Abstract
Oxidative stress and cellular response mechanisms such as NRF2-mediated antioxidant responses play differential roles in healthy and diseased cells. Constant generation and elimination of high levels of reactive oxygen species is a hallmark of many cancer cell types; this phenomenon is not observed during steady state of healthy cells. Manipulation of NRF2 transcriptional activity and the cellular redox homeostasis therefore has potential to be therapeutically exploitable for cancer therapy by preferentially targeting cancer cells for induction of oxidative stress. We found that the NRF2 inhibitor brusatol triggered increased oxidative stress while compromising viability and proliferation of multiple myeloma cells. Using a repurposing approach we discovered that the Cdc7/CDK9 inhibitor PHA-767491 is also a potent inhibitor of NRF2 transcriptional activity. The molecule was identified by high throughput screening of a library of about 5,900 drug-like molecules. Screening assays included two cell-based assays using HepG2 hepatocellular carcinoma cells: a) A NRF2 nuclear translocation assay, and b) A NRF2 luciferase reporter assay. Validation assays were performed in multiple myeloma cells and included detection of mitochondrial superoxide levels and MTS assays. We found that PHA-767491 treatment of multiple myeloma cells was associated with inhibition of nuclear translocation of NRF2, increased mitochondrial superoxide levels and inhibition of cell growth. Our findings suggest that PHA-767491 is a promising drug candidate for cancer therapy with NRF2 inhibitory potency contributing to its anti-cancer properties.
Keywords: Small molecule inhibitor, high-throughput screening, drug development, reactive oxygen species, Nrf2, cancer
Introduction
Oxidative stress and cellular response mechanisms such nuclear factor, erythroid 2 like 2 (NFE2L2, also known as NRF2)-mediated antioxidant responses play differential roles in healthy and diseased cells[1-3]. Oxidative stress triggers release of Nrf2 from Keap1 and subsequent nuclear translocation of NRF2 where it binds to the antioxidant response element (ARE), an enhancer sequence that regulates cytoprotective genes involved in the oxidative stress response[4-6]. Chemical induction of oxidative stress and subsequent NRF2 activation can be accomplished with numerous agents including tert-butylhydroquinone (t-BHQ)[7, 8] and pyocyanin[9, 10]. In addition to its role as a response mechanism to exogenous stimuli NRF2 plays an important role in the redox homeostasis of cancer cells. Constant generation and elimination of reactive oxygen species (ROS) is a hallmark of many cancer cell types including multiple myeloma (MM)[11, 12], lung cancer[11, 12], and others; this phenomenon is not observed during steady state of healthy cells. In contrast to healthy cells, cancer cells often maintain high levels of ROS to gain a survival advantage[13] and it is because of oxidative stress defense mechanisms such as NRF2 that cancer cells are able to avoid unrestrained accumulation of ROS. Without the cytoprotective safety net provided by NRF2 and other factors, however, the redox state of many cancer cells would be significantly altered, leading to decreased growth factor receptor signaling, genetic instability or even outright cytotoxicity. NRF2 activity in cancer cells has been associated with cancer cell survival, progression, and resistance to chemotherapy[14-16]. Oncogenic genes that have been have been shown to induce NRF2 expression in cancer cells include KRAS, BRAF, and MYC[17]. In the setting of cancer NRF2 initially became of interest as a therapeutic target with the goal to induce its activity for chemoprevention[18], taking advantage of the cytoprotective properties of NRF2 in an attempt to prevent progression of premalignant cells. However, due to its oncogenic potential, NRF2 has also emerged as a promising target in patients with established cancers (especially, but not limited to those with constitutive NRF2 activity), hypothesizing that NRF2 inhibition could work as a strategy to compromise cancer cell survival and sensitize cancer cells to chemotherapy[19-24]. Inhibition of NRF2 activity is expected to result in accumulation of detrimental ROS levels in cancer cells characterized by a high ROS turnover, while sparing healthy cells where basal ROS generation is minimal. Only a few compounds with NRF2 inhibitory potency have been reported in the literature including ascorbic acid, brusatol, trigonelline, chrysin, apigenin, and luteolin[22]. Studies exploring the potential anti-cancer properties of these molecules revealed that brusatol treatment increased the efficacy of chemotherapy in a preclinical model of lung cancer[24] and trigonelline enhanced apoptosis of pancreatic cancer cell lines[25]. However, no NRF2 inhibitor drug has been developed for clinical use to date. The goal of our study was therefore to identify potential NRF2 inhibitor drug candidates from a collection of 5,879 known bioactive molecules by employing a combined high-throughput and high-content screening approach. Moreover, we sought to investigate if ROS-mediated mechanisms mediated by our molecules of interest could be exploited for the treatment of multiple myeloma (MM) cells.
Materials and Methods
Cells
ARE/HepG2/Luciferase reporter cells were purchased from BPS Bioscience (San Diego, CA). The human multiple myeloma cell lines MM.1S and U266 were obtained from ATCC and L363 cells were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ), Department of Human and Animal Cell Cultures (Braunschweig, Germany). HepG2 cells were cultured in MEM medium supplemented with 10% FBS, 1% non-essential amino acids, 1 mM Na-pyruvate, 1% Penicillin/Streptomycin and 600 μg/ml of Geneticin (Life Technologies) in a humidified low oxygen (5% O2) environment with 5% CO2 at 37°C. Multiple myeloma cell lines were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (GIBCO BRL, Gaithersburg, MD), 1% l-glutamine, and penicillin-streptomycin in a humidified environment of 5% CO2 at 37°C. All cell lines were underwent mycoplasma testing at our center in 2015. Cell lines were used for experiments after no more than four passages after thawing.
Antibodies and flow cytometry
A NRF2-AF488 antibody was obtained from Abcam (Cambridge, MA). Cell viability was determined by DAPI (Thermo Fisher Scientific) staining. Flow cytometry was performed with a Fortessa II cytometer (BD Biosciences) and analyzed with FlowJo (TreeStar Software).
Small molecule compounds
We screened a collection of 5,879 known bioactive small molecule compounds and FDA-approved drugs combined from eight libraries, including 1,280 molecules from the LOPAC1280 library (Sigma Aldrich, St. Louis, MO), 1,280 compounds from the Prestwick Chemical Library 2013 (Prestwick Chemicals, Washington, DC), 240 compounds from the Prestwick Chemical Library 2006 (Prestwick Chemicals), 993 compounds from the Selleck Bioactive Compound Library (Selleck Chemicals, Houston, TX), 1,272 compounds from the Tocriscreen Plus Library (Tocris Biosciences, Bristol, UK), 727 compounds from the NIH Clinical Collection (Evotec Inc, Princeton, NJ), 61 compounds from a Signalosome Library (AG Scientific, Cayman Chemical, LC Laboratories, Sigma-Aldrich, Proteinkinase.de), 26 compounds from the Sequoia Library (Sequoia Research Products, Pangbourne, UK).
NRF2 inhibitor high-throughput screen
Compound screening was performed using a 384 well pate format. ARE/Nrf2 transcriptional activity was determined with a luminometer (EnSpire, Perkin Elmer, Waltham, MA) in compound and DMSO-treated HepG2 ARE reporter cells that were stimulated with the known NRF2 inducer tert-butylhydroquinone t-BHQ (Sigma-Aldrich). 7,000 HepG2 cells per well were seeded in 384 well plates and cultured for 24 hours before applying compounds. 200μM or 500μM t-BHQ was added to induce NRF2 translocation into the nucleus 30 minutes after compounds were added and the cells were incubated for 24 hours in a low oxygen (5%) environment. One-Step Luciferase Assay System (BPS Bioscience) was added to the cells 24 hours after incubation and plates were read on an EnSpire plate reader (Perkin Elmer). NRF2 inhibition was expressed as Z-scores that were normalized to the mean of all compound-treated wells. The primary screen was performed with a compound concentration of 5μM and included four data points per compound (200μM or 500μM t-BHQ, two independent runs for each condition). For a compound to be considered a hit at least two out of the four Z-scores had to be lower than −2.0. To rule out false positive results we assessed cell density and morphology of cells treated with molecules of interest to be able to exclude overtly toxic compounds.
NRF2 nuclear translocation high-content screen
2,000 HepG2 cells per well were seeded in 384 well plates and cultured for 24 hours before applying compounds. 200μM or 500μM t-BHQ was added to induce NRF2 translocation into the nucleus 30 minutes after compounds were added and cells were incubated for two hours. Cells were fixed with 4% PFA for 30 minutes followed by permeabilizing/blocking (2% BSA, 0.1% Triton X-100) for 30 minutes. Hoechst 33342 (4ug/mL, Invitrogen, Carlsbad, CA) and a NRF2-Alexa Fluor 488 (1:1000) antibody were used to stain nucleus and NRF2 protein. Plates were imaged with an INCell 6000 (GE Healthcare) and analyzed using Columbus image analysis software (Perkin Elmer). 300,000 MM.1S cells per well were seeded in 96 well plates, DMSO, 2 mM N-Acetyl-L-Cysteine (Sigma-Aldrich) or compounds were added and incubated for 2 hours and then fixed with 4% PFA for 30 minutes followed by permeabilizing/blocking (2% BSA, 0.1% Triton X-100) for 30 minutes. Hoechst 33342 (4ug/mL) and a NRF2-Alexa Fluor 488 antibody (1:1000) were used to stain nucleus and NRF2 protein, before imaging and analysis as described above. NRF2 inhibition was expressed as Z-scores.
MTS assay
Relative cell growth was measured by CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay (Promega) per manufacturer’s instructions. Briefly, tetrazolium compound [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; MTS] was added to the culture media and the conversion of MTS into formazon was measured by the amount of 490nm absorbance using a Synergy HTX plate reader (Biotek Instruments, Inc., Winooski VT).
Detection of mitochondrial superoxide
Cells were seeded into 96 well plates and treated with compounds of interest or DMSO for 24 hours and the levels of mitochondrial superoxide was determined with the MitoSOX Red Mitochondrial Superoxide Indicator, per manufacturer’s instructions (Life Technologies). The fluorescence intensities of cells after treatment were detected by flow cytometry (Fortessa II, BD Bioscience).
Statistics
Data are presented as mean ± SEM. The nonparametric Mann-Whitney U test was used for comparisons between two experimental groups. A P value of less than 0.05 was considered statistically significant.
Z-score calculation: groups of data were normalized to either compounds or DMSO control in primary and confirmation screen, respectively. Median and mad were used for the calculation according to the following formula: x.z-score = (x – median(x[subset]))/mad(x[subset])[26].
Results
NRF2 is a promising therapeutic target in cancer cells
NF-κB and NRF2 represent families of transcription factors that are found ubiquitously in mammalian cells with important roles in inflammation, cellular stress and cancer[20, 27-29]. We hypothesized that inhibition of the NF-κB and/or NRF2-mediated antioxidant response will alter the redox state of cancer cells and have therapeutic potential for cancer treatment. Importantly, a given cancer type could be sensitive to this mechanism of action without the need for mutation or amplification of the target gene as long as a) the cancer cells generate high levels of ROS, and b) the target gene is expressed and contributes to ROS elimination. The potential of NRF2 as a therapeutic target is underscored by its prevalence in a wide range of cancers including both hematologic malignancies and solid tumors (Supplementary Fig. 1). Of note, the majority of tumor samples did not show any NRF2 mutations, and if NRF2 gene mutations were detected they were almost exclusively missense mutations. NRF2 gene amplifications were found in up to seven percent of the tested samples in half of the tumor types (data not shown) but NRF2 deletions were limited to only six tumor types and were overall very rare (0.2 percent of all tested tumor samples, data not shown).
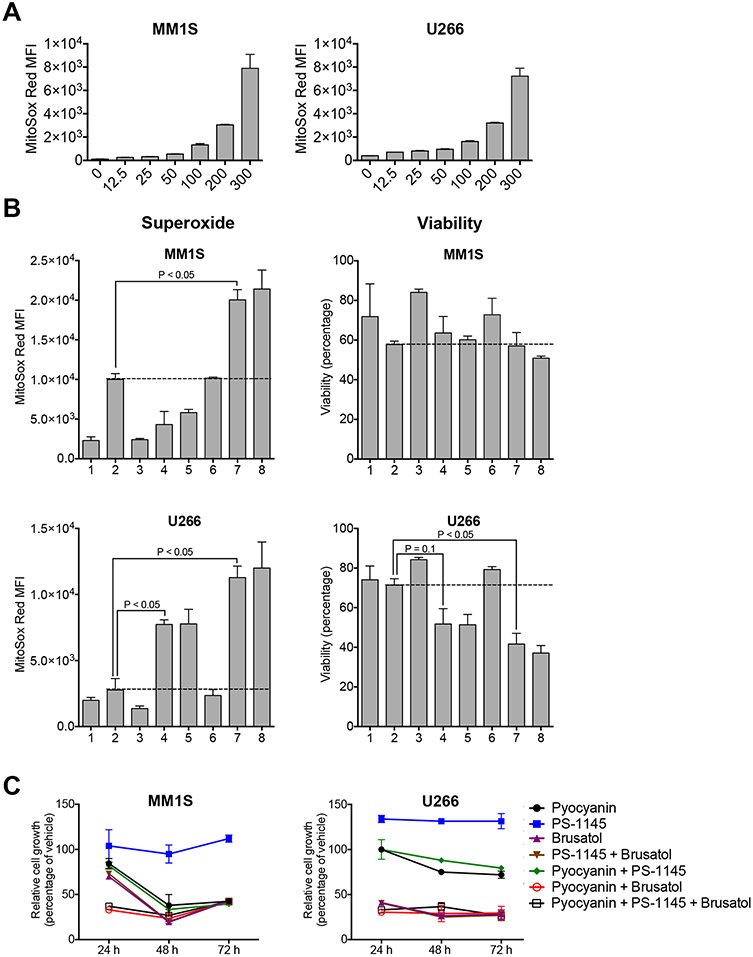
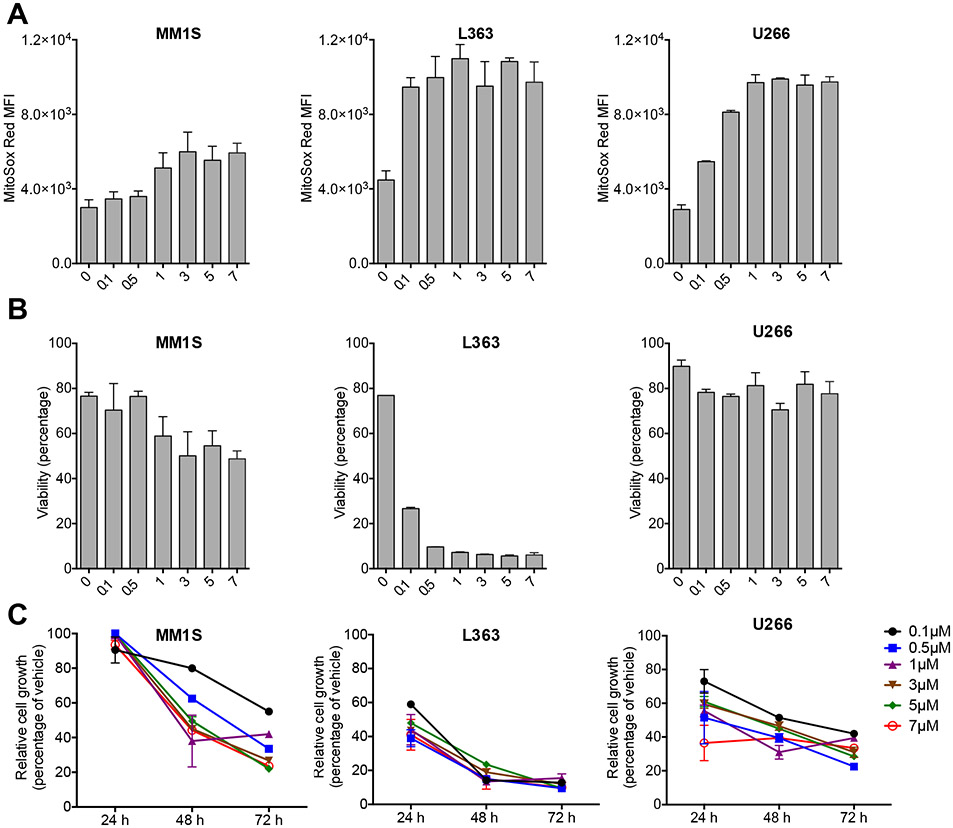
Next, we compared the effect of both NF-κB and NRF2 inhibition on the redox state and viability of MM cells. Pyocyanin treatment resulted in efficient and dose-dependent induction of mitochondrial superoxide in MM.1S and U266 MM cells (Fig. 1a), indicating that MM cells should be useful tools to test therapeutic approaches based on redox modulation. We cultured MM.1S and U266 cells under the following conditions: 1.) Baseline (empty vehicle); 2.) Oxidative stress induction with pyocyanin; 3.) NF-κB inhibition with the IKK inhibitor PS-1145[30]; 4.) NRF2 inhibition with brusatol[23]; 5.) Combination of PS-1145 with brusatol; 6.) Combination of pyocyanin with PS-1145; 7.) Combination of pyocyanin with brusatol; 8.) Combination of pyocyanin with PS-1145 and brusatol. We found that NRF2 inhibition but not NF-κB inhibition led to increased superoxide levels and decreased viability of U266 cells, and combining pyocyanin with brusatol had additive effects in both MM.1S and U266 cells (Fig. 1b). Moreover, single agent NRF2 inhibitor treatment efficiently suppressed growth of MM cells (Fig. 1c). Additional dose titration studies confirmed a reproducible, robust and dose-dependent effect of NRF2 inhibition on the superoxide levels, viability and growth rate of MM cells (Fig. 2).
Figure 1: NRF2 inhibition is more effective than NF-κB inhibition for oxidative stress induction in multiple myeloma cells.
A: MM.1S and U266 multiple myeloma cells were incubated for 24 hours in the presence of serial dilutions (ranging from 0 to 300μM) of Pyocyanin. Mitochondrial superoxide was labeled with MitoSOX Red and analyzed by flow cytometry. Mean and SEM are presented. B: MM.1S and U266 cells were incubated for 24 hours under the following conditions: 1: vehicle (negative control); 2: Pyocyanin 200μM (positive control); 3: PS-1145 5μM; 4: Brusatol 5μM; 5: PS-1145 + Brusatol; 6: Pyocyanin + PS-1145; 7: Pyocyanin + Brusatol; 8: Pyocyanin + PS-1145 + Brusatol. Mitochondrial superoxide levels (MitoSox Red MFI) and viability (percentage of DAPI-negative viable cells) were analyzed by flow cytometry. Mean and SEM are presented. Red lines indicate positive control levels in the presence of Pyocyanin. C: MM.1S and U266 cells were cultured for 72 hours under the conditions described in B. Cell growth was analyzed after 24, 48 and 72 hours by MTS assay. Mean and SEM of relative cell growth (percentage of vehicle control) are presented.
Figure 2: NRF2 inhibition efficiently induces oxidative stress and compromises viability and cell growth of multiple myeloma cells.
A: MM.1S, L363 and U266 multiple myeloma cells were incubated for 24 hours in the presence of serial dilutions (ranging from 0 to 7μM) of Brusatol. Mitochondrial superoxide levels were analyzed by flow cytometry. Mean and SEM are presented. B: MM.1S, L363 and U266 cells were incubated for 24 hours as described in A. Viability (percentage of DAPI-negative viable cells) was analyzed by flow cytometry. Mean and SEM are presented. C: MM.1S, L363 and U266 cells were cultured for 72 hours under the conditions described in A. Cell growth was analyzed after 24, 48 and 72 hours by MTS assay. Mean and SEM of relative cell growth (percentage of vehicle control) are presented.
Discovery of NRF2 inhibitor candidates through drug repurposing
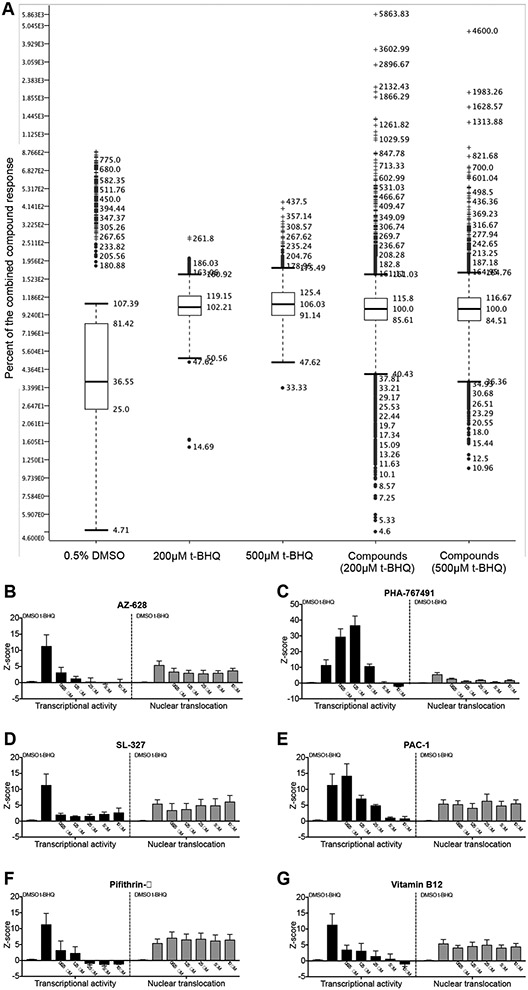
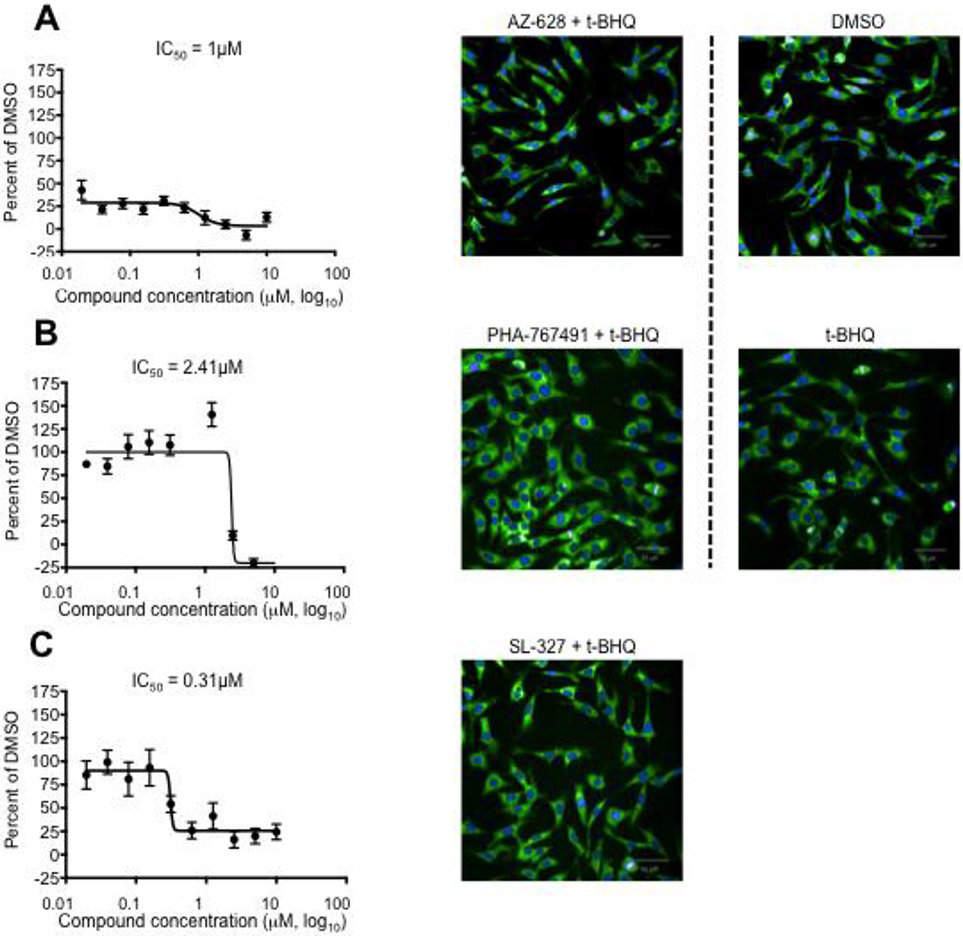
Given the potential of NRF2 inhibition for cancer therapy and the lack of NRF2 inhibitor drugs in clinical practice we set out to identify small molecule NRF2 inhibitor drug candidates by repurposing, a drug development approach that allows to speed up preclinical development since only established compounds with drug-like physicochemical properties and favorable toxicology and pharmacokinetics are included in the screen. Specifically, we performed high-throughput screening (HTS) of 5,879 compounds with known bioactivities using a luciferase assay based on an ARE/NRF2 transcriptional reporter cell line (Supplementary Table 1). We compared DMSO-treated cells (baseline) and t-BHQ-treated cells where NRF2 transcriptional activity is expected to be induced (positive control) with t-BHQ + 5uM compound-treated cells (Fig. 3a). Potential hits will be identified by significantly decreased luminescence intensity compared to t-BHQ + empty vehicle-treated control cells. In addition to reflecting true inhibition of NRF2 activity, decreased luminescence intensity could also be due to cell loss or non-specific cytotoxicity. To rule out the latter we assessed cell density and morphology using an automated microscopic imaging platform (data not shown). The initial screen demonstrated robust induction of NRF2 transcriptional activity of HepG2 cells by t-BHQ and allowed us to select a list of 30 potent and non-toxic NRF2 inhibitor molecules of interest with Z-scores ranging from −2.004 to −3.506 (mean: – 2.527). We confirmed the potency of those compounds using the same transcriptional reporter assay to analyze the effect of five compound concentrations (0.625uM, 1.25uM, 2.5uM, 5uM, 10uM) (Fig. 3b-g (left panels) and data not shown). While the luciferase reporter assay is a convenient screening assay it should be emphasized that the generated data are merely descriptive and cannot be used to support a mechanistic hypothesis with regard to NRF2 inhibition. Possible mechanisms of action include degradation of cytoplasmic NRF2, inhibition of NRF2 DNA binding and inhibition of NRF2 nuclear translocation. We assessed the latter by performing additional high-content screening (HCS) using an automated immunofluorescence assay (Supplementary Table 1) to analyze the ratio of cytoplasmic to nuclear NRF2 (Fig. 3b-g (right panels) and data not shown). The addition of imaging as an independent orthogonal assay does not only offer mechanistic insight but it improves the overall specificity by increasing the likelihood to differentiate false positive from true positive results. Based on the combined transcriptional activity and nuclear translocation data and the expected safety and side effect profile of the tested 30 compounds we were able to further shorten the list of molecules of interest to the following six potential NRF2 inhibitor drug candidates: AZ-628[31], PHA-767491 [32, 33], SL-327[34], PAC-1[35], pifithrin-α[36], and vitamin B12 (Table 1). It should be noted that differences in the ratio of cytoplasmic to nuclear NRF2 were relatively subtle (compared to differences in NRF2 transcriptional activity measured by bioluminescence) but nevertheless reproducible, revealing that three of our six molecules of interest affected nuclear translocation of NRF2 (PHA-767491, AZ-628, SL-327). Furthermore, treatment of MM.1S cells with SL-327 resulted in the anticipated inhibitory effect on nuclear translocation of NRF2, even though the effect was less pronounced than the effect of treatment with the ROS scavenger N-acetyl cysteine (Supplementary Fig. 2). In order to determine the compound concentration required for 50 percent NRF2 inhibition in vitro (IC50) we generated dose response curves for our six molecules of interest based on data generated with the luciferase reporter assay (Fig. 4a-c (left panels) and Supplementary Fig. 3). Fluorescence microscopy confirmed that cell density and morphology was not negatively affected by the tested compounds (Fig. 4a-c (right panels) and data not shown), confirming that we were able to demonstrate inhibition of NRF2 activity at a dose range that was not toxic to HepG2 cells.
Figure 3: Identification of repurposed NRF2 inhibitor compounds by a combined HTS and HCS approach.
A: ARE/HepG2/Luciferase reporter cells were incubated for 24 hours in the presence of 0.5% DMSO, 200μM t-BHQ, 500μM t-BHQ and 5μM of 5,879 compounds + 200μM t-BHQ or 5μM of 5,879 compounds + 500μM t-BHQ. ARE/Nrf2 transcriptional activity was analyzed by measuring bioluminescence of the reporter cells using an EnSpire plate reader. Alterations in bioluminescence signal intensity were expressed as Z-scores and normalized to the measurements obtained from compound-treated cells. Box plots of combined data from two independent experiments are presented. B–G, left panels (transcriptional activity): ARE/HepG2/Luciferase reporter cells were incubated for 24 hours in the presence of 0.5% DMSO, 200μM t-BHQ, or compounds (0.625μM, 1.25μM, 2.5μM, 5μM and 10μM) + 200μM t-BHQ. ARE/NRF2 transcriptional activity was analyzed by bioluminescence and mean + SEM of Z-scores from two independent experiments are presented. B–G, right panels (nuclear translocation): ARE/HepG2/Luciferase reporter cells were incubated for 2 hours in the presence of 0.5% DMSO, 200μM t-BHQ, or compounds (0.625μM, 1.25μM, 2.5μM, 5μM and 10μM) + 200μM t-BHQ. Intracellular NRF2 was detected with a NRF2-specific AF-488 conjugated antibody and nuclei were labeled with Hoechst 33342. Nuclear translocation of NRF2 was quantified using a confocal imaging platform followed by image analysis with Columbus imaging analysis software. Mean and SEM of Z-scores from two independent experiments are presented.
Table 1:
Candidates for NRF2 inhibitor drugs identified by small molecule repurposing.
| Molecule | IC50 in HepG2 ARE/NRF2 reporter assay |
Known bioactivity |
|---|---|---|
| Pifithrin-α | 0.16μM | p53 inhibition |
| SL-327 | 0.31μM | MEK inhibition |
| Vitamin B12 | 0.62μM | Vitamin B12 |
| AZ-628 | 1μM | RAF inhibition |
| PHA-767491 | 2.41μM | Cdc7/Cdk9 kinase inhibition |
| PAC-1 | 2.7μM | Procaspase activation |
Figure 4: AZ-628, PHA-767491 and SL-327 inhibit NRF2 activity in HepG2 cells.
ARE/HepG2/Luciferase reporter cells were incubated for 24 hours in the presence of 200μM t-BHQ and ten-fold serial dilutions of compounds of interest (20nM to 10μM). 0.5% DMSO was used as control. ARE/NRF2 transcriptional activity was analyzed by bioluminescence. Left panels present mean and SEM of relative transcriptional activities (percentage of DMSO control). Microscopic images show density and morphology of HepG2 cells stained with an anti-NRF2-AF488 antibody and Hoechst 33342 under negative control conditions (0.5% DMSO), positive control conditions (200μM t-BHQ), and experimental conditions (5μM compound of interest + 200μM t-BHQ). A: AZ-628. B: PHA-76749. C: SL-327.
The NRF2 inhibitor PHA-767491 induces superoxide and inhibits growth of multiple myeloma cells
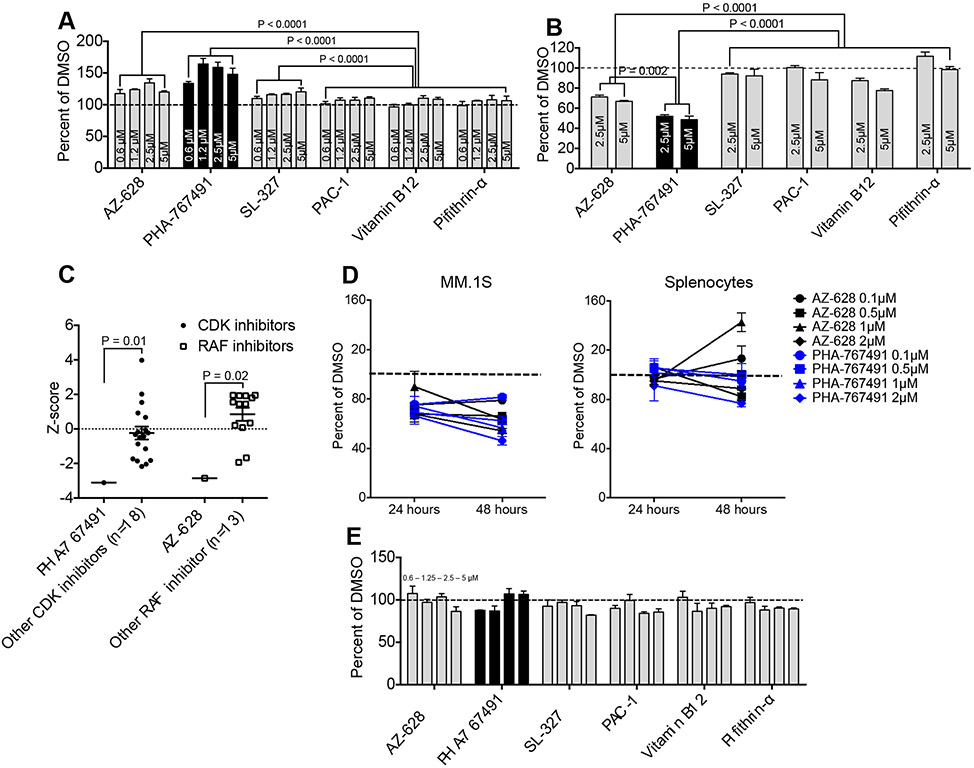
In order to show true promise for cancer therapy a NRF2 inhibitor drug candidate would have to be able to modulate the redox balance of cancer cells in the absence of chemical NRF2 inducers such as t-BHQ, similar to what we were able to demonstrate with the known NRF2 inhibitor brusatol (Fig. 2). We therefore analyzed mitochondrial superoxide levels in MM.1S cells that were treated with DMSO or two-fold serial dilutions of our molecules or interest. We found that PHA-767491 was the most potent superoxide inducer of the six drug candidates, followed by AZ-628 and SL-327 (Fig. 5a). MTS assays revealed that treatment of MM.1S cells with the leading superoxide inducers (PHA-767491 and AZ-628) resulted in significant inhibition of cell growth (Fig. 5b), consistent with the concept of a threshold effect of ROS-mediated cytotoxicity. Of note, PHA-767491 and AZ-628 have previously been reported as potential as cancer therapeutics based on their capacity to inhibit Cdc7/CDK9[32, 33] or RAF[31], respectively. Given that RAF is upstream of NRF2 and implicated in its induction we hypothesized that the observed NRF2 inhibitory effect of AZ-628 may be a secondary effect of RAF inhibition. We therefore compared inhibition of NRF2 transcriptional activity by AZ-628 with other RAF inhibitors (as well as by PHA-767491 versus other CDK inhibitors), confirming that the NRF2 inhibitory potency of PHA-767491 and AZ-628 was significantly superior to the mean potency of the tested reference molecules (Fig. 5c). This suggests that NRF2 inhibition is a unique feature of our identified molecules and not simply a secondary effect of an already known bioactivity. Additional MTS assay-based in vitro efficacy studies comparing MM.1S cells and normal mouse splenocytes demonstrated a dose-dependent and cancer cell-selective effect on cell growth of both PHA-767491 and AZ-628 (Fig. 5d). Consistent with these results we did not observe any meaningful effects of our molecules of interest on superoxide levels of normal splenocytes, reaffirming the notion that NRF2 inhibition is not expected to alter the redox homeostasis of normal cells during steady state conditions (Fig. 5e).
Figure 5: NRF2 inhibitors induce increased oxidative stress and decreased growth of multiple myeloma cells.
A: MM.1S cells were incubated for 24 hours in the presence of 0.6, 1.25, 2.5 and 5μM of NRF2 inhibitor molecules of interest. DMSO was used as control. Mitochondrial superoxide was labeled with MitoSOX Red and analyzed by flow cytometry. Mean and SEM are presented. Dotted line represents the baseline. B: MM.1S were cultured for 48 hours in the presence of empty vehicle or NRF2 inhibitor compounds of interest (2.5μM and 5μM). Cell growth was analyzed by MTS assay. Mean and SEM of relative cell growth (percentage of vehicle) are presented. Dotted line represents baseline cell growth (100 percent of vehicle). C: ARE/HepG2/Luciferase reporter cells were incubated for 24 hours in the presence of 200μM t-BHQ and 5μM of various CDK inhibitors or RAF inhibitors. ARE/NRF2 transcriptional activity was analyzed by bioluminescence. Scatter plot depicts Z-scores of PHA-767491 versus all other CDK inhibitors (n=18) and of AZ-628 versus all other RAF inhibitors (n=13). D: MM.1S and splenocytes from 8 week-old female C57BL/6 mice (Jackson Laboratories) were cultured for 48 hours in the presence of empty vehicle (DMSO) or 0.1, 0.5, 1 and 2μM of AZ-628 or PHA-767491. Cell growth was analyzed after 24 and 48 hours by MTS assay. Mean and SEM of relative cell growth (percentage of vehicle) are presented. Dotted line represents baseline cell growth. E: Splenocytes from 8 week-old female C57BL/6 mice (Jackson Laboratories) were incubated for 24 hours in the presence of 0.6, 1.25, 2.5 and 5μM of NRF2 inhibitor molecules of interest. DMSO was used as control. Mitochondrial superoxide was labeled with MitoSOX Red and analyzed by flow cytometry. Mean and SEM are presented. Dotted line represents the baseline.
Discussion
NRF2-mediated activation of the antioxidant response element is a key component of the cellular oxidative stress response while NF-κB is known to play a lesser role. The ambivalent interactions between ROS and the NF-κB can at least in part be attributed to the fact that ROS have been implicated in inhibition of NF-κB activity through oxidation of cysteine residues resulting in thiol modifications of DNA binding domains of NF-κB subunits, among others[37, 38]. Similar interactions have not been reported for the NRF2 pathway where ROS exclusively serve as inducers of transcriptional activity. Targeting NRF2 should therefore be an efficient approach for the development of therapeutic strategies designed to exploit ROS-mediated effects to treat cancer. NRF2 is a particularly attractive therapeutic target in this setting since it is expressed in a wide range of cancers but gene alterations are relatively infrequent and limited to mostly amplifications and missense mutations instead of deletions or nonsense mutations. This indicates that while cancers with the highest NRF2 expression levels may be particularly sensitive to NRF2 inhibition, the concept of NRF2 inhibition has potential to be applicable to a great variety of malignancies.
This study focused on multiple myeloma cells to demonstrate feasibility for NRF2 inhibition as an effective and safe therapeutic strategy. Our findings revealed that NRF2 inhibition modulates the redox homeostasis of MM cells resulting in increased oxidative stress and dose-dependent inhibition of cell growth, which is consistent with the threshold concept of ROS-mediated cytotoxicity[2, 3]. Furthermore, therapeutic effects of NRF2 inhibition in the setting of cancer have been associated with modulation of redox switches [15] and with regulation of serine biosynthesis[39, 40], implying that the anti-cancer effects of NRF2 inhibition may be complex and could involve a diverse set of mechanistic foundations. Importantly, the effects of NRF2 inhibition on ROS levels and cell growth/viability in our studies were selective to cancer cells while sparing normal control cells. This observation is in line with our previous findings regarding NF-κB inhibition in lymphoma cells[41] and supportive of mechanistic models predicting that inhibition of ROS elimination can result in cytotoxicity in cancer cells well before healthy cells will be affected[2, 3].
Since NRF2 inhibition has significant therapeutic potential and given the lack of NRF2 inhibitor drugs in clinical practice we sought to identify candidates for an NRF2 inhibitor drug by repurposing of drug-like small molecules with known bioactivity. Our three-step screening approach (summarized in Supplementary Fig. 4) resulted in the identification of PHA-767491 as our primary drug candidate while AZ-628 was selected as backup molecule. Both compounds inhibited nuclear translocation and transcriptional activity of NRF2 in a dose-dependent fashion, resulting in increased superoxide levels and inhibited cell growth of MM cells. Efficacy testing in additional cancer cell types was beyond the scope of this study but we anticipate that our NRF2 inhibitor drug candidates are characterized by broad anti-cancer activity, which will likely be enhanced by the previously described mechanisms of action of PHA-767491 and AZ-628 through Cdc7/CDK9 inhibition[32, 33] and RAF inhibition[31], respectively.
Taken together, our findings indicate that NRF2 inhibition mediated by PHA-767491 is robust, specific, and at least in part due to inhibition of nuclear translocation of NRF2. On the other hand it is evident that our drug candidates possess anti-cancer properties independent from NRF2 inhibition. Additional studies will be needed to decipher the differential contributions of NRF2 inhibition versus inhibition of other pathways, but it would be reasonable to expect that combined inhibition of Cdc7/CDK9 and NRF2 will result in additive effects and therapeutic synergy. Of note, the same dose range of PHA-767491 that was found to be effective in our study was previously reported to inhibit proliferation of human cancer cells via Cdc7/CDK9 inhibition[32, 33], suggesting that PHA-767491-mediated effects on cancer cell survival and proliferation may well be due to its combined net inhibitory activity on both Cdc7/CDK9 and NRF2. Investigations into relationships between multiple bioactivities and efficacy of PHA-767491 should therefore ideally be pursued in parallel with translational studies furthering clinical drug development.
Supplementary Material
Acknowledgements
This research was supported by K08CA160659 and grants from Leukemia and Lymphoma Society (6465-15) and Hyundai Hope on Wheels.
Footnotes
Conflict of Interest: The authors do not have any conflicts of interest to declare.
All studies involving animals were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) protocol 16-07-007 of Memorial Sloan Kettering Cancer Center. Memorial Sloan Kettering Cancer Center is designated as an institution with Public Health Services (PHS) approved Animal Welfare Assurance (A3311-01).
This article does not contain any studies with human participants performed by any of the authors.
References
- 1.Lee JM, Johnson JA (2004) An important role of Nrf2-ARE pathway in the cellular defense mechanism. Journal of biochemistry and molecular biology 37: 139–143 [DOI] [PubMed] [Google Scholar]
- 2.Gorrini C, Harris IS, Mak TW (2013) Modulation of oxidative stress as an anticancer strategy. Nature reviews Drug discovery 12: 931–947. DOI 10.1038/nrd4002 [DOI] [PubMed] [Google Scholar]
- 3.Trachootham D, Alexandre J, Huang P (2009) Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nature reviews Drug discovery 8: 579–591. DOI 10.1038/nrd2803 [DOI] [PubMed] [Google Scholar]
- 4.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD (2003) Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. The Biochemical journal 374: 337–348. DOI 10.1042/bj20030754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rushmore TH, Morton MR, Pickett CB (1991) The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. The Journal of biological chemistry 266: 11632–11639 [PubMed] [Google Scholar]
- 6.He X, Ma Q (2009) NRF2 cysteine residues are critical for oxidant/electrophile-sensing, Kelch-like ECH-associated protein-1-dependent ubiquitination-proteasomal degradation, and transcription activation. Molecular pharmacology 76: 1265–1278. DOI 10.1124/mol.109.058453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imhoff BR, Hansen JM (2010) Tert-butylhydroquinone induces mitochondrial oxidative stress causing Nrf2 activation. Cell Biol Toxicol 26: 541–551. DOI 10.1007/s10565-010-9162-6 [DOI] [PubMed] [Google Scholar]
- 8.Gharavi N, Haggarty S, El-Kadi AO (2007) Chemoprotective and carcinogenic effects of tert-butylhydroquinone and its metabolites. Curr Drug Metab 8: 1–7 [DOI] [PubMed] [Google Scholar]
- 9.Muller M (2002) Pyocyanin induces oxidative stress in human endothelial cells and modulates the glutathione redox cycle. Free radical biology & medicine 33: 1527–1533 [DOI] [PubMed] [Google Scholar]
- 10.Liu K, Wang X, Sha K, Zhang F, Xiong F, Wang X, Chen J, Li J, Churilov LP, Chen S, Wang Y, Huang N (2017) Nuclear protein HMGN2 attenuates pyocyanin-induced oxidative stress via Nrf2 signaling and inhibits Pseudomonas aeruginosa internalization in A549 cells. Free radical biology & medicine 108: 404–417. DOI 10.1016/j.freeradbiomed.2017.04.007 [DOI] [PubMed] [Google Scholar]
- 11.Nerini-Molteni S, Ferrarini M, Cozza S, Caligaris-Cappio F, Sitia R (2008) Redox homeostasis modulates the sensitivity of myeloma cells to bortezomib. British journal of haematology 141: 494–503. DOI 10.1111/j.1365-2141.2008.07066.x [DOI] [PubMed] [Google Scholar]
- 12.Esme H, Cemek M, Sezer M, Saglam H, Demir A, Melek H, Unlu M (2008) High levels of oxidative stress in patients with advanced lung cancer. Respirology (Carlton, Vic) 13: 112–116. DOI 10.1111/j.1440-1843.2007.01212.x [DOI] [PubMed] [Google Scholar]
- 13.Moloney JN, Cotter TG (2017) ROS signalling in the biology of cancer. Seminars in cell & developmental biology. DOI 10.1016/j.semcdb.2017.05.023 [DOI] [PubMed] [Google Scholar]
- 14.Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL (2013) The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox biology 1: 45–49. DOI 10.1016/j.redox.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chio II, Jafarnejad SM, Ponz-Sarvise M, Park Y, Rivera K, Palm W, Wilson J, Sangar V, Hao Y, Ohlund D, Wright K, Filippini D, Lee EJ, Da Silva B, Schoepfer C, Wilkinson JE, Buscaglia JM, DeNicola GM, Tiriac H, Hammell M, Crawford HC, Schmidt EE, Thompson CB, Pappin DJ, Sonenberg N, Tuveson DA (2016) NRF2 Promotes Tumor Maintenance by Modulating mRNA Translation in Pancreatic Cancer. Cell 166: 963–976. DOI 10.1016/j.cell.2016.06.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Syu JP, Chi JT, Kung HN (2016) Nrf2 is the key to chemotherapy resistance in MCF7 breast cancer cells under hypoxia. Oncotarget 7: 14659–14672. DOI 10.18632/oncotarget.7406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, Scrimieri F, Winter JM, Hruban RH, Iacobuzio-Donahue C, Kern SE, Blair IA, Tuveson DA (2011) Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475: 106–109. DOI 10.1038/nature10189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwak MK, Kensler TW (2010) Targeting NRF2 signaling for cancer chemoprevention. Toxicology and applied pharmacology 244: 66–76. DOI 10.1016/j.taap.2009.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, Wong PK, Zhang DD (2008) Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 29: 1235–1243. DOI 10.1093/carcin/bgn095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaramillo MC, Zhang DD (2013) The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes & development 27: 2179–2191. DOI 10.1101/gad.225680.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao S, Wang S, Moghaddam SJ, Ooi A, Chapman E, Wong PK, Zhang DD (2014) Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer research 74: 7430–7441. DOI 10.1158/0008-5472.can-14-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.No JH, Kim YB, Song YS (2014) Targeting nrf2 signaling to combat chemoresistance. Journal of cancer prevention 19: 111–117. DOI 10.15430/jcp.2014.19.2.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, Toppin HA, Zhang DD (2011) Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proceedings of the National Academy of Sciences of the United States of America 108: 1433–1438. DOI 10.1073/pnas.1014275108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olayanju A, Copple IM, Bryan HK, Edge GT, Sison RL, Wong MW, Lai ZQ, Lin ZX, Dunn K, Sanderson CM, Alghanem AF, Cross MJ, Ellis EC, Ingelman-Sundberg M, Malik HZ, Kitteringham NR, Goldring CE, Park BK (2015) Brusatol provokes a rapid and transient inhibition of Nrf2 signaling and sensitizes mammalian cells to chemical toxicity-implications for therapeutic targeting of Nrf2. Free radical biology & medicine 78: 202–212. DOI 10.1016/j.freeradbiomed.2014.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arlt A, Sebens S, Krebs S, Geismann C, Grossmann M, Kruse ML, Schreiber S, Schafer H (2013) Inhibition of the Nrf2 transcription factor by the alkaloid trigonelline renders pancreatic cancer cells more susceptible to apoptosis through decreased proteasomal gene expression and proteasome activity. Oncogene 32: 4825–4835. DOI 10.1038/onc.2012.493 [DOI] [PubMed] [Google Scholar]
- 26.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R (2006) Statistical practice in high-throughput screening data analysis. Nature biotechnology 24: 167–175. DOI 10.1038/nbt1186 [DOI] [PubMed] [Google Scholar]
- 27.Lawrence T (2009) The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harbor perspectives in biology 1: a001651. DOI 10.1101/cshperspect.a001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espinosa-Diez C, Miguel V, Mennerich D, Kietzmann T, Sanchez-Perez P, Cadenas S, Lamas S (2015) Antioxidant responses and cellular adjustments to oxidative stress. Redox biology 6: 183–197. DOI 10.1016/j.redox.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiDonato JA, Mercurio F, Karin M (2012) NF-kappaB and the link between inflammation and cancer. Immunol Rev 246: 379–400. DOI 10.1111/j.1600-065X.2012.01099.x [DOI] [PubMed] [Google Scholar]
- 30.Hideshima T, Chauhan D, Richardson P, Mitsiades C, Mitsiades N, Hayashi T, Munshi N, Dang L, Castro A, Palombella V, Adams J, Anderson KC (2002) NF-kappa B as a therapeutic target in multiple myeloma. The Journal of biological chemistry 277: 16639–16647. DOI 10.1074/jbc.M200360200 [DOI] [PubMed] [Google Scholar]
- 31.Khazak V, Astsaturov I, Serebriiskii IG, Golemis EA (2007) Selective Raf inhibition in cancer therapy. Expert opinion on therapeutic targets 11: 1587–1609. DOI 10.1517/14728222.11.12.1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montagnoli A, Valsasina B, Croci V, Menichincheri M, Rainoldi S, Marchesi V, Tibolla M, Tenca P, Brotherton D, Albanese C, Patton V, Alzani R, Ciavolella A, Sola F, Molinari A, Volpi D, Avanzi N, Fiorentini F, Cattoni M, Healy S, Ballinari D, Pesenti E, Isacchi A, Moll J, Bensimon A, Vanotti E, Santocanale C (2008) A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nature chemical biology 4: 357–365. DOI 10.1038/nchembio.90 [DOI] [PubMed] [Google Scholar]
- 33.Natoni A, Murillo LS, Kliszczak AE, Catherwood MA, Montagnoli A, Samali A, O'Dwyer M, Santocanale C (2011) Mechanisms of action of a dual Cdc7/Cdk9 kinase inhibitor against quiescent and proliferating CLL cells. Mol Cancer Ther 10: 1624–1634. DOI 10.1158/1535-7163.mct-10-1119 [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Wang H, Xu L, Rozanski DJ, Sugawara T, Chan PH, Trzaskos JM, Feuerstein GZ (2003) Significant neuroprotection against ischemic brain injury by inhibition of the MEK1 protein kinase in mice: exploration of potential mechanism associated with apoptosis. The Journal of pharmacology and experimental therapeutics 304: 172–178. DOI 10.1124/jpet.102.040246 [DOI] [PubMed] [Google Scholar]
- 35.Putt KS, Chen GW, Pearson JM, Sandhorst JS, Hoagland MS, Kwon JT, Hwang SK, Jin H, Churchwell MI, Cho MH, Doerge DR, Helferich WG, Hergenrother PJ (2006) Small-molecule activation of procaspase-3 to caspase-3 as a personalized anticancer strategy. Nature chemical biology 2: 543–550. DOI 10.1038/nchembio814 [DOI] [PubMed] [Google Scholar]
- 36.Strom E, Sathe S, Komarov PG, Chernova OB, Pavlovska I, Shyshynova I, Bosykh DA, Burdelya LG, Macklis RM, Skaliter R, Komarova EA, Gudkov AV (2006) Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nature chemical biology 2: 474–479. DOI 10.1038/nchembio809 [DOI] [PubMed] [Google Scholar]
- 37.Morgan MJ, Liu ZG (2011) Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell research 21: 103–115. DOI 10.1038/cr.2010.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bubici C, Papa S, Dean K, Franzoso G (2006) Mutual cross-talk between reactive oxygen species and nuclear factor-kappa B: molecular basis and biological significance. Oncogene 25: 6731–6748. DOI 10.1038/sj.onc.1209936 [DOI] [PubMed] [Google Scholar]
- 39.DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, Wistuba II, Minna JD, DeBerardinis RJ, Cantley LC (2015) NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nature genetics 47: 1475–1481. DOI 10.1038/ng.3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amelio I, Cutruzzola F, Antonov A, Agostini M, Melino G (2014) Serine and glycine metabolism in cancer. Trends in biochemical sciences 39: 191–198. DOI 10.1016/j.tibs.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shono Y, Tuckett AZ, Liou HC, Doubrovina E, Derenzini E, Ouk S, Tsai JJ, Smith OM, Levy ER, Kreines FM, Ziegler CG, Scallion MI, Doubrovin M, Heller G, Younes A, O'Reilly RJ, van den Brink MR, Zakrzewski JL (2016) Characterization of a c-Rel Inhibitor That Mediates Anticancer Properties in Hematologic Malignancies by Blocking NF-kappaB-Controlled Oxidative Stress Responses. Cancer research 76: 377–389. DOI 10.1158/0008-5472.can-14-2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.