Abstract
While pluripotent stem cell-derived kidney organoids are now being used to model renal disease, the proximal nephron remains immature with limited evidence for key functional solute channels. This may reflect early mispatterning of the nephrogenic mesenchyme and/or insufficient maturation. Here we show that enhanced specification to metanephric nephron progenitors results in elongated and radially aligned proximalised nephrons with distinct S1 - S3 proximal tubule cell types. Such PT-enhanced organoids possess improved albumin and organic cation uptake, appropriate KIM-1 upregulation in response to cisplatin, and improved expression of SARS-CoV-2 entry factors resulting in increased viral replication. The striking proximo-distal orientation of nephrons resulted from localized WNT antagonism originating from the organoid stromal core. PT-enhanced organoids represent an improved model to study inherited and acquired proximal tubular disease as well as drug and viral responses.
Subject terms: Stem-cell differentiation, Induced pluripotent stem cells, Regeneration, SARS-CoV-2, Differentiation
Proximal nephron in pluripotent stem cell derived kidney organoids are immature with limited support for functional solute channels. Vanslambrouck et al report improved metanephric specification, generating enhanced kidney organoids with superior proximal tubules, spatially arranged nephrons, and applications for disease research, and drug screening.
Introduction
The proximal tubules (PTs) of the kidney represent a highly specialised portion of the nephron performing the bulk of kidney reabsorption and secretion. This occurs via three distinct functional and anatomical segments: the convoluted (S1 and S2) and the straight (S3) segments that traverse the cortico-medullary boundary, with S1 exhibiting the highest capacity for solute, sodium, amino acid, and fluid transport1. Their unique roles and high metabolic activity render the PTs acutely vulnerable to toxins and metabolic stress2. As such, accurately patterned and segmented PTs would represent a critical tool for drug development, toxicology research, and studies of PT dysfunction.
We and others have established robust protocols for the directed differentiation of human pluripotent stem cells to kidney3–7. While these organoids display a remarkable transcriptional similarity to the developing human kidney8–11, their nephron patterning and segmentation remain immature, more closely resembling human trimester 1 fetal tissue6. PT maturation and functional segmentation are particularly underdeveloped. Despite possessing nuclear HNF4A (responsible for driving early proximal patterning ref. 12) and apical CUBILIN-MEGALIN complex expression, organoid PTs lack a range of functional solute channels that define each PT subsegment11,13. Expression levels of the principle water transporting channel, AQP1, the organic anion transporters (OATs), and the organic cation transporters (OCTs) are all low13.
Such suboptimal PT maturation may represent inappropriate anteroposterior/mediolateral patterning, suboptimal maintenance of progenitor identity or incomplete maturation. In response to distinct temporospatial signalling, the permanent (metanephric) kidney arises during human embryogenesis as the final of three embryonic excretory organs, developing sequentially from specific rostrocaudal regions of the intermediate mesoderm14. Metanephric development, commencing during weeks 4−5 of gestation, is preceded by the formation of two more rostral transient organs; the pronephros (human gestation weeks 3–4) and the mesonephros (human gestation week 4–10)15. While the mammalian pronephros is highly rudimentary, mesonephric nephrons also arise via MET and show similar patterning and segmentation to early metanephric nephron. However, the mesonephros possesses less definitive distal tubule segments and regresses around week 816–18.
Using fluorescent reporter lines and lineage tracing in human kidney organoids, we have confirmed both the presence of a SIX2+ nephron progenitor population and the contribution of these cells to nephrogenesis via MET in kidney organoids19,20. However, the possibility exists that we are modelling mesonephric rather than metanephric nephrogenesis, potentially contributing to poor PT patterning and maturation (reviewed in ref. 21). It is also possible that suboptimal maintenance of progenitor identity during iPSC differentiation in vitro limits nephron maturation. Several media have been described that are able to support the maintenance of isolated nephron progenitors in vitro22–25. While each media contains low levels of canonical WNT activity and FGF2/9, distinct differences in nephron patterning result from the inclusion of a variety of TGFβ superfamily agonists (BMP4, BMP7, Activin A) and antagonists (A83-01, LDN193189), NOTCH inhibition (DAPT), and other growth factors (TGFα, IGF1/2, LIF). The inclusion of LDN193189 (inhibitor of BMP receptor-mediated SMAD1/5/8) supported tubular patterning but not formation of glomeruli22. In contrast, the addition of LIF and either dual-SMAD inhibition (LDN193189 and A83-01) or NOTCH inhibition (DAPT) resulted in the formation of nephrons with podocytes but different nephron morphologies23,25. Finally, while proximodistal nephron patterning in mouse has previously been shown to be influenced by relative Wnt, Bmp, and Notch signalling in mouse26, these data suggest that distinct nephron progenitor states may show varying competence for different nephron segments, or that distinct SIX2 populations give rise to different regions of the nephron.
Here we show that patterning to a posterior metanephric SIX2+ nephron progenitor population by extending the duration of mesodermal patterning, while simultaneously enhancing nephron progenitor expansion, specifies progenitors with improved metanephric identity without influencing anteroposterior/mediolateral patterning. These progenitors form strongly proximalised, elongated, and spatially aligned nephrons. The PTs within these nephrons display distinct segmentation into S1, S2 and S3 cell types, upregulation of key solute channels and transporters, and functional uptake of albumin and organic cations. Treatment with cisplatin upregulates Kidney Injury Marker-1 (KIM-1), while increased expression of key viral entry factors enables improved SARS-CoV-2 infection and replication compared to standard protocols. Notably, striking nephron alignment results from localised WNT antagonism, supporting a role for WNT gradients in human nephron proximodistal patterning. Taken together, this study suggests a requirement for optimal nephron progenitor commitment for appropriate PT identity. Such PT-enhanced kidney organoids represent a model of the human proximal nephrons with likely applications for infectious and genetic disease research, drug development, and nephrotoxicity evaluation.
Results
Delayed nephron induction supports nephron progenitors
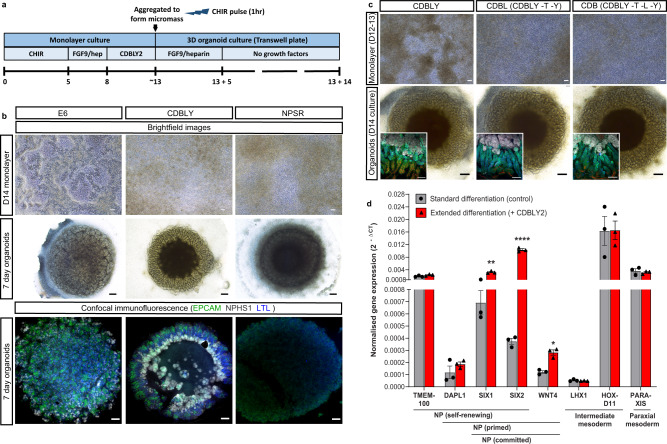
As noted previously, optimisation of nephron progenitor maintenance in vitro has been investigated by a range of studies using murine and human pluripotent stem cell-derived nephron progenitors22,23,25. While all studies reported maintenance of nephron progenitors, variations were evident with respect to the final patterning of resulting nephrons following induction. Given the clear influence that initial differentiation conditions and timing can have on nephron progenitor survival and subsequent nephron patterning, we hypothesised that expanding our nephron progenitor population whilst delaying nephron initiation may create a more metanephric population leading to organoids with improved patterning and PT maturation. We have previously shown that SIX2 expression is not detected until day 10 of pluripotent stem cell differentiation19. Hence, the initial monolayer differentiation phase was prolonged to between 12–14 days, along with culture in either of two previously defined nephron progenitor (NP) maintenance media, NPSR23 and CDBLY25 from day 7, which represents the point of intermediate mesoderm commitment6,27 (Fig. 1a). Compared to control media (TeSR-E6; E6), both NPSR and CDBLY prevented spontaneous epithelialisation of the monolayer (Fig. 1b). However, very little epithelialisation and poor nephron commitment was observed after culture in NPSR (Fig. 1b). In contrast, CDBLY preserved the nephron-forming capacity of the progenitor cells following their formation into a micromass and induction of nephrogenesis with a pulse of canonical WNT signalling) (Fig. 1b). Nephrons of these organoids were also observed to surround a stromal core region that stained positive for markers of kidney stroma MEIS1/2/3 and SNAI2 (SLUG) (Supplementary Fig. 1a)28. Upon prolonged organoid culture (>14 days), portions of this core region formed patches of Alcian blue-positive cartilage (Supplementary Fig. 1B).
Fig. 1. Extended monolayer culture in CDBLY supports nephron progenitors and preserves nephrogenic capacity.
a Schematic depicting the extended differentiation protocol in CDBLY2. b Brightfield and confocal immunofluorescence images of extended monolayer differentiations in E6, CDBLY and NPSR, and resulting organoids. Immunofluorescence depicts nephrons (EPCAM; green), podocytes of glomeruli (NPHS1; grey), and proximal tubules (LTL; blue). Scale bars represent 100 µm (monolayers) and 200 µm (organoids). c Brightfield images of extended monolayer differentiations using CDBLY variations and resulting organoids, with inset confocal immunofluorescence images highlighting organoid nephron alignment and patterning. Immunofluorescence depicts nephrons (EPCAM; green), podocytes of glomeruli (NPHS1; grey), proximal tubules (HNF4A; blue), and Loop of Henle (SLC12A1; red). Scale bars represent 100 µm (monolayer brightfields and organoid immunofluorescence) and 200 µm (organoid brightfields). d qRT-PCR analysis of standard and extended monolayer differentiations. Error bars represent SEM from n = 3 biological replicates. Statistical significance was determined using an unpaired t test. Asterisks represent two-tailed P values adjusted for multiple comparisons using the Holm–Sidak method, alpha = 0.05 (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 [SIX1: P = 0.002059; SIX2: P = 0.000075; WNT4: P = 0.017459; GATA3: P = 0.001246]). Source data are provided as a Source data file.
The prevention of spontaneous differentiation while preserving the nephrogenic capacity of the NP cells was found to be primarily a response to the presence of CDB (CHIR, DAPT, BMP7), with omission of LIF, Y27632, as well as the basal media component TGFα, found to produce a similar result with respect to growth, morphology and nephron segmentation compared to CDBLY (Fig. 1c). The inhibition of monolayer epithelialisation with preserved nephrogenic capacity was found to be consistent at monolayer differentiation lengths tested (10, 12, 13 and 14 days) (Supplementary Fig. 1C). However, a monolayer differentiation length of 12–13 days produced more consistent nephrogenesis between experiments, with 14 days observed to cause frequent detachment of the differentiating monolayer. Subsequent studies proceeded using prolonged culture in CDBLY noting the inclusion of an increased concentration of BMP7 (10 ng/mL; CDBLY2) which improved the consistency of nephrogenesis between organoids compared to standard CDBLY (5 ng/mL BMP7) (Supplementary Fig. 1D). This modified differentiation protocol is detailed in Fig. 1a.
Quantitative RT-PCR (qRT-PCR) of the extended monolayer differentiations in CDBLY2 confirmed an improved metanephric gene expression profile compared to standard differentiations performed in parallel (7-day protocol in E619,29) (Fig. 1d). Extended CDBLY2 monolayers showed a significant increase in SIX1/SIX2 (self-renewing to committed NPs) and WNT4 (primed to committed NPs), while DAPL1 (self-renewing and primed NPs) was increased without significance and no change was observed in TMEM100 (self-renewing NPs). This suggested that the extended protocol promotes a primed/committed, rather than self-renewing, NP population30–32. Extended differentiation in CDBLY2 was not found to alter mediolateral patterning, with no change in paraxial mesodermal marker PARAXIS and unchanged or increased expression of intermediate mesoderm markers HOXD11 and LHX117 (Fig. 1d).
Extended culture induces SIX2-derived proximalised nephrons
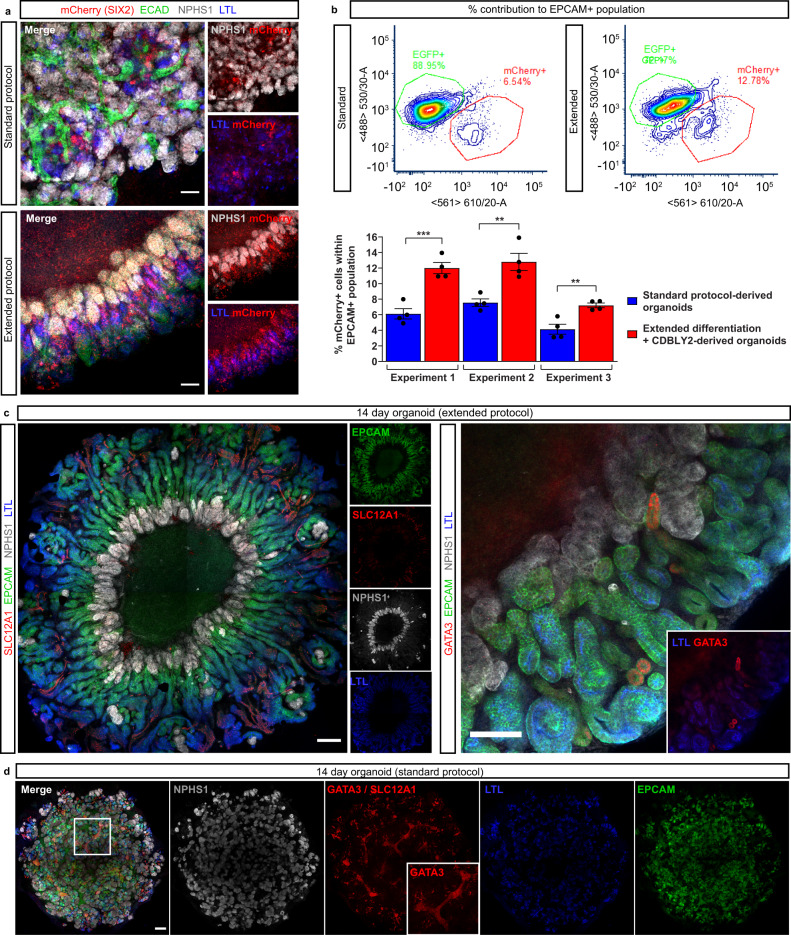
Lineage tracing studies in mouse have shown that nephrons are derived entirely from Six2+ nephron progenitors33, with histological studies suggesting a similar developmental process in human33–35. Using a SIX2Cre/Cre:GAPDHdual lineage tracing line, in which SIX2 expression induces a permanent GFP/mCherry switch, we have previously shown that kidney organoid nephrons contain cells derived from SIX2+, but also SIX2-, progenitor cells, resulting in a chimeric appearance19. To confirm and compare the competence of the metanephric progenitor-enriched monolayer differentiation to contribute to nephron formation, organoids were generated from both our standard protocol and the extended differentiation protocol using the SIX2Cre/Cre:GAPDHdual lineage tracing line. Immunofluorescence re-confirmed the chimeric contribution of SIX2+ and SIX2- progenitor-derived cells to standard organoid nephrons as shown previously19 (Fig. 2a). However, confocal imaging suggested a larger contribution of SIX2+ cells to proximal nephrons in organoids derived from the extended protocol compared to the standard protocol (7 days differentiation, cultured in E6)19, including contribution to NPHS1+ podocytes, LTL+ PTs, and to a lesser extent E-CADHERIN+ distal tubules (Fig. 2a). To quantitatively compare the contributions SIX2-derived cells to nephrons, dissociated SIX2Cre/Cre:GAPDHdual standard and extended organoids (expressing endogenous SIX2-mCherry) were co-stained with EPCAM to mark both proximal and distal nephron epithelium, then analysed via flow cytometry (Fig. 2b, Supplementary Fig. 1E). In agreement with confocal imaging, SIX2-derived cell contribution to EPCAM+ nephrons was significantly higher in organoids derived from the metanephric progenitor-enriched extended monolayers compared to those derived from the standard 7 day protocol in E6 media, suggesting improved metanephric identity of prolonged monolayers exposed to CDBLY2 (Fig. 2b).
Fig. 2. Extended monolayer culture in CDBLY2 increases SIX2+ progenitor contribution to nephrons and proximalisation.
a Confocal immunofluorescence of D7 + 14 (standard protocol) and D13 + 14 (extended protocol) organoids derived from the SIX2Cre/Cre:GAPDHdual lineage tracing iPSC line. Images depict merged and separated channels showing lineage-traced SIX2+ cells (mCherry; red), distal tubules (ECAD; green), podocytes (NPHS1; grey) and proximal tubules (LTL; blue). Scale bars represent 100 µm. b Flow cytometry of SIX2Cre/Cre:GAPDHdual lineage tracing organoids derived from extended (13 day + CDBLY2) and standard (7 day + E6 media) differentiations depicting SIX2-derived mCherry + (red) cell contribution to the EPCAM + (nephron) population. Flow plots shown in top panels are representative of the replicates across multiple experiments. Percentage mCherry contributions from flow cytometry are depicted in bar graph (bottom panel), where error bars represent SEM from n = 4 biological replicates across 3 independent experiments. Statistical significance was determined using an unpaired t test. Asterisks represent two-tailed P values adjusted for multiple comparisons using the Holm–Sidak method, alpha = 0.05 (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 [Experiment 1: P = 0.000924; Experiment 2: P = 0.004899; Experiment 3: P = 0.004845]). Source data are provided as a Source data file. c Confocal immunofluorescence of D13 + 14 organoids demonstrating (left panel) aligned nephron morphology with nephron segmentation makers (nephron epithelium [EPCAM; green], distal tubule/Loop of Henle [SLC12A1; red], proximal tubules [LTL; blue], and podocytes [NPHS1; grey]) and (right panel) the presence of few GATA3 + connecting segment/ureteric epithelium structures (red), co-stained for nephron epithelium (EPCAM; green), podocytes (NPHS1; grey), and proximal tubules (LTL; blue). Inset in right panel shows GATA3 and LTL alone. Scale bars in (i) and (ii) represent 200 µm and 100 µm, respectively. d Confocal immunofluorescence of a D7 + 14 (standard) organoid depicting homogenous distribution of podocytes (NPHS1; grey), proximal tubules (LTL; blue), and nephron epithelium (EPCAM; green), as well as the presence of extended segments of centralised GATA3 + (red [nuclear]) connecting segment/ureteric epithelium (also highlighted in insets). Scale bar represents 200 µm.
The segmentation of nephrons within organoids derived from the extended protocol was examined using a range of markers for podocytes, proximal, and distal tubules, revealing distinct proximo-distal segmentation (Fig. 2c). In contrast to the standard protocol which produced organoids with a branching GATA3 + epithelium (Fig. 2d), extended protocol-derived organoids possessed few structures expressing the ureteric epithelium marker GATA3 (Fig. 2c, right panel). The distribution of glomeruli, marked by NPHS1 + podocytes, also differed between protocols, with extended protocol-derived organoids possessing a central ring of glomeruli and elongated PTs radiating outwards that starkly opposed the more homogenous distribution of these structures in standard organoids (Fig. 2c, d). This unique organoid morphology was observed in organoids derived from 6 different iPSC lines with or without gene editing and from male or female iPSC sources (3 examples evidenced in Supplementary Fig. 1F).
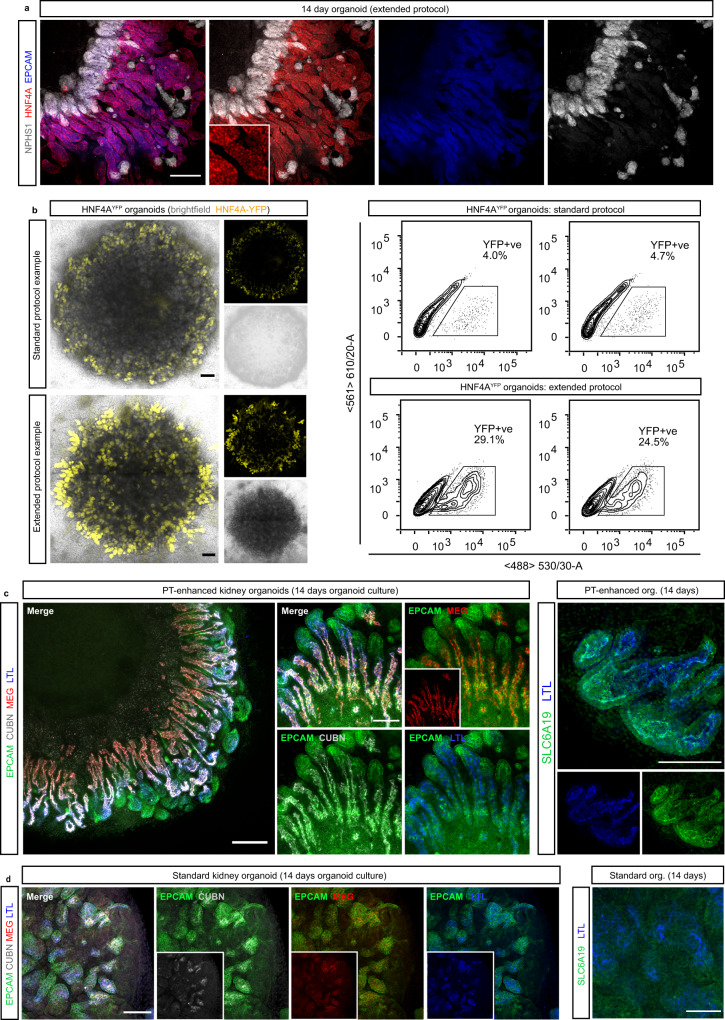
In addition to differences in the segmentation of nephrons, organoids derived via extended differentiation in CDBLY2 appeared to possess a larger proportion of PT expressing LTL and HNF4A compared to standard organoids (Figs. 2c [left panel], d, and 3a). To quantify and compare the proportion of PT cells in organoids derived from these two protocols, organoids were generated using the HNF4AYFP iPSC reporter line which reports the formation of PT20 (Fig. 3b). Flow cytometry revealed up to 6.2 times higher average proportions of HNF4AYFP+ PT cells in organoids derived from the extended monolayer protocol compared to the standard protocol (Fig. 3b, Supplementary Fig. 1E), confirming the use of extended monolayer differentiation combined with progenitor-supportive media, CDBLY2, as an effective method of generating proximal tubule-enhanced (PT-enhanced) kidney organoids.
Fig. 3. Enhanced organoids express mature and appropriately localised PT transporter proteins and transcription factors.
a Confocal immunofluorescence of HNF4A protein expression (red) in EPCAM-positive PTs (blue) of a day 14 organoid derived from extended differentiation of iPSCs in CDBLY2. NPHS1 (grey) marks podocytes of the glomeruli. Inset depicts higher magnification of HNF4A-expressing PT segments emphasising nuclear localisation. Scale bar represents 100 µm. b Left panel shows live confocal microscopy of 2 representative standard and extended protocol-derived organoids (also shown in right panel) generated using the HNF4AYFP fluorescent iPSC reporter line (PCS-201-010/HNF4AYFP). YFP (yellow) marks PTs. Transmitted light channel (brightfield) is shown as merged and separate images. Scale bars represent 200 µm. Right panel of (b) shows flow cytometry plots of the 2 representative HNF4AYFP organoids from experiment depicted in left panel, derived from standard and extended protocols. c, d Confocal immunofluorescence of PT-enhanced (D13 + 14; c) and standard (D7 + 14; d) organoids showing PT markers within EPCAM + (green) nephrons, including LTL (blue in all panels), CUBILIN (CUBN; grey [left panels of c, d]), MEGALIN (MEG; red [left panels of c, d]), and SLC6A19 (green [right panel of c]). Inset in (c) depicts MEG alone. Insets in (d) depict CUBN, MEG, and LTL. Scale bars represent 200 µm (c, left panel) and 100 µm (c [right panel] and d [all panels]).
PT-enhanced organoids show improved proximal tubule maturation
To establish the level of PT maturation within enhanced organoids, the expression and cellular localisation of functionally important brush border membrane proteins and markers, characteristic of mature PTs, were assessed via immunofluorescence (Fig. 3c). Within LTL-positive PTs, enhanced organoids showed strong expression of the protein transport complex CUBILIN-MEGALIN (CUBN-MEG) and neutral amino acid transporter SLC6A19, with all transporters displaying a highly-specific apical brush border membrane localisation (Fig. 3c). In contrast, the PTs of standard organoids possessed weaker and diffuse staining of the CUBN-MEG complex (Fig. 3d, left panel). Furthermore, the majority of standard organoids lacked SLC6A19 expression, with staining observed in just one of three independent experiments (representative images in Fig. 3d [right panel] and Supplementary Fig. 2A). Additional information regarding the maturity of PT brush border membranes was afforded by high-resolution imaging of LTL binding. LTL is a fucose-specific lectin widely used in the kidney field owing to its high-affinity binding to α-linked L fucose-containing oligosaccharides of glycoconjugates that abundantly line the brush border membrane of kidney PT cells36. High-resolution imaging of PTs within enhanced organoids showed LTL binding was highly restricted to the apical membrane where it co-localised with SLC6A19, a characteristic of correctly polarised, mature PT brush-border membranes (Supplementary Fig. 2A). In contrast, the PTs of standard organoids possessed LTL staining that was not highly apically-restricted and instead diffuse throughout the PT, even in the instance where apical SLC6A19 was detected (Fig. 2d [right panel] and Supplementary Fig. 2A). Taken together, these data suggested a more immature PT phenotype and suboptimal brush border membrane development in standard compared to enhanced organoids.
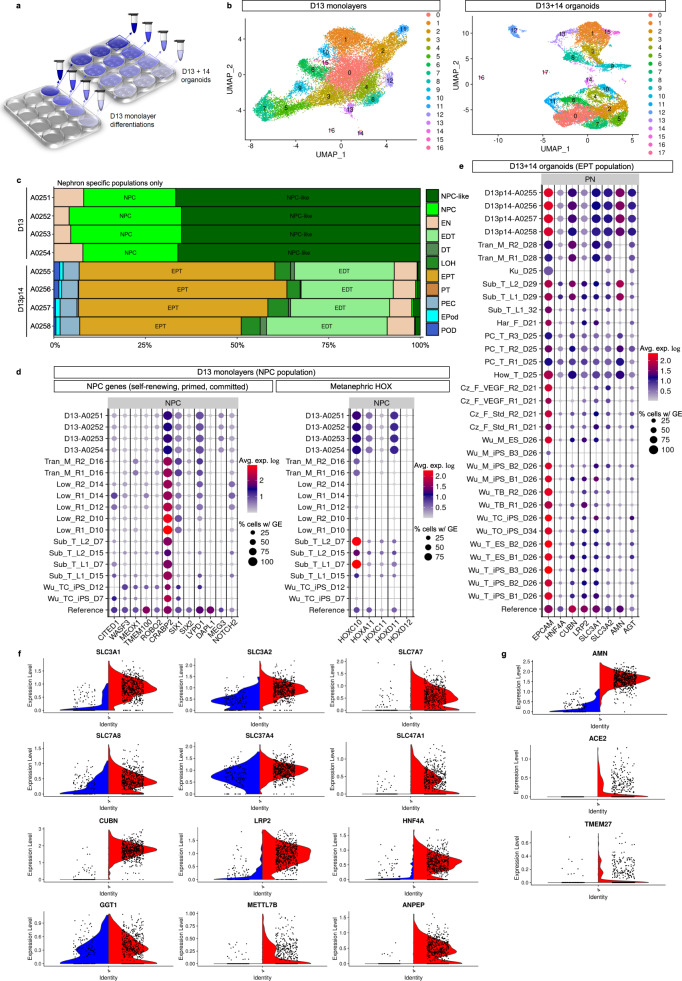
To provide a more comprehensive comparison with existing kidney organoid differentiation protocols, as well as to gain a deeper insight into the complexity and maturity of cells derived from the extended protocol, multiplexed single-cell RNA sequencing (scRNAseq) with antibody-based cell barcoding was performed on both monolayer (day 13) and resulting PT-enhanced organoids (Fig. 4). To account for variation, libraries were created from 4 separate differentiated monolayers representing distinct starting pools of iPSCs (CRL1502.C32) that were used to generate 4 separate batches of organoids (Fig. 4a). Cells from the 4 replicates (both at day 13 [D13] monolayer stage, prior to organoid formation, and day 14 of organoid culture [D13 + 14]) were barcoded using hashing antibodies before being pooled. This approach produced a single library for each timepoint (sample) which could be later deconvoluted to retrieve replicate information.
Fig. 4. Single cell transcriptional profiling (scRNAseq) shows improved specification, patterning and maturation of proximal tubules and their progenitors.
a Schematic depicting experimental design and profiled samples. Multiple organoids (2.5 × 105 cells per organoid) were generated from each of the 4 replicate differentiated cell monolayers at D13. The remaining portion of cells from each replicate monolayer were barcoded and pooled for generation of the D13 monolayer library. The resulting organoids were cultured for 14 days before being harvested and pooled within replicate wells, making 4 cell suspensions. These 4 suspensions were individually barcoded for later de-replication and pooled into a single-cell suspension for generation of the D13 + 14 organoid library. b UMAP plots of D13 and D13 + 14 samples (pooled replicates) identifying 16 and 17 distinct cell clusters, respectively. c ComparePlots depicting proportions of kidney cell types (nephron-specific populations only) in D13 and D13 + 14 sample replicates as classified by DevKidCC. Population abbreviations: nephron progenitor cell (NPC), early nephron (EN), early distal tubule (EDT), DT (distal tubule), loop of Henle (LOH), early proximal tubule (EPT), proximal tubule (PT), parietal epithelial cell (PEC), early podocyte (EPod), podocyte (Pod). d DevKidCC dot plots comparing the expression of gene signatures for (left plot) self-renewing (SIX1, SIX2, CITED1, WASF3, DAPL1, MEOX1, TMEM100, ROBO2, CRABP2), committed (SIX1, SIX2, LYPD1), and primed (DAPL1, NOTCH2, MEG3) NPC subsets, as well as (right plot) metanephric HOX genes, within the D13 monolayer NPC population to that of published stem cell-derived kidney datasets and a mixed (week 11–18) human fetal kidney reference dataset30,38,82. Comparisons were made to published monolayer and early nephrogenic datasets10,11,37,38 as outlined previously13. e DevKidCC dot plot comparing the expression of proximal nephron (PN) gene signatures within the EPT population of PT-enhanced D13 + 14 organoids to that of published stem cell-derived kidney organoid datasets40–42 and the mixed week 11–18 fetal kidney reference dataset30,38,82 as outlined previously13. f, g Violin plots in (f, g) compare PT-specific gene expression of PT-enhanced organoids (red, right on each plot) with our existing standard organoid dataset of equivalent line and age (blue, left)19. Genes encoding auxiliary proteins are shown in (g).
The resulting D13 and D13 + 14 pooled replicate libraries resolved 19,956 and 15,852 individual cell transcriptomes per timepoint, respectively. UMAP plots showed the resolution of distinct clusters for both D13 monolayers and resulting PT-enhanced (D13 + 14) organoids (Fig. 4b). Gene expression analyses confirmed the expression of a range of markers for mesenchymal cell states pre-kidney organogenesis in D13 monolayers, as well as markers of proximodistal patterning, stroma, and endothelium in D13 + 14 organoids (Supplementary Fig. 2BC and Supplementary Data 1–2). To enable unbiased comparisons of kidney cell types and gene expression levels between D13/D13 + 14 samples, published stem cell-derived, and reference human kidney datasets, datasets were analysed using the DevKidCC package13. DevKidCC enables robust classification of novel developing human or stem cell-derived kidney organoid datasets without the need for integration or prior dimensional reduction or clustering. Using the ComparePlot function, kidney cell proportions in D13 and D13 + 14 samples were directly compared, confirming distinct differences in cell populations yet consistency between the 4 replicates within each sample (Fig. 4c and Supplementary Fig. 3A). As anticipated, over 90% of cells within the D13 monolayer differentiations were classified as nephron progenitor cells (NPC) or NPC-like, with a small contribution of cells classified as early nephron (EN) (Fig. 4c). In contrast, D13 + 14 organoids possessed a range of proximal, distal, and renal corpuscle cell types. Early proximal tubule (EPT) formed the largest proportion of organoid nephron cell types (51% average across 4 samples), while two replicates possessed a small (<5%) fraction of maturing PT cells. By contrast, previous studies of the standard organoid protocol show on average <25% EPT and no PT6.
DevKidCC was next used to compare cell type-specific markers in D13/D13 + 14 samples to published stem cell-derived and reference human fetal kidney datasets (Fig. 4d, e). Analysis of the NPC population within D13 samples confirmed strong gene signatures for committed NPCs (SIX1, SIX2, and LYPD1) and the metanephric HOX code (HOXC10/11, HOXA11 and HOXD11) compared to relevant published monolayer and nephrogenic-stage differentiations10,11,37,38 that better emulated the mixed reference dataset of human fetal kidneys (weeks 11, 13, 16, 18)30,38,39. PT-enhanced organoids derived from these D13 monolayer differentiations possessed high and abundant expression of a range of proximal nephron markers in their EPT population (Fig. 4e). These included genes encoding several membrane proteins critical for PT transport of proteins and amino acids (CUBN, LRP2, SLC3A1 and SLC3A2), as well as auxiliary proteins and transcription factors required for transporter regulation and functionality, such as AMN, AGT and HNF4A. This gene signature showed remarkable congruence to reference human fetal kidney and improved PT identity compared to existing published kidney organoid datasets40–42 (Fig. 4e).
An important anatomical feature of the mature PT is its segmentation into functionally and morphologically distinct regions defined as the S1/S2 convoluted tubule segments and the S3 straight segment. In addition to differences in proliferation characteristics and protein synthesis, the convoluted and straight segments display distinct differences in solute handling to accommodate the declining concentration of solutes as the ultrafiltrate passes through the nephron1,43. As such, early S1–S2 convoluted segments express low-affinity/high-capacity transporters, with a gradual transition to high-affinity/low-capacity transporters in the later S3 straight segment44–46. To determine whether the PTs of enhanced organoids show evidence of this segmentation, PT clusters from the 4 integrated D13 + 14 replicate datasets were isolated and re-clustered, resolving 4740 PT cells across 6 distinct clusters (Supplementary Fig. 3B). The PT population was analysed for the expression of segment-specific PT markers with critical functional roles, including solute carriers for ions (SLC34A1/NPT247 expressed in S1 > S2), glucose (SLC2A2/GLUT2 and SLC5A2/SGLT2 expressed in S1 > S2; SLC2A1/GLUT1 and SLC5A1/SGLT1 expressed in S2 < S348–50), amino acids (SLC7A9/b(0,+)AT transporter of cystine, aspartate, and glutamate expressed in S1/S2 > S351, and cationic drugs/toxins (SLC47A1/MATE1 expressed in S1/S2 > S352), as well as AKAP12 (involved in cell cycle regulation, expressed in S2 < S353 and GPX3 (glutathione peroxidase; secreted antioxidant synthesised in S1/S2 > S343) (Supplementary Fig. 3C). UMAP plots revealed the largely opposing distributions of cells expressing S1 > S2 and S2 > S3 gene signatures (Supplementary Fig. 3C). Cells expressing S1 > S2 convoluted PT markers (SLC34A1/MATE1, SLC2A2/GLUT2 and SLC5A2/SGLT2) were predominantly located in clusters 0, 3, and the lower portion of cluster 4, whereas cells expressing S2 < S3 straight PT markers (AKAP12, SLC2A1/GLUT1 and SLC5A1/SGLT1) were primarily within clusters 1, 2, and the upper portion of cluster 4. When analysed for markers that exhibit a gradient of expression along the length of the nephron (S1/S2 > S3), UMAP plots for each gene revealed a similar graded expression pattern, with a higher concentration of positive cells within the S1 > S2 cluster (0) and decreasing in prevalence within S2 < S3 clusters (0, 2) (Supplementary Fig. 3C). Together this suggested that, despite the low expression of some markers indicating PT immaturity, the PTs of enhanced kidney organoids show evidence of separation into the 3 anatomically distinct PT segments.
Comparison between organoids is confounded by the inherent variability of different organoid protocols, technical variables, and individual cell line characteristics. To minimise potential bias when comparing cell maturation, PT-enhanced organoid scRNAseq data were compared to an existing standard organoid dataset derived from the same iPSC line and of equivalent organoid age19. Libraries from the PT-enhanced and standard organoid samples resolved 6737 and 1879 cells, respectively. Datasets were integrated prior to quality control measures to enable direct comparison of PT maturation and UMAP plots confirmed the resolution of distinct kidney cell clusters for both samples (Supplementary Fig. 3D). Violin plots of the PT cluster alone in integrated datasets confirmed that the PT-enhanced organoid dataset possessed higher and more abundant expression of genes critical for PT functionality compared to the standard organoid (Fig. 4f, g). Examples included genes encoding membrane transporters CUBILIN/CUBN and MEGALIN/LRP2 (important for protein uptake54, heavy-chain subunit solute carriers rBAT/SLC3A1 and 4F2/SLC3A2 (required for heteromer formation and amino acid transport by SLC7 family members55, light-chain subunit solute carriers y+LAT-1/SLC7A7 and LAT2/SLC7A8 (responsible for regulating intracellular amino acid pool via basolateral efflux of basic and neutral amino acids for transport systems y + L and L, respectively56,57, and solute carriers critical for PT metabolism and drug transport (G6PT1/SLC37A4 and MATE1/SLC47A158 (Fig. 4f). Several auxiliary proteins essential for correct apical localisation and transporter functionality also showed higher expression in the PT-enhanced dataset, including AMN (AMNIONLESS), ACE2 and TMEM27 (COLLECTRIN)55,59–61 (Fig. 4g). Expression of genes encoding drug transporters SLC22A2 (OCT2) and SLC22A6 (OAT1) were low in both conditions but increased in PT-enhanced compared to standard organoids (Supplementary Fig. 3E).
To investigate PT maturation further, an unbiased ToppFun GO Molecular Function analysis was performed on genes that were significantly differentially expressed within the PT cluster of PT-enhanced compared to standard organoids (945 input genes). This analysis revealed key differences in genes involved in cell metabolism (Supplementary Fig. 3F). PT-enhanced organoid cells within the PT cluster showed increased expression of genes related to fatty acid metabolism and its regulation, such as PPARG, FABP3, PRKAA2 and FAT1 (Supplementary Fig. 3G). Given the known reliance of mature PT cells on fatty acid metabolism in vivo (reviewed in1, this gene signature was suggestive of a more mature metabolic profile in enhanced compared to standard organoid PT cells.
Together, these comprehensive scRNAseq analyses confirmed an increased abundance and relative maturation of PT within this extended protocol. Analyses of D13 monolayers suggest this higher-order PT patterning arises from improved NPC identity at the point of metanephric specification.
Localised WNT antagonism drives radial nephron alignment
Of interest was the characteristic radial patterning observed in all PT-enhanced organoids, where tubules align with their glomeruli towards the centre of the organoid, surrounding a central core region, and distal SLC12A1 + segments towards the organoid periphery (refer to Fig. 2c). This orientation was suggestive of a directional patterning cue emanating from the core region, shown earlier to express stroma marker proteins MEIS1/2/3 and SNAI2 (Supplementary Fig. 1A). Previous studies have not only suggested a role of interstitial/stromal populations in nephron differentiation28,62, but have also indicated proximo-distal patterning is controlled by Wnt/β-catenin signalling along the nephron axis, with lower WNT signalling leading to improved formation and maturation of the proximal nephron26. In agreement with this, WNT inhibition has been observed to promote podocyte commitment in PSC cultures63. These findings suggested that the central core of PT-enhanced organoids may possess stromal populations influencing nephron patterning and/or express a localised WNT antagonist leading to directional signalling cues.
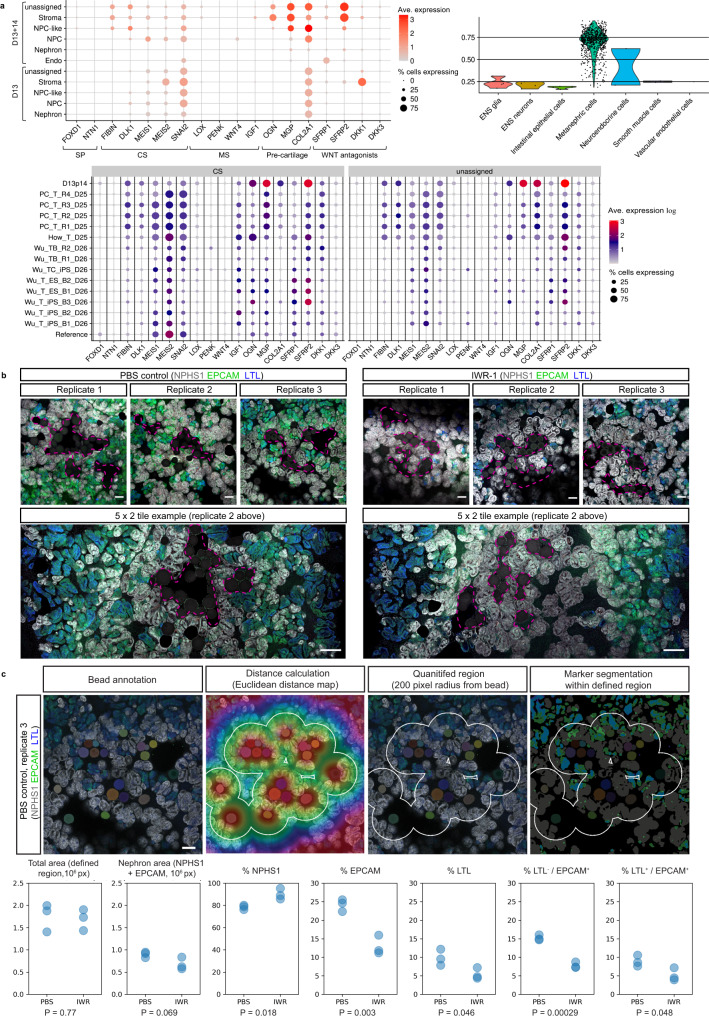
PT-enhanced scRNAseq datasets classified by DevKidCC were re-analysed to examine the stromal populations at greater depth. In addition to nephron-related and endothelial populations, previous classification of D13 + 14 organoids identified 48.2% of cells as stroma (enriched for CRABP1, COL3A1, COL1A1, COL1A2 and CXCL12) and 23.8% of cells as unassigned but similarly enriched for collagens (e.g., COL2A1 and COL9A1) (Supplementary Fig. 3A). Further analyses of D13 + 14 populations for defined markers of stromal zones curated in mouse kidney28 revealed the stromal cells of PT-enhanced organoids were most like those of kidney cortex (Fig. 5a, top left plot). High expression of cortical stroma (CS) markers, including FIBN, DLK1, MEIS1/2 and SNAI2, were observed predominantly in the unassigned, stroma, and NPC-like subsets, while medullary stroma and stromal progenitor markers were largely absent (Fig. 5a, top left plot). Unassigned and stroma clusters also highly expressed the WNT antagonist Secreted Frizzled-Related Protein-2 (SFRP2) and developing cartilage markers (ONG, MGP and COL2A1) that been previously identified in mouse kidney stromal cells (ONG and MGP)64 and nephrogenic mesenchyme (COL2A1)65,66 (Fig. 5a, top left plot). When compared to standard organoid datasets derived from a range of relevant published protocols, D13 + 14 PT-enhanced organoids possessed a similar cortical stroma gene signature to several datasets, but notably higher expression of the WNT antagonist, SFRP2, and pre-cartilage markers, within cortical stroma and unassigned populations (Fig. 5a, bottom plot).
Fig. 5. WNT signalling gradient influences nephron alignment and directionality.
a Analyses of scRNAseq datasets for PT-enhanced D13 + 14 organoid replicates (top left and bottom panels) and D13 monolayer replicates (top left and right panels) as classified by DevKidCC. Top left dot plot depicts expression of stroma compartment markers28 and WNT antagonists in D13 + 14 and D13 samples. DevKidCC’s DotPlotCompare (bottom panel) shows the comparison of D13 + 14 to other relevant published datasets (cortical stroma [CS] and unassigned populations only). Dot colour and size in dot plots represent unscaled gene expression and percentage of cells expressing each gene, respectively. Violin plot (top right) depicts the similarity scores for all unassigned cells within D13 monolayer replicates as predicted by the Azimuth label transfer method (https://azimuth.hubmapconsortium.org/) with the human developmental reference dataset67, where cells are grouped by population with the highest similarity score. Population abbreviations in all panels: nephron progenitor cell (NPC), endothelium (Endo), stroma progenitor (SP), cortical stroma (CS), medullary stroma (MS), enteric nervous system (ENS). b Confocal immunofluorescence images of replicate standard organoids bioprinted in a patch conformation and in contact with either agarose beads soaked in PBS (control; left panel) or in 10 µM IWR-1 (right panel). Clusters of beads are outlined with magenta dashed lines. Organoids are stained with markers of epithelium (EPCAM; green), proximal tubule (LTL; blue), and podocytes of the glomeruli (NPHS1; grey). Scale bars represent 100 µm (all top panels) and 200 µm (bottom tile panels). c Top panel shows and example image from (b, left panel [PBS control, replicate 3]) illustrating the image annotation approach used to segment and quantify the proportion of nephron structures (NPHS1 + [grey], EPCAM + [green], and LTL + [blue]) within a defined region 200 pixels from any bead (white outline). Solid colours represent masks for beads and nephrons. Scale bar represents 200 µm. Bottom panel of (c) shows quantification of PBS control and IWR-1 treated organoid images from (b) using approach illustrated in top panel images of (c), with n = 3 replicates per condition. Total area and nephron area values are shown in pixels (106 px). Percentage (%) of each structure (NPHS1+, EPCAM+, LTL+) are shown as a proportion of the total nephron area. P values were calculated using a two-sided t-test without adjustment for multiple comparisons and are indicated below each plot. Source data are provided as a Source data file.
D13 monolayers were similarly re-analysed to determine at which stage of the differentiation protocol (monolayer or 3D culture) stroma and pre-cartilage subtypes appear. Previously shown to contain just 0.9% stromal cells (Supplementary Fig. 3A), analysis of the D13 sample following DevKidCC classification confirmed a lack of stromal progenitor (SP) and medullary stroma (MS) zone markers, while expression of cortical stroma (CS) and pre-cartilage markers were limited (Fig. 5a, top left plot). This suggested that these definitive CS and pre-cartilage populations arise during the organoid culture period, but possibly from precursor NPC-like and/or unclassified cell populations in the D13 monolayer owing to their dominance in the differentiations (83%) (Supplementary Fig. 3A). Indeed, the NPC-like population in D13 monolayers showed a high similarity to the NPC population without key NPC markers (e.g. PAX8 and SIX2), while Azimuth label transfer method using a human developmental reference dataset67 still classified the majority (~75%) of D13 monolayer cells as metanephric despite 52.3% being unclassified by DevKidCC (Supplementary Fig. 3A, Fig. 5a [top right plot]).
The cortical stromal gene expression, notably including the WNT antagonist SFRP2, suggested that the central core region of PT-enhanced organoids may control WNT pathway-mediated nephron patterning, in turn driving the observed radial alignment. To functionally test this hypothesis, a WNT signalling gradient was recreated using agarose beads soaked in the tankyrase inhibitor, IWR-1 (10 µM), which antagonises canonical WNT/β-catenin pathway activity68 (Fig. 5b). Following the 7 day (standard) differentiation protocol, iPSC-derived kidney progenitors were bioprinted and cultured to create rectangular patch organoids69. At 5 days of organoid culture (D7 + 5), by which time renal vesicle formation had occurred, IWR-1-soaked or control (PBS-soaked) beads were added to the centre of the organoids where they made contact with the early epithelial structures (Supplementary Fig. 4A). After 9 days of organoid culture, organoids with IWR-1-soaked beads exhibited visible differences in the morphology of structures surrounding the beads compared to controls with PBS-soaked beads (Supplementary Fig. 4B). This became more apparent when these organoids were stained via immunofluorescence (Fig. 5b). In control organoids with PBS-soaked beads, beads were in contact with a mixture of proximal and distal EPCAM-positive nephron epithelium, as well as NPHS1-positive podocytes of glomeruli (Fig. 5b, left panel). In contrast, IWR-1-soaked beads were predominantly surrounded by glomeruli, with few distal structures (LTL-negative/EPCAM-positive) visible overall (Fig. 5b, right panel). These observations were confirmed by image quantification, showing that the percentage of NPHS1 + podocytes (glomeruli) was significantly higher in the region adjacent to IWR-1-soaked beads compared to PBS-soaked control beads (Fig. 5c and Supplementary Table 1).
Taken together, these analyses supported establishment of a gradient arising from centralised WNT antagonism as responsible for the nephron directionality and alignment in PT-enhanced organoids.
Functionally enhanced organoid nephrons enable drug screening
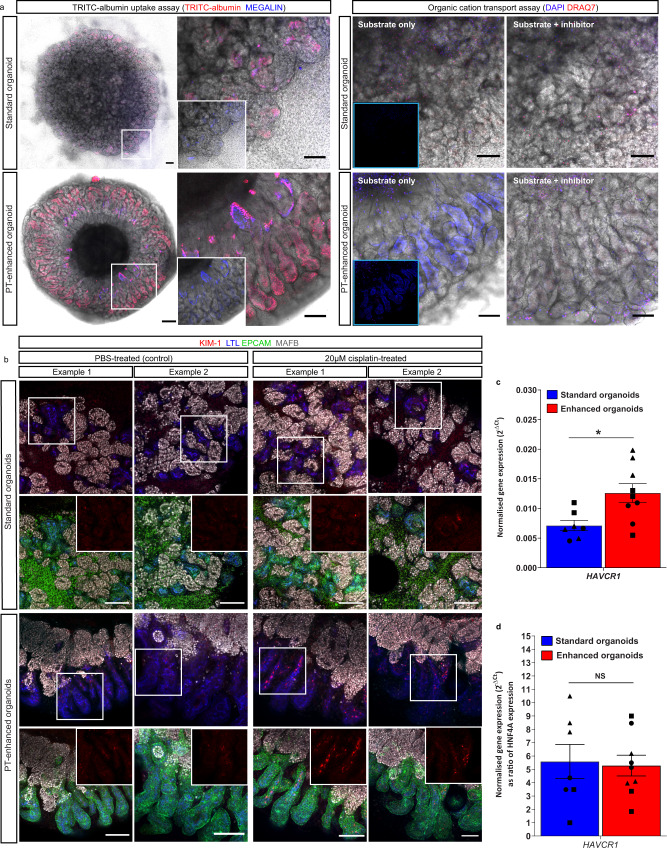
The strong expression and apical cellular localisation of transporters in PT-enhanced organoids was suggestive of nephron functionality. To test this, we firstly performed multiple substrate uptake assays specific to PTs in both standard and PT-enhanced kidney organoids (Fig. 6a). While standard organoids showed evidence of uptake of fluorescently labelled albumin (TRITC-albumin) into MEG-positive PTs (indicative of MEG-CUBN transport function), this uptake was visibly higher in PT-enhanced organoids, with large portions of elongated PTs displaying high-intensity TRITC-albumin fluorescence (Fig. 6a, left panel). In addition, PTs of enhanced organoids demonstrated robust uptake of 4′,6-diamidino-2-phenylindole (DAPI), which is an effective probe for evaluation of the PT-specific SLC47 family of organic cation/H+ antiporters, MATE-1 (Multidrug and Toxin Extrusion Protein 1) and MATE2-K (Multidrug and Toxin Extrusion Protein 2 K)70 (Fig. 6a, right panel). The uptake of DAPI by PT cells was successfully inhibited via pre-treatment of organoids with the cation transporter inhibitor Cimetidine, supporting the specificity of transport activity, while the absence of DRAQ7 staining excluded the possibility of DAPI uptake in PTs due to cell death (Fig. 6a, right panel, bottom images ). In contrast, standard organoids showed no uptake of DAPI, suggesting functional immaturity of these same drug transporters (Fig. 6a, right panel, top images).
Fig. 6. Enhanced organoids possess functional PT transporters and appropriate injury response.
a Live confocal images of standard and PT-enhanced organoids (transmitted light and fluorescence overlays) depicting (left panel) uptake of TRITC-albumin (red) into MEGALIN-positive PTs (blue) and (right panel) uptake of DAPI (blue; surrogate for organic cation transport capacity). White boxed areas on left images of left panel are shown as higher magnification on right (insets depict MEGALIN staining alone). Organic cation uptake image set (right panel) depicts organoids exposed to substrate alone (DAPI; blue, left images) or a combination of substrate/DAPI + inhibitor/Cimetidine (right images). Dead cells (right panel) are labelled with DRAQ7 (red). Right panel insets depict blue channel only without brightfield overlay. Scale bars represent 200 µm (whole organoid images in left panel) and 100 µm (right images in left panel, all images in right panel). b Confocal immunofluorescence of representative D7 + 14 (standard; top panels) and D13 + 14 (PT-enhanced; bottom panels) line-matched organoids following 24 h treatment with E6 media containing either (ii) 20 µM cisplatin or (i) an equivalent volume of PBS. Images depict KIM-1-expressing cells (red) in LTL + proximal tubules (blue) with nephron epithelium co-stained with EPCAM (green). Insets of bottom row images for standard and PT-enhanced organoids show KIM-1 staining (red channel) alone from white boxed regions in top row images. Scale bars in all images represent 100 µm. c, d qRT-PCR analyses depicting KIM-1 gene (HAVCR1) expression in standard (blue) and PT-enhanced (red) organoids from experiments shown in (b). HAVCR1 gene expression values are normalised to the expression of housekeeping gene GAPDH and depicted both with (d) and without (c) compensation for differences in PT proportion (expressed as a ratio of HNF4A). Error bars represent SEM from n = 8 (control) and n = 9 (cisplatin-treated) biological replicates across 3 replicate experiments as indicated. Statistical significance was determined using an unpaired t test. Asterisk represents two-tailed P value (*P ≤ 0.05 [P = 0.014837]) adjusted for multiple comparisons using the Holm–Sidak method alpha = 0.05. NS non-significant. Source data are provided as a Source data file.
Having established albumin and organic cation transport capacity in PT-enhanced organoids, we next assessed their response to nephrotoxic insult (Fig. 6b–d). Several recent studies have explored the suitability of kidney organoids as a human-relevant model of cisplatin-induced nephrotoxicity3,4,6, a common complication that limits usage of this chemotherapeutic agent71,72. The biomarker KIM-1 is sensitive for early detection of PT injury in humans and animals73–77 and has been shown to increase in response to cisplatin in kidney organoids, despite conflicting reports regarding its PT-specificity4,29,78. This discrepancy may arise from immature expression of the predominant cisplatin transporters, particularly SLC22A2/OCT278, combined with heterogeneity in cisplatin uptake mechanisms. Re-analysis of our PT-enhanced and existing standard organoid scRNAseq datasets19 revealed higher expression of the majority of cisplatin influx and efflux transporters in enhanced compared to standard organoid PT cells (Supplementary Fig. 4C), suggestive of cisplatin transport capacity. This included SLC22A2/OCT2, previously reported to show low expression in kidney organoids78. To confirm the functionality of these transporters and appropriate injury response by PTs, iPSC line-matched D7 + 14 (standard) and D13 + 14 (enhanced) organoids were derived from monolayer differentiations across 3 independent experiments. Organoids were exposed to 20 µM cisplatin for 24 h prior to assessment for expression of KIM1 protein its corresponding gene, HAVCR1. Immunofluorescence revealed an upregulation of KIM-1 protein expression within LTL-positive PTs of both standard and enhanced organoids compared to PBS-treated controls (Fig. 6b). This was supported by a significant increase in KIM-1 gene (HAVCR1) expression in PT-enhanced organoids compared to standard organoids (Fig. 6c). Also noteworthy was the similar HAVCR1 expression levels in standard and PT-enhanced organoids when gene level was expressed relative to the absolute amount of PT in each organoid (marked by HNF4A). This suggested that the levels of HAVCR1 upregulation may be dictated by proximal tubule proportion (Fig. 6d). However, in both standard and PT-enhanced organoids, HAVCR1 expression was significantly increased compared to control organoids (Supplementary Fig. 4D).
Improved organoid PTs increase SARS-CoV-2 research suitability
Kidney organoids have previously proven useful to model inherited, early-onset kidney disease3,5,40,79–85. More recently, organoids have been successfully applied to understanding the pathogenesis of the infectious respiratory disease COVID-19, with SARS-CoV-2 viral infection and replication being achieved in a range of stem cell-derived tissues86–90. Driven by the occurrence of AKI in COVID-19 patients91–94, a handful of studies have explored kidney organoids as a potential model of COVID-1995,96. While it is still debated whether kidney damage results from direct viral infection or a combination of inflammatory responses and drug nephrotoxicity (reviewed in ref. 97, human PTs show high expression of the key SARS-CoV-2 receptor ACE255,98 and evidence of viral infection99–105.
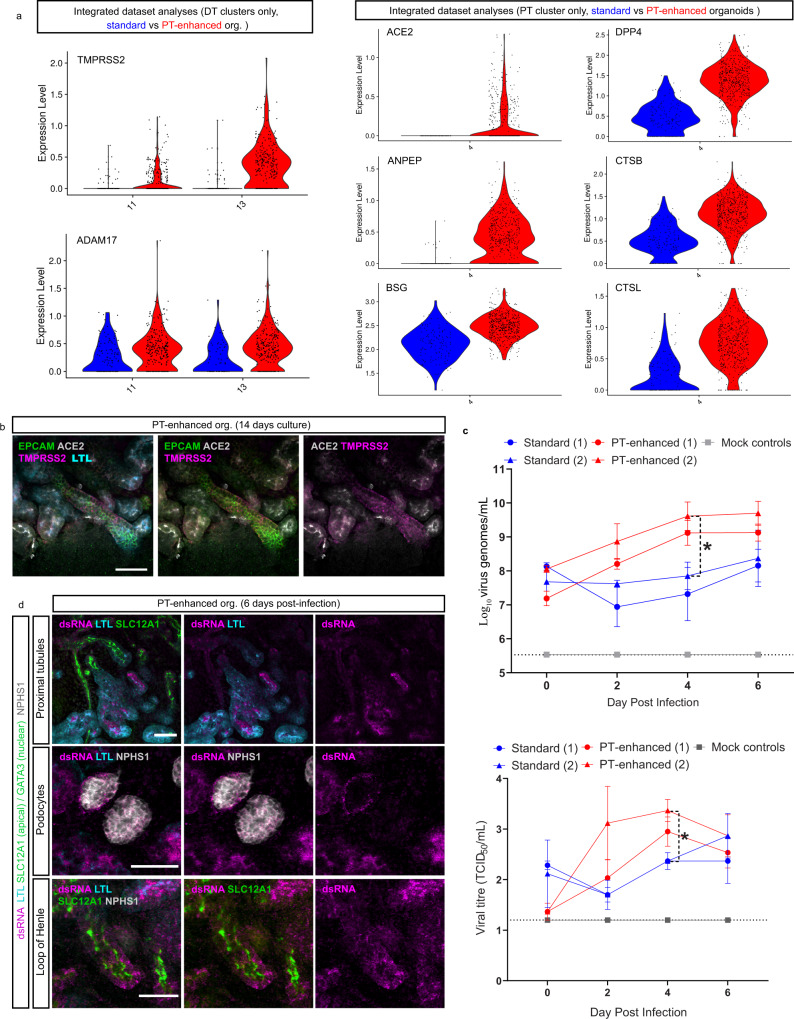
Given the high proportion of PT in enhanced organoids, we investigated their suitability as a model of SARS-CoV-2 infection and pathogenesis. Comprehensive analysis of scRNAseq data from >15,800 D13 + 14 organoid cells revealed expression levels and cellular localisation of a range of entry factors (receptors, proteases and binding proteins) previously implicated in SARS-CoV-2 infection106,107 predominantly across proximal, distal, and endothelial clusters (Supplementary Fig. 5A). DevKidCC was used to broadly compare viral entry factor expression within the PT-enhanced D13 + 14 (D13p14) dataset with that of relevant published kidney organoid datasets and human fetal kidney (Supplementary 5B). This revealed higher and/or more abundant expression of several SARS-CoV-2 entry factors in the D13 + 14 dataset, most strikingly in the PT population where the gene signature of PT-enhanced organoids more closely matched that of the human fetal kidney reference dataset (Supplementary Fig. 5B). To minimise the impacts of experimental variation, entry factor gene expression was compared in line- and age-matched standard and PT-enhanced organoid datasets following isolation of the relevant clusters according to entry factor expression patterns, as depicted in Supplementary Fig. 5A (Fig. 7a). SARS-CoV-2 entry factors of the proximal and distal tubular segments showed increased expression levels and abundance in PT-enhanced organoids compared to our existing standard organoid dataset (Fig. 7a). The two most frequently reported viral entry factors in literature, ACE2/ACE2 and TMPRSS2/TMPRSS298, were confirmed to be expressed at both a gene- and protein-level in proximal and distal nephron compartments, respectively (Fig. 7a, b), supporting previous reports in vivo and in kidney organoids55,59,86,95,96.
Fig. 7. PT-enhanced organoids show improved SARS-CoV-2 entry factor expression, infectivity, and viral replication.
a scRNAseq analyses comparing the expression of SARS-CoV-2 entry factors in PT-enhanced organoids (red) and our existing standard organoid dataset (line- and age-matched)19. Violin plots compare expression of genes within integrated datasets from which distal tubule (DT [left panel]) and proximal tubule (PT [right panel]) clusters have been isolated according to the biodistribution of the entry factors. b Confocal immunofluorescence of ACE2 (grey) and TMPRSS2 (magenta) demonstrating protein localisation in PT-enhanced kidney organoids. Nephron epithelium and proximal tubules are stained with EPCAM (green) and LTL (cyan), respectively. Scale bars represent 50 µm. c Line plots depict representative experiments, (1) and (2), replicated using the same iPSC line and culture conditions, for both PT-enhanced (red lines) and standard (blue lines) conditions. Mock-infected (grey) line is representative of all mock results across the same 4 independent experiments. Dotted line represents lower limit of detection (LOD). Error bars represent SEM from n = 3 individual wells of organoids (3 organoids per well) at each timepoint (individual values depicted in Supplementary Fig. 7D). Top plot displays qRT-PCR for SARS-CoV-2 viral envelope (E) gene (genome copies per mL) in the same culture media samples as depicted in bottom plot. Statistical significance was determined using a one-way ANOVA with Tukey’s multiple comparisons test. Asterisk (*) represents two-tailed P value ≤ 0.05 for PT-enhanced (2) vs. Standard (2) (P = 0.0297). Bottom plot shows viral titre as determined by Vero cell assays (Median Tissue Culture Infectious Dose; TCID50) of culture media sampled from SARS-CoV-2 infected standard and PT-enhanced organoids, as well as mock-infected organoids. Statistical significance was determined using a one-way ANOVA with Tukey’s multiple comparisons test. Asterisk (*) represents two-tailed P value ≤ 0.05 for PT-enhanced (2) vs. Standard (1) and PT-enhanced vs. Standard (2) (P = 0.0457). Source data are provided as a Source data file. d Confocal immunofluorescence of PT-enhanced organoids 6 days post-infection indicating viral dsRNA (magenta) localisation, co-stained for PTs (LTL; cyan), Loop of Henle (SLC12A1; apical green), and podocytes (NPHS1; grey). Scale bars represent 50 µm.
Apical ACE2 expression was also identified in epithelial cells lining the initial portion of Bowman’s capsule transitioning from the S1 segment of the PT (Supplementary Fig. 6A). Previous studies in mice have identified these transitionary cells as cuboidal and intermediate parietal epithelial cells (cuPECs and iPECs), making up the most proximal part of the PT prior to transitioning to flat PECs that line Bowmans’s capsule108,109. Accordingly, high ACE2 gene expression correlated with a subset of cells co-expressing general PEC markers with a cuPEC/iPEC-specific profile (PAX8 + , AKAP12+, PROM1−) (Supplementary Fig. 6B). This region also partly coincided with the SLC34A1Hi/HNF4A+/SLC36A2+ population marking early (S1) PT cells58,110 (Supplementary Fig. 6C), which, along with LTL-positivity of the early Bowmans capsule epithelium (Supplementary Fig. 6A), agreed with the known S1-PEC transitionary phenotype reported for cPECs and iPECs108. However, ACE2 was absent from podocytes (cluster 12; Supplementary Figs. 5A and 6B, C). These expression patterns were further supported by analyses of human fetal kidney, with expression of SARS-CoV-2 entry factors exhibiting a highly similar expression pattern to our extended kidney organoids, including low levels of ACE2 in human fetal kidney PECs (Supplementary Fig. 7A–C).
Having confirmed the expression of viral entry factors, PT-enhanced and standard organoids were assessed for infectivity following incubation with SARS-CoV-2. Viral infection of kidney organoids was confirmed by visualisation of GFP-expressing SARS-CoV-2 reporter virus (marking replicating virus)111 in combination with immunofluorescence staining for the spike protein (S; the transmembrane protein responsible for host cell binding and viral entry) (Supplementary Fig. 6D). To confirm the presence of viral genome, culture media from standard and PT-enhanced organoids were harvested every second day post-infection for qRT-PCR of SARS-CoV-2 viral envelope gene expression (E; genome copies per mL) (Fig. 7c and Supplementary Fig. 7D [left panels in both]) and virus titration in Vero cells to calculate median Tissue Culture Infectious Dose (TCID50) (Fig. 7c and Supplementary Fig. 7D [right panels in both]). Infectious virus was detected earlier in PT-enhanced compared to standard organoids (at 2 days post-infection) across independent experiments replicated using the same iPSC line and organoid conditions. In both instances, infectious virus levels reached significance at 4 days post-infection (P = 0.0297 and P = 0.0457, respectively) (Fig. 7c and Supplementary Fig. 7D).
To determine the kidney cell types targeted by SARS-CoV-2 in PT-enhanced organoids, infected organoids were analysed via immunofluorescence for double-stranded RNA (dsRNA) and nephron-specific markers 6 days post-infection (Fig. 7d). In agreement with scRNAseq analyses of ACE2 receptor expression (Supplementary Fig. 5A, B, Supplementary Fig. 6B, and Supplementary Fig. 7A–C), infected organoids showed dsRNA predominantly in LTL-positive PTs, as well as Bowman’s capsule surrounding NPHS1-positive podocytes (undetectable in podocytes themselves) and some detection in SLC12A1-positive Loops of Henle (Fig. 7d). The specificity of this staining was confirmed by immunofluorescence of uninfected control organoids, which showed no staining for dsRNA (Supplementary Fig. 7E). Despite their infection, tubular epithelium in organoids exposed to SARS-CoV-2 retained key characteristics such as apically-restricted LTL and SLC12A1, as well as membrane-bound EPCAM staining (Fig. 7d, Supplementary Fig. 7F [top panel]). However, upregulation of KIM-1 was observed in infected organoids and found to be significantly higher than mock (uninfected) control organoids at a gene level, complementing results of previous publications (Supplementary Fig. 7F)112,113.
Discussion
The utility of human PSC-derived kidney organoids as accurate models for disease research applications will rely upon their nephron maturation and functionality. To date, proximal tubules characterised within kidney organoids have lacked significant evidence of functional solute transport. In this study, we have shown that prolonged maintenance and delayed epithelialisation of the nephron progenitor population improved PT maturation and functionality compared to standard organoid protocols. Critically, this approach promoted development of distinct S1, S2 and S3 cell populations within the PT, a feature not previously identified in a kidney organoid. The application of DevKidCC in the current study enabled an unbiased and quantitative transcriptional comparison to previous published kidney organoid and human fetal kidney datasets, providing a reliable readout of cell identity and maturation and minimising the caveats associated with comparing restricted marker panels13.
Treatment strategies for coronavirus infections, including SARS-CoV and MERS-CoV, are still in their infancy with progress reliant upon an improved understanding of virus biology and interaction with host factors114. Despite the rapid accumulation of information on SARS-CoV-2, findings have often been conflicting or challenging to interpret, including reported heterogeneity in the expression of viral entry factors and the correlation between expression levels and disease outcome115–117. The utility of kidney organoids to study such aspects of infection has been illustrated by recent studies, including the demonstration of reduced infectivity following blocking of the ACE2 receptor95,96. PT-enhanced organoids show clear apical ACE2 protein staining of the proximal tubules via immunofluorescence, together with ACE2 expression within the PT cell clusters. Previous reports have shown ACE2 protein in organoids derived from separate protocols96,113 and ACE2/SARS-CoV-2 interaction was previously detected in the cell membrane protein fraction extracted from kidney organoids96,113. Compared to line- and age-matched standard organoids, PT-enhanced organoids exhibited improved expression of a range of previously identified viral entry factors compared to standard organoids, translating to superior infectivity. Along with their robust response to the nephrotoxic chemotherapeutic cisplatin, these findings underscore the advantage of organoids with enhanced PT patterning and functionality for disease modelling and drug screening.
Despite the clear demonstration of apical ACE2 protein expression in PT-enhanced organoids, ACE2 gene expression levels were lower than expected for both standard and enhanced organoid conditions. While it is known that protein levels are imperfectly correlated with mRNA abundance, the impact of “drop-out” effects arising from lowly expressed genes, transcriptional bursting, lower capture rates of certain cell types, or inadequate sequencing depth, cannot be discounted and remains a consideration when applying scRNAseq approaches118. Regardless, when compared more broadly to other organoid protocols, PT-enhanced organoids displayed wider expression of a range of entry factors that better emulated human fetal kidney in the PT population. These entry factor expression levels translated to higher virus replication levels in PT-enhanced organoids, determined by both dsRNA quantification and infectious viral genome copies across multiple timepoints, replicates, and independent experiments. Previous kidney organoid studies have reported podocyte SARS-CoV-2 infection using stem cell-derived kidney models113,119. In contrast, we saw limited viral entry factor expression and no evidence of ACE2/ACE2 within podocytes of PT-enhanced organoids and human fetal kidney. It is possible that reports of podocyte infection reflected viral entry in more immature podocytes or parietal cells, given the reported variation in genuine podocyte gene expression arising from the use of different cellular models/formats81,119. In addition, while previous transcriptional profiling of infected organoids claimed the presence of virus within most cell populations113, no viral entry factor expression was observed in any cell cluster within that study. Here again we conclude that PT-enhanced organoids represent a more accurate model of the mature nephron.
It remains to be seen whether the enhanced PT development in these organoids results from improved nephron progenitor expansion or sufficient time to form a more metanephric nephron progenitor population. Transcriptional profiling of day 13 monolayers exposed to CDBLY2 showed a high proportion of nephron progenitors with a significant increase in nephron progenitor gene expression (SIX1, LYPD1) and metanephric HOX ortholog expression (HOX11A/C/D) in comparison to other relevant published scRNAseq datasets. One unique feature critical to the overall outcome of this modified protocol included the addition of nephron progenitor maintenance media that prolongs low-level canonical WNT signalling (CHIR), suppresses NOTCH signalling (DAPT), and increases BMP7 activity (BMP7)25. Inclusion of these factors agreed with mouse studies which have shown a requirement for Notch to initiate nephron progenitor commitment and nephron formation, as well as demonstration that Notch2 supports proximal nephron patterning120,121. In addition, low levels of canonical Wnt activity and Bmp/BMP signalling via MAPK and PI3K pathways have been proposed to support nephron progenitor survival22,122–126. Despite containing both low CHIR and BMP7, the alternate nephron progenitor maintenance media NPSR was unable to support subsequent nephron formation in the resulting organoids, possibly due to the inclusion of BMP and TGFβ receptor inhibitors (dual inhibition of SMAD1/5/8 and SMAD2/3)23, which may maintain a less competent nephron progenitor population127.
The influence of timing on protocol outcome also cannot be discounted. Recent studies of the relative timing of PSC differentiation suggest that development and maturation in vitro is influenced by a predetermined species-specific biological clock. This has been elegantly demonstrated by Matsuda et al., showing that the markedly different paces of differentiation exhibited by mouse and human PSCs can be attributed to biochemical rate variations that influence the segmentation clock128. Indeed, brain organoids require months in culture to develop specific neural subtypes, akin to human gestation129,130. While our PT-enhanced kidney organoid protocol already shows considerable improvements in maturation after only 3–4 weeks, there is likely room for additional improvements including the timing of growth factor exposure and optimisation of metabolic conditions beyond the monolayer differentiation phase.
Despite enhancing PT development, this protocol faces some limitations with respect to nephron patterning and off-target populations. While providing a powerful model of PT function, reduced patterning to distal tubular segments highlights the challenge of simultaneously generating all kidney cell types in a single protocol, as previously described in mouse (refs. 3, 4, 7, 131). In addition, the formation of pre-cartilage cells is problematic for any potential clinical application, albeit not unique to this approach. Cartilage development has been observed in organoids from several protocols following transplantation132–134. In PT-enhanced organoids, this may represent a side-effect of prolonged BMP signalling that could potentially be suppressed through timed SMAD1/5/8 inhibition. The presence of central pre-cartilage within the cortical stroma population of the organoid core resulted in strong central WNT antagonism (SFRP2) that contributed to the striking nephron alignment observed. The establishment of a sink and source of WNT activity along the length of the tubule, driving nephron directionality, is in agreement with our current understanding of proximodistal patterning during mouse development26, while the cortical stroma population likely supports and promotes the proximal nephron development62. Interestingly, while standard organoids develop regions of cartilage post transplantation, they do not display this characteristic nephron spatial arrangement either before or after transplant. It is possible that this core is the result of altered biophysical parameters. We have previously shown that higher density standard organoids favour the development of a central unpatterned core, whereas a bioprinted sheet does not69. Such observations indicate that an interplay between cell deposition density and the patterning of the mesodermal population in the enhanced protocol facilitated the strong centralised source of WNT antagonism. Together this suggests an approach to further control the spatial organisation of bioengineered tissue through manipulation of signalling gradients.
In conclusion, we describe here a protocol that enabled improved patterning and maturation of proximal tubules within kidney organoids. These show significant advantages for modelling an appropriate damage response following drug-induced injury and SARS-CoV-2 infection, underscoring the utility of this approach as a platform to model a range of proximal tubular disease states.
Methods
iPSC lines and maintenance
Non-reporter iPSC lines used in this study include CRL1502.C32 (cell line derived from WS1 CRL-1502TM female fibroblasts from ATCC)6,135 and PB010/MCRIi010-A (derived from peripheral blood mononuclear cells of healthy adult individuals136). Reporter iPSC lines used in this study include CRL-2429/SIX2Cre/Cre:GAPDHdual and PCS-201-010/HNF4AYFP (derived from human foreskin fibroblasts [CCD-1112Sk/CRL-2429TM, ATCC] and primary dermal fibroblasts [PCS-201-101TM, ATCC], respectively, with simultaneous reprogramming and reporter line generation performed in-house. Reporter lines are available for distribution from the Washington University Kidney Translational Research Centre, St Louise, MO, accessible via the ReBuilding a Kidney Reporter Cell Line database [https://www.rebuildingakidney.org/chaise/recordset/#2/Cell_Line:Reporter_Cell_Line@sort(RID)] and identified as “SIX2:Cre/GAPDH:Dual” and “HNF4A:YFP”, respectively)20. All iPSC lines were maintained and expanded at 37 °C, 5% CO2 and 5% O2 in Essential 8 medium (Thermo Fisher Scientific, Waltham, MA) on Matrigel- (BioStrategy, Victoria, Australia) coated plates with daily media changes and passaged every 2–3 days at 80% confluency via 3 min incubation in EDTA in 1X PBS137.
Directed differentiation and kidney organoid generation
Monolayer differentiation
For standard protocol-derived organoids (manually made and bioprinted), differentiation of iPSC lines and organoid culture was performed as follows using published methodology19,29. The day prior to differentiation, iPSCs at day 2 post-passage were dissociated to near-single-cell suspensions via a 3 min incubation in TrypLE (Thermo Fisher Scientific) and seeded onto Laminin-521-coated (BioLamina, Sundbyberg, Sweden) 12-well plates before commencement of differentiation 24 h later via addition of 1 mL TeSR-E6 medium (StemCell Technologies) supplemented with CHIR99021 (R&D Systems). The concentration of Laminin-521 used to coat 12-well plates, initial iPSC seeding density within 12-well plates, and CHIR99021 concentration and duration of exposure varied according to the iPSC line used (CRL1502.C32, CRL-2429/SIX2Cre/Cre:GAPDHdual and PB010/MCRIi010-A were seeded at 25,000 cells/well and exposed to 6 µM CHIR for 5 days; PCS-201-010/HNF4AYFP was seeded at 40,000 cells/well and exposed to 6 µM CHIR for 4 days; CRL1502.C32, CRL-2429/SIX2Cre/Cre:GAPDHdual were seeded with 20 µL/mL Laminin-521; PB010/MCRIi010-A and PCS-201-010/HNF4AYFP were seeded with 40 µL/mL Laminin-521). CHIR medium was refreshed every second day. Following this initial CHIR exposure, monolayers were exposed to 1 mL/well of TeSR-E6 medium supplemented with 200 ng/mL FGF9 (R&D Systems) and 1 µg/mL Heparin (Sigma Aldrich, St Louise, MO), with medium refreshed every second day until day 7 of differentiation.
For extended monolayer differentiation of iPSCs prior PT-enhanced organoid generation, Laminin-521 concentrations, iPSC seeding density, plate formats, and cell handling conditions were as stated for standard conditions above. However, iPSCs were subjected to prolonged (5 day) initial monolayer differentiation in TeSR-E6 supplemented with 6 µM CHIR, refreshed every second day. At day 5 of monolayer differentiation, cells were exposed to 200 ng/mL FGF9 and 1 µg/mL Heparin until day 8, refreshing the medium every second day. At day 8, the monolayer was exposed to 1 mL/well nephron progenitor maintenance medium, NPSR or CDBLY23,25, refreshing these media daily. Final PT-enhanced organoid conditions utilised CDBLY2, containing 2X concentration of BMP7. PT-enhanced organoid generation proceeded at day 10–14 of monolayer differentiation.
Organoid generation
Standard organoids (manually generated and bioprinted) and PT-enhanced organoids (manually generated only) were made at days 7 or 10–14 of monolayer differentiation, respectively. Cell monolayers were dissociated to single-cell suspensions via 3 min incubation in 1 mL/well of TrypLE. After neutralisation using 5 mL of TeSR-E6 containing 2% FBS (Hyclone, Logan, UT), cell suspensions were centrifuged (3 min at 1200 rpm), resuspended in 2 mL of TeSR-E6/2% FBS, and cell number counted. For manually generated organoids, 250,000 cells were aliquoted into a single 1.5 mL Eppendorf for each organoid to be generated and the total volume reconstituted to 200 µL per tube with TeSR-E6/2% FBS. Eppendorfs were centrifuged 3 times (3 min each at 1800rpm), rotating the tubes 180° between spins. Using a wide-bore 200 µL pipette tip, organoids were transferred to 0.4 µm Transwell polyester membranes in 6-well plates (Corning Incorporated, Corning, NY). For standard organoids bioprinted as patches, cell pellets from dissociated and centrifuged monolayers were resuspended to a density of 200,000 cells/µL for 3D extrusion-based bioprinting onto Transwell membranes of 6-well plates using the NovoGen MMX bioprinter following published methodology69.
Following generation, standard and PT-enhanced organoids were exposed to 1 mL/well of TeSR-E6 supplemented with 5 µM CHIR for 1 h at 37 °C, 5% CO2 and 5% O2. After incubation, CHIR pulse medium was replaced with TeSR-E6 medium supplemented with 200 ng/mL FGF9 and 1 µg/mL Heparin, refreshing medium on day 3 post-organoid generation. Organoid medium was replaced with TeSR-E6 medium without growth factors on day 5 post-organoid generation and refreshed every second day until organoid harvest.
Immunofluorescence and confocal microscopy
For immunofluorescence, organoids were fixed and stained using the following published methodology20. Following removal from the Transwell membrane and transfer to 48-well plates, organoids were fixed in 4% paraformaldehyde (Sigma Aldrich) for 20 min on ice and washed for 15 min in 3 changes of PBS. Overnight blocking and staining incubations were performed at 4 °C on a rocking platform. Blocking solution consisted of 10% donkey serum and 0.3% Triton-X-100 diluted in PBS (Sigma Aldrich). Staining solutions consisted of the antibodies detailed in Table 1, diluted in 0.3% Triton-X-100/PBS. Primary antibodies were probed with Alexa Fluor-conjugated fluorescent secondary antibodies (Life Technologies) diluted 1:500. Organoids were washed in 3 changes of PBS for 3 h between primary and secondary antibody incubations, as well as after the final secondary antibody incubation. Imaging was performed on the ZEISS LSM 780 confocal microscope (Carl Zeiss, Oberkochen, Germany) with acquisition and processing performed using ZEISS ZEN Black software (version 2.3 SP1) (Zeiss Microscopy, Thornwood, NY) and Fiji ImageJ (version: 2.1.0/1.53c)138.
Table 1.
Antibodies used in immunofluorescence studies
| Specificity | Host species | Dilution range | Manufacturer and identifier |
|---|---|---|---|
| ACE2 | Rabbit polyclonal IgG | 1:300 | Abcam (ab15348) Lot GR3333640-R |
| CUBILIN (Y-20) | Goat polyclonal IgG | 1:300 | Santa Cruz Biotechnology (sc-20607) Lot J1012 |
| dsRNA | Mouse monoclonal IgG2a, Kappa | 1:300 | Absolute Antibody (Ab01299-2.0) clone J2, Lot T2007A26 |
| ECADHERIN | Mouse monoclonal IgG2a, Kappa | 1:300 | BD Biosciences (610181) clone 36 |
| EpCAM (Alexa488 or Alexa647 conjugate) | Mouse monoclonal IgG2a, Kappa | 1:300 | BioLegend (324210 and 324212) clone 9C4, Lot B352438 and B314235 |
| GATA3 | Goat polyclonal IgG or rabbit monoclonal IgG | 1:300 | R&D Systems (AF2605) and Cell Signalling Technology (5852S clone D13C9. Lot UZQ0219011) |
| GFP | Chicken polyclonal IgY | 1:200–1:300 | Abcam (ab13970) Lot GR3361051-11 |
| HNF4A | Mouse monoclonal IgG2a | 1:300 | Invitrogen (MA1-199) clone K9218, Lot WI3366551 |
| KIM-1 | Goat polyclonal IgG | 1:300 | R&D Systems (AF1750), Lot JTB0821051 |
| mCherry (RFP) | Rabbit polyclonal IgG | 1:300–1:400 | MBL Medical & Biological Laboratories Co. Ltd. (PM005), Lot 048 |
| MEGALIN | Rabbit polyclonal IgG | 1:300 | Novus Biologicals (NBP2-39033), Lot R91329 |
| NEPHRIN | Sheep polyclonal IgG | 1:300 | R&D Systems (AF4269), Lot ZMU0221031 |
| Proximal tubule brush border membrane | Lotus tetragonobulus lectin (LTL), biotinylated | 1:300–1:500 | Vector Laboratories (B-1325-2) |
| TMPRSS2 | Mouse monoclonal IgG1, Kappa | 1:300 | Merck (MABF2158) clone P5H9-A3, Lot 351024 |
| S2 subunit of SARS-CoV-2 spike protein (stain: Sin2774) | Rabbit monoclonal IgG1 | 1:300 | GeneTex (GTX632604) clone HL1002, Lot 44405 |
| SLC6A19 | Chicken | 1:100–1:200 | Aves Laboratories (custom antibody) |
| SLC12A1 | Rabbit polyclonal IgG | 1:300–1:400 | Proteintech (18970-1-AP), Lot 00045810 |
Flow cytometry
Flow cytometry of reporter line-derived organoids using endogenous fluorescence was performed and analysed following published methodology and as follows20. Following Accutase (StemCell Technologies) dissociation of the organoids at 37 °C (1 mL Accutase per 6 organoids for 15 min, pipetting every 3 min), the enzymes were inactivated with TeSR-E6/2% FBS and by placing the cell suspension on ice for 2 min. Dissociated organoid cells were passed through 40 µm and 70 µm cell strainers with additional TeSR-E6/2% FBS prior to centrifugation (1500 rpm for 3 min) and resuspension in 100–500 µL PBS containing 1% FBS (FACS wash). To determine the contribution of SIX2-mCherry + cells to EPCAM + populations in organoids derived from the SIX2Cre lineage tracing iPSC line, cells were stained using directly conjugated anti-EPCAM Alexa Fluor-647 antibody (see Table 1) diluted 1:100 in 100 µL of FACS wash for every 5 × 105 cells. Following 30 min incubation on ice, stained cells were washed 3 times in 2 mL FACS wash via centrifugation prior to flow cytometry. Flow cytometry was performed using the BD LSR Fortessa X-20 Cell Analyzer (BD Biosciences, CA) with analyses performed using FACsDiva versions 8.0.1 and 9.0.1 (BD Biosciences) and FlowLogic software version 8.6 (Inivai).
Histology
For Alcian Blue detection of cartilage, organoids were fixed in 4% PFA as described above and processed for routine paraffin embedding using the Excelsior AS Tissue Processor (rapid biopsy setting; Thermo Fisher Scientific). Samples were embedded in wax and 5 µm sections cut using a Waterfall HM325 microtome (Thermo Fisher Scientific). Sections were dewaxed, hydrated through graded alcohols to running water, then covered with Alcian Blue Solution (1% Alcian blue in 3% acetic acid, pH 2.5). After 10 min, sections were washed in tap water for 2 min and counterstained for 7 min in Nuclear Fast Red stain (0.1% Nuclear Fast Red [Sigma Aldrich] and 5% ammonium potassium sulfate in water). Following staining, sections were dehydrated in graded alcohols, cleared in Safsolvent (Bacto Laboratories, NSW, Australia), and coverslipped. Images were acquired on a Zeiss Axio Imager A2 with Zeiss Zen software (Zeiss Microscopy, Thornwood, NY).
Real-time quantitative reverse transcription PCR (qRT-PCR)
RNA extraction, cDNA synthesis and quantitative RT-PCR (qRT-PCR) were performed using the Bioline Isolate II Mini/Micro RNA Extraction Kit, SensiFAST cDNA Synthesis Kit and the SensiFAST SYBR Lo-ROX Kit (Bioline, NSW, Australia), respectively, as per manufacturer’s instructions. Each qRT-PCR reaction was performed in triplicate using the primer pairs detailed in Table 2. RT-PCR data were collected using the Applied Biosystems 7500 Sequence Detection Software (version 1.5.1) installed on the Applied Biosystems 7500 Real Time PCR System. Data were graphed and analysed in Prism 8 (GraphPad).
Table 2.
Forward and reverse primers used for qRT-PCR
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| DAPL1 | CTCGGAAAGGGGGACATCCT | AGTTGAGCTTCTCCAGTGCG |
| GAPDH | CTCTCTGCTCCTCCTGTTCGA | TGAGCGATGTGGCTCGGCT |
| GATA3 | GCCCCTCATTAAGCCCAAG | TTGTGGTGGTCTGACAGTTCG |
| HAVCR1 | GTTCCTCCAATGCCTTTGCC | CGGTGTCATTCCCATCTGTTG |
| HNF4A | ACCCTCGTCGACATGGACA | GCCTTCTGATGGGGACGTG |
| HOXD11 | GCCAGTGTGCTGTCGTTCCC | CTTCCTACAGACCCCGCCGT |
| LHX1 | CGTCATTCAGGTCTGGTTCC | CCCGTAGTACTCGCTCTGGT |
| PARAXIS | GGGGGTGGCCGTCGT | CAGGCTGAATGGATCCTCAC |
| SIX1 | AAAGGGAAGGAGAACAAGGATAG | GGAGCCTACATGATTACTGGG |
| SIX2 | TCCTGGTCCCTCCGTATGTA | TAGGGGCAGATAGACCACCA |
| TMEM-100 | CAGGCGTTGCTGTTTCTTGT | CAGGGTGAAAGCTCGGAGAG |
| WNT4 | AACTGCTCCACACTCGACTC | TGACCACTGGAAGCCCTGT |
Gene names are italicised.
Single-cell RNA sequencing (scRNAseq) and dataset generation
The D13 + 12 dataset was generated using the CRL-2429/SIX2Cre/Cre:GAPDHdual iPSC line. The D13 and D13 + 14 organoids were generated using the CRL1502.C32 with four replicates per timepoint, where each replicate was derived from an independent well. Cells were dissociated using Accutase as described above and following published methodology69. For the D13 and D13 + 14 samples, replicates were multiplexed following the method of Soeckius et al.139. Cells were stained for 20 min on ice with 1 µg (1:50 dilution) of BioLegend TotalSeq-A anti-human hashtag oligo antibody (BioLegend TotalSeq-A0251 to A0258). Cells were washed 3 times then pooled at equal ratios for sequencing. A single library was generated for each suspension/condition, composed of equally sized pools of each replicate (Set 1–4). Libraries were generated following the standard 10x Chromium Next GEM Single Cell 3ʹ Reagent Kits v3.1 protocol except that superloading of the 10x device was performed with ~30k cells. Hash tag oligo (HTO) libraries were generated following the BioLegend manufacturer protocol. Single-cell RNA sequencing data were collected with NovaSeq Control Software for the Illumina NovaSeq 6000 sequencing system.
10x mRNA libraries were demultiplexed using CellRanger (version 3.1.0) to generate matrices of UMI counts per cell. HTO libraries were demultiplexed using Cite-seq-count (1.4.3) to generate matrices of HTO counts per cell barcode. All data were loaded into Seurat (3.1.4) and HTO libraries were matched to mRNA libraries. Seurat was used to normalise HTO counts and determine cut-offs to assign HTO identity per cell using the HTODemux function with the positive.quantile parameter set at 0.99. HTO doublet and unassigned cells were removed, as were cells with mitochondrial content greater than 35% accounting for the increased metabolic activity of renal epithelium140, number of genes per cell greater than 500 and the number of UMIs less than 100,000, to obtain filtered datasets (D13 replicates: 3694 cells [A0251], 3545 cells [A0252], 3785 cells [A0253], 3641 cells [A0254]; D13 + 14 replicates: 3415 cells [A0255], 2350 cells [A0256], 2904 cells [A0257], 2578 cells [A0258]). The combined datasets contained a median of 3915 genes expressed per cell, with a median of 16352 UMI counts per cell.
Analysis of scRNAseq datasets
Data was normalised using the SCTransform method141 including the regression of cell cycle scores. A 30 component Principal Component Analysis (PCA) was performed, followed by Uniform Manifold Approximation and Projection (UMAP) using these PCA components. Seurat’s graph-based clustering approach was used to identify, with resolutions of 0.7 (D13) and 0.5 (D13 + 14) chosen for downstream analysis. Marker analysis was performed using the Seurat FindMarkers function, using student’s t-test, limited to positive markers (i.e. increased expression within a cluster) above 0.25 log fold-change expressed in at least 10% of cells within a cluster. Marker lists were exported and cluster identities were determined by comparison with published human single-cell data19 or Gene ontology analysis using ToppFun (https://toppgene.cchmc.org/enrichment.jsp; version 2021-Mar-29 15:59/#31/639dfe36cbad55ba69e8255099537d5c2fcb9483). The PT cluster was isolated and re-analysed as above to further investigate any subpopulations.
The D13 + 12 dataset was integrated with an age- and line-matched published dataset19 using the anchor-based method within Seurat142,143. This integrated dataset was analysed as above, isolating the PT cluster and comparing gene expression of cells from both samples within this population.
For DevKidCC analyses, The D13 and D13p14 samples were analysed using DevKidCC (version 0.0.3); a hierarchical set of machine-learning binary classifiers trained on a human fetal kidney reference dataset. The classified dataset was then compared to relevant existing single-cell organoid datasets using the DotPlotCompare function.
For Azimuth analyses, cells were uploaded to the online Azimuth portal at https://app.azimuth.hubmapconsortium.org/app/human-fetus and instructions were followed as per the website for the analysis.
Agarose bead-mediated morphogen signalling assay
Standard organoids bioprinted as patches were generated and cultured as described above prior to the addition of morphogen-soaked beads at 5 days post-organoid generation (D7 + 5)69. The day before bead addition, 100 µL of Affi-Gel Blue Gel 100–200 mesh crosslinked agarose beads (Bio-Rad Laboratories, Hercules, CA), were washed 3 times in PBS via centrifugation. Washed beads were resuspended in 100 µL of PBS (control) or 10 µM IWR-1 (stock reconstituted according to manufacturer’s instructions; Sigma Aldrich) and incubated for 1 h at room temperature prior to overnight storage at 4 °C. On day 7 + 5, suspensions were agitated to resuspend beads and 0.3 µL was added to the centre of each patch organoid with the aid of a P2 pipette and dissecting microscope (Leica Microsystems, Wetzlar, Germany). Organoid media (TeSR-E6 [STEMCELL Technologies, Vancouver Canada]) was refreshed every second day prior to harvest at D7 + 9 for immunofluorescence.
Quantification of tissue patterning changes in response to IWR-1 soaked beads
Tissue patterning within the radius of beads was quantified using custom Python (version 3.10.2) scripts, with method as follows. Images (n = 3 per condition, IWR soaked and control) were loaded as Numpy (version 1.22.1)144 arrays using the Czifile library (version 2019.7.2) and masks of bead location were generated by manually segmenting each bead using the Napari (version 0.4.13) labels layer feature. Nephron segments were segmented by applying a gaussian filter to each channel (sigma of 5 pixels) followed by Otsu thresholding (for NPHS1 staining) or multi-otsu thresholding (for LTL, EPCAM) using the second threshold value. All processing was implemented using functions in Scikit-image (version 0.19.2)145. The distance of each pixel in the image from the bead edge was calculated using the Euclidian distance transform in Scipy (version 1.7.3)146. These values were used to define the total region within 200 pixels of the bead surface, including the beads themselves. The percentage of pixels assigned to each nephron marker as a proportion of total nephron tissue (defined by the total pixels that were segmented as NPHS1 or EPCAM positive), within the 200 pixel region of each image was then calculated. Scipy was used to conduct t-tests, Matplotlib (version 3.5.1) was used to generate plots and Napari was used to generate composite images.
Cisplatin toxicity assay
D13 + 14 PT-enhanced organoids were exposed through the basolateral compartment of the Transwell tissue culture plate (Corning Incorporated) to 1 mL per well of 20 µM Cisplatin (Accord Healthcare, Durham, NC), or an equivalent volume of PBS, in TeSR-E6 for 24 h (37 °C, 5% CO2 and 5% O2). Following incubation, organoids within Transwells were washed with PBS and harvested for flow cytometry as described above.
Fluorescent substrate uptake assays
For albumin uptake assays, D13 + 14 enhanced and D7 + 14 standard organoids (triplicate wells per condition) were incubated in TeSR-E6 containing 1:1000 TRITC albumin (10 mg/mL stock, Sigma Aldrich) and 1:500 anti-MEGALIN/LRP2 (pre-incubated with neat alpaca Nano-secondary Alexa Fluor 647 secondary antibody [Proteintech, Rosemont, IL]) via the basolateral compartment of the Transwell. Control organoids were incubated in secondary antibody alone. After overnight incubation (37 °C, 5% CO2 and 5% O2), plates containing organoids were washed in at least 3 changes of Hanks’ Balanced Salt Solution (HBSS; Thermo Fisher Scientific) for 30 min and live-imaged immediately using a ZEISS LSM 780 confocal microscope. For organic cation transport assays, D13 + 14 PT-enhanced and D7 + 14 standard organoids (triplicate wells per condition) were incubated in 4’,6-diamindino-2-phenylindole substrate (DAPI; 1:1000 [Thermo Fisher Scientific]) with 1:500 DRAQ7 dead cell label (Thermo Fisher Scientific) diluted in TeSR-E6 for 1 h (37 °C, 5% CO2 and 5% O2). Control organoids were pre-incubated for 15 min in 100 µM Cimetidine inhibitor (Sigma Aldrich) prior to incubation for 1 h in TeSR-E6 containing both inhibitor, substrate, and dead cell label (1:1000 DAPI, 1:500 DRAQ7, 100 µM Cimetidine). Following incubation, substrate and substrate + inhibitor solutions were replaced with HBSS and live-imaged immediately using a ZEISS LSM 780 confocal microscope.
Viral infection assays
Standard and PT-enhanced organoids grown on Transwells were infected with 104 tissue-culture infectious dose 50 (TCID50) of SARS-CoV-2 (Australia/VIC01/2020) in TeSR-E6 media added above the Transwell for 3 h (virus titration experiments) or below the Transwell with a drop ontop of the organoid for 1 h (virus localisation experiments). Following incubation (37 °C and 5% CO2), the viral inoculum was removed and replaced with 1 mL of plain TeSR-E6 medium beneath the Transwell as for typical organoid culture29. Culture medium was collected on days 0, 2, 4 and 6 post-infection for viral titer quantification and replaced with fresh medium. Median TCID50 in supernatants were determined, as detailed below, by 10-fold serial dilution in Vero cells (#CCL-81, ATCC) and calculated using the Reed and Muench method. Organoids were harvested at 6 days post-infection and fixed with 4% PFA fixation for immunofluorescence.
Infectious virus titration (median tissue culture infectious dose assay; TCID50)
Viral titrations were performed on confluent monolayers of Vero cells in 96-well plates. Wells were washed with plain minimum essential media (MEM) and replaced with 180 µl of infection media (MEM, 50 U/ml Penicillin, 50 µg/ml Streptomycin, 2 mM GlutaMax, 15 mM HEPES and 1 µg/ml TPCK-treated Trypsin). 20 µl of the samples to be titred were added to four wells and 10-fold serial dilutions were made. Plates were incubated at 37 °C and 5% CO2. Four days post-infection, SARS-CoV-2-induced cytopathic effect was assessed by microscopy.
RT-qPCR for SARS-CoV-2 genome
RNA was extracted from supernatant culture media using the QIAamp 96 Virus QIAcube HT Kit (Qiagen). E-gene expression was determined using the SensiFAST Probe No-Rox One Step Kit (Bioline) and the following primers/probes: Fwd: 5′-ACAGGTACGTTAATAGTTAATAGCGT-3′, Rev: ATATTGCAGCAGTACGCACACA and Probe: FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ. Viral genome copies were interpolated using a standard curve generated by using a plasmid vector containing the E-gene.
Statistics and reproducibility
The results depicted in this manuscript are representative of the similar observations and analyses made across multiple independent experiments, biological replicates, and technical replicates. Sample sizes were based on even group distribution and no statistical method was used to predetermine sample size owing to the exploratory nature of the study. Each experiment included a minimum of 3 biological replicates per condition across multiple experiments. Biological replicates were classed as monolayer differentiations or organoids derived from separate wells. Assortment of well-formed organoids of equivalent morphology into experimental groups was randomized for all experiments. Investigators were not blinded to allocation during analyses and outcome assessment owing to the automated processes used to collect and analyse results. Minimal data were excluded from the study. Organoids that failed to form kidney structures were omitted from downstream analyses without affecting sample size. For scRNASeq studies, cells were excluded based on poor QC scores and barcode readouts that indicate empty cells or multiplets. For qRT-PCR experiments, a technical replicate or sample was excluded if amplification failed. For SARS-CoV-2 infection experiments, one experimental replicate was excluded from downstream analyses due to a failure to infect.
To ensure robustness and reproducibility, experiments were replicated multiple times as either stated in the Figure Legend or as follows. Differentiation condition comparisons (Fig. 1b, c, Supplementary Fig. 1C, D) using CDBLY and standard E6 conditions were replicated in more than 5 independent experiments, including NPSR and CDBLY modifications (exposure and composition) in 2 independent experiments, with between 3 and 8 organoids generated per condition (depending on monolayer cell counts). Successful generation of organoids from multiple iPSC lines using the extended differentiation protocol (Supplementary Fig. 1F) was replicated in more than 5 independent experiments, with more variability in morphology notable in PCS-201-010/HNF4αYFP. Lineage tracing of SIX2-mCherry cells in PT-enhanced organoids (Fig. 2a, b) was performed across 4 independent experiments of 3 or 4 biological replicates. Observations of radially aligned and proximilised nephrons in PT-enhanced organoids compared to standard organoids (Fig. 2c, d) have been made across more than 10 independent experiments. Improved PT marker protein expression in PT-enhanced organoids compared to standard organoids (Fig. 3) has been assessed and observed in 3 independent experiments, with stromal marker analyses (Supplementary Fig. 1A) performed in 3 organoids matched to one these same experiments. Alcian Blue cartilage staining (Supplementary Fig. 1B) was performed and similarly detected in 4 separate PT-enhanced organoids across 3 independent experiments. PT brush border membrane characteristics (Supplementary Fig. 2A) of PT-enhanced organoids compared to standard organoids were replicated in more than 3 independent experiments, with apical SLC6A19 expression observed in standard organoids derived from 1 out of 3 independent experiments. Fluorescent substrate uptake assays for PT-enhanced and standard organoids (Fig. 6a) included 3 individual wells of 3 organoids per substrate/control condition and uptake was assessed in 4 independent experiments (2 per organoid protocol) with identical results. IWR-1 soaked agarose beads (Supplementary Fig. 4A, B) were added to organoids in 4 independent experiments, with influences on organoid morphology observed in 3 of these experiments (1 independent experiment excluded from downstream analyses owing to failed organoid differentiation). SARS-CoV-2 entry factor detection via immunofluorescence (Fig. 7b, Supplementary Fig. 6A) has been replicated in 3 independent experiments. Viral protein detection (Fig. 7C, Supplementary Fig. 6D) has been performed in 3 independent experiments with variation in viral protein intensity and abundance owing to variable infection levels between individual organoids and organoid batches.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This research was supported by the National Health and Medical Research Council (GNT1156440 [M.H.L., J.M.V., K.S.T., S.B.W., K.L., E.G., M.S.]), the National Institutes of Health (UH3DK107344; M.H.L., J.M.V., S.E.H.) and the Victorian State Government (DJPR/COVID-19 [MHL, KS, J.M.V., S.M., J.N., R.R.]). M.H.L. is an NHMRC Senior Principal Research Fellow (GNT1136085). We acknowledge the Stafford Fox Medical Research Foundation MCRI genome editing facility for the generation of all pluripotent stem cell lines. We thank Maelle Le Moing and the Murdoch Children’s Research Institute Translational Genomics Unit for 10x single-cell and hash-tag oligo library preparation and sequencing, and bulk-RNAseq sequencing; Matthew Burton and the Murdoch Children’s Research Institute Microscopy Core; Professor John Rasko and Dr Charles Bailey for providing the SLC6A19 antibody; Dr Dad Abu-Bonsrah for providing the dsRNA antibody; Professor Ralph Baric for providing the GFP-tagged SARS-CoV-2.
Author contributions
J.M.V., M.H.L. and K.S. contributed to experimental design and planning. J.M.V., K.S.T., E.G., R.R., J.N., S.M., M.S. and S.E.H. performed experiments and developed reagents and methods. S.B.W. and J.M.V. performed bioinformatics analyses. K.L. performed image analyses. J.M.V., M.H.L. and S.B.W. contributed to manuscript preparation. J.M.V. and M.H.L. wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks Daniel Batlle and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Data availability
The transcriptional profiling datasets generated in this study have been deposited in GEO under accession code GSE184928. The raw data from scRNAseq and immunofluorescence image analyses have been deposited in the Github repository [https://github.com/KidneyRegeneration/Vanslambrouck2022]. Raw and processed data from qRT-PCR, TCID50 assays, and image analyses are provided in the Source data file. Source data are provided with this paper.
Code availability
Code for scRNAseq and image analyses are available through the Github repository (https://github.com/KidneyRegeneration/Vanslambrouck2022; 10.5281/zenodo.7021393)147.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sean B. Wilson, Ker Sin Tan.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-33623-z.
References
- 1.Zhuo JL, Li XC. Proximal nephron. Compr. Physiol. 2013;3:1079–1123. doi: 10.1002/cphy.c110061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirita Y, Wu H, Uchimura K, Wilson PC, Humphreys BD. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc. Natl Acad. Sci. USA. 2020;117:15874–15883. doi: 10.1073/pnas.2005477117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freedman BS, et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 2015;6:8715. doi: 10.1038/ncomms9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morizane R, et al. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 2015;33:1193–1200. doi: 10.1038/nbt.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taguchi A, Nishinakamura R. Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell. 2017;21:730–746 e736. doi: 10.1016/j.stem.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Takasato M, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- 7.Toyohara T, et al. Cell therapy using human induced pluripotent stem cell-derived renal progenitors ameliorates acute kidney injury in mice. Stem Cells Transl. Med. 2015;4:980–992. doi: 10.5966/sctm.2014-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Combes, A. N. et al. Single cell analysis of the developing mouse kidney provides deeper insight into marker gene expression and ligand-receptor crosstalk. Development146, 10.1242/dev.178673 (2019). [DOI] [PubMed]
- 9.Howden SE, et al. Plasticity of distal nephron epithelia from human kidney organoids enables the induction of ureteric tip and stalk. Cell Stem Cell. 2021;28:671–684.e676. doi: 10.1016/j.stem.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subramanian A, et al. Single cell census of human kidney organoids shows reproducibility and diminished off-target cells after transplantation. Nat. Commun. 2019;10:5462. doi: 10.1038/s41467-019-13382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu H, et al. Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell. 2018;23:869–881.e868. doi: 10.1016/j.stem.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marable SS, Chung E, Park JS. Hnf4a is required for the development of Cdh6-expressing progenitors into proximal tubules in the mouse kidney. J. Am. Soc. Nephrol. 2020;31:2543–2558. doi: 10.1681/ASN.2020020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson SB, et al. DevKidCC allows for robust classification and direct comparisons of kidney organoid datasets. Genome Med. 2022;14:19. doi: 10.1186/s13073-022-01023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dressler GR. Advances in early kidney specification, development and patterning. Development. 2009;136:3863–3874. doi: 10.1242/dev.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bakker BS, van den Hoff MJB, Vize PD, Oostra RJ. The pronephros; a fresh perspective. Integr. Comp. Biol. 2019;59:29–47. doi: 10.1093/icb/icz001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgas KM, Chiu HS, Lesieur E, Rumballe BA, Little MH. Expression of metanephric nephron-patterning genes in differentiating mesonephric tubules. Dev. Dyn. 2011;240:1600–1612. doi: 10.1002/dvdy.22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mugford JW, Sipila P, Kobayashi A, Behringer RR, McMahon AP. Hoxd11 specifies a program of metanephric kidney development within the intermediate mesoderm of the mouse embryo. Dev. Biol. 2008;319:396–405. doi: 10.1016/j.ydbio.2008.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiedemann K, Welling LW, Basto P. Structural and functional comparison of mesonephric and metanephric proximal tubules. Pediatr. Nephrol. 1987;1:297–305. doi: 10.1007/BF00849227. [DOI] [PubMed] [Google Scholar]
- 19.Howden, S. E., Vanslambrouck, J. M., Wilson, S. B., Tan, K. S. & Little, M. H. Reporter-based fate mapping in human kidney organoids confirms nephron lineage relationships and reveals synchronous nephron formation. EMBO Rep. 10.15252/embr.201847483 (2019). [DOI] [PMC free article] [PubMed]
- 20.Vanslambrouck JM, et al. A toolbox to characterize human induced pluripotent stem cell-derived kidney cell types and organoids. J. Am. Soc. Nephrol. 2019;30:1811–1823. doi: 10.1681/ASN.2019030303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Little MH, Combes AN. Kidney organoids: accurate models or fortunate accidents. Genes Dev. 2019;33:1319–1345. doi: 10.1101/gad.329573.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown AC, Muthukrishnan SD, Oxburgh L. A synthetic niche for nephron progenitor cells. Develop. Cell. 2015;34:229–241. doi: 10.1016/j.devcel.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, et al. 3D Culture supports long-term expansion of mouse and human nephrogenic progenitors. Cell Stem Cell. 2016;19:516–529. doi: 10.1016/j.stem.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanigawa S, Sharma N, Hall MD, Nishinakamura R, Perantoni AO. Preferential propagation of competent SIX2+ nephronic progenitors by LIF/ROCKi treatment of the metanephric mesenchyme. Stem Cell Rep. 2015;5:435–447. doi: 10.1016/j.stemcr.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanigawa S, Taguchi A, Sharma N, Perantoni AO, Nishinakamura R. Selective in vitro propagation of nephron progenitors derived from embryos and pluripotent stem cells. Cell Rep. 2016;15:801–813. doi: 10.1016/j.celrep.2016.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindstrom NO, et al. Integrated beta-catenin, BMP, PTEN, and Notch signalling patterns the nephron. Elife. 2015;3:e04000. doi: 10.7554/eLife.04000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takasato M, et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat. Cell Biol. 2014;16:118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- 28.England, A. R. et al. Identification and characterization of cellular heterogeneity within the developing renal interstitium. Development147, 10.1242/dev.190108 (2020). [DOI] [PMC free article] [PubMed]
- 29.Takasato M, Er PX, Chiu HS, Little MH. Generation of kidney organoids from human pluripotent stem cells. Nat. Protoc. 2016;11:1681–1692. doi: 10.1038/nprot.2016.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hochane M, et al. Single-cell transcriptomics reveals gene expression dynamics of human fetal kidney development. PLoS Biol. 2019;17:e3000152. doi: 10.1371/journal.pbio.3000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindstrom NO, et al. Progressive recruitment of mesenchymal progenitors reveals a time-dependent process of cell fate acquisition in mouse and human nephrogenesis. Develop. Cell. 2018;45:651–660.e654. doi: 10.1016/j.devcel.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindstrom NO, et al. Conserved and divergent features of mesenchymal progenitor cell types within the cortical nephrogenic niche of the human and mouse kidney. J. Am. Soc. Nephrol. 2018;29:806–824. doi: 10.1681/ASN.2017080890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi A, et al. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindstrom NO, et al. Conserved and divergent features of human and mouse kidney organogenesis. J. Am. Soc. Nephrol. 2018;29:785–805. doi: 10.1681/ASN.2017080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindstrom NO, et al. Conserved and divergent molecular and anatomic features of human and mouse nephron patterning. J. Am. Soc. Nephrol. 2018;29:825–840. doi: 10.1681/ASN.2017091036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hennigar RA, Schulte BA, Spicer SS. Heterogeneous distribution of glycoconjugates in human kidney tubules. Anat. Rec. 1985;211:376–390. doi: 10.1002/ar.1092110403. [DOI] [PubMed] [Google Scholar]
- 37.Low JH, et al. Generation of human PSC-derived kidney organoids with patterned nephron segments and a de novo vascular network. Cell Stem Cell. 2019;25:373–387.e379. doi: 10.1016/j.stem.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran T, et al. In vivo developmental trajectories of human podocyte inform in vitro differentiation of pluripotent stem cell-derived podocytes. Develop. Cell. 2019;50:102–116.e106. doi: 10.1016/j.devcel.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holloway EM, et al. Differentiation of human intestinal organoids with endogenous vascular endothelial cells. Develop. Cell. 2020;54:516–528.e517. doi: 10.1016/j.devcel.2020.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Czerniecki SM, et al. High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell. 2018;22:929–940.e924. doi: 10.1016/j.stem.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harder, J. L. et al. Organoid single cell profiling identifies a transcriptional signature of glomerular disease. JCI Insight4, 10.1172/jci.insight.122697 (2019). [DOI] [PMC free article] [PubMed]
- 42.Kumar, S. V. et al. Kidney micro-organoids in suspension culture as a scalable source of human pluripotent stem cell-derived kidney cells. Development146, 10.1242/dev.172361 (2019). [DOI] [PMC free article] [PubMed]
- 43.Avissar N, et al. Human kidney proximal tubules are the main source of plasma glutathione peroxidase. Am. J. Physiol. 1994;266:C367–C375. doi: 10.1152/ajpcell.1994.266.2.C367. [DOI] [PubMed] [Google Scholar]
- 44.Palacin M, Fernandez E, Chillaron J, Zorzano A. The amino acid transport system b(o,+) and cystinuria. Mol. Membr. Biol. 2001;18:21–26. doi: 10.1080/09687680010028771. [DOI] [PubMed] [Google Scholar]
- 45.Schuh CD, et al. Combined structural and functional imaging of the kidney reveals major axial differences in proximal tubule endocytosis. J. Am. Soc. Nephrol. 2018;29:2696–2712. doi: 10.1681/ASN.2018050522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verrey F, et al. Novel renal amino acid transporters. Annu. Rev. Physiol. 2005;67:557–572. doi: 10.1146/annurev.physiol.67.031103.153949. [DOI] [PubMed] [Google Scholar]
- 47.Fenollar-Ferrer C, et al. Identification of the first sodium binding site of the phosphate cotransporter NaPi-IIa (SLC34A1) Biophys. J. 2015;108:2465–2480. doi: 10.1016/j.bpj.2015.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hummel CS, et al. Glucose transport by human renal Na+/D-glucose cotransporters SGLT1 and SGLT2. Am. J. Physiol. Cell Physiol. 2011;300:C14–C21. doi: 10.1152/ajpcell.00388.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rahmoune H, et al. Glucose transporters in human renal proximal tubular cells isolated from the urine of patients with non-insulin-dependent diabetes. Diabetes. 2005;54:3427–3434. doi: 10.2337/diabetes.54.12.3427. [DOI] [PubMed] [Google Scholar]
- 50.Wood IS, Trayhurn P. Glucose transporters (GLUT and SGLT): expanded families of sugar transport proteins. Br. J. Nutr. 2003;89:3–9. doi: 10.1079/BJN2002763. [DOI] [PubMed] [Google Scholar]
- 51.Nagamori S, et al. Novel cystine transporter in renal proximal tubule identified as a missing partner of cystinuria-related plasma membrane protein rBAT/SLC3A1. Proc. Natl Acad. Sci. USA. 2016;113:775–780. doi: 10.1073/pnas.1519959113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otsuka M, et al. A human transporter protein that mediates the final excretion step for toxic organic cations. Proc. Natl Acad. Sci. USA. 2005;102:17923–17928. doi: 10.1073/pnas.0506483102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vogetseder A, et al. Proliferation capacity of the renal proximal tubule involves the bulk of differentiated epithelial cells. Am. J. Physiol. Cell Physiol. 2008;294:C22–C28. doi: 10.1152/ajpcell.00227.2007. [DOI] [PubMed] [Google Scholar]
- 54.Nielsen R, Christensen EI, Birn H. Megalin and cubilin in proximal tubule protein reabsorption: from experimental models to human disease. Kidney Int. 2016;89:58–67. doi: 10.1016/j.kint.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 55.Kowalczuk S, et al. A protein complex in the brush-border membrane explains a Hartnup disorder allele. FASEB J. 2008;22:2880–2887. doi: 10.1096/fj.08-107300. [DOI] [PubMed] [Google Scholar]
- 56.Kanai Y, et al. Transport properties of a system y+L neutral and basic amino acid transporter. Insights into the mechanisms of substrate recognition. J. Biol. Chem. 2000;275:20787–20793. doi: 10.1074/jbc.M000634200. [DOI] [PubMed] [Google Scholar]
- 57.Verrey F. System L: heteromeric exchangers of large, neutral amino acids involved in directional transport. Pflug. Arch. 2003;445:529–533. doi: 10.1007/s00424-002-0973-z. [DOI] [PubMed] [Google Scholar]
- 58.Lee JW, Chou CL, Knepper MA. Deep sequencing in microdissected renal tubules identifies nephron segment-specific transcriptomes. J. Am. Soc. Nephrol. 2015;26:2669–2677. doi: 10.1681/ASN.2014111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Camargo SM, et al. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology. 2009;136:872–882. doi: 10.1053/j.gastro.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fyfe JC, et al. The functional cobalamin (vitamin B12)-intrinsic factor receptor is a novel complex of cubilin and amnionless. Blood. 2004;103:1573–1579. doi: 10.1182/blood-2003-08-2852. [DOI] [PubMed] [Google Scholar]
- 61.Ahuja R, et al. Interactions of cubilin with megalin and the product of the amnionless gene (AMN): effect on its stability. Biochem. J. 2008;410:301–308. doi: 10.1042/BJ20070919. [DOI] [PubMed] [Google Scholar]
- 62.Das A, et al. Stromal-epithelial crosstalk regulates kidney progenitor cell differentiation. Nat. Cell Biol. 2013;15:1035–1044. doi: 10.1038/ncb2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshimura Y, et al. Manipulation of nephron-patterning signals enables selective induction of podocytes from human pluripotent stem cells. J. Am. Soc. Nephrol. 2019;30:304–321. doi: 10.1681/ASN.2018070747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanigawa S, et al. Generation of the organotypic kidney structure by integrating pluripotent stem cell-derived renal stroma. Nat. Commun. 2022;13:611. doi: 10.1038/s41467-022-28226-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Menon, R. et al. Single-cell analysis of progenitor cell dynamics and lineage specification in the human fetal kidney. Development145, 10.1242/dev.164038 (2018). [DOI] [PMC free article] [PubMed]
- 66.Zhu Y, Oganesian A, Keene DR, Sandell LJ. Type IIA procollagen containing the cysteine-rich amino propeptide is deposited in the extracellular matrix of prechondrogenic tissue and binds to TGF-beta1 and BMP-2. J. Cell Biol. 1999;144:1069–1080. doi: 10.1083/jcb.144.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cao, J. et al. A human cell atlas of fetal gene expression. Science370, 10.1126/science.aba7721 (2020). [DOI] [PMC free article] [PubMed]
- 68.Gunaydin H, Gu Y, Huang X. Novel binding mode of a potent and selective tankyrase inhibitor. PLoS ONE. 2012;7:e33740. doi: 10.1371/journal.pone.0033740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lawlor KT, et al. Cellular extrusion bioprinting improves kidney organoid reproducibility and conformation. Nat. Mater. 2021;20:260–271. doi: 10.1038/s41563-020-00853-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yasujima T, Ohta KY, Inoue K, Ishimaru M, Yuasa H. Evaluation of 4’,6-diamidino-2-phenylindole as a fluorescent probe substrate for rapid assays of the functionality of human multidrug and toxin extrusion proteins. Drug Metab. Dispos. 2010;38:715–721. doi: 10.1124/dmd.109.030221. [DOI] [PubMed] [Google Scholar]
- 71.Ozkok A, Edelstein CL. Pathophysiology of cisplatin-induced acute kidney injury. Biomed. Res. Int. 2014;2014:967826. doi: 10.1155/2014/967826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao X, Panichpisal K, Kurtzman N, Nugent K. Cisplatin nephrotoxicity: a review. Am. J. Med. Sci. 2007;334:115–124. doi: 10.1097/MAJ.0b013e31812dfe1e. [DOI] [PubMed] [Google Scholar]
- 73.Abdelsalam M, et al. Urinary biomarkers for early detection of platinum based drugs induced nephrotoxicity. BMC Nephrol. 2018;19:219. doi: 10.1186/s12882-018-1022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chiusolo A, et al. Kidney injury molecule-1 expression in rat proximal tubule after treatment with segment-specific nephrotoxicants: a tool for early screening of potential kidney toxicity. Toxicol. Pathol. 2010;38:338–345. doi: 10.1177/0192623310362244. [DOI] [PubMed] [Google Scholar]
- 75.Sasaki D, et al. Comparison of the course of biomarker changes and kidney injury in a rat model of drug-induced acute kidney injury. Biomarkers. 2011;16:553–566. doi: 10.3109/1354750X.2011.613123. [DOI] [PubMed] [Google Scholar]
- 76.Shinke H, et al. Urinary kidney injury molecule-1 and monocyte chemotactic protein-1 are noninvasive biomarkers of cisplatin-induced nephrotoxicity in lung cancer patients. Cancer Chemother. Pharm. 2015;76:989–996. doi: 10.1007/s00280-015-2880-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vaidya VS, et al. Kidney injury molecule-1 outperforms traditional biomarkers of kidney injury in preclinical biomarker qualification studies. Nat. Biotechnol. 2010;28:478–485. doi: 10.1038/nbt.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Digby JLM, Vanichapol T, Przepiorski A, Davidson AJ, Sander V. Evaluation of cisplatin-induced injury in human kidney organoids. Am. J. Physiol. Ren. Physiol. 2020;318:F971–F978. doi: 10.1152/ajprenal.00597.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cruz NM, et al. Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease. Nat. Mater. 2017;16:1112–1119. doi: 10.1038/nmat4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Forbes TA, et al. Patient-iPSC-derived kidney organoids show functional validation of a ciliopathic renal phenotype and reveal underlying pathogenetic mechanisms. Am. J. Hum. Genet. 2018;102:816–831. doi: 10.1016/j.ajhg.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hale LJ, et al. 3D organoid-derived human glomeruli for personalised podocyte disease modelling and drug screening. Nat. Commun. 2018;9:5167. doi: 10.1038/s41467-018-07594-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hollywood JA, et al. Use of human induced pluripotent stem cells and kidney organoids to develop a cysteamine/mTOR inhibition combination therapy for cystinosis. J. Am. Soc. Nephrol. 2020;31:962–982. doi: 10.1681/ASN.2019070712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mae S, et al. Monitoring and robust induction of nephrogenic intermediate mesoderm from human pluripotent stem cells. Nat. Commun. 2013;4:1367. doi: 10.1038/ncomms2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Przepiorski A, et al. A simple bioreactor-based method to generate kidney organoids from pluripotent stem cells. Stem Cell Rep. 2018;11:470–484. doi: 10.1016/j.stemcr.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tanigawa S, et al. Organoids from nephrotic disease-derived iPSCs identify impaired NEPHRIN localization and slit diaphragm formation in kidney podocytes. Stem Cell Rep. 2018;11:727–740. doi: 10.1016/j.stemcr.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han L, et al. Cell transcriptomic atlas of the non-human primate Macaca fascicularis. Nature. 2022;604:723–731. doi: 10.1038/s41586-022-04587-3. [DOI] [PubMed] [Google Scholar]
- 87.Marchiano S, et al. SARS-CoV-2 infects human pluripotent stem cell-derived cardiomyocytes, impairing electrical and mechanical function. Stem Cell Rep. 2021;16:478–492. doi: 10.1016/j.stemcr.2021.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mills RJ, et al. BET inhibition blocks inflammation-induced cardiac dysfunction and SARS-CoV-2 infection. Cell. 2021;184:2167–2182 e2122. doi: 10.1016/j.cell.2021.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sharma A, et al. Human iPSC-derived cardiomyocytes are susceptible to SARS-CoV-2 infection. Cell Rep. Med. 2020;1:100052. doi: 10.1016/j.xcrm.2020.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tiwari SK, Wang S, Smith D, Carlin AF, Rana TM. Revealing tissue-specific SARS-CoV-2 infection and host responses using human stem cell-derived lung and cerebral organoids. Stem Cell Rep. 2021;16:437–445. doi: 10.1016/j.stemcr.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kunutsor SK, Laukkanen JA. Renal complications in COVID-19: a systematic review and meta-analysis. Ann. Med. 2020;52:345–353. doi: 10.1080/07853890.2020.1790643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang X, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Monteil V, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e907. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wysocki, J. et al. A novel soluble ACE2 variant with prolonged duration of action neutralizes SARS-CoV-2 infection in human kidney organoids. J. Am. Soc. Nephrol.10.1681/ASN.2020101537 (2021). [DOI] [PMC free article] [PubMed]
- 97.Motavalli R, et al. The lethal internal face of the coronaviruses: kidney tropism of the SARS, MERS, and COVID19 viruses. IUBMB Life. 2021;73:1005–1015. doi: 10.1002/iub.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Braun F, et al. SARS-CoV-2 renal tropism associates with acute kidney injury. Lancet. 2020;396:597–598. doi: 10.1016/S0140-6736(20)31759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Farkash EA, Wilson AM, Jentzen JM. Ultrastructural evidence for direct renal infection with SARS-CoV-2. J. Am. Soc. Nephrol. 2020;31:1683–1687. doi: 10.1681/ASN.2020040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kissling S, et al. Collapsing glomerulopathy in a COVID-19 patient. Kidney Int. 2020;98:228–231. doi: 10.1016/j.kint.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Puelles VG, et al. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Su H, et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98:219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Werion A, et al. SARS-CoV-2 causes a specific dysfunction of the kidney proximal tubule. Kidney Int. 2020;98:1296–1307. doi: 10.1016/j.kint.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hanley B, et al. Histopathological findings and viral tropism in UK patients with severe fatal COVID-19: a post-mortem study. Lancet Microbe. 2020;1:e245–e253. doi: 10.1016/S2666-5247(20)30115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Amraei, R. et al. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2. ACS Cent Sci.7, 1156–1165 (2021). [DOI] [PMC free article] [PubMed]
- 107.Singh M, Bansal V, Feschotte C. A single-cell RNA expression map of human coronavirus entry factors. Cell Rep. 2020;32:108175. doi: 10.1016/j.celrep.2020.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kuppe C, et al. Novel parietal epithelial cell subpopulations contribute to focal segmental glomerulosclerosis and glomerular tip lesions. Kidney Int. 2019;96:80–93. doi: 10.1016/j.kint.2019.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang M. Not your usual parietal cell. Nat. Rev. Nephrol. 2019;15:318. doi: 10.1038/s41581-019-0141-8. [DOI] [PubMed] [Google Scholar]
- 110.Broer S, et al. Iminoglycinuria and hyperglycinuria are discrete human phenotypes resulting from complex mutations in proline and glycine transporters. J. Clin. Invest. 2008;118:3881–3892. doi: 10.1172/JCI36625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hou YJ, et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446 e414. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen Z, et al. SARS-CoV-2 causes acute kidney injury by directly infecting renal tubules. Front. Cell Dev. Biol. 2021;9:664868. doi: 10.3389/fcell.2021.664868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jansen J, et al. SARS-CoV-2 infects the human kidney and drives fibrosis in kidney organoids. Cell Stem Cell. 2022;29:217–231 e218. doi: 10.1016/j.stem.2021.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.V’Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zlacka J, Stebelova K, Zeman M, Herichova I. Interactions of renin-angiotensin system and COVID-19: the importance of daily rhythms in ACE2, ADAM17 and TMPRSS2 expression. Physiol. Res. 2021;70:S177–S194. doi: 10.33549/physiolres.934754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Muus C, et al. Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics. Nat. Med. 2021;27:546–559. doi: 10.1038/s41591-020-01227-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qiu P. Embracing the dropouts in single-cell RNA-seq analysis. Nat. Commun. 2020;11:1169. doi: 10.1038/s41467-020-14976-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kalejaiye TD, et al. SARS-CoV-2 employ BSG/CD147 and ACE2 receptors to directly infect human induced pluripotent stem cell-derived kidney podocytes. Front. Cell Dev. Biol. 2022;10:855340. doi: 10.3389/fcell.2022.855340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chung E, Deacon P, Park JS. Notch is required for the formation of all nephron segments and primes nephron progenitors for differentiation. Development. 2017;144:4530–4539. doi: 10.1242/dev.156661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Surendran K, et al. The contribution of Notch1 to nephron segmentation in the developing kidney is revealed in a sensitized Notch2 background and can be augmented by reducing Mint dosage. Dev. Biol. 2010;337:386–395. doi: 10.1016/j.ydbio.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Karner CM, et al. Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development. 2011;138:1247–1257. doi: 10.1242/dev.057646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Park JS, Valerius MT, McMahon AP. Wnt/beta-catenin signaling regulates nephron induction during mouse kidney development. Development. 2007;134:2533–2539. doi: 10.1242/dev.006155. [DOI] [PubMed] [Google Scholar]
- 124.Blank U, Brown A, Adams DC, Karolak MJ, Oxburgh L. BMP7 promotes proliferation of nephron progenitor cells via a JNK-dependent mechanism. Development. 2009;136:3557–3566. doi: 10.1242/dev.036335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lindstrom NO, Carragher NO, Hohenstein P. The PI3K pathway balances self-renewal and differentiation of nephron progenitor cells through beta-catenin signaling. Stem. Cell Rep. 2015;4:551–560. doi: 10.1016/j.stemcr.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Muthukrishnan SD, Yang X, Friesel R, Oxburgh L. Concurrent BMP7 and FGF9 signalling governs AP-1 function to promote self-renewal of nephron progenitor cells. Nat. Commun. 2015;6:10027. doi: 10.1038/ncomms10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tanigawa S, et al. Activin is superior to BMP7 for efficient maintenance of human iPSC-derived nephron progenitors. Stem Cell Rep. 2019;13:322–337. doi: 10.1016/j.stemcr.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Matsuda M, et al. Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science. 2020;369:1450–1455. doi: 10.1126/science.aba7668. [DOI] [PubMed] [Google Scholar]
- 129.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Velasco S, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Taguchi A, et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14:53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 132.Bantounas I, Silajdzic E, Woolf AS, Kimber SJ. Formation of mature nephrons by implantation of human pluripotent stem cell-derived progenitors into mice. Methods Mol. Biol. 2020;2067:309–322. doi: 10.1007/978-1-4939-9841-8_19. [DOI] [PubMed] [Google Scholar]
- 133.Nam SA, et al. Graft immaturity and safety concerns in transplanted human kidney organoids. Exp. Mol. Med. 2019;51:1–13. doi: 10.1038/s12276-019-0336-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.van den Berg CW, et al. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell Rep. 2018;10:751–765. doi: 10.1016/j.stemcr.2018.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Briggs JA, et al. Integration-free induced pluripotent stem cells model genetic and neural developmental features of down syndrome etiology. Stem Cells. 2013;31:467–478. doi: 10.1002/stem.1297. [DOI] [PubMed] [Google Scholar]
- 136.Vlahos K, et al. Generation of iPSC lines from peripheral blood mononuclear cells from 5 healthy adults. Stem Cell Res. 2019;34:101380. doi: 10.1016/j.scr.2018.101380. [DOI] [PubMed] [Google Scholar]
- 137.Chen G, et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stoeckius M, et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018;19:224. doi: 10.1186/s13059-018-1603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ransick A, et al. Single-cell profiling reveals sex, lineage, and regional diversity in the mouse kidney. Develop. Cell. 2019;51:399–413 e397. doi: 10.1016/j.devcel.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Harris CR, et al. Array programming with NumPy. Nature. 2020;585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.van der Walt S, et al. scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Virtanen P, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vanslambrouck, J. et al. Enhanced metanephric specification to functional proximal tubule enables toxicity screening and infectious disease modelling in kidney organoids. KidneyRegeneration/Vanslambrouck2022, 10.5281/zenodo.7021393 (2022). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The transcriptional profiling datasets generated in this study have been deposited in GEO under accession code GSE184928. The raw data from scRNAseq and immunofluorescence image analyses have been deposited in the Github repository [https://github.com/KidneyRegeneration/Vanslambrouck2022]. Raw and processed data from qRT-PCR, TCID50 assays, and image analyses are provided in the Source data file. Source data are provided with this paper.
Code for scRNAseq and image analyses are available through the Github repository (https://github.com/KidneyRegeneration/Vanslambrouck2022; 10.5281/zenodo.7021393)147.