Abstract
Proteins of the LuxR family detect the presence of N-acylhomoserine lactones (AHLs) and regulate transcription accordingly. When AHLs are synthesized by the same species that detects them, the system allows a bacterium to measure the population density of its own species, a phenomenon known as quorum sensing. The sdiA genes of Escherichia coli and Salmonella enterica serovar Typhimurium are predicted to encode LuxR homologs. However, these species do not appear to synthesize AHLs or any other molecule detected by SdiA. It has previously been demonstrated that overexpression of sdiA results in the activation of the ftsQAZ locus in E. coli and four other loci in Salmonella serovar Typhimurium. Here we report that transcriptional fusions to these five loci fall into two classes. The first class requires overexpression of sdiA for activation. The second class responds to sdiA expressed from its natural position in the chromosome if the appropriate AHLs are added to the culture. The only member of the second class is a series of Prck-luxCDABE fusions in Salmonella serovar Typhimurium. SdiA responds with highest sensitivity to AHLs that have a keto modification at the third carbon and an acyl chain length of 6 or 8 (half-maximal response between 1 and 5 nM). Growth of Salmonella in proximity to species known to synthesize these AHLs results in sdiA-dependent activation of the Prck-luxCDABE fusions. SdiA appears to be the first AHL receptor discovered that detects signals emanating exclusively from other species.
Numerous species of gram-negative bacteria use N-acylhomoserine lactone (AHL) signals to monitor their own population density (for reviews see references 14, 39, and 48). The prototypical example is the Vibrio fischeri LuxR/LuxI system (10, 12, 35). Proteins of the LuxR type have a domain for binding AHL and a second domain for binding DNA (44), while proteins of the LuxI type catalyze the final step in AHL synthesis (21, 26, 34, 36, 41, 51). A variety of AHL signaling molecules have been discovered. These differ primarily in acyl chain length and the nature of the substituents at the C-3 position. Each LuxI homolog makes a specific AHL, although many LuxI enzymes also make lesser amounts of related AHL molecules. In addition to the LuxI family of AHL synthases, a second type (LuxLM) has been described for Vibrio harveyi and V. fischeri (2, 17). These enzymes use biosynthetic substrates similar to those used by the LuxI family (22). More recently, a third type of AHL synthase has been proposed, although enzymatic studies are required to confirm this (32).
In a widely accepted model for quorum sensing, each bacterial cell in a population produces AHL. As the population density increases, the concentration of AHL also increases. Above a threshold concentration, representing a “quorum” of bacterial cells, the LuxR homolog binds AHL and activates transcription of target genes. One of the target genes is often the luxI homolog, which results in a positive feedback loop of increased AHL synthesis. Many bacterial behaviors have been shown to be regulated in a population density-dependent manner by AHLs, including plasmid conjugal transfer, protein secretion, exoenzyme and cytotoxin synthesis, antibiotic synthesis, capsular exopolysaccharide synthesis, biofilm formation, twitching motility, and swarming motility (9).
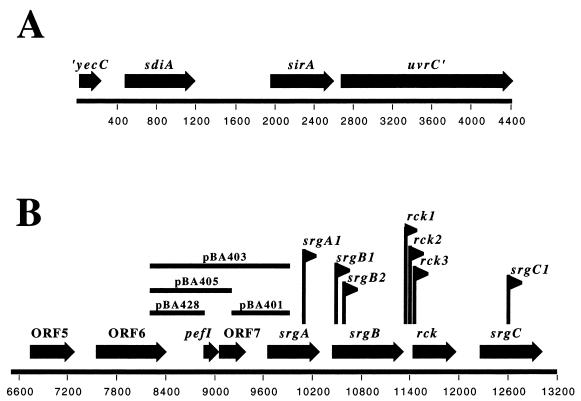
Escherichia coli and Salmonella enterica serovar Typhimurium encode a single luxR homolog named sdiA (1, 53). The genomic organizations of the sdiA region are identical in the two species (Fig. 1A). Upstream of sdiA is an uncharacterized open reading frame (ORF) named yecC which is similar to the ATP binding component of ABC transporters (1, 3). Downstream of sdiA is a gene named uvrY in E. coli and sirA in Salmonella serovar Typhimurium (27, 29). Further downstream is the uvrC gene, which encodes a DNA repair enzyme. Despite the name, uvrY plays no role in DNA repair but instead encodes a transcription factor of the FixJ family that controls virulence functions in all γ-proteobacterial pathogens examined to date (see references in reference 18). Our recent searches of genome databases suggest that orthologs of uvrY and uvrC are present in all members of the γ-Proteobacteria, while sdiA is located directly upstream only in Escherichia, Salmonella, and Klebsiella spp. Furthermore, it is striking that although these three genera possess a copy of sdiA, they are not known to synthesize the AHLs that are typically detected by LuxR homologs (reference 48 and this report). In fact, there are no AHL synthase genes (luxI or luxLM homologs) in any of the available genome sequences for these organisms. Therefore, Escherichia, Salmonella, and Klebsiella appear to be unusual with regard to quorum sensing in that they encode a putative AHL receptor, SdiA, but not an AHL synthase.
FIG. 1.
ORF maps of the sdiA (A) and rck (B) regions of the Salmonella serovar Typhimurium genome derived from GenBank accession numbers U88651 (A) and L08613 (B). Numbers along the bottom of each map represent nucleotide positions.
The E. coli sdiA gene was initially isolated as a regulator of the ftsQp2 promoter upstream of the ftsQAZ operon (53). However, this 5- to 13-fold up-regulation required that sdiA be expressed from a low-copy-number plasmid (pSC101 origin). SdiA expressed from its natural position in the chromosome had only a marginal effect on ftsQp2 expression (40% higher expression in the wild type than in the sdiA mutant) (53). It was later demonstrated that E. coli SdiA activates ftsQ in response to AHL (43). This was a twofold stimulation using a strain in which sdiA was expressed from a plasmid. No experiments comparing a wild-type strain to an sdiA mutant with respect to detection of AHL and activation of ftsQ were reported (43). It was also reported that SdiA responds to an unidentified compound present in spent E. coli culture supernatants (43), although this effect was later explained by growth rate differences in fresh medium compared to spent medium (16).
A study with E. coli O157:H7 (enterohemorrhagic E. coli [EHEC]) recently determined that expression of sdiA from a plasmid causes repression of both motility and virulence factor expression (30). A second study, performed with E. coli K-12, also found that sdiA expressed from a plasmid causes repression of motility gene expression (54). Neither study reported the phenotype of a wild-type strain compared to that of an isogenic sdiA mutant. It was also reported that an SdiA-dependent ligand could be removed from the EHEC culture supernatant using immobilized SdiA as an affinity matrix (30).
In Salmonella serovar Typhimurium, a genetic screen was performed to identify genes regulated by SdiA (1). The sdiA gene was expressed under the control of the araBAD promoter on a multicopy plasmid (pJVR2) and placed in a Salmonella serovar Typhimurium sdiA mutant strain so that expression of sdiA was dependent on the presence of arabinose. Random lacZY transcriptional fusions (MudJ transposon insertions) were created in this strain and screened for a difference in expression on plates containing glucose versus arabinose. Ten MudJ fusions that respond to plasmid-borne sdiA but not to a vector control were identified. Although these fusions were responsive to sdiA overexpression, they were not active when sdiA was expressed from its natural position in the chromosome (1). This suggested that any putative ligand detected by SdiA was not present in pure cultures of Salmonella serovar Typhimurium and that overexpression of sdiA somehow bypasses the requirement for a ligand (1).
Seven of the 10 sdiA-responsive MudJ insertions are located within four genes on the 90-kb virulence plasmid of Salmonella serovar Typhimurium (srgA, srgB, rck, and srgC [Fig. 1B]). Sequence analysis suggests that srgA (sdiA-regulated gene) encodes a dsbA homolog, srgB encodes a putative lipoprotein, rck (resistance to complement killing) encodes a small outer membrane protein, and srgC encodes a putative araC type transcriptional regulator (13). The functions of srgA, srgB, and srgC are not known. However, rck has been studied in detail and appears to form an 8-stranded β-barrel in the outer membrane (7). The rck gene is not expressed well from the 90-kb plasmid of Salmonella serovar Typhimurium, but when expressed from a heterologous promoter in a rough background of either Salmonella serovar Typhimurium or E. coli, rck confers resistance to human complement (4, 20, 23, 24). The rough background is required because lipopoysaccharide is a redundant complement resistance factor. Rck also confers adhesiveness to epithelial cells and/or extracellular matrix when expressed from a plasmid in E. coli K-12 (7, 8, 24). The requirement for a heterologous promoter in these experiments is consistent with our observation that the rck promoter is sdiA dependent and is not active in pure culture. Given the lack of an obvious AHL synthase gene in the E. coli or Salmonella serovar Typhimurium genome, we hypothesized that SdiA is used to detect only the signals of other bacterial species (1). In this report we test and confirm this hypothesis.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids used in this study are listed in Tables 1 and 2. Bacteria were grown in Luria-Bertani (LB) medium or on LB plates containing 1.5% agar (EM Science) unless otherwise indicated. AB mannitol plates were used for growth of Agrobacterium tumefaciens as described previously (6). For filter disk assays the reporter strains were grown in 3 ml of LB soft agar (0.75% agar) overlaid on a standard LB plate. Tetracycline, chloramphenicol, ampicillin, and kanamycin were used at 20, 30, 100, and 60 μg/ml when appropriate. Glucose and arabinose were used at a final concentration of 0.2% unless otherwise indicated. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used at a final concentration of 80 μg/ml. AHLs with even-numbered side chains of 4 to 12 C atoms and with or without a 3-oxo group were synthesized according to the work of Chhabra et al. (5). The unsubstituted AHLs are abbreviated as C4, C6, C8, C10, or C12 based on acyl chain length. Those AHLs with a 3-oxo modification are abbreviated as oxoC4, oxoC6, oxoC8, oxoC10, or oxoC12. AHLs were added to media as dilutions from a 10 mM stock solution in acetonitrile. The final concentration of acetonitrile was always less than 1% and had no effect on the growth or AHL response of the reporter. All incubations were carried out at 37°C unless otherwise stated.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Strains | ||
| 14028 | Wild-type Salmonella serovar Typhimurium | American Type Culture Collection |
| BA612 | 14028 sdiA1::mTn3 (Ampr) | 1 |
| UT481 | E. coli K-12 Δlac | 53 |
| WX2 | UT481 sdiA::kan | 53 |
| BA1101 | 14028 srg-5::MudJ | 1 |
| BA1102 | 14028 srgB2::MudJ | 1 |
| BA1103 | 14028 srgA1::MudJ | 1 |
| BA1104 | 14028 rck2::MudJ | 1 |
| BA1105 | 14028 rck3::MudJ | 1 |
| BA1107 | 14028 srgB1::MudJ | 1 |
| BA1109 | 14028 srgC1::MudJ | 1 |
| BA1110 | 14028 srg-6::MudJ | 1 |
| BA1111 | 14028 rck1::MudJ | 1 |
| BA1112 | 14028 srg-7::MudJ | 1 |
| BA1303 | 14028 srgA1::MudJ sdiA::mTn3 | 1 |
| Plasmids | ||
| pWSK29 | pSC101 cloning vector (Ampr) | 52 |
| pWSK129 | pSC101 cloning vector (Kanr) | 52 |
| pBA306 | pWSK29 Salmonella serovar Typhimurium sdiA+ (Ampr) | 1 |
| pBM1 | pWSK129 Salmonella serovar Typhimurium sdiA+ (Kanr) | This study |
| pCX39 | Mini-F′ carrying ftsQAZ promoter 2 (sdiA sensitive) fusion to galK′-lacZYA; Ampr | 53 |
| pJVR2 | sdiA under control of araBAD promoter; pACYC origin; Cmr | 1 |
| pBA321 | sdiA under control of araBAD promoter; ColE1 origin; Kanr | This study |
| pSB401 | luxR+luxI::luxCDABE; pACYC origin; Tetr | 57 |
| pBA401 | pSB401ΔEcoRI carrying ORF7-srgA intergenic region | This study |
| pBA403 | pSB401ΔEcoRI carrying ORF6-pefI intergenic region, pefI, and ORF7 | This study |
| pBA405 | pSB401ΔEcoRI carrying ORF6-pefI intergenic region and pefI | This study |
| pBA428 | pSB401ΔEcoRI carrying ORF6-pefI intergenic region | This study |
TABLE 2.
Strains tested for activation of the reporter strain 14028/pBA428
| Strain | Greatest fold induction | Sourcea | AHL(s) produced | Reference(s)d |
|---|---|---|---|---|
| Agrobacterium tumefaciens ATCC 23308 | None | DCC | C6, oxoC8 | 15, 59 |
| Citrobacter freundii strain 849 | None | DCC | None detected | |
| Chromobacterium violaceum ATCC 31532 | 10b | Simon Swift | C6 | 33 |
| Escherichia coli O157:H7 ATCC 35150 | None | DCC | None detected | |
| Escherichia coli O157:H3 strain 317 | None | DCC | None detected | |
| Escherichia coli O157:H7 ATCC 43895 | None | DCC | None detected | |
| Escherichia coli O157:H12 Strain 316 | None | DCC | None detected | |
| Escherichia coli O157:H7 ATCC 4388 | None | DCC | None detected | |
| Escherichia coli O157:H7 ATCC 700927 | None | ATCC | None detected | |
| Escherichia coli MG1655 | None | EGSC | None detected | |
| Hafnia alvei 1058 | 8 | IIISC | Unknown | |
| Klebsiella pneumoniae subsp. ozaenae ATCC 11296 | None | Valley Stewart | None detected | |
| Klebsiella oxytoca ATCC 13182 | None | Valley Stewart | None detected | |
| Klebsiella pneumoniae subsp. pneumoniae ATCC 13883 | None | Valley Stewart | None detected | |
| Klebsiella pneumoniae subsp. rhinoscleromatis ATCC 13884 | None | Valley Stewart | None detected | |
| Klebsiella terrigena ATCC 33257 | None | Valley Stewart | None detected | |
| Klebsiella planticola ATCC 33531 | None | Valley Stewart | None detected | |
| Klebsiella ornithinolytica JCM6096 | None | Valley Stewart | None detected | |
| Klebsiella pneumoniae CDC 2665-69 | None | DCC | None detected | |
| Providencia alkafaciens CDC 671-66 | None | DCC | None detected | |
| Proteus vulgaris ATCC 12454 | None | DCC | None detected | |
| Pseudomonas aeruginosa PAO1 | 3.0b | Dieter Haas | C4, C6, oxoC6, oxoC8, oxoC10, oxoC12 | 28, 37, 38, 42, 56 |
| Pseudomonas aeruginosa PAK | 2.5b | Shouguang Jin | C4, C6, oxoC6, oxoC8, oxoC10, oxoC12 | 28, 37, 38, 42, 56 |
| Salmonella serovar Gallinarum ATCC 9184 | None | ATCC | None detected | |
| Salmonella serovar Pullorum ATCC 9120 | None | ATCC | None detected | |
| Salmonella serovar Typhi ATCC 19430 | None | ATCC | None detected | |
| Salmonella serovar Typhi ATCC 33458 | None | ATCC | None detected | |
| Salmonella serovar Typhimurium 14028 | None | Laboratory collection | None detected | |
| Salmonella serovar Typhimurium SL1344 | None | Laboratory collection | None detected | |
| Salmonella serovar Typhimurium SR-11 | None | Laboratory collection | None detected | |
| Shigella flexneri | None | DCC | None detected | |
| Vibrio fischeri ESRI | 11c | Karen Visick | C6, oxoC6, C8 | 11 |
| Yersinia enterocolitica 10460 | 13 | Steve Atkinson | C6, oxoC6 | 50 |
| Yersinia enterocolitica 10460 yenI::kan | None | Steve Atkinson | None detected | 50 |
DCC, Ohio State University Department of Microbiology Culture Collection; EGSC, E. coli Genetic Stock Center; ATCC, American Type Culture Collection; IIISC, Institute of Infections and Immunity Strain Collection, Nottingham, United Kingdom.
Test strain incubated for 16 h at 28°C prior to cross-streaking with reporter strains and incubation at 37°C for 10 h.
Test strain incubated for 16 h at 24°C prior to cross-streaking with reporter strains and incubation at 37°C for 10 h.
References refer to identification of particular AHLs in a species, not a specific strain.
Plasmid constructions.
To identify the promoter region of the rck operon, episomal transcriptional fusions were constructed (Fig. 1B). The putative promoter regions were amplified using Pfu Turbo DNA polymerase (Stratagene) with Salmonella serovar Typhimurium strain ATCC 14028 genomic DNA as the template. Each primer had an EcoRI site in the 5′ end. The resulting DNA fragments were gel purified using Qiagen gel extraction columns and cloned into the SrfI site of pCR-script Amp (Stratagene). One PCR product that gave rise to pBA428 was instead cloned into pCR-Blunt-II-Topo, which has EcoRI sites in the vector flanking the cloning site. The EcoRI fragment of each clone was removed, gel purified, and ligated into pSB401 that had been digested with EcoRI and exposed to phosphatase. pSB401 is a reporter vector containing a p15A origin of replication, a tetracycline resistance marker, and a promoterless luxCDABE operon from Photorhabdus luminescens (57). Upstream of the luciferase operon is an EcoRI fragment containing luxR and the luxI promoter from V. fischeri. This EcoRI fragment was removed and replaced with regions of DNA hypothesized to encode the rck promoter (see Fig. 1B). According to the numbering system of GenBank accession number L08613, which is used in Fig. 1B, pBA401 contains nucleotides 9276 to 9976, pBA403 contains nucleotides 8178 to 9976, pBA405 contains nucleotides 8178 to 9300, and pBA428 contains nucleotides 8178 to 8910. pBM1 was constructed by cloning the sdiA-containing PstI fragment of pBA306 (1) into the PstI site of pWSK129 (52). Orientation of inserts was determined by restriction mapping. One clone containing the sdiA gene oriented opposite the lac promoter of pWSK129 was saved and named pBM1. pBA321 was constructed by digesting pJVR2 (1) with XbaI and SacI, gel purifying the sdiA-containing fragment, and ligating it to pBAD18-Kn (19) that had been digested with XbaI and SacI. Plasmids were introduced into the appropriate strains using electroporation with a Bio-Rad Gene Pulser II.
Assay of luciferase activity.
Luciferase activity was measured in liquid culture using a Turner Designs TD-20/20 luminometer. The optical density of the culture at 550 nm was measured using either a Spectronic 20D+ or a Beckman DU-64 spectrophotometer. All luminometer samples consisted of 10 μl of liquid culture placed into a 12- by 75-mm polystyrene tube. The samples were oxygenated by “ratcheting” the sample tube across a tube rack prior to insertion into the luminometer.
Expression of luciferase activity in soft agar plates was imaged and quantified using a C2400–32 intensified charge-coupled device camera with an Argus 20 image processor (Hamamatsu Photonics) or a Luminograph LB980 photon video camera (E. G. & G. Berthold). Images were captured with a Macintosh G4 computer and Adobe Photoshop 5.0 software.
For microplate format dose response assays, a logarithmic dilution series of each AHL was first made in LB medium containing the appropriate antibiotics (final volume, 100 μl) from a 10 mM stock solution in acetonitrile. The sensor strain was then diluted 1:100 from an overnight culture into LB broth with 0.6% agar, and 100 μl was added to the test microplate wells to give a final concentration of 0.3% agar. Luciferase expression was quantified after 6 h using a Victor2 1420 multilabel counter (Wallac).
RESULTS
Identification of the rck promoter region.
Previously, seven MudJ fusions clustering in or around the rck gene on the Salmonella serovar Typhimurium virulence plasmid were isolated based on their transcriptional responsiveness to sdiA overexpression (1) (Fig. 1B). To test whether these MudJ insertions were disrupting a single operon, the polarity of each MudJ insertion on the downstream genes was assessed using Northern hybridization. A MudJ insertion in each of the four genes was placed into a strain that allows arabinose-dependent overexpression of sdiA (BA612/pJVR2). RNA was isolated from each strain after growth in either glucose or arabinose. Northern blot analysis using probes specific to each individual gene confirmed that each MudJ insertion is polar on the transcription of each of the downstream genes and that expression is dependent on arabinose (data not shown). This demonstrates that srgA, srgB, rck, and srgC are expressed as a polycistronic transcript with no detectable internal start sites.
To identify the promoter region of the rck operon, plasmid-based transcriptional fusions to individual fragments of DNA found upstream of srgA were constructed. The resulting constructs form transcriptional fusions between the DNA of interest and the luxCDABE operon of P. luminescens. Each reporter plasmid (pBA401, pBA403, pBA405, and pBA428) (Fig. 1B) was then placed into an arabinose-conditional sdiA strain (BA612/pBA321). The luminescence resulting from each construct was compared during growth in glucose to that during growth in arabinose. A fragment containing the DNA region between ORF7 and srgA was not responsive to sdiA overexpression (pBA401 [data not shown]). However, all fragments that contained the region between ORF6 and pefI were responsive to sdiA overexpression (data not shown for pBA403 and pBA405; data for the smallest plasmid, pBA428, are shown in Fig. 2). No fragment was responsive when cloned in the opposite orientation with respect to the luxCDABE genes (data not shown). In total, this demonstrates that the promoter for the rck operon is unidirectional and lies between ORF6 and pefI (Fig. 1B and 2). The results also indicate that pefI and ORF7 are previously unrecognized members of the rck operon. Both of these genes are homologous to transcription factors.
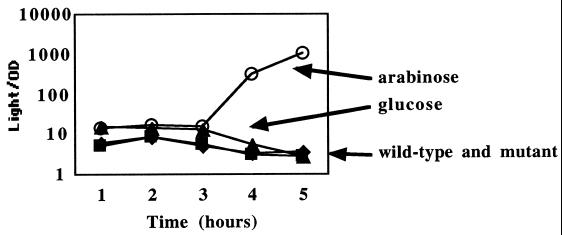
FIG. 2.
Expression of the Prck-luxCDABE fusion, pBA428, in various strain backgrounds. Levels of expression are identical in the wild-type (14028/pBA428 [⧫]) and sdiA mutant (BA612/pBA428 [▪]) backgrounds. In a strain where sdiA is plasmid borne under the control of the arabinose-dependent araBAD promoter (BA612/pBA428/pBA321), Prck-luxCDABE expression is high in the presence of 0.2% arabinose (▴) and low in the presence of 0.2% glucose (53). Results shown are means of results from triplicate cultures. Error bars representing standard deviations were smaller than the symbols used and are therefore not shown.
Testing of fusions for a response to chemically synthesized AHLs.
All previous reports on sdiA have utilized plasmid-based expression of sdiA to obtain regulatory effects greater than 40% on any particular transcriptional fusion (1, 43, 53). We hypothesized that SdiA is used to detect AHLs synthesized by other species of bacteria and that overexpression of sdiA bypasses the requirement for AHL (1). To test this hypothesis, we screened all previously identified sdiA-regulated transcriptional fusions for a response to chemically synthesized AHLs in the absence of sdiA overexpression. The fusion collection consists of a plasmid-based E. coli P2ftsQ-lacZYA fusion (pCX39 [53]), 3 plasmid-based Salmonella serovar Typhimurium Prck-luxCDABE fusions (pBA403, pBA405, and pBA428 [this study]), and 10 Salmonella serovar Typhimurium MudJ insertion mutations, of which 3 are uncharacterized and 7 are located within the rck operon (1). Each fusion was tested using a filter disk assay. Individual reporter strains were placed in a soft agar layer on top of an LB plate. For the lacZ fusions X-Gal was included in the plates at 80 μg/ml. This relatively high concentration of X-Gal was chosen because it allowed perceptible detection of the background level of β-galactosidase activity coming from the MudJ fusions. Any further increase in β-galactosidase activity would then be readily detected visually. Filter disks impregnated with 100 pmol of synthetic AHLs were placed on top of each plate. At various times during incubation at 37°C the plates were examined for either blue halos surrounding the filter disks or, in the case of the luxCDABE fusions, luminescence surrounding the filter disks. This methodology was chosen because a concentration gradient of AHL is formed as the AHL diffuses away from the filter disk, so that all concentrations can be tested simultaneously.
The E. coli P2ftsQ-lacZYA fusion, pCX39, failed to respond to AHL when present in either E. coli or Salmonella serovar Typhimurium (data not shown). All of the Salmonella serovar Typhimurium MudJ fusions also failed to respond to AHL (data not shown). This includes the MudJ insertions that lie within the rck operon. Only the Prck-luxCDABE fusions pBA403, pBA405, and pBA428 responded to any of the AHLs (Fig. 3). The response did not occur in the isogenic sdiA mutant control and is therefore sdiA dependent. AHLs with a 3-oxo modification and acyl chain lengths of 6 or 8 induced the greatest luminescence (Fig. 3). Of the three Prck-luxCDABE reporter plasmids, pBA428 contains the smallest promoter region and was chosen for further study of sdiA-dependent responses to AHL.
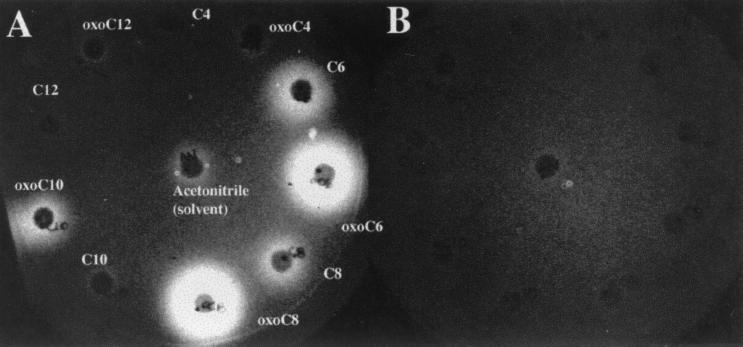
FIG. 3.
Testing of individual chemically synthesized AHLs for activation of a Prck-luxCDABE fusion in either a wild-type background (14028/pBA405) (A) or the isogenic sdiA mutant background (BA612/pBA405) (B) using a filter disk assay. A soft agar overlay containing the appropriate reporter strain was poured onto an LB agar plate and allowed to harden at room temperature for 2 h. Filter disks impregnated with 100 pmol of the indicated AHL were then placed onto the overlay, and the plate was incubated at 37°C for 8 h. Similar results were obtained using reporter plasmids pBA403 and pBA428 (data not shown).
Quantification of the sdiA-dependent response of Salmonella serovar Typhimurium to AHLs.
Dose-response experiments were performed to quantify the response of SdiA to each synthetic AHL using the 14028/pBA428 reporter (Fig. 4). It was determined that Salmonella serovar Typhimurium does not respond well to AHLs in liquid LB medium but responds best in LB semisolid agar (0.3, 0.75, or 1.5% agar). In liquid cultures sdiA-dependent responses were 3- to 5-fold, while on semisolid agar the responses reached 14- to 17-fold (data not shown). Therefore, we chose to grow the reporter strain in 0.3% agar in 96-well plates for the dose-response experiments for which results are shown. The plates were incubated at 37°C for 6 h to reach optimal response levels. The Prck-luxCDABE reporter responded to all of the AHLs tested at concentrations greater than 1 μM (Fig. 4). The two most potent AHLs tested were oxoC6 and oxoC8, which exhibited half-maximal responses between 1 and 5 nM (Fig. 4). Detectable concentrations in this range are similar to values obtained for other LuxR homolog reporter systems, suggesting that these are physiologically relevant detection limits (15, 31, 37, 58, 59).
FIG. 4.
Quantitation of Salmonella serovar Typhimurium responses to chemically synthesized AHLs. The reporter strain 14028/pBA428 was incubated in LB broth with 0.3% agar containing varying amounts of each AHL. The responses of triplicate cultures were measured after 6 h of incubation at 37°C. Error bars are not shown for clarity, but standard deviations did not exceed 32% of any value. This experiment is representative of experiments performed on four separate occasions.
Salmonella serovar Typhimurium responds in an sdiA-dependent manner to the presence of AHL-synthesizing bacterial species.
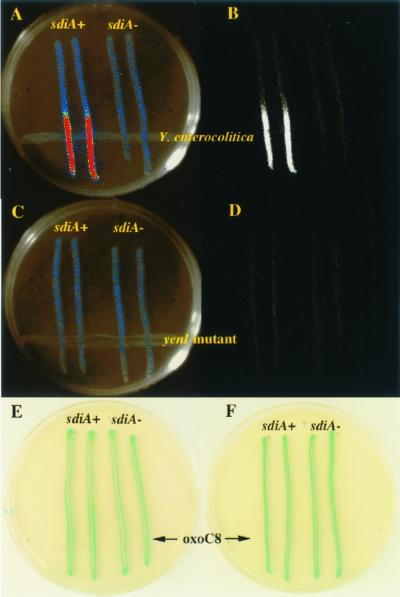
Numerous gram-negative bacterial species are known to synthesize the AHLs that are most readily detected by SdiA (oxoC6 and oxoC8). Therefore, we tested the ability of the Salmonella serovar Typhimurium reporter strain (14028/pBA428) and an isogenic sdiA mutant (BA612/pBA428) to respond to the presence of other bacterial species on an LB agar plate using a cross-streak assay. Several species that elicit sdiA-dependent responses were identified (Table 2 and Fig. 5A and B). The best responses were obtained with Yersinia enterocolitica and Hafnia alvei (induction ratios, 8- to 13-fold). It is not known what AHLs are synthesized by Hafnia, but AHL type activity has been detected from this genus before (49). Y. enterocolitica synthesizes both C6 and oxoC6, which are consistent with the specificity preferences of SdiA (50). An isogenic yenI mutant of Y. enterocolitica that lacks AHL synthetic capabilities was unable to elicit a response from the Salmonella serovar Typhimurium Prck-luxCDABE reporter (Fig. 5C and D). It is noteworthy that at a distance from the test species, the wild-type and sdiA mutant reporters produce equal levels of background luminescence. These results demonstrate that Salmonella serovar Typhimurium can detect the physical proximity of other species in an sdiA-dependent manner.
FIG. 5.
Detection of Y. enterocolitica by Salmonella serovar Typhimurium. Y. enterocolitica strain NCTC 10460 is struck across the bottom of the plate (A and B). The wild-type Salmonella serovar Typhimurium reporter strain 14028/pBA428 is struck in duplicate perpendicular to the Y. enterocolitica on the left side of the plate (A through D). The isogenic sdiA mutant, BA612/pBA428, is struck in duplicate on the right side of the plate (A through D). Raw luminescence data are shown in panels B and D, while the pseudocolored version of the same data is shown in panels A and C. The luminescence of the wild-type reporter strain near the Y. enterocolitica strain is 13-fold greater than that of the sdiA mutant control (A and B). No response is obtained using an isogenic yenI::kan mutant of Y. enterocolitica (C and D). These data show that SdiA expressed from its natural position in the chromosome of Salmonella serovar Typhimurium can detect the physical proximity of other species that are capable of synthesizing AHLs. (E) MudJ insertions in the rck operon fail to respond to AHL. The wild-type Salmonella serovar Typhimurium reporter strain BA1103 (14028 srgA1::MudJ) is struck in duplicate on the left side of the LB kanamycin–X-Gal plate, while the isogenic sdiA mutant, BA1303, is struck in duplicate on the right side of the plate. Across the bottom is 20 μl of 10 μM oxoC8. All other Salmonella serovar Typhimurium MudJ insertions previously shown to respond to sdiA overexpression also failed to respond when present in the wild-type 14028 background in this assay (data not shown). (F) The E. coli P2ftsQ-lacZYA reporter fails to respond to AHL. The wild-type E. coli reporter strain UT481/pCX39 is struck in duplicate on the left side of the LB ampicillin–X-Gal plate, while the isogenic sdiA mutant, WX2/pCX39, is struck in duplicate on the right side of the plate. Across the bottom is 20 μl of 10 μM oxoC8. The E. coli P2ftsQ-lacZYA reporter and the Salmonella serovar Typhimurium MudJ insertions also failed to respond in cross-streak assays against Y. enterocolitica, in filter disk assays with synthetic AHL, and in liquid cultures supplemented with synthetic AHL (data not shown).
A. tumefaciens, Chromobacterium violaceum, and V. fischeri are known to produce the AHLs that sdiA most readily detects but failed to elicit a response from 14028/pBA428 using the standard cross-streak assay. However, by preincubation of the test strain on the plate for 16 h at lower temperatures, responses to both C. violaceum and V. fischeri were obtained (Table 2). A. tumefaciens was tested on both LB plates and AB mannitol plates at 22, 30, and 37°C. However, this species failed to stimulate the Salmonella serovar Typhimurium reporter strain under any condition (data not shown). This is probably due to the observation that the A. tumefaciens quorum-sensing system is activated by plant-derived octopine compounds which were not present in the assay (15).
Optimal assay conditions for SdiA and AI-2.
To date, we have found that the maximal response from 14028/pBA428 is 14- to 18-fold using a cross-streak assay at 37°C in which chemically synthesized oxoC8 is spread across the plate, rather than any particular bacterial species (20 μl of 10 μM oxoC8 [data not shown]). Using this optimal assay the P2ftsQ-lacZYA reporter (pCX39) and the Salmonella serovar Typhimurium MudJ insertions all failed to respond to AHL (Fig. 5E and F).
AI-2 is a molecule produced by numerous bacterial species, including E. coli and Salmonella serovar Typhimurium, that induces V. harveyi luminescence genes (45). E. coli and Salmonella serovar Typhimurium were found to produce maximal levels of AI-2 during the exponential phase of growth in agitated LB broth containing 0.5% glucose (46). To determine if SdiA can respond to AI-2 production, the luminescence of the Prck-luxCDABE reporter (pBA428) was measured during growth under these conditions (in the absence of any exogenous AHL). Under these conditions, which are optimal for AI-2 production, the wild-type and sdiA mutant strains (14028/pBA428 and BA612/pBA428, respectively), produced identical levels of background luminescence throughout the growth curve (data not shown). This indicates that SdiA does not respond to AI-2.
Expression of sdiA from pSC101-based vectors results in SdiA activity in the absence of ligand.
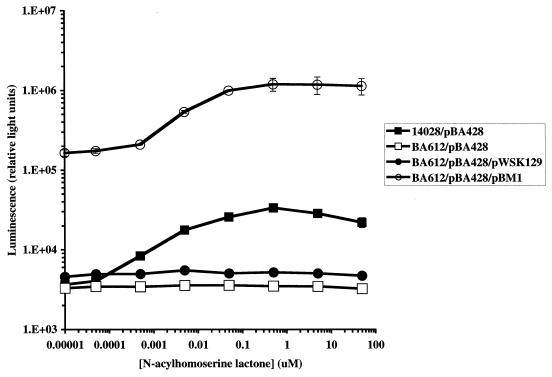
The first studies of sdiA that were performed in E. coli used low-copy-number pSC101-based vectors to express sdiA (43, 53). However, because sdiA-dependent activation of the ftsQ reporter (pCX39) does not occur in a wild-type background (even in the presence of AHL), we tested the possibility that the expression of sdiA from a low-copy-number vector may result in artificial activation (i.e., activation in the absence of ligand). Because only the Prck-luxCDABE fusions in Salmonella serovar Typhimurium respond to both conditions, the responses of the pBA428 reporter to chromosomal and plasmid-borne sdiA were compared. Expression of sdiA from a low-copy-number vector resulted in greater activation of the rck promoter than addition of AHL to a chromosomal copy of sdiA (Fig. 6). However, plasmid-borne sdiA still responded to AHL by further increasing the activity of the rck promoter. The concentration of oxoC8 resulting in half-maximal activation was between 1 and 5 nM regardless of whether sdiA was plasmid borne (Fig. 6).
FIG. 6.
Comparison of Prck-luxCDABE (pBA428) activation when sdiA is expressed from its natural position in the Salmonella serovar Typhimurium chromosome and when it is expressed from a pSC101-derived plasmid (pBM1). Strains were incubated for 6 h at 37°C in LB broth with 0.3% agar containing various concentrations of AHL before measurement of luminescence. Error bars represent standard deviations of triplicate samples.
Members of the Escherichia, Salmonella, and Klebsiella genera do not activate AHL biosensors.
Members of the Escherichia, Salmonella, and Klebsiella genera are known to contain sdiA orthologs in their genomes. To test the hypothesis that SdiA does not detect signals emanating from the same species that contain sdiA, representatives of these three genera were tested for activation of the pBA428 reporter using the cross-streak assay (on an LB plate at 37°C). All seven type species of Klebsiella, six different strains of E. coli O157, the E. coli K-12 strain MG1655, a strain of Shigella flexneri, three strains of Salmonella serovar Typhimurium, two strains of Salmonella enterica serovar Typhi, and one strain each of Salmonella enterica serovar Gallinarum and Salmonella enterica serovar Pullorum were tested. All strains tested negative for the ability to elicit a response from the Salmonella serovar Typhimurium Prck-luxCDABE reporter under the conditions employed (Table 2).
The above strains were also tested for the ability to activate other AHL reporter systems. In cross-streak assays on LB plates at 37°C, all of the Escherichia, Klebsiella, and Salmonella strains failed to activate a LasR reporter (JM109/pSB1075 [57]), an AhyR reporter (DH5α/pSB536 [47]), and a LuxR reporter (JM109/pSB401 [57]) (data not shown). Therefore, these particular members of the Escherichia, Salmonella, and Klebsiella genera do not produce quantities of AHL that are detectable with these reporter systems and growth conditions.
DISCUSSION
Despite decades of research on E. coli and Salmonella serovar Typhimurium, microbiologists are still unable to assign, or even convincingly predict, functions for approximately 30% of the ORFs in the E. coli genome. A further 30% of the ORFs are predicted to encode proteins that belong to recognized families, but their specific functions remain undetermined (3). It seems likely that the functions of many of these genes are not observable using pure cultures. In nature, such bacteria do not normally exist as pure cultures and a percentage of their genetic capacity is almost certainly involved with “mixed-community” interactions.
In this report we have determined that Salmonella serovar Typhimurium can respond to the physical proximity of other bacterial species in an sdiA-dependent manner. This response requires that the other bacterial species be able to synthesize AHLs. Salmonella serovar Typhimurium detects chemically synthesized AHLs at nanomolar concentrations in an sdiA-dependent manner but fails to synthesize detectable quantities of these molecules under the conditions tested. SdiA also does not respond to AI-2 or any other molecule present in its own culture supernatant. Therefore, sdiA appears to encode a receptor that exclusively detects the signal molecules of other species.
This is the first study in which significant SdiA activity has been detected without the use of plasmid-based expression. In fact, expression of sdiA from even a low-copy-number plasmid vector leads to higher levels of rck promoter activation than does addition of AHL to a wild-type cell. Studies of other LuxR homologs indicate that binding of AHL promotes dimerization or some higher form of oligomerization, which is required for promoter activation (40, 44, 55, 60). Therefore, our working model for SdiA is that in pure culture (in the absence of AHL) SdiA is found primarily as a monomer in the cell. Because monomers and dimers are in equilibrium, a small percentage of the SdiA in a cell may be found as dimers even in the absence of AHL. When sdiA is expressed from a plasmid, the concentration of the SdiA monomer is elevated and there is a corresponding increase in the equilibrium concentration of the dimer. The number of dimers in this situation appears to exceed the number achieved with chromosomal expression of sdiA even in the presence of AHL (based on the level of reporter activation shown in Fig. 6), although the equilbrium still greatly favors the monomeric form. When AHL is added to cells with plasmid-borne sdiA, a high number of monomers in the cell are converted to dimers, with a corresponding increase in reporter activity (Fig. 6). Figure 6 also demonstrates that the half-maximal response to oxoC8 occurs between 1 and 5 nM regardless of whether sdiA is expressed from the chromosome or from a plasmid.
It is unclear why the ftsQ locus and the three uncharacterized loci in Salmonella serovar Typhimurium respond to plasmid-borne sdiA but fail to respond to chromosomal sdiA and AHL. The question arises whether these loci ever respond to SdiA and AHL in nature. It is possible that these are only weakly regulated or indirectly regulated promoters that never respond significantly in nature. Alternatively, it is possible that environmental conditions exist that increase the expression of sdiA from the chromosome and/or remove competitive regulatory influences. The regulation of genes that respond to plasmid-borne sdiA, and the regulation of sdiA itself, must be studied in natural environments to address these questions.
It is also not clear why the plasmid-based Prck-luxCDABE fusions respond to chromosomal sdiA and AHL, but the MudJ insertions in the rck operon do not. The explanation cannot be a difference in sensitivity between the luxCDABE fusions and the lacZ fusions, because in Fig. 5E and F the background levels of lacZ activity are clearly observed (by including high levels of X-Gal in the plate), yet there is no increase in activity near the AHL cross-streak. (Even twofold differences in β-galactosidase activity are easily detected visually.) We have also eliminated the polarity of the MudJ insertions as a viable explanation (unpublished data). Given that there are three putative transcription factors encoded within the rck operon, there may not be a simple answer. Regardless, the sdiA-dependent activation of the Prck-luxCDABE fusions reported here is the first demonstration of a phenotype for sdiA expressed from its natural position in the chromosome.
It has been hypothesized that quorum sensing may be used by pathogenic species to prevent the expression of virulence factors until the population of bacteria has increased to a point at which victory is ensured. In the commonly studied quorum-sensing systems, the areas of colonization have only low populations of other bacterial species (e.g., the squid light organ for V. fischeri, plant wound sites for Erwinia and Pseudomonas species, human lung tissues for P. aeruginosa). In contrast, the intestinal environment has microbial population densities reaching 1011 cells/g of fecal contents (reviewed in reference 25). Given that Salmonella serovar Typhimurium is most often associated with the intestinal environment, we hypothesize that instead of using LuxR homologs to gauge the population density of its own species, Salmonella serovar Typhimurium (and possibly E. coli and Klebsiella species as well) appears to have dispensed with any AHL synthase genes it may have had in the past and instead uses SdiA to detect the AHLs produced by other species of bacteria. If this is true, then SdiA is technically not a “quorum sensor” but instead is strictly an AHL receptor. Interestingly, there are no reports of AHLs in the intestinal environment, and no members of the normal intestinal flora have been reported to synthesize AHLs. We are currently examining the intestinal environment for the presence of AHLs and screening individual members of the normal flora for AHL production.
ACKNOWLEDGMENTS
We thank many researchers for invaluable suggestions, strains, and/or critical reading of the manuscript. These include Shougang Jin, Karen Visick, Valley Stewart, Dietz Bauer, Steve Atkinson, Paul Williams, Eb Pesci, Brian Hanzelka, Linda Kenney, and Tina Henkin. B.M.M.A. thanks Chris Contag and the members of his laboratory, especially Nick Olomu, Michael Bachmann, and Blythe Bartos, for allowing him to use their laboratory and instrumentation during the preliminary stages of this work.
This work was supported by startup funds and an institutional seed grant from The Ohio State University and an Institutional Research Grant (IRG-98-278-01) from the American Cancer Society (to B.M.M.A.), NIH grant R01AI22933 (to F.H.), and United Kingdom Medical Research Council Programme Grant G9632750 and Cooperative Group Grant G9800920 (to S.S.).
REFERENCES
- 1.Ahmer B M M, van Reeuwijk J, Timmers C D, Valentine P J, Heffron F. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J Bacteriol. 1998;180:1185–1193. doi: 10.1128/jb.180.5.1185-1193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler B L, Wright M, Showalter R E, Silverman M R. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 3.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Bliska J B, Falkow S. Bacterial resistance to complement killing mediated by the Ail protein of Yersinia enterocolitica. Proc Natl Acad Sci USA. 1992;89:3561–3565. doi: 10.1073/pnas.89.8.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chhabra S R, Stead P, Bainton N J, Salmond G P, Stewart G S, Williams P, Bycroft B W. Autoregulation of carbapenem biosynthesis in Erwinia carotovora by analogues of N-(3-oxohexanoyl)-l-homoserine lactone. J Antibiot. 1993;46:441–454. doi: 10.7164/antibiotics.46.441. [DOI] [PubMed] [Google Scholar]
- 6.Chilton M D, Currier T C, Farrand S K, Bendich A J, Gordon M P, Nester E W. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci USA. 1974;71:3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cirillo D M, Heffernan E J, Wu L, Harwood J, Fierer J, Guiney D G. Identification of a domain in Rck, a product of the Salmonella typhimurium virulence plasmid, required for both serum resistance and cell invasion. Infect Immun. 1996;64:2019–2023. doi: 10.1128/iai.64.6.2019-2023.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crago A M, Koronakis V. Binding of extracellular matrix laminin to Escherichia coli expressing the Salmonella outer membrane proteins Rck and PagC. FEMS Microbiol Lett. 1999;176:495–501. doi: 10.1111/j.1574-6968.1999.tb13703.x. [DOI] [PubMed] [Google Scholar]
- 9.Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: ASM Press; 1999. [Google Scholar]
- 10.Eberhard A. Inhibition and activation of bacterial luciferase synthesis. J Bacteriol. 1972;109:1101–1105. doi: 10.1128/jb.109.3.1101-1105.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eberhard A, Burlingame A L, Eberhard C, Kenyon G L, Nealson K H, Oppenheimer N J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 12.Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich M J, Kinsey N E, Vila J, Kadner R J. Nucleotide sequence of a 13.9-kb segment of the 90-kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 14.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 15.Fuqua W C, Winans S C. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia L J, Shang L H, Rothfield L I. An extracellular factor regulates expression of sdiA, a transcriptional activator of cell division genes in Escherichia coli. J Bacteriol. 1996;178:2742–2748. doi: 10.1128/jb.178.10.2742-2748.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilson L, Kuo A, Dunlap P V. AinS and a new family of autoinducer synthesis proteins. J Bacteriol. 1995;177:6946–6951. doi: 10.1128/jb.177.23.6946-6951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodier R I, Ahmer B M M. SirA orthologs affect both motility and virulence. J Bacteriol. 2001;183:2249–2258. doi: 10.1128/JB.183.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hackett J, Wyk P, Reeves P, Mathan V. Mediation of serum resistance in Salmonella typhimurium by an 11-kilodalton polypeptide encoded by the cryptic plasmid. J Infect Dis. 1987;155:540–549. doi: 10.1093/infdis/155.3.540. [DOI] [PubMed] [Google Scholar]
- 21.Hanzelka B L, Greenberg E P. Quorum sensing in Vibrio fischeri: evidence that S-adenosylmethionine is the amino acid substrate for autoinducer synthesis. J Bacteriol. 1996;178:5291–5294. doi: 10.1128/jb.178.17.5291-5294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanzelka B L, Parsek M R, Val D L, Dunlap P V, Cronan J E, Greenberg E P. Acylhomoserine lactone synthase activity of the Vibrio fischeri AinS protein. J Bacteriol. 1999;181:5766–5770. doi: 10.1128/jb.181.18.5766-5770.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heffernan E J, Reed S, Hackett J, Fierer J, Roudier C, Guiney D. Mechanism of resistance to complement-mediated killing of bacteria encoded by the Salmonella typhimurium virulence plasmid gene rck. J Clin Investig. 1992;90:953–964. doi: 10.1172/JCI115972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heffernan E J, Wu L, Louie J, Okamoto S, Fierer J, Guiney D G. Specificity of the complement resistance and cell association phenotypes encoded by the outer membrane protein genes rck from Salmonella typhimurium and ail from Yersinia enterocolitica. Infect Immun. 1994;62:5183–5186. doi: 10.1128/iai.62.11.5183-5186.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hooper L V, Bry L, Falk P G, Gordon J I. Host-microbial symbiosis in the mammalian intestine: exploring an internal ecosystem. Bioessays. 1998;20:336–343. doi: 10.1002/(SICI)1521-1878(199804)20:4<336::AID-BIES10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Jiang Y, Camara M, Chhabra S R, Hardie K R, Bycroft B W, Lazdunski A, Salmond G P, Stewart G S, Williams P. In vitro biosynthesis of the Pseudomonas aeruginosa quorum-sensing signal molecule N-butanoyl-l-homoserine lactone. Mol Microbiol. 1998;28:193–203. doi: 10.1046/j.1365-2958.1998.00789.x. [DOI] [PubMed] [Google Scholar]
- 27.Johnston C, Pegues D A, Hueck C J, Lee A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 28.Jones S, Yu B, Bainton N J, Birdsall M, Bycroft B W, Chhabra S R, Cox A J, Golby P, Reeves P J, Stephens S, et al. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 1993;12:2477–2482. doi: 10.1002/j.1460-2075.1993.tb05902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kahn D, Ditta G. Modular structure of FixJ: homology of the transcriptional activator domain with the −35 binding domain of sigma factors. Mol Microbiol. 1991;5:987–997. doi: 10.1111/j.1365-2958.1991.tb00774.x. [DOI] [PubMed] [Google Scholar]
- 30.Kanamaru K, Kanamaru K, Tatsuno I, Tobe T, Sasakawa C. SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 2000;38:805–816. doi: 10.1046/j.1365-2958.2000.02171.x. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan H B, Greenberg E P. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol. 1985;163:1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laue B E, Jiang Y, Chhabra S R, Jacob S, Stewart G S, Hardman A, Downie J A, O'Gara F, Williams P. The biocontrol strain Pseudomonas fluorescens F113 produces the Rhizobium small bacteriocin, N-(3-hydroxy-7-cis-tetradecenoyl)homoserine lactone, via HdtS, a putative novel N-acylhomoserine lactone synthase. Microbiology. 2000;146:2469–2480. doi: 10.1099/00221287-146-10-2469. [DOI] [PubMed] [Google Scholar]
- 33.McClean K H, Winson M K, Fish L, Taylor A, Chhabra S R, Camara M, Daykin M, Lamb J H, Swift S, Bycroft B W, Stewart G S, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 34.More M I, Finger L D, Stryker J L, Fuqua C, Eberhard A, Winans S C. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 35.Nealson K H, Platt T, Hastings J W. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsek M R, Val D L, Hanzelka B L, Cronan J E, Jr, Greenberg E P. Acyl homoserine-lactone quorum-sensing signal generation. Proc Natl Acad Sci USA. 1999;96:4360–4365. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson J P, Passador L, Iglewski B H, Greenberg E P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierson L S, Wood D W, von Bodman S B. Quorum sensing in plant-associated bacteria. In: Dunny G M, Winans S, editors. Cell-cell signaling in bacteria. Washington, D.C.: ASM Press; 1999. pp. 101–115. [Google Scholar]
- 40.Qin Y, Luo Z Q, Smyth A J, Gao P, von Bodman S B, Farrand S K. Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm. EMBO J. 2000;19:5212–5221. doi: 10.1093/emboj/19.19.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaefer A L, Val D L, Hanzelka B L, Cronan J E, Greenberg E P. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci USA. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw P D, Ping G, Daly S L, Cha C, Cronan J E, Jr, Rinehart K L, Farrand S K. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sitnikov D M, Schineller J B, Baldwin T O. Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc Natl Acad Sci USA. 1996;93:336–341. doi: 10.1073/pnas.93.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens A M, Greenberg E P. Transcriptional activation by LuxR. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: ASM Press; 1999. pp. 231–242. [Google Scholar]
- 45.Surette M G, Bassler B L. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Surette M G, Bassler B L. Regulation of autoinducer production in Salmonella typhimurium. Mol Microbiol. 1999;31:585–595. doi: 10.1046/j.1365-2958.1999.01199.x. [DOI] [PubMed] [Google Scholar]
- 47.Swift S, Karlyshev A V, Fish L, Durant E L, Winson M K, Chhabra S R, Williams P, Macintyre S, Stewart G S. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J Bacteriol. 1997;179:5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swift S, Williams P, Stewart G S A B. N-Acylhomoserine lactones and quorum sensing in proteobacteria. In: Dunny G M, Winans S, editors. Cell-cell signaling in bacteria. Washington, D.C.: ASM Press; 1999. pp. 291–313. [Google Scholar]
- 49.Swift S, Winson M K, Chan P F, Bainton N J, Birdsall M, Reeves P J, Rees C E, Chhabra S R, Hill P J, Throup J P, et al. A novel strategy for the isolation of luxI homologues: evidence for the widespread distribution of a LuxR:LuxI superfamily in enteric bacteria. Mol Microbiol. 1993;10:511–520. doi: 10.1111/j.1365-2958.1993.tb00923.x. [DOI] [PubMed] [Google Scholar]
- 50.Throup J P, Camara M, Briggs G S, Winson M K, Chhabra S R, Bycroft B W, Williams P, Stewart G S. Characterisation of the yenI/yenR locus from Yersinia enterocolitica mediating the synthesis of two N-acylhomoserine lactone signal molecules. Mol Microbiol. 1995;17:345–356. doi: 10.1111/j.1365-2958.1995.mmi_17020345.x. [DOI] [PubMed] [Google Scholar]
- 51.Val D L, Cronan J E., Jr In vivo evidence that S-adenosylmethionine and fatty acid synthesis intermediates are the substrates for the LuxI family of autoinducer synthases. J Bacteriol. 1998;180:2644–2651. doi: 10.1128/jb.180.10.2644-2651.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 53.Wang X D, de Boer P A J, Rothfield L I. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 1991;10:3363–3372. doi: 10.1002/j.1460-2075.1991.tb04900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei Y, Lee J M, Smulski D R, LaRossa R A. Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:2265–2272. doi: 10.1128/JB.183.7.2265-2272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welch M, Todd D E, Whitehead N A, McGowan S J, Bycroft B W, Salmond G P. N-Acyl homoserine lactone binding to the CarR receptor determines quorum-sensing specificity in Erwinia. EMBO J. 2000;19:631–641. doi: 10.1093/emboj/19.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winson M K, Camara M, Latifi A, Foglino M, Chhabra S R, Daykin M, Bally M, Chapon V, Salmond G P, Bycroft B W, et al. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winson M K, Swift S, Fish L, Throup J P, Jorgensen F, Chhabra S R, Bycroft B W, Williams P, Stewart G S. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol Lett. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- 58.Wood D W, Gong F, Daykin M M, Williams P, Pierson L S., III N-Acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. J Bacteriol. 1997;179:7663–7670. doi: 10.1128/jb.179.24.7663-7670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang L, Murphy P J, Kerr A, Tate M E. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature. 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]
- 60.Zhu J, Winans S C. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc Natl Acad Sci USA. 2001;98:1507–1512. doi: 10.1073/pnas.98.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]