Abstract
Methanothermobacter wolfeii (formerly Methanobacterium wolfei), a thermophilic methanoarchaeon whose cultures lyse upon hydrogen starvation, carries a defective prophage called ΨM100 on its chromosome. The nucleotide sequence of ΨM100 and its flanking regions was established and compared to that of the previously sequenced phage ΨM2 of Methanothermobacter marburgensis (formerly Methanobacterium thermoautotrophicum Marburg). The ΨM100 genome extends over 28,798 bp, and its borders are defined by flanking 21-bp direct repeats of a pure-AT sequence, which very likely forms the core of the putative attachment site where the crossing over occurred during integration. A large fragment of 2,793 bp, IFa, apparently inserted into ΨM100 but is absent in the genome of ΨM2. The remaining part of the ΨM100 genome showed 70.8% nucleotide sequence identity to the whole genome of ΨM2. Thirty-four open reading frames (ORFs) on the forward strand and one ORF on the reverse strand were identified in the ΨM100 genome. Comparison of ΨM100-encoded ORFs to those encoded by phage ΨM2 and to other known protein sequences permitted the assignment of putative functions to some ORFs. The ORF28 protein of ΨM100 was identified as the previously known autolytic enzyme pseudomurein endoisopeptidase PeiW produced by M. wolfeii.
Cultures of the thermophilic methanoarchaeon Methanothermobacter wolfeii (formerly Methanobacterium wolfei) (20, 21) spontaneously lyse upon hydrogen limitation (10). However, no phage-like particles have been detected in the culture supernatant or autolysate. A lytic enzyme was purified from the autolysate, and this enzyme has been shown to be a pseudomurein endoisopeptidase (9). M. wolfeii proved insensitive to the virulent phage ΨM1 and its deletion mutant ΨM2 of Methanothermobacter marburgensis (formerly Methanobacterium thermoautotrophicum) Marburg (P. Pfister, unpublished observation). The results of Southern hybridization suggested that there is a prophage in the chromosome of M. wolfeii, ΨM100, which is homologous to phages ΨM1 and ΨM2 (14, 19; P. Pfister, unpublished data). The approximate location of ΨM100 in the M. wolfeii chromosome was determined, and most of the prophage was shown to be located on a ca. 30-kb NotI-NheI fragment (19). Since attempts to detect free phage particles had failed, ΨM100 was assumed to be defective. In contrast, infection of M. marburgensis Marburg with phages ΨM1 and ΨM2 consistently led to lysis of the host. The complete nucleotide sequence of ΨM2 was established, and some of its open reading frames (ORFs) and the proteins they encode were characterized (15). In order to explore the relationships between the defective prophage ΨM100, the autolysis phenomenon, and the ΨM1 and ΨM2 phages, we determined and analyzed the sequence of ΨM100 and its flanking regions.
Cloning, PCR amplification, and nucleotide sequence determination of ΨM100 and its flanking regions.
Portions of ΨM100 and its flanking regions were obtained from the M. wolfeii chromosome as overlapping clones and PCR fragments (data not shown). The methods used included the following: shotgun cloning, construction, and screening of a SuperCos1-based cosmid library; screening of a λ-ZAP Express genomic library (7); PCR amplification of either nonclonable portions or portions that were difficult to clone on both sides of ΨM100; and PCR amplification of the regions across the restriction sites of fragments obtained by shotgun cloning. The nucleotide sequences of both strands were determined by primer walking and then assembled.
Properties of ΨM100 DNA.
The border of ΨM100 is defined by the flanking 21-bp AT-only direct repeats, which probably represent the core of the putative attachment site (see below). The length of the ΨM100 genome extends over 28,798 bp with an overall GC content of 45.4%. This is somewhat lower than the 48.3% GC determined for M. wolfeii by a melting point analysis (11). Sequence alignment of ΨM100 and ΨM2 (Fig. 1) reveals that, other than point mutations and small deletions or insertions, there is a large fragment of 2,793 bp, IFa, inserted into ΨM100 between coordinates 5272 and 8066. The GC content of the IFa element is 33.4%, which is significantly lower than those of ΨM100 and ΨM100 without IFa (46.7%). Like ΨM2, the GC content of the ΨM100 sequence is not evenly distributed, with five low-GC (<40%) DNA regions of at least 300 nucleotides (nt) extending over parts of ORFs orf6, orf28, orf29, and orf30 and over the entire ORFs orf5, the IFa-encoded orfB, -C, and -D, and the putative attachment sites attL and attR (Fig. 1).
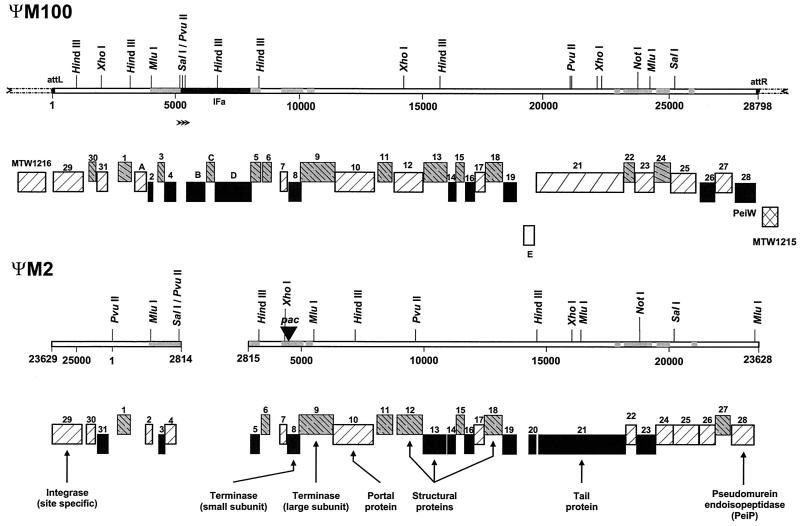
FIG. 1.
Schematic representation of the defective prophage with its flanking regions and comparison to that of phage ΨM2. The ORFs are represented by boxes numbered as in Table 1, and their vertical placement indicates the gene location in one of the six possible reading frames. Homologous ORFs carry the same numbers, and the assigned functions for some ORFs of ΨM2 are reported. MTW1215 and MTW1216 represent the two ORFs encoded by the chromosome sequences flanking ΨM100. orfA to orfE are unique to ΨM100. The experimentally determined pac locus for ΨM2 is shown as a solid black triangle. The IFa fragment (black bar) and the DNA regions with at least 200 bp with >98% identity in ΨM100 and ΨM2 (grey bars) are indicated. Three arrowheads represent three contiguous copies of a direct repeat of 125 nt in the vicinity of the left end of the IFa fragment.
Some direct repeats are clustered and prominent in their length and locations. For example, there are three contiguous copies of a direct repeat of 125 nt in the region between coordinates 5081 and 5455, and a short copy is duplicated between coordinates 5456 and 5481. This is apparently due to the insertion of IFa, since there are only two direct repeats of 67 nt in the corresponding region of ΨM2, which, based on its similarity to oriC of Escherichia coli, was proposed to be the most-probable replication origin (Pfister, unpublished). Moreover, the direct repeats do not overlap any coding regions of ΨM100 and ΨM2.
After the IFa element was removed, the remaining region of ΨM100 (26,005 bp) could be aligned with that of ΨM2 (26,111 bp) over their full length with the GAP program of the Genetics Computer Group (GCG). It shows 70.8% identity in a 26,752-nt overlap. Eight stretches of sequence longer than 200 bp in ΨM100 and ΨM2, including the regions harboring the putative origin of replication and the experimentally determined pac site (8), have >98% identity (Fig. 1).
Coding capacity of ΨM100 and comparison to that of ΨM2.
Putative ORFs were defined based on the following assumptions: an ORF should code for a polypeptide of at least 90 amino acids (aa) and be preceded by a ribosome binding site with at least 45% identity to the proposed consensus sequence 5′-AGGAGGTGATC-3′ (2). Two exceptions were made; these exceptions were orf2, encoding a 85-aa polypeptide with a homologue of 95 aa in ΨM2, and orf29, encoding a putative integrase, the ribosome binding site of which has only 36% identity to the consensus sequence (as in ΨM2). Consequently, 34 genes were identified on the forward strand and 1 gene was identified on the reverse strand (Fig. 1 and Table 1).
TABLE 1.
General features of the putative ORFs encoded by ΨM100 and comparison to the corresponding ORFs of ΨM2 and to other proteins in databases
| ORF | Fa | Start/end | Size (aa) | Motifb | Related
ORF in ΨM2
|
Other related protein in database
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF | Size (aa)c | Proposed function | % aa identityd | Name | Accession no.e | Size (aa) | % aa identity | |||||
| orf29 | 2 | 65/1273 | 402 | orf29 | 402 | Integrase or recombinase | 63.8 (406) | Integrase or recombinase RipX (Bacillus subtilis) | sp P46352 | 296 | 28.0 (416) | |
| orf30 | 1 | 1489/1791 | 100 | orf30 | 124 | 44.8 (137) | Hypothetical protein (Emericella nidulans mitochondrion) | sp P03883 | 228 | 24.2 (230) | ||
| orf31 | 2 | 1826/2278 | 150 | orf31 | 150 | 39.9 (153) | ||||||
| orf1 | 1 | 2578/3102 | 174 | orf1 | 177 | 73.0 (178) | ||||||
| orfA | 2 | 3287/3754 | 155 | NA | NA | |||||||
| orf2 | 3 | 3828/4085 | 85 | orf2 | 95 | 95.3 (95) | ||||||
| orf3 | 1 | 4324/4608 | 94 | orf3 | 94 | 98.9 (94) | ||||||
| orf4 | 3 | 4605/5069 | 154 | HTH | orf4 | 154 | 100 (154) | |||||
| orfB | 3 | 5577/6335 | 252 | NA | NA | Hypothetical protein MTH315 (Methanothermobacter thermautotrophicus ΔH) | pir D69140 | 254 | 62.9 (258) | |||
| orfC | 1 | 6385/6708 | 107 | NA | NA | Hypothetical protein MTH316 (Methanothermobacter thermautotrophicus ΔH) | pir E69140 | 114 | 56.9 (119) | |||
| orfD | 3 | 6720/8198 | 492 | NA | NA | |||||||
| orf5 | 1 | 8176/8565 | 129 | orf5 | 127 | 89.1 (129) | Hypothetical protein APE1779 (Aeropyrum pernix K1) | pir A72562 | 158 | 26.4 (158) | ||
| orf6 | 1 | 8665/9000 | 111 | HTH | orf6 | 117 | 66.7 (119) | ORF6 of plasmid pME2200 (Methanothermobacter marburgensis ZH3) | gb AAF00129 | 135 | 40.5 (136) | |
| orf7 | 2 | 9389/9661 | 90 | orf7 | 90 | 97.8 (90) | ||||||
| orf8 | 3 | 9702/10223 | 173 | HTH | orf8 | 173 | Small terminase subunit | 100 (173) | Prophage PBSX small terminase subunit (Bacillus subtilis) | sp P39785 | 265 | 21.5 (276) |
| orf9 | 1 | 10168/11571 | 467 | ATP or GTP binding motif A | orf9 | 468 | Large terminase subunit | 91.5 (468) | Unknown protein of bacteriophage Felix 01 | gb AAC24136.1 | 533 | 36.0 (540) |
| orf10 | 2 | 11564/13186 | 540 | orf10 | 542 | Portal protein | 78.2 (542) | Portal protein of temperate phage HP1 (Haemophilus influenzae) | sp P51717 | 345 | 24.9 (561) | |
| orf11 | 1 | 13327/13932 | 201 | orf11 | 216 | 60.3 (220) | ||||||
| orf12 | 2 | 14003/15223 | 406 | orf12 | 351 | Structural protein | 55.8 (416) | Unknown protein of a cryptic prophage (Bacillus subtilis) | sp P45920 | 322 | 26.0 (424) | |
| orf13 | 1 | 15238/16182 | 314 | orf13 | 315 | Structural protein | 57.1 (321) | Unknown protein of a cryptic prophage (Bacillus subtilis) | sp P45921 | 311 | 30.2 (327) | |
| orf14 | 3 | 16254/16574 | 106 | orf14 | 110 | 53.9 (114) | ||||||
| orf15 | 1 | 16561/16902 | 113 | orf15 | 112 | 41.1 (114) | ||||||
| orf16 | 3 | 16908/17309 | 133 | orf16 | 133 | 63.9 (133) | Unknown protein of phage SPP1 (Bacillus subtilis) | pir T42287 | 144 | 31.0 (148) | ||
| orf17 | 2 | 17294/17710 | 138 | orf17 | 139 | 54.0 (140) | ||||||
| orf18 | 1 | 17707/18423 | 238 | orf18 | 243 | Structural protein | 64.4 (245) | |||||
| orf19 | 3 | 18435/18986 | 183 | orf19 | 189 | 59.2 (193) | ||||||
| orfE | 6 | 19613/19185 | 142 | NA | NA | Hypothetical protein MTH1518 (Methanothermobacter thermautotrophicus ΔH) | pir E69069 | 133 | 28.1 (147) | |||
| orf21 | 2 | 19679/23152 | 1,157 | orf21 | 1,186 | Tail protein | 49.8 (1,222) | Hypothetical protein M151.4 (Caenorhabditis elegans) | pir T32297 | 1,034 | 20.0 (1,686) | |
| orf22 | 1 | 23149/23583 | 144 | ATP or GTP binding motif A | orf22 | 144 | ATP or GTP binding protein | 100 (144) | ||||
| orf23 | 2 | 23588/24373 | 261 | orf23 | 268 | 92.7 (268) | ||||||
| orf24 | 1 | 24370/25068 | 232 | orf24 | 232 | 98.3 (232) | ||||||
| orf25 | 2 | 25070/26101 | 343 | orf25 | 354 | 86.2 (357) | ||||||
| orf26 | 3 | 26280/26912 | 210 | orf26 | 216 | 81.8 (217) | ||||||
| orf27 | 2 | 26909/27631 | 240 | orf27 | 206 | 41.9 (248) | ORFA of phage ΨM1 element DR1 (Methanothermobacter marburgensis Marburg) | gb AAC27071 | 217 | 38.5 (249) | ||
| orf28 | 3 | 27780/28634 | 284 | orf28 | 305 | Pseudomurein endoisopeptidase PeiP | 53.4 (306) | Conserved hypothetical protein MTH412 (Methanothermobacter thermautotrophicus ΔH) | pir F69153 | 583 | 20.0 (602) | |
F, reading frame in which an ORF was defined.
HTH, helix-turn-helix motif.
NA, not available.
The percentages of amino acid (aa) identity were calculated using the GAP program of GCG. The numbers in parentheses are the sizes (in amino acids) of the alignment overlap. NA, not available.
The database sources are shown before the accession numbers as follows: sp, SWISSPROT; pir, PIR database; gb, GenBank.
Except for orfA and the IFa-encoded orfB, -C, -D, and -E, all other ORFs are similar to their counterparts encoded by phage ΨM2. The ORFs of ΨM100 and ΨM2 exhibit similarities in size, location in the genome, and sequence identity, which ranges from 39.9 to 100% at both the nucleotide and amino acid levels. The homologue of ΨM2 orf20 is missing in ΨM100 (Fig. 1 and Table 1). The ORFA protein (155 aa) apparently has no homologue of similar length in ΨM2. The nucleotide sequence of ΨM2 encodes a peptide of 86 aa, homologous to the N terminus of ORFA (probability calculated using BLAST, 10−24). Sequence alignment suggests that the C-terminal part of ORFA was created by insertion of three noncontiguous DNA stretches into the ΨM100 genome. Database searches with BLAST at the National Center for Biotechnology Information (1) also revealed that 16 of the proposed proteins are similar to other protein sequences (Table 1).
Integration site of ΨM100 in the chromosome of M. wolfeii.
Screening for ORFs encoded by regions of the M. wolfeii chromosome flanking ΨM100 revealed two genes encoding MTW1216 on the forward strand upstream of the putative attL site and MTW1215 on the reverse strand downstream of the putative attR site (Fig. 1). The two host genes share high similarity to their counterparts (i.e., mth1215 and mth1216) in the Methanobacterium thermautotrophicus ΔH genome (18); therefore, the same numbers were used to designate the M. wolfeii homologues. At the amino acid level, MTW1216 exhibits 77% identity to MTH1216 (pantothenate metabolism flavoprotein) and MTW1215 is 81.9% identical to MTH1215 (fibrillarin-like pre-rRNA processing factor). MTW1215 and MTW1216 are transcribed convergently, as are MTH1215 and MTH1216. The MTH1215 gene in the M. thermautotrophicus ΔH genome is located at nucleotides 1117886 to 1118560 on the direct strand, and the gene encoding MTH1216 is located at nucleotides 1119977 to 1118817 on the complementary strand (18). Remarkably, integration of ΨM100 occurred exactly within the intergenic regions between these two host genes and between prophage genes orf28 (peiW) and orf29. Neither the host genes nor ORFs of the defective prophage were disrupted in terms of their transcription potential. Thus, the integration mode of ΨM100 resembles that of the major integration pattern of coliphage λ (4).
Putative attachment sites of ΨM100.
Sequence analysis revealed that ΨM100 is flanked by direct repeats of a 21-bp pure-AT nucleotide sequence. This sequence is very likely the core of putative attachment sites where the crossing-over occurred during integration and/or excision. Therefore, the flanking regions of the core might be defined as hybrid attL and attR sites. Conversely, the sequences of attP [(pro)phage-encoded attachment site] and attB [chromosomal attachment site] can be derived, although their exact lengths remain to be determined (Fig. 2A). In the region between orf28 and orf29 of phage ΨM2 (i.e., in the putative attP site of phage ΨM2) and between MTH1216 and MTH1215 of M. thermautotrophicus ΔH (i.e., in the putative attB site), similar AT-rich sequences with four and three mismatches, respectively, were identified (Fig. 2B). Interestingly, one stretch of sequence which is 100% identical to the core sequence but located in a different context was present in another region of the M. thermautotrophicus ΔH genome (Fig. 2B). It is not known whether additional core-like sequences occur in the M. wolfeii genome. For phage λ, the core of the attachment sites consists of 15-bp AT-rich sequences (3). There are some similarities between the sequences of the λ and ΨM100 cores (Fig. 2C).
FIG. 2.
(A) Sequences of the attR, attL, attP, attB, and the N-terminal part of the putative integrase ORF29. Sequences originating from the M. wolfeii chromosome are shown in italics. The crossed lines represent the sequences of attP and attB derived from the sequences of attR and attL. The core of the putative attachment sites (grey box), the stop codon of MTW1215 (CTA on the reverse strand) (grey box), and the start codon of ORF29 (ATG on the forward strand) (black box) are indicated. The numbers −38∼38 and 39∼85 indicate the coordinates of the prophage ΨM100 sequence. (B) DNA sequences of the corresponding regions in the phage ΨM2 and M. thermautotrophicus ΔH genomes as well as of another region in the M. thermautotrophicus ΔH genome. The numbers refer to the coordinates of the sequences in the phage ΨM2 or M. thermautotrophicus ΔH genome. (C) DNA sequence of the core of coliphage λ attachment sites. In panels B and C, nucleotides identical to those of the core of the ΨM100 attachment sites are also highlighted.
Experimental identification of the autolytic enzyme pseudomurein endoisopeptidase PeiW.
The deduced N-terminal sequence of the protein encoded by orf28 of prophage ΨM100 is identical to that of the experimentally determined N-terminal sequence (18 aa) of the autolytic enzyme produced by M. wolfeii (15). This strongly suggests that the ORF28 protein is the pseudomurein endoisopeptidase PeiW that has been known for more than 10 years (9). The predicted molecular mass (33.4 kDa) is consistent with the observed 33 kDa for the autolytic enzyme (9).
The structural gene of PeiW was cloned and successfully overexpressed in E. coli BL21(DE3) grown under aerobic conditions. The cell wall-degrading activity of PeiW was confirmed in the activity assay described previously (15; data not shown).
Conclusions.
The complete nucleotide sequences of phage ΨM2 of M. marburgensis and of the defective prophage ΨM100 of M. wolfeii and its flanking regions now allow us to make a thorough comparison between these two elements. In contrast to two other archaeal defective prophages in Halobacterium halobium (17) and in Sulfolobus sp. strain B12 (16), the sequence of ΨM100 contains all the information necessary for synthesis of structural proteins homologous to those of ΨM2. In addition to small insertions, deletions, and point mutations, downstream of the putative replication origin, ΨM100 carries the inserted fragment IFa of 2,793 bp, which apparently originated from another source. The IFa element has none of the distinct characteristics usually found in IS sequences (13). It encodes three putative ORFs, of which two short ones (i.e., orfB and orfC) are similar to hypothetical ORFs of M. thermautotrophicus ΔH. The largest ORF, orfD, has no homologues in the database, although PropSearch at the EMBL (6) yields some hits related to DNA metabolism. One of these hits is the potential transposase for IS1151 of Clostridium perfringens (5). Using the GAP program of GCG, the two proteins show 18% identity in a 522-aa overlap. Only the 5′ part of the IFa DNA sequence, which is a duplication of the sequence harboring the putative origin of replication, shows traces of terminal redundancy, possibly derived from the packaging mechanism of phage ΨM2 (8). Therefore, the rearrangement due to the insertion of this IFa in the DNA of ΨM100 might have led to this defect. However, there is no evidence explaining the order of insertion of IFa into the ΨM100 genome and integration of the ΨM100 ancestor(s), if any, into the M. wolfeii chromosome. Alternatively, ΨM100 might be defective due to the lack of a functional excisionase, which is usually required to excise (pro)- phage genomes from their host chromosomes, while ΨM2 might recruit the putative excisionase function of the ORF5 encoded by plasmid pME2001 present in the host strain (12).
Together with the finding that the ORF29 proteins of both ΨM2 and ΨM100 are highly similar to members of the Int family of site-specific recombinases, the presence of a 21-bp pure-AT direct repeat in the flanking regions of ΨM100 supports the view that ΨM100 is really a ΨM2-related prophage. Such direct repeats are typically derived from integrative recombination of phage DNA with the host chromosome (3). Since genes encoding site-specific integrases are not found in genomes of virulent phages, phages ΨM1 and ΨM2 might be temperate phages.
Nucleotide sequence accession number.
The sequence is available in GenBank under accession number AF301375.
Acknowledgments
We thank R. Hedderich for providing the λ-ZAP Express genomic library of M. wolfeii and U. Kües for providing E. coli strain NM554 and cosmid SuperCos1.
This work was supported in part by grant 3100-50593.97 from the Swiss National Foundation for Scientific Research.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Brown J W, Daniels C J, Reeve J N. Gene structure, organization, and expression in archaebacteria. Crit Rev Microbiol. 1989;16:287–338. doi: 10.3109/10408418909105479. [DOI] [PubMed] [Google Scholar]
- 3.Campbell A M. Chromosomal insertion sites for phages and plasmids. J Bacteriol. 1992;174:7495–7499. doi: 10.1128/jb.174.23.7495-7499.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell A M. Thirty years ago in genetics: prophage insertion into bacterial chromosomes. Genetics. 1993;133:433–437. doi: 10.1093/genetics/133.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daube G, Simon P, Kaeckenbeeck A. IS1151, an IS-like element of Clostridium perfringens. Nucleic Acids Res. 1993;21:352. doi: 10.1093/nar/21.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hobohm U, Sander C. A sequence property approach to searching protein databases. J Mol Biol. 1995;251:390–399. doi: 10.1006/jmbi.1995.0442. [DOI] [PubMed] [Google Scholar]
- 7.Hochheimer A, Hedderich R, Thauer R K. The formylmethanofuran dehydrogenase isoenzymes in Methanobacterium wolfei and Methanobacterium thermoautotrophicum: induction of the molybdenum isoenzyme by molybdate and constitutive synthesis of the tungsten isoenzyme. Arch Microbiol. 1998;170:389–393. doi: 10.1007/s002030050658. [DOI] [PubMed] [Google Scholar]
- 8.Jordan M, Meile L, Leisinger T. Organization of Methanobacterium thermoautotrophicumbacteriophage ΨM1 DNA. Mol Gen Genet. 1989;220:161–164. doi: 10.1007/BF00260872. [DOI] [PubMed] [Google Scholar]
- 9.Kiener A, König H, Winter J, Leisinger T. Purification and use of Methanobacterium wolfei pseudomurein endopeptidase for lysis of Methanobacterium thermoautotrophicum. J Bacteriol. 1987;169:1010–1016. doi: 10.1128/jb.169.3.1010-1016.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.König H, Semmler R, Lerch C, Winter J. Evidence for the occurrence of autolytic enzymes in Methanobacterium wolfei. Arch Microbiol. 1985;141:177–180. [Google Scholar]
- 11.Kotelnikova S V, Obraztsova A Y, Blotevogel K-H, Popov I N. Taxonomic analysis of thermophilic strains of the genus Methanobacterium: reclassification of Methanobacterium thermoalcaliphilum as a synonym of Methanobacterium thermoautotrophicum. Int J Syst Bacteriol. 1993;43:591–596. [Google Scholar]
- 12.Luo Y, Leisinger T, Wasserfallen A. Comparative sequence analysis of plasmids pME2001 and pME2200 of Methanothermobacter marburgensisstrains Marburg and ZH3. Plasmid. 2001;45:18–30. doi: 10.1006/plas.2000.1493. [DOI] [PubMed] [Google Scholar]
- 13.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meile L, Jenal U, Studer D, Jordan M, Leisinger T. Characterization of ΨM1: a virulent phage of Methanobacterium thermoautotrophicumMarburg. Arch Microbiol. 1989;152:105–110. [Google Scholar]
- 15.Pfister P, Wasserfallen A, Stettler R, Leisinger T. Molecular analysis of Methanobacteriumphage ΨM2. Mol Microbiol. 1998;30:233–244. doi: 10.1046/j.1365-2958.1998.01073.x. [DOI] [PubMed] [Google Scholar]
- 16.Reiter W D, Palm P. Identification and characterization of a defective SSV1 genome integrated into a tRNA gene in the archaebacterium Sulfolobussp. B12. Mol Gen Genet. 1990;221:65–71. doi: 10.1007/BF00280369. [DOI] [PubMed] [Google Scholar]
- 17.Schnabel H. An immune strain of Halobacterium halobiumcarries the invertible L segment of phage ΦH as a plasmid. Proc Natl Acad Sci USA. 1984;81:1017–1020. doi: 10.1073/pnas.81.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicumΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stettler R, Thurner C, Stax D, Meile L, Leisinger T. Evidence for a defective prophage on the chromosome of Methanobacterium wolfei. FEMS Microbiol Lett. 1995;132:85–89. doi: 10.1111/j.1574-6968.1995.tb07815.x. [DOI] [PubMed] [Google Scholar]
- 20.Wasserfallen A, Nölling J, Pfister P, Reeve J, Conway de Macario E. Phylogenetic analysis of 18 thermophilic Methanobacterium isolates supports the proposals to create a new genus, Methanothermobacter gen. nov., and to reclassify several isolates in three species, Methanothermobacter thermautotrophicus comb. nov., Methanothermobacter wolfeii comb. nov., and Methanothermobacter marburgensissp. nov. Int J Syst Evol Microbiol. 2000;50:43–53. doi: 10.1099/00207713-50-1-43. [DOI] [PubMed] [Google Scholar]
- 21.Winter J, Lerp C, Zabel H-P, Wildenauer F X, König H, Schindler F. Methanobacterium wolfeisp. nov., a new tungsten-requiring, thermophilic, autotrophic methanogen. Syst Appl Microbiol. 1984;5:457–466. [Google Scholar]