Significance
How parental epigenomes influence offspring development and health is poorly understood. Here, we used the model organism Caenorhabditis elegans to elucidate the role of sperm-inherited histone marking in shaping offspring gene expression. We found that absence of the conserved repressive mark H3K27me3 from the sperm genome caused up-regulation from sperm alleles in offspring somatic and germline tissues, and that tissue context determined which genes were prone to up-regulation. Our findings in worms mirror findings in mammalian cells and implicate a conserved mechanism for chromatin-based regulation between worms and mammals. Finally, we showed that the sperm-inherited pattern of gene marking and gene up-regulation were transmitted to grandoffspring, demonstrating that H3K27me3 marking can serve as a bona fide transgenerational epigenetic carrier in C. elegans.
Keywords: epigenetic inheritance, gene regulation, H3K27me3, C. elegans, transgenerational
Abstract
The transmission of chromatin states from parent cells to daughter cells preserves cell-specific transcriptional states and thus cell identity through cell division. The mechanism that underpins this process is not fully understood. The role that chromatin states serve in transmitting gene expression information across generations via sperm and oocytes is even less understood. Here, we utilized a model in which Caenorhabditis elegans sperm and oocyte alleles were inherited in different states of the repressive mark H3K27me3. This resulted in the alleles achieving different transcriptional states within the nuclei of offspring. Using this model, we showed that sperm alleles inherited without H3K27me3 were sensitive to up-regulation in offspring somatic and germline tissues, and tissue context determined which genes were up-regulated. We found that the subset of sperm alleles that were up-regulated in offspring germlines retained the H3K27me3(−) state and were transmitted to grandoffspring as H3K27me3(−) and up-regulated epialleles, demonstrating that H3K27me3 can serve as a transgenerational epigenetic carrier in C. elegans.
An area of intense interest is regulation of development by epigenetic mechanisms involving factors beyond the DNA code. For all animals, each generation emerges from the union of two germ cells, an oocyte and a sperm, that in some species carry a transcriptional memory of the germ tissue from which they arose (1–3). During embryogenesis, this germline memory must be “forgotten” in somatic lineages and either “remembered” or re-established in the nascent germ lineage. Transgenerational epigenetic inheritance, defined as spanning at least three generations, requires that some epigenetic memory survive reprogramming events that occur during embryogenesis and germline development (4, 5). Studies of humans suggest that various lifestyle and dietary conditions can alter epigenetic information carried in sperm or oocytes, and epigenetic mechanisms underlie links between conditions experienced by parents and the health outcomes of their offspring (6, 7). However, studies of humans are limited in their ability to identify the mechanisms that underlie these processes. Studies in model organisms are illuminating how epigenetic inheritance can shape the development and health of future generations.
A major challenge in the field of epigenetic inheritance is demonstrating a causal link between changes to the parental epigenome carried in the sperm or oocyte and changes in offspring gene expression and development. Epigenetic information can be carried in different forms. DNA methylation, histone modifications, and noncoding RNAs (ncRNAs) are the three most heavily studied epigenetic carriers (4). After transmission to the embryo, DNA methylation and histone modifications reside at the loci whose transcription they regulate, and so are referred to as cis epigenetic regulators. In contrast, ncRNAs can diffuse through the nucleoplasm and affect transcription from multiple genomic loci, and so are referred to as trans epigenetic regulators. This feature of ncRNAs has allowed researchers to inject sperm-derived ncRNAs into fertilized wild-type oocytes and demonstrate their role in shaping embryonic gene expression and development (8–12). Demonstrating a similar role for cis regulators requires a different strategy. A unique feature of epigenetic information carried in cis is that it can regulate expression in an allele-specific manner, resulting in different transcriptional outputs from sperm-inherited and oocyte-inherited alleles within a single nucleus. This feature is what led to the discovery of parental imprinting in which the sperm and oocyte contribute different patterns of DNA methylation that drive different transcriptional states of sperm versus oocyte alleles (13).
We developed a paradigm in which we could exploit the cis regulatory feature of histone modifications to test whether sperm-inherited patterns of histone marking directly influence transcription in offspring (14). In that study, we showed that in Caenorhabditis elegans, parental alleles inherited with different states of the repressive histone mark H3K27me3 (histone H3 trimethylated on Lys-27) could achieve different transcriptional outputs in the germline of offspring. We attributed these transcriptional differences to the differential inheritance of H3K27me3 on parental alleles, since C. elegans lacks canonical DNA methylation on cytosines. Importantly, these findings demonstrated that histone modifications, or “marks,” inherited on gamete genomes directly influence offspring transcription in cis and thus represent a bona fide intergenerational epigenetic carrier. Recent studies in mice have shown that changes to sperm-inherited H3K4me3 impact offspring gene expression and development (15). These studies demonstrate a conserved role for sperm-inherited histone marks in the epigenetic transmission of gene expression information across generations. However, our mechanistic understanding of this process is limited.
To transmit information across generations requires that gamete-inherited patterns of histone marking are maintained through cell division. We have a strong foundation for understanding how histone modifications are transmitted from parent chromosomes to newly replicated daughter chromosomes during cell division (16–19) and how those parent-cell-derived modifications help reestablish the chromatin landscape in daughter cells (20, 21). This process is probably best understood for the conserved mark of repression H3K27me3 and the enzyme that generates it polycomb repressive complex 2 (PRC2). For instance, histones bearing H3K27me3 have been shown to maintain their genomic positioning when transferred from parent chromosomes to daughter chromosomes (22) where their presence can stimulate PRC2 to generate H3K27me3 on unmarked neighboring histones (20, 21, 23) and thus restore parental patterns of H3K27me3. It is thought that preserving H3K27me3 domains is an important factor in maintaining gene repression and cell identity through cell division. This model is supported by in vivo studies that demonstrated a causal role for the mark itself in maintaining gene repression (24–26). Despite this, controversies remain over the importance of parental templating of H3K27me3 in propagating H3K27me3 landscapes through cell division and in maintaining gene repression (24, 27). Indeed, recent studies have demonstrated that PRC2 can reestablish appropriate patterns of H3K27me3 in the absence of preexisting H3K27me3 (28, 29). These controversies highlight our limited understanding of this process even in a relatively simple paradigm in which chromatin landscapes participate in the maintenance of transcriptional programs through cell division. The situation is more challenging when cell- and tissue-specific transcriptional programs and chromatin landscapes are being established during development.
To investigate the mechanisms through which gamete-inherited histone marks influence transcriptional programs in offspring, we developed a model in which we could assess sperm and oocyte allele-specific transcription patterns in worms that were genetically engineered to inherit the sperm genome lacking H3K27me3 (14). Our initial analysis indicated that the H3K27me3(−) state of sperm-inherited chromosomes was maintained in all cells during early embryogenesis, followed by de novo appearance of H3K27me3 immunostaining on the sperm chromosomes as cell lineages launched their transcriptional programs (14, 30). Focusing on the germline of offspring revealed that inheritance of the sperm genome lacking H3K27me3 resulted in the up-regulation of some tissue-inappropriate transcripts specifically from sperm alleles (14). These findings suggested that in the germline of offspring, while the H3K27me3 landscape was largely reestablished on the H3K27me3(−) sperm-inherited genome, some sperm-inherited alleles escaped H3K27me3 repression. These findings also highlighted several burning questions in the field that our genetic model was uniquely poised to address: 1) In different cellular contexts, are different subsets of genes sensitive to gamete-inherited H3K27me3 states? 2) How are gamete-inherited H3K27me3 states preserved or rewritten at the gene level in mature tissues? and 3) Does inheritance of a gamete genome lacking H3K27me3 influence transcription in grandoffspring as well as in offspring, in other words transgenerationally? In the present study, we found that inheriting the sperm genome lacking H3K27me3 resulted in up-regulated genes in each tissue context analyzed, many of which were up-regulated specifically from the sperm allele. Up-regulated genes were mainly tissue-specific, indicating that cellular context in offspring dictated which genes were sensitive to the absence of parentally inherited H3K27me3. Our analysis of transcript and chromatin profiles in germlines revealed that genes that were up-regulated in offspring maintained the inherited H3K27me3(−) state of sperm alleles, while genes that were not up-regulated reestablished H3K27me3 on the sperm allele, consistent with preexisting H3K27me3 not being required to direct de novo H3K27me3 deposition (28, 29). Notably, we found that for genes up-regulated in offspring germlines, the H3K27me3(−) and up-regulated state of sperm alleles was transmitted to grandoffspring germlines. Our findings indicate that in C. elegans, the H3K27me3 status of a gene can be inherited in a Mendelian fashion, in which allele-specific H3K27me3 states or “epialleles” are packaged into sperm and oocyte and delivered to the next generation where they drive allele-specific transcription patterns. This exemplifies that H3K27me3 marking can transmit epigenetic information transgenerationally.
Results
In K27me3 M+P− Worms, Sperm Alleles Were Up-Regulated in Both Soma and Germline.
To generate F1 offspring worms that inherited a sperm genome lacking H3K27me3, we mated wild-type mothers with fathers that lacked a critical component of PRC2, the enzyme that generates H3K27me3 (Fig. 1A). We refer to these F1 offspring as K27me3 M+P− and their genetically identical controls as K27me3 M+P+ (M+ for maternal H3K27me3(+), P− or P+ for paternal H3K27me3(−) or H3K27me3(+), respectively). To distinguish transcript reads that emanated from sperm versus oocyte alleles, we created these F1 worms using parents whose genomes can be distinguished by single nucleotide polymorphisms (SNPs). We achieved this by using mothers from the Hawaii (CB4856) background and fathers from the Bristol (N2) background. As a result, we could assign SNP-containing RNA sequences to having originated from the sperm or oocyte allele.
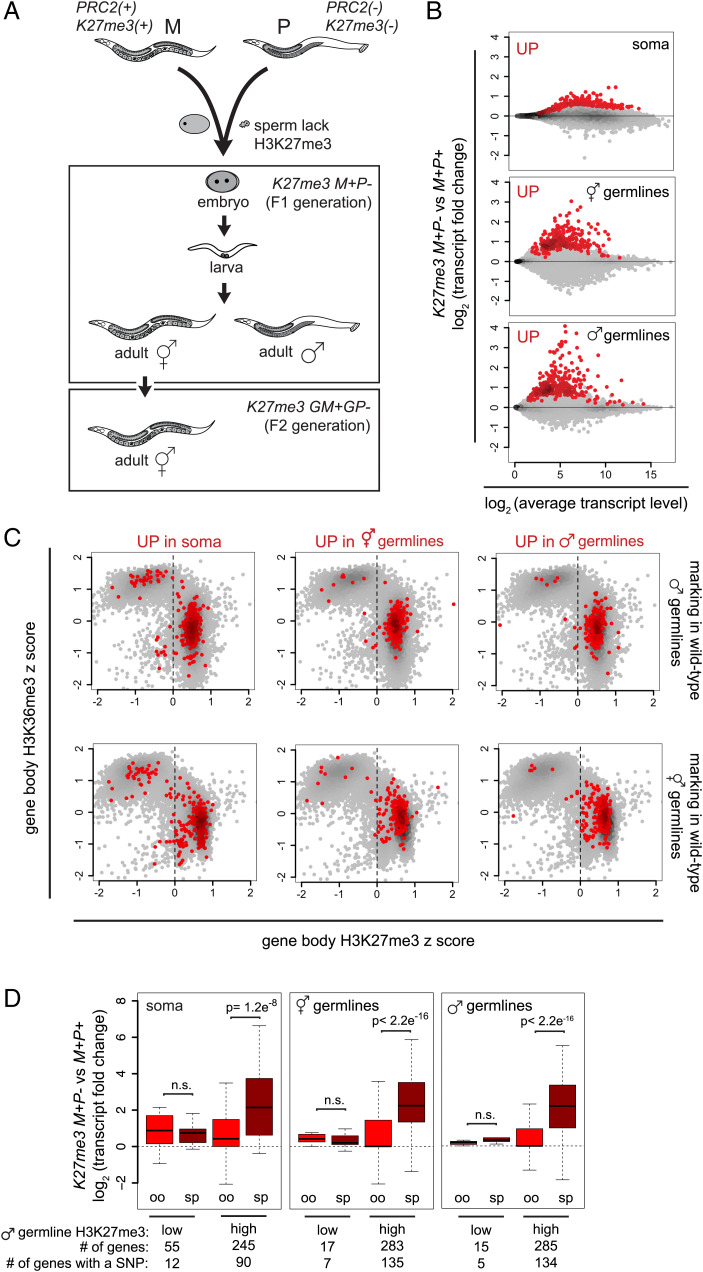
Fig. 1.
In K27me3 M+P− offspring, different tissues up-regulate genes specifically from sperm-inherited alleles. (A) Diagram showing crosses between PRC2(+) K27me3(+) maternal (M) parents and mes-3/mes-3 PRC2(−) K27me3(−) paternal (P) parents that generated K27me3 M+P− (maternal and paternal) F1 offspring and K27me3 GM+GP− (grandmaternal and grandpaternal) F2 grandoffspring. The Upper Box displays different tissue contexts in the F1 generation, including newly hatched L1 larvae (source of soma), adult hermaphrodites (source of oogenic germlines), and adult males (source of spermatogenic germlines). The Lower Box displays the F2 generation. (B) MA plots of K27me3 M+P− vs. K27me3 M+P+ L1s, which are >99% somatic cells, oogenic hermaphrodite germlines, and spermatogenic male germlines, showing the top 300 up-regulated genes in red. SI Appendix, Fig. S1A includes MA plots showing significantly up-regulated and down-regulated genes. (C) Scatterplots of H3K27me3 vs. H3K36me3 gene body enrichment on genes in “wild-type” (him-8) male germlines (Upper Plots) and in “wild-type” (K27me3 M+P+) hermaphrodite germlines (Lower Plots) with the top 300 up-regulated genes in different F1 tissue contexts in red. Positive z scores indicate gene body enrichment of H3K27me3 or H3K36me3 relative to all gene bodies. Dashed lines separate genes with “low” and “high” H3K27me3 marking. (D) Boxplots of oocyte (oo) and sperm (sp) allele-specific changes in transcript levels for the top 300 up-regulated genes with low or high H3K27me3 marking in wild-type male germlines and containing a SNP. SI Appendix, Fig. S1B includes boxplot analysis of significantly up-regulated genes. Boxplots show the median (horizontal line), the 25th and 75th percentiles (boxes), and whiskers extending to the most extreme data point, which is no more than 1.5 times the interquartile range. P values were calculated using Student’s t test (two-tailed).
Our previous comparison of transcript levels in the germline of K27me3 M+P− and K27me3 M+P+ hermaphrodite offspring revealed that, although the entire sperm genome was inherited lacking H3K27me3, only a subset of sperm alleles was up-regulated in K27me3 M+P− germlines. We wondered if different F1 tissues also selectively up-regulate sperm alleles. We tested this by comparing patterns of gene misexpression in three different tissues in K27me3 M+P− F1 offspring: adult hermaphrodite oogenic germlines, adult male spermatogenic germlines, and larval somatic cells (Fig. 1A). Newly hatched L1 larvae consist of greater than 99% somatic cells (556 somatic cells and two germ cells). Thus, transcripts from L1 larvae represent gene expression from various somatic tissues. We also investigated if genes up-regulated in F1s are typically those that are marked by H3K27me3 in the germline of wild-type fathers. To assess this, we used CUT&RUN (cleavage under targets & release using nuclease) to analyze the distribution of H3K27me3 and the active mark H3K36me3 in wild-type spermatogenic germlines (dissected from adult males). CUT&RUN is a high-resolution chromatin analysis technique that requires fewer cells than traditional chromatin immunoprecipitation (31, 32), allowing us to interrogate the chromatin landscapes of hand-dissected germlines.
We focused on analyzing up-regulated genes, since our goal was to understand what drives some genes and not others to become up-regulated when the entire sperm genome was inherited lacking repressive H3K27me3. To compare similar-sized sets of up-regulated genes across K27me3 M+P− tissues, we selected the 300 up-regulated genes with the lowest adjusted P values from each tissue (Fig. 1B). We found that in each F1 tissue context, the vast majority of up-regulated genes was enriched for repressive H3K27me3 and depleted of the active mark H3K36me3 in wild-type spermatogenic and also oogenic germlines (Fig. 1C). This suggests that genes prone to up-regulation in K27me3 M+P− worms were those typically inherited with repressive H3K27me3 on both sperm and oocyte alleles. In K27me3 M+P− worms, those genes were inherited in different H3K27me3 states from sperm and oocyte, which we predicted would lead to differential sensitivity of sperm and oocyte alleles to up-regulation. Indeed, we observed that in each K27me3 M+P− tissue analyzed, genes that are normally packaged with high levels of H3K27me3 (z score >0) in father’s spermatogenic germline were preferentially up-regulated in cis from the sperm-inherited allele, and genes that are normally packaged with low levels of H3K27me3 (z score <0) in father’s spermatogenic germline were modestly up-regulated from both sperm- and oocyte-inherited alleles (Fig. 1D and SI Appendix, Fig. S1B). The latter may reflect a secondary effect of inheriting the sperm genome lacking H3K27me3. Our results show that different tissues in K27me3 M+P− F1s selectively up-regulated sperm alleles.
Because the X chromosomes in wild-type germlines have higher levels of H3K27me3 than the autosomes, we analyzed X genes separately. We observed that in K27me3 M+P− offspring more X-linked genes had a negative log2(fold change) than expected based on the transcriptome distribution (χ2 P < 2.2e−16 for both germline tissues and 2.9e−13 for larval soma) (SI Appendix, Fig. S2B). This is likely explained by our observation that in K27me3 M+P− embryos, the sperm-inherited X chromosome acquired H3K27me3 during the early divisions, while the sperm-inherited autosomes remained H3K27me3(−) through many rounds of cell division (SI Appendix, Fig. S2 A–C). Indeed, none of the up-regulated genes in K27me3 M+P− hermaphrodite and male germlines are X-linked.
In K27me3 M+P− Worms, Sperm Alleles Were Up-Regulated in a Manner Dependent on Tissue Context.
Having established that different K27me3 M+P− F1 tissues displayed gene up-regulation when the sperm genome was inherited lacking H3K27me3, we investigated whether different tissues displayed up-regulation of similar or different sets of genes. We found that there was very little overlap in top up-regulated genes between soma and germline (Fig. 2A and SI Appendix, Fig. S1C), highlighting the importance of cellular context.
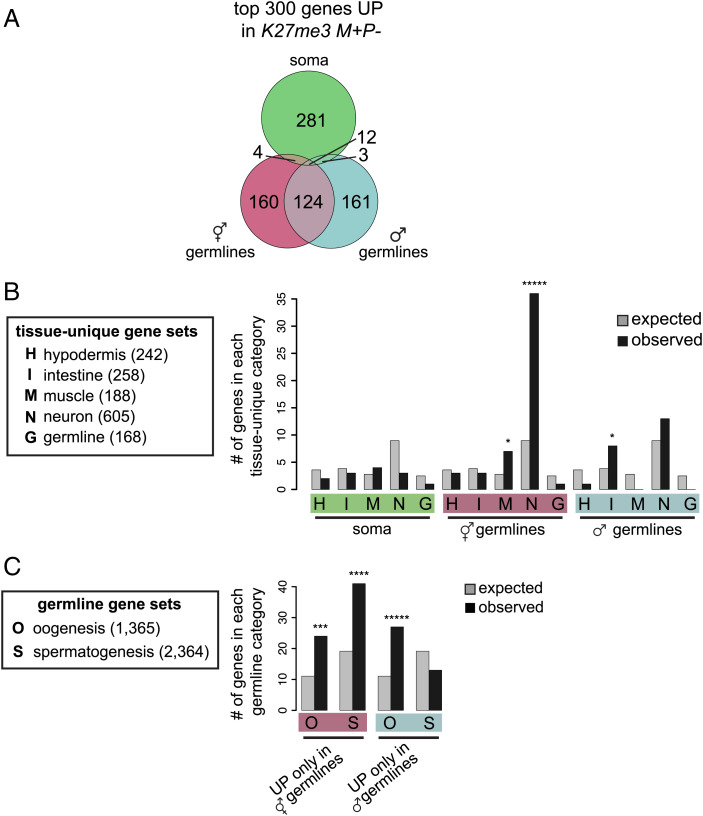
Fig. 2.
K27me3 M+P− worms up-regulate genes in a tissue-specific manner. (A) Venn diagram showing overlap of the top 300 up-regulated genes in each of the three F1 tissues. SI Appendix, Fig. S1C includes a Venn diagram showing overlap of significantly up-regulated genes. (B) Histograms showing expected (gray bars) and observed (black bars) numbers of up-regulated genes that overlap with tissue-unique genes identified in (33). (C) Histograms showing expected (gray bars) and observed (black bars) numbers of up-regulated genes that overlap with oogenesis or spermatogenesis genes identified in (34). The number of genes in each gene set is in parentheses. Gene set enrichments were calculated using hypergeometric tests. P values are: *<0.05, ***< 0.0005, ****<0.00005, and *****<0.000005.
To explore the identity of up-regulated genes in each K27me3 M+P− F1 tissue context, we determined whether the top 300 up-regulated genes are enriched for genes whose expression is typically restricted to particular tissues (Fig. 2B). We found that genes up-regulated in germline samples were enriched for particular soma-specific genes (1, 33). Hermaphrodite germlines mainly up-regulated neuron-specific genes and some muscle-specific genes, while male germlines up-regulated intestine-specific genes (Fig. 2B). Although up-regulated genes in both germline contexts were not enriched for our very select set of germline-specific genes (168 genes expressed in germline but not soma), we observed enrichment of the larger sets of germline-expressed genes that are mostly restricted to either oogenic or spermatogenic germlines (34). Specifically, genes up-regulated in K27me3 M+P− hermaphrodite oogenic germlines were enriched for spermatogenesis genes, while genes up-regulated in K27me3 M+P− male spermatogenic germlines were enriched for oogenesis genes (Fig. 2C). We conclude that gamete-inherited patterns of H3K27me3 differentially influence gene expression patterns in various offspring tissues, and in the germline, inherited H3K27me3 marking is important for preventing transcription of somatic and sex-inappropriate genes.
In the Germline of K27me3 M+P− Worms, Genes That Were Up-Regulated Retained the H3K27me3(−) State of Sperm Alleles, While Genes That Were Not Up-Regulated Regained H3K27me3 on the Sperm Allele.
Our previous immunostaining revealed that when the sperm genome was inherited without H3K27me3, sperm chromosomes retained the H3K27me3(−) state in the germ lineage until germ cells started proliferating during larval development, after which sperm-inherited chromosomes acquired H3K27me3 immunostaining (14, 30). To analyze the distribution of H3K27me3 at high resolution in the germline of K27me3 M+P− adults, we performed CUT&RUN analysis. We first assessed wild-type patterns of H3K27me3 and H3K36me3 in adult hermaphrodite germlines, by calculating z scores for H3K27me3 and H3K36me3 reads that mapped to gene bodies in “wild-type” K27me3 M+P+ germlines. Consistent with H3K27me3 marking repressed genes and H3K36me3 marking transcribed genes, we found that in the germline, H3K27me3 was enriched over genes that are silent and H3K36me3 was enriched over genes that are expressed (e.g., Fig. 3A).
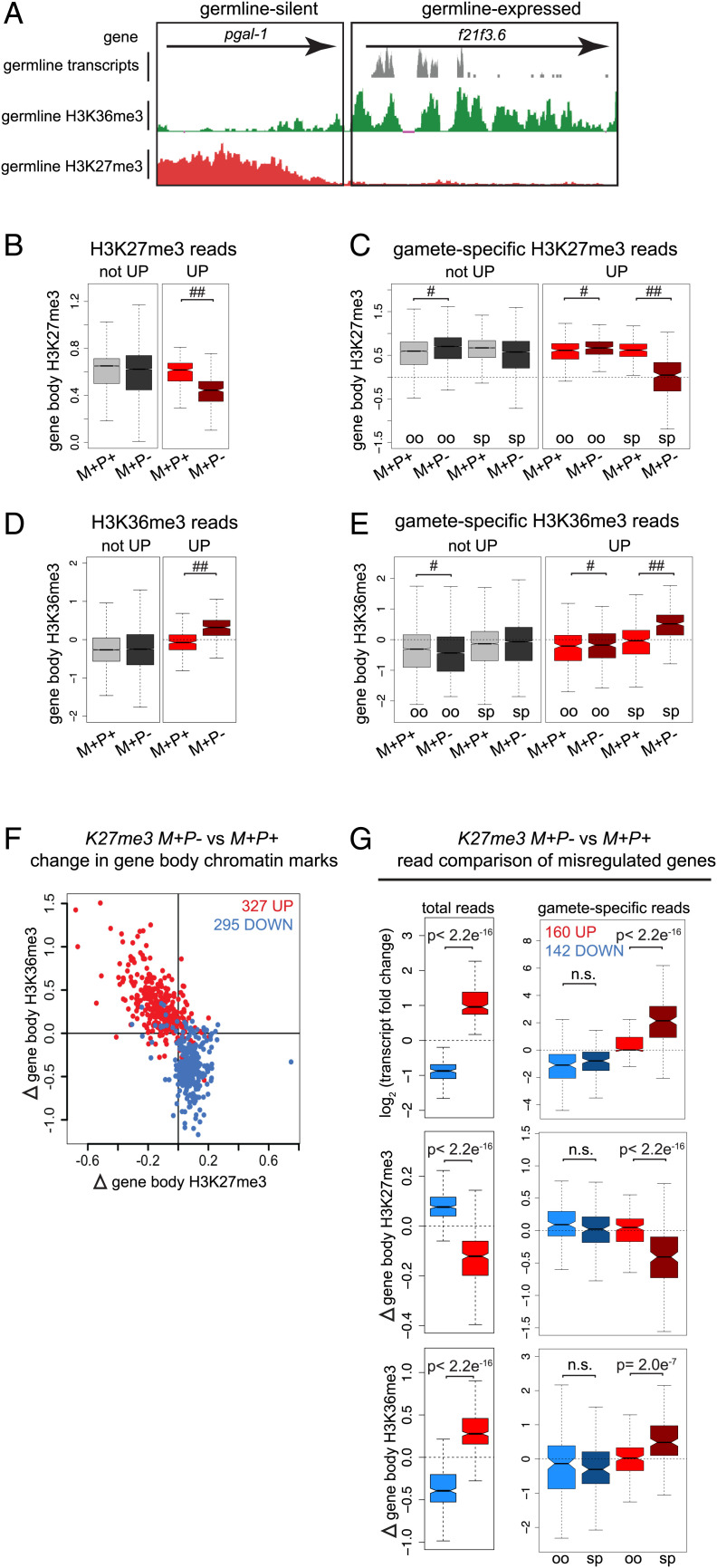
Fig. 3.
Genes that are up-regulated in K27me3 M+P− germ cells retain the H3K27me3(−) state of sperm alleles, while genes that are not up-regulated become de novo methylated to reestablish wild-type levels of H3K27me3. (A) UCSC genome browser images showing transcript reads (gray; scale 0–200), H3K36me3 (green; scale 0–150), and H3K27me3 (red; scale 0–30) CUT&RUN sequencing reads from “wild-type” (K27me3 M+P+) hermaphrodite germlines for a germline-silent gene and a germline-expressed gene. (B, D) Boxplots of gene body (B) H3K27me3 or (D) H3K36me3 z scores in K27me3 M+P+ and K27me3 M+P− germlines for genes that are H3K27me3(+) in K27me3 M+P+ germlines and either UP (310 genes) or not UP (9,318 genes) in K27me3 M+P− germlines. (C, E) Boxplots of gamete-specific gene body (C) H3K27me3 or (E) H3K36m3 z scores in K27me3 M+P+ and K27me3 M+P− germlines for genes that are H3K27me3(+) in K27me3 M+P+ germlines and either UP or not UP in K27me3 M+P− germlines. Of 310 UP genes, 153 have at least one gene-associated SNP. Of 9,318 not-UP genes, 3,780 have at least one gene-associated SNP. #P value <0.01 and absolute(FC) <1.1. ##P value <0.01 and absolute(FC) >1.1. (F) Scatterplot of K27me3 M+P− vs. K27me3 M+P+ change in gene body H3K27me3 vs. H3K36me3 in hermaphrodite germlines for significantly up-regulated genes (red) and down-regulated genes (blue). (G) Boxplots of transcript fold change, H3K27me3 change, and H3K36me3 change for significantly up-regulated genes (red) and down-regulated genes (blue) in K27me3 M+P− vs. K27me3 M+P+ germlines. Left Plots: changes based on all RNA-seq reads across a transcript or CUT&RUN reads across a gene body. Right Plots: changes based on allele-specific (SNP-containing) reads. Of 327 up-regulated genes, 160 have at least one gene-associated SNP. Of 295 down-regulated genes, 142 have at least one gene-associated SNP. Oocyte alleles (oo). Sperm alleles (sp). Boxplots show the median (horizontal line at waist), the 25th and 75th percentiles (boxes), and whiskers extending to the most extreme data point, which is no more than 1.5 times the interquartile range. The waist indicates the 95% confidence interval for the medians. P values were calculated using Student’s t test (two-tailed).
To investigate whether all or only some sperm alleles acquired H3K27me3 in K27me3 M+P− hermaphrodite germlines, we analyzed CUT&RUN reads containing an allele-specific SNP, which allowed us to determine whether the reads emanated from the sperm- and/or oocyte-inherited allele. To assess persistence of the inherited H3K27me3(−) state versus reversion to a H3K27me3(+) state, we focused on genes that are normally H3K27me3(+) in “wild-type” K27me3 M+P+ germlines. We found that genes that were up-regulated in K27me3 M+P− germlines remained depleted of H3K27me3 compared to K27me3 M+P+ control germlines, and genes that were not up-regulated in K27me3 M+P− germlines had acquired H3K27me3 over their gene bodies at levels comparable to K27me3 M+P+ controls (Fig. 3B). Incorporating SNP analysis revealed that restoration of H3K27me3 in K27me3 M+P− germlines was due to de novo deposition of H3K27me3 on sperm alleles of genes that were not up-regulated; sperm alleles of up-regulated genes did not acquire H3K27me3 (Fig. 3C). H3K36me3 enrichment mirrored H3K27me3 depletion (Fig. 3 D and E).
To assess whether gene expression changes coincided with changes in histone marking more generally, we analyzed misregulated genes for changes in H3K27me3 and H3K36me3 in germlines from adult K27me3 M+P− and K27me3 M+P+ hermaphrodites. We found that up-regulated genes had increased levels of H3K36me3 and reduced levels of H3K27me3 across their gene bodies in K27me3 M+P− compared to K27me3 M+P+ germlines, and the reverse was true for down-regulated genes (Fig. 3F). Both the increase in H3K36me3 and the decrease in H3K27me3 on up-regulated genes reflected a shift toward a more euchromatic chromatin state, while the opposite changes on down-regulated genes reflected a shift toward a more heterochromatic chromatin state.
To determine whether allele-specific changes in transcript levels were associated with allele-specific changes in chromatin state, we analyzed transcript and CUT&RUN reads containing an allele-specific SNP. This analysis confirmed our previous findings that up-regulated genes were primarily due to increased transcript levels from sperm alleles, while down-regulated genes were similarly down-regulated from both sperm and oocyte alleles (Fig. 3G, Upper Panels) (14). Notably, our allele-specific CUT&RUN analysis demonstrated that allele-specific changes in transcript levels were associated with allele-specific changes in chromatin state (Fig. 3G, Lower Panels). These findings indicate that, although much of the H3K27me3(+) landscape had been reestablished on the sperm-inherited genome in the germlines of adult K27me3 M+P− hermaphrodites, the inherited H3K27me3(−) state was retained on the subset of sperm alleles that was up-regulated. This raised the exciting possibility that altered chromatin and transcriptional states can be transmitted to and influence gene expression in the next generation.
K27me3 M+P− Worms Transmitted the H3K27me3(−) and Up-Regulated State of Sperm Alleles to the Next Generation.
To test whether inheriting a sperm genome lacking H3K27me3 has transgenerational (to grandoffspring) impacts on transcription, we repeated profiling of the germlines of K27me3 M+P− hermaphrodites (F1s) and also profiled the germlines of their genetically identical hermaphrodite offspring (F2s), which we refer to as K27me3 GM+GP− (GM+ for grandmaternal H3K27me3(+); GP− for grandpaternal H3K27me3(−)) (Figs. 1A and 4A). K27me3 M+P− (and K27me3 M+P+) F1s inherited the oocyte genome with the mother’s background genotype (Hawaii CB4856) and the sperm genome with the father’s background genotype (Bristol N2). Therefore, for every SNP-containing gene, K27me3 M+P− nuclei contained an oocyte-inherited allele from an H3K27me3(+) mother and a sperm-inherited allele from an H3K27me3(−) father. When K27me3 M+P− F1 hermaphrodite worms produced gametes, the maternal and paternal alleles were independently assorted into their sperm and oocytes, resulting in a multitude of allelic combinations inherited by their offspring (F2s) (Fig. 4A). We reasoned that if the H3K27me3(−) state of sperm-inherited alleles persisted transgenerationally, then at the population level of F2 worms, we should observe transcript level and chromatin state differences between the GP+ and GP− grandpaternal alleles regardless of whether those alleles were packaged into sperm or oocyte and whether they were inherited in 0, 1, or 2 copies by any individual F2 worm (Fig. 4A).
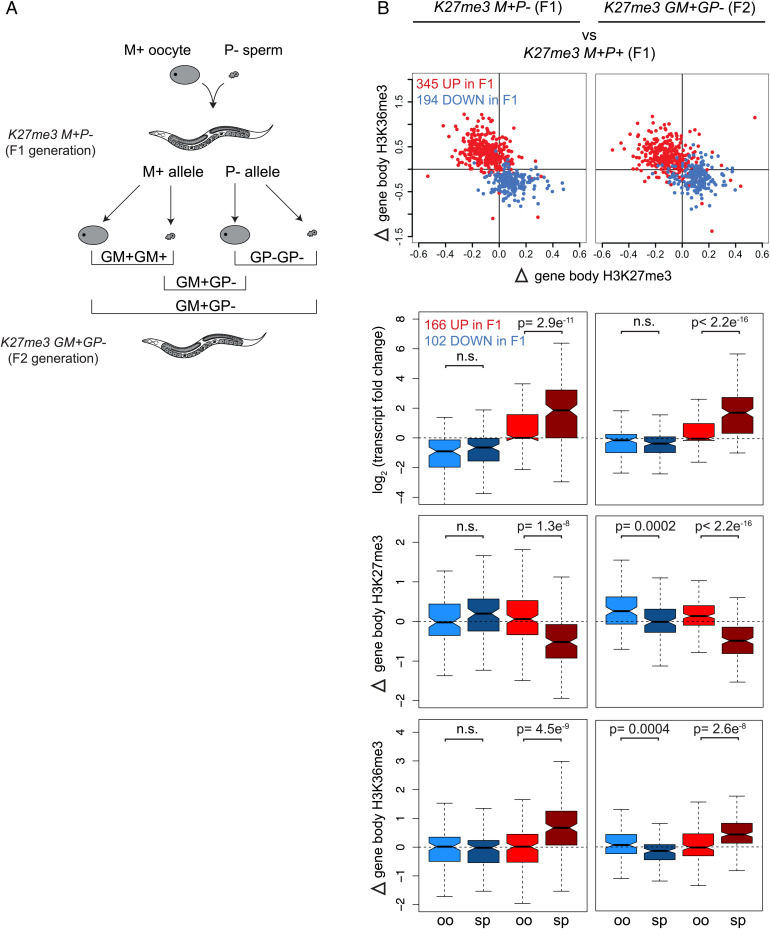
Fig. 4.
Transcriptional states and chromatin state shifts of genes misregulated in K27me3 M+P− germ cells persist in K27me3 GM+GP− germ cells of the next generation. (A) Diagram showing independent assortment of parental alleles (M+ from the oocyte and P− from the sperm) into F1 offspring sperm and oocytes to yield three possible grandoffspring epigenotypes: GM+GM+ in which the H3K27me3(+) grandmaternal allele is inherited from both sperm and oocyte, GP−GP− in which the H3K27me3(−) grandpaternal allele is inherited from both sperm and oocyte, and GM+GP− in which a H3K27me3(+) grandmaternal and H3K27me3(−) grandpaternal allele are each inherited, one from each gamete. (B) Scatterplots of K27me3 M+P− vs. K27me3 M+P+ (Left Panels) and K27me3 GM+GP− vs. K27me3 M+P+ (Right Panels) transcript and chromatin changes in hermaphrodite germ cells for genes significantly up-regulated (red) and down-regulated (blue) in F1s. Scatterplots show gene body H3K27me3 vs. H3K36me3 (top row, similar to Fig. 3F). Boxplots show oocyte (oo) and sperm (sp) allele-specific transcript changes and chromatin state shifts (lower three rows, similar to Fig. 3G). Of 345 genes up-regulated in F1s, 166 have at least one gene-associated SNP. Of 194 genes down-regulated in F1s, 102 have at least one gene-associated SNP. P values were calculated using Student’s t test (two-tailed).
Focusing on genes that were significantly misexpressed in K27me3 M+P− (F1) hermaphrodite germlines (Fig. 4B, Left Panels), we compared the transcript level and chromatin state changes of those genes in newly isolated germlines of K27me3 M+P− (F1) (Fig. 4B, Left Panels) and in the germlines of their K27me3 GM+GP− (F2) hermaphrodite offspring (Fig. 4B, Right Panels) to K27me3 M+P+ (F1) germlines. As shown in Fig. 3F, this new analysis of F1 germlines demonstrated that genes that were up-regulated in F1 germlines had reduced H3K27me3 and increased H3K36me3, representing a shift toward a more euchromatic chromatin state; the opposite was true for genes that were down-regulated in F1 germlines (Fig. 4B, Left). These changes in gene expression and chromatin state persisted in the germlines of the next generation (F2s) (Fig. 4B, Right). Strikingly, these changes were driven by changes in transcript levels and histone marking of sperm alleles specifically (Fig. 4B, Right). For example, H3K27me3(−) sperm-inherited alleles up-regulated in cis in K27me3 M+P− (F1) germlines continued to be up-regulated in cis in the germlines of K27me3 GM+GP− (F2) grandoffspring. Chromatin state analysis of those genes indicated that the euchromatically shifted chromatin state of sperm alleles also persisted in cis. These findings suggest that, whether repackaged in sperm or oocyte of the F1 generation, the H3K27me3(−) state of sperm-inherited alleles was maintained in the next generation, where it continued to sensitize those alleles to transcriptional up-regulation. We note that our analysis was of the same tissue context (i.e., adult hermaphrodite germlines from K27me3 M+P− F1s and K27me3 GM+GP− F2s), which likely predisposes them to up-regulate a similar set of genes. These findings raise the possibility that as long as germline health and therefore reproduction are not compromised, cis up-regulation of ancestrally inherited H3K27me3(−) alleles could persist from hermaphrodite parent to hermaphrodite offspring indefinitely.
Discussion
The major take-homes from our study of K27me3 M+P− F1 offspring that inherited the sperm genome lacking the repressive mark H3K27me3 are: 1) different tissues (both soma and germline) up-regulated genes specifically from sperm alleles, 2) different tissues up-regulated different sets of genes, revealing the importance of tissue context, 3) in the germline, up-regulated genes retained the inherited H3K27me3(−) state while genes not up-regulated regained H3K27me3, and 4) K27me3 M+P− F1 offspring transmitted the inherited H3K27me3(−) state and the resulting up-regulated state of sperm alleles to the next generation, so transgenerationally.
Model for How Tissue Context Dictates Gene Up-Regulation.
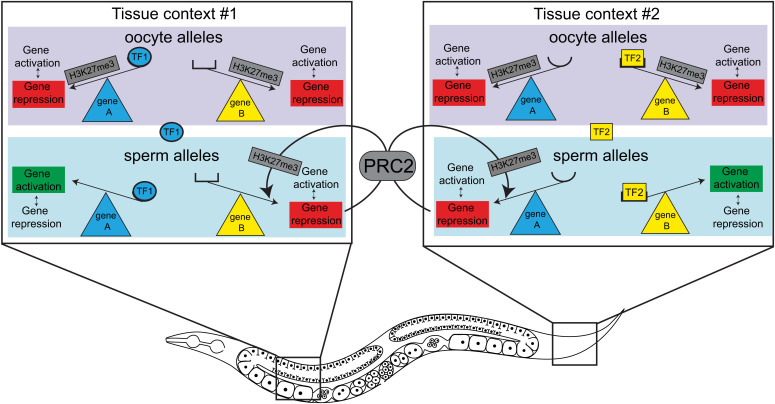
We hypothesize that gamete-inherited H3K27me3 prevents lowly expressed or weak transcriptional activators from activating their targets in various tissues in offspring. In the case of K27me3 M+P− cells, weak activators can activate their targets on H3K27me3(−) sperm alleles but are prevented from doing so on H3K27me3(+) oocyte alleles (Fig. 5). This model predicts that the transcription factor milieu of a particular cell dictates which genes will be up-regulated when H3K27me3 is absent, which echoes recent findings in mammalian cells (28). This model is consistent with a recent study in C. elegans that found that upon global loss of the repressive histone mark H3K9me, the tissue-specific transcription factor environment determines which genes are sensitive to up-regulation (35). Once sperm alleles are up-regulated, our model invokes that they are poor substrates for de novo methylation of H3K27. Thus, our model captures the bidirectional relationship between chromatin state and transcription.
Fig. 5.
Model for tissue-context-specific transcriptional up-regulation of sperm alleles when the sperm genome is inherited lacking H3K27me3. Boxes represent nuclei from two different tissue contexts. Tissue context #1 (Left Box) expresses transcriptional activator TF1 (blue oval), which can activate its target, gene A, when gene A is inherited lacking H3K27me3, as occurs for the sperm allele in K27me3 M+P− worms. The active state of gene A opposes de novo deposition of H3K27me3 by PRC2 on the sperm allele of gene A. Tissue context #1 does not express transcriptional activators for gene B and thus does not activate gene B regardless of its H3K27me3 state. The repressed state of gene B allows PRC2 to reestablish H3K27me3 on the sperm allele of gene B. Tissue context #2 (Right Box) expresses transcriptional activator TF2 (yellow rectangle), which can activate its target, gene B, when gene B is inherited lacking H3K27me3, as occurs for the sperm allele in K27me3 M+P− worms. The active state of gene B opposes de novo deposition of H3K27me3 by PRC2 on the sperm allele of gene B. Tissue context #2 does not express transcriptional activators for gene A and thus does not activate gene A regardless of its H3K27me3 state. The repressed state of gene A allows PRC2 to reestablish H3K27me3 on the sperm allele of gene A.
Evidence for a Conserved Mechanism of Histone-Based Gene Regulation across Generations and through Cell Division.
In mammals, the development of mature sperm involves the replacement of most histones with nonhistone proteins, resulting in a limited, but developmentally important, set of loci that are packaged with histones bearing chromatin marks (36, 37). Unlike mammals, C. elegans sperm retain histones genome-wide (2). This feature makes C. elegans a powerful model for investigating the mechanism through which sperm-inherited histone marking is preserved (or rewritten) in embryos and how such marking influences transcription and development in offspring. Using this system, we showed that sperm-inherited histone marks directly influence offspring transcription and development and elucidated a mechanism through which gamete-inherited states of H3K27me3 are preserved in early C. elegans embryos [(14, 30, 38) and this study]. Despite the reduced transmission of sperm-inherited histones in mammals, recent findings suggest that histone-based processes may underlie at least some epigenetic inheritance in mammals as well (15, 39, 40).
A recent study investigated mechanisms of H3K27me3-mediated transcriptional memory across cell divisions in cultured mammalian cells (28). By comparing different cultured cell types after transient disruption of PRC2 activity, Holoch et al. (28) found that cellular context was important in dictating which genes reestablished H3K27me3 de novo and which remained H3K27me3(−) through cell divisions and were prone to up-regulation. After fusing cells with different epigenetic states, they found that those different states persisted through many cell divisions and regulated gene expression in cis. The similarities between our findings in worms and those of Holoch et al. (28) in cultured mammalian cells strongly support a conserved mechanism for PRC2-mediated gene regulation and epigenetic memory between worms and mammals.
K27me3 M+P− Worms Up-Regulate Genes in Somatic Tissues.
Given that the 1-cell embryo inherits chromatin states established in the parental germline, somatic tissues must overcome germline-appropriate patterns of H3K27me3 that mark some loci critical for somatic development. This predicts that gamete-inherited H3K27me3 may not have strong barrier function in developing somatic cells. However, our findings indicate that in nascent somatic tissues, gamete-inherited H3K27me3 influenced transcription patterns and that the absence of H3K27me3 sensitized genes to up-regulation. Our model predicts that different genes are up-regulated in different somatic tissues. In this study, we analyzed newly hatched L1 larvae as they are >99% somatic cells. However, larvae are a mix of several somatic tissues (e.g., intestine, muscle, hypodermis, and neuron); we do not know which genes were up-regulated in which somatic tissues. We observed fewer significantly up-regulated genes in K27me3 M+P− L1 samples compared to adult germline samples. This may be due to up-regulation of genes by only a small percentage of cells in L1s (e.g., only intestinal cells but no other somatic cells). Our findings that gamete-inherited chromatin states can impact transcription in somatic tissues have important implications for how inherited chromatin states can influence offspring development and health.
K27me3 M+P− Hermaphrodite Germlines Up-Regulate Neuronal Genes.
We observed that loss of H3K27me3 from the sperm-inherited genome primed hermaphrodite germ cells to misexpress neuronal genes [(14) and this study] and in a sensitized background resulted in germ cells transforming toward neurons (14). Thus, H3K27me3 is particularly important to keep neuronal genes silent during germline development. We speculate that neuronal genes are particularly susceptible to up-regulation because some neuronal transcriptional activators are normally expressed in the hermaphrodite germline (e.g., die-1 and nsy-7) (41) and others become expressed when their promoter lacks an H3K27me3 barrier (Fig. 5); these may up-regulate a neuronal program, leading to a germ-toward-neuron conversion. This model is consistent with observations that loss of PRC2 and other chromatin regulators allows ectopic expression of a neuronal transcription factor to drive germ cells toward a neuronal fate (42–44).
Up-Regulation of Genes in the Germline of K27me3 M+P− Worms Can Be Passed across Generations.
Here, we showed that alterations to the sperm epigenome influenced transcription across two generations of germ cells. We found that sperm-inherited alleles that became up-regulated in the germline of K27me3 M+P− offspring continued to be up-regulated in the germline of the next generation. Most sperm-inherited alleles in the germline of K27me3 M+P− worms reestablished H3K27me3 and never became up-regulated. However, sperm-inherited alleles that did become up-regulated maintained an H3K27me3(−) state in K27me3 M+P− germ cells and in the germ cells of their offspring. Both the F1 and F2 generations analyzed were hermaphrodite germlines and thus represent the same tissue context. We hypothesize that the genes sensitive to up-regulation when the H3K27me3(+) state is absent in the F1 generation are similarly sensitive to up-regulation in the F2 generation. We did not analyze further generations for the persistence of the H3K27me3(−) state and transcriptional up-regulation of sperm-inherited alleles. However, we speculate that, so long as transmission occurs from hermaphrodite parent to hermaphrodite offspring, this allele-specific pattern could persist for numerous generations.
Passage of the H3K27me3(−) State of Sperm Alleles across Generations As Epialleles.
In Mendelian genetics, an F1 “hybrid” has parents with different versions of the same gene. Typically, one parent is mutant and does not produce functional gene product, while the other parent is wild type. In this way, hybrids have one allele that produces product and the other that does not. The offspring of hybrid organisms, F2s, can inherit various combinations of alleles, affording this generation a range of genotypes and potential phenotypes. In this study, we present an epigenetic paradigm that functions similarly to Mendelian inheritance. The major difference is that differences in chromatin state (epialleles) rather than DNA sequence (alleles) determine whether gene product is produced or not. Consistent with Mendelian inheritance, these chromatin states and resulting expression states are preserved in the F2 generation. Because of this, genetically identical F2s can have a range of phenotypes, those that inherit both alleles in the same state, either “off” or “on,” and those that inherit alleles in different states. In the K27me3 M+P− paradigm, many genes were inherited in this manner and therefore represent a wide spectrum of potential “epigenotypes.” We hypothesize that these epigenotypes produce the spectrum of germline health observed in the F2 generation, in which worms range from sterile and lacking a germline to fertile with a well-proliferated germline (14). This model provides one way in which epigenetic changes to the parental genome can rapidly create epigenetic variation in an otherwise genetically identical population (45). We speculate that this ability is particularly advantageous for species that primarily reproduce clonally, such as C. elegans. A further advantage of epialleles may be rapid responsiveness to environmental conditions, enabling animals to adapt to changing habitats, diets, and climates.
Materials and Methods
C. elegans Growth Conditions.
All strains (SI Appendix includes worm strains used) were maintained on nematode growth medium agar plates seeded with E. coli OP50 and maintained at 20 °C except for fem-2 containing strains, which were maintained at 15 °C. To generate females, fem-2 containing strains were raised at 25 °C until the L4 stage.
Collection of K27me3 M+P− and K27me3 M+P+ Soma (L1 Larvae) and Hermaphrodite and Male Germlines.
Feminized fem-2 (M+) (CB4856 background) hermaphrodites were mated to either mes-3; him-8 (P−) or mes-3/hT2-GFP;him-8 (P+) (N2 background) males to generate genetically identical mes-3/+; fem-2/+; him-8/+ K27me3 M+P- and K27me3 M+P+ offspring, respectively.
To collect K27me3 M+P− and M+P+ L1 larvae (source of somatic tissue), gravid mothers were transferred to fresh plates seeded with OP50 and allowed to lay eggs overnight at 20 °C. The next day, late-stage embryos along with some bacteria were transferred to 50 μL drops of S Basal (5.85 g NaCl, 1 g K2HPO4, 6 g KH2PO4, 1 mL cholesterol [5 mg/mL in ethanol], water to 1 L) on gelatin chrom alum-coated slides. After 2 h at room temperature, newly hatched K27me3 M+P− and K27me3 M+P+ (no GFP balancer) L1 larvae were transferred to 300 μL of ice-cold TRIzol reagent (Life Technologies) and stored at −80 °C for later RNA-sequencing library preparation. This process of transferring embryos to S Basal, incubating 2 h, and transferring newly hatched L1 larvae to TRizol was repeated throughout the day to reach a final collection of ∼100 newly hatched larvae per replicate. Three biological replicates were collected for each genotype from parental crosses set up on different days.
To collect K27me3 M+P− and M+P+ F1 germlines, hermaphrodite and male K27me3 M+P− and M+P+ (no GFP balancer) worms were picked at the L4 stage and grown 1 d at 20 °C to day 1 adults from which distal germlines (from the distal tip to the bend) were dissected. Approximately 35 distal germlines were dissected in egg buffer (25 mM Hepes pH 7.4, 118 mM NaCl, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2) with 1 mM levamisole, 0.5% Tween after which they were transferred to 300 μL of ice-cold TRIzol and stored at −80 °C for later RNA-sequencing library preparation. For CUT&RUN-sequencing library preparation, distal germlines were transferred to a drop of 100 μL GWB (20 mM Hepes pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 0.05% BSA, protease inhibitor tablet [Roche]), then to a 1.5 mL tube on ice with 1 mL GWB. This process was repeated until each tube contained enough germlines for each sample: 16 for H3K27me3 and 50 for H3K36me3. Four biological replicates were collected per genotype for RNA-sequencing library construction, and two biological replicates were collected per genotype per histone mark for CUT&RUN-sequencing library construction. Each biological replicate was from parental crosses set up on different days.
Collection of Germlines from K27me3 M+P− and K27me3 M+P+ (F1) Worms and Their Genetically Identical K27me3 GM+GP− (F2) Offspring.
Feminized fem-2 (M+) (CB4856 background) hermaphrodites were mated to either glh-1::GFP mes-3 (P−) or glh-1::GFP mes-3/hT2-GFP (P+) (N2 background) males to generate genetically identical glh-1::GFP mes-3/+ K27me3 M+P− and M+P+ hermaphrodite offspring (F1). Four F1 hermaphrodite offspring were picked at the L4 stage and allowed to self-fertilize to generate genetically identical glh-1::GFP mes-3/+ K27me3 GM+GP− hermaphrodite grandoffspring (F2). Approximately 150 F1 offspring and F2 grandoffspring were picked at the L4 stage and grown 1 d at 20 °C to day 1 adults from which distal germlines were dissected for RNA-sequencing and CUT&RUN-sequencing library preparation. The glh-1::GFP reporter gene is closely linked to mes-3 and was used to assess the mes-3 genotype of F2 worms. Worms carrying the glh-1::GFP reporters contain fluorescent germ granules in their germline. The level of germline fluorescence as viewed on a fluorescent microscope indicated the number of mes-3 mutant alleles in each worm. The accuracy of our visual assessment was confirmed by genotyping individual worms via PCR for mutant and wild-type copies of mes-3. 100% of tested worms (n = 45) confirmed the visual assessments. Three biological replicates per genotype per generation were collected for library construction.
Collection of Germlines from PRC2(−) Mutant and Wild-Type Fathers.
PRC2(−) mutant male germlines were collected from homozygous mutant mes-3; him-8 males generated from heterozygous mes-3/hT2-GFP; him-8 hermaphrodite mothers. Wild-type male germlines were collected from him-8 males. L4 males were selected and allowed to mature 1 d at 20 °C to day 1 adults. Three biological replicates per genotype were collected for RNA-sequencing and H3K27me3 and H3K36me3 CUT&RUN-sequencing library construction.
RNA Sequencing.
SI Appendix includes details. In brief, total RNA from germlines or L1s was extracted using TRIzol reagent and chloroform followed by isopropyl alcohol precipitation. The RNA pellet was resuspended in RNase-free water, and RNA concentration was measured and RNA quality assessed. Total RNA was polyA selected, and libraries were prepared. Sequencing libraries prepared from L1 larvae were sequenced on the Illumina NextSeq platform to acquire paired-end 75 bp reads. All other libraries were sequenced on the Illumina NovaSeq platform to acquire paired-end 50 bp reads.
CUT&RUN of Hand-Dissected Germlines.
Immediately following germline collection, 10 μL of activated ConA beads (Bangs Laboratory cat. no. BP531) were added to each tube containing germlines in GWB (31, 32). ConA beads were activated by washing twice with 1.5 mL binding buffer (31, 32), then resuspended in 10 μL binding buffer. The beads/germlines were washed via a quick spin in a microcentrifuge, placed on a magnet stand for 30 seconds, and the supernatant was removed. The activated beads/germlines were incubated 5 min at room temperature with gentle rotation, then washed twice with 1 mL GWB. Primary antibodies (1 μg rabbit anti-H3K27me3 [Cell Signaling Technology] or 0.5 μg mouse anti-H3K36me3 [Wako]) were added to 50 μL antibody buffer (GWB with 2 mM EDTA and 0.1% digitonin). Diluted antibodies were added to each sample and incubated at 4 °C overnight with gentle rotation. Samples were washed twice for 5 min in 1 mL GWB-D (GWB with 0.1% digitonin) with gentle rotation. H3K36me3 samples required an additional incubation with rabbit anti-mouse IgG (0.5 μg [Abcam] in 50 μL GWB-D) at 4 °C for 1 h with gentle rotation. Samples were washed twice for 5 min with 1 mL GWB-D, and the beads resuspended in 50 μL GWB-D. 2.5 μL of 1:10 dilution of pA-MNase (gift from Steve Henikoff’s laboratory) were added to each sample. Tubes were rotated for 1 h at room temperature, then washed three times for 10 min with 1 mL GWB-D. GWB-D (100 μL) was added to each tube and then incubated in a cold metal block for 5 min. pA-MNase digestion was initiated by adding 2 μL 100 mM CaCl2 to each tube and incubating in a cold block for 30 min. pA-MNase digestion was stopped by adding 100 μL G2XSTOP buffer (31, 32). The samples were mixed, incubated at 37 °C for 30 min, spun down for 5 min at 4 °C at full speed, and placed on a magnet stand. The supernatant was transferred to a new 1.5 mL tube containing 2 μL 10% SDS and 2.5 μL Proteinase K (20 mg/mL) and incubated for 10 min at 70 °C. Each sample was phenol chloroform extracted, ethanol precipitated overnight at −80 °C, and the pellet was resuspended in 13 μL water. A total of 2 μL of each sample was run on a High Sensitivity TapeStation to check for cutting, and the remaining 10 μL were used to make libraries with the Ovation Ultralow System V2 kit (Tecan Genomics). Replicate 1 of K27me3 M+P− germline H3K36me3 and H3K27me3 libraries were sequenced on the Illumina NextSeq platform (paired-end 75 bp reads). All other CUT&RUN libraries were sequenced on the Illumina NovaSeq platform (paired-end 50 bp reads).
Analysis of RNA Sequencing Data.
SI Appendix includes details. In brief, all reads were mapped to the Bristol (N2) genome version WS220. Reads longer than 50 bp were trimmed to 50 bp. For reads per kilobase of transcript per million mapped reads (RPKM) and reads per single nucleotide polymorphism per million (RPSM) calculations, 50 bp reads were mapped to the WS220 genome twice: once allowing 1 mismatch (1mm) and a second time allowing 0 mismatches (0 mm). For all reads, RPKM was calculated by scaling total reads to 1 million, then dividing by transcript length in kb (union of all exons). For single nucleotide polymorphism (SNP) reads, RPSM was calculated by scaling total SNP reads to 1 million, then dividing by the number of SNPs per transcript. For differential expression analysis, reads were mapped using TopHat2 for paired-end reads. Reads per transcript or gene body were counted using htseq and a gtf file specifying transcript or gene body or SNP locations per transcript or gene body. Significantly misexpressed genes were determined using the differential analysis program DESeq2 from 50 bp reads. Genes with a multiple hypothesis adjusted P value <0.05 were considered significant. The top 300 up-regulated genes per F1 tissue context were selected based on having a positive fold change and the lowest P values in K27me3 M+P− samples relative to their K27me3 M+P+ controls. We defined 6,664 germline-silent genes (RPKM of transcript reads = 0 in K27me3 M+P+ germlines) and 5,931 germline-expressed genes (RPKM of transcript reads >5 in K27me3 M+P+ germlines). To identify genes that failed to reestablish H3K27me3 in K27me3 M+P− hermaphrodite germlines, genes were first selected that were H3K27me3-enriched (H3K27me3 gene body z score >0.2) in K27me3 M+P+ hermaphrodite germlines. This set of genes was split into two categories: UP (transcriptionally up-regulated) in K27me3 M+P− vs. M+P+ germlines (log2 fold change > 0 and adjusted P value < 0.05) and not UP (not transcriptionally up-regulated) (all other H3K27me3-enriched genes). This yielded 310 and 9,318 H3K27me3-enriched UP and not UP genes, respectively. To perform gene set enrichment analysis on the top 300 up-regulated genes in different F1 tissue contexts, the hypergeometric test (phyper in R) was performed using previously identified tissue-unique gene sets (33), hermaphrodite germline- and male germline-specific gene sets (34), and a germline-specific gene set (1).
Analysis of CUT&RUN Sequencing Data.
Reads longer than 50 bp were trimmed to 50 bp. 50 bp single-end (R1 only) reads were mapped (tophat2 with bowtie2) to the Bristol (N2) genome version WS220 twice: once allowing 1 mismatch (1 mm) and a second time allowing 0 mismatches (0 mm). Reads mapping anywhere in the gene body were counted using htseq and a gtf file specifying the entire gene body interval from start to end (exons and introns). To assess gene body enrichment of H3K27me3- and H3K36me3-based CUT&RUN reads, gene reads per million reads were calculated by dividing gene body CUT&RUN read counts (exonic and intronic) by gene length in kb, then scaling to 1 million total reads. To calculate gene body z scores, log2 transformed gene body reads (plus a pseudocount of 1) were scaled using the R package “scale.” For each gene within a sample, this package subtracts the log2 (gene body reads +1) mean, then divides by the log2 (gene body reads +1) SD. To calculate the change in gene body H3K36me3 or H3K27me3 in K27me3 M+P– vs. M+P+, the z score for K27me3 M+P+ was subtracted from the z score for K27me3 M+P−. To visualize H3K27me3 and H3K36me3 enrichment over genes, mapped CUT&RUN reads were scaled to 10 million and loaded into the University of California Santa Cruz (UCSC) genome browser.
SNP Assessment.
SI Appendix includes details. In brief, annotated SNPs of the Hawaiian CB4856 genome compared to the Bristol N2 WS220 genome were downloaded from WormBase. 50 bp CUT&RUN-seq and RNA-seq reads from K27me3 M+P−, K27me3 M+P+, and K27me3 GM+GP− worms were mapped to the WS220 Bristol genome two ways: 1) allowing 1 mismatch, and 2) allowing 0 mismatches. Reads that overlapped with 1 of the 78,780 gene-associated SNPs and that were also determined to reliably detect SNPs between wild-type Bristol and Hawaii genomes were selected. These SNP-overlapping reads were assigned to having originated from a Bristol (oocyte-inherited) genome or Hawaii (sperm-inherited) genome. Reads that mapped to the Bristol genome under both mapping parameters (allowing 0 or 1 mismatch) mapped as expected for having originated from a Bristol genome and were assigned to the oocyte allele. Reads that only mapped to the Bristol genome when allowing 1 mismatch mapped as expected for having originated from a Hawaii genome and were assigned to the sperm allele. Total SNP-overlapping reads (Bristol + Hawaii reads) were scaled to 1 million. SNP-overlapping reads per SNP per million SNP reads (RPSM) was calculated for each gene by dividing the total SNP-overlapping reads per gene by the number of SNPs per gene. Log2 (fold change), SNP-specific z scores, and change in gene body H3K27me3 or H3K36me3 in K27me3 M+P− vs. M+P+ were calculated as described in SI Appendix.
Analysis of the Paternal X Chromosome in K27me3 M+P− Embryos in a plk-1 Background for SI Appendix, Fig. S2.
SI Appendix includes generation and staining of K27me3 M+P− embryos to evaluate the H3K27me3 status of the sperm-inherited X chromosome.
Supplementary Material
Acknowledgments
We thank past and present members of the Strome lab for stimulating discussions and for contributing to a happy and supportive lab environment. We thank Dustin Updike for sharing the glh-1::GFP strain and Steve Henikoff for providing reagents for and advice on CUT&RUN. This work was supported by NIH (T32GM008646 and F31GM120882 to K.R.K. and R01GM34059 to S.S.). Some strains were provided by the CGC, which is funded by NIH Office of Infrastructure Programs (P40 OD010440). Portions of this manuscript were developed from the dissertation of K.R.K.
Footnotes
Reviewers: S.M.G., Fondation ISREC; and A.B., Stanford University School of Medicine.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2209471119/-/DCSupplemental.
Data, Materials, and Software Availability
All sequencing data generated in this study were deposited in the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) under accession GSE205113, and are accessible upon publication at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE205113, (46).
References
- 1.Rechtsteiner A., et al. , The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 6, e1001091 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tabuchi T. M., et al. , Caenorhabditis elegans sperm carry a histone-based epigenetic memory of both spermatogenesis and oogenesis. Nat. Commun. 9, 4310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu S.-F., Zhang H., Cairns B. R., Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 21, 578–589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fitz-James M. H., Cavalli G., Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet. 23, 325–341 (2022). [DOI] [PubMed] [Google Scholar]
- 5.Özdemir I., Steiner F. A., Transmission of chromatin states across generations in C. elegans. Semin. Cell Dev. Biol. 127, 133–141 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Pembrey M., Saffery R., Bygren L. O.; Network in Epigenetic Epidemiology; Network in Epigenetic Epidemiology, Human transgenerational responses to early-life experience: Potential impact on development, health and biomedical research. J. Med. Genet. 51, 563–572 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vågerö D., Pinger P. R., Aronsson V., van den Berg G. J., Paternal grandfather’s access to food predicts all-cause and cancer mortality in grandsons. Nat. Commun. 9, 5124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gapp K., et al. , Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 17, 667–669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q., et al. , Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Sharma U., et al. , Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grandjean V., et al. , RNA-mediated paternal heredity of diet-induced obesity and metabolic disorders. Sci. Rep. 5, 18193 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conine C. C., Sun F., Song L., Rivera-Pérez J. A., Rando O. J., Small RNAs gained during epididymal transit of sperm are essential for embryonic development in mice. Dev. Cell 46, 470–480.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson-Smith A. C., Genomic imprinting: The emergence of an epigenetic paradigm. Nat. Rev. Genet. 12, 565–575 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Kaneshiro K. R., Rechtsteiner A., Strome S., Sperm-inherited H3K27me3 impacts offspring transcription and development in C. elegans. Nat. Commun. 10, 1271 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lismer A., Siklenka K., Lafleur C., Dumeaux V., Kimmins S., Sperm histone H3 lysine 4 trimethylation is altered in a genetic mouse model of transgenerational epigenetic inheritance. Nucleic Acids Res. 48, 11380–11393 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petryk N., et al. , MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361, 1389–1392 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Alabert C., et al. , Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 29, 585–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reverón-Gómez N., et al. , Accurate recycling of parental histones reproduces the histone modification landscape during DNA replication. Mol. Cell 72, 239–249.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu C., et al. , A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 361, 1386–1389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margueron R., et al. , Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poepsel S., Kasinath V., Nogales E., Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol. 25, 154–162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Escobar T. M., et al. , Active and repressed chromatin domains exhibit distinct nucleosome segregation during DNA replication. Cell 179, 953–963.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen K. H., et al. , A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 10, 1291–1300 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Coleman R. T., Struhl G., Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science 356, eaai8236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pengelly A. R., Copur Ö., Jäckle H., Herzig A., Müller J., A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339, 698–699 (2013). [DOI] [PubMed] [Google Scholar]
- 26.McKay D. J., et al. , Interrogating the function of metazoan histones using engineered gene clusters. Dev. Cell 32, 373–386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laprell F., Finkl K., Müller J., Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science 356, 85–88 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Holoch D., et al. , A cis-acting mechanism mediates transcriptional memory at Polycomb target genes in mammals. Nat. Genet. 53, 1686–1697 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Højfeldt J. W., et al. , Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat. Struct. Mol. Biol. 25, 225–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaydos L. J., Wang W., Strome S., Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science 345, 1515–1518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skene P. J., Henikoff S., An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, e21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skene P. J., Henikoff J. G., Henikoff S., Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006–1019 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Kaletsky R., et al. , Transcriptome analysis of adult Caenorhabditis elegans cells reveals tissue-specific gene and isoform expression. PLoS Genet. 14, e1007559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzur Y. B., et al. , Spatiotemporal gene expression analysis of the Caenorhabditis elegans germline uncovers a syncytial expression switch. Genetics 210, 587–605 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Methot S. P., et al. , H3K9me selectively blocks transcription factor activity and ensures differentiated tissue integrity. Nat. Cell Biol. 23, 1163–1175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brykczynska U., et al. , Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 17, 679–687 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Hammoud S. S., et al. , Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, 473–478 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaydos L. J., Rechtsteiner A., Egelhofer T. A., Carroll C. R., Strome S., Antagonism between MES-4 and Polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Rep. 2, 1169–1177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lismer A., et al. , Histone H3 lysine 4 trimethylation in sperm is transmitted to the embryo and associated with diet-induced phenotypes in the offspring. Dev. Cell 56, 671–686.e6 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Inoue A., Jiang L., Lu F., Suzuki T., Zhang Y., Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hobert O., “Neurogenesis in the nematode Caenorhabditis elegans” in Patterning and Cell Type Specification in the Developing CNS and PNS (Elsevier, 2013), pp. 609–626. [Google Scholar]
- 42.Tursun B., Patel T., Kratsios P., Hobert O., Direct conversion of C. elegans germ cells into specific neuron types. Science 331, 304–308 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel T., Tursun B., Rahe D. P., Hobert O., Removal of Polycomb repressive complex 2 makes C. elegans germ cells susceptible to direct conversion into specific somatic cell types. Cell Rep. 2, 1178–1186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hajduskova M., et al. , MRG-1/MRG15 is a barrier for germ cell to neuron reprogramming in C. elegans. Genetics 211, 121–139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarutani Y., Takayama S., Monoallelic gene expression and its mechanisms. Curr. Opin. Plant Biol. 14, 608–613 (2011). [DOI] [PubMed] [Google Scholar]
- 46.K. R. Kaneshiro et al., Data for “Sperm-inherited H3K27me3 epialleles are transmitted transgenerationally in cis.” NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE205113. Deposited on 31 May 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data generated in this study were deposited in the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) under accession GSE205113, and are accessible upon publication at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE205113, (46).