Significance
The fibroblast growth factor 19 (FGF19)–FGF receptor 4 (FGFR4)–coreceptor klotho β (KLB) signaling axis has been implicated as a therapeutic target in 20 to 30% of hepatocellular carcinomas. However, patient responses to FGFR4-selective inhibitors were unsatisfactory in recent clinical trials. In this study, we reveal the redundancy of FGFR3 and FGFR4 as mechanisms of de novo resistance to FGFR4-selective inhibitors, supporting the clinical evaluation of pan-FGFR inhibitors for treating FGF19-positive hepatocellular carcinoma.
Keywords: FGFR inhibitor, HCC, FGFR4, FGF19, KLB
Abstract
Aberrant fibroblast growth factor 19 (FGF19) signaling mediated by its receptor, FGF receptor 4 (FGFR4), and coreceptor, klotho β (KLB), is a driver of hepatocellular carcinoma (HCC). Several potent FGFR4-selective inhibitors have been developed but have exhibited limited efficacy in HCC clinical trials. Here, by using HCC cell line models from the Cancer Cell Line Encyclopedia (CCLE) and the Liver Cancer Model Repository (LIMORE), we show that selective FGFR4 inactivation was not sufficient to inhibit cancer cell proliferation and tumor growth in FGF19-positive HCC. Moreover, genetic inactivation of KLB in these HCC cells resulted in a fitness defect more severe than that resulting from inactivation of FGFR4. By a combination of biochemical and genetic approaches, we found that KLB associated with FGFR3 and FGFR4 to mediate the prosurvival functions of FGF19. KLB mutants defective in interacting with FGFR3 or FGFR4 could not support the growth or survival of HCC cells. Genome-wide CRISPR loss-of-function screening revealed that FGFR3 restricted the activity of FGFR4-selective inhibitors in inducing cell death; the pan-FGFR inhibitor erdafitinib displayed superior potency than FGFR4-selective inhibitors in suppressing the growth and survival of FGF19-positive HCC cells. Among FGF19-positive HCC cases from The Cancer Genome Atlas (TCGA), FGFR3 is prevalently coexpressed with FGFR4 and KLB, suggesting that FGFR redundancy may be a common mechanism underlying the de novo resistance to FGFR4 inhibitors. Our study provides a rationale for clinical testing of pan-FGFR inhibitors as a treatment strategy for FGF19-positive HCC.
Fibroblast growth factor (FGF) receptors 1 to 4 (FGFR1, FGFR2, FGFR3, and FGFR4) are receptor tyrosine kinases that orchestrate diverse biological processes such as embryonic development, cellular proliferation, metabolic homeostasis, and tissue repair (1). In the presence of FGFs, FGFR kinase activity is activated, leading to the activation of phospholipase Cγ, PI3K-AKT, and RAS-MAPK signaling pathways (2). Genetic alterations (amplification, gene fusion, or point mutation) in FGFRs are observed in 5 to 10% of all human cancers, and this frequency increases to 10 to 30% in urothelial carcinoma and intrahepatic cholangiocarcinoma (3, 4). Recently, several FGFR inhibitors have demonstrated therapeutic benefit and gained regulatory approval in treating metastatic urothelial carcinoma and unresectable cholangiocarcinoma with FGFR alterations (5–7). In addition, many inhibitors that target one or more FGFRs are currently under clinical evaluation for treating FGFR-altered cancers.
Among the FGFR family, FGFR4 is an emerging target for hepatocellular carcinoma (HCC). Under physiological conditions, FGFR4 participates in the gut-liver crosstalk to regulate bile acid synthesis (8). Bile acids are synthesized in the liver and stored in the gallbladder in the fasting state. After meal ingestion, bile acids are released into the duodenum to facilitate fat absorption. A majority of bile acids are reabsorbed into the ileum, where they trigger the production of endocrine FGF19. Via portal circulation, FGF19 reaches the liver and binds to its receptor, FGFR4, and coreceptor, klotho β (KLB), on hepatocytes to repress bile acid synthesis (9, 10). Thus, the physiological function of FGFR4 is to control bile acid homeostasis via feedback regulation (11).
Whereas normal hepatocytes do not express FGF19, aberrant expression of FGF19 occurs in ∼20% of patients with HCC via gene amplification or epigenetic activation (12). In contrast, FGFR4 and KLB are consistently expressed in normal liver and HCC. As a result, HCC-derived FGF19 functions in an autocrine fashion to activate FGFR4 to promote HCC initiation and progression. Indeed, preclinical studies showed that FGFR4 inactivation blocks tumor initiation in a mouse HCC model driven by transgenic overexpression of FGF19 (13, 14).
FGFR4 possesses a unique cysteine residue (Cys552) in its kinase pocket, not found in FGFR1, FGFR2, or FGFR3, thus enabling the development of potent and selective FGFR4 inhibitors covalently targeting this cysteine (15–17). Two FGFR4-selective inhibitors, BLU-554 and FGF-401, have been evaluated in phase 1/2 clinical trials in patients with HCC whose tumors expressed FGF19. Although BLU-554 and FGF-401 were well tolerated, the overall response rates were 7 to 17%, mostly consisting of partial responses (ClinicalTrials.gov identifiers: NCT02508467 and NCT02325739) (18). In order to prioritize treatments that provide clinically meaningful benefit to patients, it is important to understand the mechanisms responsible for the intrinsic resistance to FGFR4 inhibitors in FGF19-positive HCC.
Results
FGFR4 Inhibition Is Not Sufficient for Cell Death Induction in FGF19-Positive HCC Cells.
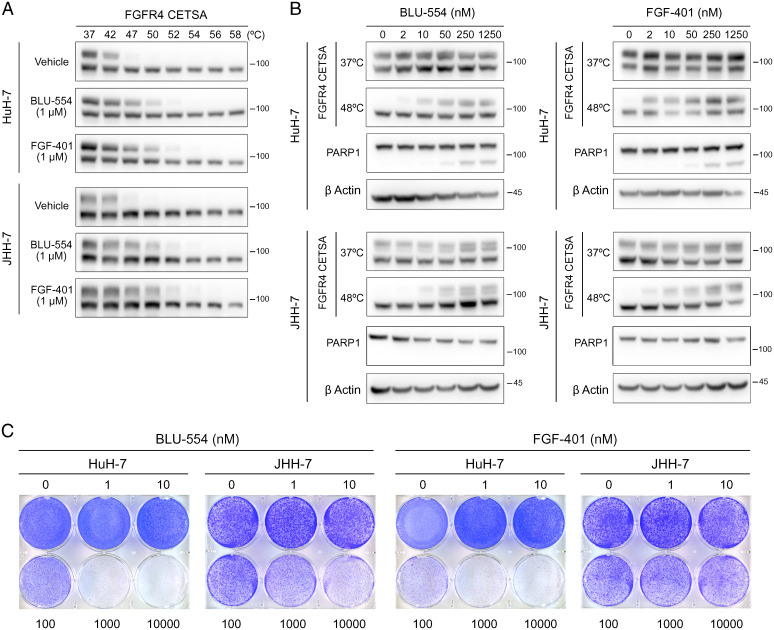
We first examined the in vitro pharmacodynamic properties of BLU-554 and FGF-401 in two HCC cell lines, HuH-7 and JHH-7, commonly used as models of FGF19-positive HCC. We used the cellular thermal shift assay (CETSA) to reveal inhibitor engagement of FGFR4 in intact cells. CETSA is based on the phenomenon that heating induces protein unfolding and aggregation, while inhibitor binding can shift the temperatures at which target proteins unfold and aggregate in the cellular milieu. Aggregated proteins disappear from cell lysate after centrifugation (19). We treated HuH-7 and JHH-7 with vehicle, 1 μM BLU-554, or 1 μM FGF-401 for 12 h, heated cells at temperatures ranging from 37 to 58 °C, and quantified soluble FGFR4 by Western blotting. Two prominent bands of FGFR4 were observed: the faster migrating band was thermostable, whereas the slower migrating band was thermolabile, with an estimated melting temperature (Tm) of ∼42 °C. We focused on the latter species to probe FGFR4 inhibitor engagement. BLU-554 treatment increased the Tm of FGFR4 by ∼5 °C in HuH-7 and JHH-7 cells, respectively. Similarly, FGF-401 treatment also increased the Tm of FGFR4 by ∼8 °C in HuH-7 and JHH-7 cells (Fig. 1A). In contrast, the melting temperature of β actin was not altered by inhibitor treatment (SI Appendix, Fig. S1A). Next, we performed CETSA by treating HuH-7 and JHH-7 cells with different concentrations of BLU-554 or FGF-401 for 12 h, heating cells at 48 °C, and then measuring the amounts of soluble FGFR4. Consistent with their reported in vitro potencies in inhibiting FGFR4 kinase activity (SI Appendix, Fig. S1B), 10 nM BLU-554 or 2 nM FGF-401 was sufficient to stabilize FGFR4 in HuH-7 cells. An approximately fivefold higher concentration of BLU-554 (50 nM) or FGF-401 (10 nM) was required to stabilize FGFR4 in JHH-7 cells (Fig. 1 B and C). These results provide evidence that BLU-554 and FGF-401 potently inhibit FGFR4 in intact HCC cells.
Fig. 1.
FGFR4 inhibition is not sufficient to trigger cell death in HuH-7 or JHH-7. (A) CETSA for in-cell target engagement by selective FGFR4 inhibitors. Western blots showing thermostable FGFR4 following heat shock at indicated temperatures in HuH-7 and JHH-7 cells treated with vehicle (DMSO), BLU-554 (1 μM), or FGF-401 (1 μM) for 12 h. (B) CETSA and PARP1 cleavage in HuH-7 and JHH-7 cells treated with indicated concentrations of BLU-554 or FGF-401 for 12 (CETSA) or 48 h (PARP1 cleavage). (C) Crystal violet staining of HuH-7 and JHH-7 cells treated with indicated concentrations of BLU-554 or FGF-401 for 6 d.
To explore the cellular outcomes of FGFR4 inhibition, we examined poly (ADP ribose) polymerase-1 (PARP1) cleavage as a marker for apoptosis in HuH-7 and JHH-7 cells treated with BLU-554 or FGF-401 for 48 h. In HuH-7 cells, 250 nM BLU-554 or FGF-401 was required to induce PARP1 cleavage. In JHH-7 cells, as high as 1.25 μM BLU-554 or FGF-401 failed to induce PARP1 cleavage (Fig. 1 B and C). Similar results were observed by crystal violet staining of viable cells after BLU-554 or FGF-401 treatment (Fig. 1D). Taken together, these observations reveal that the anticancer activity of selective FGFR4 inhibitors in HCC requires concentrations at lease 25-fold higher than the concentration sufficient for FGFR4 inhibition.
Genetic Inactivation of KLB Results in a Fitness Defect More Severe Than That Resulting From Inactivation of FGFR4.
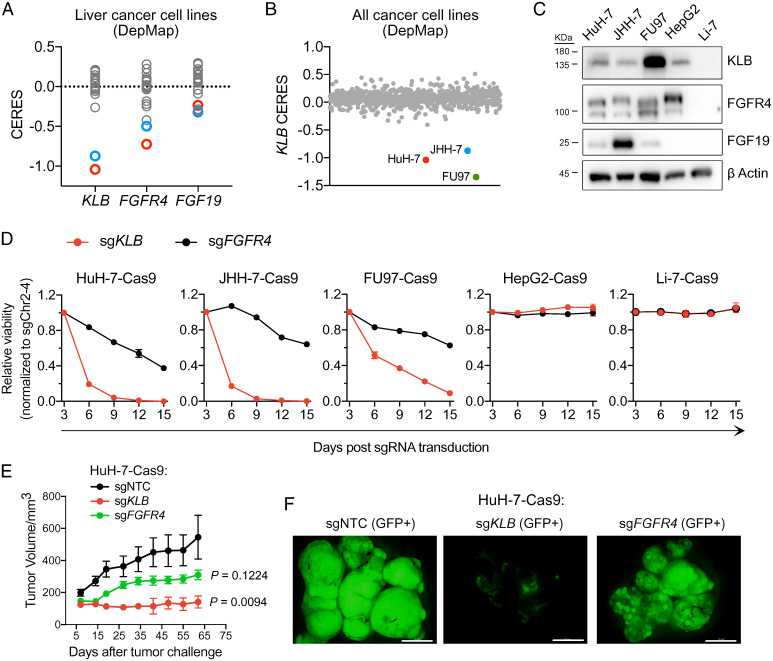
The lack of potent anticancer activity upon selective FGFR4 inhibition in FGF19-positive HCC cell lines prompted us to evaluate their genetic dependencies on FGFR4, FGF19, and KLB. CRISPR loss-of-function screens have enabled the identification of genes essential for diverse human cancer cell lines in the Cancer Cell Line Encyclopedia (CCLE), resulting in the cancer dependency map (DepMap) (20, 21). Among 22 HCC cell lines in the DepMap, HuH-7 and JHH-7 displayed lower CERES scores (reflecting higher likelihood of essentiality) for FGFR4 and KLB than the remaining 20 cell lines. In contrast, FGF19 did not score as an essential gene in HuH-7 or JHH-7 (Fig. 2A). We interpreted the lack of FGF19 dependency as an artifact of pooled CRISPR screening; FGF19-knockout cells could be rescued by paracrine FGF19 secreted by other cells in the population.
Fig. 2.
CRISPR inactivation of KLB displays a more severe fitness defect than that seen with inactivation of FGFR4 in FGF19-positive cancer cell lines. (A) CERES scores of KLB, FGFR4, and FGF19 in 22 liver cancer cell lines tested in CRISPR-Cas9 loss-of-viability screens (DepMap). (B) KLB CERES scores of 807 cell lines in DepMap. (C) Western blots showing expression levels of KLB, FGFR4, and FGF19 in indicated cell lines. (D) Viability effects after CRISPR inactivation of KLB or FGFR4 in indicated cell lines normalized to control (sgChr2-4). (E) Tumor growth curve after CRISPR inactivation of KLB or FGFR4 in HuH-7 cells xenografted subcutaneously into nude mice. Data are mean ± SEM, with n = 5 mice per group. Comparisons with the control group (sgNTC) were performed with ordinary one-way ANOVA (F [2, 12] = 5.904, P = 0.0164) followed by Dunnett multiple comparison test (sgKLB vs. sgNTC: P = 0.0094; sgFGFR4 vs. sgNTC: P = 0.1224). (F) Fluorescence images of dissected tumors on day 62 of E (scale bar, 1 cm).
Notably, KLB CERES scores of HuH-7 and JHH-7 were lower than their FGFR4 CERES scores (Fig. 2A). By examining KLB CERES score across all 807 cell lines in the DepMap, we found that KLB was essential in only three cell lines: HuH-7, JHH-7, and a gastric cancer cell line, FU97 (Fig. 2B). Using Western blotting, we confirmed that HuH-7, JHH-7, and FU97 expressed FGF19, FGFR4, and KLB (Fig. 2C). However, these cell lines were predicted to be less sensitive to CRISPR inactivation of FGFR4 than to that of KLB. To experimentally validate the DepMap prediction, we identified single guide RNAs (sgRNAs) that could efficiently deplete FGFR4 and KLB (SI Appendix, Fig. S2A) and then devised a competitive cell growth assay (SI Appendix, Fig. S2B) to quantify the effects of knocking out FGFR4 versus KLB on the fitness of three FGF19-positive cell lines (HuH-7, JHH-7, and FU97) and two FGF19-negative HCC cell lines (HepG2 and Li-7). After stably transducing these cell lines with lentiviral Cas9, we infected them with lentiviruses coexpressing an sgRNA and a green fluorescent protein (GFP) marker. We then monitored the percentages of GFP-positive cells over the course of 2 wk. An sgRNA targeting an intergenic region (sgChr2-4) was included as a control for data normalization (22). Consistent with the DepMap prediction, Cas9-expressing HuH-7, JHH-7, and FU97 cells infected with sgKLB were depleted more rapidly than cells infected with sgFGFR4. Neither sgKLB nor sgFGFR4 resulted in a detectable fitness drop in HepG2 or Li-7 (Fig. 2D).
We further extended the competitive cell growth assay from in vitro to in vivo by transplanting control, FGFR4-knockout, or KLB-knockout HuH-7 cells onto the dorsal flanks of nude mice. Tumors derived from FGFR4-knockout HuH-7 cells grew slower than tumors derived from control HuH-7 cells, but the differences were not statistically significant. In contrast, KLB knockout significantly inhibited tumor growth compared with control (Fig. 2E). Because our sgRNA vector also encoded GFP, we further imaged dissected tumors for the presence of GFP-positive cells and found that KLB-knockout cells were depleted from the tumors, whereas control or FGFR4-knockout cells were retained (Fig. 2F). In conclusion, CRISPR inactivation of KLB resulted in a fitness defect more pronounced than that of FGFR4 in FGF19-positive cancer cell lines.
KLB Associates With Multiple FGFRs to Mediate FGF19 Signaling.
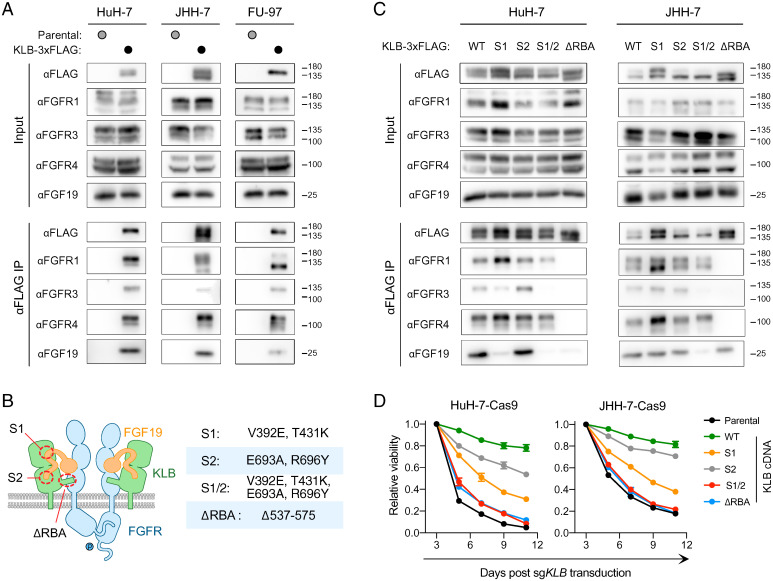
The differences in the phenotypic outcomes of FGFR4 versus KLB inactivation suggest that KLB may have FGFR4-independent functions. To reveal such functions, we performed KLB immunoprecipitation followed by mass spectrometry (IP-MS) to identify proteins interacting with KLB. We expressed KLB-3xFLAG driven by a doxycycline-inducible promoter in HuH-7 cells and performed immunoprecipitation with an anti-FLAG antibody (SI Appendix, Fig. S3A). Mass spectrometry revealed that KLB associated with FGFR1, FGFR3, and FGFR4. Among the three FGFRs in complex with KLB, the number of peptides derived from FGFR4 was the highest, whereas the number of peptides derived from FGFR1 was the lowest (SI Appendix, Fig. S3B). To corroborate our IP-MS findings, we performed KLB-3xFLAG IP followed by Western blotting (IP-WB) in HuH-7, JHH-7, and FU97. In all three cell lines, we confirmed the coimmunoprecipitation of FGFR1, FGFR3, and FGFR4 with KLB. In addition, IP-WB revealed FGF19 enrichment in the KLB complex (Fig. 3A). Taken together, these results demonstrate that KLB associates with multiple FGFRs in FGF19-positive cell lines.
Fig. 3.
KLB associates with multiple FGFRs to mediate FGF19 signaling. (A) Coimmunoprecipitation of FGFR1, FGFR3, and FGFR4 with KLB-3×FLAG in HuH-7, JHH-7, and FU97 cells. (B) A cartoon model of the FGF19-FGFR-KLB complex with indicated mutations introduced to perturb complex formation. (C) Effects of indicated KLB mutations on the coimmunoprecipitation of FGFR1, FGFR3, and FGFR4 with KLB-3×FLAG in HuH-7 and JHH-7 cells. (D) Effects of indicated KLB mutations on the viability of HuH-7 and JHH-7 cells after CRISPR inactivation of endogenous KLB. WT, wild type.
KLB is a transmembrane protein consisting of two extracellular glycoside hydrolase-like domains, a transmembrane helix, and a short intracellular tail (23). Previous structural biology studies revealed that KLB interacts with the C-terminal tail of FGF19 via two sites, S1 and S2 (24). We mutated key residues in these two sites alone or in combination. In addition, KLB interacts with FGFR via a protruding loop known as the receptor binding arm (RBA) (25), which was deleted from KLB (ΔRBA) to impair its ability to interact with FGFRs (Fig. 3B).
We introduced these KLB mutants tagged with C-terminal 3xFLAG into HuH-7 cells and JHH-7 cells and then used IP-WB to examine whether these mutants were able to associate with FGFRs and FGF19. KLB S1 mutant exhibited reduced binding to FGF19 in HuH-7 cells but not in JHH-7 cells. KLB S2 mutant was indistinguishable from wild-type KLB in its ability to coimmunoprecipitate FGFRs and FGF19. Combining S1 and S2 mutations (S1/2) prevented KLB binding to FGF19 and FGFR3 in both HuH-7 and JHH-7 cells. KLB ΔRBA mutant lost the ability to coimmunoprecipitate FGFR1, FGFR3, and FGFR4 in both HuH-7 and JHH-7 cells. Moreover, KLB ΔRBA mutant failed to coimmunoprecipitate FGF19 in HuH-7 cells but not in JHH-7 cells (Fig. 3C). Taken together, we generated a series of KLB mutations that impaired the formation of the FGF19-FGFR-KLB complex to varying extents.
To examine the functions of these KLB mutants, we stably expressed sgRNA-resistant forms of these KLB mutants in HuH-7 and JHH-7 cells and tested whether these mutants could rescue the fitness defect resulting from CRISPR inactivation of endogenous KLB. Whereas expression of wild-type KLB rescued the viability of KLB-knockout cells, expression of KLB mutants exhibited a range of responses. ΔRBA and S1/2 mutants were unable to rescue KLB-knockout cells, whereas S1 and S2 mutants partially rescued KLB-knockout cells (Fig. 3D). The differential ability of KLB mutants to rescue KLB-knockout cells correlated with their ability to associate with FGFRs and FGF19 (Fig. 3 C and D). Taken together, the above structure-function analyses of KLB suggest that the essential function of KLB in FGF19-positive HCC requires its interaction with FGFRs and FGF19.
FGFR3 Reduces the Anticancer Activity of FGFR4 Inhibitors in FGF19-Positive HCC.
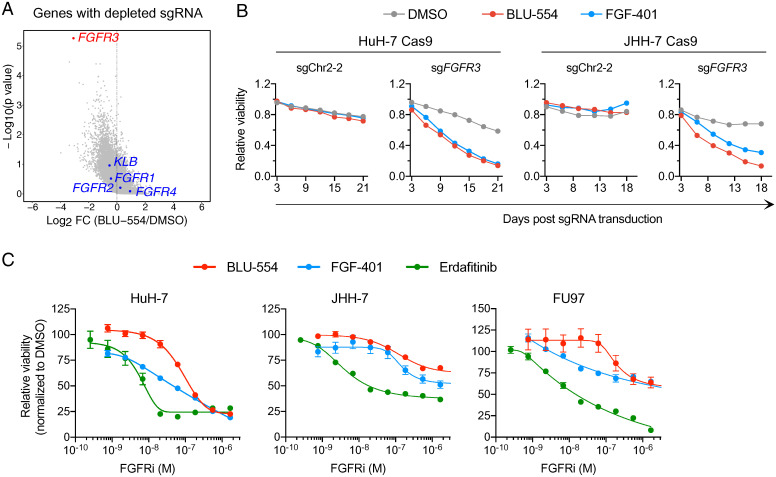
The observation that multiple FGFRs interact with KLB to mediate oncogenic FGF19 signaling suggests that FGFR redundancy may limit the anticancer activity of selective FGFR4 inhibitors. To test this hypothesis in an unbiased manner, we performed pooled genome-scale CRISPR-Cas9 knockout screening (26) by treating sgRNA-transduced HuH-7–Cas9 cells with either vehicle or a sublethal concentration (40 nM) of BLU-554 for 3 wk. Afterward, we isolated genomic DNAs and performed next-generation sequencing to quantify the abundance of each sgRNA in surviving cells. MAGeCK (Model-based Analysis of Genome-wide CRISPR/Cas9 Knockout) (27) algorithm ranked FGFR3 as the top 1 depleted gene in BLU-554–treated cells relative to vehicle-treated cells, whereas FGFR1, FGFR2, and FGFR4 were not significantly depleted (Fig. 4A and Dataset S1).
Fig. 4.
Inactivation of FGFR3 sensitizes HuH-7 and JHH-7 cells to selective FGFR4 inhibitors (FGFR4i). (A) Scatterplot depicting log2-transformed average fold change (FC) of sgRNA abundance (BLU-554 divided by DMSO) and log10-transformed P value computed by MaGeCK. (B) Viability effects after CRISPR inactivation of FGFR3 in HuH-7 and JHH-7 cells in the presence of vehicle (DMSO), BLU-554 (50 nM), or FGF-401 (10 nM). (C) Measurements of IC50 of FGFR inhibitors on the viability of HuH-7, JHH-7, and FU97 cells. Cells were treated with indicated concentrations of FGFR inhibitors for 3 (HuH-7 and JHH-7) and 6 d (FU97) before viability assessment.
Based on the CRISPR screen results, we predicted that FGFR3-knockout cells would be more sensitive to FGFR4 inhibitors. To test this prediction, we measured the viability of control (sgChr2-2) or FGFR3-knockout HuH-7 and JHH-7 cells in the presence of vehicle, BLU-554, or FGF-401. Compared with control cells, FGFR3-knockout cells were indeed more rapidly depleted in the presence of BLU-554 or FGF-401 (Fig. 4B). Erdafitinib is a pan-FGFR inhibitor approved by the US Food and Drug Administration (FDA) to treat locally advanced or metastatic urothelial carcinoma with FGFR2/3 alterations (5, 28, 29). Erdafitinib inhibited all four FGFRs with half maximal inhibitory concentrations (IC50s) of 1 to 6 nM in vitro (SI Appendix, Fig. S1B), which are comparable to its cellular potency (IC50 = 5.77 nM in HuH-7 cells and 3.65 nM in JHH-7 cells). In contrast, BLU-554 and FGF-401 were 14- to 38-fold less potent than erdafitinib (Fig. 4C). In FU97 cells, erdafitinib inhibited cell viability by 92% inhibition. In contrast, BLU-554 and FGF-401 could inhibit FU97 cell viability by only 36% (Fig. 4C). These results suggest that pan-FGFR inhibitors such as erdafitinib may offer greater therapeutic benefit to patients with FGF19-positive HCC by overcoming FGFR redundancy.
Multiple FGFRs Redundantly Promote FGF19-Positive HCC Tumor Growth.
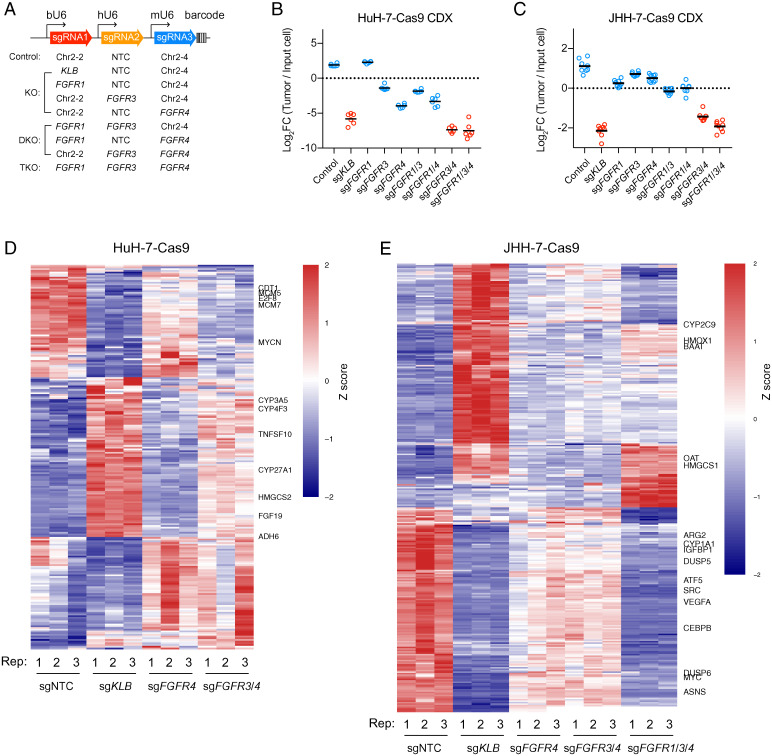
To evaluate the roles of FGFR redundancy in driving HCC tumor growth, we used a polycistronic sgRNA lentiviral vector that has been optimized for triple gene targeting; three sgRNAs could be expressed simultaneously under bovine, human, and mouse U6 promoters, respectively (Fig. 5A) (30). The vector also harbored a set of barcode sequences for decoding sgRNA representation. We generated a combination of constructs targeting one, two, or three FGFRs and then transduced these sgRNA constructs into HuH-7–Cas9 and JHH-7–Cas9 cells. Transduced cells were transplanted subcutaneously into nude mice. Afterward, tumors were harvested, and their sgRNA representations were analyzed by barcode sequencing.
Fig. 5.
FGFR redundancy in HuH-7 and JHH-7 revealed by in vivo tumor growth and transcriptome analyses. (A) Barcoded polycistronic vectors expressing indicated combinations of sgRNAs driven by bovine, human, and mouse U6 promoters. (B and C) Relative abundance (log2 transformed) of HuH-7–Cas9 and JHH-7–Cas9 cells harboring indicated combinations of sgRNAs from subcutaneous cell line–derived tumor xenografts (CDX) normalized to input cells. Bars represent averages from n = 6 mice (HuH-7–Cas9) or n = 7 mice (JHH-7–Cas9). Comparisons among groups were performed with ordinary one-way ANOVA (FHuH-7 [8, 45] = 247.5; FJHH-7 [8, 63] = 206; P < 0.000001) followed by Tukey multiple comparison test (P values in SI Appendix, Tables S1 and S2). (D and E) Heat map showing differentially expressed genes in HuH-7–Cas9 and JHH-7–Cas9 cells after CRISPR inactivation of indicated genes relative to control (sgNTC). The genes were filtered as described in Materials and Methods. Row-wise z score transformation was performed. DKO, double knockout; FC, fold change; KO, knockout; TKO, triple knockout.
In HuH-7 tumors, compared with control cells, FGFR3- or FGFR4-knockout cells were depleted by 10- and 57-fold respectively, whereas KLB-knockout cells were depleted by 206-fold. Dual knockout of FGFR3 and FGFR4 resulted in 607-fold of depletion (Fig. 5B and SI Appendix, Table S1). In JHH-7 tumors, single knockout of FGFRs did not cause significant depletion, whereas KLB knockout resulted in 9.5-fold of depletion. FGFR3 and FGFR4 double knockout led to 5.8-fold of depletion. FGFR1, FGFR3, and FGFR4 triple knockout resulted in 8.2-fold of depletion (Fig. 5C and SI Appendix, Table S2). Thus, multiple FGFRs redundantly promote FGF19-positive tumor growth.
We further performed RNA sequencing of HuH-7 and JHH-7 cells after CRISPR inactivation of KLB or various combinations of FGFR1, FGFR3, and FGFR4 (Dataset S2). Gene ontology analyses revealed that KLB inactivation in HuH-7 and JHH-7 cells up-regulated genes involved in steroid and sterol metabolic processes, consistent with the known endocrine function of FGF19 signaling (11). KLB inactivation down-regulated genes involved in DNA replication and response to nutrient levels in HuH-7 and JHH-7 cells, respectively (SI Appendix, Fig. S4 A and B). We further generated heat maps of differentially expressed genes to visualize the impact of FGFR redundancy in regulating gene expression. In HuH-7 cells, FGFR3 and FGFR4 dual inactivation resulted in a gene expression profile similar to that resulting from KLB inactivation, which was dissimilar to that resulting from FGFR4 inactivation (Fig. 5D). In JHH-7 cells, FGFR1, FGFR3, and FGFR4 triple inactivation, but not FGFR4 single inactivation, displayed a gene expression profile similar to that resulting from KLB inactivation (Fig. 5E). Taken together, transcriptome analyses further support the existence of FGFR redundancy in shaping the gene expression programs of FGF19-positive HCC.
Prevalent Coexpression of FGFR3 and FGFR4 in FGF19-Positive HCC.
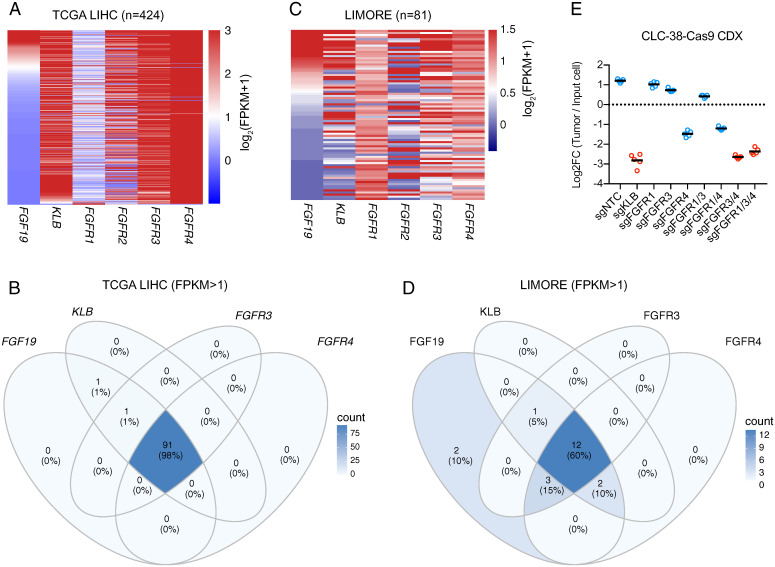
To examine whether FGFR redundancy could be observed in human patients, we queried the RNA expression data of 424 hepatocellular carcinoma cases from the Cancer Genome Atlas (TCGA) (Fig. 6A). Approximately 20% of these cases expressed FGF19 mRNA, based on a cutoff of 1 fragment per kilobase transcript per million mapped reads (FPKM). Nighty-eight percent of these FGF19-positive HCC tumors coexpressed FGFR3, FGFR4, and KLB mRNAs (Fig. 6 A and B). We further analyzed genetic alterations of FGF19, KLB, and FGFR1/2/3/4 in these cases and found that only FGF19 amplification was enriched (13% of cases) in this patient cohort (SI Appendix, Fig. S5).
Fig. 6.
Prevalent coexpression of FGFR3 and FGFR4 in FGF19-positive HCC cases and cell lines. (A) Heat map depicting RNA expression levels of indicated genes in HCC cases from the TCGA LIHC dataset. (B) Venn diagram indicating TCGA LIHC cases coexpressing indicated genes (FPKM >1). (C) Heat map depicting RNA expression levels of indicated genes from the LIMORE cell line collection. (D) Venn diagram indicating LIMORE cell lines coexpressing indicated genes (FPKM >1). (E) Relative abundance (log2 transformed) of CLC38-Cas9 cells harboring indicated combinations of sgRNAs from subcutaneous tumor xenografts normalized to input cells. Bars represent averages from n = 5 nude mice. Comparisons among groups were performed with ordinary one-way ANOVA (F [8, 36] = 588.9; P < 0.000001) followed by Tukey multiple comparison test (P values in SI Appendix, Table S3).
We further queried the RNA expression data of the Liver Cancer Model Repository (LIMORE), which is composed of 81 human liver cancer cell line models (31), and also observed the coexpression of FGFR3, FGFR4, and KLB mRNAs in 60% of FGF19-positive cell lines (Fig. 6 C and D). Most cell lines in LIMORE were unique to the collection, and their genetic dependencies have not been profiled, providing us an opportunity to test the generalizability of our findings. We identified three cell lines (CLC-4, CLC-37, and CLC-38) that expressed FGF19, KLB, and FGFR4 mRNAs (SI Appendix, Fig. S6A). Among the three cell lines, CLC-4 was not tumorigenic, whereas CLC-37 did not express detectable FGFR3 or FGFR4 protein by Western blotting (SI Appendix, Fig. S6B). We therefore performed a xenograft experiment with the remaining cell line, CLC-38. In CLC-38 tumors, inactivation of KLB and FGFR4 resulted in 16- and 6.4-fold of depletion, respectively. Dual inactivation of FGFR3 and FGFR4 resulted in 14.4-fold of depletion (Fig. 6E and SI Appendix, Table S3). Taken together, FGFR redundancy in mediating FGF19 signaling may be a general phenomenon in human patients with HCC.
Discussion
The rationale for developing FGFR4-selective inhibitors to treat FGF19-dependent HCC is that FGFR4 is the sole receptor mediating the tumorigenic function of FGF19. This premise is largely based on experiments conducted in mouse models. Mice lacking either Fgf15 (the mouse ortholog of human FGF19) or Fgfr4 fail to maintain bile acid homeostasis and have increased bile acid pool relative to wild-type mice (8, 9). Furthermore, while ectopic expression of human FGF19 in skeletal muscle promotes HCC in mice, breeding these transgenic mice with Fgfr4-knockout mice prevented HCC development (13, 14). In our study, we used multiple FGF19-positive cell lines to demonstrate that FGFR3 and FGFR4 were functionally redundant in mediating the oncogenic function of FGF19. One possible explanation for the apparent discrepancy between our study and prior studies is the different modes of FGF19 (Fgf15) signaling. In studies conducted in mice, FGF19 (Fgf15) is secreted by distant tissues as an endocrine factor, whereas in human HCC cell lines, FGF19 is constitutively expressed as an autocrine factor. Therefore, HCC cells may experience constant high local concentrations of FGF19 that can activate both FGFR4 and FGFR3 in complex with KLB. The ability of FGFR3 to mediate FGF19 (Fgf15) signaling has previously been hinted in the study of gallbladder filling with bile acids. Fgf15-knockout mice showed a more pronounced reduction in gallbladder volume than Fgfr4-knockout mice. Moreover, FGF19 administration increased gallbladder volume in FGFR4-knockout mice. The fact that FGFR3 is the most abundantly expressed FGFR in the gallbladder suggests that FGFR3 may mediate the gallbladder filling activity of FGF19 (32).
Although FGFR3 and FGFR4 are redundant, they are not functionally equivalent in FGF19-positive HCC. Phenotypically, FGFR4 knockout caused a mild fitness defect, whereas FGFR3 knockout did not cause a measurable fitness defect in HCC cells. Interestingly, KLB mutants that displayed weakened interaction with FGF19 also displayed weakened interaction with FGFR3, whereas their interaction with FGFR4 was maintained (Fig. 3C). These results suggest that the KLB-FGFR4 complex can form in the absence of FGF19 binding, whereas KLB-FGFR3 complex formation requires FGF19 binding. In addition to FGF19, other FGF ligands can activate FGFR3 and/or FGFR4 in a KLB-dependent or -independent manner. Whether FGF ligand redundancy can drive resistance to FGFR4-specific inhibitors in FGF19-positive HCC warrants further investigation.
Current FGFR inhibitors in clinical use or in development can be categorized into pan-FGFR inhibitors, FGFR1/2/3 inhibitors, and selective FGFR4 inhibitors (33). Recent studies of adaptive resistance to FGFR inhibitors in FGFR-altered triple-negative breast cancer and cholangiocarcinoma revealed that feedback activation of mTORC1, YAP, or EGFR signaling limits FGFR inhibitor efficacy (34, 35). Our observation that FGFR3 inactivation sensitized FGF19-positive HCC to selective FGFR4 inhibitors provides a potential mechanism of de novo resistance to FGFR4-selective inhibitors in FGF19-positive HCC, which can be overcome by pan-FGFR inhibitors. A previous study observed synergistic antitumor activity between multikinase inhibitors (lenvatinib, sorafenib, and regorafenib) and a selective FGFR4 inhibitor (BLU-9931) in FGF19-positive HCC cell lines (36). In light of our findings, such a synergy could be explained by FGFR redundancy. Taken together, the redundancy of growth signaling pathways could be a common mechanism driving intrinsic or adaptive resistance to FGFR inhibitors.
Materials and Methods
Cell Lines and Cell Culture.
All cell lines were grown under standard conditions in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco), except for Li-7 and CLC-38, which were grown in RPMI-1640 medium (Gibco). Culture medium was supplemented with 10% fetal bovine serum (FBS) (Gibco), 2 mM glutamine (Invitrogen), 100 U/mL penicillin (Gibco), and 0.1 mg/mL streptomycin (Gibco). For CLC-38, 40 ng/mL EGF (Sino Biological), 1× Insulin-Transferrin-Selenium (ITS, Thermo Fisher Scientific), and 10 μM Y27632 (TargetMol) were added to the culture medium (31). All cell lines were confirmed to be mycoplasma free by PCR. Cas9-expressing stable cell lines were constructed by lentivirus infection followed by 50 μg/mL blasticidin (InvivoGen) selection. HuH-7 and JHH-7 cell lines stably expressing exogenous KLB were constructed by lentivirus infection followed by 5 μg/mL puromycin (InvivoGen) selection.
Mice and Tumor Xenograft.
For subcutaneous transplantation, 8-wk-old NU/NU nude female mice, which were housed in the specific pathogen-free (SPF) unit of the National Institute of Biological Sciences, Beijing, China, were used. All animal experimental procedures were approved by the Experimental Animal Welfare Ethical Committee of the National Institute of Biological Sciences, Beijing, China. For tumor inoculation into mice, cultured HCC cells were trypsinized, washed three times with Dulbecco’s phosphate-buffered saline (DPBS), and resuspended in DPBS. Five million HuH-7 cells, 5 million CLC-38 cells, or 1 million JHH-7 cells were injected subcutaneously in 125 μL of PBS into the left flank of recipient mice. Tumor growth was monitored by caliper measurement to determine growth. Tumor volume was calculated as ([length × width2] × 3.14)/6. At the end point of the experiment, tumors were dissected, and GFP fluorescence images were taken using Eclipse Ni-E Microscope (Nikon Instruments, Inc.). For in vivo competitive tumor growth assay, Cas9-expressing stable cell lines were separately infected with different barcoded polycistronic sgRNAs lentivirus, equally mixed, and then subcutaneously implanted into mice. Tumors were harvested 1 mo later, and the relative abundance of indicated combinations of sgRNAs was measured by barcode sequencing.
Generation of Single sgRNA and Barcoded Polycistronic Vectors.
A single sgRNA lentiviral vector harboring an hU6-sgRNA cassette and an SFFV-mNeonGreen-P2A-ZsGreen1 (referred to as GFP thereafter) cassette was constructed by a standard molecular cloning technique. Annealed sgRNA oligos were inserted into the vector digested by BsmBI. Polycistronic vectors were generated by inserting bU6-sgRNA and mU6-sgRNA cassettes derived from pMJ114 (bovine U6) and pMJ179 (mouse U6) (Addgene: 85995 and 85996) into the single sgRNA vector as previously described (30). A set of 20-nucleotide barcode sequences were then inserted, resulting in a collection of barcoded polycistronic sgRNA vectors.
Western Blotting.
Standard sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting procedures were used, with the following modifications. For preparation of total lysates, cells were washed with DPBS to remove residual medium and then lysed in 20 mM Hepes-NaOH (pH 8.0), 10 mM NaCl, 2 mM MgCl 2, and 1% SDS freshly supplemented with 0.5 unit/mL benzonase (Yeasen) and 1× cOmplete Mini EDTA-free protease inhibitor mixture (Roche). Protein concentrations of the resulting lysates were quantified by the BCA method. Between 30 and 60 μg proteins were resolved on SDS-PAGE and transferred to nitrocellulose membranes with a pore size of 0.5 mm. Membranes were blocked in 5% nonfat milk phosphate-buffered saline with Tween 20 (PBST) (0.1% vol/vol Tween 20) at room temperature for 30 min before blotting with antibodies. Primary antibodies and horseradish peroxidase (HRP)–linked secondary antibodies were diluted in 5% nonfat milk PBST. Membranes were incubated with primary antibodies at 4 °C overnight and then washed with PBST at room temperature three times. Afterward, membranes were further incubated with secondary antibodies at room temperature for 2 h and then washed with PBST at room temperature three times. M5 HiPer ECL Western HRP Substrate (Mei5bio) was used for the detection of HRP enzymatic activity. Western blot images were taken with the Fusion FX (Vilber) imager. The following antibodies were used: anti-Flag–HRP (Sigma-Aldrich, A8592; 1:5,000), anti-FGFR1 (Cell Signaling Technology, 9740; 1:1,000), anti-FGFR3 (Cell Signaling Technology, 4574; 1:1,000), anti-FGFR4 (Cell Signaling Technology, 8562; 1:5,000), anti-FGF19 (ABclonal, A6589; 1:2,000), anti-KLB (R&D Systems, MAB5889; 1:1,000), anti-PARP1 (Cell Signaling Technology, 9542; 1:5,000), anti–β actin–HRP (Huaxingbio, HX1827; 1:10,000).
CETSA.
HuH-7 or JHH-7 cells were plated in cell culture plates. After overnight attachment, cells were treated with indicated concentrations of FGFR4 inhibitors for 12 h. Cells were scraped off plates, washed twice with DPBS, and then resuspended in DPBS. Cell suspensions were equally divided into several 0.2-mL PCR tubes, heated at designated temperature for 3 min, and then incubated at 25 °C for 3 min in a thermal cycler. Afterward, heat-treated cell suspensions were transferred into 1.5-mL microcentrifuge tubes and supplemented with protease inhibitor. Cells were lysed by three freeze-thaw cycles, and the resulting lysates were centrifuged at 20,000 g for 10 min at 4 °C to pellet cell debris together with aggregated proteins. The supernatant was collected for Western blotting.
Crystal Violet Staining.
For overnight attachment, 100,000 HuH-7 or JHH-7 cells per well were plated in a six-well plate. Afterward, cells were treated with indicated concentrations of FGFR4 inhibitors. Six days later, cells were washed with DPBS, fixed for 10 min in 10% formalin at room temperature, and then stained for 20 min with 0.05% crystal violet (Yeasen). Plates were washed and imaged by Fusion FX (Vilber).
Competitive Cell Growth Assay.
Cas9-transduced cell lines were infected with single sgRNA lentivirus at a low multiplicity of infection (0.2∼0.4). The infected and uninfected cell mixtures were then passaged every 3 d, and the percentage of transduced GFP-positive cells was measured by flow cytometry.
Measurements of Inhibitor IC50 on Cell Viability.
BLU-554 (Targetmol), FGF-401 (Targetmol), and erdafitinib (Targetmol) were prepared as 10-mM stocks in dimethyl sulfoxide (DMSO). In 100 μL culture medium per well, 3,000 HuH-7 cells, 3,000 JHH-7 cells, or 1,000 FU97 cells were plated in 96-well flat clear-bottom white polystyrene tissue culture (TC)–treated microplates (Corning). After overnight attachment, cells were dosed with a serial dilution of compounds. Cell survival was measured 3 (HuH-7 and JHH-7) or 6 d (FU97) later using the CellTiter-Glo Luminescent Cell Viability Assay Kit (Promega) following vendor instructions. Luminescence was recorded by an EnVison multimode plate reader (PerkinElmer). IC50 was determined with GraphPad Prism using baseline correction (by normalizing to DMSO control), the asymmetric (four-parameter) equation, and least squares fit.
Genome-Wide CRISPR Screen.
The human CRISPR Brunello library (26) (77,441 sgRNAs consisting of an average of 4 sgRNAs per gene and 1,000 nontargeting control sgRNAs) was transduced into HuH-7–Cas9 at a low multiplicity of infection (0.2∼0.3) and a coverage of ∼400 cells per sgRNA. After 2 w of puromycin (2 μg/mL) selection, transduced cells were treated with vehicle DMSO (0.04%) or BLU-554 (40 nM) for 3 w. Library preparation for sequencing was carried out in PCR performed on genomic DNA isolated from cells. Sequencing reads were analyzed by MAGeCK (27) to determine relative sgRNA abundance.
KLB-3×FLAG IP-MS.
Anti-FLAG–conjugated beads were prepared as previously described (37). HuH-7 cell lines stably expressing doxycycline-inducible KLB-3×Flag were cultured in the presence of doxycycline with three different doses (0, 4, and 20 ng/mL) for 2 d. Cells were detached from plates by scraping, washed in DPBS, and frozen in liquid nitrogen. Frozen cells were pulverized using a mixer mill MM 400 (Retsch) with two rounds of 1-min ball milling at 30 Hz. Per experiment, 500 mg cell powder was suspended with 2 mL lysis buffer (25 mM Hepes [pH 7.4] and 10 mM KCl), supplemented with 1× cOmplete Mini EDTA-free protease inhibitor mixture (Roche), and incubated on ice for 5 min. The resulting lysates were centrifuged at 15,000 g at 4 °C for 5 min. Afterward, pellets containing membrane fractions were washed with lysis buffer three times and then solubilized with 2 mL IP buffer (25 mM Hepes [pH 7.4], 300 mM NaCl, 1% wt/wt lauryl maltose neopentyl glycol [LMNG], and 0.1% wt/wt cholesteryl hemisuccinate [CHS]), supplemented with 1× cOmplete Mini EDTA-free protease inhibitor mixture (Roche), and incubated on ice for 5 min. The resulting lysates were centrifuged at 15,000 g for 5 min at 4 °C. Afterward, 30 μL anti-FLAG–conjugated magnetic beads were mixed with clarified lysates on a rotating platform at 4 °C for 15 min, followed by three washes with washing buffer (25 mM Hepes [pH 7.4], 300 mM NaCl, 0.1% wt/wt LMNG, and 0.01% wt/wt CHS). Bound proteins were eluted from magnetic beads with 30 μL 1-mg/mL 3× FLAG peptide (Sigma-Aldrich) with agitation at 4 °C for 30 min. The final eluted proteins were subjected to in-gel digestion following protein identification by MS as described (38).
RNA Sequencing.
Total RNA was isolated from in vitro cultured cells using TRNzol (Tiangen). RNA sequencing library construction and sequencing were performed by Berry Genomics. Sequencing reads were aligned to the Homo sapiens GRCh38 reference transcriptome to obtain raw read counts, and the differential expression genes were analyzed using DESeq2 (39) followed by filtering for average FPKM greater than 5, log2-transformed fold changes greater than 1 and 1.5, and adjusted P values less than 0.05 and 0.01 for HuH-7 and JHH-7, respectively. Heat maps were generated with pheatmap in R. Gene ontology analysis of differentially expressed genes was performed with clusterProfiler (40).
TCGA and LIMORE Analyses.
RNA sequencing data of TCGA liver HCC (LIHC) cases were downloaded from the University of California Santa Cruz Xena browser (41). RNA sequencing data of LIMORE were downloaded from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) (accession: GSE97098). Heat map and Venn diagram were generated with pheatmap and ggVennDiagram in R. A cutoff of 1 FPKM was used to specify FGF19-, FGFR-, and KLB-positive cases. Genetic alteration analysis was performed with cBioPortal (42, 43).
Statistical Analyses.
Comparisons among groups were performed with ordinary one-way ANOVA followed by post hoc Tukey or Dunnett multiple comparison corrections. Statistical analyses were conducted with GraphPad Prism.
Supplementary Material
Acknowledgments
We thank Shuang Li for help with the in vitro competitive growth assay. The research reported here was supported by the Beijing Municipal Commission of Science and Technology (Z201100005320010 and Z211100003321009) (T.H.) and intramural funding from the National Institute of Biological Sciences, Beijing, and Tsinghua Institute of Multidisciplinary Biomedical Research (T.H.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2208844119/-/DCSupplemental.
Data Availability
RNA sequencing data have been deposited in GEO (GSE208350) (44). All study data are included in the article and/or supporting information.
References
- 1.Ornitz D. M., Itoh N., The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 4, 215–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemmon M. A., Schlessinger J., Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hatlen M. A., et al. , Acquired on-target clinical resistance validates FGFR4 as a driver of hepatocellular carcinoma. Cancer Discov. 9, 1686–1695 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Katoh M., Fibroblast growth factor receptors as treatment targets in clinical oncology. Nat. Rev. Clin. Oncol. 16, 105–122 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Loriot Y., et al. ; BLC2001 Study Group, Erdafitinib in locally advanced or metastatic urothelial carcinoma. N. Engl. J. Med. 381, 338–348 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Abou-Alfa G. K., et al. , Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: A multicentre, open-label, phase 2 study. Lancet Oncol. 21, 671–684 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Javle M., et al. , Infigratinib (BGJ398) in previously treated patients with advanced or metastatic cholangiocarcinoma with FGFR2 fusions or rearrangements: Mature results from a multicentre, open-label, single-arm, phase 2 study. Lancet Gastroenterol. Hepatol. 6, 803–815 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Yu C., et al. , Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J. Biol. Chem. 275, 15482–15489 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Inagaki T., et al. , Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2, 217–225 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Ito S., et al. , Impaired negative feedback suppression of bile acid synthesis in mice lacking betaKlotho. J. Clin. Invest. 115, 2202–2208 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gadaleta R. M., Moschetta A., Metabolic Messengers: Fibroblast growth factor 15/19. Nat. Metab. 1, 588–594 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Sawey E. T., et al. , Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell 19, 347–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholes K., et al. , A mouse model of hepatocellular carcinoma: Ectopic expression of fibroblast growth factor 19 in skeletal muscle of transgenic mice. Am. J. Pathol. 160, 2295–2307 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.French D. M., et al. , Targeting FGFR4 inhibits hepatocellular carcinoma in preclinical mouse models. PLoS One 7, e36713 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagel M., et al. , First selective small molecule inhibitor of FGFR4 for the treatment of hepatocellular carcinomas with an activated FGFR4 signaling pathway. Cancer Discov. 5, 424–437 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Fairhurst R. A., et al. , Discovery of roblitinib (FGF401) as a reversible-covalent inhibitor of the kinase activity of fibroblast growth factor receptor 4. J. Med. Chem. 63, 12542–12573 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Joshi J. J., et al. , H3B-6527 is a potent and selective inhibitor of FGFR4 in FGF19-driven hepatocellular carcinoma. Cancer Res. 77, 6999–7013 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Kim R. D., et al. , First-in-human phase I study of fisogatinib (BLU-554) validates aberrant FGF19 signaling as a driver event in hepatocellular carcinoma. Cancer Discov. 9, 1696–1707 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Martinez Molina D., et al. , Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Tsherniak A., et al. , Defining a cancer dependency map. Cell 170, 564–576.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyers R. M., et al. , Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan E. M., et al. , WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature 568, 551–556 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuro-O M., The Klotho proteins in health and disease. Nat. Rev. Nephrol. 15, 27–44 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Lee S., et al. , Structures of β-klotho reveal a ‘zip code’-like mechanism for endocrine FGF signalling. Nature 553, 501–505 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen G., et al. , α-klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling. Nature 553, 461–466 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanson K. R., et al. , Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 9, 5416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W., et al. , MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karkera J. D., et al. , Oncogenic characterization and pharmacologic sensitivity of activating fibroblast growth factor receptor (FGFR) genetic alterations to the selective FGFR inhibitor erdafitinib. Mol. Cancer Ther. 16, 1717–1726 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Perera T. P. S., et al. , Discovery and pharmacological characterization of JNJ-42756493 (Erdafitinib), a functionally selective small-molecule FGFR family inhibitor. Mol. Cancer Ther. 16, 1010–1020 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Adamson B., et al. , A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882 e1821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu Z., et al. , A pharmacogenomic landscape in human liver cancers. Cancer Cell 36, 179–193.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi M., et al. , Identification of a hormonal basis for gallbladder filling. Nat. Med. 12, 1253–1255 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Facchinetti F., et al. , Facts and new hopes on selective FGFR inhibitors in solid tumors. Clin. Cancer Res. 26, 764–774 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y., et al. , FGFR-inhibitor-mediated dismissal of SWI/SNF complexes from YAP-dependent enhancers induces adaptive therapeutic resistance. Nat. Cell Biol. 23, 1187–1198 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Wu Q., et al. , EGFR inhibition potentiates FGFR inhibitor therapy and overcomes resistance in FGFR2 fusion-positive cholangiocarcinoma. Cancer Discov. 12, 1378–1395 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanzaki H., et al. , The impact of FGF19/FGFR4 signaling inhibition in antitumor activity of multi-kinase inhibitors in hepatocellular carcinoma. Sci. Rep. 11, 5303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lv L., et al. , Discovery of a molecular glue promoting CDK12-DDB1 interaction to trigger cyclin K degradation. eLife 9, e59994 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han T., et al. , Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 356, eaal3755 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu G., Wang L. G., Han Y., He Q. Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldman M. J., et al. , Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38, 675–678 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cerami E., et al. , The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao J., et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao Z., Cui Y., Xu X. and Han T., FGFR redundancy limits the efficacy of FGFR4-selective inhibitors in hepatocellular carcinoma. GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE208350. Deposited 17 July 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data have been deposited in GEO (GSE208350) (44). All study data are included in the article and/or supporting information.