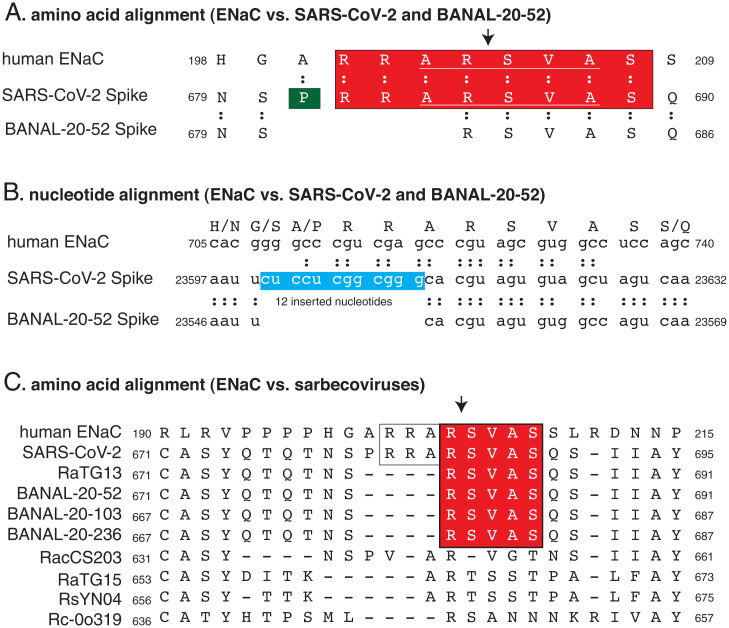
Harrison and Sachs (1) make a serious accusation against scientists at the University of North Carolina (UNC) and the Wuhan Institute of Virology (WIV) based on an eight-amino-acid sequence similarity between the furin cleavage site (FCS) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike and one of the FCSs of human amiloride-sensitive epithelial sodium channel α subunit (ENaC) (2). Both proteins have the sequence RRARSVAS (Fig. 1A). Harrison and Sachs cite work on rat ENaC from UNC (3, 4) and suggest that the UNC and WIV coronavirologists may have mimicked human ENaC FCS to make SARS-CoV-2 more infectious for lung epithelia.
Fig. 1.
Alignment of the human ENaC with sarbecovirus Spikes. (A) Amino acid alignment of ENaC with Spike proteins of SARS-CoV-2 and BANAL-20-52. (B) Nucleotide alignment of the gene for ENaC with SARS-CoV-2 and BANAL-20-52 Spike genes. One of two possible out-of-frame insertions is shown. (C) Amino acid alignment of ENaC with the S1/S2 junction of selected sarbecovirus spikes. Arrows denote the sites of cleavage in the proteins.
Numerous features of SARS-CoV-2 FCS demonstrate that it was not engineered to mimic human ENaC:
-
•
Alignment of the nucleotide sequence of the SARS-CoV-2 Spike gene with the closest known coronavirus Spike gene from Laotian bat coronavirus BANAL-20-52 (5) clearly shows that four extra amino acids (PRRA), not eight, were added to the SARS-CoV-2 Spike protein (Fig. 1B).
-
•
There was an insertion of 12 nucleotides into the Spike gene (Fig. 1B, box) (6). This nucleotide insertion is out of frame (6, 7).
-
•
The insertion adds a proline not present in ENaC.
-
•
Except for one codon (cgu that encodes arginine 685), each of the codons for RRARSVAS is different in human ENaC and SARS-CoV-2 (Fig. 1B).
-
•
Five of eight amino acids (RSVAS; underlined in Fig. 1A, red box in Fig. 1C) in or near the ENaC FCS sequence shared with SARS-Cov-2 Spike are present in Spikes of sarbecoviruses, such as BANAL-20-52. It would be illogical to use the FCS from ENac rather than from a FCS of another coronavirus.
Harrison and Sachs’s (1) claim that alignment of sarbecovirus Spike amino acid sequences illustrates“the unusual nature of the [SARS-CoV-2] FCS” is misleading. FCSs are common in coronaviruses, and present in representatives of four out of five betacoronavirus subgenuses (8). The highly variable nature of the S1/S2 junction is easily ascertained by inspecting a precise alignment of sarbecovirus Spikes (Fig. 1C).
After commenting about the “unusual nature” of the SARS-CoV-2 FCS, Harrison and Sachs (1) then argue the opposite. With regard to our earlier publication (7), they write, “In fact, the assertion that the FCS in SARS-CoV-2 has an unusual, nonstandard amino acid sequence is false.” We made no such assertion. Rather, we noted that the SARS-CoV-2 FCS is “suboptimal.” We also noted, correctly, that placing the insertion out of frame would be “an unusual and needlessly complex feat of genetic engineering.”
The immediate proximal ancestor of SARS-CoV-2 did not come directly from a bat to a human, but first evolved in an intermediate host. Two related lineages of SARS-CoV-2—lineage A and lineage B—first infected humans via the wildlife trade at the Huanan Market in Wuhan (9, 10). For the ENaC hypothesis to be true, UNC or WIV researchers would have had to possess the direct SARS-CoV-2 progenitor isolated from another animal—not a bat.
Harrison and Sachs (1) allege that scientists at NIH and elsewhere, including myself and colleagues, conspired to suppress theories of a laboratory origin of SARS-CoV-2. This is false. A possible laboratory origin of SARS-CoV-2 was discussed in our earlier publications (6, 7).
Footnotes
Competing interest statement: R.F.G. is the cofounder of Zalgen Labs, a biotechnology company developing countermeasures to emerging viruses.
References
- 1.Harrison N. L., Sachs J. D., A call for an independent inquiry into the origin of the SARS-CoV-2 virus. Proc. Natl. Acad. Sci. U.S.A. 119, e2202769119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand P., Puranik A., Aravamudan M., Venkatakrishnan A. J., Soundararajan V., SARS-CoV-2 strategically mimics proteolytic activation of human ENaC. eLife 9, e58603 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.García-Caballero A., Dang Y., He H., Stutts M. J., ENaC proteolytic regulation by channel-activating protease 2. J. Gen. Physiol. 132, 521–535 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kota P., Gentzsch M., Dang Y. L., Boucher R. C., Stutts M. J., The N terminus of α-ENaC mediates ENaC cleavage and activation by furin. J. Gen. Physiol. 150, 1179–1187 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Temmam S., et al. , Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature 604, 330–336 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Andersen K. G., Rambaut A., Lipkin W. I., Holmes E. C., Garry R. F., The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes E. C., et al. , The origins of SARS-CoV-2: A critical review. Cell 184, 4848–4856 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y., Zhao S., Furin cleavage sites naturally occur in coronaviruses. Stem Cell Res. 50, 102115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pekar J. E., et al. , The molecular epidemiology of multiple zoonotic origins of SARS-CoV-2. Science 377, 960–966 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worobey M., et al. , The Huanan Seafood Wholesale Market in Wuhan was the early epicenter of the COVID-19 pandemic. Science 377, 951–959 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]