Significance
Levels of the orphan G protein–coupled receptor GPR3 are elevated in a subset of patients with Alzheimer’s disease (AD). Our group previously showed that genetic deletion of Gpr3 attenuates amyloid-β (Aβ) pathology in multiple AD mouse models, highlighting the therapeutic potential of GPR3 as a drug target for AD. However, Gpr3-deficient mice display several adverse phenotypes, including anxiety-like behavior and cognitive deficits. Here, we genetically modified Gpr3 in naive mice and in a preclinical AD mouse model and demonstrated that biased GPR3 signaling reduces AD pathology and induces glial activation in the absence of an effect on basal anxiety levels or cognitive function. Thus, biased GPR3 therapeutics are potentially neuroprotective and a safer avenue for therapeutic intervention in AD.
Keywords: G protein–coupled receptor, arrestin, biased signaling, Alzheimer’s disease, amyloid plaques
Abstract
Biased G protein–coupled receptor (GPCR) ligands, which preferentially activate G protein or β-arrestin signaling pathways, are leading to the development of drugs with superior efficacy and reduced side effects in heart disease, pain management, and neuropsychiatric disorders. Although GPCRs are implicated in the pathophysiology of Alzheimer’s disease (AD), biased GPCR signaling is a largely unexplored area of investigation in AD. Our previous work demonstrated that GPR3-mediated β-arrestin signaling modulates amyloid-β (Aβ) generation in vitro and that Gpr3 deficiency ameliorates Aβ pathology in vivo. However, Gpr3-deficient mice display several adverse phenotypes, including elevated anxiety-like behavior, reduced fertility, and memory impairment, which are potentially associated with impaired G protein signaling. Here, we generated a G protein–biased GPR3 mouse model to investigate the physiological and pathophysiological consequences of selective elimination of GPR3-mediated β-arrestin signaling in vivo. In contrast to Gpr3-deficient mice, G protein–biased GPR3 mice do not display elevated anxiety levels, reduced fertility, or cognitive impairment. We further determined that G protein–biased signaling reduces soluble Aβ levels and leads to a decrease in the area and compaction of amyloid plaques in the preclinical AppNL-G-F AD mouse model. The changes in amyloid pathology are accompanied by robust microglial and astrocytic hypertrophy, which suggest a protective glial response that may limit amyloid plaque development in G protein–biased GPR3 AD mice. Collectively, these studies indicate that GPR3-mediated G protein and β-arrestin signaling produce discrete and separable effects and provide proof of concept for the development of safer GPCR-targeting therapeutics with more directed pharmacological action for AD.
G protein-coupled receptors (GPCRs) are the most successful class of drug targets (1). However, more than 90% of agents that enter phase 1 trials fail to achieve U.S. Food and Drug Administration approval, primarily because of safety concerns or lack of efficacy (2). GPCRs classically activate G proteins (e.g., Gs, Gi/o, Gq, G12) and β-arrestins (e.g., βarr1, βarr2) to mediate distinct cellular and physiological effects (3). Biased GPCR ligands preferentially activate G protein- or β-arrestin-mediated signaling pathways and present opportunities to fine-tune physiology and develop more selective and safer therapeutics in the absence of on-target side effects (4). Accordingly, biased GPCR ligands are currently being tested in various therapeutic areas, including cardiovascular disease, pain management, chronic inflammation, and neuropsychiatric disorders (5). Surprisingly, although several GPCRs have been implicated in the pathophysiology of Alzheimer’s disease (AD) (6–8), biased GPCR signaling is an unexplored area of therapeutic intervention for AD.
AD is a progressive neurodegenerative disease characterized by extracellular deposition of amyloid-β (Aβ) in amyloid plaques and intracellular aggregation and accumulation of tau in neurofibrillary tangles (9). The pathological accumulation of Aβ and tau is accompanied by neurotoxicity, cell death, and neuroinflammation; the latter includes alterations in the morphology and transcription expression profile of microglia, astrocytes, and oligodendrocytes (10). Recent studies indicate that Aβ is sufficient to drive a neuroinflammatory response (11). Glial activation, in turn, may be neuroprotective, leading to increased Aβ clearance (12). Nevertheless, the mechanisms involved in the crosstalk between Aβ pathology and neuroinflammation are still poorly understood. In AD, GPCRs are associated with multiple stages of amyloid precursor protein (APP) proteolysis by the α-, β-, and γ-secretases, regulation of Aβ-mediated toxicity and Aβ degradation, tau pathobiology, neuroinflammation, and memory deficits (6–8, 13, 14). Accordingly, there is an intimate association between GPCRs and AD pathogenesis, highlighting the potential of GPCRs as drug targets of the multifactorial nature of AD.
We previously identified the orphan GPCR GPR3 as a key modulator of γ-secretase activity and determined that GPR3 levels are elevated in a subset of patients with AD (15, 16). Genetic deletion of Gpr3 leads to a significant reduction in Aβ pathology and alleviation of the cognitive deficits in AD transgenic mice (15, 17). However, in contrast to the beneficial effects on Aβ pathogenesis, Gpr3−/− mice also display several adverse phenotypes, including elevated anxiety-like and depression-like behavior (18), increased sensitivity to neuropathic pain (19), reduced neuronal survival following brain ischemia (20), higher responsiveness to cocaine reward (21), and reduced fertility (22). These observations are clinically significant as they indicate that total inhibition of GPR3 signaling may attenuate Aβ pathology to the detriment of other AD-related clinical symptoms (e.g., anxiety). Importantly, the in vivo contribution of G protein–biased signaling to the physiological and pathophysiological regulation of GPR3 has not been previously tested but is necessary to provide insight into the therapeutic potential and putative safety of biased GPR3, and GPCR, signaling to treat patients with AD.
Here, we hypothesized that the generation of a G protein–biased GPR3 mouse model, which maintains G protein signaling while eliminating βarr2 signaling, would lower Aβ levels while preserving the beneficial physiological effects of G protein signaling. Accordingly, we find that, in contrast to Gpr3 knockout (KO) mice, G protein–biased GPR3 (HA-Ala) mice are not anxious, display intact fertility, and maintain cognitive function with age, demonstrating that G protein, but not β-arrestin, signaling is involved in the maintenance of these physiological functions. To investigate the effect of G protein–biased GPR3 signaling on AD pathogenesis, we determined that G protein–biased signaling leads to a reduction in Aβ generation in primary neuronal cultures and Aβ levels in mouse brain samples from the AppNL-G-F AD mouse model. Concomitant with reduced Aβ levels, G protein–biased signaling in AD mice alters the neuroinflammatory state of microglia and astrocytes, which is in accordance with the observed reduction in amyloid plaque area and the increase in amyloid plaque compaction. Taken together, these results demonstrate that biased GPR3 signaling is a safer and more selective therapeutic intervention for AD that maintains the vital physiological functions of GPR3 while targeting Aβ pathology and neuroinflammation.
Results
Two Phosphorylation Clusters in the C Terminus of GPR3 Differentially Affect βarr2 Recruitment.
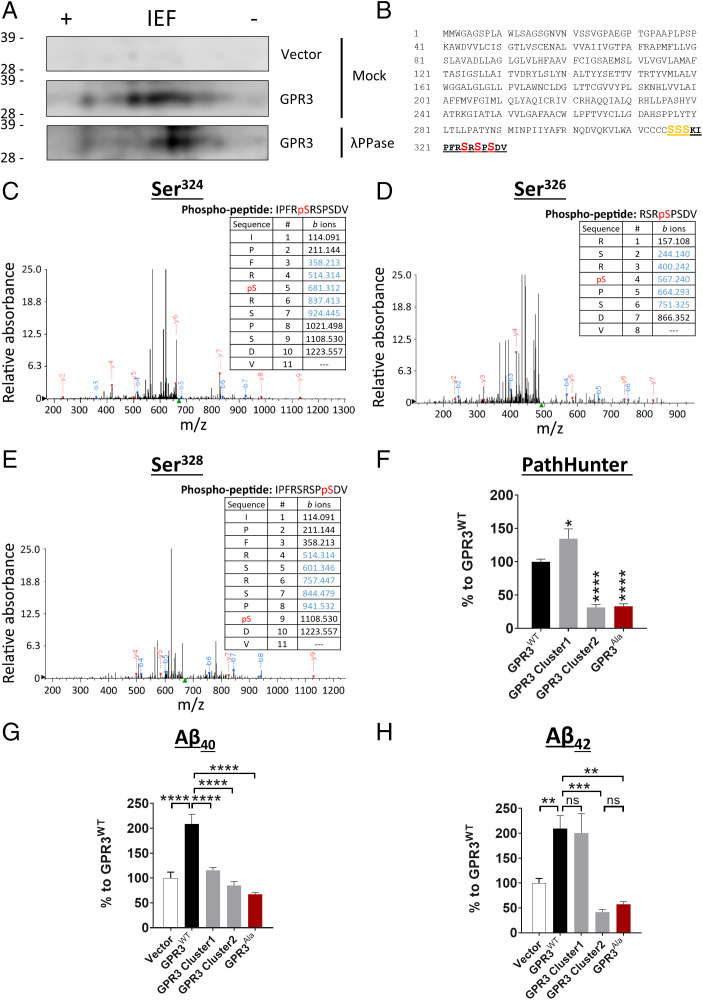
To initially determine whether GPR3 is phosphorylated, we expressed GPR3 in human embryonic kidney 293 cells (HEK293) and treated cell lysate samples with λ-phosphatase to remove the phosphate groups from serine, threonine, and tyrosine residues (23). We then analyzed the samples by two-dimensional electrophoresis and observed a shift in GPR3WT to the right (a more basic protein) after λ-phosphatase treatment, indicating that GPR3 is indeed phosphorylated (Fig. 1A).
Fig. 1.
The phosphorylation status of the GPR3 C terminus dictates βarr2 recruitment and Aβ generation. (A) Cell lysates from HEK293 cells, overexpressing empty vector or HA-tagged human GPR3, were immunoprecipitated with an HA antibody and subjected to mock or λ phosphatase (λPPase) treatment. Two-dimensional electrophoresis analysis with an isoelectric focusing (IEF) strip and sodium dodecyl sulfate–polyacrylamide gel electrophoresis (indicates that GPR3 is phosphorylated on C-terminal serine residues. (B) Schematic representation of the protein sequence of human GPR3 indicates the potential C-terminal phosphorylation sites. The peptide sequence identified by mass spectrometry analysis is highlighted in bold and underlined. The consistently identified phosphorylated residues Ser324, Ser326, and Ser328 are highlighted in red. The putative phosphorylated residues Ser316, Ser317, and Ser318 are highlighted in orange. n = 3 independent experiments. (C–E) Representative mass spectra and fragmentation tables show the three phosphorylated residues; detected b and y ions are indicated in blue and red, respectively. (F–H) Alanine mutagenesis of both cluster 1 and 2 serine residues shows a robust reduction in βarr2 recruitment to GPR3 (F) and Aβ levels (G and H). Vector condition refers to cells transfected with an empty control plasmid without GPR3. (F-H) P < 0.0001 by one-way ANOVA. Data are presented as mean ± SEM. ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by one-way ANOVA with Tukey’s post hoc test.
To identify the specific GPR3 phosphorylation sites, we immunopurified GPR3 from HEK293 cell lysates and analyzed the samples by liquid chromatography–tandem mass spectrometry. We consistently identified phosphorylated serine residues in the C terminus of GPR3 (Fig. 1 B–E). The distribution of the phosphorylation sites is arranged in two distinct clusters, a putative phosphorylated cluster 1 and a confirmed phosphorylated cluster 2 (Fig. 1B), consistent with previous findings for other GPCRs (24, 25). To determine whether the two phosphorylation clusters dictate different signaling outcomes, we performed alanine mutagenesis of the serine residues in each phosphorylation cluster and determined the effect on βarr2 signaling and Aβ generation. Surprisingly, mutagenesis of each cluster differentially affects βarr2 recruitment to GPR3 while similarly lowering Aβ levels (Fig. 1 F–H). In contrast, mutagenesis of both clusters (GPR3Ala) drastically diminishes βarr2 recruitment (∼70%) (Fig. 1F) and Aβ40 and Aβ42 generation (∼70%) (Fig. 1 G and H). Taken together, these results demonstrate that GPR3 is phosphorylated at the C terminus and identify two serine clusters that differentially contribute to βarr2 recruitment but exhibit similar effects on Aβ generation.
The GPR3Ala Mutant Is a G Protein–Biased Receptor.
GPCR-mediated mitogen-activated protein kinase (MAPK) activation can be regulated by G proteins (26) and β-arrestins (3). Given the drastic reduction in βarr2 recruitment to the GPR3Ala mutant (Fig. 1F), we sought to determine whether alanine mutagenesis of GPR3 generates a G protein–biased receptor (i.e., a receptor that maintains G protein signaling in the absence of coupling to βarr2). Expression of both GPR3WT and GPR3Ala shows a similar increase in phosphorylation of ERK and JNK relative to vector control–transfected cells (SI Appendix, Fig. S1 A–C). In contrast, phosphorylation of P38 is not regulated by GPR3 (SI Appendix, Fig. S1D). These results indicate that expression of GPR3 induces MAPK activation and, importantly, that alanine mutagenesis of GPR3 does not affect GPR3-dependent MAPK signaling.
To further investigate whether the GPR3Ala mutant affects G protein signaling, we expressed GPR3WT or GPR3Ala in a CHO-K1 cell line, which stably expresses βarr2, and treated the cells with a Gs (NF449) (27), Gi (pertussis toxin, PTX) (28), or Gq (FR900359) protein inhibitor (29). Pharmacological inhibition of Gs and Gi, but not Gq, significantly reduces ERK phosphorylation levels in GPR3WT- and GPR3Ala-transfected cells relative to vector control–transfected cells (SI Appendix, Fig. S1 E and F). These results indicate that GPR3 couples to Gs and Gi to mediate ERK phosphorylation and are in accordance with previous findings showing that GPR3 can couple to Gs and Gi (30) and, most recently, that constitutively active GPR3 couples primarily to Gs (31). Significantly, inhibition of G protein signaling does not affect the recruitment of βarr2 to GPR3 (SI Appendix, Fig. S1G). These results demonstrate that the GPR3Ala mutant specifically disrupts βarr2 recruitment while maintaining GPR3-mediated G protein signaling. Furthermore, expression of GPR3Ala leads to an increase in cyclic adenosine monophosphate (cAMP) levels relative to GPR3WT (SI Appendix, Fig. S1H), providing further validation of the G protein bias of the GPR3Ala mutant. Finally, we do not observe a change in the membrane localization of GPR3WT or GPR3Ala, indicating that alanine mutagenesis does not affect the cell surface expression of GPR3 (SI Appendix, Fig. S1 I and J). Taken together, these results indicate that the GPR3Ala mutant is a G protein–biased receptor and provide significant support for investigation of the physiological relevance of biasing GPR3 signaling in vivo.
CRISPR/Cas9 Genome Editing Generates a G Protein–Biased GPR3 Mouse Model.
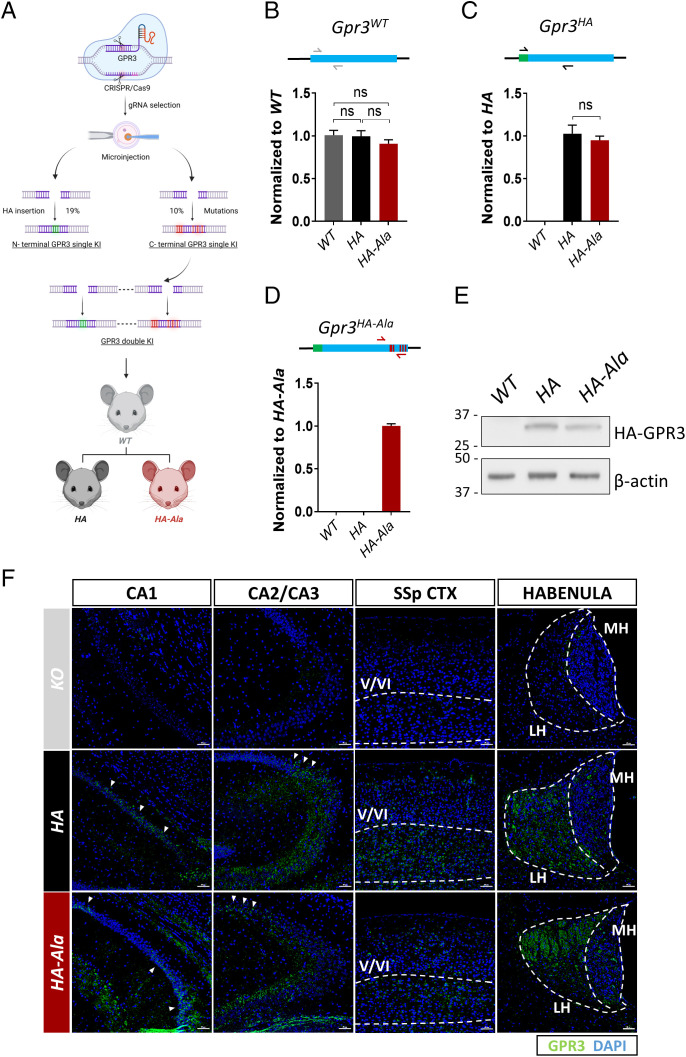
To date, no study has investigated the putative benefit of G protein–biased GPR3 signaling in vivo in modulating disease-associated phenotypes such as Aβ generation. Therefore, we used a CRISPR/Cas9 genome editing strategy to mutate the six serine/threonine residues in the GPR3 C terminus to phosphorylation-deficient alanine residues (S316A, T317A, S318A, S324A, S326A, and S328A) in naive mice. The Thr317 residue in murine GPR3 is similar to the human Ser317 residue. We also introduced an HA tag in the N terminus of GPR3 to determine the expression and localization of the GPR3 protein in vivo (Fig. 2A). The 15 highest-ranking potential off targets in the founder lines were excluded by Sanger sequencing analysis. Importantly, insertion of the HA tag and alanine mutagenesis of the endogenous Gpr3 gene were successful and do not affect Gpr3 messenger RNA (mRNA) levels in Gpr3+/+ (WT), Gpr3HA/HA (HA), or Gpr3HA-Ala/HA-Ala (HA-Ala) mice (Fig. 2 B–D). Accordingly, GPR3 protein levels are similar in HA and HA-Ala mice (Fig. 2E). Moreover, we established that the receptor is expressed in the CA1, CA2, and CA3 regions of the hippocampus, layers V and VI of the cortex, and the habenula in both mouse strains (Fig. 2F), which is consistent with previous reports on Gpr3 mRNA localization (18, 32, 33). These results demonstrate the successful generation of a G protein–biased GPR3 mouse model in the absence of effects on GPR3 mRNA and protein levels and reveal GPR3 protein localization in the brain.
Fig. 2.
An in vivo CRISPR/Cas9 gene-editing strategy was used to generate the G protein–biased GPR3 mouse model. (A) The schematic diagram shows the workflow to generate the two mouse models. The single KI HA contains 2xHA insertions in the N terminus of Gpr3. The double KI HA-Ala mouse contains 2xHA insertions at the N terminus of Gpr3 and serine–alanine mutations (S316A, T317A, S318A, S324A, S326A, S328A) in the C terminus of Gpr3. The double KI mouse model was generated from the embryos from F2 C terminus single KI mice. The success rates to obtain single KI mice are indicated in the graph. (B–D) The mRNA levels of Gpr3 in WT, HA, and HA-Ala male mice (n = 6 mice/genotype) at 4 mo of age were analyzed by quantitative PCR with three different sets of primers. (B) Gray arrows indicate endogenous Gpr3-specific primers, (C) black arrows indicate HA-GPR3-specific primers, and (D) red arrows indicate HA-GPR3 with the alanine mutation–specific primers. The results indicate that HA and HA-Ala mice express physiological levels of Gpr3. As a control, WT Gpr3 expression is undetectable with the HA- and HA-Ala-specific primers (C and D). HA-GPR3 is undetectable with HA-Ala-specific primers (D). Data were analyzed via one-way ANOVA (P = 0.42 [B], P < 0.0001 [C and D]) and are presented as mean ± SEM. ns, not significant. (E) Representative immunoblot analysis using an antibody to HA indicates that HA-GPR3 is expressed in the mouse cortex of HA and HA-Ala. (F) Representative images from the somatosensory parietal (SSp) cortex (CTX), hippocampal regions (CA1, CA2, and CA3), and habenula of KO (Top Panels), HA (Middle Panels), and HA-Ala (Bottom Panels) mice brains stained with anti-HA (GPR3, green) and DAPI (nuclei, blue). Dashed lines indicate layers V/VI of the cortex and medial (MH) and lateral (LH) habenula. Arrows indicate regions of the CA rich in GPR3 expression. Detection of GPR3 through HA-specific antibodies confirms localization of GPR3 in the cortex and hippocampus, with no region-specific differences between GPR3 WT and Ala mutant. Scale bars = 50 µm. Schematic (A) created with BioRender.com.
G Protein–Biased GPR3 Mice Maintain cAMP Levels and Do Not Display Cognitive or Behavioral Deficits.
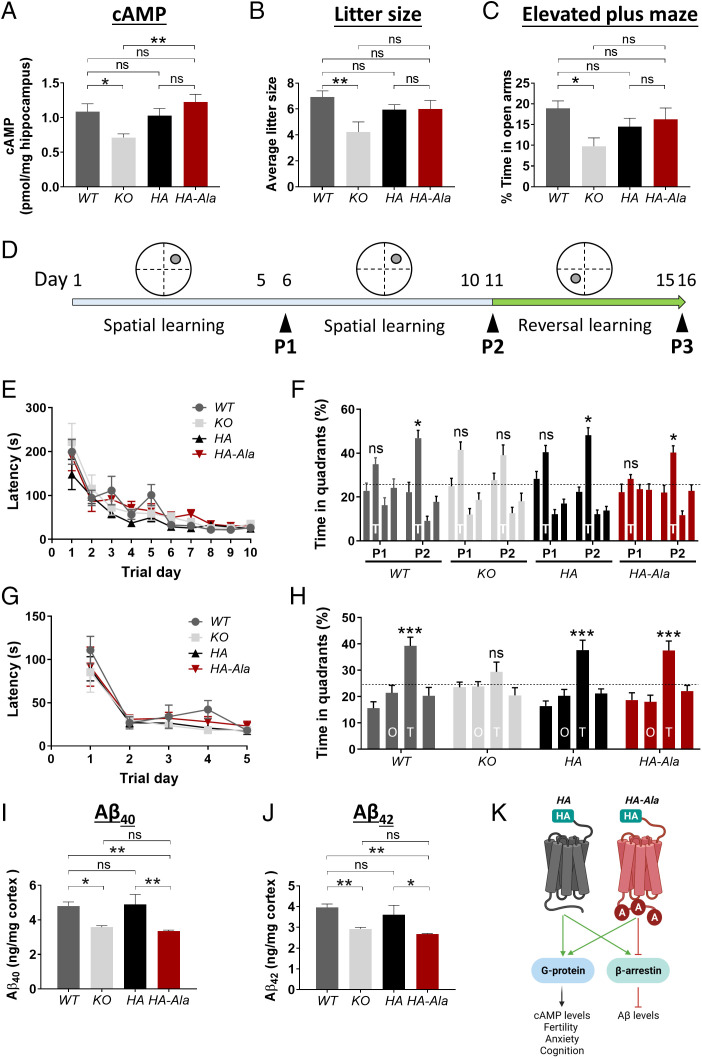
Expression of GPR3 activates G protein signaling, which leads to an elevation in cAMP levels (34, 35). Therefore, we measured cAMP levels in the hippocampus of WT, KO, HA, and HA-Ala mice. Levels of cAMP are reduced in KO mice but are similar in HA-Ala, HA, and WT mice (Fig. 3A). These results indicate that G protein signaling is not affected in G protein–biased GPR3 mice.
Fig. 3.
G protein–biased GPR3 mice display reduced Aβ levels and intact cognitive function. (A) cAMP levels were assessed by ELISA in male WT, KO, HA, and HA-Ala mice at 4 mo of age. cAMP levels are reduced in KO mice and unaffected in HA-Ala mice (n = 7–9 mice/genotype; P = 0.0041). (B) Average litter size is reduced in KO (mice relative to WT, HA, and HA-Ala mice at 3–6 mo of age; n = 7–15 mice/genotype; P = 0.0137). (C) Anxiety levels were assessed by the elevated plus maze behavioral paradigm in WT, KO, HA, and HA-Ala at 4 mo of age. The results indicate that KO mice display elevated anxiety (n = 14–21 male and female mice/genotype; P = 0.0445). (D) Schematic of the MWM behavioral paradigm illustrates the study design. P1, P2, and P3 indicate the probe trials. (E–H) Spatial learning and reference memory (E), spatial memory and retrieval (F), reversal learning (G), and reversal memory (H) were determined in WT, KO, HA, and HA-Ala mice (n = 10 male and female mice/genotype). T in (F) represents the target quadrant; O and T in (H) represent the opposite quadrant and new target quadrant, respectively. The establishment of spatial memory, reversal memory, and memory retrieval were detected in WT, HA, HA-Ala, but not in KO mice. (I and J) Endogenous Aβ40 (I) and Aβ42 (J) are reduced in male KO and HA-Ala mice relative to control WT and HA mice at 4 mo of age (n = 8–10 mice/genotype; Aβ40, P = 0.0004; Aβ42, P = 0.0006). For all datasets, data are presented as mean ± SEM. ns, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001 are determined by one-way ANOVA with a Tukey’s post hoc test. (K) The schematic diagram summarizes the physiological function of G protein–biased GPR3-mediated signaling in the HA and HA-Ala mouse models. Schematic created with BioRender.com.
Genetic deletion of Gpr3 (KO) results in several adverse phenotypes, including an increase in anxiety-like and depression-like behaviors (18) and reduced fertility (22). The increased anxiety-like and depression-like behaviors, which are elevated in patients with AD (36), and reduced fertility are reported to be due to reduced G protein signaling (22). We observed a reduction in the average litter size, an indication of reduced fertility, in KO, but not in HA or HA-Ala, relative to WT mice (Fig. 3B). Using the elevated plus maze behavioral paradigm, we observe an elevation in anxiety-like behavior in KO mice (Fig. 3C). However, importantly, HA-Ala mice do not display an anxiogenic phenotype (Fig. 3C). These results demonstrate that G protein–biased GPR3 mice do not display characteristic deficits associated with the absence of G protein signaling, which are observed in the KO animals.
To further establish whether G protein–biased GPR3 signaling affects hippocampal-dependent learning and memory, we used a battery of behavioral tests to assess cognitive function. We determined that WT, KO, HA, and HA-Ala mice have intact short-term and working memory by using the novel arm recognition and Y-maze behavioral paradigms (SI Appendix, Fig. S2). To assess the involvement of G protein–biased signaling in spatial learning and reference memory, we used the Morris water maze (MWM) behavioral paradigm (37) (Fig. 3D). Although all four genotypes display similar spatial learning (Fig. 3E), the KO mice fail to locate the target quadrant during the second probe trial (P2) (Fig. 3F), indicating that G protein signaling is essential for spatial memory and memory retrieval. To investigate cognitive flexibility, which is a correlate of executive function in humans (38), we tested spatial reversal learning and reference memory (39) by moving the platform to the opposite quadrant (Fig. 3D). All four genotypes display an intact reversal learning (Fig. 3G). However, similar to spatial memory and memory retrieval, only KO mice exhibit a deficit in spatial reversal memory and memory retrieval (Fig. 3H). All four genotypes display intact locomotor function (SI Appendix, Fig. S3). These studies demonstrate that GPR3-mediated G protein signaling is necessary for cognitive flexibility, specifically in the integration of spatial reversal memory and memory retrieval, which is compromised in KO mice but preserved in G protein–biased GPR3 mice.
Endogenous Aβ Levels Are Reduced in the G Protein–Biased GPR3 Mouse.
To investigate whether endogenous murine Aβ generation is affected in G protein–biased GPR3 mice, we analyzed Aβ40 and Aβ42 levels by enzyme-linked immunosorbent assay (ELISA). HA-Ala mice display reduced Aβ40 and Aβ42 levels, similar to KO mice (Fig. 3 I and J). These findings show that selective perturbation of β-arrestin signaling reduces endogenous murine Aβ generation. Immunoblot analysis of full-length APP, βarr1/2, and the γ-secretase subunits indicates that protein levels are not altered in HA-Ala mice in comparison to WT, KO, and HA mice (SI Appendix, Fig. S4). Taken together, these studies provide in vivo proof of concept for a putative pathophysiological role of biased GPR3 signaling in AD (Fig. 3K).
G Protein–Biased GPR3 Mice Display Reduced Pathology in a Preclinical AD Mouse Model.
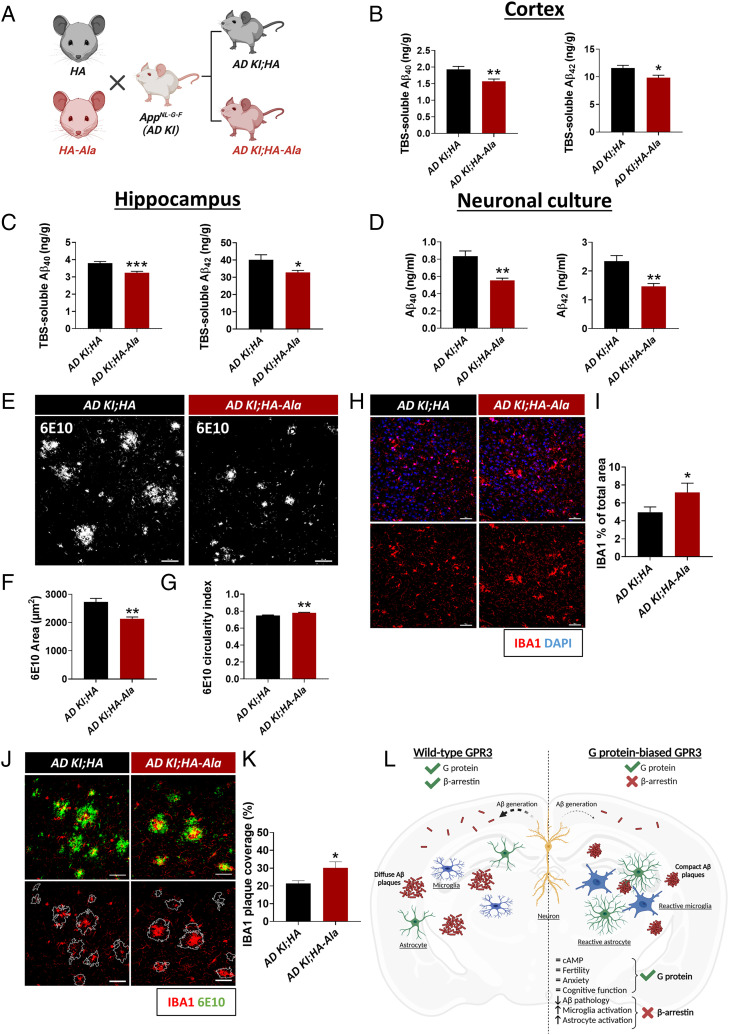
Given the physiological benefits of G protein–biased GPR3 signaling in naive mice, we sought to establish the putative impact of G protein–biased GPR3 signaling on AD pathogenesis. We used the App knock-in AD mouse model AppNL-G-F (AD KI), which has the humanized Aβ sequence containing the Swedish “NL” (KM670NL), Iberian “F” (I716F), and Arctic “G” (E693G) mutations (40). We crossed the AD KI mice with the HA (AppNL-G-F/NL-G-F;Gpr3HA/HA, hereafter referred to as AD KI;HA) and HA-Ala (AppNL-G-F/NL-G-F;Gpr3HA-Ala/HA-Ala, hereafter referred to as AD KI;HA-Ala) mice (Fig. 4A). The AD KI;HA and AD KI;HA-Ala mouse models display similar patterns of GPR3 expression and region-specific distribution (SI Appendix, Fig. S5). Significantly, Tris-buffered saline (TBS)-soluble Aβ40 and Aβ42 levels are reduced in both the cortex (Fig. 4B) and hippocampus (Fig. 4C) of AD KI;HA-Ala mice relative to AD KI;HA mice at 6 mo of age. Aβ40 and Aβ42 generation is also significantly reduced in neuronal cultures from AD KI;HA-Ala mice relative to AD KI;HA mice (Fig. 4D). These studies show that neuronal GPR3-mediated β-arrestin signaling is necessary for the modulation of Aβ generation endogenously and in an AD mouse model.
Fig. 4.
G protein–biased GPR3 AD mice display reduced Aβ pathology and increased microglial and astrocytic activation. (A) AD KI mice were crossed with the HA (AD KI;HA) and HA-Ala (AD KI;HA-Ala) mice. (B and C) TBS-soluble Aβ40 and Aβ42 are reduced in cortex and hippocampus of AD KI;HA-Ala mice relative to AD KI;HA mice at 6 mo of age (n = 12–16 male and female mice/genotype; Aβ40, P = 0.004; Aβ42, P = 0.013) (B) and hippocampus (Aβ40, P = 0.0004; Aβ42, P = 0.018) (C). (D) Aβ40 and Aβ42 levels are reduced in AD KI;HA-Ala relative to AD KI;HA neuronal cultures (n = 6 P0 mice/genotype; Aβ40, P = 0.0014; Aβ42, P = 0.0028). (E) Representative confocal immunofluorescence images show the cortex of AD KI;HA (Left Panel) and AD KI; HA-Ala (Right Panel) mice immunolabeled for Aβ plaques with an anti-APP antibody (6E10; white). (F) Aβ plaque area (in µm2) is reduced in AD KI;HA-Ala relative to AD KI;HA mice (P = 0.0027). (G) Amyloid plaque circularity index (scale of 0–1, with 1 being the most circular), as a measure of amyloid plaque compaction, indicates increased circularity of amyloid plaques in AD KI;HA-Ala relative to AD KI;HA mice (n = 6 male and female mice/genotype; P = 0.0027). (H) Representative confocal immunofluorescence images from the cortex indicate an increase in the percentage area occupied by IBA1+ cells in AD KI; HA-Ala (I) relative to AD KI;HA mice (n = 6 male and female mice/genotype). (J) Representative confocal immunofluorescence images show the cortex of AD KI;HA (Left Panel) and AD KI;HA-Ala (Right Panel) mice immunolabeled for microglia (IBA1; red) and Aβ plaques (6E10; green). (K) The amyloid plaque area covered by IBA1+ cells is increased in AD KI;HA-Ala relative to AD KI;HA mice. Data are presented as mean ± SEM, ns, not significant; *P < 0.05, **P < 0.01, and **P < 0.001 by unpaired Student’s t test. Scale bars = 10 µm. (L) Schematic diagram depicts the physiological and pathophysiological phenotypes of wild-type GPR3 and G protein–biased GPR3 mice in AD. Schematics (A and L) created with BioRender.com.
Neuroinflammation is a complex response to brain insult that involves the activation of glial cells, which accompanies the accumulation of AD pathology (41). In addition to the decrease in Aβ generation, we observe a decrease in the amyloid plaque area and an increase in the circularity of amyloid plaques in AD KI;HA-Ala relative to AD KI;HA mice (Fig. 4 E–G), which is indicative of amyloid plaque compaction. In accordance with these findings, we observe an increase in the total area occupied by microglia (Fig. 4 H and I) and in the amyloid plaque area occupied by microglia in the brains of AD KI;HA-Ala relative to AD KI;HA mice (Fig. 4 J and K), which is indicative of elevated microglia activation and recruitment to amyloid plaques in G protein–biased GPR3 AD mice. In addition, we observed an increase in the total area occupied by astrocytes (SI Appendix, Fig. S6 A and B) in the absence of a change in the amyloid plaque area occupied by astrocytes (SI Appendix, Fig. S6 C and D). Importantly, we do not observe a difference in glial reactivity in nontransgenic mouse brains (i.e., WT, HA, and, HA-Ala mice) (SI Appendix, Figs. S7 and S8). Taken together, these results demonstrate that G protein–biased GPR3 signaling in an AD mouse model specifically attenuates Aβ accumulation and leads to the activation and hypertrophy of astrocytes and microglia, with the latter actively restricting the development of amyloid plaques.
Discussion
Biased GPCR signaling (i.e., the preferential activation of G protein- or β-arrestin-mediated signaling pathways) is a comparatively new area of GPCR investigation that is transforming the conceptualization of GPCR signaling and its application for disease therapeutics. Here, we show that mutagenesis of C-terminal GPR3 phosphorylation disrupts the interaction with βarr2 while maintaining G protein signaling (i.e., cAMP levels), effectively generating an in vivo G protein–biased GPR3 model. We show that G protein–biased GPR3 mice do not display the memory deficits and elevated anxiety-like behavior observed in Gpr3−/− mice. Moreover, we demonstrate that G protein–biased GPR3 signaling decreases endogenous murine Aβ generation, providing exciting premise for the investigation of biased GPR3 signaling as a therapeutic avenue for AD. In this regard, we determined that biased GPR3 signaling in an AD mouse model lowers soluble Aβ levels and leads to a decrease in the area and an increase in the compaction of amyloid plaques. The changes in amyloid pathology correspond with the hypertrophy of microglia and astrocytes, suggesting a protective glial response that probably restricts the growth of amyloid plaques in G protein–biased GPR3 AD mice. Collectively, this study provides pioneering insight into how biased GPCR signaling can modify AD pathogenesis in vivo in a safer and more effective manner.
GPR3-mediated G protein signaling is responsible for the production of cAMP (42). cAMP signaling is critically involved in fertility, affective disorders (e.g., anxiety and depression), synapse growth, long-term memory formation, and memory consolidation (18, 22, 43, 44), which highlights the importance of maintaining this signaling pathway. Accordingly, we show that Gpr3−/− mice have reduced cAMP levels, decreased litter size (which is indicative of compromised fertility), memory deficits, and exacerbated anxiety-like behavior. All of these phenotypes are absent in G protein–biased GPR3 mice. Importantly, high anxiety levels are implicated in preclinical AD (45–47) through exacerbation of the Aβ-driven deterioration of global cognition, verbal memory, and executive function (48). Therefore, it is imperative for AD therapeutic interventions to avoid side effects that may worsen psychocognitive behavior. Notably, our findings strongly indicate that biased GPR3 signaling may potentially combat the highly complex and multifactorial nature of AD without contributing to the deterioration of behavioral physiology.
Several decades of AD therapeutic research have focused on Aβ-reducing agents (e.g., secretase inhibitors, aggregation blockers, and immunotherapies) (49). However, many safety concerns have halted the approval of broad secretase inhibitors to counteract Aβ pathology in AD (50–52). We showed that G protein–biased GPR3 signaling consistently reduces Aβ levels and pathology in neuronal models, nontransgenic murine brains, and an AD mouse model. Additionally, we previously demonstrated that GPR3 modulates Aβ generation via βarr2- and γ-secretase-dependent pathways in vitro (15, 16). Collectively, our studies show a neuron-specific decrease in γ-secretase-mediated Aβ generation upon disruption of GPR3–βarr2 signaling in vivo. These findings present a more specific strategy to modulate γ-secretase activity in the putative absence of side effects and toxicity associated with broad γ-secretase inhibitors (50–52).
Aberrant accumulation of Aβ is sufficient to drive a neuroinflammatory response in AD brains (11). Given that microglia and astrocytes are involved in Aβ clearance through active phagocytosis and degradation of toxic Aβ species (53), glial activation may be neuroprotective in AD (12). Here, we observed a reduction in the area and an increase in the compaction of amyloid plaques that is paralleled by an increase in the area covered by microglia and astrocytes in G protein–biased GPR3 AD mice. These findings suggest that glial cells may undergo a protective and accelerated response to slow or limit the Aβ burden in G protein–biased GPR3 AD mice. Importantly, disruption of microglial function can directly prevent amyloid plaque compaction (54–56) and drive neurotoxicity (54, 57–60). These studies support the putative beneficial nature of increased microglial hypertrophy and amyloid plaque coverage observed in our model. Future studies will help determine the causal relationship between biased GPR3 signaling, reactive gliosis, and Aβ pathogenesis. Nonetheless, the fact that the G protein–biased GPR3 mice display changes in both neuronal-driven Aβ generation and glial activation opens interesting avenues to explore the putative cell-type-specific functions of GPR3.
Here, we demonstrated that biased GPR3 signaling is protective against the generation and accumulation of Aβ pathology and that this effect is dependent on the phosphorylation status of the receptor. We showed that GPR3 is phosphorylated at the C terminus in two distinct serine clusters (clusters 1 and 2) that differentially regulate βarr2 recruitment, but both lead to a decrease in Aβ40 and Aβ42 generation. Specific GPCR phosphorylation barcodes have been shown to lead to distinct β-arrestin conformations that affect both GPCRs and activation of distinct downstream signaling pathways (24, 25, 61–73). For example, key phosphorylation sites in the rhodopsin C terminus act as “inhibitory sites,” interrupting β-arrestin binding, or as “modulator sites,” changing the global conformation and activation state of β-arrestin (73). Therefore, differential phosphorylation of serine clusters in the C terminus of GPR3 may induce specific conformations of βarr2, changing its availability to interact with secondary binding partners such G proteins, APP, and the γ-secretase complex subunits upstream of Aβ generation. Nevertheless, further investigation will be necessary to determine whether in vivo disruption of GPR3 phosphorylation may affect βarr2-independent signaling outcomes.
There is a growing body of evidence supporting the clinical use of biased GPCR signaling as a safer approach for therapeutic intervention. This strategy has been recently explored for the neurotensin receptor 1 in the treatment of addictive behaviors, the μ-opioid receptor in pain management, the muscarinic 1 receptor in prion disease, and the dopamine receptor D2 in Parkinson’s disease (61, 74–78). GPR3 is the first GPCR investigated to show the effects of biased GPCR signaling on AD pathogenesis. Although GPR3 is an orphan GPCR, the use of intracellular pepducins (i.e., small membrane-permeable peptides that mimic the C terminus of GPCRs) has been reported to be a readily translational strategy to bias GPCR signaling from the inside of cell without the need for an exogenous ligand (79). This observation combined with the proven efficacy of biased signaling of other GPCRs in ameliorating pathogenic phenotypes provides an exciting premise for the future application of GPR3-biased signaling in AD.
Materials and Methods
Mice.
B6.129P2-Gpr3tm1Dgen/Mmnc mice (Gpr3−/−; stock no. 011623-UNC) were obtained from the Mutant Mouse Resource and Research Center at the University of North Carolina at Chapel Hill, an NIH-funded strain repository, and were donated to the Mutant Mouse Resource and Research Center by Deltagen. The AppNL-G-F mice (40) were a gift from Dr. Takaomi C. Saido (RIKEN Brain Science Institute, Saitama, Japan). Mice were maintained in a specific-pathogen-free animal facility and group housed in individually ventilated microisolater cages under a 12-h light/dark cycle and had ad libitum access to food and water. All mice used in the study were on a C57BL/6J genetic background. Gpr3−/− and AppNL-G-F mice were backcrossed with WT mice for six generations before experimentation. All mice were bred to homozygosity unless otherwise specified.
In-House Generation of Mouse Models.
The Gpr3 mouse models HA and HA-Ala were generated in house through CRISPR/Cas9-mediated genome editing on an inbred C57BL/6J genetic background by introducing 2xHA tags in the N terminus of GPR3 or mutating serine and threonine residues in the C terminus of GPR3 to alanine residues. Potential off targets of individual single-guided RNA were excluded by Sanger sequencing. Detailed methods of the generation of mouse models are included in SI Appendix.
Mass Spectrometry.
Immunoprecipitated HA-GPR3 from 4 mg of total cell extracts was separated on a 4–12% Bis-Tris gel (Thermo Fisher Scientific). The gel was washed with double-distilled water (ddH2O) for 10 min and fixed with fixation buffer (50% methanol and 10% acetic acid) for 20 min. After fixation, the gel was washed three times with ddH2O for 10 min and stained with Bio-Safe Coomassie blue reagent (Bio-Rad) for 1 h. The gel was destained with ddH2O for 30 min. The target gel band was excised and analyzed by the Taplin Biological Mass Spectrometry Facility at Harvard Medical School to investigate phosphorylation of GPR3.
PathHunter Assay.
The CHO-K1 βarr2 cells (DiscoverX) were seeded on 96-well plates at a cell density of 20,000 cells per well. One day after seeding, cells were cotransfected with the GPR3 mutants and human APP-C99 with X-tremeGENE HP DNA transfection reagent (MilliporeSigma). Two days after seeding, the culture medium was replaced with serum-free Dulbecco’s modified Eagle medium overnight. The interaction between GPR3 and βarr2 was measured in cells with a PathHunter β-arrestin assay from DiscoverX according to the manufacturer’s protocol. The culture media were collected for Aβ ELISA analysis as previously described (16).
In Vitro and In Vivo cAMP Measurements.
For In Vitro cAMP.
The CHO-K1 βarr2 cells were seeded in six-well plates at a cell density of 500,000 cells per well. One day after seeding, cells were cotransfected with the GPR3 mutants and human APP-C99 with X-tremeGENE HP DNA Transfection Reagent (MilliporeSigma). Two days after seeding, the culture medium was replaced with serum-free Dulbecco’s modified Eagle medium overnight. Intracellular cAMP levels were then measured in cells with a cAMP Parameter Assay Kit (R&D Systems) according to the manufacturer’s protocol.
For In Vivo cAMP.
Mice were transcardially perfused with ice-cold phosphate-buffered saline. Mouse hippocampi were dissected and snap-frozen in liquid nitrogen. The tissue was homogenized with 0.1 M HCl, and endogenous cAMP levels were measured with a Direct cAMP ELISA kit (Enzo Life Sciences) according to the manufacturer’s protocol.
Aβ ELISA.
Aβ40 and Aβ42 levels were determined by standard sandwich ELISA with end-specific antibodies provided by Janssen Pharmaceuticals as previously described (17). Briefly, 96-well plates were coated and incubated overnight with monoclonal antibodies JRFcAb40/28 and JRFcAb42/26, which recognize the C terminus of Aβ species terminating at amino acid 40 or 42, respectively. Horseradish peroxidase–conjugated JRFAbN/25 and JRF/fAB/2 were used as the detection antibodies for human Aβ or murine Aβ, respectively. The culture supernatants from GPR3-transfected CHO-K1 βarr2 cells and primary neuronal cultures were subjected to Aβ40 and Aβ42 ELISA. Alternately, mouse cortices were extracted in a 0.4% diethylamine/50 mM NaCl solution containing complete protease inhibitors (MilliporeSigma). The homogenates were ultracentrifuged at 4 °C for 1 h at 100,000 × g. A 10% neutralization buffer (0.5 M Tris⋅HCl, pH 6.8) was added to the supernatant. The supernatant was subjected to the analysis of the endogenous murine Aβ40 and Aβ42 ELISA. Extraction of human Aβ40 and Aβ42 with TBS from the cortices and hippocampi of AppNL-G-F mice was performed as previously described (40). Immunoblot analysis of mouse brain homogenates was performed with 4–20% Mini-PROTEAN TGX Precast Gels (Bio-Rad). Immunodetection was performed with horseradish peroxidase–coupled secondary antibodies (Bio-Rad) and the chemiluminescent detection reagent Renaissance (PerkinElmer Life Sciences).
Statistical Analysis.
Data are presented as mean ± SEM unless otherwise noted. Statistical significance was determined in GraphPad Prism 7 software. Statistical analyses used for each study are noted in the figure legends. A two-tailed Student’s t test was used for the comparison of two means with one independent variable. A one-way ANOVA followed by a Tukey’s post hoc test was used for the comparison of multiple means with one independent variable. A two-way repeated-measures ANOVA followed by a Tukey’s post hoc test was used for the comparison of multiple means in MWM studies to determine whether there is significant time or genotype effect during training. Statistical significance is noted when P < 0.05, and the degree of significance is reported with *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, respectively.
Supplementary Material
Acknowledgments
We greatly appreciate the support and advice of Dr. Bart De Strooper (VIB and KU Leuven, Leuven, Belgium; UK Dementia Research Institute and University College London, London, United Kingdom) and the gift of the PS1-NTF, APP, and NCT antibodies. We thank Dr. Marc Mercken (Johnson & Johnson Pharmaceuticals R&D, Beerse, Belgium) for the Aβ antibodies. We are grateful to Katrien Horré, Elke Vandewyer, and Leen Wolfs (KU Leuven, Leuven, Belgium) for technical support. We thank Sean-Paul Gerard Williams and Gabi Little (University of Pittsburgh, Pittsburgh, PA) for technical support. We thank Dr. Stacey J. Sukoff Rizzo for discussions regarding the behavioral studies (Aging Institute, University of Pittsburgh, Pittsburgh, PA).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2204828119/-/DCSupplemental.
Data, Materials, and Software Availability
Additional materials and methods and other study data are included in the SI Appendix. All other data are included in the article and/or supporting information.
References
- 1.Hauser A. S., Attwood M. M., Rask-Andersen M., Schiöth H. B., Gloriam D. E., Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dowden H., Munro J., Trends in clinical success rates and therapeutic focus. Nat. Rev. Drug Discov. 18, 495–496 (2019). [DOI] [PubMed] [Google Scholar]
- 3.DeWire S. M., Ahn S., Lefkowitz R. J., Shenoy S. K., β-arrestins and cell signaling. Annu. Rev. Physiol. 69, 483–510 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Wisler J. W., Rockman H. A., Lefkowitz R. J., Biased G protein-coupled receptor signaling: Changing the paradigm of drug discovery. Circulation 137, 2315–2317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slosky L. M., Caron M. G., Barak L. S., Biased allosteric modulators: New frontiers in GPCR drug discovery. Trends Pharmacol. Sci. 42, 283–299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thathiah A., De Strooper B., The role of G protein-coupled receptors in the pathology of Alzheimer’s disease. Nat. Rev. Neurosci. 12, 73–87 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Zhao J., Deng Y., Jiang Z., Qing H., G protein-coupled receptors (GPCRs) in Alzheimer’s disease: A focus on BACE1 related GPCRs. Front. Aging Neurosci. 8, 58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y., Todd N., Thathiah A., The role of GPCRs in neurodegenerative diseases: Avenues for therapeutic intervention. Curr. Opin. Pharmacol. 32, 96–110 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Duyckaerts C., Delatour B., Potier M.-C., Classification and basic pathology of Alzheimer disease. Acta Neuropathol. 118, 5–36 (2009). [DOI] [PubMed] [Google Scholar]
- 10.De Strooper B., Karran E., The cellular phase of Alzheimer’s disease. Cell 164, 603–615 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Sierksma A., et al. , Novel Alzheimer risk genes determine the microglia response to amyloid-β but not to TAU pathology. EMBO Mol. Med. 12, e10606 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deczkowska A., et al. , Disease-associated microglia: A universal immune sensor of neurodegeneration. Cell 173, 1073–1081 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Chidambaram H., Chinnathambi S., G-protein coupled receptors and tau-different roles in Alzheimer’s disease. Neuroscience 438, 198–214 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Azam S., et al. , G-protein-coupled receptors in CNS: A potential therapeutic target for intervention in neurodegenerative disorders and associated cognitive deficits. Cells 9, E506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thathiah A., et al. , The orphan G protein-coupled receptor 3 modulates amyloid-beta peptide generation in neurons. Science 323, 946–951 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Thathiah A., et al. , β-arrestin 2 regulates Aβ generation and γ-secretase activity in Alzheimer’s disease. Nat. Med. 19, 43–49 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Huang Y., et al. , Loss of GPR3 reduces the amyloid plaque burden and improves memory in Alzheimer’s disease mouse models. Sci. Transl. Med. 7, 309ra164 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Valverde O., et al. , GPR3 receptor, a novel actor in the emotional-like responses. PLoS One 4, e4704 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz-Medina J., Ledent C., Valverde O., GPR3 orphan receptor is involved in neuropathic pain after peripheral nerve injury and regulates morphine-induced antinociception. Neuropharmacology 61, 43–50 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Tanaka S., et al. , Developmental expression of GPR3 in rodent cerebellar granule neurons is associated with cell survival and protects neurons from various apoptotic stimuli. Neurobiol. Dis. 68, 215–227 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Tourino C., et al. , The orphan receptor GPR3 modulates the early phases of cocaine reinforcement. Br. J. Pharmacol. 167, 892–904 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freudzon L., et al. , Regulation of meiotic prophase arrest in mouse oocytes by GPR3, a constitutive activator of the Gs G protein. J. Cell Biol. 171, 255–265 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhuo S., Clemens J. C., Hakes D. J., Barford D., Dixon J. E., Expression, purification, crystallization, and biochemical characterization of a recombinant protein phosphatase. J. Biol. Chem. 268, 17754–17761 (1993). [PubMed] [Google Scholar]
- 24.Prihandoko R., et al. , Distinct phosphorylation clusters determine the signaling outcome of free fatty acid receptor 4/G protein-coupled receptor 120. Mol. Pharmacol. 89, 505–520 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Bouzo-Lorenzo M., et al. , Distinct phosphorylation sites on the ghrelin receptor, GHSR1a, establish a code that determines the functions of ß-arrestins. Sci. Rep. 6, 22495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldsmith Z. G., Dhanasekaran D. N., G protein regulation of MAPK networks. Oncogene 26, 3122–3142 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Hohenegger M., et al. , Gsalpha-selective G protein antagonists. Proc. Natl. Acad. Sci. U.S.A. 95, 346–351 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milligan G., Kostenis E., Heterotrimeric G-proteins: A short history. Br. J. Pharmacol. 147 (suppl. 1), S46–S55 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrage R., et al. , The experimental power of FR900359 to study Gq-regulated biological processes. Nat. Commun. 6, 10156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uhlenbrock K., Gassenhuber H., Kostenis E., Sphingosine 1-phosphate is a ligand of the human gpr3, gpr6 and gpr12 family of constitutively active G protein-coupled receptors. Cell. Signal. 14, 941–953 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Sveidahl Johansen O., et al. , Lipolysis drives expression of the constitutively active receptor GPR3 to induce adipose thermogenesis. Cell 184, 3502–3518.e33 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyagi T., Tanaka S., Hide I., Shirafuji T., Sakai N., The subcellular dynamics of the Gs-linked receptor GPR3 contribute to the local activation of PKA in cerebellar granular neurons. PLoS One 11, e0147466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikawa F., et al. , Detailed neuronal distribution of GPR3 and its co-expression with EF-hand calcium-binding proteins in the mouse central nervous system. Brain Res. 1750, 147166 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Eggerickx D., et al. , Molecular cloning of an orphan G-protein-coupled receptor that constitutively activates adenylate cyclase. Biochem. J. 309, 837–843 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinckley M., Vaccari S., Horner K., Chen R., Conti M., The G-protein-coupled receptors GPR3 and GPR12 are involved in cAMP signaling and maintenance of meiotic arrest in rodent oocytes. Dev. Biol. 287, 249–261 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Mendez M. F., Degenerative dementias: Alterations of emotions and mood disorders. Handb. Clin. Neurol. 183, 261–281 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Inostroza M., et al. , Hippocampal-dependent spatial memory in the water maze is preserved in an experimental model of temporal lobe epilepsy in rats. PLoS One 6, e22372 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guarino A., et al. , Executive functions in alzheimer disease: A systematic review. Front. Aging Neurosci. 10, 437 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah D., et al. , Spatial reversal learning defect coincides with hypersynchronous telencephalic BOLD functional connectivity in APPNL-F/NL-F knock-in mice. Sci. Rep. 8, 6264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito T., et al. , Single App knock-in mouse models of Alzheimer’s disease. Nat. Neurosci. 17, 661–663 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Heneka M. T., et al. , Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 14, 388–405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang C.-R., et al. , The G protein coupled receptor 3 is involved in cAMP and cGMP signaling and maintenance of meiotic arrest in porcine oocytes. PLoS One 7, e38807 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrot M., et al. , Regulation of anxiety and initiation of sexual behavior by CREB in the nucleus accumbens. Proc. Natl. Acad. Sci. U.S.A. 102, 8357–8362 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kandel E. R., The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol. Brain 5, 14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donovan N. J., et al. , Subjective cognitive concerns and neuropsychiatric predictors of progression to the early clinical stages of Alzheimer disease. Am. J. Geriatr. Psychiatry 22, 1642–1651 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geda Y. E., et al. , Baseline neuropsychiatric symptoms and the risk of incident mild cognitive impairment: A population-based study. Am. J. Psychiatry 171, 572–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steenland K., et al. , Late-life depression as a risk factor for mild cognitive impairment or Alzheimer’s disease in 30 US Alzheimer’s disease centers. J. Alzheimers Dis. 31, 265–275 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pietrzak R. H., et al. ; Australian Imaging, Biomarkers, and Lifestyle Research Group, Amyloid-β, anxiety, and cognitive decline in preclinical Alzheimer disease: A multicenter, prospective cohort study. JAMA Psychiatry 72, 284–291 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Panza F., Lozupone M., Logroscino G., Imbimbo B. P., A critical appraisal of amyloid-β-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 15, 73–88 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Doody R. S., et al. ; Alzheimer’s Disease Cooperative Study Steering Committee; Semagacestat Study Group, A phase 3 trial of semagacestat for treatment of Alzheimer’s disease. N. Engl. J. Med. 369, 341–350 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Fleisher A. S., et al. , Phase 2 safety trial targeting amyloid beta production with a gamma-secretase inhibitor in Alzheimer disease. Arch. Neurol. 65, 1031–1038 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gijsen H. J. M., Mercken M., γ-secretase modulators: Can we combine potency with safety? Int. J. Alzheimers Dis. 2012, 295207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song W. M., Colonna M., The identity and function of microglia in neurodegeneration. Nat. Immunol. 19, 1048–1058 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Yuan P., et al. , TREM2 haplodeficiency in mice and humans impairs the microglia barrier function leading to decreased amyloid compaction and severe axonal dystrophy. Neuron 90, 724–739 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y., et al. , TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160, 1061–1071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ulland T. K., et al. , TREM2 maintains microglial metabolic fitness in Alzheimer’s disease. Cell 170, 649–663.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Condello C., Yuan P., Schain A., Grutzendler J., Microglia constitute a barrier that prevents neurotoxic protofibrillar Aβ42 hotspots around plaques. Nat. Commun. 6, 6176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., et al. , TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213, 667–675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mazaheri F., et al. , TREM2 deficiency impairs chemotaxis and microglial responses to neuronal injury. EMBO Rep. 18, 1186–1198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meilandt W. J., et al. , Trem2 deletion reduces late-stage amyloid plaque accumulation, elevates the Aβ42:Aβ40 ratio, and exacerbates axonal dystrophy and dendritic spine loss in the PS2APP Alzheimer’s mouse model. J. Neurosci. 40, 1956–1974 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bradley S. J., et al. , Biased M1-muscarinic-receptor-mutant mice inform the design of next-generation drugs. Nat. Chem. Biol. 16, 240–249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Butcher A. J., et al. , Differential G-protein-coupled receptor phosphorylation provides evidence for a signaling bar code. J. Biol. Chem. 286, 11506–11518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nobles K. N., et al. , Distinct phosphorylation sites on the β(2)-adrenergic receptor establish a barcode that encodes differential functions of β-arrestin. Sci. Signal. 4, ra51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poulin B., et al. , The M3-muscarinic receptor regulates learning and memory in a receptor phosphorylation/arrestin-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 107, 9440–9445 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bradley S. J., et al. , Mapping physiological G protein-coupled receptor signaling pathways reveals a role for receptor phosphorylation in airway contraction. Proc. Natl. Acad. Sci. U.S.A. 113, 4524–4529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kliewer A., et al. , Phosphorylation-deficient G-protein-biased μ-opioid receptors improve analgesia and diminish tolerance but worsen opioid side effects. Nat. Commun. 10, 367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mann A., et al. , Agonist-induced phosphorylation bar code and differential post-activation signaling of the delta opioid receptor revealed by phosphosite-specific antibodies. Sci. Rep. 10, 8585 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Latorraca N. R., et al. , How GPCR phosphorylation patterns orchestrate arrestin-mediated signaling. Cell 183, 1813–1825.e18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou X. E., et al. , Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors. Cell 170, 457–469.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baidya M., et al. , Key phosphorylation sites in GPCRs orchestrate the contribution of β-Arrestin 1 in ERK1/2 activation. EMBO Rep. 21, e49886 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He Q.-T., et al. , Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2. Nat. Commun. 12, 2396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sente A., et al. , Molecular mechanism of modulating arrestin conformation by GPCR phosphorylation. Nat. Struct. Mol. Biol. 25, 538–545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mayer D., et al. , Distinct G protein-coupled receptor phosphorylation motifs modulate arrestin affinity and activation and global conformation. Nat. Commun. 10, 1261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Slosky L. M., et al. , β-arrestin-biased allosteric modulator of NTSR1 selectively attenuates addictive behaviors. Cell 181, 1364–1379.e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grim T. W., et al. , A G protein signaling-biased agonist at the μ-opioid receptor reverses morphine tolerance while preventing morphine withdrawal. Neuropsychopharmacology 45, 416–425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scarpa M., et al. , Biased M1 muscarinic receptor mutant mice show accelerated progression of prion neurodegenerative disease. Proc. Natl. Acad. Sci. U.S.A. 118, e2107389118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu J., et al. , Drd2 biased agonist prevents neurodegeneration against NLRP3 inflammasome in Parkinson’s disease model via a β-arrestin2-biased mechanism. Brain Behav. Immun. 90, 259–271 (2020). [DOI] [PubMed] [Google Scholar]
- 78.Brown A. J. H., et al. , From structure to clinic: Design of a muscarinic M1 receptor agonist with potential to treatment of Alzheimer’s disease. Cell 184, 5886–5901.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shukla A. K., Biasing GPCR signaling from inside. Sci. Signal. 7, pe3 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional materials and methods and other study data are included in the SI Appendix. All other data are included in the article and/or supporting information.