Significance
Multiple sclerosis is a disease of the central nervous system with an autoimmune and neurodegenerative component. While numerous drugs are available to ameliorate autoimmunity, just some drugs so far have demonstrated effects suggestive of neuroprotection. In this study, we demonstrate that the sphingosine-1 phosphate receptor modulator siponimod exerts protective effects, which are independent of lymphocytes and regulated by the sphingosine-1 phosphate receptor 5 subtypes.
Keywords: multiple sclerosis, siponimod, cuprizone, oligodendrocyte, S1PR5
Abstract
Multiple sclerosis (MS), an autoimmune-driven, inflammatory demyelinating disease of the central nervous system (CNS), causes irreversible accumulation of neurological deficits to a variable extent. Although there are potent disease-modifying agents for its initial relapsing–remitting phase, immunosuppressive therapies show limited efficacy in secondary progressive MS (SPMS). Although modulation of sphingosine-1 phosphate receptors has proven beneficial during SPMS, the underlying mechanisms are poorly understood. In this project, we followed the hypothesis that siponimod, a sphingosine-1 phosphate receptor modulator, exerts protective effects by direct modulation of glia cell function (i.e., either astrocytes, microglia, or oligodendrocytes). To this end, we used the toxin-mediated, nonautoimmune MS animal model of cuprizone (Cup) intoxication. On the histological level, siponimod ameliorated cuprizone-induced oligodendrocyte degeneration, demyelination, and axonal injury. Protective effects were evident as well using GE180 translocator protein 18-kDa (TSPO) imaging with positron emission tomography (PET)/computed tomography (CT) imaging or next generation sequencing (NGS). Siponimod also ameliorated the cuprizone-induced pathologies in Rag1-deficient mice, demonstrating that the protection is independent of T and B cell modulation. Proinflammatory responses in primary mixed astrocytes/microglia cell cultures were not modulated by siponimod, suggesting that other cell types than microglia and astrocytes are targeted. Of note, siponimod completely lost its protective effects in S1pr5-deficient mice, suggesting direct protection of degenerating oligodendrocytes. Our study demonstrates that siponimod exerts protective effects in the brain in a S1PR5-dependent manner. This finding is not just relevant in the context of MS but in other neuropathologies as well, characterized by a degeneration of the axon–myelin unit.
Multiple sclerosis (MS), an inflammatory disease of the central nervous system (CNS), initially presents with a relapsing–remitting disease course. During this early stage of the disease, leukocytes cross the blood–brain barrier to drive the formation of focal demyelinating plaques. Disease-modifying agents that modulate or suppress the peripheral immune system or the recruitment of peripheral immune cells into the CNS provide significant therapeutic benefits during relapsing–remitting MS (RRMS). The majority of individuals with RRMS ultimately enter a secondary progressive disease stage, clinically characterized by the progressive accumulation of neurological deficits, a process that is essentially independent of inflammatory-driven relapses. The cellular and molecular basis for this transition and the ongoing disease progression is unclear, and the role of inflammation during the secondary progressive disease stage is a subject of intense and controversial debate. Although the extent of axonal damage correlates positively with the density of intracerebral lymphocytes (1), and histological correlates of lymphoid tissue with a high B cell density are found in the meninges of deceased progressive MS patients (2–4), it is entirely unclear why classical immunosuppressive drugs, such as natalizumab, lose their effectiveness in the progressive stage of the disease (5).
For patients with progressive MS, either secondary progressive MS (SPMS) or primary progressive MS (PPMS), there are only a few approved treatment options available. In 2020, siponimod (trade name Mayzent) received European Union approval for the treatment of adults with SPMS with disease activity demonstrated by clinical relapses or imaging of inflammatory activity. The sphingosine-1 phosphate receptor modulator (S1P modulator) siponimod selectively binds to two of the five G protein–coupled receptors for S1P: the S1P receptor 1 (S1PR1) and the S1P receptor 5 (S1PR5) (6). Functionally, siponimod prevents the egress of lymphocytes from the lymph nodes and, thus, reduces the recirculation of T cells into the CNS, resulting in an amelioration of the inflammatory attacks driven by the peripheral immune systems (7). In addition, S1PR1 and S1PR5 are expressed by cells of the CNS, as demonstrated by several groups (8–11). On a cellular level, S1PR1 is predominantly expressed by astrocytes and microglia, whereas cells of the oligodendrocytic lineage are the major cell type in the CNS expressing S1PR5. So far, it is unknown to what extent the reported beneficial effects of siponimod in SPMS patients are mediated by direct modulation of glial cells.
In this project, we followed the hypothesis that siponimod exerts protective effects by direct modulation of glia cell function (i.e., either astrocytes, microglia, or oligodendrocytes). To this end, we used the cuprizone (Cup) model of demyelination, which is a toxin-mediated MS animal model independent of autoimmune processes (12–14).
Results
Siponimod Exerts Long-Lasting Immunosuppressive Effects.
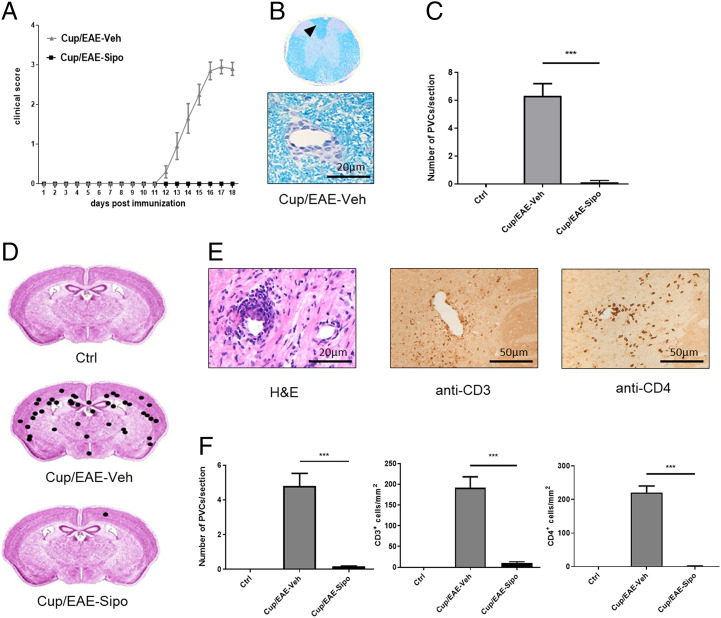
In an initial step, we aimed to verify the reported antiinflammatory effects of siponimod (15, 16) and at the same time investigated potential prolonged immunosuppressive potencies. To this end, Cup/experimental autoimmune encephalomyelitis (EAE) was induced in C57BL/6 wild-type mice (17) and the animals were treated with either vehicle (Veh) or siponimod (Sip) solution during the initial 3-wk cuprizone intoxication period (i.e., groups B and C in Fig. 1). Myelin oligodendrocyte glycoprotein (MOG)-EAE was induced at the end of week 5. Ten out of 10 vehicle-treated mice developed tail and partial hindlimb paralysis, whereas no motor-behavioral deficits were observed in the eight siponimod-treated mice (Fig. 2A). In line with this finding, Luxol fast blue-periodic acid Schiff (LFB/PAS) stains of spinal cord tissues demonstrated multifocal inflammatory demyelination exclusively in vehicle- but not siponimod-treated mice (Fig. 2 B and C). Beyond, multifocal perivascular cuffs (PVCs) and high CD3+ and CD4+ cell numbers were found in the forebrain of vehicle-treated, but not siponimod-treated mice (Fig. 2 D–F).
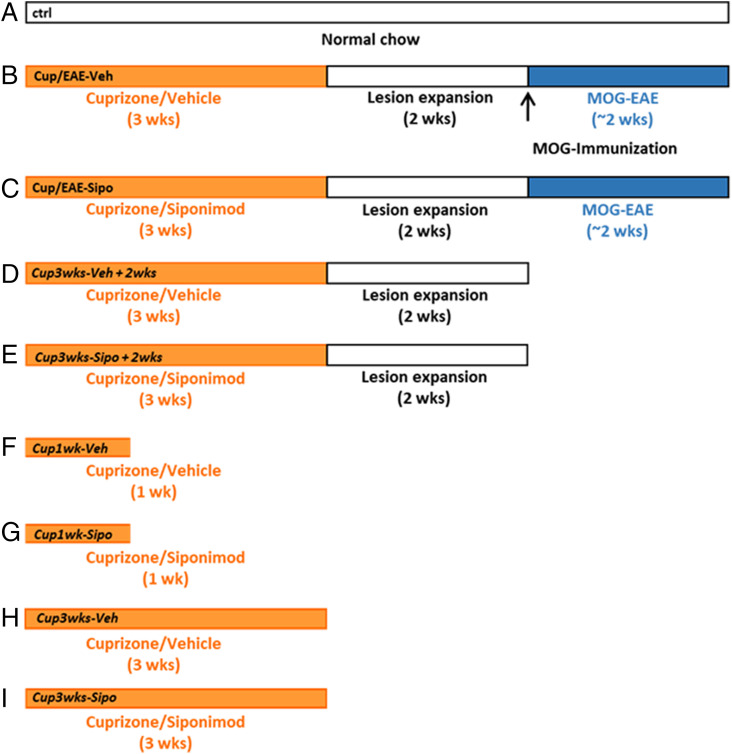
Fig. 1.
Schematic depicting the experimental setup. A (control); B (Cup/EAE-Veh); C (Cup/EAE-Sipo); D (Cup3wks-Veh+2wks); E (Cup3wks-Sipo+2wks); F (Cup1wk-Veh); G (Cup1wk-Sipo); H (Cup3wks-Veh); I (Cup3wks-Sipo).
Fig. 2.
Siponimod ameliorates the disease course and inflammatory demyelination in the Cup/EAE model. (A) Clinical scores for Cup/EAE-vehicle (gray triangles; n = 10) and Cup/EAE-siponimod (black squares; n = 8) groups. (B) Representative LFB/PAS stained section of the spinal cord. Arrowhead indicates an inflammatory PVC shown beneath in higher magnification. (C) Quantification of PVCs in spinal cord sections in control (n = 3), Cup/EAE-vehicle (n = 9) and Cup/EAE-siponimod (n = 8) mice. Differences were determined using one-way ANOVA followed by Tukey's multiple comparisons test. (D) Cumulative spatial distribution of PVCs in the forebrain of control (n = 3), Cup/EAE-vehicle (n = 10), and Cup/EAE-siponimod (n = 8) groups. (E) Histological presentation of a representative forebrain PVC, visualized by H&E stain, anti-CD3, and anti-CD4 immunohistochemistry. (F) Numbers of PVCs in the entire brain as well as CD3+ and CD4+ lymphocyte densities in the corpus callosum (CC). Differences were determined using one-way ANOVA followed by Tukey's multiple comparisons test. Data are shown as mean ± SEM. ***P < .001.
Siponimod Ameliorates Toxin-Induced Demyelination.
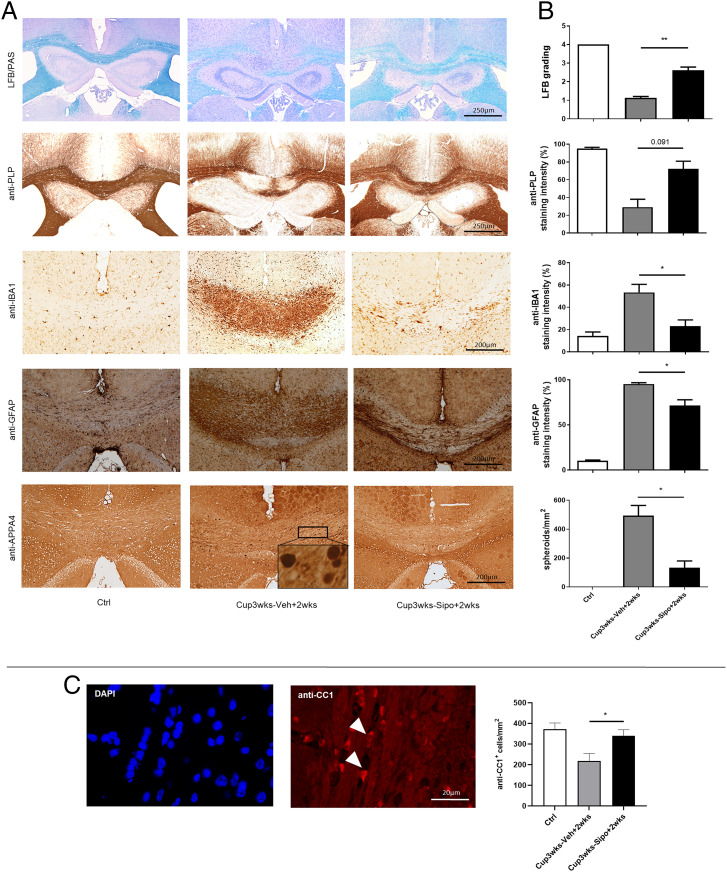
While these observed antiinflammatory effects of siponimod have been expected based on its modulatory function on S1PR1 receptors (6) and its antiinflammatory activity in clinical trials (18), beneficial effects by direct modulation of brain cell function are not well understood. To investigate this, we used the cuprizone model in which metabolic oligodendrocyte stress results in oligodendrocyte degeneration, demyelination, gliosis, and acute axonal injury (19–21). Mice were intoxicated with cuprizone for 3 wk and treated with either vehicle or siponimod during the cuprizone intoxication period. The brains were analyzed at the end of week 5 after autonomous lesion progression (i.e., groups D and E in Fig. 1). As shown in Fig. 3A, vehicle-treated mice demonstrated severe demyelination in the midline of the corpus callosum (CC) (i.e., loss of LFB and anti-proteolipid protein [PLP] staining intensities), paralleled by pronounced activation of anti-IBA1+ microglia and anti-GFAP+ astrocytes. Furthermore, anti-APP A4 stains revealed numerous stained spheroids, indicative of acute axonal injury. Signs of demyelination, glia activation, and acute axonal injury were less severe in siponimod-treated mice (Fig. 3 A and B). Furthermore, we processed adjacent sections for anti-CC1 immunofluorescence double stains to be able to identify mature oligodendrocytes. Significantly higher anti-CC1+ mature oligodendrocyte numbers were found in siponimod-treated compared to vehicle-treated mice (Fig. 3C, vehicle 218.7 ± 35.66 cells/mm2 versus siponimod 340.1 ± 29.22 cells/mm2).
Fig. 3.
Siponimod ameliorates cuprizone-induced pathologies. (A) Representative LFB/PAS, anti-PLP, anti-IBA1, anti-GFAP and anti-APP A4 stains of the medial CC from control (n = 3), Cup3wks-Veh+2wks (n = 8) and Cup3wks-Sipo+2wks (n = 9) treated cuprizone-intoxicated mice. (B) Evaluation of the extent of demyelination (LFB/PAS and anti-PLP), microgliosis (anti-IBA1), astrocytosis (anti-GFAP) and acute axonal injury (anti-APP A4). Statistical analyzes were performed by Kruskal–Wallis test followed by Dunn’s multiple comparison test (C) Representative anti-CC1 immunofluorescence staining of the medial CC. Arrowheads indicate the localization of anti-CC1+ cells. (D) Quantification of CC1+ cell numbers. Statistical analyzes were performed by Kruskal–Wallis test followed by Dunn’s multiple comparison test. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01.
To verify these results in a second independent experiment, and to evaluate the dose dependency of siponimod treatment, separate cohorts of mice were treated daily with 0.315 mg/kg, 3.125 mg/kg, or 15.5 mg/kg siponimod solution during the 3-wk cuprizone intoxication period, and the brains were histologically evaluated. As demonstrated in SI Appendix, Fig. S1, the protective effect clearly followed a dose-dependent manner, with moderate protective effects in the 0.315 mg/kg and the most pronounced protective effects in the 15.5 mg/kg treatment group, respectively. In the 0.315 mg/kg treatment group, we found a loss of LFB and anti-PLP staining intensities, which is indicative of demyelination. Demyelination in low-dose–treated mice was paralleled by microglia activation (anti-IBA1) and acute axonal injury (anti-APP A4 and anti-vGLUT1). Such pathologies were less severe in the 3.125 mg/kg-treated and almost absent in the 15.5 mg/kg-treated mice.
Protective Siponimod Effects Can Be Visualized by PET/CT Imaging and Is Evident in Next Generation Sequencing Experiments.
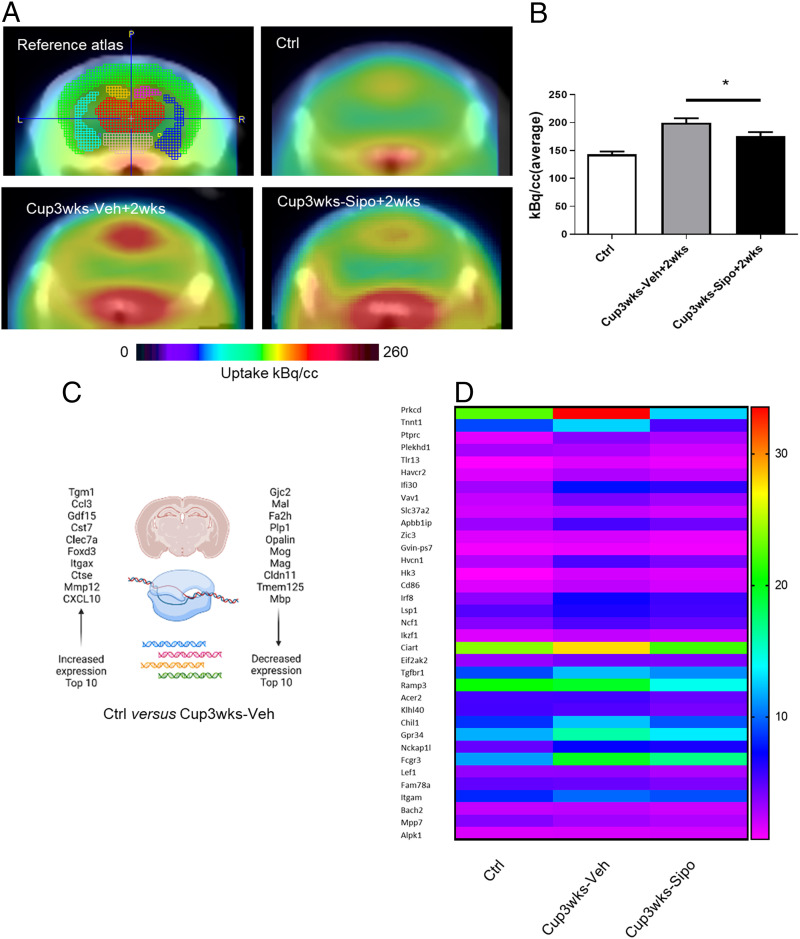
These first results suggest that siponimod can ameliorate metabolic oligodendrocyte injury in the cuprizone mouse model. Positron emission tomography (PET) is a sensitive method for visualizing the activation of microglia and astrocytes in living animals (22). To assess whether the protective effects of siponimod can be visualized by PET/computed tomography (CT) imaging, mice (i.e., groups D and E in Fig. 1) were injected with [18F]-GE180 and the binding/accumulation of the radioligand was analyzed. In line with previous results (22), we found a region-dependent increase of [18F]-GE180 uptake in cuprizone-treated mice (Fig. 4A). A visual interpretation of the translocator protein (TSPO) μPET scans indicated increased tracer uptake, particularly in the midline of the white matter tract corpus callosum. Statistical comparison between the vehicle- and siponimod-treated groups showed significant higher tracer uptake in vehicle (199.5 ± 8.2 kBq; mean ± SEM) compared to siponimod-treated mice (176 ± 6.9 kBq; P = 0.037; Fig. 4B). These results are well in line with the reduced gliosis observed in siponimod-treated cuprizone mice.
Fig. 4.
Protective effects of siponimod are evident by [18F]F-GE180 PET/CT measurements and next generation sequencing studies. (A) Schematic illustration of the evaluated areas and cumulative heat map illustrating radioligand uptake in control (n = 6), Cup3wks-Veh+2wks (n = 4) and Cup3wks-Sipo+2wks (n = 4) mice. The color codes demonstrate the standardized radioligand uptake levels relative to the control group. (B) Quantification of radioligand uptake. Statistical analyzes were performed by Mann–Whitney test as indicated. (C) Top 10 up-regulated and down-regulated genes in Ctrl versus Cup3wks-Veh mice. (D) Heat-map illustrating expression levels of the top 35 genes with lower expression in Cup3wks-Sipo versus Cup3wks-Veh mice. Control groups (Ctrl) are shown in addition. n = 5 animals per experimental group. Data are shown as mean ± SEM. *P < 0.05.
Next, we performed next generation sequencing (NGS) experiments with mixed white/gray matter tissues isolated from control, Cup3wks-Veh–, and Cup3wks-Sipo– treated mice (i.e., groups H and I in Fig. 1). A total of 1,418 genes were found to be regulated in cuprizone-vehicle versus control mice (821 up-regulated and 597 down-regulated). Among those genes that were found to be most profoundly up-regulated (Dataset S1, marked in red and Fig. 4C) were classical proinflammatory cytokines such as Ccl3 (5.3-log2-fold up-regulated) or Cxcl10 (3.8-log2-fold up-regulated). These chemokines are well known to promote neuroinflammatory responses. Beyond, microglia-related genes such as Cst7 (5.3-log2-fold up-regulated) or Itgax (5.0-log2-fold up-regulated) were among those genes most profoundly induced. On the other hand, among those genes that were found to be most profoundly down-regulated (Dataset S1, marked in green and Fig. 4C) were found oligodendrocyte-enriched transcripts among Fa2h (−2.8-log2-fold down-regulated), Plp1 (−2.7-log2-fold down-regulated) or Mog (−2.7-log2-fold down-regulated). Interestingly, the expression of S1pr5, which is well known to be expressed by mature oligodendrocytes, was also found to be significantly reduced (−2.1-log2-fold down-regulated). This finding demonstrates the validity of our NGS experiment.
Next, we were interested in differentially expressed genes in vehicle- versus siponimod-treated groups. We found that 87 genes showed lower expression (Dataset S2, marked in green) whereas 192 genes showed higher expression (Dataset S2, marked in red) in siponimod-treated compared to vehicle-treated mice. Although not applicable for all of the lower expressed genes in siponimod-treated versus vehicle-treated mice, many of those genes were low expressed in control mice, high expressed in vehicle-treated, and low/intermediate expressed in siponimod-treated mice (see heat map in Fig. 4D). For example protein kinase c delta (Prkcd), Transforming Growth Factor Beta-Receptor I (Tgfbr1), or Integrin α-M (Itgam), which are known to be expressed by activated microglia cells followed this pattern. When we applied the PANTHER overrepresentation test (23) for the 87 genes that were lower expressed in siponimod- versus vehicle-treated mice, the gene ontology (GO) biological process terms lymphocyte activation (6.2-fold enrichment), negative regulation of multicellular organismal process (3.7-fold enrichment), and positive regulation of metabolic process (2.4-fold enrichment) were found to be overrepresented. No overrepresentations were found for the GO “molecular function” or “cellular component” categories. When we performed the same analysis with the 192 genes that were higher expressed in siponimod-treated compared to vehicle-treated mice, the GO biological process terms “regulation of neurotransmitter receptor activity” (11.9 fold enrichment), “synaptic vesicle cycle” (8.3-fold enrichment), and “regulation of synapse organization” (5.7-fold enrichment) were found to be overrepresented. For the GO cellular component the terms “smooth endoplasmic reticulum” (16.6-fold enrichment), “dendritic shaft” (11.9-fold enrichment), and “excitatory synapse” (10.9-fold enrichment) were found to be overrepresented. No overrepresentations were found for the GO molecular function category. In summary, siponimod impacts cuprizone-induced demyelinating effects on the transcriptome level as well.
Protective Siponimod Effects Are Independent of T Cells.
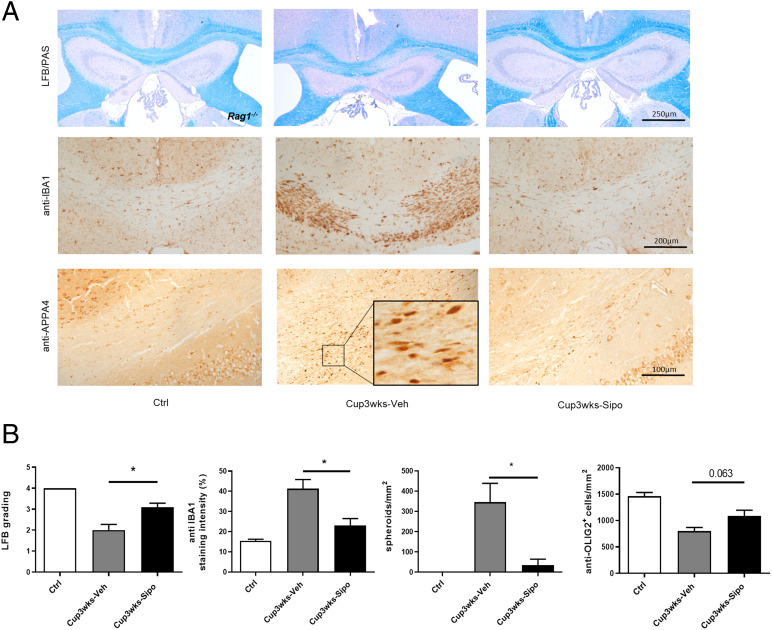
Although the results of functional studies, using Rag1−/− mice with no functional B and T cells, suggest that adaptive immunity is not mediating the cuprizone-induced pathology (14, 24), our group recently demonstrated that lymphocytes, in particular CD8+ cytotoxic T cells, populate the demyelinated corpus callosum in the cuprizone model, while displaying an activated phenotype (25). Of note, densities of the recruited T cells and the composition of lymphocytic infiltrates in cuprizone-intoxicated mice were found to be comparable to those found in progressive MS lesions. Comparably, our NGS-based enrichment analysis suggests that siponimod might exert its protective effects in the cuprizone model via modulation of lymphocyte function (see above). To formally exclude that the observed protective effects of siponimod in the cuprizone model are due to a modulation of T cell function, we used Rag1−/− mice, which lack functional B and T cells (26). In these experiments, the mice were killed after a 3-wk cuprizone intoxication period (i.e., groups H and I in Fig. 1). As demonstrated in Fig. 5, vehicle-treated Rag1−/− mice displayed the typical histopathological alterations of cuprizone-intoxicated mice (i.e., demyelination, microgliosis, acute axonal injury, and loss of oligodendrocytes), whereas siponimod-treated Rag1−/− mice were almost completely protected from the cuprizone-mediated insult. Beyond, densities of CD3+ lymphocytes and CD4+ Th cells were found to be increased in vehicle- and siponimod-treated wild-type mice (SI Appendix, Fig. S2). These data strongly suggest that the protective siponimod effects are independent of T and B cells, but are mediated by direct modulation of brain cell function.
Fig. 5.
Siponimod ameliorates cuprizone-induced pathologies in Rag1−/− mice. (A) Representative LFB/PAS, anti-IBA1, and anti-APP A4 stains of the medial CC from control (n = 3), Cup3wks-Veh (n = 5) and Cup3wks-Sipo (n = 5) Rag1−/− mice. (B) Evaluation of the extent of demyelination (LFB/PAS), microgliosis (anti-IBA1), acute axonal injury (anti-APP A4) and pan-oligodendrocyte densities (anti-OLIG2). Statistical analyzes were performed by Mann–Whitney test between the Cup3wks-Veh and Cup3wks-Sipo groups. Data are shown as mean ± SEM. *P < 0.05.
Siponimod Does Not Modulate Astrocyte/Microglia Function In Vitro.
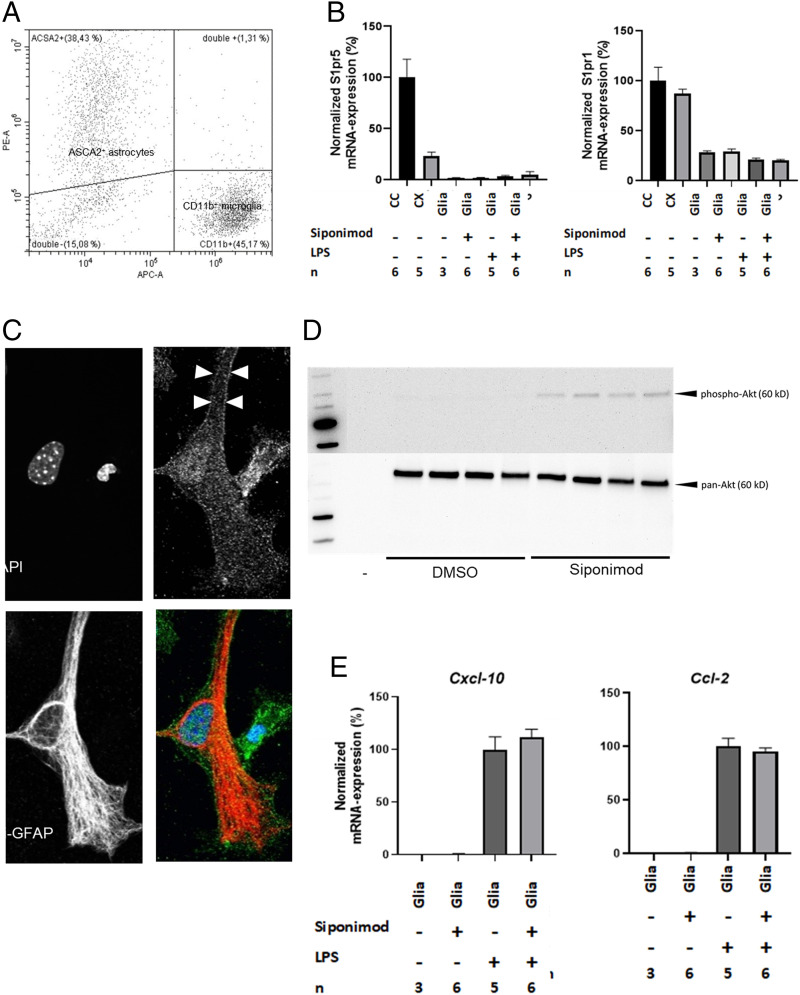
Siponimod interacts with S1PR1 and S1PR5. In the CNS, S1PR1 predominantly regulates astrocyte (27–29) and microglia (30) function, whereas S1PR5 is predominantly expressed by cells of the oligodendrocyte lineage (31). Early during cuprizone-induced demyelination, astrocytes and microglia are activated (19) and are believed to secrete a variety of proinflammatory cytokines and chemokines, thus orchestrating the progression of demyelination in this model (32). To investigate a potential modulatory function of astrocytes/microglia by siponimod, we prepared mixed glia cell cultures (CD11b+ microglia/ASCA2+ astrocyte ratio 1:1.2; see Fig. 6A) and investigated the potential of siponimod to modulate proinflammatory responses in this in vitro model. In a first step, we examined the expression of S1pr5 and S1pr1 mRNA isolated from mixed glia cell cultures and mouse brain tissues. As expected, the expression of S1pr5 was low in the isolated cortex but high in the oligodendrocyte-rich isolated white matter tract corpus callosum. The lowest S1pr5 expression levels were found in the mixed astrocyte/microglia cultures (Fig. 6 B, Left). In contrast, S1pr1 expression was evident in isolated brain tissues and the mixed glial cell cultures (Fig. 6 B, Right). The mean Ct values for S1pr1 in the cortex were 24.1 ± 0.12 (mean ± SEM) and in the nontreated mixed glia cell cultures, 26.1 ± 0.23. Beyond, neither stimulation of cultured astrocytes with lipopolysaccharides (LPS) nor siponimod treatment induced or repressed S1pr5 or S1pr1 expression levels. The expression of S1PR1 protein was verified by immunocytochemistry, demonstrating a punctate expression at the cell surface of GFAP+ cells (see arrowheads in Fig. 6C). To verify that the expressed receptors are functionally active in these cells, mixed glia cell cultures were serum starved for 4 h, then treated with siponimod (1 μM) for 60 min and the samples prepared for Western blotting experiments. In line with a previous report (33), the treatment of the mixed glia cell cultures with siponimod induced AKT phosphorylation (Fig. 6D).
Fig. 6.
Effects of siponimod in mixed glia cell cultures. (A) Flow cytometry analysis of ACSA-2+ astrocytes and CD11b+ microglia in mixed glia cultures. (B) Qualitative relative expression of S1pr5 and S1pr1 in tissues isolated from control mice (corpus callosum [CC] and cortex [CX]) as well as mixed glia cell cultures exposed to LPS (100 ng/mL) and/or siponimod (1 µM). Data are normalized to the mean expression in the CC. No statistical comparison was performed. (C) Representative double stain against GFAP (red) and S1PR1 (green) in the mixed glia culture. (D) Protein levels of total AKT and phosphorylated AKT in mixed glia cultures treated with DMSO (vehicle solution) or siponimod (n = 4). (E) Qualitative relative expression of Cxcl-10 and Ccl-2 in mixed glia cell cultures. Data are shown as mean ± SEM.
Next, the mixed glia cell cultures were stimulated with either LPS, TNFα, or IFNγ, and cytokine release into the cell culture supernatant was analyzed in vehicle- or siponimod-treated experimental groups by Bio-Plex protein arrays. As expected, low cytokine levels were observed in control cultures, whereas cytokine levels increased in a stimulus-specific manner (Dataset S3). For example, IL10 levels were significantly increased in LPS-, but not TNFα- or IFNγ-exposed cultures. In contrast, all three applied stimuli increased the levels of the chemokine RANTES/CCL5. Of note, siponimod treatment did not decrease cytokines release in cultured glia cells. In contrast, the concentration of some cytokines, such as MCP-1, IL12-p40, or G-CSF, was found to be significantly higher in siponimod-exposed cultures (see # sign in Dataset S3). In line with this finding on the protein level, gene expression studies showed expression induction of the two chemokines Cxcl10 and Ccl2 by LPS treatment with no effects of siponimod on chemokine expression induction (Fig. 6E).
Siponimod Protects Oligodendrocytes in a S1PR5-Dependent Manner.
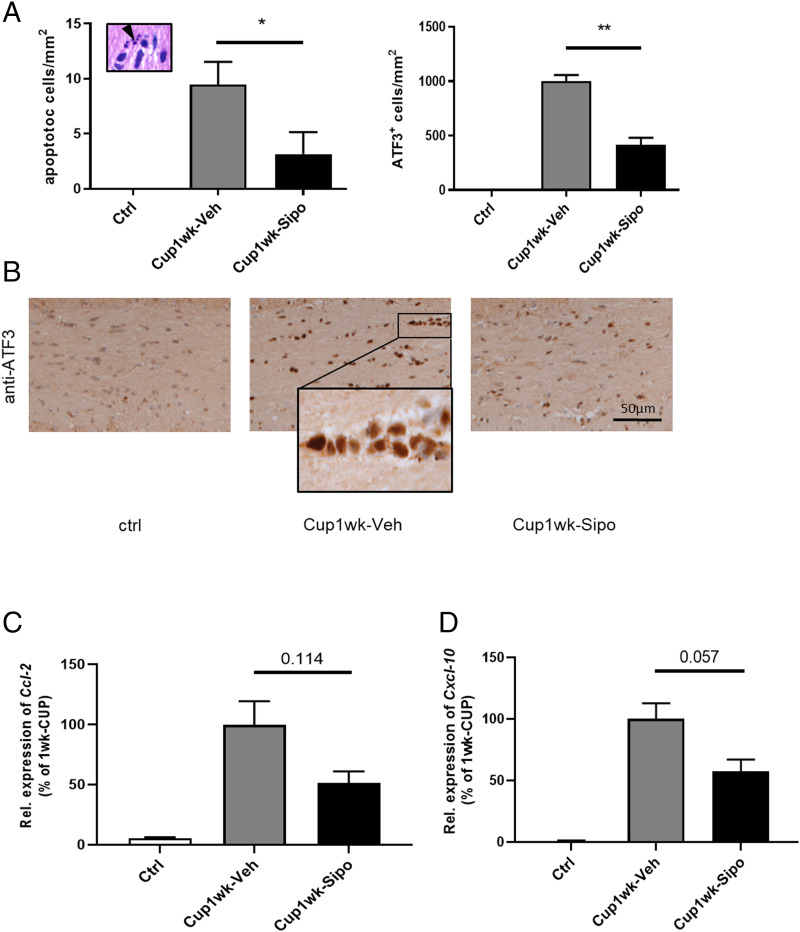
Next, we asked whether the observed protective effects of siponimod might be due to protection of oligodendrocytes. To this end, we first intoxicated mice for 1 wk with cuprizone and investigated whether or not the cuprizone-induced oligodendrocyte degeneration is ameliorated by siponimod. As demonstrated in Fig. 7, densities of apoptotic cells were by trend lower in siponimod- compared to vehicle-treated mice (vehicle 9.46 ± 2.08 versus siponimod 3.15 ± 2.00 apoptotic cells; P = 0.124). Beyond, densities of ATF3+ cells, which have been shown to mark stressed oligodendrocytes in this model (34), were significantly reduced by siponimod. This early protective effect of siponimod was paralleled by a trend of reduced expression induction of the two proinflammatory chemokines Ccl2 and Cxcl10.
Fig. 7.
Siponimod ameliorates cuprizone-induced oligodendrocyte degeneration. (A) Quantification of apoptotic cells in H&E (Left) or anti-ATF3 (Right) stained sections of control (n = 3), Cup1wk-Veh (n = 6) and Cup1wk-Sipo (n = 6) mice. Statistical analyzes were performed by Mann–Whitney test as indicated. (C and D) Relative Ccl2 and Cxcl10 mRNA expression levels in the forebrain from control, Cup1wk -vehicle (n = 4), and Cup1wk-siponimod (n = 4) mice. Statistical analyzes were performed by Mann–Whitney test as indicated. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01.
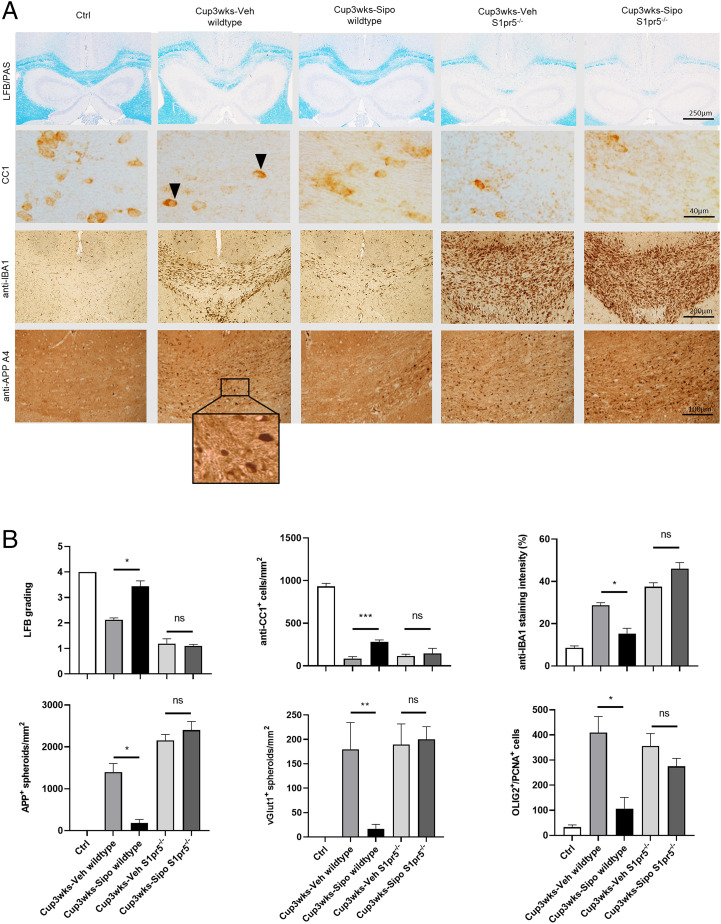
It is well known that oligodendrocytes express high levels of S1PR5 (31, 35). To test whether S1PR5 is involved in siponimod-mediated protection in the cuprizone model, wild-type and S1pr5−/− mice were intoxicated with cuprizone for 3 wk (i.e., groups H and I in Fig. 1), and the level of demyelination, glia activation, and oligodendrocyte loss was analyzed via immunohistochemistry. As demonstrated in Fig. 8, vehicle-treated wild-type mice showed myelin integrity loss, glia activation, and acute axonal injury after cuprizone intoxication. In line with our previous experiments, cuprizone-induced pathologies were significantly less severe in siponimod-treated wild-type mice. Of note, the protective effects of siponimod were virtually absent in S1pr5−/− mice. The parameters myelin densities (LFB; 1.15 ± 0.15 versus 1.1 ± 0.6 arbitrary units [AU]), mature oligodendrocytes (CC1; 117 ± 19.63 versus 146. ± 59.43 cells/mm2), microglia cells (anti-IBA1; 37.5 ± 1.9 versus 46.0 ± 2.91 optical density), acute axonal injury (anti-APP A4; 2,153 ± 14 versus 2,402 ± 20 spheroids/mm2, and anti-vGLUT1; 189.4 ± 42.35 versus 200.3 ± 25.82 spheroids/mm2; see SI Appendix, Fig. S3 for representative images of anti-vGLUT1 stains) were comparable in vehicle- and siponimod-treated cuprizone-intoxicated S1pr5−/− mice, demonstrating that siponimod mediates its protective effects by the S1PR5 receptor. Finally, there was an increase of OLIG2+/PCNA+ proliferating oligodendrocyte progenitor cells after 3 wk of cuprizone intoxication, and this increase was ameliorated by siponimod in wild-type but not S1pr5−/− mice (Fig. 8B and SI Appendix, Fig. S4). This observation demonstrates that the observed beneficial effects of siponimod in this study are not mediated by induction of myelin regeneration pathways but rather due to protection of oligodendrocytes.
Fig. 8.
Protective siponimod effects are absent in S1pr5−/− mice. (A) Representative LFB/PAS, anti-CC1, anti-IBA1 and anti-APP A4 stains of the medial CC from control (n = 4), Cup3wks-Veh-wildtype (n = 4), Cup3wks-Sipo-wildtype (n = 4), Cup3wks-Veh-S1pr5−/− (n = 5) and Cup3wks-Sipo-S1pr5−/− (n = 5) mice. Arrowheads highlight CC1+ mature oligodendrocytes. (B) Evaluation of the histological results of one experiment. The results have been verified in a second independent experiment (i.e., absence of protective siponimod effects in S1pr5−/− mice; see SI Appendix, Fig. S6). Statistical analyzes were performed by two separate Mann–Whitney tests as indicated. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. ns, nonsignificant.
Discussion
In this study, we were able to demonstrate that 1) siponimod exerts long-lasting immunosuppressive effects in an EAE model with severe forebrain involvement (i.e., in the Cup/EAE model); 2) in addition, siponimod exerts protective effects, which are independent of their immunomodulatory function; and 3) these protective effects are mediated via S1PR5 receptors.
In a first step, we reinvestigated the well-known immunosuppressive functions of siponimod (15, 36, 37) in the Cup/EAE model, which combines the toxin-mediated cuprizone model with the autoimmune MS model EAE to provoke peripheral immune cell recruitment into the forebrain (17). Even 2 wk after the last siponimod dosing (see Fig. 1 for the experimental setup) active MOG35–55 immunization in C57BL/6 mice did not result in EAE disease development. In line with this observation, a recent study demonstrated that the formation of meningeal inflammatory infiltrates, which have been described in postmortem MS tissues, is reduced by siponimod (37). Future studies have to show the underlying mechanisms of this prolonged immunosuppression.
While the role of S1PR1 signaling in regulating T cell migration is well established (38, 39), direct effects within the CNS are less clear. To study the potential direct effects of siponimod in the CNS, we used the cuprizone demyelination model. In this model, oral ingestion of the copper chelator cuprizone induces reproducible oligodendrocyte degeneration followed by region-specific demyelination and axonal injury (40). Lymphocytes are believed to play no functional role in this model (14). During the 3-wk cuprizone intoxication period, mice were treated with either vehicle or 3.125 mg/kg siponimod and their brains were histologically analyzed after an additional 2 wk of autonomous lesion progression (17). This dosing was chosen because it has been shown to ameliorate EAE in rats (15). Protective effects of siponimod were not just evident at week 5 but also at earlier time points such as weeks 1 and 3.
Of note, there are reports as well suggesting that peripheral immune cells might impact on the cuprizone-induced CNS pathologies. For example, it has been suggested that circulating CXCR2+ neutrophils are important for cuprizone-induced demyelination (41). We and others observed the recruitment of CD8+ lymphocytes into the CNS of cuprizone-intoxicated mice (25, 42). Despite the recruitment of peripheral immune cells into the CNS of cuprizone-intoxicated mice, the observed protective effects of siponimod in this study are likely to be mediated by a direct modulatory function of neuronal cells (i.e., cells within the CNS and not peripheral immune cells). First, protective effects were observed as well in Rag1−/− mice, which lack functional B and T cells. Second, in line with previous reports, siponimod concentrations were found to be increased in the brain parenchyma (∼650 nM; see SI Appendix, Fig. S5) (43). Third, the early recruitment of lymphocytes into the CNS of cuprizone-intoxicated mice was not ameliorated by siponimod. The results of a number of previous studies support the view of a direct protective effect of siponimod in the CNS. In EAE, siponimod prevented the degeneration of synapses after intracerebroventricular infusion (16). In slice cultures, where the CNS and the peripheral immune system are virtually uncoupled, siponimod attenuated lysophosphotidyl choline–mediated demyelination (33), whereas in vivo siponimod increased myelin basic protein levels after lysophosphotidyl choline–induced focal demyelination (44). Dietrich et al. recently demonstrated that siponimod can support remyelination, presumably by modulating the polarization of microglia cells (45).
To understand which receptor subtype mediated the protective effects of siponimod, we used mice deficient for S1pr5. While pronounced protective effects of siponimod were observed in wild-type mice, any protection was virtually absent in S1pr5−/− mice. Beyond, the secretion of proinflammatory mediators was not ameliorated by siponimod in mixed glia cell cultures where S1pr1 expression was found to be high, but S1pr5 expression is low (compare Fig. 6). Kim et al. (46) recently showed that the nonselective S1PR modulator FTY720 (fingolimod) reduced cuprizone-induced oligodendrocyte apoptosis and preserved myelin protein levels after 3 wk of cuprizone intoxication. Similar effects were observed when the authors used the specific S1PR1 modulator CYM5542 (46). Protective effects have been described as well by another group using the S1PR modulator ponesimod, which was recently approved for treating RRMS patients (47). Finally, it has been shown that ozanimod, a S1P1/5 modulator recently approved in the United States for the treatment of MS, attenuates ex vivo EAE glutamatergic synaptic alterations (48). Of note, electrophysiological studies with selective S1PR1 (AUY954) and S1PR5 (A971432) agonists suggested that S1PR1 modulation is the main driver of the antiexcitotoxic activity mediated by ozanimod. We cannot formally rule out that the observed protective effects of siponimod in our experiments are mediated by modulatory effects of both S1PR1 and S1PR5. Due to the essential function of S1PR1 for vascular maturation, S1pr1−/− mice exhibit embryonic hemorrhage leading to intrauterine death (49). Experiments using mice carrying the floxed S1pr1 alleles (50) would be required to address this interesting point in the future. However, the complete absence of protective siponimod effects in S1pr5−/− mice strongly suggests that this receptor subtype is critically involved.
This study is not without limitations. Several important differences exist between the cuprizone model of demyelination and the development of inflammatory lesions in MS patients. Maybe most importantly, in contrast to MS, T and B cells are not relevant for cuprizone‐induced demyelination. This is best documented by the finding that RAG‐deficient mice are fully susceptible to cuprizone‐induced demyelination and oligodendrocyte pathology (14). Furthermore, although the exact mode of action of cuprizone intoxication is not well understood, it is believed that the copper‐chelation action of cuprizone inhibits mitochondrial enzymes of the respiratory chain, leading to oxidative and endoplasmic reticulum stress responses (51, 52) and consequently to primary oligodendrocyte apoptosis. This mode of action is artificial and most likely not the underlying cause of oligodendrocyte and myelin degeneration in the CNS of MS patients.
Nevertheless, in summary, our study demonstrates that siponimod exerts protective effects in the brain in an S1PR5-dependent manner, which is independent of a T and B cell modulation. This finding is not just relevant in the context of MS but other neuropathologies as well characterized by degeneration of the axon–myelin unit.
Materials and Methods
Animals and Experimental Groups.
C57BL/6 female mice at 10 wk (wks) of age were obtained from Janvier Labs. Breeding pairs of mice homozygous for the Rag1tm1Mom mutation (Rag1−/−), lacking mature B and T cells (26), were kindly provided by Sven G. Meuth, Heinrich Heine University, Düsseldorf, Germany). Breeding pairs of mice lacking the S1pr5 receptor (S1pr5−/−) were kindly provided by Thierry Walzer, Centre International de Recherche en Infectiologie, Lyon, France. Both strains were bred in our animal facility and subsequently used for the experiments. All experimental procedures were approved by the review boards for the care of animal subjects of the district government (Regierung Oberbayern, reference no. 55.2-154-2532-73-15 and Regierung Mecklenburg-Vorpommern, reference no. 7221.3-1-001/19).
A maximum of five animals were housed per cage (814 cm2 area). Animals were kept under standard laboratory conditions (12 h light/dark cycle, controlled temperature 23 °C ± 2 °C and 50% ± 5% humidity) with access to food and water ad libitum. The mice were allowed to accommodate to the environment for at least 1 wk prior to the beginning of the experiments. Body weights of mice were controlled once per week. The mice were randomly assigned to the experimental groups (see Fig. 1 for a schematic illustration). Detailed descriptions of the experimental groups are given in SI Appendix, Materials and Methods.
Siponimod was provided as powder formulation from Novartis Pharma AG, Basel, Switzerland and dissolved in a 1% carboxymethy cellulose sodium solution (Sigma-Aldrich; order no. 4888-500gr). If not stated otherwise, the mice were treated with a siponimod concentration of ∼3.1 mg/kg. For dose-dependency studies, ∼0.31 mg/kg (low dose [LD]) and ∼15.5 mg/kg (high dose [HD]) were additionally used.
Cuprizone Intoxication and EAE Induction.
Cuprizone intoxication and active EAE induction were performed as previously published by our group (17, 53). Detailed descriptions are given in SI Appendix, Materials and Methods.
Tissue Sampling.
For the histological and immunohistochemical studies, the preparation of tissues was performed as previously described in detail (54). In brief, mice were transcardially perfused with cold phosphate-buffered saline (PBS), followed by a 3.7% formalin solution (pH 7.4). After overnight postfixation in the same fixative, the brains were dissected and embedded in paraffin, and coronal 5-μm-thick sections were prepared for immunohistochemical stains. The coronal slices were analyzed at the level R265 according to the mouse brain atlas published by Sidman et al. (http://www.hms.harvard.edu/research/brain/atlas.html). For EAE studies, spinal cord sections were analyzed, which were, prior to paraffin-embedding, decalcified by incubating the vertebral column samples in a 10% ethylenediaminetetraacetic acid (EDTA) solution for 35 d at 22 °C. For gene expression or next generation sequencing studies, mice were transcardially perfused with ice-cold PBS to avoid contamination with blood cells; and the brains were removed and manually dissected using a stereomicroscopic approach. In this study, the corpus callosum and overlying neocortex between the levels 215 and 355 according to Sidman et al. were sampled and further processed. Hippocampal and other subcortical areas were discarded.
Histology and Immunohistochemistry.
For histological evaluations, randomly selected brain sections containing the region of interest (ROI) were stained with standard LFB/PAS to determine demyelination, and hematoxylin & eosin (H&E) to 1) evaluate the number and spatial distribution of perivascular infiltrates within the forebrain or 2) to visualize apoptotic bodies in the corpus callosum, following established protocols in our laboratory. Spinal cord sections were stained with LFB/PAS to evaluate inflammatory demyelination within the white matter as described in detail elsewhere (55). Immunohistochemistry was performed using the following primary antibodies: Anti-ionized calcium-binding adaptor molecule 1 (IBA1) to visualize microglia and macrophages, anti-myelin PLP to target myelin proteins, anti-adenomatous polyposis coli (APC) clone CC1 (APC/CC1) to target mature oligodendrocytes, anti-activating transcription factor 3 (ATF3) as a stress marker protein, anti-glial fibrillary acidic protein (GFAP) to study the extent of astrocyte activation, anti-amyloid precursor protein (APP) or anti-vesicular glutamate transporter 1 (vGLUT1) to detect acutely damaged axons, anti-CD3 and anti-CD4 to label T lymphocytes and Th cells, respectively, and anti-oligodendrocyte transcription factor 2 (OLIG2) to detect the nuclei of oligodendrocytes (pan-oligodendrocyte marker). OLIG2/PCNA double labelings were performed to visualize proliferating oligodendrocyte progenitor cells. Order numbers, suppliers, applied concentrations, and the performed heat-induced epitope retrieval methods are given in SI Appendix, Table S1. The stains were conducted following established protocols. Detailed descriptions are given in SI Appendix, Materials and Methods. Beyond, detailed information regarding the methods applied for the histological evaluations can be found in SI Appendix, Materials and Methods.
Immunofluorescence Double Labeling.
To specifically label proliferating oligodendrocyte progenitor cells, immunofluorescence double labeling experiments using anti-OLIG2 and anti-PCNA antibodies were performed as previously described by our laboratory (56). Unspecific secondary antibody binding to the tissue itself was excluded by performing negative controls by incubating sections with each of the fluorescent secondary antibodies alone. Stained and processed sections were documented with the microscope working station Leica DM6 B. Order numbers, suppliers, applied concentrations, and the performed heat-induced epitope retrieval method are given in SI Appendix, Table S1.
PET/CT.
PET imaging to visualize activated glia cells, in particular microglia, was performed as previously described (22, 57) with minor modifications (SI Appendix, Materials and Methods).
Mixed Glia Cell Cultures.
Primary astrocyte cultures were prepared from the brains of P1–P3 C57BL/6 mice as previously described with some modifications (58). Flow cytometry analyses have been performed to analyze the relative proportion of astrocytes and microglia (SI Appendix, Materials and Methods).
Immunocytochemistry.
Immunofluorescence stains of the cultured cells were performed to analyze S1PR1 and S1PR5 expression. Detailed descriptions are given in SI Appendix, Materials and Methods.
Protein Analyses.
To analyze siponimod-induced signaling cascades in mixed glia cultures, cells were treated after 4 h of serum starvation with either vehicle (0.01% dimethyl sulfoxide [DMSO] in starving medium) or siponimod (1 µM in 0.01% DMSO in starving medium) solution for 60 min. Western blotting experiments were performed to analyze the activation of the Akt signaling pathway (see SI Appendix, Materials and Methods for a detailed description).
To analyze whether siponimod can modulate cytokine secretion in stimulated mixed glia cell cultures, cells were seeded in poly-L-lysine–coated six-well plates and stimulated with either lipopolysaccharides (LPS) (100 ng/mL, Sigma-Aldrich, L4391-1 MG), interferon gamma (IFNγ) (100 U/mL, Peprotech, 315-05), or tumor necrosis factor alpha (TNFα) (100 U/mL, Peprotech, 315-01A) for 24 h. A total of 2,000 µL of medium was applied per well. The cells in control groups were pretreated with either vehicle (DMSO) or siponimod solution (1 µM) for 60 min. After the indicated stimulation/treatment period, 1,000 µL of the cell supernatant was harvested, snap frozen in liquid nitrogen, and cytokine concentrations were analyzed by applying the Bio-Plex multiplex platform and using the Bio-Plex Pro Mouse Cytokine 23-Plex Assay M60009RDPD (Bio-Rad). Results are shown as concentration in picogram/milliliter after standard curve calibration.
RT-PCR.
To analyze S1pr1 and S1pr5 as well as cytokine mRNA expression levels in mixed glia cell cultures and isolated brain tissues, gene expression levels were semiquantified by qRT-PCR (Bio-Rad), using SensiMix Plus SYBR and fluorescein (Quantace, Bioline) with a standardized protocol as described previously by our group (52). The primer sequences and individual annealing temperatures are given in SI Appendix, Table S1.
Next Generation Sequencing.
NGS experiments were performed with mixed corpus callosum/neocortical tissues isolated from control (n = 5), Cup3wks-Veh (n = 5) and Cup3wks-Sipo (3.125 mg/kg) (n = 5) mice. After mechanical dissection, tissues were snap frozen in liquid nitrogen and afterward homogenized using the Precellys tissue homogenizer (VWR, Bertin Technologies). RNA isolation was performed using the RNeasy Lipid Tissue Mini Kit (Qiagen) strictly following the manufacturer’s instructions.
Quality checks of the prepared RNA samples were performed with the TapeStation 4200 using the Agilent RNA ScreenTape Assay Kit (Agilent). Mean RNA integrity numbers (RINs) were about 8.5 with lowest RIN ∼7.9. RNA quantification was performed using the Quantus RNA System (Promega). According to the manufacturer’s instructions, all cDNA libraries were generated from 1 μg total RNA using the TrueSeq Stranded Total RNA Library Preparation Kit with the Ribo-Zero Gold Kit (Illumina Inc.). The quality and quantity of the cDNA libraries were assessed using the 4200 TapeStation (D1000 screen tape assay) and the Quantus Fluorometer, respectively. All samples were sequenced on a NextSEq. 500 sequencing system with the following run parameters: 76 cycles for read 1, 6 cycles for index 1, and 76 cycles for read 2.
RNA-Sequencing Primary Analysis.
Generation of the fastq files and removal of adapters were completed using the Illumina demultiplexing and conversion software bcl2fastq (https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html). Subsequently, RNA-Sequencing (RNA-Seq) primary analysis was performed with the RNA-Seq pipeline of nf.core v3.0 using standard parameters. For instance, quality control of the sequenced transcripts was done with online routine fastqc v0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc); library complexity was measured with preseq v2.0.3 (59). Adapter and quality trimming was performed using TrimGalore v.0.6.6 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Subsequently, the reads were aligned to the mm10 reference genome sequence using STAR v2.6.1d (60) with default parameters. For quantification, reads were counted with the quasi-mapping quantification tool salmon v1.4.0 (61).
Statistical Evaluations.
Differences between groups were statistically tested using GraphPad Prism 5. In all cases, the Kolmogorov–Smirnov test was applied to test for normal data distribution. To compare multiple groups, one-way analysis of variance (ANOVA) was used with Tukey’s posttest for the normally distributed and Kruskal–Wallis test with Dunn’s multiple comparision test for not normally distributed data. The Bonferroni correction was applied for NGS-enrichment analyses. All data are given as arithmetic means ± SEM. Significance levels are indicated as *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgments
This work was supported by Novartis Pharma AG (M.K.), intramural funding (FORUN program, Rostock University Medical Center), and the Deutsche Forschungsgemeinschaft, Grant/Award No. KI 1469/8-1. We thank Prof. Dr. Walter Thierry for providing the S1pr5−/− mouse line as well as Joanna Foerster, Susann Lehmann, Frauke Winzer, Robin Piecha, and Antje Schümann for their excellent and valuable technical assistance. We thank Dr. Marc Bigaud (Novartis Basel, Switzerland) for critical feedback and the siponimod level measurements in isolated brain tissues.
Footnotes
Competing interest statement: This work was supported by Novartis Pharma AG (M.K.), intramural funding (FORUN program, Rostock University Medical Center [S.J.]), and the Deutsche Forschungsgemeinschaft, Grant/Award No. KI 1469/8-1.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2204509119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information. Raw data of the NGS experiments have been uploaded to the GEO website (GSE213374) (62).
References
- 1.Frischer J. M., et al. , The relation between inflammation and neurodegeneration in multiple sclerosis brains. Brain 132, 1175–1189 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howell O. W., et al. , Meningeal inflammation is widespread and linked to cortical pathology in multiple sclerosis. Brain 134, 2755–2771 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Serafini B., Rosicarelli B., Magliozzi R., Stigliano E., Aloisi F., Detection of ectopic B-cell follicles with germinal centers in the meninges of patients with secondary progressive multiple sclerosis. Brain Pathol. 14, 164–174 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magliozzi R., et al. , Meningeal B-cell follicles in secondary progressive multiple sclerosis associate with early onset of disease and severe cortical pathology. Brain 130, 1089–1104 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Kapoor R., et al. ; ASCEND investigators, Effect of natalizumab on disease progression in secondary progressive multiple sclerosis (ASCEND): A phase 3, randomised, double-blind, placebo-controlled trial with an open-label extension. Lancet Neurol. 17, 405–415 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Behrangi N., Fischbach F., Kipp M., Mechanism of Siponimod: Anti-inflammatory and neuroprotective mode of action. Cells 8, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGinley M. P., Cohen J. A., Sphingosine 1-phosphate receptor modulators in multiple sclerosis and other conditions. Lancet 398, 1184–1194 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Novgorodov A. S., El-Alwani M., Bielawski J., Obeid L. M., Gudz T. I., Activation of sphingosine-1-phosphate receptor S1P5 inhibits oligodendrocyte progenitor migration. FASEB J. 21, 1503–1514 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Van Doorn R., et al. , Sphingosine 1-phosphate receptor 1 and 3 are upregulated in multiple sclerosis lesions. Glia 58, 1465–1476 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Janssen S., et al. , Effect of FTY720-phosphate on the expression of inflammation-associated molecules in astrocytes in vitro. Mol. Med. Rep. 12, 6171–6177 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Nishimura H., Akiyama T., Irei I., Hamazaki S., Sadahira Y., Cellular localization of sphingosine-1-phosphate receptor 1 expression in the human central nervous system. J. Histochem. Cytochem. 58, 847–856 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zendedel A., Beyer C., Kipp M., Cuprizone-induced demyelination as a tool to study remyelination and axonal protection. J. Mol. Neurosci. 51, 567–572 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Kipp M., Clarner T., Dang J., Copray S., Beyer C., The cuprizone animal model: New insights into an old story. Acta Neuropathol. 118, 723–736 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Hiremath M. M., Chen V. S., Suzuki K., Ting J. P., Matsushima G. K., MHC class II exacerbates demyelination in vivo independently of T cells. J. Neuroimmunol. 203, 23–32 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gergely P., et al. , The selective sphingosine 1-phosphate receptor modulator BAF312 redirects lymphocyte distribution and has species-specific effects on heart rate. Br. J. Pharmacol. 167, 1035–1047 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentile A., et al. , Siponimod (BAF312) prevents synaptic neurodegeneration in experimental multiple sclerosis. J. Neuroinflammation 13, 207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheld M., et al. , Neurodegeneration triggers peripheral immune cell recruitment into the forebrain. J. Neurosci. 36, 1410–1415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kappos L., et al. ; EXPAND Clinical Investigators, Siponimod versus placebo in secondary progressive multiple sclerosis (EXPAND): A double-blind, randomised, phase 3 study. Lancet 391, 1263–1273 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Gudi V., Gingele S., Skripuletz T., Stangel M., Glial response during cuprizone-induced de- and remyelination in the CNS: Lessons learned. Front. Cell. Neurosci. 8, 73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skripuletz T., Gudi V., Hackstette D., Stangel M., De- and remyelination in the CNS white and grey matter induced by cuprizone: The old, the new, and the unexpected. Histol. Histopathol. 26, 1585–1597 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Praet J., Guglielmetti C., Berneman Z., Van der Linden A., Ponsaerts P., Cellular and molecular neuropathology of the cuprizone mouse model: Clinical relevance for multiple sclerosis. Neurosci. Biobehav. Rev. 47, 485–505 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Nack A., et al. , Expression of translocator protein and [18F]-GE180 ligand uptake in multiple sclerosis animal models. Cells 8, 94 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P. D., PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnett H. A., et al. , TNF alpha promotes proliferation of oligodendrocyte progenitors and remyelination. Nat. Neurosci. 4, 1116–1122 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Kaddatz H., et al. , Cuprizone-induced demyelination triggers a CD8-pronounced T cell recruitment. Glia 69, 925–942 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Mombaerts P., et al. , RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992). [DOI] [PubMed] [Google Scholar]
- 27.Rothhammer V., et al. , Sphingosine 1-phosphate receptor modulation suppresses pathogenic astrocyte activation and chronic progressive CNS inflammation. Proc. Natl. Acad. Sci. U.S.A. 114, 2012–2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi J. W., et al. , FTY720 (fingolimod) efficacy in an animal model of multiple sclerosis requires astrocyte sphingosine 1-phosphate receptor 1 (S1P1) modulation. Proc. Natl. Acad. Sci. U.S.A. 108, 751–756 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colombo E., et al. , Siponimod (BAF312) activates Nrf2 while hampering NFκB in Human astrocytes, and protects from astrocyte-induced neurodegeneration. Front. Immunol. 11, 635 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaire B. P., et al. , Identification of sphingosine 1-Phosphate Receptor Subtype 1 (S1P1) as a pathogenic factor in transient focal cerebral ischemia. Mol. Neurobiol. 55, 2320–2332 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Jaillard C., et al. , Edg8/S1P5: An oligodendroglial receptor with dual function on process retraction and cell survival. J. Neurosci. 25, 1459–1469 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zamora N. N., Cheli V. T., Santiago González D. A., Wan R., Paez P. M., Deletion of voltage-gated calcium channels in astrocytes during demyelination reduces brain inflammation and promotes myelin regeneration in mice. J. Neurosci. 40, 3332–3347 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Sullivan C., Schubart A., Mir A. K., Dev K. K., The dual S1PR1/S1PR5 drug BAF312 (Siponimod) attenuates demyelination in organotypic slice cultures. J. Neuroinflammation 13, 31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldberg J., et al. , Short-term cuprizone feeding induces selective amino acid deprivation with concomitant activation of an integrated stress response in oligodendrocytes. Cell. Mol. Neurobiol. 33, 1087–1098 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miron V. E., Hall J. A., Kennedy T. E., Soliven B., Antel J. P., Cyclical and dose-dependent responses of adult human mature oligodendrocytes to fingolimod. Am. J. Pathol. 173, 1143–1152 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis N. D., et al. , Circulating monocytes are reduced by sphingosine-1-phosphate receptor modulators independently of S1P3. J. Immun. 190, 3533–3540 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Brand R. M., et al. , Siponimod inhibits the formation of meningeal ectopic lymphoid tissue in experimental autoimmune encephalomyelitis. Neurol. Neuroimmunol. Neuroinflamm. 9, e1117 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baeyens A. A. L., Schwab S. R., Finding a Way Out: S1P Signaling and Immune Cell Migration. Annu. Rev. Immunol. 38, 759–784 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Matloubian M., et al. , Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 427, 355–360 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Rühling S., et al. , Visualization of the breakdown of the axonal transport machinery: A comparative ultrastructural and immunohistochemical approach. Mol. Neurobiol. 56, 3984–3998 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Liu L., et al. , CXCR2-positive neutrophils are essential for cuprizone-induced demyelination: Relevance to multiple sclerosis. Nat. Neurosci. 13, 319–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Almuslehi M. S. M., Sen M. K., Shortland P. J., Mahns D. A., Coorssen J. R., CD8 T-cell recruitment into the central nervous system of cuprizone-fed mice: Relevance to modeling the etiology of multiple sclerosis. Front. Cell. Neurosci. 14, 43 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bigaud M., et al. , Siponimod (BAF312) penetrates, distributes, and acts in the central nervous system: Preclinical insights. Mult. Scler. J. Exp. Transl. Clin. 7, 20552173211049168 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson S. J., Giovannoni G., Baker D., Fingolimod modulates microglial activation to augment markers of remyelination. J. Neuroinflammation 8, 76 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dietrich M., et al. , Increased remyelination and proregenerative microglia under siponimod therapy in mechanistic models. Neurol. Neuroimmunol. Neuroinflamm. 9, e1161 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S., et al. , Functional antagonism of sphingosine-1-phosphate receptor 1 prevents cuprizone-induced demyelination. Glia 66, 654–669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kihara Y., et al. , Ponesimod inhibits astrocyte-mediated neuroinflammation and protects against cingulum demyelination via S1P1 -selective modulation. FASEB J. 36, e22132 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Musella A., et al. , Central modulation of selective sphingosine-1-phosphate receptor 1 ameliorates experimental multiple sclerosis. Cells 9, 1290 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y., et al. , Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J. Clin. Invest. 106, 951–961 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allende M. L., Yamashita T., Proia R. L., G-protein-coupled receptor S1P1 acts within endothelial cells to regulate vascular maturation. Blood 102, 3665–3667 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Teske N., et al. , Chemical hypoxia-induced integrated stress response activation in oligodendrocytes is mediated by the transcription factor nuclear factor (erythroid-derived 2)-like 2 (NRF2). J. Neurochem. 144, 285–301 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Fischbach F., et al. , Cuprizone-induced graded oligodendrocyte vulnerability is regulated by the transcription factor DNA damage-inducible transcript 3. Glia 67, 263–276 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Rüther B. J., et al. , Combination of cuprizone and experimental autoimmune encephalomyelitis to study inflammatory brain lesion formation and progression. Glia 65, 1900–1913 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Kipp M., Kiessling M. C., Hochstrasser T., Roggenkamp C., Schmitz C., Design-based stereology for evaluation of histological parameters. J. Mol. Neurosci. 61, 325–342 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Nyamoya S., et al. , Laquinimod supports remyelination in non-supportive environments. Cells 8, 1363 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baertling F., et al. , ADAM12 is expressed by astrocytes during experimental demyelination. Brain Res. 1326, 1–14 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Mann T., et al. , [18F]fallypride-PET/CT analysis of the dopamine D2/D3 receptor in the hemiparkinsonian rat brain following intrastriatal botulinum neurotoxin A injection. Molecules 23, 587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kipp M., et al. , Brain-region-specific astroglial responses in vitro after LPS exposure. J. Mol. Neurosci. 35, 235–243 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Daley T., Smith A. D., Predicting the molecular complexity of sequencing libraries. Nat. Methods 10, 325–327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.N. Behrangi et al., Data from Siponimod ameliorates metabolic oligodendrocyte injury via the sphingosine-1 phosphate receptor 5. Gene Expression Omnibus. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE213374. Deposited 16 September 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. Raw data of the NGS experiments have been uploaded to the GEO website (GSE213374) (62).