Abstract
Intervening sequences (IVSs) in the rrl genes for 23S rRNA are transcribed but later removed by RNase III without religation during RNA processing, leading to fragmented rRNA. We examined about 240 strains of the family Enterobacteriaceae for presence of IVSs using PCR. No IVSs were detected in strains belonging to Escherichia, Shigella, Enterobacter, Erwinia, Ewingella, Hafnia, Kluyvera, Morganella, Pantoea, or Serratia. Previously unreported IVSs were detected in Klebsiella oxytoca, Citrobacter amalonaticus, and Providencia stuartii; previously reported IVSs are in species of Salmonella, Proteus, Providencia, and Yersinia. The sporadic distribution of IVSs indicates lateral genetic transfer of IVSs.
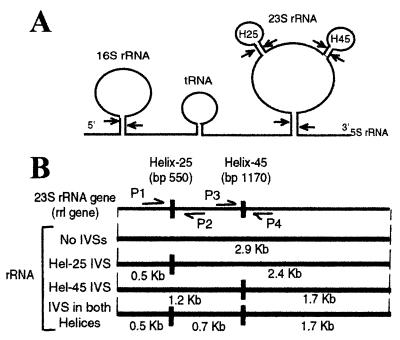
Fragmented 23S rRNA has been detected in Rhodopseudomonas (12, 16), Anacystis (4), and Salmonella (31). This fragmentation was shown to be due to intervening sequences (IVSs) in the rrl genes (3, 5); these IVSs are excised without religation during rRNA processing by RNase III (3). In Salmonella, IVSs appear at two sites in the proposed secondary structure of the rRNA, at helix 25 and helix 45, where the tetraloops are replaced by an elongated stem-loop structure of about 110 bp (Fig. 1).
FIG. 1.
(A) Processing of the rRNA transcript, showing removal of IVSs. A typical 30S rRNA transcribed from an rrn operon is shown with 16S, spacer tRNA, 23S, and 5S rRNAs. The positions of helix 25 IVS (H25) at bp 550 and helix 45 IVS (H45) at bp 1170 are also shown as stem-loop structures on the 23S rRNA. The arrows indicate sites of cleavage by RNase III (an enzyme responsible for primary processing of the nascent 30S rRNA in E. coli and S. enterica serovar Typhimurium). (B) rrl gene for 23S rRNA, indicating positions of helix 25 (about bp 550) and helix 45 (about bp 1170). Also indicated are primers used for PCR amplification of the helix 25 region (P1 and P2) and the helix 45 region (P3 and P4). The rRNA fragmentation patterns expected from excision of IVSs are also shown. The presence of 2.4- and 0.5-kb fragments indicates that an rrl gene contains an IVS in helix 25 only, 1.7 and 1.2-kb fragments indicate an IVS in helix 45 only, and 1.7-, 0.7-, and 0.5-kb fragments indicate IVSs in both helices. Mature 23S rRNA that lacks IVSs is 2.9 kb.
In spite of the overall conservation of rrn operons (32), IVSs are remarkable both in their sporadic distribution among bacteria (28) and in sequence variability. IVSs have been observed in 14 different genera of bacteria, including Rhodopseudomonas (5), Agrobacterium (8), Brucella (8), and Rhizobium (26) (alpha group of Proteobacteria, defined by 16S rRNA sequence) (32), Coxiella (1), Actinobacillus (7), Haemophilus (29), Salmonella (3, 17) (21), Proteus (20), Providencia (20), and Yersinia (27) (gamma group of Proteobacteria), and Campylobacter (10) and Helicobacter (9) (epsilon group of Proteobacteria), as well as Leptospira (23) (spirochetes).
IVSs which show less than 90% nucleotide identity within the family Enterobacteriaceae were placed in different families; those families which have near identity of sequences in the stem region of the IVS were grouped into superfamilies, but families in the same superfamily often have major variation in the loop region of the IVS (22). Superfamily I consists of helix 25 IVSs in families A, B, and C in Salmonella and family D in Proteus (18, 20, 22). Superfamily II, also in helix 25, comprises families E and G in Proteus and family F in Providencia (20). Superfamily III in helix 45 contains six families, M and O in Salmonella (22), N and P in Yersinia groups II and I, respectively (27), Q in Providencia (20), and R in Proteus (20).
The function of IVSs is not known. Strains of Salmonella containing IVSs with a deficiency of RNase III in which the IVSs are not excised still grow, but at reduced rates; the growth reduction is probably due to RNase III catalysis of other important reactions, not to the unexcised IVSs (19). In addition, strains of Escherichia coli (which normally do not contain IVSs) carrying a plasmid with an IVS-containing Salmonella enterica serovar Typhimurium rrl gene have fragmented 23S rRNA but grow at normal rate (6).
We examined about 240 strains of Enterobacteriaceae for the possession of IVSs, using PCR with whole genomic DNA as a template to amplify the helix 25 and helix 45 regions of the rrl gene. Bacterial strains (Table 1) were grown as reported earlier (21, 22). Enzymes, chemicals, and PCR primers have been described (18, 21). PCR methods and electrophoresis (21, 22, 24) and pulsed-field gel electrophoresis methods (14, 15) were also as previously described. DNA from PCR amplification was cycle sequenced by University of Calgary Core DNA Services (18)
TABLE 1.
Strains of Enterobacteriaceae in which IVSs were detected by PCR and in which rRNA fragmentation was observed
| Species | Strain | No. of genes with IVSs ina
|
|
|---|---|---|---|
| Helix 25 | Helix 45 | ||
| S. enterica serovar Typhimurium LT2 | SGSC1412b | 2 | 6 |
| E. coli K-12 | SAB1332c | 0 | 0 |
| C. amalonaticus | SA5617d | 1 | 0 |
| SA5664d | 0 | 3 | |
| SA5689d | 0 | 1 | |
| SA5690d | 0 | 4 | |
| K. oxytoca | SA5362e | 4 | 0 |
| SA5363e | 1 | 0 | |
| SA5364e | 2 | 0 | |
| SA5365e | 1 | 0 | |
| ATCC 13182fg | 2 | 0 | |
| P. stuartii | ATCC 33672fh | 5 | 0 |
| SA5625d | 4 | 0 | |
| SA5626d | 2 | 0 | |
| SA5627d | 3 | 0 | |
| SA5628d | 4 | 0 | |
| SA5639d | 2 | 0 | |
| P. rettgeri | SA5573d | 7 | 0 |
| SA5574d | 7 | 1 | |
The number of rrn genes (out of the total of seven) evaluated to contain IVSs (according to PCR data) and to contain sites leading to rRNA fragmentation (see the text for details).
From L. Lilleengen via J. Lederberg, stored in the Salmonella Genetic Stock Centre as strain SGSC1412.
From the Salmonella Genetic Stock Centre.
From P. Gibb, Calgary Laboratory Services, Calgary, Alberta, Canada.
From F. Liang, Calgary Regional Health Authority, Calgary, Alberta, Canada.
From the American Type Culture Collection.
K. oxytoca ATCC 13182 can be found in the Salmonella Genetic Stock Centre as SA5925.
P. stuartii ATCC 33672 can be found in the Salmonella Genetic Stock Centre as SA5582.
The negative control used here, E. coli K-12 (Fig. 2, lane 1), is known to lack IVSs in both helices 25 and 45 (3, 18); as expected, it shows for each helix only one fast-running band, which is 731 bp. Organisms in which the amplicons from PCR of genomic DNA for both helix 25 and helix 45 were indistinguishable from those of E. coli K-12 and which therefore do not contain detectable IVSs in these regions are as follows (data not shown): Citrobacter freundii (11 strains), Citrobacter koseri (23 strains), four species of Enterobacter (32 strains), three species of Erwinia (4 strains), Hafnia alvei (19 strains), Kluyvera (1 strain), Morganella morganii (19 strains), 3 species of Serratia (34 strains), Shigella sonnei (1 strain), and Yersinia frederiksenii (1 strain). These strains are listed in Fig. 3; some of the results are from earlier studies (see the figure legend). Ewingella americana (4 strains) and Pantoea agglomerans (7 strains) are enteric bacteria not shown in Fig. 3; they also lack IVSs.
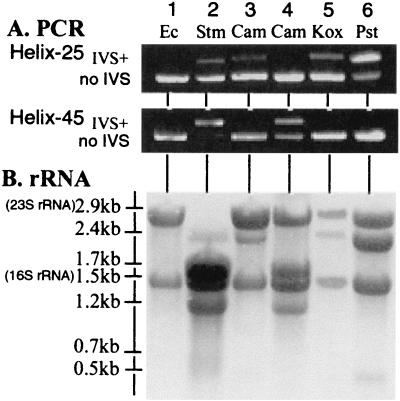
FIG. 2.
PCR amplification of IVSs and rRNA fragmentation in strains of enteric bacteria. Ec, E. coli K-12; Stm, S. enterica serovar Typhimurium LT2; Cam, C. amalonaticus SA5617 (lane 3) and SA5664 (lane 4); Kox, K. oxytoca SA5365; Pst, P. stuartii ATCC 33672. (A) Agarose gel containing PCR products from amplification of regions of rrl genes containing IVSs in helix 25 or helix 45; after PCR they were separated by electrophoresis in a gel containing ethidium bromide and visualized under UV light. (B) rRNA was blotted on a Hybond-N+ membrane and stained with methylene blue.
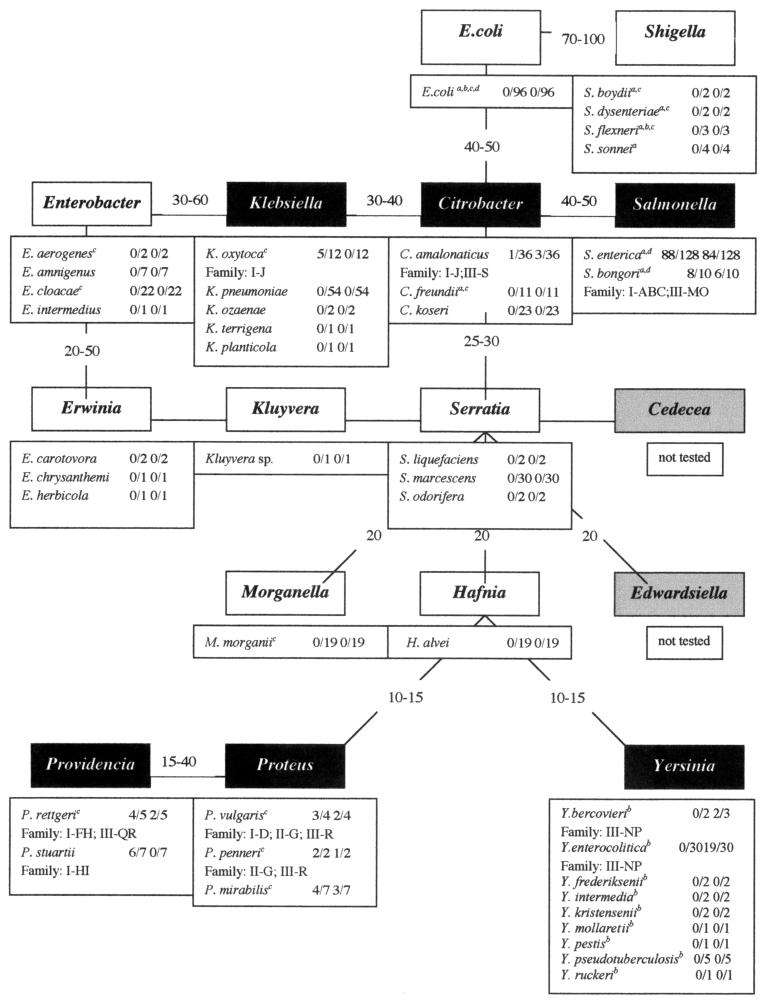
FIG. 3.
Distribution of IVSs and rRNA fragmentation in genera of the Enterobacteriaceae. The percentages of DNA-DNA reassociation, which are shown between the genera, come from Bergey's Manual of Systematic Bacteriology (11). Genera in which IVSs were not detected are shown in white boxes; genera in which some strains have IVSs and fragmented rRNA are shown in black boxes. The boxes below the genera show the species tested by PCR for the possession of IVSs; all strains which revealed IVSs were tested for rRNA fragmentation also. The numerators of the first and second fractions after the species show the number of strains which have IVSs in helix 25 and helix 45, respectively; the denominator is the total number of strains tested. For species that contain IVSs, the superfamily and family of IVSs are shown. The results are from this report except where indicated, as follows: a, reference 28; b, reference 27; c, reference 20; d, reference 22.
The positive control, S. enterica serovar Typhimurium LT2 (Fig. 2, lane 2), which contains IVSs in both helices 25 and 45 (18), yields two bands for each helix, as expected. Enteric bacteria usually have seven rrl genes, each of which may or may not have IVSs. Comparisons of the intensity of the slower-running band with the faster-running band estimate the number of the genes which have IVSs (18); the data indicate that helix 25 contains IVSs in two of the seven rrl genes and helix 45 contains IVSs in six rrl genes, as previously reported (18) (Table 1).
Some of the strains of Klebsiella oxytoca, Citrobacter amalonaticus, Providencia stuartii, and Providencia rettgeri yielded amplicons which were larger than the E. coli K-12 standard (Fig. 2, lanes 3 to 6), indicating that some of the rrl genes must possess IVSs. The number of rrl genes containing an IVS was estimated by the intensity of the bands that contain an IVS compared with the intensity of the bands without an IVS. Thirty-six strains of C. amalonaticus were tested by PCR; only one strain has one IVS in helix 25 (Fig. 2, lane 3), while three strains possess from one to four IVSs in helix 45 (Fig. 2, lane 4; Table 1). Of 12 strains of K. oxytoca, five possess one to four IVSs in helix 25, while none have IVSs in helix 45 (Fig. 2, lane 5). Of seven strains of P. stuartii, six have IVSs in helix 25 (Table 1; Fig. 2, lane 6). None of the strains of P. stuartii contain IVSs in helix 45, while one lacks IVSs in both helices. Of two strains of P. rettgeri, both have IVSs in helix 25 in all seven rrl genes, while one also has an IVS in helix 45 in only one rrl gene (data not shown); IVSs in this species were previously reported (20).
rRNA was extracted with phenol; the fragments were separated by gel electrophoresis and blotted on a Hybond-N+ membrane (Fig. 2B). E. coli K-12 (Fig. 2, lane 1), which does not contain IVSs, yields intact 23S rRNA (2.9 kb) and 16S rRNA (1.5 kb) as expected. S. enterica serovar Typhimurium (Fig. 2, lane 2) is known to contain IVSs in both helices; as expected, fragments of 2.4, 1.7, 1.2, 0.7, and 0.5 kb but not of 2.9 kb (intact 23S rRNA) were present, indicating that IVSs were excised from IVS-containing rRNA resulting from transcription of all seven rrl genes. The number of rrl genes containing IVSs was determined by comparing the intensity of the bands resulting from the fragmented rRNA with that of the 16S rRNA band. S. enterica serovar Typhimurium contains IVSs in helix 25 in two rrl genes, concluded from a light 2.4-kb band, and IVSs in helix 45 in six rrl genes, concluded from the dark 1.7- and 1.2-kb bands; these data are consistent with earlier conclusions (18). C. amalonaticus SA5617 (Fig. 2, lane 3) contains an IVS in helix 25 in one rrl gene according to the PCR data; the observation of a 2.9-kb band (intact 23S rRNA) and a light band at 2.4 kb indicates that this IVS in helix 25 results in rRNA fragmentation. The band expected at 0.5 kb is not seen because the concentration is too low. C. amalonaticus SA5664 (Fig. 2, lane 4) has IVSs in helix 45 according to the PCR data; the rRNA data show intact 23S rRNA and also fragments of 1.7 and 1.2 kb, indicating fragmentation at the IVSs in helix 45 in three rrl genes. Five out of 12 strains of K. oxytoca have one to four IVSs in helix 25 and also show rRNA fragmentation to yield 2.4- and 0.5-kb fragments (Table 1; Fig. 3); representative data for SA5365 in Fig. 2B, lane 5, indicate fragmentation at one helix 25 site. P. stuartii ATCC 33672 (lane 6) has IVSs in helix 25 in five rrl genes according to PCR data, and the rRNA data indicate that all these IVSs lead to fragmentation to 2.4- and 0.5-kb fragments; of seven strains tested (Fig. 3), six show rRNA fragmentation (Table 1).
IVS sequences cannot be determined by using genomic DNA unless all seven rrn operons have identical IVSs. Thus, individual rrn operons were isolated by digestion of genomic DNA in agarose by the endonuclease I-CeuI and PCR amplified as described earlier (13, 18, 20, 22). The nucleotide sequences of the DNA were determined by the University of Calgary Core DNA services. The percent similarity of DNA was calculated using DNASIS. IVSs were identified as members of the same family if they had more than 90% nucleotide identity. Then CLUSTAL X was used to align the sequences (Fig. 4). IVSs in helix 25 were placed in superfamily I or II based on the sequences in the stem region of the stem-loop structure of the RNA, which is predicted by the mfold program (33); all IVSs in superfamily II are in Proteus spp. (20), and these are not further discussed here.
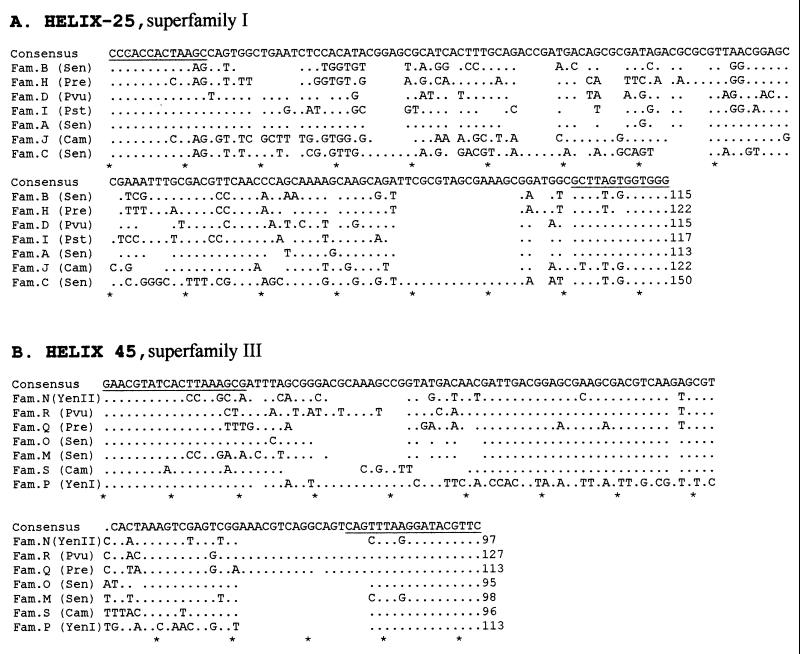
FIG. 4.
Alignment of IVSs using CLUSTAL X version 1.6 for the Macintosh. The alignment was imported into and modified in Word 98. Conserved bases are shown by dots, and deletions are shown by spaces. The consensus sequence is shown at the top of the alignment, and each 10th base is marked by an asterisk at the bottom. The number of bases on the IVS is at the end of each sequence. The bases which are underlined are postulated to be part of the primary stem, as described in the stem-loop structure postulated for the IVS based on secondary structure (22, 33). (A) Helix 25 IVSs from S. enterica (Sen) (families A, B, and C), Proteus vulgaris (Pvu) (family D), P. rettgeri (Pre) (family H), P. stuartii (Pst) (family I), and C. amalonaticus (Cam) and K. oxytoca (Kox) (family J). (B) Helix 45 IVSs from S. enterica (Sen) (families M and O), Yersinia enterocolitica (Yen) (families N and P), Proteus vulgaris (Pvu) (family R), P. rettgeri (Pre) (family Q), and C. amalonaticus (Cam) (family S).
Three new families of IVSs have been detected in helix 25, all in superfamily I. Family H in P. rettgeri (in two strains, where it was present in all 14 rrl genes) and in P. stuartii (in four rrl genes) resembles family B (Salmonella) in nucleotide identity (78 to 83%) and is close in CLUSTAL alignment (Fig. 4). Family I, also found in P. stuartii, resembles family A of Salmonella (76% nucleotide identity) and family D of Proteus (75% identity). The third new family is J, which was found in only one rrl gene of one strain of C. amalonaticus but also in nine genes of five strains of K. oxytoca; the sequences of all these IVSs are 98% identical. In helix 45, only one new family was found, family S, which is in superfamily III; it is in eight rrl genes in three strains of C. amalonaticus. It resembles family O of Salmonella in nucleotide identity (88 to 89%).
IVSs are sporadically distributed in Enterobacteriaceae. Fourteen of the 16 genera classified in Bergey's Manual of Systematic Bacteriology (11) were examined, and IVSs were detected in either helix 25 or helix 45 in six of these (Fig. 3); two genera (Cedecea and Edwardsiella) were not tested. The relationships of genera illustrated in Fig. 3 had been determined by numerous criteria but especially DNA-DNA hybridization (11). The genera that possess IVSs belong to three clusters: Salmonella, Citrobacter, and Klebsiella; Proteus and Providencia; and Yersinia (Fig. 3). More recently, many genera of the Enterobacteriaceae have been classified based on 16S rRNA sequences (30); most of the genera in Fig. 3 are placed in similar locations, but many new genera, especially of plant pathogens, were added.
IVSs are found very frequently in some genera but are uncommon in others. Salmonella and Proteus possess IVSs in more than half of the strains in both helices; Providencia has IVSs in helix 25 in six out of seven strains, though it has no IVSs in helix 45. Yersinia possesses IVSs only in two species and only in helix 45, but in each of these species over half the strains have IVSs (Fig. 3). However, IVSs are present only infrequently in other strains, such as those of Citrobacter and Klebsiella. For example, C. amalonaticus possesses IVSs in only one strain in one rrl gene in helix 25 and in three strains in helix 45 among 36 strains tested, while 11 strains of C. freundii and 23 strains of C. koseri have no IVSs. Klebsiella possesses IVSs in only one species in only 5 of 12 strains; other species, such as Klebsiella pneumoniae, have no IVSs in 54 strains. IVSs are certainly rare or absent in E. coli, since 96 strains, largely from the E. coli reference set, lacked IVSs in earlier tests (21) and in this study.
Family S in helix 45 of C. amalonaticus resembles family O in Salmonella (89% identity); the stem region of all helix 45 IVSs are similar (Fig. 4), so this is defined as a new family in superfamily III. Thus, IVSs in genera in which they were previously undescribed usually resemble known IVSs, suggesting that the major range of IVS variability in the Enterobacteriaceae has now been described.
IVSs in enteric bacteria are all inserted in helix 25 and/or in helix 45, but not at other locations in the DNA, according to earlier studies (17, 20–22) and to the work in this report. However, in other groups, such as Rhizobium, they also appear in other regions of the rRNA (25).
The sizes of IVSs in all genera of bacteria range from 90 bp (27) to 600 bp (23). IVSs in enteric bacteria fall into three different clusters, superfamily I and II in helix 25 and superfamily III in helix 45, based on sequences in the stem region. The loop regions have diverged much more, so that relationships are obscured.
Earlier studies on nucleotide sequences have supported the idea that IVSs must be exchanged between different genera by lateral transfer (20, 22, 23, 27). This report confirms and extends this theme; for example, the IVSs in helix 25 in 10 sequenced genes in six strains of the two genera represented by C. amalonaticus and K. oxytoca share 98% or more nucleotide identity. They are all members of family J (Fig. 4A) and thus are very closely related, indicating a recent lateral transfer between the genera; most IVS sequences have much less than 98% identity, even within genera.
IVSs in members of the six genera of Enterobacteriaceae show sequence homology with each other (Fig. 4) (22), but no significant homology between IVSs from Enterobacteriaceae and any other sequences, including IVSs from other bacteria, was detected by BLAST searches (2).
Acknowledgments
We appreciate the assistance of Kanti Pabbaraju and Renee Brost in providing instruction and help with numerous technical aspects of the work.
The work was supported by an operating grant from the Natural Sciences and Engineering Research Council of Canada and by grants AI34829-09 and AI43283 from the Allergy and Infectious Diseases Research Institute of the National Institutes of Health.
REFERENCES
- 1.Afseth G, Mo Y-Y, Mallavia L P. Characterization of the 23S and 5S rRNA genes of Coxiella burnetii and identification of an intervening sequences within the 23S rRNA gene. J Bacteriol. 1995;177:2946–2949. doi: 10.1128/jb.177.10.2946-2949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgin A B, Parodos K, Lane D J, Pace N R. The excision of intervening sequences from Salmonella 23S ribosomal RNA. Cell. 1990;60:405–414. doi: 10.1016/0092-8674(90)90592-3. [DOI] [PubMed] [Google Scholar]
- 4.Doolittle W F. Postmaturational cleavage of 23S ribosomal ribonucleic acid and its metabolic control in the blue-green alga Anacystis nidulans. J Bacteriol. 1973;113:1256–1263. doi: 10.1128/jb.113.3.1256-1263.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dryden S C, Kaplan S. Localization and structural analysis of the ribosomal RNA operons of Rhodobacter sphaeroides. Nucleic Acids Res. 1990;18:7267–7277. doi: 10.1093/nar/18.24.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregory S T, O'Connor M, Dahlberg A E. Functional Escherichia coli 23S rRNAs containing processed and unprocessed intervening sequences from Salmonella typhimurium. Nucleic Acids Res. 1998;24:4918–4923. doi: 10.1093/nar/24.24.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haraszthy V I, Sunday G J, Bobek L A, Motley T S, Preus H, Zambon J J. Identification and analysis of the gap region in the 23S ribosomal RNA from Actinobacillus actinomycetemcomitans. J Dent Res. 1992;71:1561–1568. doi: 10.1177/00220345920710090401. [DOI] [PubMed] [Google Scholar]
- 8.Hsu D, Zee Y C, Ingraham J, Shih L M. Diversity of cleavage patterns of Salmonella 23S rRNA. J Gen Microbiol. 1992;138:199–203. doi: 10.1099/00221287-138-1-199. [DOI] [PubMed] [Google Scholar]
- 9.Hurtado A, Clewley J P, Linton D, Owen R J, Stanley J. Sequence similarities between large subunit ribosomal RNA gene intervening sequences from different Helicobacter species. Gene. 1997;194:69–75. doi: 10.1016/s0378-1119(97)00158-3. [DOI] [PubMed] [Google Scholar]
- 10.Konkel M E, Marconi R T, Mead D J, Cieplak W., Jr Identification and characterization of an intervening sequence within the 23S ribosomal RNA genes of Campylobacter jejuni. Mol Microbiol. 1994;14:235–241. doi: 10.1111/j.1365-2958.1994.tb01284.x. [DOI] [PubMed] [Google Scholar]
- 11.Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. [Google Scholar]
- 12.Lessie T G. The atypical ribosomal RNA complement of Rhodopseudomonas spheroides. J Gen Biol. 1965;39:311–320. doi: 10.1099/00221287-39-3-311. [DOI] [PubMed] [Google Scholar]
- 13.Liu S-L, Hessel A, Sanderson K E. Genomic mapping with I-CeuI, an intron-encoded endonuclease, specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc Natl Acad Sci USA. 1993;90:6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S-L, Hessel A, Sanderson K E. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella typhimurium LT2 determined by double digestion, end labelling, and pulsed-field gel electrophoresis. J Bacteriol. 1993;175:4104–4120. doi: 10.1128/jb.175.13.4104-4120.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S-L, Sanderson K E. I-CeuI reveals conservation of the genome of independent strains of Salmonella typhimurium. J Bacteriol. 1995;177:3355–3357. doi: 10.1128/jb.177.11.3355-3357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrs B, Kaplan S. 23S precursor ribosomal RNA of Rhodopseudomonas spheroides. J Mol Biol. 1970;49:297–317. doi: 10.1016/0022-2836(70)90247-0. [DOI] [PubMed] [Google Scholar]
- 17.Mattatall N R, Daines D A, Liu S-L, Sanderson K E. Salmonella typhi contains identical intervening sequences in all seven rrl genes. J Bacteriol. 1996;178:5323–5326. doi: 10.1128/jb.178.17.5323-5326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattatall N R, Sanderson K E. Salmonella typhimurium LT2 possesses three distinct 23S rRNA intervening sequences. J Bacteriol. 1996;178:2272–2278. doi: 10.1128/jb.178.8.2272-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattatall N R, Sanderson K E. RNase III deficient Salmonella typhimurium LT2 contains intervening sequences (IVSs) in its 23S rRNA. FEMS Microbiol Lett. 1998;159:179–185. doi: 10.1111/j.1574-6968.1998.tb12858.x. [DOI] [PubMed] [Google Scholar]
- 20.Miller W L, Pabbaraju K, Sanderson K E. Fragmentation of 23S ribosomal RNA in strains of Proteus and Providencia results from intervening sequences (IVSs) in the rrn (rRNA) genes. J Bacteriol. 2000;182:1109–1117. doi: 10.1128/jb.182.4.1109-1117.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pabbaraju K, Miller W L, Sanderson K E. Distribution of intervening sequences in the genes for 23S rRNA and rRNA fragmentation among strains of the Salmonella Reference Collection B (SARB) and SARC sets. J Bacteriol. 2000;182:1923–1929. doi: 10.1128/jb.182.7.1923-1929.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pabbaraju K, Sanderson K E. Sequence diversity of intervening sequences (IVSs) in the ribosomal RNA in Salmonella spp. Gene. 2000;253:55–66. doi: 10.1016/s0378-1119(00)00239-0. [DOI] [PubMed] [Google Scholar]
- 23.Ralph D, McClelland M. Phylogenetic evidence for horizontal transfer of an intervening sequence between species in a spirochete genus. J Bacteriol. 1994;176:5982–5987. doi: 10.1128/jb.176.19.5982-5987.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning; a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Selenska-Pobell S, Doring H. Sequences around the fragmentation sites of the large subunit ribosomal RNA in the family Rhizobiaceae. 23S-like rRNAs in Rhizobiaceae. Antonie Leeuwenhoek. 1998;73:55–67. doi: 10.1023/a:1000540023194. [DOI] [PubMed] [Google Scholar]
- 26.Selenska-Pobell S, Evguenieva-Hackenberg E. Fragmentations of the large-subunit rRNA in the family Rhizobiaceae. J Bacteriol. 1995;177:6993–6998. doi: 10.1128/jb.177.23.6993-6998.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skurnik M, Toivanen P. Intervening sequences (IVSs), in the 23S ribosomal RNA genes of pathogenic Yersinia enterocolitica strains. The IVSs in Y. enterocolitica and Salmonella typhimurium have a common origin. Mol Microbiol. 1991;5:585–593. doi: 10.1111/j.1365-2958.1991.tb00729.x. [DOI] [PubMed] [Google Scholar]
- 28.Smith N H, Crichton P B, Old D C, Higgins C F. Ribosomal-RNA patterns of Escherichia coli, Salmonella typhimurium and related Enterobacteriaceae. J Med Microbiol. 1988;25:223–228. doi: 10.1099/00222615-26-3-223. [DOI] [PubMed] [Google Scholar]
- 29.Song X M, Forsgren A, Janson H. Fragmentation heterogeneity of 23S ribosomal RNA in Haemophilus species. Gene. 1999;230:287–293. doi: 10.1016/s0378-1119(99)00063-3. [DOI] [PubMed] [Google Scholar]
- 30.Sproer C, Mendrock U, Swiderski J, Lang E, Stackebrandt E. The phylogenetic position of Serratia, Buttiauxella and some other genera of the family Enterobacteriaceae. Int J Syst Bacteriol. 1999;49:1433–1438. doi: 10.1099/00207713-49-4-1433. [DOI] [PubMed] [Google Scholar]
- 31.Winkler M E. Ribosomal ribonucleic acid isolated from Salmonella typhimurium: absence of the intact 23S species. J Bacteriol. 1979;139:842–849. doi: 10.1128/jb.139.3.842-849.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuker M, Mathews D H, Turner D H. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide in RNA biochemistry and biotechnology. NATO ASI Ser. 1999. pp. 11–43. [Google Scholar]