 cells from tolerant mice are more susceptible to Treg suppression than memory TCR75 cells from rejector mice, consistent with their lower expression of Satb1 and in contrast to the ability of memory T cells to resist Treg suppression (38).
Discussion
There is accumulating evidence that certain inflammatory signals can enable Tconvs to escape suppression by Tregs (39). Conversely, using a mouse model of cardiac transplantation, we demonstrate that alloreactive TCR75 Tconvs from tolerant mice display heightened susceptibility to Treg suppression when compared with memory Tconvs from rejector mice, correlating with down-regulation of the transcription factor Satb1. This was consistent with Satb1-deficient Tconvs having enhanced susceptibility to Treg suppression in vitro and in vivo, and cell-intrinsic changes, such as reduced CD25 expression, potentially enabling CD25hi Tregs to scavenge IL-2 away from Tconvs more effectively. Addition of low-dose IL-2 further increased susceptibility of Satb1−/−, but not control Tconvs, to Treg suppression in vitro as well as in vivo, resulting in spontaneous long-term acceptance of very immunogenic fully mismatched skin allografts in the absence of a tolerogenic regimen. Thus, responsiveness of Tconvs to Treg suppression can be tuned and is associated with transplant survival.
T cell dysfunction can either be cell-intrinsic such as the hyporesponsiveness described in states of T cell exhaustion or T cell anergy or can be cell-extrinsic imparted by Tregs or other inhibitory cell types. Whereas exhaustion refers to T cells that were productively activated initially but remained chronically stimulated because of lack of antigen clearance, anergy refers to T cells receiving improper initial activation such as upon antigen encounter in the absence of costimulation. Alloreactive CD4+ T cells from tolerant mice in our model combine both features, as they initially encounter alloantigen during blockade of CD40/CD154 costimulation but also receive long-term alloantigen stimulation in the form of a persisting allograft. Both long-term alloantigen exposure and anti-CD154 treatment are necessary for their acquisition of a dysfunctional phenotype (23). The transcription factors promoting T cell dysfunction are incompletely understood. In CD8+ T cells, TOX has been suggested by several investigators to drive the program of cell-intrinsic T cell exhaustion (28–31). Similarly, following murine pregnancy, fetus-reactive CD8+ T cells were recently described to express a transcriptional profile resembling that of exhausted T cells in tumor and chronic infection settings, including up-regulation of TOX (40). Interestingly, fetus-reactive CD8+ T cells expressed reduced levels of Satb1, similarly to our alloreactive CD4+ T cells from tolerant mice, although their susceptibility to Treg suppression was not tested (40). The transcriptional profile of exhausted CD4+ T cells is distinct from that of CD8+ T cells and depends on dose and duration of antigen exposure (24, 41). Our results suggest that tolerant alloreactive T cells develop cell-extrinsic dysfunction by remaining sensitive to Treg suppression, unlike their memory counterparts from mice that have rejected their allograft. Whether part of their transcriptional program also drives features of cell-intrinsic dysfunction remains to be investigated but it does not include significant up-regulation of Tox.
The mechanisms by which Tregs mediate their suppressive functions still remain incompletely understood and a better understanding is necessary to harness their full potential to treat autoimmunity, graft-versus-host disease following hematopoietic stem cell transplantation, or to promote solid organ transplant tolerance (42, 43). Such mechanisms need to intersect with susceptibility of effector Tconvs to suppression by Tregs. Indeed, Tconvs have been reported to develop resistance to Treg suppression in several autoimmune diseases, both in mice and humans (39), which might be a contributing factor to the development of autoimmunity, or a consequence of autoimmune-accompanying inflammation. Strong costimulation via CD28, signals from tumor necrosis factor receptor family members, Toll-like receptors and inflammatory cytokines are pathways by which Tconvs can resist Treg suppression (42, 44, 45) and which converge onto the PI3K signaling pathway (45). Intriguingly, the PI3K/Akt pathway has been shown to phosphorylate Satb1 and protect it from apoptotic cleavage in a tumor cell line (46). This suggested link between T cell activation and Satb1 expression is consistent with our observation that tacrolimus and costimulation blockade inhibit Satb1 up-regulation following alloreactive T cell activation.
The role of IL-2 scavenging for Tconv suppression remains controversial (42). A recent study using high resolution imaging of lymph nodes in vivo was able to visualize local production of IL-2 by activated T cells enabling adjacent Treg proliferation and suppressor function in microdomains that counteracted CD28 costimulation and IL-2 signaling in Tconvs (47). Thus, levels of IL-2 and responsiveness to IL-2 appear crucial for the dynamic interactions between Tconvs and Tregs such that lower expression of Satb1 in tolerant CD4+ T cells and therefore of Satb1-controlled CD25 may tip the balance toward tolerance by maximizing the IL-2 scavenging potential of CD25hi Tregs away from CD25lo Tconvs.
TCR75, pKd:I-Ab-reactive, OTII and 2W-reactive CD4+ T cells all recognize graft antigens indirectly presented on host major histocompatibility complex (MHC) class II. Whether Tconvs that recognize donor MHC class II directly can also develop increased susceptibility to Treg suppression in tolerant hosts remains to be determined. Whereas donor APCs that accompany the graft die rapidly after transplantation (48), they may, before their death, shed exosomes containing intact donor-MHC class II that could perpetuate semidirect stimulation of direct alloreactive CD4+ Tconvs by exosome-decorated host APCs. Whether this form of activation could induce/maintain increased susceptibility of direct Tconvs to Treg suppression in tolerant hosts remains to be investigated.
Our data carry potential implications outside of transplantation settings. For example, exhausted T cells in human ovarian cancer and in a mouse model of LCMV infection were reported to express lower levels of Satb1 (24, 32) and may therefore be more susceptible to Treg suppression, creating an additional hurdle to achieving tumor or viral control. Importantly, mouse and human Satb1 share 99% homology in amino acid sequence and Satb1-deficiency in postthymic Tregs does not impair their suppressor function (49). Thus, therapeutic targeting of Satb-1, a molecule largely restricted to lymphocytes postembryonic development, might affect Tconvs without reducing Treg suppressive function, making this an attractive strategy in settings of autoimmunity and transplantation. Moreover, because Treg-adoptive transfer therapy is already in clinical trials for organ transplant patients (50), our data support a multifaceted approach where therapeutically targeting Satb1 may synergize with Treg-inducing or Treg transfer therapies to promote transplantation tolerance.
Materials and Methods
Mice.
C57BL/6 (B6) and BALB/c (B/c) mice were purchased from Envigo, RMS, Inc. OTII, 129/J and CD45.1+ congenic C57BL/6J mice were purchased from Jackson Laboratories. Rag−/− and P14Rag−/− mice were obtained from Taconic Biosciences. P14 (Vβ8.1+/8.2+) CD8+ T cells are specific for an LCMV viral peptide presented by H2-Db on B6 APCs. TCR75 CD4+ TCR-transgenic mice, a gift from R. Pat Bucy, were bred with B6 and Rag−/− CD45.1+ mice on a B6 background. TCR75 (Vβ8.3+) CD4+ T cells recognize the B/c derived Kd54-68 allopeptide presented by I-Ab on B6 APCs. CD4-Cre Satb1fl/fl mice were provided by Jose Conejo-Garcia and Foxp3-GFP mice were obtained from Alexander Rudensky. OTII (Vα2/Vβ5) CD4+ T cells are specific for OVA323-339. OTII mice were bred with B6 and Rag−/− CD45.1+ mice on a B6 background. 2W-OVA (membrane-bound OVA) mice on a B6 background, were previously described (51). B6 and B/c mice were crossed once to generate CB6F1 hybrids. Antigen-presenting cells (APCs) from CB6F1 mice are H-2b/Kd+ and can directly present antigen to TCR75 T cells. Mice at 2 to 5 mo of age were used in the study and were housed in specific pathogen-free conditions. Animals were used in accordance with the University of Chicago’s Institutional Animal Care and Use Committee, according to the NIH guidelines for animal use.
Organ Transplantation and Tolerance Induction.
A mouse model of heterotopic heart transplantation was used as previously described (19). The day of heart allograft rejection was defined as the day of complete cessation of heartbeat. For induction of tolerance, heart recipients received an intravenous injection of splenocytes from 1/4 of donor spleen on day 0 and were treated with 600 μg of anti-CD154 (MR1) blocking mAb (Bio-X-cell) on days 0 (intravenous), 7 (intraperitoneal), and 14 (intraperitoneal) postheart transplantation. Skin transplantation was performed following previously described methods (52). B/c or 2W-OVA-B6 tail skin allografts were transplanted on the flank of B6 recipients. The rejection of skin allografts was defined as greater than 80% necrosis. For some experiments, memory against donor B/c alloantigens was generated via an intraperitoneal injection of B/c DST.
Adoptive T Cell Transfers.
A total of 105 naïve TCR75 T cells were injected intravenously on day −1 into B6 mice. To calculate the number of naïve TCR75 T cells for adoptive transfer, 1 × 106 spleen cells were stained for CD45.1, CD4, Vβ8.3, and CD44 and the percentage of CD44loCD45.1+CD4+Vβ8.3+ TCR75 T cells was estimated. B6 mice subsequently received B/c hearts on day 0 and were either left untreated, or the tolerance protocol was initiated. For experiments involving skin graft tolerance, 105 naïve OTII T cells were used for seeding, and tolerance was induced to 2W-OVA-B6 skin grafts as described. In a different set of experiments, P14Rag−/− mice were transferred on day −1 with 4.5 × 105 sorted naïve (CD127hi CD44lo CD25− Nrp1−) littermate control or Satb1−/− Tconvs alone or with 2.25 × 105 sorted GFP+ Tregs from FoxP3-GFP+ mice followed by transplantation of B/c skin allografts on day 0. All adoptive T cell transfers were done via retro-orbital intravenous route.
Tissue Preparation.
Spleens were mechanically dissociated. Cell suspensions were red blood cell (RBC) lysed before downstream processing. Heart tissues were first perfused with heparin (Sigma)-containing Hank's balanced salt solution (HBSS) (1:1,000) using an 18-gauge needle followed by rinsing with heparin-containing HBSS in a Petri dish to eliminate contaminating blood. Hearts were then digested with 400 U/mL collagenase IV (Sigma) as described previously (19). Graft-infiltrating cells were not from contaminating blood as mean percentage of TCR75 cells within infiltrating CD4+ cells was 14.3%, whereas that in spleen was 0.67% (SI Appendix, Fig. 1B).
Cell Enrichment and Sorting.
Splenic CD4+ T cells were purified and subsequently sorted for naïve (CD127hi CD44lo CD25− Nrp1−) CD4+ Tconvs from wildtype (WT) or CD4-Cre Satb1fl/fl mice or sorted for GFP+ Tregs from Foxp3-GFP (Satb1-competent) mice. In some assays, CD4+ T cells from spleens of TCR75 mice were enriched and sorted for naïve (CD44lo CD25−) CD4+ T cells. Around 70 to 90% of naïve CD4+ T cells from TCR75 mice are Vβ8.3+ that can recognize a Kd allopeptide. The naïve T cells were then used in cell culture assays. In some experiments, spleen, lymph nodes (LN) and heart allografts were harvested at day 3 ± 1, day 12 ± 2 or day ≥35 post heart-transplantation. TCR75 cells were enriched using biotin-labeled αCD45.1 (A20, eBioscience) and streptavidin magnetic beads (Miltenyi Biotec). Enriched TCR75 cells were either analyzed by flow cytometry or sorted for CD4+Vβ8.3+ for RNA sequencing. OTII T cells were similarly enriched and sorted day ≥35 post skin-transplantation. In other assays, where TCR75Rag−/− mice were the source of cells, enriched TCR75 T cells were sorted only for CD4+ and used for in vitro cultures. CD4+ T cells were purified using a commercial kit (Miltenyi Biotec). Cell sorting was performed on FACSAria II, FACSAria III, or FACSAria Fusion (BD Biosciences).
RNA Sequencing.
A minimum of 4 × 103 TCR75 (CD45.1+CD4+Vβ8.3+) T cells were sorted from spleen and heart allograft of tolerant or rejecting mice at day ≥35 posttransplantation for RNA-seq. Briefly, total RNA was extracted (RNeasy mini and micro kit, Qiagen) and used to generate full-length cDNA and a sequencing library using the SMART-seq2 method (53). Sequencing was conducted on HisEq. 4000 (Illumina), in a single end 50-bp read length run. The raw sequences were aligned to mouse reference genome GRCm38 using STAR (version 2.5.3a) (54). Aligned sequences were quantified (featureCounts) (55) followed by normalization with the trimmed mean of m-values method (56–59). Differential gene expression (adj P ≤ 0.05) was calculated using quasi-likelihood F-test in edgeR (51–53). Normalized log2 transformed expression values in count per million (CPM) were used to generate a Principal Component Analysis (PCA) plot. The CPM values were scaled for heat maps visualization. PCA plot and heat maps were generated in R (version 3.4.1). The pathway analysis on differentially expressed genes was performed in IPA software between May–June 2020 (version 52912811, Qiagen). Mouse transcription factors were extracted from the Riken database (genome.gsc.riken.jp/TFdb/htdocs/tf_list.html).
Cell Culture.
Sorted naïve control, Satb1−/− Tconvs and TCR75 T cells were labeled with 5 µM (Invitrogen) prior to stimulation. For stimulation, naïve Tconvs were activated with 0.1 µg/mL of soluble αCD3ε mAb (145-2C11; Bio-X-cell) in presence of RBC-lysed splenocytes from Rag−/− mice at 1:1 (T cell:Rag−/− splenocyte) ratio. For activation of TCR75 T cells, either 0.1 or 1 µg/mL of soluble αCD3ε was used. In some experiments, Tconvs were activated with 5 μg/mL of soluble anti-CD3ε and cultured with a titration of Tacrolimus (Prograf; Astellas) 0 to 100,000 ng/mL for 48 h. In Treg suppression assays, graded numbers of sorted GFP+ Tregs from Foxp3-GFP mice were added to the cultures, in the presence or absence of a dose titration of exogenous human IL-2 (teceleukin, Tecin). In CFSE dilution assays, either all proliferating cells (if overall proliferation was <50%) or the cells in division >2 (if overall proliferation was >50%) were included in the analysis for significance. In some experiments, sorted naïve CD4+ T cells from TCR75 mice were cocultured with splenocytes from CB6F1 mice and 50 μg/mL anti-CD154 (MR1) was added to the cultures. Prior to use, CB6F1 splenocytes were T-cell depleted using anti-CD90.2 (30-H12, BioLegend) and rabbit complement (Pel-Freeze Biologicals) as described previously (60) and then stimulated overnight with lipopolysaccharide (10 µg/mL, Sigma). Cultured CD4+ T cells were analyzed by flow cytometry at day 3. Culture supernatants were tested for IL-2 by ELISA (BioLegend) at day 1.
Flow Cytometry.
Cells were stained with a fixable live/dead aqua stain (Invitrogen) and with fluorochrome conjugated antibodies to Thy1.2 (53-2.1), CD4 (RM4-5), CD8 (53-6.7), CD44 (IM7), CD127 (A7R34), CD25 (PC61.5), Nrp1 (3E12), Vβ8.3 (1B3.3), CD45.1 (A20), Satb1 (14/Satb1), and Foxp3 (JFK-16s). For experiments involving tetramer staining, splenocytes were either CD4-enriched (Miltenyi Biotec) or sorted for CD4+ T cells. A total of 4 × 106 cells were stained with either phycoerythrin-coupled 2W(EAQGALANWAVDSA):I-Ab (to identify endogenous polyclonal CD4+ T cells reactive to the 2W donor peptide indirectly presented by host I-Ab) or pKd(ASDFPEYWEEQTQRAKSD):I-Ab (to identify endogenous polyclonal CD4+ T cells reactive to a peptide from donor Kd indirectly presented by host I-Ab) (both obtained from the NIH tetramer core facility) for 1 h at room temperature in a dark water bath immediately after live/dead staining and prior to surface staining. The Mouse Vβ TCR screening panel (BD Biosciences) was used to stain 14 TCR Vβ families. Proliferation of CD4+ Tconvs was measured by CFSE (Invitrogen) dilution. Intracellular staining was performed using FoxP3/Transcription Factor Staining Buffer (eBioscience). Cells were acquired on LSR Fortessa (BD Biosciences) or Aurora (Cytek Biosciences) and data were analyzed using Flow Jo software (version 9 or 10; TreeStar). Antibodies were purchased from BD Biosciences, eBioscience or BioLegend.
In Vivo IL-2 Treatment.
A group of P14Rag−/− mice that were adoptively transferred with naïve control or Satb1−/− Tconvs and GFP+ Tregs on day −1 and received B/c skin grafts on day 0, were treated intraperitoneally with 35,000 IU of recombinant human IL-2 (teceleukin, Tecin) daily from day 0 to 12.
Statistics.
One-way or two-way ANOVA with pairwise comparisons or unpaired or paired Student’s t test were used as appropriate. Allograft median survival time (MST) was calculated (Kaplan-Meier analysis) and compared using log-rank (Mantel-Cox) test. P values < 0.05 were considered significant. Data analyses were performed using GraphPad Prism 8.0.
Supplementary Material
Acknowledgments
This work was supported by NIH grants to J.R.C.-.G. (R01CA240434) and to A.S.C. and M.L.A. (P01AI-97113). J.B.A. received NIH predoctoral training grant (T32 AI7090-41). J.M.F. received a Katen fellowship (Katen Scholars Program, University of Chicago, 2018). We thank members of the Gajewski, Sperling, Chong and Alegre labs for helpful comments on the project, and the staff of the Flow Cytometry (RRID: SCR_017760), Genomics (RRID:SCR_019196) and Animal Research Center (RRID:SCR_021806C) core facilities at the University of Chicago for their technical assistance.
Footnotes
Competing interest statement: J.R.C.G. has financial agreements with Compass Therapeutics, Anixa Biosciences (stock options, consulting fees and sponsored research) and Leidos (consulting fees). P.K.G. now works at AstraZeneca. J.A. now works at Kite Pharma.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2205062119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
References
- 1.Krieger N. R., Yin D. P., Fathman C. G., CD4+ but not CD8+ cells are essential for allorejection. J. Exp. Med. 184, 2013–2018 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pietra B. A., Wiseman A., Bolwerk A., Rizeq M., Gill R. G., CD4 T cell-mediated cardiac allograft rejection requires donor but not host MHC class II. J. Clin. Invest. 106, 1003–1010 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu Z., et al. , Primed macrophages directly and specifically reject allografts. Cell. Mol. Immunol. 17, 237–246 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matesic D., et al. , Type 2 immune deviation has differential effects on alloreactive CD4+ and CD8+ T cells. J. Immunol. 161, 5236–5244 (1998). [PubMed] [Google Scholar]
- 5.Yuan X., et al. , A novel role of CD4 Th17 cells in mediating cardiac allograft rejection and vasculopathy. J. Exp. Med. 205, 3133–3144 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee R. S., Grusby M. J., Glimcher L. H., Winn H. J., Auchincloss H. Jr., Indirect recognition by helper cells can induce donor-specific cytotoxic T lymphocytes in vivo. J. Exp. Med. 179, 865–872 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grazia T. J., et al. , Acute cardiac allograft rejection by directly cytotoxic CD4 T cells: Parallel requirements for Fas and perforin. Transplantation 89, 33–39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conlon T. M., et al. , Germinal center alloantibody responses are mediated exclusively by indirect-pathway CD4 T follicular helper cells. J. Immunol. 188, 2643–2652 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawai T., et al. , HLA-mismatched renal transplantation without maintenance immunosuppression. N. Engl. J. Med. 358, 353–361 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scandling J. D., et al. , Tolerance and chimerism after renal and hematopoietic-cell transplantation. N. Engl. J. Med. 358, 362–368 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Brouard S., et al. , The natural history of clinical operational tolerance after kidney transplantation through twenty-seven cases. Am. J. Transplant. 12, 3296–3307 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Leventhal J., et al. , Chimerism and tolerance without GVHD or engraftment syndrome in HLA-mismatched combined kidney and hematopoietic stem cell transplantation. Sci. Transl. Med. 4, 124ra28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawai T., et al. , Long-term results in recipients of combined HLA-mismatched kidney and bone marrow transplantation without maintenance immunosuppression. Am. J. Transplant. 14, 1599–1611 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niimi M., et al. , The role of the CD40 pathway in alloantigen-induced hyporesponsiveness in vivo. J. Immunol. 161, 5331–5337 (1998). [PubMed] [Google Scholar]
- 15.You S., et al. , Induction of allograft tolerance by monoclonal CD3 antibodies: A matter of timing. Am. J. Transplant. 12, 2909–2919 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Goto R., You S., Zaitsu M., Chatenoud L., Wood K. J., Delayed anti-CD3 therapy results in depletion of alloreactive T cells and the dominance of Foxp3+ CD4+ graft infiltrating cells. Am. J. Transplant. 13, 1655–1664 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarraj B., et al. , Impaired selectin-dependent leukocyte recruitment induces T-cell exhaustion and prevents chronic allograft vasculopathy and rejection. Proc. Natl. Acad. Sci. U.S.A. 111, 12145–12150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H., et al. , Ablation of interferon regulatory factor 4 in T cells induces “memory” of transplant tolerance that is irreversible by immune checkpoint blockade. Am. J. Transplant. 19, 884–893 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller M. L., et al. , Spontaneous restoration of transplantation tolerance after acute rejection. Nat. Commun. 6, 7566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta P. K., McIntosh C. M., Chong A. S., Alegre M.-L., The pursuit of transplantation tolerance: New mechanistic insights. Cell. Mol. Immunol. 16, 324–333 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guiducci C., Valzasina B., Dislich H., Colombo M. P., CD40/CD40L interaction regulates CD4+CD25+ T reg homeostasis through dendritic cell-produced IL-2. Eur. J. Immunol. 35, 557–567 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Honjo K., Xu Xy., Bucy R. P., CD4+ T-cell receptor transgenic T cells alone can reject vascularized heart transplants through the indirect pathway of alloantigen recognition. Transplantation 77, 452–455 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Miller M. L., et al. , Resilience of T cell-intrinsic dysfunction in transplantation tolerance. Proc. Natl. Acad. Sci. U.S.A. 116, 23682–23690 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford A., et al. , Molecular and transcriptional basis of CD4+ T cell dysfunction during chronic infection. Immunity 40, 289–302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mueller D. L., Jenkins M. K., Molecular mechanisms underlying functional T-cell unresponsiveness. Curr. Opin. Immunol. 7, 375–381 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Besançon A., et al. , The induction and maintenance of transplant tolerance engages both regulatory and anergic CD4+ T cells. Front. Immunol. 8, 218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J., et al. , Ablation of transcription factor IRF4 promotes transplant acceptance by driving allogenic CD4+ T cell dysfunction. Immunity 47, 1114–1128.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan O., et al. , TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott A. C., et al. , TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seo H., et al. , TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl. Acad. Sci. U.S.A. 116, 12410–12415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alfei F., et al. , TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Stephen T. L., et al. , SATB1 expression governs epigenetic repression of PD-1 in tumor-reactive T cells. Immunity 46, 51–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickinson L. A., Joh T., Kohwi Y., Kohwi-Shigematsu T., A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell 70, 631–645 (1992). [DOI] [PubMed] [Google Scholar]
- 34.Kakugawa K., et al. , Essential roles of SATB1 in specifying T lymphocyte subsets. Cell Rep. 19, 1176–1188 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Yasui D., Miyano M., Cai S., Varga-Weisz P., Kohwi-Shigematsu T., SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419, 641–645 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Krieg C., Létourneau S., Pantaleo G., Boyman O., Improved IL-2 immunotherapy by selective stimulation of IL-2 receptors on lymphocytes and endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 107, 11906–11911 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonnet B., et al. , Low-dose IL-2 induces regulatory T cell-mediated control of experimental food allergy. J. Immunol. 197, 188–198 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Yang J., et al. , Allograft rejection mediated by memory T cells is resistant to regulation. Proc. Natl. Acad. Sci. U.S.A. 104, 19954–19959 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mercadante E. R., Lorenz U. M., Breaking free of control: How conventional T cells overcome regulatory T cell suppression. Front. Immunol. 7, 193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis E. L., et al. , NFAT-dependent and -independent exhaustion circuits program maternal CD8 T cell hypofunction in pregnancy. J. Exp. Med. 219, e20201599 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trefzer A., et al. , Dynamic adoption of anergy by antigen-exhausted CD4+ T cells. Cell Rep. 34, 108748 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Shevach E. M., Foxp3+ T regulatory cells: Still many unanswered questions-a perspective after 20 years of study. Front. Immunol. 9, 1048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korn T., et al. , Myelin-specific regulatory T cells accumulate in the CNS but fail to control autoimmune inflammation. Nat. Med. 13, 423–431 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sojka D. K., Hughson A., Sukiennicki T. L., Fowell D. J., Early kinetic window of target T cell susceptibility to CD25+ regulatory T cell activity. J. Immunol. 175, 7274–7280 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Wehrens E. J., et al. , Anti-tumor necrosis factor α targets protein kinase B/c-Akt-induced resistance of effector cells to suppression in juvenile idiopathic arthritis. Arthritis Rheum. 65, 3279–3284 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Chen B., et al. , Akt-signal integration is involved in the differentiation of embryonal carcinoma cells. PLoS One 8, e64877 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong H. S., et al. , A local regulatory T cell feedback circuit maintains immune homeostasis by pruning self-activated T cells. Cell 184, 3981–3997.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ali J. M., et al. , Diversity of the CD4 T cell alloresponse: The short and the long of it. Cell Rep. 14, 1232–1245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beyer M., et al. , Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat. Immunol. 12, 898–907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atif M., Conti F., Gorochov G., Oo Y. H., Miyara M., Regulatory T cells in solid organ transplantation. Clin. Transl. Immunology 9, e01099 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moon J. J., et al. , Quantitative impact of thymic selection on Foxp3+ and Foxp3- subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc. Natl. Acad. Sci. U.S.A. 108, 14602–14607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kellersmann R., Zhong R., “Surgical technique for skin transplantation in mice” in Organtransplantation in Rats and Mice: Microsurgical Techniques and Immunological Principles, Timmermann W., Gassel H.-J., Ulrichs K., Zhong R., Thiede A., Eds. (Springer, 1998), pp. 151–154. [Google Scholar]
- 53.Picelli S., et al. , Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Robinson M. D., Oshlack A., A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lund S. P., Nettleton D., McCarthy D. J., Smyth G. K., Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat. Appl. Genet. Mol. Biol. 11 1544–6115 (2012). [DOI] [PubMed] [Google Scholar]
- 59.McCarthy D. J., Chen Y., Smyth G. K., Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dittel B. N., Depletion of specific cell populations by complement depletion. J. Vis. Exp. 36, 1487 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.