Abstract
The DnaK chaperone of Escherichia coli is known to interact with the J domains of DnaJ, CbpA, and DjlA. By constructing multiple mutants, we found that the djlA gene was essential for bacterial growth above 37°C in the absence of dnaJ. This essentiality depended upon the J domain of DjlA but not upon the normal membrane location of DjlA.
The Hsp70 (DnaK) family of molecular chaperones and their various cochaperone cohorts are central components of the cellular machines that assist a plethora of biological processes involving protein folding, translocation, disaggregation, and protein targeting for degradation (reviewed in reference 2). All of these functions depend upon the ATP-dependent association of Hsp70 with short hydrophobic sequences present in its various substrate proteins. The intrinsically weak ATPase activity of Hsp70 necessitates the requirement and recruitment of protein partners that regulate its ATPase cycle. Some of these proteins, such as the members of the DnaJ (Hsp40) cochaperone family, specifically stimulate the ATP hydrolysis step, while others, such as GrpE, modulate the ADP-ATP exchange process (2, 14). All members of the DnaJ cochaperone protein family invariably contain the so-called J domain, an approximately 70-amino-acid signature sequence that interacts with the ATPase domain of Hsp70 and stimulates its ATPase activity (8, 9, 11, 14). This fact implies that the substrate specificities of the DnaJ family members primarily depend upon DnaJ's other domains and/or upon its cellular localization (11).
Escherichia coli possesses three genes, dnaK, hscA, and ybeW, that code for members of the Hsp70 family and six genes, dnaJ, cbpA, djlA, hscB, ybeS, and ybeV, whose products share significant, or partial, sequence similarity with the canonical DnaJ J domain. DnaJ, CbpA, and DjlA share high sequence similarity in their J domains, and have been shown to act as bona fide cochaperones for DnaK (7, 14, 22, 23) (Fig. 1A). In contrast, the hscB gene product, Hsc20, was shown to specifically function as a cochaperone for Hsc66, the gene product of hscA (19). To date, no information is available about the two putative J domain-containing genes ybeS and ybeV, located immediately upstream of ybeW, whose gene product is Hsc62 (25). The predicted translation products of ybeS and ybeV show relatively low sequence identity with the DnaJ J domain (about 20%), and both possess a His Pro Glu tripeptide in place of the highly conserved His Pro Asp motif characteristic of all known functional J domains (11).
FIG. 1.
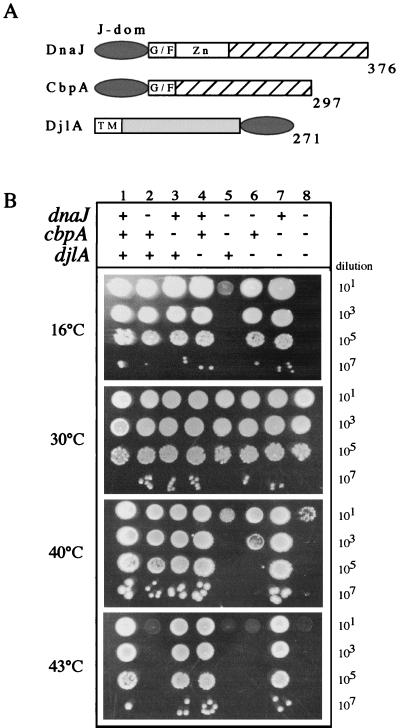
(A) Schematic representation of the E. coli DnaJ protein family. J-dom, J domain; G/F, glycine- and phenylalanine-rich region; Zn, zinc finger domain; TM, transmembrane domain. The cross-hatched boxes indicate the conserved region in DnaJ and CbpA, the light-gray box indicates a central region of unknown function, and the darker-gray ovals indicate the J domain. The numbers refer to the amino acid residues of each protein. (B) Effects of combinations of dnaJ, cbpA, and djlA mutations on E. coli growth. Representative growth on LB agar plates following overnight incubation at various temperatures is shown. + indicates the presence of the wild-type gene, and − indicates the null mutant.
The DnaJ protein has been extensively studied and shown to be a key regulator of DnaK's activities, both in vivo and in vitro (14). DnaJ is a 41-kDa cytoplasmic protein that possesses four distinct domains: an N-terminal J domain essential for stimulation of DnaK's ATPase activity, a glycine- and phenylalanine-rich region of unknown function, a zinc finger domain, and a C-terminal domain thought to bind and present specific substrates to DnaK (2, 11, 16). A strain carrying a deletion in the dnaJ gene displays various phenotypes, such as failure to replicate bacteriophage λ, inability to efficiently replicate mini-F and P1 plasmids, loss of cellular motility, and temperature sensitivity for growth above 42°C (10, 17, 18, 20, 27).
The 33-kDa CbpA (curved binding protein A) was originally isolated for its ability to bind curved synthetic oligonucleotides (21). CbpA is 39% identical to DnaJ, although it entirely lacks the zinc finger domain (Fig. 1A). CbpA is poorly expressed during the exponential phase of bacterial growth but is up-regulated upon entry into stationary phase or during phosphate starvation, both responses being dependent upon the ςs transcription factor (24). A cbpA deletion mutant exhibits no apparent growth phenotype, but the dnaJ cbpA double mutant is hypersensitive for growth below 16°C and above 37°C, a phenotype resembling that produced by a dnaK deletion mutation alone (3, 22). Multicopy expression of CbpA efficiently suppresses the phenotypes of a dnaJ null mutation, indicating that CbpA, under certain circumstances, can behave like a functional homolog of DnaJ (21, 23).
DjlA is a 30-kDa type III membrane protein with a single N-terminal transmembrane domain and with the remainder of the protein oriented towards the cytoplasm (6, 7). Apart from its C-terminal J domain, DjlA possesses no appreciable homology with either DnaJ or CbpA. DjlA was recently recognized as a bona fide DnaK cochaperone, since it can stimulate DnaK's ATPase and assist DnaK in the reactivation of denatured luciferase in vitro (7). Various in vivo approaches showed that, in contrast to CbpA, DjlA cannot complement bacteriophage λ growth in a dnaJ null background or bacterial growth above 39°C and below 16°C in the dnaJ cbpA null background (5, 12). A specific cellular role for DjlA has been difficult to pinpoint since deletion of the djlA gene results in no apparent growth phenotype. Nevertheless, overexpression of DjlA increases sensitivity to some drugs, such as novobiocin and the anticalmodulin W7. DjlA overexpression is highly toxic in minimal media and can trigger the synthesis of the colanic acid capsule (1, 5, 12, 26). DjlA-mediated capsule induction requires both interaction with DnaK and membrane localization (5, 7, 12, 26).
The dnaJ djlA double mutant is sensitive for bacterial growth at high temperatures.
To better understand the cellular role of DjlA and explore the functional harmony between the three J domain partners of DnaK, we constructed multiple bacterial strains lacking various combinations of dnaJ, cbpA, and djlA by bacteriophage P1-mediated generalized transduction at 30°C (15) and tested them for growth at various temperatures. The strains used in this study are listed in Table 1. P1 was grown on strains CU247 (dnaJ cbpA) and WKG15 (djlA), and these lysates served as donors in the transduction experiments. Growth on Luria-Bertani (LB) agar plates of bacterial strains carrying various combinations of null mutations is shown in Fig. 1B. The reproducibility of the phenotypes was assessed by constructing the same mutant combinations in various E. coli genetic backgrounds (data not shown). The growth properties of the dnaJ, cbpA, and djlA single null mutants, as well as those of the dnaJ cbpA double mutant, have already been described elsewhere (12, 21, 22). The following original observations have been made in this study: (i) both the djlA and cbpA genes can be deleted in the presence of DnaJ without any apparent effect on bacterial growth, (ii) the dnaJ cbpA djlA triple mutant is viable at 30°C and exhibits no additional growth defects on LB agar plates when compared to those of the double dnaJ cbpA mutant, and (iii) in a manner analogous to that of the dnaJ cbpA double mutant, the double djlA dnaJ mutant is unable to grow at high temperatures. However, in contrast to that of the double dnaJ cbpA mutant, growth of the double dnaJ djlA mutant is not affected at 16°C (Fig. 1B).
TABLE 1.
E. coli strains, bacteriophages, and plasmids
| Strain, bacteriophage, or plasmid | Genotype | Reference or source |
|---|---|---|
| Strains | ||
| MC4100 | Δ(argF-lac)U169 araD139 rpsL150 deoC1 relA1 ptsF25 flbB5501 rbsR F− λ− | 4 |
| CU247 | MC4100 ΔcbpA::KanrdnaJ::Tn10-42(Tetr) | 22 |
| WKG15 | MC4100 ΔdjlA::ΩSpcr | 12 |
| GP108 | MC4100 dnaJ::Tn10-42(Tetr) | This work |
| GP109 | MC4100 ΔcbpA::Kanr | This work |
| GP110 | MC4100 ΔdjlA::ΩSpcrdnaJ::Tn10-42(Tetr) | This work |
| GP111 | MC4100 ΔdjlA::ΩSpcr ΔcbpA::Kanr | This work |
| GP112 | MC4100 dnaJ::Tn10-42(Tetr) ΔcbpA::Kanr | This work |
| GP113 | MC4100 ΔdjlA::ΩSpcrdnaJ::Tn10-42(Tetr) ΔcbpA::Kanr | This work |
| Bacteriophages | ||
| λcI | Clear plaque former | Laboratory collection |
| λcI dnaJ+ | dnaJ+ transducing phage | Laboratory collection |
| Plasmids | ||
| pSE380 | ColE1 Ptrc Ampr | Invitrogen |
| pWKG90 | dnaJ+ | 13 |
| pCU60 | cbpA+ | 22 |
| pWKG54 | djlA(H233Q) | 12 |
| pWKG52 | djlAΔTMD | 12 |
| pGP134 | djlAΔTMD in pSE380 | This work |
| pGP135 | djlAΔTMD (H233Q) in pSE380 | This work |
| pGP136 | cbpA in pSE380 | This work |
| pGP137 | dnaJ in pSE380 | This work |
The considerable growth defect of the dnaJ djlA double mutant at temperatures above 37°C reveals the first phenotype associated with the loss of DjlA. The absence of both DnaJ and DjlA proteins in the dnaJ djlA double mutant was confirmed by immunoblotting using anti-DjlA and anti-DnaJ antibodies (data not shown). Furthermore, growth at high temperatures could be restored following the introduction of low-copy-number plasmids expressing either DnaJ or DjlA from their native promoter sequences (data not shown). These results reveal that, under standard laboratory conditions, either cbpA or djlA can be deleted without any apparent effect on bacterial viability. In contrast, in a dnaJ null background, cbpA or djlA becomes essential for bacterial growth at high temperatures.
A functional J domain, but not DjlA's cellular localization, is essential for bacterial growth in a strain lacking dnaJ.
Next, we asked whether DjlA's membrane localization and/or its interaction with DnaK was required for growth in the dnaJ null mutant background. To answer this question, a djlA fragment lacking the region encoding the N-terminal 31-amino-acid transmembrane domain (djlAΔTM) and a mutated gene encoding an H233Q mutation in the J domain, known to abolish productive interaction with DnaK (djlAΔTM-H233Q) (7, 12), were PCR amplified from plasmids pWKG52 and pWKG54, respectively, using primer A (5′-CCGCCATGGATAAAGCCCGTAGCCGTAAA-3′), which introduces an NcoI N-terminal site (italicized), and primer B (5′-CCGGGATCCTCATTTAAACCCTTTCTGCTGCTT-3′), which introduces a BamHI C-terminal site (italicized). The PCR fragments were digested by NcoI and BamHI and cloned into NcoI- and BamHI-digested pSE380, resulting in plasmids pGP134 (djlAΔTM) and pGP135 (djlAΔTM-H233Q). The constructs were sequence verified, and protein expression was assessed by semiquantitative immunoblotting using anti-DjlA antibodies. It was found that the expression level of both proteins was approximately 40 times higher than the expression level of chromosomally encoded DjlA (data not shown). As a positive control, full-length wild-type dnaJ from pWKG90 (13) was digested by NcoI and BglII and also cloned into NcoI- and BglII-digested pSE380, resulting in plasmid pGP137 (Table 1). Since high expression of the full-length DjlA containing the transmembrane domain is very toxic for the cell (5, 12), we could not use its corresponding construct as a control in this assay.
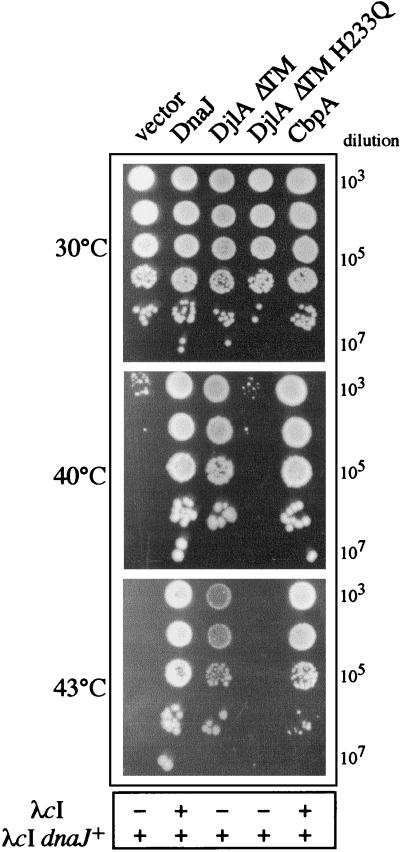
As expected, the plasmid expressing wild-type DnaJ, but not the plasmid vector alone, complemented the growth of the dnaJ djlA double mutant (Fig. 2, first two lanes). A plasmid expressing a truncated form of DjlA, which lacks the transmembrane domain (DjlAΔTM) also complemented the temperature-sensitive-phenotype of the dnaJ djlA double mutant (Fig. 2, third lane). However, the point mutation in the DjlA J domain (DjlAΔTM H233Q) abolished all complementation (Fig. 2, fourth lane). Furthermore, no complementation was observed with the DjlA J domain alone, suggesting that the J domain and the central domain of DjlA are essential for DjlA function in the absence of DnaJ (data not shown). Taken together, these results indicate that, in the absence of DnaJ, the transmembrane domain of DjlA is not critical for E. coli growth but that DjlA-DnaK interaction is.
FIG. 2.
Complementation of the bacterial temperature-sensitive phenotype and the block of bacteriophage λ growth of the dnaJ djlA double mutant. Strain GP110 (dnaJ djlA double mutant) was transformed with the pSE380-based constructs carrying the genes indicated at the top of the figure. Fresh transformants were grown overnight at 30°C, serially diluted, and spotted on LB agar plates containing 100 μg of ampicillin per ml at the indicated temperatures. Bacteriophage λ growth was measured as described previously (7). + indicates an average plaque size and an efficiency of plating of approximately 1.0 compared to that of the wild type, and − indicates no plaque formation (<10−5).
In agreement with previous findings with the dnaJ cbpA double mutant (5), neither DjlA nor DjlAΔTM could complement bacteriophage λ growth on the dnaJ djlA double mutant (Fig. 2) or bacteriophage P1 growth (data not shown). In addition, as observed for the dnaJ cbpA double mutant (5), DjlAΔTM could complement the growth defect of the dnaJ cbpA djlA triple mutant, but only up to 39°C (data not shown).
CbpA overexpression can fully complement the lack of both DjlA and DnaJ.
Previous work had shown that multicopy expression of CbpA could complement the bacterial growth defect phenotype of either a single dnaJ mutant or a double dnaJ cbpA mutant (21, 22). We asked whether CbpA could complement the lack of DjlA in the double dnaJ djlA mutant. To do so, cbpA DNA was first PCR amplified from plasmid pCU60 using primer C, 5′-GGGAATTCACCATGGAATTAAAGGATTAT-3′, which introduces an N-terminal NcoI site, and primer D, 5′-GGGGATCCAGATCTTATGCTTTCCCCCAAT-3′, which introduces a C-terminal BamHI site. The PCR fragment was digested by NcoI and BamHI and cloned into NcoI- and BamHI-digested pSE380, yielding pGP136. The construct was sequence verified and tested for functionality in the dnaJ cbpA mutant (data not shown). Plasmid pGP136 was then transformed into the double dnaJ djlA mutant and tested for its effect on bacterial growth (Fig. 2). The results clearly show (Fig. 2, compare the second and fifth lanes) that multicopy expression of CbpA can fully complement the lack of both DjlA and DnaJ at all temperatures tested. The same observation was made for the dnaJ cbpA djlA triple mutant (data not shown). These data support and extend the idea that CbpA is a functional homolog of DnaJ.
Concluding remarks.
This study sheds light on functional overlaps among the three DnaJ family members of E. coli by examining both the synthetic phenotypes using various combinations of null mutations and the extent of multicopy suppression in various genetic backgrounds. Our results reveal a surprising functional redundancy between DnaJ and either CbpA or DjlA. In the absence of DnaJ, E. coli requires the presence of either CbpA or DjlA to sustain growth at elevated temperatures. It is unknown how this is accomplished mechanistically. Although CbpA may functionally replace DnaJ by virtue of its overall domain similarity and substantial sequence homology, it is less clear how DjlA can perform the same task. Since the only common region of sequence homology shared by CbpA and DjlA is their J domain, there may be additional, but sequence-unrelated, substrate interaction regions in the two proteins that may assume a role(s) analogous to that normally provided by DnaJ.
Acknowledgments
We thank C. Ueguchi for the gift of CU247 and pCU60 and I. B. Holland and D. J. Clarke for the gift of anti-DjlA antibody.
This work was supported by Swiss National Science Foundation grant 31.47283-96 and the Canton of Geneva.
Footnotes
Present address: Division des Maladies Infectieuses, Hôpital Universitaire de Genève, 1211 Genève 4, Switzerland.
REFERENCES
- 1.Bernard S, Clarke D J, Chen M X, Holland I B, Jacq A. Increased sensitivity of Escherichia coli to novobiocin, EDTA and the anticalmodulin drug W7 following overproduction of DjlA requires a functional transmembrane domain. Mol Gen Genet. 1998;259:645–655. doi: 10.1007/pl00008629. [DOI] [PubMed] [Google Scholar]
- 2.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 3.Bukau B, Walker G C. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J Bacteriol. 1989;171:2337–2346. doi: 10.1128/jb.171.5.2337-2346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using phage λ and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 5.Clarke D J, Holland I B, Jacq A. Point mutations in the transmembrane domain of DjlA, membrane-linked DnaJ-like protein, abolish its function in promoting colanic acid production via the Rcs signal transduction pathway. Mol Microbiol. 1997;25:933–944. doi: 10.1111/j.1365-2958.1997.mmi528.x. [DOI] [PubMed] [Google Scholar]
- 6.Clarke D J, Jacq A, Holland I B. A novel DnaJ-like protein in Escherichia coli inserts into the cytoplasmic membrane with a type III topology. Mol Microbiol. 1996;20:1273–1286. doi: 10.1111/j.1365-2958.1996.tb02646.x. [DOI] [PubMed] [Google Scholar]
- 7.Genevaux P, Wawrzynow A, Zylicz M, Georgopoulos C, Kelley W L. DjlA is a third DnaK co-chaperone of Escherichia coli, and DjlA-mediated induction of colanic acid capsule requires DjlA-DnaK interaction. J Biol Chem. 2001;276:7906–7912. doi: 10.1074/jbc.M003855200. [DOI] [PubMed] [Google Scholar]
- 8.Greene M K, Maskos K, Landry S J. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci USA. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karzai A W, McMacken R. A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J Biol Chem. 1996;271:11236–11246. doi: 10.1074/jbc.271.19.11236. [DOI] [PubMed] [Google Scholar]
- 10.Kawasaki Y, Wada C, Yura T. Roles of Escherichia coli heat shock proteins DnaK, DnaJ and GrpE in mini-F plasmid replication. Mol Gen Genet. 1990;220:277–282. doi: 10.1007/BF00260494. [DOI] [PubMed] [Google Scholar]
- 11.Kelley W L. The J-domain family and the recruitment of chaperone power. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 12.Kelley W L, Georgopoulos C. Positive control of the two-component RcsC/B signal transduction network by DjlA: a member of the DnaJ family of molecular chaperones in Escherichia coli. Mol Microbiol. 1997;25:913–931. doi: 10.1111/j.1365-2958.1997.mmi527.x. [DOI] [PubMed] [Google Scholar]
- 13.Kelley W L, Georgopoulos C. The T/t common exon of simian virus 40: JC and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone. Proc Natl Acad Sci USA. 1997;94:3679–3684. doi: 10.1073/pnas.94.8.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. The Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate DnaK's ATPase activity. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 16.Rudiger S, Schneider-Mergener J, Bukau B. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sell S M, Eisen C, Ang D, Zylicz M, Georgopoulos C. The isolation and characterization of dnaJ null mutants of Escherichia coli. J Bacteriol. 1990;172:4827–4835. doi: 10.1128/jb.172.9.4827-4835.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi W, Zhou Y, Wild J, Adler J, Gross C A. DnaK, DnaJ, and GrpE are required for flagellum synthesis in Escherichia coli. J Bacteriol. 1992;174:6256–6263. doi: 10.1128/jb.174.19.6256-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silberg J J, Hoff K G, Vickery L E. The Hsc66/Hsc20 chaperone system in Escherichia coli: chaperone activity and interactions with the DnaK/DnaJ/GrpE system. J Bacteriol. 1998;180:6617–6624. doi: 10.1128/jb.180.24.6617-6624.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tilly K, Spence J, Georgopoulos C. Modulation of the stability of Escherichia coli heat shock regulatory factor ς32. J Bacteriol. 1989;171:1585–1589. doi: 10.1128/jb.171.3.1585-1589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueguchi C, Kakeda M, Yamada H, Mizuno T. An analogue of the DnaJ molecular chaperone in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:1054–1058. doi: 10.1073/pnas.91.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueguchi C, Shiozawa T, Kakeda M, Yamada H, Mizuno T. A study of the double mutation of dnaJ and cbpA, whose gene products function as molecular chaperones in Escherichia coli. J Bacteriol. 1995;177:3894–3896. doi: 10.1128/jb.177.13.3894-3896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wegrzyn A, Taylor K, Wegrzyn G. The cbpA chaperone gene function compensates for dnaJ in lambda plasmid replication during amino acid starvation of Escherichia coli. J Bacteriol. 1996;178:5847–5849. doi: 10.1128/jb.178.19.5847-5849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamashino T, Kakeda M, Ueguchi C, Mizuno T. An analogue of the DnaJ molecular chaperone whose expression is controlled by ςs during the stationary phase and phosphate starvation in Escherichia coli. Mol Microbiol. 1994;13:475–483. doi: 10.1111/j.1365-2958.1994.tb00442.x. [DOI] [PubMed] [Google Scholar]
- 25.Yoshimune K, Yoshimura T, Esaki N. Hsc62, a new DnaK homologue of Escherichia coli. Biochem Biophys Res Commun. 1998;250:115–118. doi: 10.1006/bbrc.1998.9255. [DOI] [PubMed] [Google Scholar]
- 26.Zuber M, Hoover T A, Court D L. Analysis of a Coxiella burnetii gene product that activates capsule synthesis in Escherichia coli: requirement for the heat shock chaperone DnaK and the two-component regulator RcsC. J Bacteriol. 1995;177:4238–4244. doi: 10.1128/jb.177.15.4238-4244.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zylicz M, Ang D, Liberek K, Georgopoulos C. Initiation of λ DNA replication with purified host- and bacteriophage-encoded proteins: the role of the DnaK, DnaJ and GrpE heat shock proteins. EMBO J. 1989;8:1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]