Significance
Rapid detection of pathogenic bacteria in clinical samples within a few minutes is the key to infectious disease control and precise medicine. Existing methods require time-consuming processes such as sample pretreatment, recognition, and signal amplification. This work establishes an ultra-rapid strategy for bacteria detection by direct bacteria morphological identification in free solution via a label-free single-particle imaging approach. It does not require any biological reagents and can directly detect bacteria in clinical samples within 10 min. We believe such a label-free single-particle imaging approach for rapid bacteria screening will have wide applications in clinical diagnosis and disease control due to its high sensitivity, simplicity, rapidity, and low cost.
Keywords: single bacteria detection, label-free, morphological identification, convection, dark field scattering microscopy
Abstract
Rapid detection of pathogenic bacteria within a few minutes is the key to control infectious disease. However, rapid detection of pathogenic bacteria in clinical samples is quite a challenging task due to the complex matrix, as well as the low abundance of bacteria in real samples. Herein, we employ a label-free single-particle imaging approach to address this challenge. By tracking the scattering intensity variation of single particles in free solution, the morphological heterogeneity can be well identified with particle size smaller than the diffraction limit, facilitating the morphological identification of single bacteria from a complex matrix in a label-free manner. Furthermore, the manipulation of convection in free solution enables the rapid screening of low-abundance bacteria in a small field of view, which significantly improves the sensitivity of single-particle detection. As a proof of concept demonstration, we are able to differentiate the group B streptococci (GBS)–positive samples within 10 min from vaginal swabs without using any biological reagents. This is the most rapid and low-cost method to the best of our knowledge. We believe that such a single-particle imaging approach will find wider applications in clinical diagnosis and disease control due to its high sensitivity, rapidity, simplicity, and low cost.
Bacterial infection (1, 2) is one of the major causes of mortality and results in an enormous economic loss every year because of the antibiotic-resistant bacteria originating from impropriate antibiotic prescription (3). Therefore, it has received considerable medical and public concerns recently (4). Rapid detection of pathogenic bacteria at the early stage is of vital importance (5) for infectious disease control as well as precision medicine, facilitating the appropriate treatment without the abuse of antibiotics. Therefore, it not only saves lives but also significantly cuts down the healthcare expense (1) originating from the disease spread of antibiotic-resistant bacteria. Unfortunately, the current bacteria detection methods such as bacterial culture methods, which are known as the gold standard, require days (6), and quantitative real-time PCRs can shorten the time to several hours, but they are too expensive for resource-limited environments (7). Other methods such as Raman spectrometry (2, 8), fluorescence (9, 10), and electrochemiluminescence (11) methods have been proposed with less time and lower cost. However, those methods identify the bacteria based on molecular recognition, and they are generally complicated and high in cost. For instance, they require the use of expensive biological reagents and require more than 1 h to pretreat the samples, such as incubation, separation, or signal amplification due to the complex matrix and low abundance of targets in clinical samples (12). Therefore, rapid detection of bacteria in clinical samples within several minutes is quite challenging.
The label-free single-particle optical imaging technique is one of the potential methods that is able to address this challenge (13, 14) due to its ultimate sensitivity and rapidity without incubation. It has been used for antimicrobial susceptibility testing with rapidity by either single bacterium motions–based (15) or free solution scattering imaging–based (16–19) methods. However, for direct bacterium detection, the challenges lie in the specificity on how to recognize the target particles, as well as the rapidity limited by its low density in a small field of view (ca. 100 × 100 μm) at a low concentration. First, the size of bacteria is in the range of submicrometers to a few micrometers, which is approximate to the optical diffraction limit, making the direct morphological identification difficult. Second, label-free single-particle imaging approaches such as surface plasmon resonance microscopy (SPRM) (20) and interference scattering microscopy (iSCAT) (21) are sensitive down to the single-protein level. However, they require the binding of particles to the interface before detection, implying a long incubation time due to the slow mass transportation at low concentrations (22, 23). Moreover, optical microscopy generally detects only a very tiny part of the sample due to its small field of view, and thus the detection of single bacteria in a large volume remains challenging. The large-volume solution phase scattering imaging method (16–19) can improve the limit of detection with a large field of view. However, the decrease of spatial resolution makes the differentiation of bacteria from the impurity particles difficult (17), which compromises the specificity.
In response to these challenges, we developed reflection-enhanced dark-field scattering microscopy (REDFSM) (24) that is able to directly identify single bacteria in free solution in an incubation-free manner without using any expensive biological reagents. By tracking the scattering intensity variation of single particles, REDFSM can directly identify the morphological heterogeneity of single particles with sizes smaller than the diffraction limit, facilitating the direct morphological identification of bacteria in a complex matrix in a label-free manner. Furthermore, the manipulation of convection in free solution enables the rapid screening of low-abundance bacteria in a small field of view, which significantly improves the sensitivity of single-particle detection, making the direct detection of ultra-low concentration bacteria in clinical samples possible.
Group B streptococci (GBS) are pathogenic bacteria that commonly exist in pregnant women. The vertical transmission of GBS from mother to child generally occurs during labor (25), which may cause a series of neonatal diseases or even death (26). Unfortunately, GBS detected by the bacterial culture method takes 2 d, which cannot reflect GBS status during labor (27). Therefore, intrapartum screening of GBS in a short time window is of vital importance for clinical use in obstetrics, as it facilitates the appropriate antibiotic therapy, reduces hospital costs, and prevents development of bacterial resistance to antimicrobials (28). Therefore, rapid detection of GBS in real samples within a few minutes is highly desirable in obstetrics. Herein, using GBS detection in vaginal swabs as an example, we demonstrate the direct screening of GBS in clinical samples within 10 min by such a label-free single-particle imaging approach without using any biological reagents, implying its promising application in rapid clinical diagnosis due to its high sensitivity, rapidity, simplicity, and low cost.
Results and Discussion
The Principle of Ultra-Rapid Bacteria Detection.
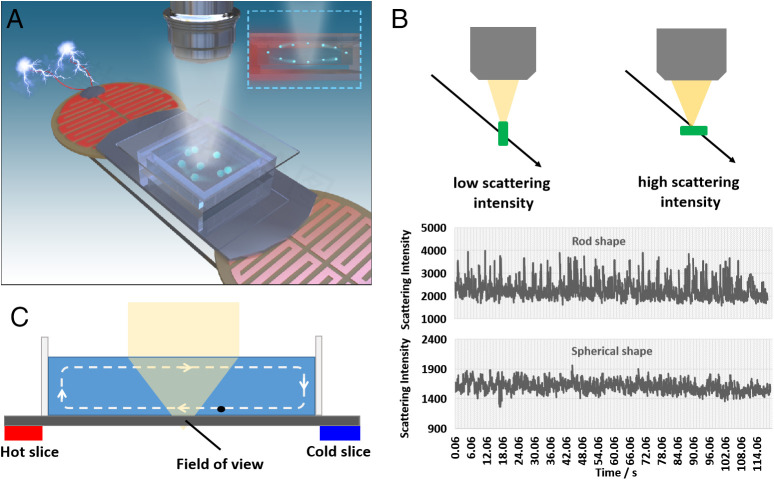
Fig. 1A demonstrates the detection principle of bacteria by a single-particle imaging approach. The sample is placed on a REDFSM with a long focus objective to enhance the depth of field (24), so that more particles can be observed in free solution. Compared with other label-free single-particle imaging methods, such as SPRM (29) or iSCAT (21), which require the binding (or approaching within a few hundred nanometers) of particles to the solid–liquid interface, the direct imaging of particles in free solution with large depth of field by dark-field scattering microscopy is much more efficient than the above interfacial imaging approaches. This is because the particle binding to the interface in a specific small field of view (ca. 100 × 100 μm) is quite a low probability event at low concentration. A polished Si chip is used to enhance the sensitivity and the depth of field by a reflection-enhanced mode as shown in our recent work (24). As the scattering intensity depends on the scattering cross-section of the particle, the fluctuation of scattering intensity in free solution reflects the morphological heterogeneity of the particle as indicated in Fig. 1B. Two electrical-heated slices are placed at the bottom of cells from one end to the other, which can induce horizontal convection in the cell as shown in Fig. 1C, so that the particles in the free solution can be screened in a small field of view.
Fig. 1.
Bacteria detection principle by a single-particle imaging approach. (A) Schematic illustration of bacteria detection by a single-particle imaging approach. (B) Particle morphological heterogeneity identification by tracking the scattering intensity fluctuation in free solution. (C) Convection induced by the hot slice for the screening of single bacteria in a small field of view.
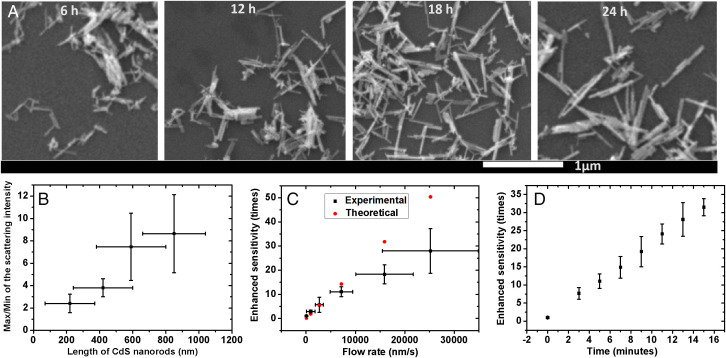
To verify whether the scattering intensity in free solution can reflect the morphological heterogeneity, CdS nanorods were synthesized by different reaction times to obtain different lengths (Fig. 2A) (30) and then dispersed in free solution to record the variation of scattering intensities. As shown in Figs. 1B and 2B, the fluctuation of scattering intensity indicated by the ratio of maximum/minimum increases with the increasing length of the nanorods, suggesting that the morphological heterogeneity is indeed related to the fluctuation of scattering intensity in free solution.
Fig. 2.
Structural heterogeneity reflected by scattering intensity and Sensitivity improved by screening time and flow rate. (A) SEM images of the CdS nanorods with different lengths by different reaction times (6, 12, 18, and 24 h). (B) Fluctuation of scattering intensity of CdS nanorods indicated by the maximum/minimum intensity value with different lengths. (C) Experimental- and theoretical-enhanced sensitivities with the increasing flow rates within 5 min. (D) Experimental-enhanced sensitivity with increasing screening times in a field of view of 150 × 150 μm with a flow rate of 7.1 μm/s. The concentration of particles is ca. 10−14 M.
The manipulation of convection to enhance the sensitivity in a small field of view was also studied. With the application of heating power on one of the heated slices, the solution at the interface flows from one end to the other. The particles move faster with increasing heating power so that the sensitivity can be improved as shown in Fig. 2C, as more particles can be observed in a small field of view, facilitating the direct screening of target particles. Correspondingly, the sensitivity can also be improved with increasing screening time as indicated in Fig. 2D. It is found that the lowest detectable concentration for one single particle (on average) in such a field of view (150 × 150 μm) is ca. 3.5 × 10−15 M (2.1 × 106 counts/mL) without convection, which is much better than the SPRM of 1.2 × 10−13 M (7.2 × 107 counts/mL) at the same field of view for 5-min counting of particles. We attribute this to the significantly different depth of field (24). With the increasing flow rate, as well as the screening time, the sensitivity can be further improved as shown in Fig. 2 C and D. For instance, we found that with a flow rate of 7.1 μm/s in the horizontal direction and a 5-min screening time, the sensitivity can be improved for ca. 11-fold in such a field of view (150 × 150 μm), which is close to the theoretical expectation of 14-fold. With increasing screening time, the sensitivity can be further improved over 30-fold for a 15-min screening. Alternatively, the sensitivity can also be further improved over 30-fold with higher heating power as shown in Fig. 2C, but a significant deviation from the theoretical expectation was observed probably due to the enhancement of turbulence in the vertical direction, resulting in the defocus of particles. The further design on the size and thickness of the cell and the optimization of heating power are likely to further improve the screening efficiency, which will be part of our future works.
The significant improvement of the sensitivity with the manipulation of convection makes the fast screening of low-abundance bacteria in clinical samples possible. As a label-free single-particle imaging approach, how to identify the bacteria from the complex matrix in clinical samples is the key to rapid detection. As indicated in Fig. 2 A and B, the direct detection of morphological heterogeneity based on the fluctuation of scattering intensity in free solution is potentially able to address the challenge with rapidity, as it neither requires staining of the sample as shown in Fig. 3E (4-h incubation) nor requires the binding of particles to the interface (extremely low probability in a smaller field of view). As a morphological identification method, the specificity significantly relies on the particle shapes and sizes in the matrix, while other interferences such as salt, small molecules, proteins, and exosomes can be eliminated due to their weak scattering intensity in free solution. Rapid detection of GBS in a vaginal swab within a few minutes, which is considered as one of the most urgent clinical diagnoses in obstetrics, is demonstrated here as a preclinical proof-of-concept study.
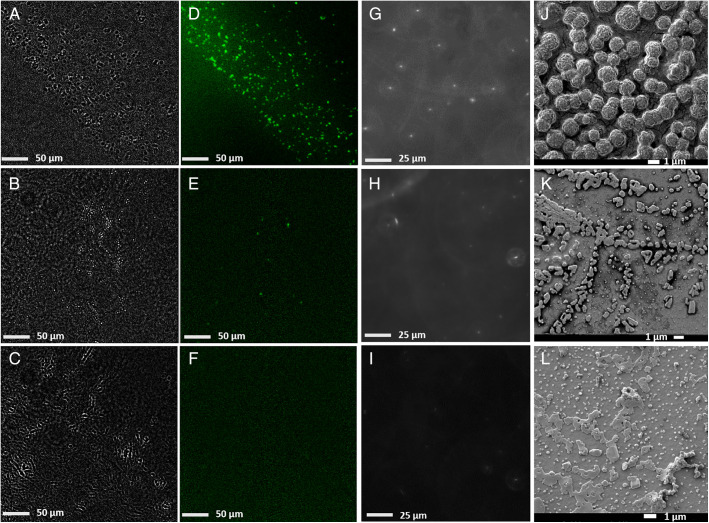
Fig. 3.
Standard GBS, GBS positive and GBS negative clinical samples imaged by different methods. (A–C) Background-subtracted bright-field, (D–F) fluorescence, and (G–I) reflection-enhanced dark-field scattering microscopy in free solution, and (J–L) SEM images of the standard GBS samples (Upper), GBS-positive clinical samples (Middle), and GBS-negative clinical samples (Lower).
The Morphological Features of GBS.
To identify the morphology of GBS and interfering particles in clinical samples, different imaging methods were employed to analyze the different samples including standard GBS samples, GBS-positive and -negative clinical samples by scanning electron microscopy (SEM), REDFSM, and fluorescence and bright-field microscopies. SEM images reveal that the morphology of GBS is generally spherical in shape with a diameter of 1.2 ± 0.2 μm, which agrees well with our expectation. Fluorescence microscopy of the stained standard GBS sample further confirms that most of the particles observed in bright-field images (Fig. 3 A and D) are indeed GBS. For GBS-negative samples, we are able to observe many more small particles with a diameter of ca. 200 nm mixed with some large aggregates with random shapes in an SEM image (Fig. 3L). No fluorescent particles were observed in the GBS-negative samples, suggesting that those particles observed in Fig. 3L are probably aggregates of proteins, exosomes, lysed cell fragments, or organelles, which are significantly different from GBS in sizes and shapes. Moreover, most of the salt, small molecules, and proteins that appeared in the dried samples in Fig. 3 K and L are likely to dissolve in free solution, which contribute negligible scattering background as shown in Fig. 3 H and I, making the morphological identification in free solution a lot easier than for the dried samples. Quite a few fluorescent particles were observed in the GBS-positive samples with a large field of view (300 × 300 μm) as indicated in Fig. 3E, but it was difficult to find a single GBS bacterium in GBS-positive samples (similar to GBS-negative samples) in SEM images probably due to the much smaller field of view for SEM, as well as the embedding of GBS in dried proteins and salt. The images shown in Fig. 3 suggest that the morphology and size of GBS are quite unique compared with the matrix and are potentially able to be recognized from the complex matrix in clinical samples.
Interferences in the Clinical Samples.
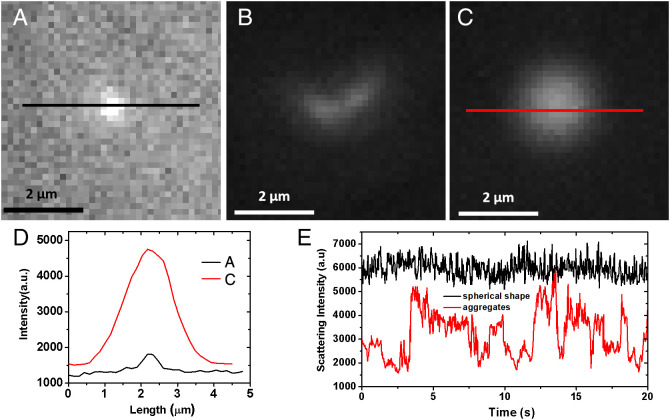
The standard GBS samples are first imaged under REDFSM in free solution as indicated in Fig. 3G and SI Appendix, Movie S1. In free solution, most of the GBS bacteria are single cells with quite a few portions of dimers or trimers, which is somewhat to our surprise, as the chain shape (frequently observed in SEM) is not found in free solution. The discrepancy could be attributed to the requirement of GBS deposition on the substrate for SEM imaging, resulting in the aggregation of GBS. The spherical shape of a single-cell GBS bacterium makes the fluctuation of scattering intensity remain in a small relative SD (RSD) of less than 15% (as shown in SI Appendix, Fig. S1) due to its homogeneous shape with weak fluctuation in scattering intensity and relatively large size (1.2 μm) with weak Brownian motion. However, in GBS-negative samples, the scattering intensity of most of the particles is quite small as shown in Fig. 4A, corresponding to the smaller particles (200 nm) observed in SEM images. Some aggregates or the fragments of cells with much larger scattering intensities close to GBS are also observed in the clinical samples (Fig. 4B and SI Appendix, Movie S2), but the scattering intensities of these particles fluctuate significantly (SI Appendix, Fig. S1) compared with GBS due to the morphological heterogeneity, as the morphologies of aggregates or fragments are much more random. Additionally, there are still quite a few particles (estimated to be 106 counts/mL, Fig. 4C) with a spherical feature and large scattering intensity that are observed in the negative samples, which indeed pose a great challenge to the morphological identification by simply tracking the scattering intensity variation. A recent work on the morphological identification of bacteria by large-volume scattering imaging also reveals the relatively low accuracy in real samples resulting from the morphological heterogeneity of particles in the matrix (31), implying that simply tracking scattering intensity might not be enough to achieve high specificity. Therefore, more parameters are needed.
Fig. 4.
Particles imaged by REDFSM in GBS negative clinical samples. (A–C) Snapshots of dark-field scattering images of different types of particles in the GBS-negative samples. (D) Scattering intensity from spherical shape particles in (A) and (C). (E) Scattering intensity curves of particles in (B) (aggregate) and (C) (spherical particle) due to the different shapes.
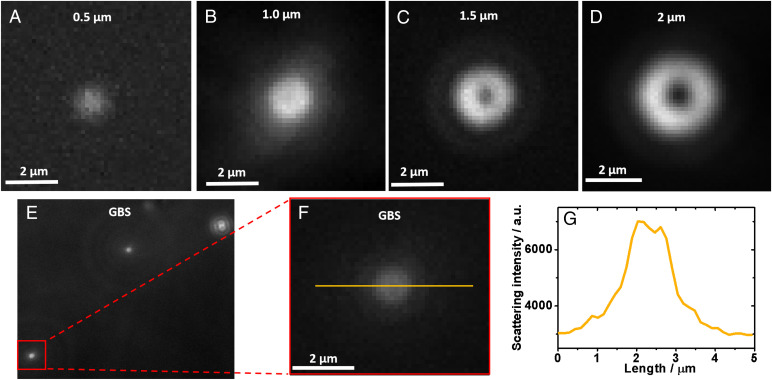
Besides morphological identification, the size of particles is another parameter that can be used for further GBS detection. However, particle size can be directly determined by Brownian motion in free solution based on the Stokes–Einstein equation (24). The manipulation of convection for particle screening makes the size determination by pure Brownian motion difficult. However, as the size of GBS is slightly larger than the diffraction limit, the light spot of a single-cell GBS bacterium imaged by REDFSM is somewhat different from the particles smaller than the diffraction limit, despite it still being hard to identify its exact shape directly. That is, the light spot is slightly larger than the critical size, and the intensity distribution of the light spot is actually not a Gaussian shape as shown in Fig. 4D, but a ring shape with a slight intensity decrease in the center (Fig. 5 F and G). This is probably because of the unique particle size of GBS (1.2 μm) that is slightly larger than the diffraction limit, in which the particle shape is about to appear. To confirm whether the feature of the light spot is related the size of particles, spherical polystyrene particles with different sizes from 0.5 to 2 μm were imaged by REDFSM as shown in Fig. 5 A–D. The center-decrease feature becomes more perceptible with the increasing size while the diameter is larger than 1 μm as shown in Fig. 5 A–D and SI Appendix, Fig. S2. In other words, a particle with a diameter smaller than 1 μm (less than or equal to the diffraction limit) displays a quasi-Gaussian light intensity distribution of the light spot, while a spherical particle larger than 1 μm possesses the center-decrease feature (ring shape) in the REDFSM image. Note that the center-decrease feature of the light spots originates from the projection of ring shape dark-field illuminator as shown in SI Appendix, Fig. S3, in which the projection of the ring shape can be well observed using larger particles. The spherical shape particle with a diameter larger than 1 μm can be a unique feature to further differentiate the GBS-positive sample as shown in SI Appendix, Movie S3 because we found that none of the organelles is spherical in shape with a size larger than 1 μm in the human cells, which guarantees the specificity in clinical sample detection. The smaller spherical shape interferences (<1 μm) with high scattering intensity observed in Fig. 4C in real samples could be the released organelles such as the lysomes according to our previous observation (24), considering their relatively high scattering cross-section. Therefore, the size-dependent feature of the light spot further improves the accuracy for direct screening of GBS bacteria in free solution. Also, we believe this method possesses better specificity than the large-volume solution scattering imaging method (31) due to the incorporation of size feature at high spatial resolution.
Fig. 5.
Scattering pattern of spherical particles with different sizes. (A–D) Reflection-enhanced dark-field scattering pattern of spherical particles with different particle sizes. (E) Scattering pattern of single GBS bacteria in free solution. (F) Zoom-in of scattering pattern of a single-cell GBS bacterium. (G) Scattering intensity distribution along the yellow line in (F).
Ultra-Rapid GBS Screening in Clinical Samples.
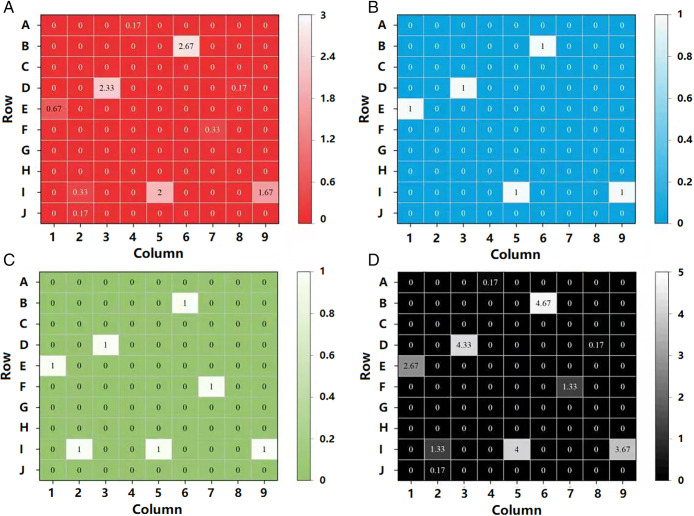
To further confirm whether the proposed method can be used for rapid detection of GBS in clinical samples, a screening of 90 vaginal swabs is carried out as displayed in Fig. 6A. As a control experiment, the samples are detected by the bacterial culture method as the gold standard to determine which samples are GBS-positive. It is found that 5 out of 90 clinical samples are GBS-positive as shown in Fig. 6B. Particles with the above features can be easily found in these five GBS-positive samples within 5 min of screening with a flow rate of 7.1 μm/s imaged by this method, but they can hardly be found in GBS-negative samples, indicating that the weak scattering intensity fluctuation and center-decrease features determined by morphology and size can be an easy way for rapid GBS screening in free solution. To further confirm the result, a small portion (1/20) of the same clinical samples are also detected by the PCR method. Besides the five positive samples, it is revealed that two more samples (I2 and F7) are found to be positive as shown in Fig. 6C. As the PCR method is considered to be more sensitive (32) than the bacterial culture method, it is hard to say whether these two samples are really false-positives, or they are actually positive but the concentration of GBS is too low to read out by the bacterial culture method. To clarify the results, we further detect these 90 samples by our method with prolonged screening time (6 × 5 min, by six repeated experiments) to further improve the sensitivity. It is revealed that two single GBS bacteria are found in these two samples for 30-min screening (0.33 counts/5 min), suggesting that these two samples are probably positive, but it is too low to be detected by the gold standard method. This result underscores the importance of developing new methodologies for bacteria detection. This result also implies that our method is probably more sensitive than the bacterial culture method with a longer screening time. It is noteworthy that only one single GBS bacterium is found in three more samples within 30 min of screening (J2, A4, and D8, 0.17 counts/5 min) as shown in Fig. 6A. Supposing these five samples are false-positive while referring to the bacterial culture method, they can be easily eliminated by setting a proper threshold of 0.5 counts/5 min as shown in the receiver operator characteristic (ROC) curves in SI Appendix, Fig. S4A and Table S1, or shorten the total screening time to ca. 10 min.
Fig. 6.
GBS detection results of 90 clinical samples indicated by three different methods. (A) Averaged frequency of single GBS bacteria detected in a 5-min screening with a flow rate of 7.1 μm/s in a field of view of 150 × 150 μm from 90 clinical samples with six repeated experiments. (B) GBS detection of these clinical samples indicated by standard bacterial culture method. (C) GBS detection of these clinical samples indicated by PCR method. (D) Merged image of the results of these three methods. Note that for the reference methods (both PCR and bacterial culture method), “1” represents the GBS-positive result, while “0” represents the GBS-negative result.
The bacterial culture method indicates a 5.6% GBS-positive rate (5/90), while the PCR method (1/20 of the sample) gives a 7.8% GBS-positive rate (7/90). With prolonged screening time, our method proposes an 11.1% GBS-positive result (10/90) within 30 min of screening. By merging the results together as shown in Fig. 6D, the GBS-positive samples determined by both reference methods are all found to be positive by our method with longer screening time. It has been reported that 10 to 30% of the women are GBS-positive (28). It seems that our method is mostly closed to this result. The PCR method used in this work possesses a nominal detection limit of 2 × 104 counts/mL, which does not match with the normal PCR detection limit due to the use of 1 out of 20 of the samples. Our method can detect one bacteria when averaged for 30-min screening when the concentration is 3.2 ×104 counts/mL, which does not completely support the result that these three samples are positive. However, the different experimental skills from person to person, as well as the stochasticity at low concentration, might result in the variation of the detection limit for each screening (see SI Appendix, Fig. S5 and Discussion for more details), so it is still difficult to say that these three samples (J2, A4, and D8) are false-positive in our method. Even if they were indeed false-positive, they can still be easily eliminated by setting a proper threshold of 0.25 counts/5 min as indicated in the ROC curve in SI Appendix, Fig. S4B and Table S2.
Further Applications in Clinical Samples.
As the sensitivity depends on the flow rate of the solution, future works on how to better manipulate the convection are likely to further improve the sensitivity. For instance, the flow rate increase up to hundreds of times (over 1 mm/s) in the horizontal direction can be simply achieved by either optical stimulation (33) or the use of an additional pump to accelerate the flow rate in a microchannel. These methods are likely to push the detection limit down to a much lower level.
Besides the spherical shape bacteria (Streptococcus), such as GBS, which have been well identified from the background, other bacteria such as Escherichia coli, which represents another major branch of pathogenic bacteria with a rod shape, have been imaged under REDFSM as shown in SI Appendix, Fig. S6. The shape and size are also quite different from the interfering particles in the background, which can also be well identified due to their unique feature.
Besides the vaginal swabs, clinical samples such as urine, saliva, and blood serum samples were all preliminarily examined as shown in SI Appendix, Fig. S7. It is found that the particle interferences from the urine and blood serum samples are also composed of exosomes, as well as a small portion of organelles and cell fragments, which are also significantly different from the bacteria in SI Appendix, Fig. S7 A and C, making the direct bacteria detection in clinical samples possible. While few bacteria can be observed in the urine samples, the preliminary study shows that spherical bacteria in the saliva samples are at a much higher level as indicated in SI Appendix, Fig. S7B. This is within our expectation, as Streptococcus represents the most abundant species in saliva. The concentrations of Streptococcus in saliva and urine clinical samples have been examined and are listed in SI Appendix, Table S3, which generally agree with the clinical expectations, implying their wide potential applications. However, Streptococcus bacteria cannot be observed in the blood serum. Interestingly, we occasionally observe a rod-shaped bacterium that swims in blood serum with a much faster rate than the Brownian motion as shown in SI Appendix, Movie S4, making the exploration of new types of bacteria in the blood possible.
Although REDFSM is able to directly identify bacteria from the background in clinical samples due to their significant different morphologies in free solution, the main challenge lies in the differentiation of bacteria with a similar shape and size. Despite this, REDFSM already has its clinical significance in disease diagnosis, as the differentiation of type rather than the subtype of bacteria is enough for the proper prescription. On the other hand, researchers have been working hard on the development of new methods to direct identification of bacteria from similar morphologies (13, 34), such as the Fourier transform light scattering method (13), to further identify the bacteria with similar shapes and sizes. With the development of these advanced imaging analysis methods, as well as the better manipulation on the convection, we believe that the single-particle imaging approach will be widely applied to the ultra-rapid detection of bacteria in clinical samples in the future.
Conclusion
In summary, we employed a label-free single-particle imaging approach to detect low concentrations of bacteria in clinical samples within several minutes. Compared with the SEM morphological imaging method that requires the deposition of a clinical sample on the surface, directly imaging the bacteria in the free solution can largely reduce the background originating from the soluble interferences such as protein, exosomes, and others. By tracking the particle motion in free solution, we are able to differentiate the morphological heterogeneity of single particles based on the variation of the scattering cross-section in the presence of Brownian motion, facilitating the detection of single bacteria from a complex matrix without using any biological reagents. Furthermore, the manipulation of convection in free solution enables the fast and facile screening of low-abundance bacteria in a small field of view, which significantly improves the sensitivity of single particle detection. It was found that GBS bacteria in clinical samples can be identified in less than 10 min (including a 2-min sample pretreatment and a 5-min screening) without using any biological reagents, which is the most rapid and low-cost method to the best of our knowledge. With further improvements in the manipulation of convection, as well as developments of advanced image recognition technology, we believe that such a single-particle imaging approach for bacteria detection will be able to find its wider applications in clinical diagnosis and infectious disease control due to its high sensitivity, rapidity, simplicity, and low cost.
Materials and Methods
The dark-field scattering images were obtained in a reflection-enhanced mode by applying a mercury light source (Nikon Intensilight C-HGFI; the irradiation intensity is ca. 10 mW/mm2) on a Nikon Ci-S reflection dark-field microscope using a polished Si chip as a reflective surface according to our previous work (24). A ×50 dark-field objective (numerical aperture [NA] of 0.60, working distance of 11 mm) was used in the experiment to collect the scattering light. The dark-field images were recorded by a scientific CMOS (complementary metal oxide semiconductor) camera (Dhyana 400BSI, Tucsen). The polished Si chips were purchased from Zhejiang Lijing Optoelectronics Technology, and the glass coverslips (no. 1, BK7, 22 × 22 mm) were purchased from Fisher Scientific. The construction of the cell was a “sandwich” structure, composed of a polished Si chip, a polydimethylsiloxane (PDMS) slice punched with a square-shaped hole (15 × 15 × 0.8 mm) in the center, and a glass coverslip on the top. Two heated slices were placed at the bottom of the cell to manipulate the convection, one of which was electrically heated to induce the horizontal convection at the bottom of the cell. A fluorescent GBS antibody (PA1-73063 from Thermo Fisher Scientific) was used to differentiate the GBS bacteria from the clinical samples. The surface topographies of the standard GBS and clinical samples were deposited on a glass coverslip and observed by SEM (JSM-7900F) and bright-field and florescence microscopies (Ti2-E, Nikon).
Monodisperse polystyrene aerosols of different sizes (500 nm, 1 μm, 1.5 μm, and 2 μm, Wuxi Regal Biotechnology) were used to obtain the size-dependent dark-field pattern of the microparticles. Deionized water (18.2 MΩ) prepared by MYZ-US10 (from Miaozhiyi Nanjing) was used throughout the experiment.
The CdS nanorods were synthesized with a solvent thermal method. Briefly, 6 mmol of Cd(NO3)2·4H2O and 18 mmol of thiourea were added into an autoclave with an inner Teflon lining which had been filled with ethylenediamine to 60% of its capacity (50 mL) and maintained at 160 °C for 6, 12, 18, and 24 h to obtain different lengths. After that, the yellow precipitate was collected by centrifugation, washed with distilled water and ethanol three times, and then dried under vacuum at 60 °C for 12 h.
A total of 90 clinical vaginal swab samples were collected from the volunteers. This study was approved by the Sir Run Run Hospital Ethics Committee (approval no. 2022-SR-001-A1). The volunteers were informed about the research project and voluntarily signed the informed consent form before the collection of samples. The swabs were immersed in 10 mL of PBS solution and vortexed for 2 min. The supernatant was divided into several aliquots for detection. A nucleic acid detection kit based on enzymatic probe isothermal amplification for GBS from Jiangsu Macro & Micro-Test Med-Tech was used as the reference method (nominal detection limit of 1,000 copies/mL). A GBS chromogenic agar plate (registration certificate for medical device no. 20152400814) from Autobio Diagnostics was used to detect the GBS in clinical samples, which is considered as the gold standard for GBS detection. The standard GBS sample was obtained by further cultivation of the known GBS colony in a GBS agar base.
A gold film with a thickness of 50 nm was coated on a glass coverslip by magnetron sputtering (Au chip) and served as the substrate for SPR imaging of particles. The plasmonic imaging was carried out using an inverted microscope (Ti2-E, Nikon) with a ×60 (NA of 1.49) oil immersion objective. Light source was a p-polarized superluminescent diode (wavelength = 680 nm, 1 mW, Qphotonics).
Supplementary Material
Acknowledgments
We thank the National Natural Science Foundation of China (Grants 22174069, 21874072, 21605078, 12005301), the Jiangsu Agricultural Science and Technology Innovation Fund (Grant CX(21)3064), the Doctoral Program of Entrepreneurship and Innovation in Jiangsu Province (Grant JSSCBS20211310), the Guangdong Basic and Applied Basic Research Foundation (Grant 2022A1515012456), and the Hospital Research Project (Grant SZ2020MS002) for financial support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2206990119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Alafeef M., Moitra P., Pan D., Nano-enabled sensing approaches for pathogenic bacterial detection. Biosens. Bioelectron. 165, 112276 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Ho C. S., et al. , Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 10, 4927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fleming-Dutra K. E., et al. , Prevalence of inappropriate antibiotic prescriptions among US Ambulatory care visits, 2010–2011. JAMA 315, 1864–1873 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Kang M., et al. , Evaluation of structure-function relationships of aggregation-induced emission luminogens for simultaneous dual applications of specific discrimination and efficient photodynamic killing of gram-positive bacteria. J. Am. Chem. Soc. 141, 16781–16789 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Zhou C., et al. , One stone, three birds: One AIEgen with three colors for fast differentiation of three pathogens. Chem. Sci. (Camb.) 11, 4730–4740 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao M., et al. , Stochastic DNA walkers in droplets for super-multiplexed bacterial phenotype detection. Angew. Chem. Int. Ed. Engl. 58, 15448–15454 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Loman N. J., et al. , High-throughput bacterial genome sequencing: An embarrassment of choice, a world of opportunity. Nat. Rev. Microbiol. 10, 599–606 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Gan Q., et al. , Culture-free detection of crop pathogens at the single-cell level by micro-Raman spectroscopy. Adv. Sci. (Weinh.) 4, 1700127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon H. Y., et al. , Development of a universal fluorescent probe for gram-positive bacteria. Angew. Chem. Int. Ed. Engl. 58, 8426–8431 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Li J., Savagatrup S., Nelson Z., Yoshinaga K., Swager T. M., Fluorescent Janus emulsions for biosensing of Listeria monocytogenes. Proc. Natl. Acad. Sci. U.S.A. 117, 11923–11930 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling J., et al. , An enzyme-free electrochemiluminescence biosensor for ultrasensitive assay of Group B Streptococci based on self-enhanced luminol complex functionalized CuMn-CeO2 nanospheres. Biosens. Bioelectron. 127, 167–173 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Chen S., et al. , Fast and ultrasensitive visual detection of exosomes in body fluids for point-of-care disease diagnosis. Anal. Chem. 93, 10372–10377 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Jo Y., et al. , Label-free identification of individual bacteria using Fourier transform light scattering. Opt. Express 23, 15792–15805 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Sevenler D., Trueb J., Ünlü M. S., Beating the reaction limits of biosensor sensitivity with dynamic tracking of single binding events. Proc. Natl. Acad. Sci. U.S.A. 116, 4129–4134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Syal K., et al. , Antimicrobial susceptibility test with plasmonic imaging and tracking of single bacterial motions on nanometer scale. ACS Nano 10, 845–852 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Yu H., et al. , Phenotypic antimicrobial susceptibility testing with deep learning video microscopy. Anal. Chem. 90, 6314–6322 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Mo M., et al. , Rapid antimicrobial susceptibility testing of patient urine samples using large volume free-solution light scattering microscopy. Anal. Chem. 91, 10164–10171 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang F., et al. , Rapid antimicrobial susceptibility testing on clinical urine samples by video-based object scattering intensity detection. Anal. Chem. 93, 7011–7021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iriya R., et al. , Rapid antibiotic susceptibility testing based on bacterial motion patterns with long short-term memory neural networks. IEEE Sens. J. 20, 4940–4950 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang P., et al. , Plasmonic scattering imaging of single proteins and binding kinetics. Nat. Methods 17, 1010–1017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor R. W., Sandoghdar V., Interferometric scattering microscopy: Seeing single nanoparticles and molecules via Rayleigh scattering. Nano Lett. 19, 4827–4835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Squires T. M., Messinger R. J., Manalis S. R., Making it stick: Convection, reaction and diffusion in surface-based biosensors. Nat. Biotechnol. 26, 417–426 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Zeng Q., et al. , Dynamic single-molecule sensing by actively tuning binding kinetics for ultrasensitive biomarker detection. Proc. Natl. Acad. Sci. U.S.A. 119, e2120379119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S., et al. , Label free imaging and deep tracking of single biological nanoparticles in free solution by reflection enhanced dark field scattering microscopy. Sens. Actuators B Chem. 355, 131317 (2022). [Google Scholar]
- 25.Puopolo K. M., Lynfield R., Cummings J. J.; Committee on Fetus and Newborn; Committee on Infectious Diseases, Management of infants at risk for group B streptococcal disease. Pediatrics 144, e20191881 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Ye X., Li Y., Fang X., Kong J., Integrated microfluidic sample-to-answer system for direct nucleic acid-based detection of group B Streptococci in clinical vaginal/anal swab samples. ACS Sens. 5, 1132–1139 (2020). [DOI] [PubMed] [Google Scholar]
- 27.El Helali N., Nguyen J. C., Ly A., Giovangrandi Y., Trinquart L., Diagnostic accuracy of a rapid real-time polymerase chain reaction assay for universal intrapartum group B streptococcus screening. Clin. Infect. Dis. 49, 417–423 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Vieira L. L., et al. , Group B Streptococcus detection in pregnant women: Comparison of qPCR assay, culture, and the Xpert GBS rapid test. BMC Pregnancy Childbirth 19, 532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S., et al. , Label-free imaging, detection, and mass measurement of single viruses by surface plasmon resonance. Proc. Natl. Acad. Sci. U.S.A. 107, 16028–16032 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang J. S., Joshi U. A., Lee J. S., Solvothermal synthesis of CdS nanowires for photocatalytic hydrogen and electricity production. J. Phys. Chem. C 111, 13280–13287 (2007). [Google Scholar]
- 31.Zhang F., et al. , Rapid detection of urinary tract infection in 10 min by tracking multiple phenotypic features in a 30 s large-volume scattering video of urine microscopy. ACS Sens. 7, 2262–2272 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin Y., et al. , Group B Streptococcus DNA copy numbers measured by digital PCR correlates with perinatal outcomes. Anal. Chem. 91, 9466–9471 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Wang P. C., et al. , Sustainable removal of nano/microplastics in water by solar energy. Chem. Eng. J. 428, 131196 (2022). [Google Scholar]
- 34.Han J., et al. , Label-free identification of bacteria species based on divergence angle distribution of light scattering patterns. Sens. Mater. 32, 2929–2940 (2020). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.