Significance
Circadian clock dysfunction is frequently associated with cancer and metabolic diseases. CRYs are core components of the circadian clock, and CRY modulation has been shown to regulate metabolic homeostasis in animal disease models. In this report, we characterized the direct and preferential interaction of a small-molecule compound SHP656 with CRY2 and identified its active R form, SHP1703. Administration of SHP1703 to patient-derived GSCs inhibited their growth in culture. Our data highlight the importance of determining the isoform selectivity of compounds, as well as the active isomer, to facilitate the understanding of individual roles of CRY isoforms in disease, and to increase the potency and efficacy of compounds.
Keywords: circadian clock, cryptochromes, small-molecule modulators, glioblastoma, X-ray crystallography
Abstract
The mammalian cryptochrome isoforms, CRY1 and CRY2, are core circadian clock regulators that work redundantly. Recent studies revealed distinct roles of these closely related homologs in clock output pathways. Isoform-selective control of CRY1 and CRY2 is critical for further understanding their redundant and distinct roles. KL001 was the first identified small-molecule CRY modulator that activates both CRY1 and CRY2. SHP656 is an orally available KL001 derivative and has shown efficacy in blood glucose control and inhibition of glioblastoma stem cell (GSC) growth in animal models. However, CRY isoform selectivity of SHP656 was uncharacterized, limiting understanding of the roles of CRY1 and CRY2. Here, we report the elucidation of CRY2 selectivity of SHP656. SHP656 lengthened cellular circadian period in a CRY2-dependent manner and selectively interacted with CRY2. By determining the X-ray crystal structure of CRY2 in complex with SHP656 and performing molecular dynamics simulations, we elucidated compound interaction mechanisms. SHP656 binding was compatible with the intrinsic CRY2 gatekeeper W417 “in” orientation and also a close “further in” conformation. Perturbation of W417 interaction with the lid loop resulted in a reduced effect of SHP656 on CRY2, supporting an important role of gatekeeper orientation in isoform selectivity. We also identified the R form of SHP656 (called SHP1703) as the active isomer. Treatment with SHP1703 effectively reduced GSC viability. Our results suggest a direct role of CRY2 in glioblastoma antitumorigenesis and provide a rationale for the selective modulation of CRY isoforms in the therapeutic treatment of glioblastoma and other circadian clock-related diseases.
The circadian clock encompasses a cell-autonomous network of molecular interactions that enables endogenous regulation of daily physiological rhythms, while allowing for inputs from environmental cues (1). The suprachiasmatic nucleus (SCN) located in the hypothalamus is the master regulator of circadian behavioral rhythms and receives light/dark signals from the retina for entrainment (2). Peripheral clocks in various tissues throughout the body receive inputs from the SCN, and locally regulate output rhythms such as metabolism in the liver (3). Core components of the circadian clock include the heterodimeric transcription factors CLOCK and BMAL1, which activate widescale gene transcription (4). Genes activated by CLOCK-BMAL1 include Period (Per1 and Per2) and Cryptochrome (Cry1 and Cry2), which encode core components of the transcription–translation negative feedback loop (TTFL). After being translated in the cytosol, PER and CRY proteins form a large complex with other regulatory proteins, and translocate to the nucleus where they repress CLOCK-BMAL1 to close the TTFL (5). PER- and CRY-mediated repression of CLOCK-BMAL1 delays the start of a new circadian cycle, and, for a new cycle to begin, PER and CRY proteins need to be degraded. Over the course of the circadian cycle, PERs are phosphorylated by casein kinase I and CK2, thereby regulating their proteasomal degradation and nuclear localization (6–9). Proteasomal degradation of CRYs is mediated by ubiquitination of the proteins via the E3 ligase FBXL3 (10–12), while its homolog FBXL21 stabilizes CRYs (13, 14).
Proper functioning of the circadian clock is important for physiological homeostasis, and its dysfunction has been associated with a variety of diseases. For example, mutations in CRY1 and CRY2 genes are known to induce sleep phase disorders in humans (15, 16), and the genetic loss of Cry1 and Cry2 in mice results in glucose intolerance (17), while Cry1 overexpression in the liver lowers blood glucose levels by inhibiting glucagon response (18). Modulation of CRY activity was enabled by the discovery of the carbazole-containing compound KL001 (Fig. 1A) as the first synthetic small molecule that targets CRY proteins (19). KL001 interacts with the FAD pockets of CRY1 and CRY2 and competes with FBXL3, thereby inhibiting proteasomal degradation (19–22). In cell-based assays, this stabilization of CRY results in repression of CLOCK-BMAL1 transcriptional activity and lengthening of the circadian period via the TTFL. KL001 also inhibits glucagon-induced gluconeogenesis in primary hepatocytes, suggesting that CRY modulation has therapeutic potential in the treatment of diabetes (19). Based on these findings, orally available KL001 derivatives were developed to facilitate in vivo studies of metabolic disease. Derivatives identified as compound 41 (23) and compound 50 (SHP656) (24) (Fig. 1A) exhibit optimal potency, lipophilicity, and toxicity characteristics, with improved bioavailability (24). These compounds improved glucose clearance in diet-induced obese mice and db/db mice, demonstrating their efficacy in animal models (23, 24). Furthermore, we recently discovered that self-renewing glioblastoma stem cells (GSCs) causing a highly malignant and lethal primary brain tumor are dependent on CLOCK and BMAL1 for their malignancy (25). Both KL001 and SHP656 suppressed patient-derived GSC growth and survival in culture, presumably through inhibition of CLOCK-BMAL1 activity via CRY stabilization, and oral administration of SHP656 prolonged the survival of mice intracranially implanted with GSCs (25). CRY-targeting molecules therefore represent a potential new direction in the treatment of metabolic disease and glioblastoma.
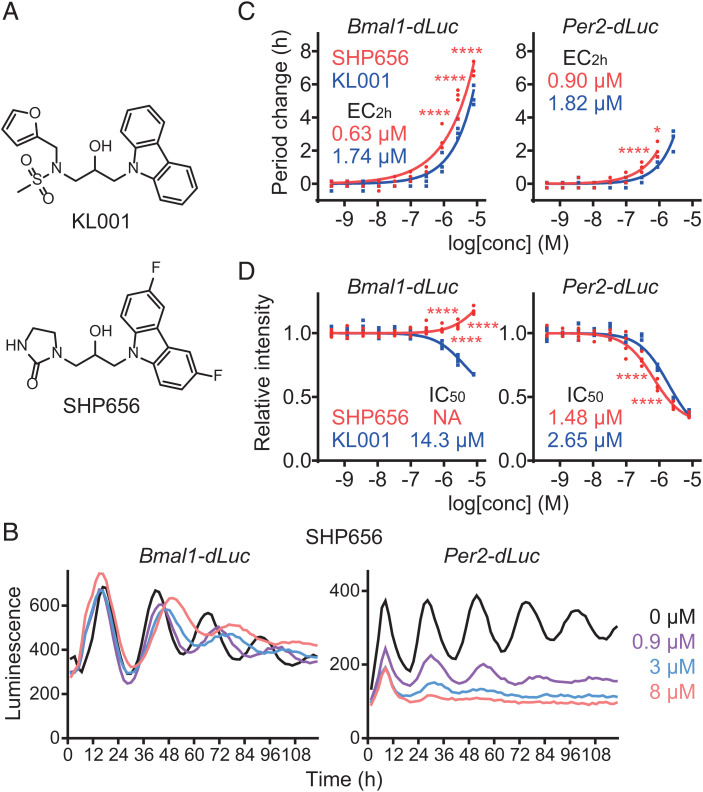
Fig. 1.
SHP656 lengthens circadian period. (A) The chemical structures of KL001 and SHP656. (B–D) Effects on circadian rhythms in Bmal1-dLuc and Per2-dLuc U2OS cells. Luminescence rhythms in the presence of various concentrations of compounds (B, mean of n = 4) and changes in period (C) and luminescence intensity (D) compared to a DMSO control are shown (n = 4 biologically independent samples). When arrhythmic, period is not plotted. Concentrations for 2-h period lengthening (EC2h) and 50% inhibition (IC50) are indicated. ****P < 0.0001; *P < 0.05 relative to KL001 by two-way ANOVA followed by Sidak’s multiple comparisons test.
CRY1 and CRY2 are highly homologous proteins and have been considered to work redundantly. However, these isoforms have been increasingly implicated in distinct regulatory roles. For example, only CRY1 can form a repressive complex with CLOCK-BMAL1 without PER2 (4, 26–28), and only CRY2 enhances degradation of a protooncogene product c-MYC by recruiting FBXL3 (29). Furthermore, emerging evidence indicates that CRY1 and CRY2 may have either protumorigenic or antitumorigenic activity in different types of cancer (30–33). Therefore, isoform-selective molecules will be useful to elucidate differential roles of CRY1 and CRY2, and enable individual control of each isoform. While in vivo efficacy of SHP656 has been reported (24, 25), its isoform selectivity was not previously characterized, partly because it is a derivative of KL001, which targets both CRY1 and CRY2 (19). To better understand the roles of CRY1 and CRY2 in glioblastoma and other disease models, it is imperative that isoform selectivity of this compound is elucidated. In this study, we unexpectedly found that SHP656 preferentially targeted CRY2. We interrogated the molecular basis of SHP656 interaction with CRY2 by determining the X-ray crystal structure of a CRY2-SHP656 complex, in combination with molecular dynamics (MD) simulations and functional analyses of mutant CRY2 proteins. While SHP656 was a racemic mixture, the crystal structure only bound the R form of SHP656, suggesting that this is the active isomer. SHP1703, which contained pure R form, showed higher activity than the S form of SHP656 in Per2 repression and a decrease in GSC viability in culture. Due to the selectivity of SHP656 and SHP1703 for CRY2, our results indicate CRY2 as a potential therapeutic target for the treatment of clock-related diseases such as glioblastoma.
Results
SHP656 Lengthens Circadian Period.
We first evaluated the circadian effect of SHP656 in human U2OS cells harboring either a Bmal1 promoter–luciferase (Bmal1-dLuc) reporter or a Per2-dLuc reporter (Fig. 1B). Treatment with SHP656 lengthened the circadian period in a dose-dependent manner with higher potency than KL001 (Fig. 1C), without affecting cell viability (SI Appendix, Fig. S1). Increasing doses of SHP656 suppressed the intensity of the Per2-dLuc reporter, but not Bmal1-dLuc (Fig. 1D). These results show that SHP656 has a circadian clock–modulating effect.
SHP656 Preferentially Targets CRY2.
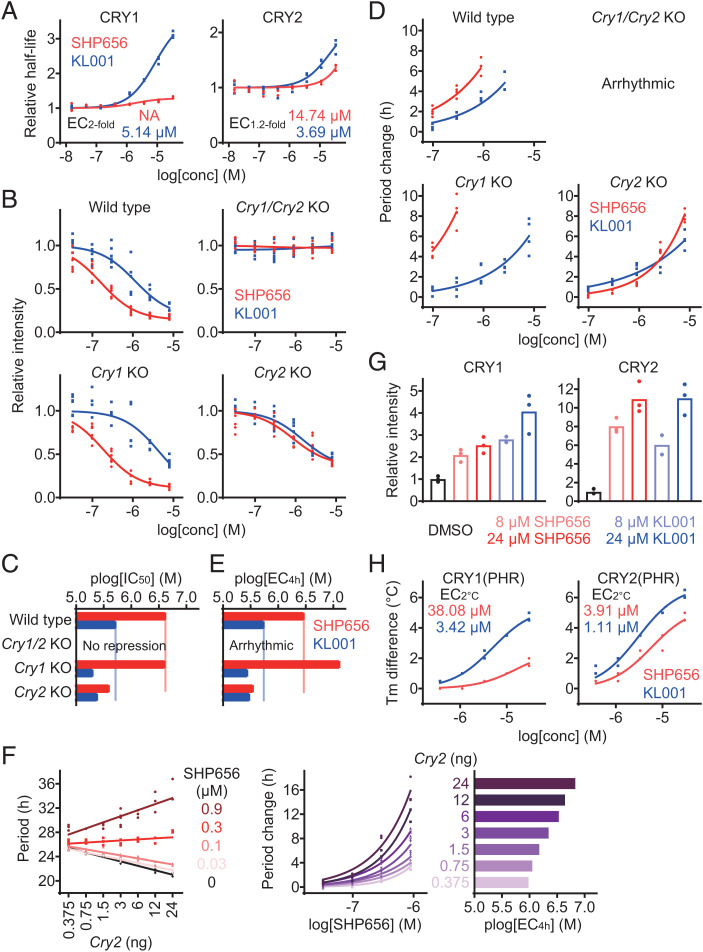
We next evaluated the effect of SHP656 on CRY stability in cell-based degradation assays using HEK293 cells harboring a CRY1-LUC or CRY2-LUC fusion protein reporter (Fig. 2A). Compared with KL001, which stabilized both CRY1 and CRY2, SHP656 showed preference for CRY2, while the overall effect was less than KL001. We further analyzed the effects of SHP656 on endogenous CRY1 and CRY2 activity, by evaluating Per2 reporter expression in fibroblasts from Per2::Luc knock-in mice. SHP656 repressed Per2::Luc reporter intensity with a higher potency than KL001 in wild-type cells in the presence of both CRY1 and CRY2 (Fig. 2 B, Top Left). This effect was CRY dependent, because no Per2 repression occurred in Cry1/Cry2 double-knockout cells (Fig. 2 B, Top Right). SHP656 was effective in Cry1 knockout cells in a manner similar to wild-type cells, consistent with its effect on CRY2 (Fig. 2 B, Bottom Left and C). In Cry2 knockout cells, SHP656 showed reduced Per2 repression activity when compared to wild-type cells (Fig. 2 B, Bottom Right and C). We also evaluated the period lengthening effect in Per2::Luc knock-in fibroblasts. SHP656 showed period lengthening in wild-type cells with greater potency than KL001 (Fig. 2 D, Top Left). In Cry1 and Cry2 knockout cells, SHP656 produced a greater and lesser period change, respectively, compared to wild-type cells (Fig. 2 D, Bottom and E). Increased sensitivity to CRY2 activation by SHP656 in Cry1 knockout cells can be a result of dosage compensation of Cry2 in Cry1 knockout cells (34). We therefore analyzed the effect of SHP656 in Cry1/Cry2 double-knockout fibroblasts rescued with different amounts of CRY2 together with a fixed amount of CRY1 (Fig. 2F). The period-lengthening effect of SHP656 was enhanced with an increasing amount of CRY2, supporting the idea that higher expression of CRY2 increases sensitivity to SHP656. These data indicate that SHP656 preferentially activates CRY2 to suppress Per2 activity and lengthen the circadian period.
Fig. 2.
SHP656 shows selectivity against CRY2. (A) Effects of SHP656 and KL001 on CRY degradation in HEK293 cells. The half-lives of CRY1-luciferase fusion protein CRY1-LUC and CRY2-LUC relative to LUC are plotted by setting a DMSO control to one (n = 3 biologically independent samples). Concentrations for twofold (EC2-fold for CRY1) or 1.2-fold (EC1.2-fold for CRY2) stabilization are indicated. (B and C) Effects on Per2::Luc knock-in reporter activity in wild-type (Top Left, B), Cry1/Cry2 double-knockout (Top Right), Cry1 knockout (Bottom Left), and Cry2 knockout (Bottom Right) fibroblasts. Changes in luminescence intensity compared to a DMSO control are shown (B, n = 6 biologically independent samples). Concentrations for 50% inhibition (plog[IC50] that represents −log[IC50]) are plotted in C. (D and E) Effects on circadian period in Per2::Luc knock-in wild-type (Top Left, D), Cry1/Cry2 double-knockout (Top Right), Cry1 knockout (Bottom Left), and Cry2 knockout (Bottom Right) fibroblasts. Changes in period compared to a DMSO control are shown (D, n = 4 biologically independent samples). When arrhythmic, period is not plotted. Concentrations for 4-h period lengthening (plog[EC4h] that represents −log[EC4h]) are shown in E. (F) Effect of SHP656 on cellular circadian period of Bmal1-Eluc reporter rhythms in Cry1/Cry2 double-knockout fibroblasts rescued with 3 ng of Cry1 plasmid together with different amounts (0.375 ng to 24 ng) of Cry2 plasmid (n = 3 to 6 biologically independent samples). Circadian period is plotted in Left. Changes in period compared to a DMSO control of each Cry2 amount are shown in Middle. Concentrations for 4-h period lengthening (plog[EC4h]) are plotted in Right. (G) Interaction with CRY proteins in HEK293T cells. The band intensities of Flag-tagged CRY1 and HA-tagged CRY2 proteins protected from thermal denaturation in cells are plotted by setting a DMSO control to one (mean of n = 3 biologically independent samples). Compound interaction induced thermal stabilization. (H) Interaction with CRY1(PHR) and CRY2(PHR) in vitro. Changes in denaturing temperatures of recombinant CRY(PHR) proteins in the presence of various concentrations of compounds compared to a DMSO control are shown (n = 3 biologically independent samples). Concentrations for 2 °C stabilization (EC2 °C) are indicated.
To evaluate the interaction of SHP656 with CRY proteins, a cellular thermal shift assay in HEK293T cells expressing CRY1-Flag and CRY2-HA was conducted (Fig. 2G). Compared to KL001, SHP656 showed a preference for CRY2 over CRY1. Furthermore, SHP656 showed a preference for recombinant CRY2(PHR) over CRY1(PHR) compared to KL001 in an in vitro thermal shift assay (Fig. 2H). Together, these data indicate that SHP656 exerts its effects by directly and preferentially interacting with CRY2.
The Binding Mode of SHP656 in CRY2 Crystal Structure.
To obtain insights into the effect of SHP656 on CRY2, we determined the X-ray crystal structure of a CRY2-SHP656 complex at a resolution of 1.95 Å (Protein Data Bank [PDB] ID: 7V8Z) (SI Appendix, Table S1). The overall structure (SI Appendix, Fig. S2A) formed canonical photolyase folds similar to published literature (20, 26, 35, 36), and the conformations of FAD pocket residues (Fig. 3A) were similar to CRY2-KL001 (PDB ID: 4MLP) and CRY1-KL001 (PDB ID: 7DLI) structures (21, 22) (Fig. 3B). Electron density was, overall, well defined, enabling accurate model building of the compound and interacting residues.
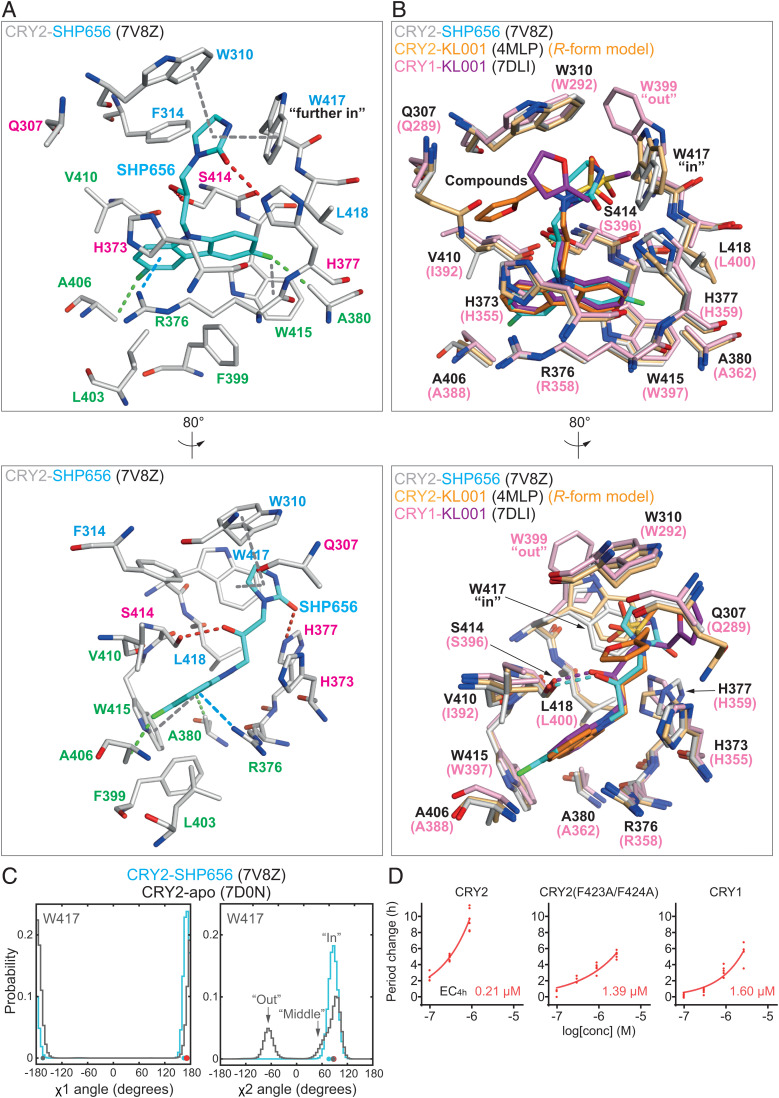
Fig. 3.
SHP656 interacts with CRY2 W417 gatekeeper. (A) Crystal structure of CRY2-SHP656 (PDB ID: 7V8Z). The binding mode of SHP656 in the FAD pocket of CRY2 is shown with a cation–π interaction in blue dashed line; C–H interactions in green dashed lines; F−π interactions, π−π stacking, and T-shaped stacking in gray dashed lines; and H-bonds in red dashed lines. Residues comprising hydrophobic region 1, the affinity region, and hydrophobic region 2 are colored blue, magenta, and green, respectively. Lower is an 80° rotated view around the y axis with respect to Upper. (B) Superposition of KL001 onto SHP656. CRY1-KL001 (magenta–purple) (PDB ID: 7DLI) and CRY2-KL001 (PDB ID: 4MLP) with the R form of KL001 (light orange–orange) were modeled into the FAD pocket of CRY2-SHP656 (white–cyan). Lower is an 80° rotated view around the y axis with respect to Upper. (C) Gatekeeper W417 χ1 and χ2 angles in MD simulations. W417 in CRY2-SHP656 (cyan) and CRY2-apo (gray) exhibited very similar conformations throughout the simulation. Major population peaks correlated closely to the χ1 and χ2 angles observed in the crystal structures (cyan and gray dots). Energy minimized CRY2-SHP656 is represented by red dots. (D) Effect of SHP656 on cellular circadian period of Bmal1-Eluc reporter rhythms in Cry1/Cry2 double-knockout fibroblasts rescued with CRY2, CRY2(F434A/F424A), or CRY1. Changes in period compared to a DMSO control are shown (n = 6 biologically independent samples). When arrhythmic, period is not plotted. Concentrations for 4-h period lengthening (EC4h) are indicated.
The 3,6-difluorocarbazole moiety of SHP656 bound to CRY2 in hydrophobic region 2 of the FAD pocket. This region comprises R376, A380, F399, L403, A406, V410, and W415, and formed a hydrophobic annulus for the difluorocarbazole. In addition to R376 C-β and C-δ hydrophobic interactions with the carbazole, the guanidinium group of R376 formed a stable cation-π interaction (Fig. 3A). These interactions were analogous to CRY2 R376 and CRY1 R358 interactions with KL001 (Fig. 3B). In addition, the difluoro functional groups in SHP656 may form unique C–H interactions with A380 and A406, or a possible F−π contact with W415 to stabilize the orientation of the carbazole moiety (Fig. 3A).
The hydroxypropyl linker that connects the difluorocarbazole and 2-imidazolidinone groups in SHP656 formed a canonical hydrogen bond with S414 in the affinity region, almost identical to CRY1 and CRY2 interactions with KL001 (Fig. 3 A and B). The R form of SHP656 bound to CRY2 in the CRY2-SHP656 structure (Fig. 3A), similar to the binding mode of CRY1-KL001 (PDB ID: 7DLI) (Fig. 3B). Furthermore, we determined that the R form of KL001 is the correct isomer in CRY2-KL001 (PDB ID: 4MLP) (22). These data suggest that the R form is the active isomer of these compounds.
In contrast to the high chemical similarity of the carbazole and hydroxypropyl linkers of SHP656 and KL001, different functional groups occupied hydrophobic region 1 of the FAD pocket: a 2-imidazolidinone (SHP656) and a methanesulfonamide (KL001) (Fig. 3 A and B). Hydrophobic region 1 is composed of residues W310, F314, W417, and L418 in CRY2, corresponding to W292, F296, W399, and L400 in CRY1. This region is considerably smaller than region 2 and provides a hydrophobic environment in which compound groups nestle. We recently identified intrinsic differences in the conformation of a key hydrophobic region 1 residue (CRY2 W417, equivalent to CRY1 W399) which was named the gatekeeper, due to distinct conformations regulating compound binding in this region (22). The 2-imidazolidinone group in SHP656 has considerably less steric bulk and less potential flexibility compared to the methanesulfonamide in KL001 and formed a hydrophobic interaction with W417 as well as an edge-to-face interaction with W310 (Fig. 3A). Compared to CRY2-apo (PDB ID: 7D0N) and CRY2-KL001, SHP656 induced a small conformational change in the gatekeeper that we designated “further in” (Fig. 3A and SI Appendix, Fig. S2B).
A space group in CRY2-SHP656 that is different from the CRY2-apo crystal structure induced different crystal packing around the lid loop, and the close proximity of a crystallographic symmetry-related molecule in CRY2-SHP656 resulted in a flipped lid loop where F428 was positioned adjacent to W417 (SI Appendix, Fig. S2 B and C). It is possible that this lid loop conformation pushed W417 further in. However, a similar lid loop conformation in CRY2-KL001 did not result in a “further in” W417 conformation, but rather the intrinsic “in” conformation as observed in CRY2-apo (SI Appendix, Fig. S2B).
In addition to stacking interactions, the carbonyl of the 2-imidazolidinone potentially formed an H-bond with H377 and may have influenced the orientation of the 2-imidazolidinone (Fig. 3A). Structural activity relationship analyses have determined that the addition of substituents at the 3 position of the 2-imidazolidinone reduced compound potency (24). Moreover, substitution of the 2-imidazolidinone for bicyclic lactams significantly reduced potency (24), most likely through steric clashes with residues that make up the constricted hydrophobic region 1. Overall, the crystal structure of CRY2-SHP656 revealed binding mechanisms that accounted for the effect of SHP656 on CRY2.
MD Simulations.
To evaluate the interaction between the gatekeeper W417 and the 2-imidazolidinone of SHP656, we performed MD simulations in solution. This approach enables the characterization of energetically favorable and predominant conformations of residues in the absence of restraints by a neighboring (crystal lattice) molecule. In the crystal structures, the χ1 and χ2 angles of W417 in CRY2-SHP656 and CRY2-apo differed by 30° and 10°, respectively. After energy minimization for MD simulations, W417 in CRY2-SHP656 adopted χ1 and χ2 angles almost identical to CRY2-apo and remained close to these values throughout the combined 600-ns simulation (Fig. 3C), indicating stabilization of W417 at the “in” conformation by SHP656. The stacking interaction between W417 and F428 was predominantly maintained, and F428 showed only a low level of χ1 and χ2 angle variability (SI Appendix, Fig. S3 A and B).
The 2-imidazolidinone group of SHP656 adopted two major orientations, designated clusters C2 and C3, and one minor orientation, C1 (SI Appendix, Fig. S3C). C2 was the most similar to the conformation in the crystal structure and also the minimized structure for the MD simulation. Throughout the simulation, the 2-imidazolidinone kept interacting distance from W417 (SI Appendix, Fig. S3D), with a mean interaction energy of −1.89 ± 0.57 kcal/mol. The potential H-bond between H377 and the carbonyl group of the 2-imidazolidinone was transient in the simulation, exhibiting an interaction energy of −0.97 ± 0.67 kcal/mol. Together, the data show that W417 “in” was the predominant conformation and energetically favorable for SHP656 binding. The “further in” conformation observed in the CRY2-SHP656 crystal structure was likely caused by crystal packing.
Lid Loop Mutation Affects SHP656 Period Length Regulation.
Interaction of the SHP656 2-imidazolidinone group with CRY2 gatekeeper W417 suggested a role of this residue in CRY2 selectivity of SHP656, because the corresponding CRY1 W399 has an “out” conformation (Fig. 3B). We recently found, in cell-based degradation assays, that a CRY2(F423A/F424A) mutant showed reversed response to CRY1- and CRY2-selective compounds KL101 and TH301, respectively, potentially by affecting W417 orientation (22). We therefore evaluated the effect of this mutation on SHP656 response. Because SHP656 showed only a small effect on CRY1 and CRY2 stability, albeit with a greater effect on CRY2 (Fig. 2A), we evaluated its effect on the circadian period of a Bmal1-Eluc reporter in Cry1/Cry2 double-knockout mouse fibroblasts rescued with CRY2 or CRY1 (Fig. 3D). SHP656 elicited a more potent period-lengthening effect in CRY2-rescued cells than CRY1-rescued cells. In contrast, CRY2(F423A/F424A) dramatically reduced SHP656 response to a level comparable to CRY1 wild type (Fig. 3D). These results indicate that CRY2 lid loop mutation interferes with SHP656 selectivity mechanisms.
R Form of SHP656 Is the Active Form.
Although SHP656 was a racemic mixture, only the R form of SHP656 bound to CRY2 in the CRY2-SHP656 crystal structure (Fig. 3A), suggesting that this is the active form. We therefore synthesized SHP1703, which contained pure R form (Fig. 4A and SI Appendix, Fig. S4A), and analyzed its effect on circadian rhythms of Bmal1-dLuc and Per2-dLuc reporters in U2OS cells (Fig. 4 B and C). As expected, SHP1703 showed higher potency than SHP656 in period lengthening and Per2 repression. In contrast, the S form of SHP656 (SHP1704) exhibited >10-fold lower potency than SHP1703 in Per2 repression (SI Appendix, Fig. S4B). Furthermore, SHP1703 had a greater stabilizing effect than SHP656 on recombinant CRY2(PHR) in an in vitro thermal shift assay and retained CRY2 selectivity (Fig. 4D). These data support that the R form is the active form.
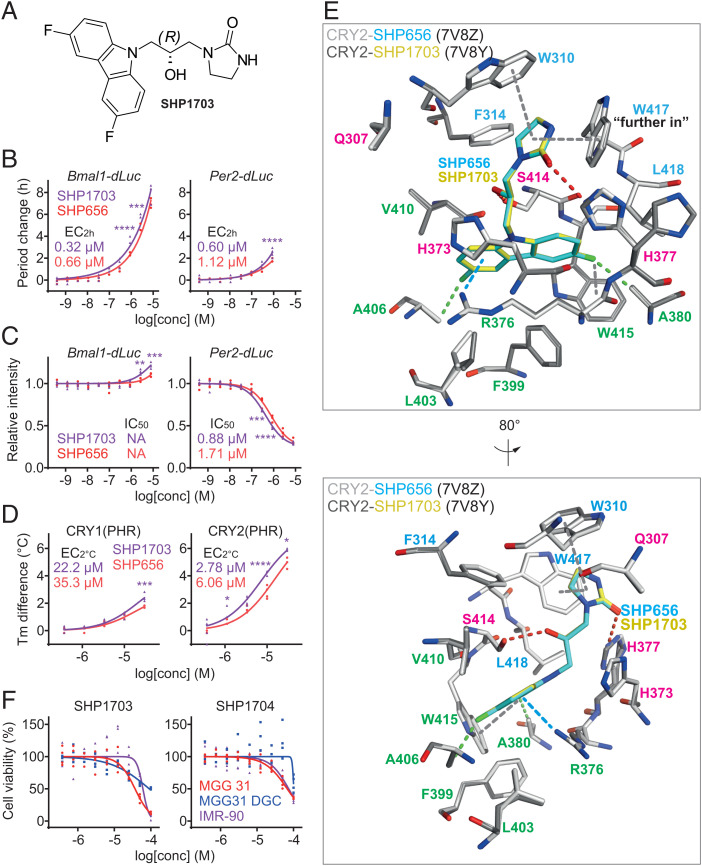
Fig. 4.
The R form of SHP656 is the active isomer. (A) The chemical structure of SHP1703, the R form of SHP656. (B and C) Effects of SHP1703 and SHP656 on circadian rhythms in Bmal1-dLuc and Per2-dLuc U2OS cells. Changes in period (B) and luminescence intensity (C) compared to a DMSO control are shown (n = 4 biologically independent samples). When arrhythmic, period is not plotted. Concentrations for 2-h period lengthening (EC2h) and 50% inhibition (IC50) are indicated. ****P < 0.0001; ***P < 0.001; **P < 0.01 relative to SHP656 by two-way ANOVA followed by Sidak’s multiple comparisons test. (D) Interaction with CRY1(PHR) and CRY2(PHR) in vitro. Changes in denaturing temperatures of recombinant CRY(PHR) proteins in the presence of various concentrations of compounds compared to a DMSO control are shown (n = 3 to 4 biologically independent samples). Concentrations for 2 °C stabilization (EC2 °C) are indicated. ****P < 0.0001; ***P < 0.001; *P < 0.05 relative to SHP656 by two-way ANOVA followed by Sidak’s multiple comparisons test. (E) Crystal structure of CRY2-SHP1703 (PDB ID: 7V8Y). Superposition of CRY2-SHP656 (white–cyan) and CRY2-SHP1703 (gray–yellow) indicated identical conformations and binding modes. A cation–π interaction is shown in blue dashed line; C–H interactions are shown in green dashed lines; F−π interactions, π−π stacking, and T-shaped stacking are shown in gray dashed lines; and H-bonds are shown in red dashed lines. Residues comprising hydrophobic region 1, the affinity region, and hydrophobic region 2 are colored blue, magenta, and green, respectively. H377 was flexible in both CRY2-SHP656 and CRY2-SHP1703 structures but was only modeled with an alternate conformation in CRY2-SHP1703. Lower is an 80° rotated view around the y axis with respect to Upper. (F) Effects of SHP1703 (Left) and SHP1704 (Right) on cell viability (n = 3 to 4 biologically independent samples). The 50% effective concentrations (EC50) are indicated in the main text. Statistical significance by two-way ANOVA followed by Tukey’s multiple comparisons test are summarized in SI Appendix, Table S2.
For further evaluation of the active form, we determined the crystal structure of CRY2-SHP1703 at a resolution of 1.9 Å (PDB ID: 7V8Y) (SI Appendix, Table S1). Superposition of CRY2-SHP656 and CRY2-SHP1703 showed almost identical conformations and interactions of compounds and residues (Fig. 4E). In CRY2-SHP1703, H377 was modeled with a split conformation. This alternate conformer was also present in CRY2-SHP656, albeit to a slightly lesser degree, and not modeled. Typically, in CRY2 structures, H377 is reasonably stable in a conformation that would position it within interacting distance of the compound (22), and would therefore be compatible with H-bond formation with the 2-imidazolidinone group. The conformation of H377 in CRY2-apo was restrained by Y431, which is positioned adjacent to H377. In CRY2-SHP656 and CRY2-SHP1703 structures, but not CRY2-apo, disulfide bonds were observed between C430 and C381, which correlated to increased flexibility of Y431 and likely facilitated the alternative H377 conformation. Overall, the chemical synthesis and crystal structures indicated that SHP656 R form is the active form.
SHP1703 Reduces Glioblastoma Cell Viability.
Given that SHP656 was shown to suppress GSC growth (25), we compared the effects of SHP1703 and SHP1704, the active and less active enantiomers of SHP656, respectively, in cell viability assays. SHP1703 reduced cell viability to a greater extent in patient-derived GSCs (MGG 31 cells, 50% effective concentration [EC50] 36.9 µM), compared to differentiated glioblastoma cells (MGG 31 DGC cells, EC50 90.6 µM) and noncancerous human lung fibroblasts (IMR-90 cells EC50 60.8 µM) (Fig. 4 F, Left). In contrast, SHP1704 had a reduced effect on all cell types (MGG 31, EC50 67.3 µM; MGG 31 DGC, EC50 101.6 µM; and IMR-90, EC50 73.9 µM) (Fig. 4 F, Right). These results indicate that CRY2-targeting activity of SHP1703 correlates with specificity against GSCs.
Discussion
We revealed selectivity of SHP656 for CRY2 over CRY1 and characterized its interaction with CRY2 both structurally and functionally. Our results provided molecular insights into the selective binding of SHP656 to CRY2 and identified the R form (SHP1703) as the active isomer. We further determined that the selective activation of CRY2 inhibits cell viability of patient-derived GSCs in culture, directly implicating CRY2 in the regulation of GSC growth and survival.
We previously identified conformational isomerism in the gatekeeper residues CRY2 W417 and CRY1 W399, where CRY2 W417 intrinsically adopts an “in” conformation and CRY1 W399 adopts an “out” conformation (22). These distinct conformations have a pronounced effect on the size and type of functional groups that can be accommodated in hydrophobic region 1. CRY1-selective compounds like KL101 and KL201 typically have functional groups with greater steric bulk that form hydrophobic or C−π stacking interactions with an “out” gatekeeper (PDB IDs: 6KX6 and 6LUE) (36, 37). Other CRY1-selective compounds TH303 and TH129 (PDB IDs: 7D1C and 7D19) have even greater steric bulk, due to the incorporation of a benzophenone moiety, which forms steric overlap with an “out” gatekeeper, forcing it to adopt a new conformation buried in the auxiliary pocket (38). Superposition of CRY1-KL101, CRY2-apo, and CRY2-TH301 onto CRY2-SHP656 showed their differential gatekeeper conformations (SI Appendix, Fig. S5A). The dimethylphenyl of KL101 severely clashes with the “in” gatekeeper of CRY2-TH301, and also the slightly rotated “further in” gatekeeper conformation of the CRY2-SHP656 crystal structure (SI Appendix, Fig. S5A). In contrast, TH301 binding to CRY2 (PDB ID: 6KX8) forms a stacking interaction with the intrinsically “in” conformation of W417 in CRY2. The 2-imidazolidinone of SHP656 has significantly less steric bulk than CRY1-selective KL101 and is similar in size to the cyclopentane group of moderately CRY2-selective TH301 (SI Appendix, Fig. S5A). Minimization of the CRY2-SHP656 structure and subsequent MD data suggested that, in the absence of crystal packing, the gatekeeper predominantly adopted a conformation close to the “in” conformation observed in CRY2-TH301 and CRY2-apo. However, a small degree of wobble around the “in” orientation could indicate that SHP656 is more tolerant of small W417 χ2-angle rotation. These data suggest that W417 “further in” was likely induced by crystal packing–mediated movement of the lid loop and reorientation of W417 to avoid steric overlap with F428. Although this potentially represents an artificial lid loop conformation, albeit a stable one, F428 in CRY2-PER2 (PDB ID: 4U8H) is also orientated toward W417, which, in turn, is rotated even further around the χ2 angle to a “middle” position (SI Appendix, Fig. S5B). If SHP656 can bind to a W417 “middle” conformation, then it may be able to bind to a CRY2-PER2 complex. Overall, the data support that SHP656 stably binds to an “in” gatekeeper conformation but can tolerate “further in,” both of which are more easily adopted in CRY2 than CRY1, suggesting a potential mechanism for CRY2 selectivity. Functional assays supported that the conformation of the gatekeeper mediated by gatekeeper–lid loop interactions is an important mechanism in CRY isoform selectivity. Because CRY2 F423A/F424A mutation was shown to reduce the response to TH301 and increase the response to KL101 (22), which requires a gatekeeper “out” conformation due to its bulky dimethylphenyl moiety in hydrophobic region 1, it is possible that the reduced SHP656 response was due to repositioning of W417 to an “out” conformation, which would subsequently lose a stacking interaction with the 2-imidazolidinone group.
We previously showed that disulfide bond formation between C430 (at the C-terminal end of the lid loop) and C381 in the FAD pocket can reposition Y431, thereby providing additional conformational freedom of H377, which is in close proximity to Y431 (22). In correlation to this observation, the C430–C381 disulfide bond in CRY2-SHP656 and CRY2-SHP1703 crystal structures resulted in disorder of the side chain of Y431, which was truncated at the β-carbon due to poorly interpretable electron density, and a split H377 conformation. This Y431 and H377 flexibility was reflected in their χ2 and χ1 angles, respectively, during CRY2-SHP656 MD simulation (SI Appendix, Fig. S6A). In contrast, our previous CRY2-apo (no disulfide bond) simulation showed H377 and Y431 to be more stable, and the major orientation of H377 positioned the side chain within H-bonding distance of the 2-imidazolidinone carbonyl (22) (SI Appendix, Fig. S6A). The greater conformational freedom of H377 in disulfide bond–containing CRY2-SHP656 may have hindered more stable hydrogen bonding to the 2-imidazolidinone carbonyl, because intermittent loss of the H377–2-imidazolidinone interaction, during MD simulation frames, correlated to transient H-bonding between H373 and the 2-imidazolidinone. In the absence of a disulfide bond, like in the CRY2-apo structure, it is likely that H377 would maintain an interaction with the 2-imidazolidinone to optimally position SHP656 in the FAD pocket of CRY2 in combination with W417 “in.” We therefore performed an additional MD simulation of CRY2-apo docked with SHP656 (CRY2-apo-SHP656 model), which supported this hypothesis. In the CRY2-apo-SHP656 model, Y431 was more stabilized than in CRY2-SHP656, and H377 maintained a position within H-bonding distance of the 2-imidazolidinone carbonyl (SI Appendix, Fig. S6A). These differences resulted in higher SHP656–H377 interaction and more favorable free energy of binding in the CRY2-apo-SHP656 model (−1.81 ± 1.14 kcal/mol), compared to CRY2-SHP656 (−0.97 ± 0.67 kcal/mol). In addition, W417 in CRY2-apo-SHP656 was more stabilized at the “in” position than CRY2-apo (SI Appendix, Fig. S6B), further supporting a gatekeeper-mediated mechanism of compound selectivity.
Progress in the development of novel therapy options for glioblastoma has largely stalled. Targeting GSCs has been especially challenging given that GSCs are resistant to both chemotherapy and radiation, which are part of the standard treatment for glioblastoma patients following maximal surgical resection. We previously showed that patient-derived GSCs (T387 GSC, EC50 15.0 µM; T3565 GSC, EC50 8.5 µM) display higher sensitivity to SHP656 treatment compared to control differentiated GSCs (T387 DGC, EC50 94.6 µM; T3565 DGC, EC50 68.4 µM) and noncancerous neural cells (NM263, EC50 55.2 µM; NM290, EC50 47.8 µM) (25). Similarly, we report here that another patient-derived GSC (MGG 31) had higher sensitivity to SHP1703 treatment compared to differentiated GSCs (MGG 31 DGCs) and IMR-90 human lung fibroblasts, while SHP1704 showed reduced effects (Fig. 4F). CRY2 selectivity of SHP656 and SHP1703 indicates that GSCs have higher sensitivity to CRY2 modulation compared to non–stem cell-like cancer cells (T387 DGC, T3565 DGC, and MGG 31 DGC) and noncancerous cells (NM263, NM290, and IMR-90). The higher dependency of GSCs on CLOCK-BMAL1 for stemness maintenance and metabolism makes them more sensitive to CRY activation compared to other cell types (25). EC50 values of SHP656 and SHP1703 for GSC viability suppression (∼10 µM or more) are much higher than those for Per2 repression in U2OS cells and mouse fibroblasts (∼1 µM or less). It is likely that viability suppression is less sensitive to the compounds than Per2 repression, because we previously found that the efficacy of KL001 in viability suppression (∼20 µM) is lower than Per2 gene repression (considerably less than 10 µM) in GSCs (25). Further investigation of in vivo properties and efficacy of SHP1703 will provide a rationale for the use of CRY-targeting compounds as a therapy paradigm for glioblastoma treatment.
In summary, we have determined that SHP656 selectively binds to CRY2. The data show that CRY2 can regulate GSC growth and survival, and demonstrate the potential of CRY2-selective activation in the treatment of glioblastoma. Future experiments utilizing CRY1-selective compounds could help elucidate overlapping and distinct roles of CRY isoforms in glioblastoma and other types of cancer as well as circadian clock–related diseases.
Materials and Methods
Cell-Based Circadian Assays.
Bmal1-dLuc and Per2-dLuc U2OS cells (39, 40) were suspended in culture medium (Dulbecco’s modified Eagle’s medium [DMEM] [11995-073, Gibco] supplemented with 10% fetal bovine serum, 0.29 mg/mL L-glutamine, 100 units/mL penicillin, and 100 µg/mL streptomycin) and plated onto a white, solid-bottom 384-well plate at 30 µL (3,000 cells) per well. After 2 d, 40 µL of explant medium containing 1 mM luciferin (DMEM [12800-017, Gibco] supplemented with 2% B27 [Gibco], 10 mM Hepes, 0.38 mg/mL sodium bicarbonate, 0.29 mg/mL L-glutamine, 100 units/mL penicillin, 100 µg/mL streptomycin, and 1 mM luciferin; pH 7.2) was dispensed into each well, followed by the application of 500 nL of compounds (final 0.7% dimethylsulfoxide [DMSO]). Luminescence was recorded every 100 min for 5 d in a microplate reader, Infinite M200Pro (Tecan).
To compare the effects of SHP1703 and SHP1704, Per2-dLuc U2OS cells were plated onto a white, solid-bottom 96-well plate at 30,000 cells per well. After 2 d, medium was replaced with 200 µL of cell culture medium supplemented with 5 µM forskolin and 1 mM luciferin, followed by the application of 1 µL of compounds (final 0.5% DMSO). Luminescence was recorded for >3 d in a microplate reader, Infinite M200 (Tecan).
Wild-type, Cry1/Cry2 double-knockout, Cry1 knockout, and Cry2 knockout fibroblasts harboring a Per2::Luc knock-in reporter (41) were plated on a white, solid-bottom 96-well plate and cultured for 2 d to reach confluency. The cells were treated with forskolin (final 10 µM) for 2 h. Then, the medium was replaced with explant medium containing various concentrations of compounds (final 0.2% DMSO) and 0.2 mM luciferin, and luminescence was recorded every 30 min for 5 d in a luminometer LumiCEC (Churitsu).
Cell Viability Assay.
Bmal1-dLuc and Per2-dLuc U2OS cells were plated by following a cell-based circadian assay protocol. After 5 d of compound application, CellTiter-Glo Reagent (Promega) was applied to each well, and luminescence corresponding to the cellular ATP level was recorded in a multimode reader, Cytation3 (BioTek).
Degradation Assay.
Stable HEK293 cells harboring a C-terminally luciferase-fused CRY1 (CRY1-LUC), CRY2-LUC, or LUC reporter (19) were suspended in culture medium and plated onto a white, solid-bottom 96-well plate at 100 µL (30,000 cells) per well. After 24 h, 1 µL of compounds (final 1% DMSO) were added to the medium. After another 24 h, the medium was supplemented with luciferin (final 0.1 mM) and Hepes-NaOH (pH7.2; final 10 mM). After 1 h, cycloheximide (final 20 µg/mL) was added, and the luminescence was recorded every 10 min for 18 h in Infinite M200Pro.
Per2::Luc Repression Assay.
Wild-type, Cry1/Cry2 double-knockout, Cry1 knockout, and Cry2 knockout fibroblasts harboring a Per2::Luc knock-in reporter were plated on a white, solid-bottom 384-well plate and cultured for 2 d to reach confluency, followed by the application of 500 nL of compounds (final 0.7% DMSO). After 2 d, the medium was replaced with BrightGlo (Promega), and luminescence was recorded in Cytation3.
Cry Rescue Assay.
Functional rescue of Cry1/Cry2 double-knockout mouse embryonic fibroblasts with CRY expression vectors was performed as described previously (42) with modifications. Cry1/Cry2 double-knockout fibroblasts (15,000 cells) were plated onto a white, solid-bottom 96-well plate. After 24 h, they were cotransfected by Fugene 6 (Promega) with 3 ng (or the indicated amount in the CRY2 dose-dependency experiment) of an expression vector for CRY or its mutant (in pMU2-P(Cry1)-FLAG-I/RRE) and 100 ng of a Bmal1-Eluc reporter vector (36). After 24 h incubation, fresh culture medium was supplied. After 2 d, the cells were treated with forskolin (final 10 µM) for 2 h. Then, the medium was replaced with explant medium containing 0.2 mM luciferin, followed by the application of 500 nL of compounds (final 0.4% DMSO). Luminescence was recorded every 36 min in Infinite M200Pro for 5 d.
Cellular Thermal Shift Assay.
HEK293T cells were reverse cotransfected on a 10-cm dish by Polyethylenimine (Polysciences) with 18 µg each of an expression vector for Flag-tagged CRY1 and HA-tagged CRY2 (36). After 2 d, the cells were suspended in serum-free DMEM supplemented with Complete Protease Inhibitor Mixture (Roche), dispensed into a PCR tube, and treated with various concentrations of compounds (final 0.7% DMSO) at 37 °C for 1 h, followed by heat treatment for 3 min. The optimized temperature for heat treatment is as follows: CRY1, 55 °C; CRY2, 49 °C. They were subjected to freeze–thaw cycles (three times) and centrifuged (18,000 × g) at 4 °C for 20 min. The supernatants were treated with sodium dodecyl sulfate (SDS) sample buffer, separated by SDS polyacrylamide gel electrophoresis, and analyzed by immunoblotting with anti-Flag-HRP (A8592, Sigma) and anti-HA-HRP (12013819001, Roche) antibodies.
Recombinant CRY Expression and Purification.
His6-MBP-CRY2(PHR) bacmid (36) was transfected into Sf9 (Spodoptera frugiperda) insect cells (Invitrogen) to produce recombinant P1 baculovirus. Sf9 cells were infected with P1 virus at a density of 1.2 × 106 cells per mL in Sf-900 II SFM medium (Gibco) (supplemented with 0.5× final penicillin-streptomycin-amphotericin) and grown at 27 °C for 4 d before collection. Cell pellets were flash-cooled in liquid nitrogen and stored at −80 °C.
His6-MBP-CRY2(PHR) cell pellets were resuspended in lysis buffer (1× phosphate-buffered saline, 50 mM NaNO3, 1% [vol/vol] glycerol, 0.1% Triton X-100, and Complete Protease Inhibitor Mixture [Roche]; pH 7.4) and sonicated on ice for 8 min. Lysates were centrifuged (19,000 × g) at 4 °C for 90 min. The supernatant was passed through a 0.45-μm filter and then loaded onto a HisTrap HP 5-mL column (GE Healthcare). The column was attached to a high-performance liquid chromatography system and washed with 100 mL of wash buffer (50 mM Tris⋅HCl, 100 mM NaCl, 10 mM NaNO3, 10% [vol/vol] glycerol, 15 mM imidazole; pH 7.5), before elution with a 20-column volume gradient of 15 mM to 500 mM imidazole in 50 mM Tris⋅HCl, 100 mM NaCl, 10 mM NaNO3, 10% (vol/vol) glycerol; pH 7.5. Pooled fractions were cleaved overnight with Tobacco Etch Virus (TEV) protease to remove the His6-MBP tag. TEV recognizes the sequence ENLYFQ/G and cuts before the Gly, leaving the additional residues Gly-Thr at the N terminus of the target protein. The protein was subsequently purified via a HiTrap Heparin HP column (GE Healthcare) with a linear gradient of 0.05 M to 1 M NaCl in 20 mM Tris⋅HCl, 50 mM NaNO3, 5% (vol/vol) glycerol, and 2 mM dithiothreitol (DTT); pH 7.5. The protein was further purified by passing the sample over amylose resin (New England Biolabs E8021) in an open chromatography column. The amylose resin was then washed with 20 mM Tris⋅HCl, 100 mM NaCl, 100 mM NaNO3, 5 mM L-Arginine, 5% (vol/vol) glycerol, and 2 mM DTT; pH 7.5, to collect all unbound (TEV-cut) CRY2(PHR) target protein. The pooled fractions were concentrated and subjected to gel filtration chromatography on a Superdex 75 16/60 column (GE Healthcare) equilibrated in 25 mM Tris⋅HCl, 200 mM NaCl, 50 mM NaNO3, 5% (vol/vol) glycerol, and 2 mM DTT; pH 7.5. Protein-containing fractions were pooled, buffer-exchanged, and concentrated to 8 mg/mL using an Amicon Ultra (Merck) concentrator (see next section), aliquoted, flash-cooled in liquid nitrogen, and stored at −80 °C.
Thermal Shift Assay.
CRY2(PHR) and CRY1(PHR) were diluted to 2 μM with DSF buffer (20 mM Hepes-NaOH, 150 mM NaCl, 2 mM DTT; pH 7.5) and dispensed into a 384-well white PCR plate (Bio-Rad) at 17 μL per well, followed by the application of 1 μL of compounds (final 5% DMSO). The mixtures were incubated at room temperature with gentle shaking for 60 min. Then 2 μL of 50× SYPRO Orange in DSF buffer (final 5× SYPRO Orange) was added, and thermal denaturation was performed using a real-time PCR detection system, CFX384 Touch (Bio-Rad).
Protein Crystallization and Structure Determination.
CRY2(PHR) was buffer-exchanged into 20 mM Hepes-NaOH, 150 mM NaCl, and 2 mM DTT; pH 7.5, concentrated to 8 mg/mL, and then mixed with 0.5 mM SHP656 or SHP1703 (final 5% DMSO). After removing insoluble materials by centrifugation, the samples were cocrystallized via hanging-drop vapor diffusion at 20 °C. Then 1 μL of CRY2(PHR)-SHP656 or CRY2(PHR)-SHP1703 complex was mixed with 1 μL of precipitant solution containing 100 mM Hepes-NaOH, 12 to 16% (wt/vol) PEG3350, 100 mM to 150 mM NaCl; pH 7.0. The crystals were cryoprotected in mother liquor plus 30% (vol/vol) ethylene glycol, and flash-cooled in liquid nitrogen.
X-ray diffraction data for CRY2-SHP656 and CRY2-SHP1703 were collected at BL17A of the Photon Factory (KEK) at a wavelength of 0.98 Å and a temperature of 100 K. The datasets were processed with DIALS (Diffraction Integration for Advanced Light Sources) and xia2 (43), and scaled with SCALA (44) in the CCP4 suite (45). The structures of CRY2-SHP656 and CRY2-SHP1703 were determined by molecular replacement (MR) with CRY2-TH301 (PDB ID: 6KX8) as a search molecule in the program Phaser (46). Fifteen residues surrounding the FAD-binding pocket were removed from the MR templates in order to reduce model bias. CRY2-SHP656 and CRY2-SHP1703 were both solved in space group P65 (1 mol/AU). Density modification was performed with the program PARROT (47), part of the CCP4 suite (45), utilizing solvent flattening and histogram matching techniques to produce electron density maps with improved phase probability distributions. Model building was performed iteratively using Coot (48) and alternated with refinement in REFMAC5 (49). Final refinement was performed in PHENIX (Python-based Hierarchical ENvironment for Integrated Xtallography) (50).
MD Simulations.
MD simulations were performed for the CRY2-SHP656 complex and for the CRY2-apo-SHP656 model complex built by aligning the crystal conformation of SHP656 onto the FAD pocket of our previous CRY2-apo model (22). Such simulations were compared to previously reported simulations of CRY2-apo (22). In the case of the CRY2-SHP656 complex, missing residues (297 and 298) were de novo modeled using the Crosslink Protein module available in Schrodinger software (Schrodinger 2019). For all simulations, restrained energy minimization and protonation states, at pH = 7, were assigned following pKa calculations with the PROPKA program available in the Protein preparation module of Schrodinger software (Schrodinger 2019). The Antechamber package was used to create parameters, General Amber force field, for the SHP656 ligand (51). The protein–ligand system was solvated using a TIP3P (Transferable Intermolecular Potential with 3 Points) water box with a minimum edge distance of 10 Å from protein atoms. The MD simulations were performed with the Amber99sb-ildn force field. After neutralization, Na+ and Cl− ions were added to maintain an ionic concentration of 0.15 M. All MD simulations were performed using GROMACS (52).
The system was initially minimized with 2,000 cycles of steepest descent followed by 8,000 cycles of a conjugate gradient. After energy minimization, the system was equilibrated in an NVT (fixed number of particle, constant volume and temperature) ensemble for 500 ps followed by 500-ps equilibration in NPT (fixed number of particle, constant pressure and temperature). Each equilibration was performed with position restraint force on backbone atoms (400 KJ⋅mol−1⋅nm−2) and sidechain atoms (40 KJ⋅mol−1⋅nm−2). The temperature was maintained at 310 K using the modified Berendsen Thermostat (V-rescale), and pressure was maintained at 1 atm using Berendsen Barostat with coupling time constants of 1 ps. The pressure of the system was further stabilized for 1 ns with the Parinello–Rahman barostat. Finally, a 200-ns production run was performed (500 ns for CRY2-apo-SHP656), without any restraints, in an NPT ensemble with the temperature maintained at 310 K and pressure at 1 bar. Three independent simulations were completed and were used for analyses.
The orientation between W417 and the ligand was monitored by following the angle defined by the W417 planar ring and the ligand 2-imidazolidinone group planar ring. All angle distributions were calculated using the cpptraj program (53). Three distribution ensembles were observed, and representative structures for each ensemble were clustered based on the rmsd of all pairs of internal distances calculated for the nonhydrogen atoms of W310, H373, H377, W417, F424, and F428, and the SHP656 ligand. An average linkage type of hierarchical agglomerative (bottom-up) algorithm was used for clustering, which was stopped when the minimum distance between clusters was greater than 2 Å (or a minimum of 10 clusters remained). The binding free energy (enthalpic term) between SHP656 and Cry2 was calculated using MMGBSA from 1,000 frames extracted from the trajectories (54).
SHP1703 Synthesis.
Compounds 1 and 2 (SI Appendix, Fig. S4A) were charged to a reactor. Anhydrous toluene, t-BuONa, t-Bu3PHBF4, and Pd2(dba)3 were added sequentially under nitrogen. After reaction completion, Celite-545 was added, and the mixture was filtered with a toluene rinse. The filtrate was distilled, and the resulting compound 3 was concentrated to dryness on a rotary evaporator. After reaction completion, Celite was added, and the mixture was filtered with a Celite pad. The reactor and filter were rinsed with toluene. The material was distilled and concentrated to dryness. Crude compound 3 was mixed with DMAc. Potassium carbonate was added under nitrogen, followed by PCy3HBF4 addition. Pd(OAc)2 was then added under nitrogen, and the reaction temperature was increased to 120 °C. After reaction completion, Celite was added, and the mixture was filtered with a Celite pad. The reactor was rinsed with the product using DMAc. The filtrates were distilled to reduce volume, and then crude compound 4 was precipitated in water/heptanes. Toluene was added to the solid to azeotrope water, and the slurry was concentrated to dryness, then slurried in EtOAc and purified on a silica plug. The solvent was removed under vacuum, yielding a white to off-white powder.
N,N-dimethylformamide was charged to the reactor, and compound 4 was added with stirring. The mixture was cooled to 0 °C, and then (S)-(+)-m-glycidyl nosylate (compound 5) was added slowly. After reaction completion, water was added to precipitate compound 6. The filter cake was dried and then recrystallized from EtOAc. Compound 6 was dissolved in EtOH and cooled to 0 °C. Ethane-1,2-diamine was added, and the reaction temperature was increased to 30 °C. After reaction completion, concentration under reduced pressure yielded compound 7.
Compound 7 was dissolved in tetrahydrofuran (THF) and cooled to 0 °C. Carbonyldiimidazole was added, and the reaction temperature was increased to 30 °C. The reaction was quenched by adding water at 0 °C to 10 °C. The mixture was concentrated under reduced pressure to remove THF, then filtered, and the filter cake was washed with water. The filter cake was dissolved in THF, dried over Na2SO4, filtered, and concentrated under reduced pressure to yield crude SHP1703. Trituration with methyl tert-butyl ether yielded pure SHP1703 as a light yellow solid.
Glioblastoma Cell Viability Assay.
IMR-90 cells were cultured in Eagle's Minimum Essential Medium (30-2003, ATCC) and 10% fetal bovine serum. MGG 31 cells were provided by Massachusetts General Hospital from resection specimens and cultured in complete Neurobasal medium (Neurobasal-A medium, minus phenol red [12349-015, Life Technologies] with 2% B27 without Vitamin A [12587-010, Life Technologies], 20 ng/mL Epidermal Growth Factor [236-EG-01M, R&D], 20 ng/mL basic Fibroblast Growth Factor [4114-TC-01M, R&D], 1 mM GlutaMAX [35050-61, Gibco], 50 units per mL penicillin-streptomycin [15140-122, Gibco], and 1 mM sodium pyruvate [11360-070, Gibco]). To induce differentiation of MGG 31, cells were cultured with 10% fetal bovine serum in complete Neurobasal medium. Cells were plated in a 96-well black, clear-bottom plate (1,000 cells per well) and treated with SHP1703 or SHP1704 for 3 d. CellTiter-Glo Luminescent Cell Viability Assay (Promega) was used to measure cell viability. Luminescence corresponding to the cellular ATP level was recorded in a microplate reader, Infinite M200Pro (Tecan).
Quantification and Statistical Analysis.
Circadian period was determined from luminescence rhythms by a curve-fitting program, MultiCycle (Actimetrics). The luminescence intensity was calculated by averaging the intensity during the entire experiment. Data from the first day were excluded from analysis, because of transient changes in luminescence upon medium exchange. Concentrations for 2- or 4-h period lengthening (EC2h or EC4h) were obtained by exponential growth fitting of dilution series data with Prism software (GraphPad Software). For reporter activity repression, the half maximal inhibitory concentrations (IC50 or log[IC50]) were obtained by sigmoidal dose–response fitting of dilution series data with Prism software. In degradation assays, half-life was obtained by one-phase exponential decay fitting with Prism software. In cellular thermal shift assays, band intensity was analyzed by ImageQuant TL software (GE Healthcare). In thermal shift assays, the first derivative of the fluorescence intensity was plotted as a function of temperature (dF/dT), and the highest peak of the curve was defined as the melting temperature. In cell viability assay, the half maximal effective concentrations (EC50) were obtained by sigmoidal dose–response fitting of dilution series data with Prism software. Statistical significance was evaluated using two-way ANOVA, followed by a Sidak’s or Tukey’s multiple comparisons test using Prism software.
Supplementary Material
Acknowledgments
We thank Dr. Yoshiki Aikawa, Dr. Kazuhiro Abe, Dr. Kunio Hirata, and Dr. Toshiya Senda for technical assistance; Dr. Hiroki R. Ueda for Cry1/Cry2 double-knockout cells and pMU2-P(Cry1)-FLAG-I/RRE-Cry1 plasmid; and Dr. Hiroaki Wakimoto and Dr. Khalid Shah for the MGG 31 cells. This work was supported, in part, by Japan Society for the Promotion of Science Grants 18H02402, 20K21269, and 21H04766; the Takeda Science Foundation; the Uehara Memorial Foundation; the Tokyo Biochemical Research Foundation; the Hitachi Global Foundation (T.H.); the Rosalie and Harold Rae Brown Center for Cancer Drug Development at the University of Southern California Norris Comprehensive Cancer Center; Synchronicity Pharma LLC; the National Cancer Institute (Grant R01CA238662-01); the National Institute of Neurological Disorders and Stroke (Grant 1F31NS120654-01); and the Charlie Teo Foundation More Data Better Tools grant (S.A.K.). The contents of this publication are solely the responsibility of the authors and do not reflect the views of the Charlie Teo Foundation, nor do they necessarily represent the official views of the National Cancer Institute, National Institute of Neurological Disorders and Stroke, or the NIH. X-ray diffraction data collection and preliminary experiments were carried out at beamlines BL44XU of SPring-8 synchrotron facility (Proposals 2019A6942 and 2019B6942) and BL-17A of Photon Factory (Proposal 2019G024). Recombinant CRY expression and beamline experiments were supported, in part, by Basis for Supporting Innovative Drug Discovery and Life Science Research from Japan Agency for Medical Research and Development Support Numbers JP19am0101074-0055 and JP19am0101071-0529.
Footnotes
Competing interest statement: S.A.K. serves on the board of Synchronicity Pharma LLC and receives financial compensation for his role.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2203936119/-/DCSupplemental.
Data, Materials, and Software Availability
The final coordinates of CRY2-SHP656 and SHP-1703 were deposited into the PDB with the accession numbers 7V8Z (55) and 7V8Y (56), respectively. All other study data are included in the article and/or SI Appendix.
References
- 1.Bass J., Lazar M. A., Circadian time signatures of fitness and disease. Science 354, 994–999 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Ralph M. R., Foster R. G., Davis F. C., Menaker M., Transplanted suprachiasmatic nucleus determines circadian period. Science 247, 975–978 (1990). [DOI] [PubMed] [Google Scholar]
- 3.Schibler U., Ripperger J., Brown S. A., Peripheral circadian oscillators in mammals: Time and food. J. Biol. Rhythms 18, 250–260 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Koike N., et al. , Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi J. S., Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maier B., et al. , A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 23, 708–718 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuchiya Y., et al. , Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci. Signal. 2, ra26 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Oshima T., et al. , Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth. Sci. Adv. 5, eaau9060 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narasimamurthy R., Virshup D. M., The phosphorylation switch that regulates ticking of the circadian clock. Mol. Cell 81, 1133–1146 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Busino L., et al. , SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316, 900–904 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Godinho S. I. H., et al. , The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316, 897–900 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Siepka S. M., et al. , Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129, 1011–1023 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirano A., et al. , FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152, 1106–1118 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Yoo S.-H., et al. , Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152, 1091–1105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patke A., et al. , Mutation of the human circadian clock gene CRY1 in familial delayed sleep phase disorder. Cell 169, 203–215.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirano A., et al. , A Cryptochrome 2 mutation yields advanced sleep phase in humans. eLife 5, e16695 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamia K. A., et al. , Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480, 552–556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang E. E., et al. , Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat. Med. 16, 1152–1156 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirota T., et al. , Identification of small molecule activators of cryptochrome. Science 337, 1094–1097 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xing W., et al. , SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496, 64–68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nangle S., Xing W., Zheng N., Crystal structure of mammalian cryptochrome in complex with a small molecule competitor of its ubiquitin ligase. Cell Res. 23, 1417–1419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller S., et al. , Structural differences in the FAD-binding pockets and lid loops of mammalian CRY1 and CRY2 for isoform-selective regulation. Proc. Natl. Acad. Sci. U.S.A. 118, e2026191118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphries P. S., et al. , Carbazole-containing sulfonamides and sulfamides: Discovery of cryptochrome modulators as antidiabetic agents. Bioorg. Med. Chem. Lett. 26, 757–760 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Humphries P. S., et al. , Carbazole-containing amides and ureas: Discovery of cryptochrome modulators as antihyperglycemic agents. Bioorg. Med. Chem. Lett. 28, 293–297 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Dong Z., et al. , Targeting glioblastoma stem cells through disruption of the circadian clock. Cancer Discov. 9, 1556–1573 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michael A. K., et al. , Formation of a repressive complex in the mammalian circadian clock is mediated by the secondary pocket of CRY1. Proc. Natl. Acad. Sci. U.S.A. 114, 1560–1565 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosensweig C., et al. , An evolutionary hotspot defines functional differences between CRYPTOCHROMES. Nat. Commun. 9, 1138 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fribourgh J. L., et al. , Dynamics at the serine loop underlie differential affinity of cryptochromes for CLOCK:BMAL1 to control circadian timing. eLife 9, e55275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huber A.-L., et al. , CRY2 and FBXL3 cooperatively degrade c-MYC. Mol. Cell 64, 774–789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia M., et al. , Circadian clock protein CRY1 prevents paclitaxel-induced senescence of bladder cancer cells by promoting p53 degradation. Oncol. Rep. 45, 1033–1043 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shafi A. A., et al. , The circadian cryptochrome, CRY1, is a pro-tumorigenic factor that rhythmically modulates DNA repair. Nat. Commun. 12, 401 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanoun M., et al. , Epigenetic silencing of the circadian clock gene CRY1 is associated with an indolent clinical course in chronic lymphocytic leukemia. PLoS One 7, e34347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan A. B., et al. , CRY2 missense mutations suppress P53 and enhance cell growth. Proc. Natl. Acad. Sci. U.S.A. 118, e2101416118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baggs J. E., et al. , Network features of the mammalian circadian clock. PLoS Biol. 7, e52 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Czarna A., et al. , Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell 153, 1394–1405 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Miller S., et al. , Isoform-selective regulation of mammalian cryptochromes. Nat. Chem. Biol. 16, 676–685 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Miller S., et al. , An isoform-selective modulator of cryptochrome 1 regulates circadian rhythms in mammals. Cell Chem. Biol. 27, 1192–1198.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Kolarski D., et al. , Photopharmacological manipulation of mammalian CRY1 for regulation of the circadian clock. J. Am. Chem. Soc. 143, 2078–2087 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirota T., et al. , A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3β. Proc. Natl. Acad. Sci. U.S.A. 105, 20746–20751 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang E. E., et al. , A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell 139, 199–210 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu A. C., et al. , Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129, 605–616 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ode K. L., et al. , Knockout-rescue embryonic stem cell-derived mouse reveals circadian-period control by quality and quantity of CRY1. Mol. Cell 65, 176–190 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Winter G., xia2: An expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010). [Google Scholar]
- 44.Evans P., Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowtan K., Recent developments in classical density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 470–478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murshudov G. N., et al. , REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Afonine P. V., et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J., Wolf R. M., Caldwell J. W., Kollman P. A., Case D. A., Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Pronk S., et al. , GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roe D. R., Cheatham T. E. III, PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Miller B. R. III, et al. , MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 8, 3314–3321 (2012). [DOI] [PubMed] [Google Scholar]
- 55.S. Miller et al., CRY2 isoform selectivity of a circadian clock modulator with antiglioblastoma efficacy. PDB. https://www.rcsb.org/structure/7V8Z. Deposited 23 August 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. Miller et al., CRY2 isoform selectivity of a circadian clock modulator with antiglioblastoma efficacy. PDB. https://www.rcsb.org/structure/7V8Y. Deposited 23 August 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The final coordinates of CRY2-SHP656 and SHP-1703 were deposited into the PDB with the accession numbers 7V8Z (55) and 7V8Y (56), respectively. All other study data are included in the article and/or SI Appendix.