Abstract
The B. subtilis ΔhelD allele rendered cells proficient in transformational recombination and moderately sensitive to methyl methanesulfonate when present in an otherwise Rec+ strain. The ΔhelD allele was introduced into rec-deficient strains representative of the α (recF strain), β (addA addB), γ (recH), ɛ (ΔrecU), and ζ (ΔrecS) epistatic groups. The ΔhelD mutation increased the sensitivity to DNA-damaging agents of addAB, ΔrecU, and ΔrecS cells, did not affect the survival of recH cells, and decreased the sensitivity of recF cells. ΔhelD also partially suppressed the DNA repair phenotype of other mutations classified within the α epistatic group, namely the recL, ΔrecO, and recR mutations. The ΔhelD allele marginally reduced plasmid transformation (three- to sevenfold) of mutations classified within the α, β, and γ epistatic groups. Altogether, these data indicate that the loss of helicase IV might stabilize recombination repair intermediates formed in the absence of recFLOR and render recFLOR, addAB, and recH cells impaired in plasmid transformation.
Genetic analysis using chromosomal and plasmid transformation allows the classification of Bacillus subtilis Rec− strains, other than recA strains, within five different epistatic groups (α, β, γ, ɛ, and ζ groups) (6, 12). Unless otherwise stated, the indicated genes and products are of B. subtilis origin. The α epistatic group activity requires the recF, recL, recO, recR, and recN genes (3–5). The recF, recL, recO, and recR strains and the Escherichia coli recF (recFEco), recOEco, and recREco strains have similar phenotypes and share indirect suppressors (e.g., a special type of recA mutant) (2, 10). It was assumed therefore that RecFLOR has its counterpart in the RecFOREco complex (6, 13). The β epistatic group activity is dependent on the addA and addB genes (3). The addA and addB genes encode different subunits of the nuclease-helicase AddAB (also termed exonuclease V or RecBCDEco for E. coli) (8, 9, 16). The γ epistatic group activity requires the recP and recH genes (6). The ɛ epistatic group activity requires the recB, recD, and recU genes (4), and the ζ group is dependent on recS, a gene sharing homology with recQ of both B. subtilis and E. coli origin (12). The function whose mutants were classified within the γ, ɛ, and ζ epistatic groups does not seem to have a counterpart in E. coli (6, 14).
Identification of the helD gene.
DNA helicases are involved in both the generation of the recombinogenic substrates and branch migration of synapsed Holliday junctions (16). RecBCDEco and the recQEco, uvrDEco, and helDEco gene products have been implicated in the RecBCDEco and RecFEco recombination pathways, respectively. The RecBCD DNA helicase is implicated in the presynaptic stage of recombination. Furthermore, it has been suggested that the presence of RecQEco, UvrDEco, or E. coli helicase IV (helD gene product, a 3′ to 5′ DNA helicase) is required for efficient recombination and repair in a recBCEco sbcB(C)Eco background (21, 23). The B. subtilis pcrA, addAB, and yvgS genes encode a DNA helicase or have a high degree of identity with bona fide DNA helicases of the SF1 family (8, 9, 16, 24, 25), and the recS and recQ gene products have identity with DNA helicases of the SF2 family, according to the nomenclature of Gorbalenya and Koonin (15). No UvrDEco, RepEco, or HelEco counterparts have been described for B. subtilis (17, 24).
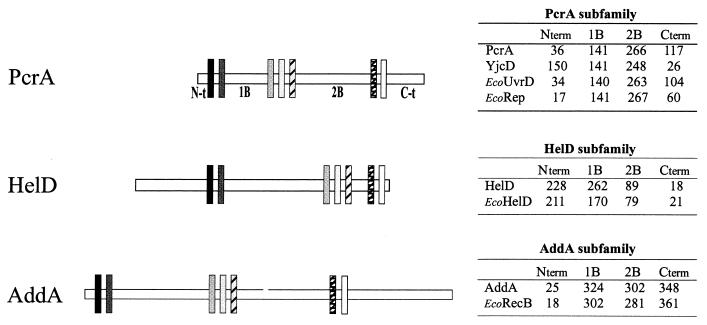
Helicases of the SF1 family share seven conserved motives scattered along the protein sequence. Structural analysis has shown that these motives come close together in the folded protein, bind ATP, and correspond to the translocating domain of these helicases, allowing the tracking of single-stranded DNA through the protein. The seven motives delimit four domains in the primary sequence of the SF1 helicases, an N-terminal domain preceding the first motive, a 1B domain between the second and third motives, a 2B domain between the fifth and sixth motives, and a C-terminal (C-t) domain after the last motive (Fig. 1). An analysis of the length of these four domains in any helicase of the SF1 family allows their classification into one of the three following groups: (i) the PcrA subfamily, helicases with a fixed 1B and 2B length (∼134 and 220 to 240 residues, respectively); (ii) the HelDEco subfamily, helicases with a long C-t and a short 2B domain; and (iii) the AddA subfamily, helicases with a long C-t domain. The four helicases of the SF1 family in B. subtilis and in E. coli were classified accordingly, as shown Fig. 1. PcrA and YjcD belonged to the first group, together with UvrDEco and RepEco. The YvgS protein belonged to the second group, together with HelDEco or E. coli helicase IV. We therefore propose to name the YvgS protein HelD. It should be noted, however, that the overall level of similarity between HelD and HelDEco is low (22% identity). AddA and RecBEco belonged to the third group. Again, these last two proteins have only 21.7% identity, but they have been shown to be functionally equivalent (8, 9).
FIG. 1.
Classification of DNA helicases of the SF1 family from B. subtilis and E. coli into three subfamilies. The seven conserved motives are shown with boxes separated by the four domains, called N-t (N-terminal), 1B, 2B, and C-t (C-terminal). The distances (in amino acid residues) separating the different clusters of motives are reported on the right part of the figure. See the text for details.
PcrA, which is able to suppress the UV sensitivity defect of a uvrDEco mutant, is an essential DNA helicase (24). A defect in RecS decreases plasmid transformation ∼100-fold when present in an otherwise Rec+ strain (12). The AddAB protein has its counterpart in the RecBCDEco enzyme (8, 9, 16). Nothing is known about the role of RecQ and helicase IV (helD gene product) in recombination and DNA repair. Here, we report the genetic analysis of a helD null allele (ΔhelD) in combination with rec function, classified within the α, β, γ, ɛ, and ζ epistatic groups. We show that the loss of helicase IV stabilized recombination repair intermediates formed in the absence of recFLOR and rendered recFLOR, addAB, and recH cells impaired in plasmid transformation. Furthermore, the fact that the ΔhelD mutation is a common partial suppressor of the recFLOR mutations further supported the classification of the recFLOR genes within the α epistatic group.
Disruption of the helD gene by use of a selectable marker.
It has been shown previously that (i) E. coli helicase IV is a DNA helicase (26), (ii) it shares several biochemical properties with the E. coli helicase II and Rep proteins (27), (iii) double helicase II (uvrDEco)-helicase IV (helDEco) deletion mutants are defective in DNA recombination and repair (20), (iv) B. subtilis possesses two genes homologous to the uvrDEco-repEco tandem, the pcrA and yjcD genes (reference 24; also see above), (v) PcrA, which suppresses the UV sensitivity defect of a uvrDEco mutant, is an essential helicase of B. subtilis, whereas a yjcD mutant does not exhibit a UV-sensitive phenotype (24). To learn whether the helD gene is involved in recombinational DNA repair, we have constructed a ΔhelD deletion strain. The helD gene was disrupted using the plasmid pMUTIN2 (details of the mutant construction are available on the Micado site [http://locus.jouy.inra.fr/cgi-bin/genmic/madbase/progs/madbase.operl]). As a result, the helD gene is disrupted starting 139 bp downstream from the start codon. The ΔhelD mutant allele was transferred into the B. subtilis chromosome of the wild type (YB886) and its isogenic rec-deficient derivatives of groups α (recF15 [BG129], recL16 [BG107], ΔrecO [BG439], and recR13 [BG127]) and β (addA5 addB72 [BG189]), γ (recH342 [BG119]), ɛ (ΔrecU [BG427]), and ζ (ΔrecS [BG425]) listed in Table 1, by a double crossing-over event as previously described (5). The presence of the desired replacement was confirmed by PCR amplification and nucleotide sequence analysis (data not shown).
TABLE 1.
Bacterial strainsa
| Single-mutant genotype (strain) | Double-mutant genotype or strain |
|---|---|
| helD1 | |
| rec+ (YB886) | NA |
| helD1 (BG400) | NA |
| recF15 (BG129) | BG477 |
| recL16 (BG107) | BG533 |
| recO1 (BG439) | BG529 |
| recR13 (BG127) | BG531 |
| addA5 addB72 (BG189) | BG483 |
| recH342 (BG119) | BG509 |
| recU1 (BG427) | BG489 |
| recS1 (BG425) | BG471 |
The isogenic background of all strains is trpC2 metB5 amyE sigB xin-1 attSPβ. Strain BG189 by construction is resistant to phleomycin. Strains BG439, BG427, and BG425 and its derivatives by construction are resistant to chloramphenicol. Strain BG400 and its derivatives by construction are resistant to erythromycin. NA, not applicable.
Interaction between ΔhelD and the functions classified within the α, β, γ, ɛ, and ζ epistatic groups.
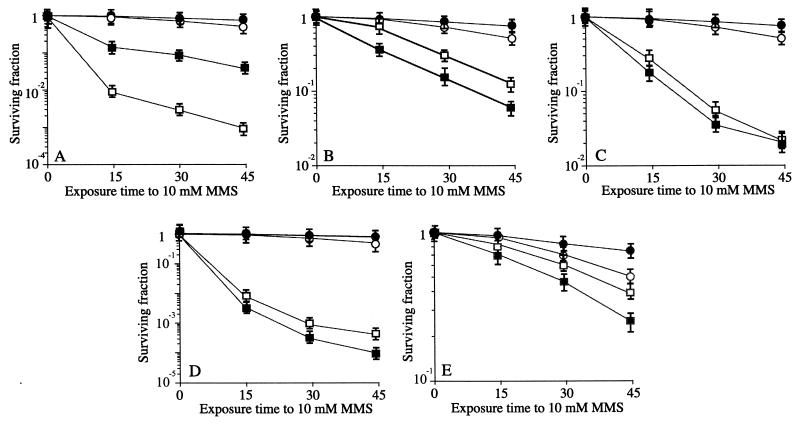
To understand the role of the helD gene product in homologous recombination, we investigated the genetic interaction between helD and the following mutant genes: recF, addA addB (addAB), recH, ΔrecU, and ΔrecS, which were selected as representatives of the five different epistatic groups (α, β, γ, ɛ, and ζ, respectively). B. subtilis (rec+) and its isogenic rec-deficient derivative strains (listed in Table 1) were exposed to the killing action of DNA-damaging agents as previously described (1). The removal of lesions from Sn2-type simple alkylating agents, such as methyl methanesulfonate (MMS), could be carried out to a different degree by adaptive response and recombination repair (1). As revealed in Fig. 2, various degrees of increased sensitivity to 10 mM MMS were observed. The ΔhelD and ΔrecS cells are slightly sensitive, the addAB and recH cells are moderately sensitive, and the recF and ΔrecU cells are very sensitive to the killing action of MMS compared to the rec+ control (1, 5, 12, 13) (Fig. 2).
FIG. 2.
Survival of B. subtilis strains following exposure to 10 mM MMS. Survival of rec+ (filled circles) and helD1 strains (empty circles) is shown. (A) Survival of recF15 cells (empty squares) and helD1 recF15 cells (filled squares). (B) Survival of addA5 addB72 cells (empty squares) and helD1 addA5 addB72 cells (filled squares). (C) Survival of recH342 cells (empty squares) and helD1 recH342 cells (filled squares). (D) Survival of recU1 cells (empty squares) and helD1 recU1 cells (filled squares). (E) Survival of recS1 cells (empty squares) and helD1 recS1 cells (filled squares). The chemical treatment of DNA repair-deficient mutant strains was performed essentially as previously described (1).
The ΔhelD mutant allele increases the MMS sensitivity of addAB, ΔrecU, and ΔrecS cells but does not affect the survival rate of recH cells (Fig. 2B to E). Previously we have shown that the ΔrecS null allele partially suppressed the sensitivity of addAB cells to MMS (12). Since ΔhelD slightly increases the sensitivity of addAB and ΔrecS cells, we have to assume that helicase IV operates at a different stage than the AddAB and RecS DNA helicases. Furthermore, if a helicase is needed at the presynaptic stage in the RecF pathway, as suggested by some experiments with E. coli (21), the fact that ΔhelD addAB and ΔhelD ΔrecS cells are more proficient in recombinational DNA repair than recF and/or ΔrecU cells (Fig. 2) suggests that helicase IV is not the only helicase responsible for presenting the DNA substrate to RecA. An alternative DNA helicase (e.g., PcrA, YjcD, RecQ, RecG or RuvAB), alone or in concerted action with a nuclease, could generate the recombinogenic substrate. Because the pcrA gene product is essential (24), the double mutant pcrA helD could not be tested. Whether any of the remaining putative helicases, encoded by yjcD, recG, recQ, or ruvAB, is involved at this stage remains to be determined.
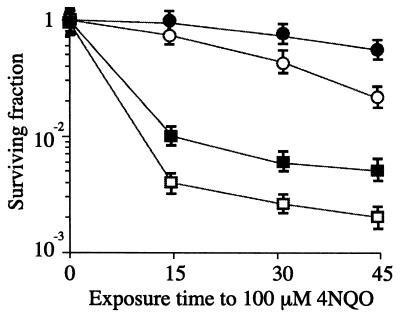
The helDEco cells do not exhibit a UV-sensitive phenotype (21). In B. subtilis the removal of purine adducts produced by 4-nitroquinoline-1-oxide (4NQO) has been reported to involve nucleotide excision repair and recombination repair (1). To analyze and interpret the involvement of the ΔhelD allele in recombinational repair, we have exposed the ΔhelD recF strain to the killing action of 100 μM 4NQO. helD cells are slightly sensitive to 4NQO (Fig. 3), but when the ΔhelD recF strain was exposed to the killing action of 4NQO a suppression of the recF defect was observed (Fig. 3).
FIG. 3.
Survival of B. subtilis strains following exposure to 100 μM 4NQO. Survival of rec+ cells (filled circles), helD1 cells (empty circles), recF15 cells (empty squares), and helD1 recF15 cells (filled squares) is shown.
From these data we can infer that (i) ΔhelD renders cells slightly sensitive to DNA-damaging agents, (ii) the helD gene product (helicase IV) contributed to different extents to the removal of DNA damage in addAB (group β), recH (group γ), ΔrecU (group ɛ), and ΔrecS (group ζ) cells, and (iii) the absence of helicase IV partially suppressed the recombinational repair deficiency of recF cells.
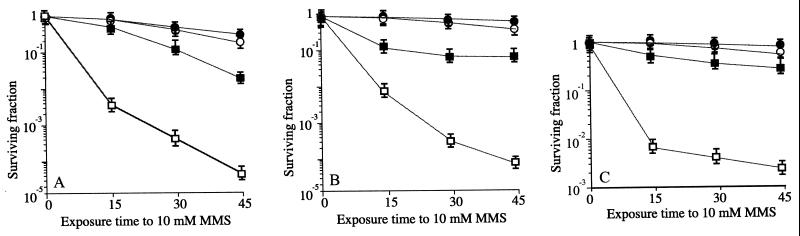
ΔhelD partially suppresses the DNA repair defects of the recF, recL, recO, and recR mutants.
Previously we have shown that suppressors of the recF defect (e.g., recA73 mutant, overexpression of ssb product) also suppressed the recL, recO, and recR defects (2, 13). To address whether the ΔhelD allele also suppressed the defect of the recL, ΔrecO, and recR mutations, the respective double mutant strains have been constructed (Table 1).
Unlike the case with recBCEco sbcB(C)Eco cells, which show a synergistic interaction between the helDEco gene and the recFEco or recOEco gene for the repair of MMS-damaged DNA (23), the ΔhelD allele suppressed the recombinational repair defect of recF, recL, recR, and ΔrecO cells (Fig. 2A, 3, and 4). Similar results were observed when the mutant cells were challenged with 100 μM 4NQO (Fig. 3 and data not shown).
FIG. 4.
Survival of B. subtilis strains following exposure to 10 mM MMS. Survival of rec+ (filled circles) and helD1 (empty circles) strains is analyzed. (A) Survival of recL16 cells (empty squares) and helD1 recL16 cells (filled squares). (B) Survival of recO1 cells (empty squares) and helD1 recO1 cells (filled squares). (C) Survival of recR13 cells (empty squares) and helD1 recR13 cells (filled squares).
The introduction of a plasmid-borne helD gene in the ΔhelD recF background resulted in a phenotype indistinguishable from that of recF cells (data not shown). It is likely, therefore, that ΔhelD is a bona fide suppressor of the recFLOR mutants and that the absence of helicase IV redirects recombinational repair into an avenue different from the RecFLOR complex.
Effect of ΔhelD on genetic recombination.
To study the effect of the ΔhelD mutation on homologous DNA recombination, we analyzed the requirement of the ΔhelD function in natural transformational recombination. Natural transformation in B. subtilis involves the transfer of naked double-stranded DNA from the media, with the subsequent degradation of one of the exogenous DNA strands by a set of competence (com) genes (11, 18, 19). Under these conditions, the level of expression of the recA and addAB genes is also increased. The recipient competent uninucleated cell takes up the other DNA strand in a linear single-stranded form (see references 11, 18, and 19 for reviews). Therefore, by the activity of the com genes, the donor single-stranded DNA is presented to the recombinational machinery in a form that it is ready for a homology search on the parental molecule. Hence, in our analysis we are studying the late presynaptic, synaptic, and postsynaptic stages but neglect the involvement of rec functions in the presentation of the substrate (early presynaptic stage) (14).
Except in recA cells where chromosomal transformation is blocked (∼10−4-fold reduction), chromosomal transformation did not change more than fourfold relative to the wild-type value when present in an otherwise wild-type strain (1, Table 2). The effect of double or triple mutations in some cases, however, is drastic. A recFLOR (epistatic group α) mutation blocks (>1,000-fold reduction) chromosomal transformation of addAB (group β) and recH (group γ) cells and reduces (>20-fold) that of ΔrecU (group ɛ) or ΔrecS (group ζ) cells (2, 6, 12, 13). A recH mutation blocks (>1,000-fold) chromosomal transformation of addAB cells and reduces (>200-fold) that of ΔrecS cells, but it does not affect chromosomal transformation of ΔrecU cells more than fourfold (5, 6, 12, 13).
TABLE 2.
Effect of the ΔhelD mutation on homologous recombination as measured by transformation of chromosomal and plasmid DNAa
| Relevant genotype | Results
|
|
|---|---|---|
| Normalized plasmid transformation | Normalized chromosomal transformation | |
| rec+ | 1 | 1 |
| ΔhelD | 0.97 | 0.96 |
| recF | 0.86 | 0.65 |
| addA addB | 0.88 | 0.63 |
| recH | 0.95 | 0.46 |
| ΔrecU | 0.02 | 0.36 |
| ΔrecS | 0.02 | 0.26 |
| recF ΔhelD | 0.22 | 0.62 |
| addA addB ΔhelD | 0.28 | 0.51 |
| recH ΔhelD | 0.13 | 0.28 |
| ΔrecU ΔhelD | 0.01 | 0.14 |
| ΔrecS ΔhelD | 0.01 | 0.10 |
Competent B. subtilis cells auxotrophic for methionine harboring the indicated mutations were transformed with chromosomal DNA from a met+ strain. The yield of met+ transformants (chromosomal transformation) and that of Nmr transformants (plasmid transformation) were corrected for DNA uptake and cell viability, and the values obtained were normalized relative to that for the rec+ strain, taken as 1 (see reference 1). Each result is the average from at least three independent experiments and is within a 10% standard error.
Plasmid establishment on transformation of B. subtilis competent cells is dependent on the degree of oligomerization of the plasmid genome and requires the RecU, RecO, and RecS products (7, 12, 13) but is independent of the RecA activity (12, 18, 19). Except in the ΔrecO, ΔrecU, and ΔrecS strains (50-fold reduction), the frequency of plasmid transformation did not change more than twofold relative to the wild-type value (12, 13). A recFLOR mutation blocks (>100-fold) plasmid transformation of addAB (group β) and recH (group γ) cells and reduces (>15-fold) that of ΔrecU or ΔrecS cells (5, 6, 12, 13). A recH mutation reduced plasmid transformation of ΔrecU and ΔrecS cells about 10-fold (12). Altogether those data indicate that the gene products classified with the α, β, γ, ɛ, and ζ epistatic groups provide overlapping activities that compensate for the effects of a single mutation.
By measuring both chromosomal (intermolecular recombination) and plasmid (intramolecular recombination) transformation we can examine different types of events. B. subtilis competent cells were transformed with 1 μg of homologous chromosomal DNA or plasmid DNA/ml to determine the transformation frequency of the Rec− mutant strains. The frequency of appearance of met+ transformants in the single Rec− strains and in certain double Rec− strains has been previously reported (1). Here, however, the experiments were performed in parallel for comparison with other strains (Table 2). We show here that ΔhelD deletion did not affect more than threefold chromosomal transformation of recF, addAB, recH, ΔrecU, and ΔrecS cells (Table 2).
The ΔhelD deletion did not affect more than twofold plasmid transformation of the highly impaired ΔrecU and ΔrecS cells, and it reduced three- to sevenfold plasmid transformation of recF, addAB, and recH cells (Table 2).
What is the role for helicase IV in DNA recombination?
The AddAB, PcrA, helicase IV, RecS, and RecQ helicases are the orthologs of the presynaptic RecBCDEco, UvrDEco, E. coli helicase IV, and RecQEco DNA helicases, respectively. However, genetic studies on mutants of these various helicases start to uncover some differences in terms of their respective roles in repair and recombination in the two hosts. PcrA is essential in B. subtilis, whereas UvrDEco is not essential under laboratory growth conditions (24). It has been shown that RecQEco is required for conjugational recombination and DNA repair in a recBCEco sbcB(C)Eco background (22), whereas the RecS helicase partially suppressed the DNA repair defect of addAB cells and was required for plasmid transformation and DNA repair in an otherwise Rec+ background (12). The uvrDEco, helDEco, recQEco, and helD cells were recombination and repair proficient when present in otherwise Rec+ cells (21, 22, this work). A helDEco mutation, which has no effect on repair and recombination in an recBCEco sbcB(C)Eco background, interacts synergistically with recFEco and recOEco in the repair of MMS-damaged DNA (21), but we show here that ΔhelD partially suppresses the DNA repair defect of recFLOR cells.
A possible way to reconcile these contrasting observations is to consider that in B. subtilis recombinational repair takes place mostly through the functions classified within the α (recFLOR) and ɛ (recB, recU, and recD) epistatic groups. To illustrate this point, one should compare the steepness of MMS curves for a recF or a recU mutant to the modest effect of MMS on survival of an addAB mutant. Furthermore, genetic studies are made possible in a RecF− Add+ background in B. subtilis which are prevented in E. coli given the preponderance of the RecBCD pathway, hiding all possible effect of mutations on the RecF pathway. Taking these data into account, we propose that in a recF background, helicase IV extends the number of gaps created upon injury to DNA, which results in a severe sensitivity to genotoxic agents. When it is absent, fewer gaps are created, which are taken in charge by the functions classified within the ɛ (RecB, RecU, and RecD) and β (AddAB) epistatic groups, and this results in the partial suppression of the repair defect. Such an effect cannot be observed in E. coli for the above-mentioned reasons (work in a recBC sbcBC background). Still, one observation made for E. coli could be reminiscent of such a role for helicase IV. It was found that the double recQEco uvrDEco mutant could not be constructed in a recBCEco sbcBCEco background, whereas the triple recQEco uvrDEco helDEco mutant was successfully constructed, albeit with difficulty (22). This suggests that somehow the absence of helicase IV in this mutant had a positive effect, possibly by reducing the amount of substrate that RecFOR had to deal with. A similar and symmetrical argument can be drawn in the case of recS (12). In conclusion, studies on recombination in B. subtilis are instructive and stimulating for the broadening of our view on recombination mechanisms in bacteria at large.
Acknowledgments
This research was supported by grant BMC2000–0548 from MCyT-DGI to J.C.A.
REFERENCES
- 1.Alonso J C, Tailor R H, Lüder G. Characterization of recombination-deficient mutants of Bacillus subtilis. J Bacteriol. 1988;170:3001–3007. doi: 10.1128/jb.170.7.3001-3007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso J C, Lüder G. Characterization of recF suppressors in Bacillus subtilis. Biochimie. 1991;73:277–280. doi: 10.1016/0300-9084(91)90213-k. [DOI] [PubMed] [Google Scholar]
- 3.Alonso J C, Lüder G, Tailor R H. Characterization of the Bacillus subtilis recombinational pathways. J Bacteriol. 1991;173:3977–3980. doi: 10.1128/jb.173.13.3977-3980.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonso J C, Lüder G, Trautner T A. Intramolecular homologous recombination in Bacillus subtilis 168. Mol Gen Genet. 1992;236:60–64. doi: 10.1007/BF00279643. [DOI] [PubMed] [Google Scholar]
- 5.Alonso J C, Stiege A C, Lüder G. Molecular analysis of the Bacillus subtilis recF function. Mol Gen Genet. 1993;239:129–136. doi: 10.1007/BF00260632. [DOI] [PubMed] [Google Scholar]
- 6.Alonso J C, Ayora S, Rojo F. Recombinación Genética en Bacillus subtilis, In: Casadesús J, editor. Microbiología y genética molecular. Huelva, Spain: Publicaciones Universidad de Huelva; 1996. pp. 229–240. [Google Scholar]
- 7.Canosi U, Iglesias A, Trautner T A. Plasmid transformation in Bacillus subtilis: effects of insertion of Bacillus subtilis DNA into plasmid pC194. Mol Gen Genet. 1981;181:434–440. doi: 10.1007/BF00428732. [DOI] [PubMed] [Google Scholar]
- 8.Chédin F, Noirot P, Biaudet V, Ehrlich S D. A five-nucleotide sequence protects DNA from exonucleolytic degradation by AddAB, the RecBCD analogue of Bacillus subtilis. Mol Microbiol. 1998;29:1369–1377. doi: 10.1046/j.1365-2958.1998.01018.x. [DOI] [PubMed] [Google Scholar]
- 9.Chédin F, Ehrlich S D, Kowalczykowsky S C. The Bacillus subtilis AddAB helicase/exonuclease is regulated by its cognate Chi sequence in vitro. J Mol Biol. 2000;298:7–20. doi: 10.1006/jmbi.2000.3556. [DOI] [PubMed] [Google Scholar]
- 10.Clark A J, Sandler S J. Homologous genetic recombination: the pieces begin to fall into place. Crit Rev Microbiol. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]
- 11.Dubnau D. Genetic exchange and homologous recombination. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 555–584. [Google Scholar]
- 12.Fernández S, Sorokin A, Alonso J C. Genetic recombination in Bacillus subtilis 168: effects of recU and recS on DNA repair and homologous recombination. J Bacteriol. 1998;180:3405–3409. doi: 10.1128/jb.180.13.3405-3409.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández S, Kobayashi Y, Ogasawara N, Alonso J C. Analysis of the Bacillus subtilis recO gene: RecO is part of RecFLOR function. Mol Gen Genet. 1999;261:567–573. doi: 10.1007/s004380051002. [DOI] [PubMed] [Google Scholar]
- 14.Fernández S, Ayora S, Alonso J C. Bacillus subtilis homologous recombination gene and products. Res Microbiol. 2000;151:481–486. doi: 10.1016/s0923-2508(00)00165-0. [DOI] [PubMed] [Google Scholar]
- 15.Gorbalenya A E, Koonin E V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 16.Kooistra J, Venema G. Cloning, sequencing and expression of Bacillus subtilis genes involved in ATP-dependent nuclease synthesis. J Bacteriol. 1991;173:3644–3655. doi: 10.1128/jb.173.12.3644-3655.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunst F, Ogasawara N, Moszer I, Albertini A M G, Alloni, Azevedo V M G, Bestero, et al. The complete genome sequence of the Gram-positive model organism Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 18.Lacks S A. Mechanisms of genetic recombination in gram-positive bacteria. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C.: American Society for Microbiology; 1988. pp. 43–86. [Google Scholar]
- 19.Lorenz M G, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matson S W. Escherichia coli DNA helicase II (uvrD gene product) catalyzes the unwinding of DNA-RNA hybrids in vitro. Proc Natl Acad Sci USA. 1989;86:4430–4434. doi: 10.1073/pnas.86.12.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendonca V M, Kaiser-Rogers K, Matson S W. Double helicase II (uvrD)-helicase IV (helD) deletion mutants are defective in the recombination pathways of Escherichia coli. J Bacteriol. 1993;175:4641–4651. doi: 10.1128/jb.175.15.4641-4651.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendonca V M, Klepin H D, Matson S W. DNA helicases in recombination and repair: construction of a ΔuvrD ΔhelD ΔrecQ mutant deficient in recombination and repair. J Bacteriol. 1995;177:1326–1335. doi: 10.1128/jb.177.5.1326-1335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendonca V M, Matson S W. Genetic analysis of ΔhelD and ΔuvrD mutation in combination with other genes in the RecF recombination pathways in Escherichia coli: suppression of a ruvB mutation by a uvrD deletion. Genetics. 1995;141:443–452. doi: 10.1093/genetics/141.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petit M A, Dervyn E, Entian K D, McGovern S, Ehrlich S D, Bruan C. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling circle replication. Mol Microbiol. 1998;29:261–273. doi: 10.1046/j.1365-2958.1998.00927.x. [DOI] [PubMed] [Google Scholar]
- 25.Subramanya H S, Bird L E, Brannigan J A, Wigle D B. Crystal structure of the Dexx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 26.Wood E R, Matson S W. The molecular cloning of the gene encoding the Escherichia coli 75-kDa helicase and the determination of its nucleotide sequence and genetic map position. J Biol Chem. 1989;264:8297–8303. [PubMed] [Google Scholar]
- 27.Yancey-Wrona J E, Wood E R, George J W, Smith K R, Matson S W. Escherichia coli Rep protein and helicase IV. Distributive single-stranded DNA-dependent ATPases that catalyze a limited unwinding reaction in vitro. Eur J Biochem. 1992;207:479–485. doi: 10.1111/j.1432-1033.1992.tb17074.x. [DOI] [PubMed] [Google Scholar]