Abstract
Human cytomegalovirus (HCMV) infection is associated with renal allograft failure. Allograft damage in animal models is accelerated by CMV‐induced T helper 17 (Th17) cell infiltrates. However, the mechanisms whereby CMV promotes Th17 cell‐mediated pathological organ inflammation are uncharacterized. Here we demonstrate that murine CMV (MCMV)‐induced intragraft Th17 cells have a Th1/17 phenotype co‐expressing IFN‐γ and/or TNF‐α, but only a minority of these cells are MCMV specific. Instead, MCMV promotes intragraft expression of CCL20 and CXCL10, which are associated with recruitment of CCR6+CXCR3+ Th17 cells. MCMV also enhances Th17 cell infiltrates after ischemia–reperfusion injury, independent of allogeneic responses. Pharmacologic inhibition of the Th17 cell signature cytokine, IL‐17A, ameliorates MCMV‐associated allograft damage without increasing intragraft viral loads or reducing MCMV‐specific Th1 cell infiltrates. Clinically, HCMV DNAemia is associated with higher serum IL‐17A among renal transplant patients with acute rejection, linking HCMV reactivation with Th17 cell cytokine expression. In summary, CMV promotes allograft damage via cytokine‐mediated Th1/17 cell recruitment, which may be pharmacologically targeted to mitigate graft injury while preserving antiviral T cell immunity.
Keywords: basic (laboratory) research/science; clinical research/practice; kidney transplantation/nephrology; rejection: acute, cytokines/cytokine receptors; chemokines/chemokine receptors; infection and infectious agents—viral: cytomegalovirus (CMV)
Short abstract
The authors show that urine cytomegalovirus infection promotes cytokine‐induced recruitment of Th1/17 cells into acutely rejecting murine renal allografts and that pharmacologic inhibition of IL‐17A ameliorates allograft damage without increasing intragraft viral loads.
Abbreviations
- ABMR
antibody‐mediated rejection
- ACR
acute cellular rejection
- AR
acute rejection
- CLIP
class II‐associated invariant chain peptide
- FOXP3
Forkhead Box P3
- HCMV
human cytomegalovirus
- IPA
ingenuity pathway analysis
- IRI
ischemia–reperfusion injury
- MCMV
murine cytomegalovirus
- OVA
ovalbumin
- PMA
phorbol 12‐myristate 13‐acetate
- RORγt
RAR‐related orphan receptor gamma t
1. INTRODUCTION
Human cytomegalovirus (HCMV) is a ubiquitous virus with seroprevalence of 50–98% for adult populations worldwide. 1 , 2 , 3 , 4 Among healthy individuals, HCMV establishes latency with intermittent asymptomatic reactivations that are controlled by antiviral T cell responses. 5 , 6 , 7 CMV‐specific T helper 1 (Th1) cells and cytotoxic CD8+ T cells are necessary for control of CMV infections in immunocompromised transplant and HIV‐infected patients, and in animal models of murine CMV (MCMV) infection. 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 HCMV can cause viral disease including pneumonitis, hepatitis, colitis, retinitis, or encephalitis in solid organ and hematopoietic cell transplant patients. 16 , 17 HCMV also increases risk for rejection and allograft failure by unknown mechanisms. 18 , 19 , 20 , 21 , 22 , 23 , 24 To prevent HCMV disease after transplant, short‐term valganciclovir prophylaxis or pre‐emptive treatment at DNAemia onset can be given. 16 Antiviral prophylaxis is associated with lower graft loss rates compared to pre‐emptive treatment, suggesting that viral reactivation contributes to graft failure. 25 However, valganciclovir frequently causes neutropenia, preventing long‐term prophylactic use, so adjunctive interventions to mitigate HCMV induced organ damage deserve further exploration. 26 , 27
In rodent renal transplant models, rat CMV and MCMV infection exacerbate allograft inflammation and accelerate fibrosis. 28 , 29 , 30 Similar to clinical observations, valganciclovir prophylaxis in murine transplants inhibits viral reactivation and improves late allograft fibrosis. 31 , 32 Recently, our group showed that T helper 17 (Th17) cells are more abundant in MCMV‐infected than uninfected allografts, and that IL‐6 inhibition reduces Th17 cell infiltrates and histopathologic allograft injury. 30 In animal models and clinical organ transplantation, Th17 cells, IL‐17A, and CCL20 are elevated in allografts, blood, and urine during acute rejection and are associated with late graft failure, but HCMV serostatus and DNAemia were not assessed in these studies. 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 Thus, a link between HCMV infection and Th17 cells in transplant rejection has not yet been described in the clinical literature.
Th17 cells have known effector functions against extracellular pathogens, mucoepithelial infections, and intracellular bacteria but are not typically thought to be essential for antiviral immune responses. 43 , 44 However, Th17 cells do promote pathological organ inflammation during experimental and clinical viral infections. 45 , 46 , 47 , 48 , 49 , 50 Mechanisms by which viruses facilitate Th17 cell activity are not well understood. Furthermore, although HCMV and MCMV‐specific Th17 cells have been described, their role in viral‐induced inflammation has not been reported. 51 , 52 In this study, we investigated the antigen specificity of MCMV‐induced Th17 cells in renal allografts, the MCMV‐induced microenvironment promoting their generation, and the impact of IL‐17A inhibition upon viral loads, T cell subsets and organ damage. The clinical relevance of animal model findings was examined among a cohort of renal transplant patients by comparing serum IL‐17A quantities between groups with and without HCMV DNAemia.
2. MATERIALS AND METHODS
2.1. Mice and virus
BALB/c, C57BL/6 (B6), C57BL/6‐IL‐17A tm1Bcgen (IL‐17A‐GFP), and B6.Cg‐Tg(TcraTcrb)425Cbn (OT‐II) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Donor and recipient mice were infected with MCMVΔm157 at 1x106 plaque forming units (pfu) at least 12 weeks prior to transplantation to clear acute systemic infection, establish donor organ infection, and generate recipient MCMV‐specific T cells, as previously described. 30 , 51 , 53 , 54
2.2. Murine renal transplantation
Renal transplantation was performed as described, using male and female BALB/c or B6 donors (D) and C57BL/6, IL‐17a‐GFP, or OT‐II recipients (R). 30 , 55 Native kidneys were retained, and recipients were treated with cyclosporine at 10 mg/kg/day, subcutaneously once daily until terminal sacrifice. 30 , 31 , 55
2.3. Intracellular cytokine staining (ICS)/flow cytometry
Splenocytes, blood, and intragraft leukocytes were isolated from mice, unstimulated or stimulated with either PMA‐ionomycin or 5 μg/ml of MCMV peptide pool (Table S1) and stained for flow cytometry as previously described (Methods S.1, S.2; Table S2). 30
2.4. RNA‐Seq analysis
Details are provided in Methods S.3. In brief, total RNA was isolated from allografts or native kidneys, cDNA libraries generated, and paired end 150 base pair sequencing performed using the Illumina HiSeq 4000. Gene expression between groups was compared using Qiagen Ingenuity Pathway Analysis using normalized read counts.
2.5. Cytokine and chemokine detection
Mouse cytokines and chemokines were quantitated in grafts and spleens using LEGENDplex panels and analyzed with LEGENDplex Data Analysis Software, V8.0 (Biolegend). 30 Cytokine and chemokine levels were depicted as picograms/gram (pg/g) of tissue.
2.6. MCMV viral load quantitation
MCMV viral loads were quantified as previously described. 30 (Methods S.4), except that DNA was extracted from blood and tissues using the Zymo Viral DNA extraction kit (Zymo Research).
2.7. In vivo IL‐17A neutralization and Tregs depletion
Mice were treated with αIL‐17A (Clone 17F3) or αCD25 (Clone PC61) antibodies to deplete IL‐17A or Tregs, respectively (Methods S.5). Kidneys, spleens, and blood were isolated for flow cytometry and histology. Histopathology was scored by a veterinary nephropathologist (R.C.) blinded to sample identity using a previously published grading scale, 30 modified to reflect contemporary rejection criteria (Table S3). 56 , 57 Scores (0–3) were assigned for 12 histopathologic criteria, for a maximum score of 36.
2.8. Human renal transplant study
A retrospective cohort study was conducted to determine the association between blood mRNA RORγt:FOXP3 ratio and acute renal allograft rejection, using clinical data and samples derived from a previously completed IRB‐approved prospective observational study conducted at PGIMER in India (Methods S.6). 58 , 59 , 60 Acute rejection (AR) cases were matched by age and sex with controls without AR. Blood RNA was analyzed by RT‐PCR for RORγt, FOXP3, and β‐actin expression (Methods S.7; Table S4). Serum cytokines were measured using the human Th1/Th2/Th17 cytometric bead array kit. Blood HCMV viral loads were quantified by DNA PCR (Methods S.8).
2.9. Statistical analysis
Patient characteristics were described using ranges and frequency distributions or mean ± standard deviation (SD) unless otherwise indicated. Continuous variables were analyzed using the two‐tailed Student's t‐test or the one‐way analysis of variance. Categorical variables were compared using the chi‐square test. Statistical analyses were performed using GraphPad Prism 8 (San Diego, CA) accepting statistical significance at p value <.05.
3. RESULTS
3.1. MCMV infection induces graft‐infiltrating Th1/17 cells
Th17 cells infiltrate both MCMV D+R− and D+R+ allografts at day 14 post‐transplantation. 30 To distinguish virus‐specific from non‐viral Th17 cells, R+ recipients with pretransplant MCMV‐specific T cell immunity were analyzed for these studies and compared to D−R− recipients without MCMV infection. To characterize events prior to day 14, D+R+ and D−R− transplants were compared at day 7. D+R+ allografts had significantly higher frequencies of Th17 cells expressing IL‐17A alone or with Th1 cytokines, IFN‐γ, and/or TNF‐α (Figure 1A–C; Figure S1, S2). IL‐17A and IFN‐γ cytokine concentrations were higher in D+R+ than D−R− grafts (Figure 1D). Of the IL‐17A expressing CD4+, CD8+, and MHCII+ cells, only Th17 cells were more abundant in D+R+ allografts (Figure 1E). D+R+ allografts had significantly higher frequencies of Th17 cells and higher quantities of IL‐17A, IFN‐γ, and TNF‐α compared to spleens (Figure 1F,G). Together, these results indicate that MCMV infection induces intragraft infiltration of Th17 cells with a Th1/17 profile, which are associated with higher intragraft quantities of IL‐17A and IFN‐γ.
FIGURE 1.
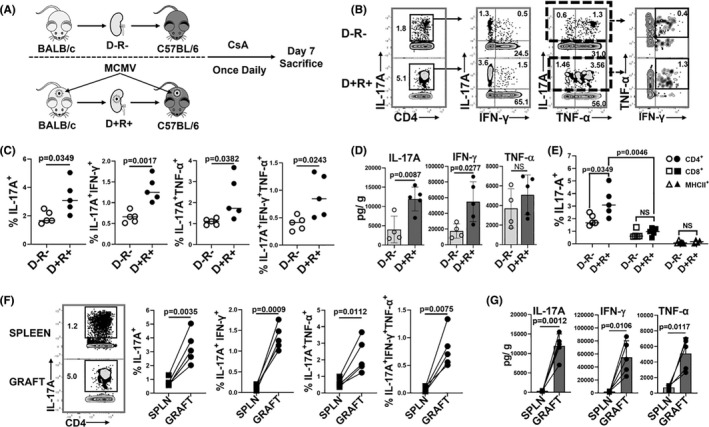
MCMV induces intragraft infiltration of Th17 cells co‐expressing Th1 cytokines. (A) Study design. Renal transplantation was performed between BALB/c and C57BL/6 mice with cyclosporine A (CsA) subcutaneous injections given daily for posttransplant immunosuppression. On day 7, splenocytes and intragraft leukocytes were analyzed by flow cytometry. (B) Representative flow cytometry plots showing frequencies of CD4+IL‐17A+ (Th17) cells and cytokine expression profiles. (C) Frequencies of cytokine‐expressing Th17 cell subsets were compared for D−R− and D+R+ allografts. (D) Intragraft quantities of IL‐17A, IFN‐γ, and TNF‐α (picogram per gram tissue, pg/g) in D−R− and D+R+ allografts. (E) Frequencies of IL‐17A expressing CD4+, CD8+, and MHC‐II+ leukocytes in D−R− and D+R+ allografts. (F) Representative flow plots and frequencies of Th17 cells in D+R+ spleens (SPLN) and allografts (GRAFT). (G) Cytokine quantities (pg/g) in D+R+ spleens and allografts. All data are represented as mean ± standard deviation (SD) and analyzed by two‐sided Student's t‐test. NS, not significant (p > .05)
3.2. MCMV antigen‐specific Th17 cell cytokine expression differs from antigen‐independent Th17 cells
To define the temporal kinetic of MCMV‐specific Th17 cells after primary infection, splenocytes from MCMV‐infected B6 mice were analyzed for MCMV‐specific Th17 cell frequencies at days 0, 7, 14, 21, and 28 (Figure 2A). MCMV‐specific Th17 cells comprised 13.55% (±4.67%) of total Th17 cells at day 7 (Figure 2B) and rapidly contracted to baseline levels from day 14 onward. In D+R+ allografts (Figure 2C–E), MCMV‐specific Th17 cells comprised only 5.48 ± 0.97% of total graft‐infiltrating Th17 cells (Figure 2D) and predominantly expressed IL‐17A alone or with IFN‐γ, but not TNF‐α (Figure 2E).
FIGURE 2.
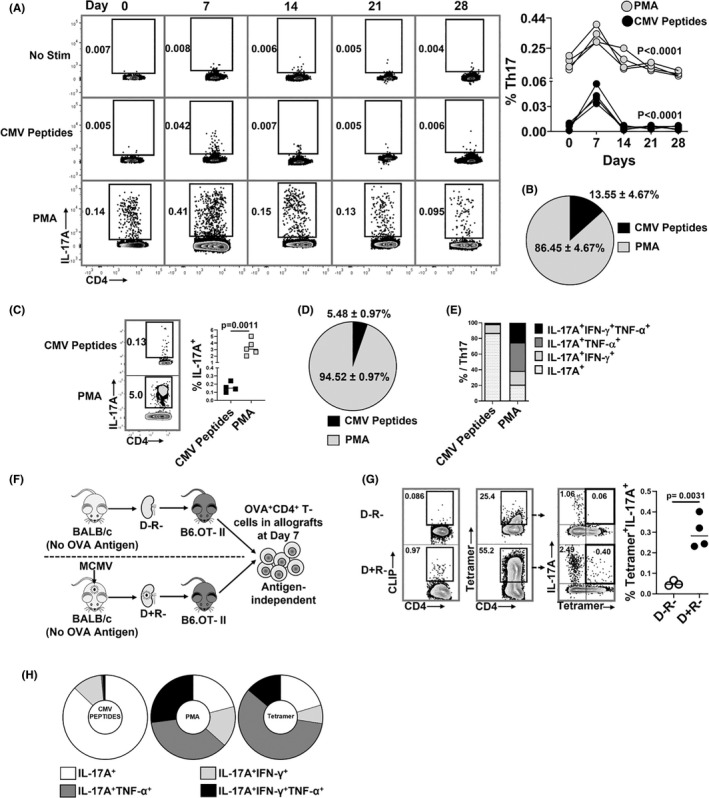
MCMV‐specific and antigen‐independent Th17 cells infiltrate virus‐infected allografts. (A,‐B) Non‐transplant B6 mice were infected with MCMV on day 0 and splenic Th17 cell frequencies were quantified at days 0, 7, 14, 21, and 28 post‐infection. Splenocytes were stimulated with either PMA or MCMV peptides and stained for IL‐17A expressing CD4+ T cells. (A) Representative flow plots showing frequencies of MCMV‐specific and PMA+ Th17 cells at indicated days; graph shows frequencies of PMA+ (gray circles) and MCMV‐specific (black circles) Th17 cells over time. (B) Pie chart shows the percentages of MCMV‐specific and PMA+ Th17 cells in non‐transplant spleens at day 7 post‐infection. (C) Representative flow plots and frequencies of CMV‐specific (CMV peptides+) and total (PMA+) Th17 cells in allografts of D+R+ transplant recipients. (D) Pie chart shows percentages of MCMV‐specific and PMA+ Th17 cells in D+R+ allografts. (E) Proportions of intragraft Th17 cells expressing IL‐17A, IFN‐γ, and/or TNF‐α, compared between Th17 cells responding to MCMV peptides or PMA in D+R+ allografts. (F) Experimental design. B6.OT‐II transgenic recipients received D− or D+ allografts lacking expression of OVA antigen, so that OVA+ Th17 cells are recruited to allografts by antigen‐independent mechanisms. (G) OVA‐specific Th17 cells were detected using I‐Ab‐OVA323–339‐APC tetramer staining, with human CLIP‐APC tetramer used as control (Figure S3). Representative flow plots show tetramer staining of CD4+ T cells derived from D−R− and D+R+ allografts. Graph shows the frequencies of OVA tetramer+ Th17 cells compared between the groups. (H) Cytokine expression profiles were compared for intragraft MCMV specific and PMA+ Th17 cells in wild‐type recipients, and for OVA tetramer+ Th17 cells from OTII recipients. Data are represented as mean ± standard deviation (SD) and are analyzed by two‐sided Student's t‐test (C,G) or one‐way ANOVA (A).
As most MCMV‐induced Th17 cells lacked viral antigen specificity, the requirement for any graft‐specific antigens was assessed using recipient B6.OTII mice, transgenic for CD4+ T cells recognizing chicken ovalbumin (OVA) antigen, transplanted with BALB/c wild‐type kidneys lacking OVA expression (Figure 2F; Figure S3). OVA tetramer+ Th17 cells were significantly higher in the D+ than D− grafts (Figure 2G), indicating that MCMV infection can induce infiltration of Th17 cells lacking specificity for any viral or allograft‐expressed antigens. Tetramer+ Th17 cells co‐expressed TNF‐α ± IFN‐γ, similar to PMA+ but not MCMV peptide+ Th17 cells (Figure 2H). In sum, MCMV infection induces infiltration of both viral antigen‐dependent and antigen‐independent Th17 cells that differ in their cytokine co‐expression profiles.
3.3. MCMV‐infected allograft microenvironment favors Th17 cell recruitment
Next, to identify MCMV‐induced microenvironmental cues promoting antigen‐independent Th17 cell infiltration, gene expression profiles of D+ allografts were compared to native MCMV‐infected kidneys by tissue RNA‐seq and Ingenuity pathway analysis (IPA). Allogeneic transplantation led to significant changes in 5502 genes (adjusted p values <.05), involved in 391 canonical pathways, with 107 differentially expressed genes in the Th17 activation pathway including transcription factors, cytokines, and cytokine receptors involved in Th17 cell differentiation and transcription (Figure 3A) and recruitment (Figure 3C), including CXCL10, CCL20, CXCR3, and CCR6. Similar findings were identified in comparisons of D− allografts and uninfected native kidneys (Figure 3B). However, D− and D+ allografts had no significantly differentially expressed Th17 pathway‐associated transcripts; therefore, transcriptional heatmaps comparing D− and D+ organs are not shown. Similarly, no differences were observed between non‐transplant MCMV‐uninfected and infected kidneys (data not shown), indicating that MCMV infection does not dysregulate native kidneys. 30 Thus, the transplant microenvironment of both D+ and D− allografts favors Th17 cell differentiation and recruitment, but differential expression was not observed at the analyzed timepoint, possibly due to insufficient sensitivity of whole‐organ RNA‐seq to discern differences in low‐abundance Th17 cell transcripts, or to kinetic differences in gene expression not captured at a single timepoint.
FIGURE 3.
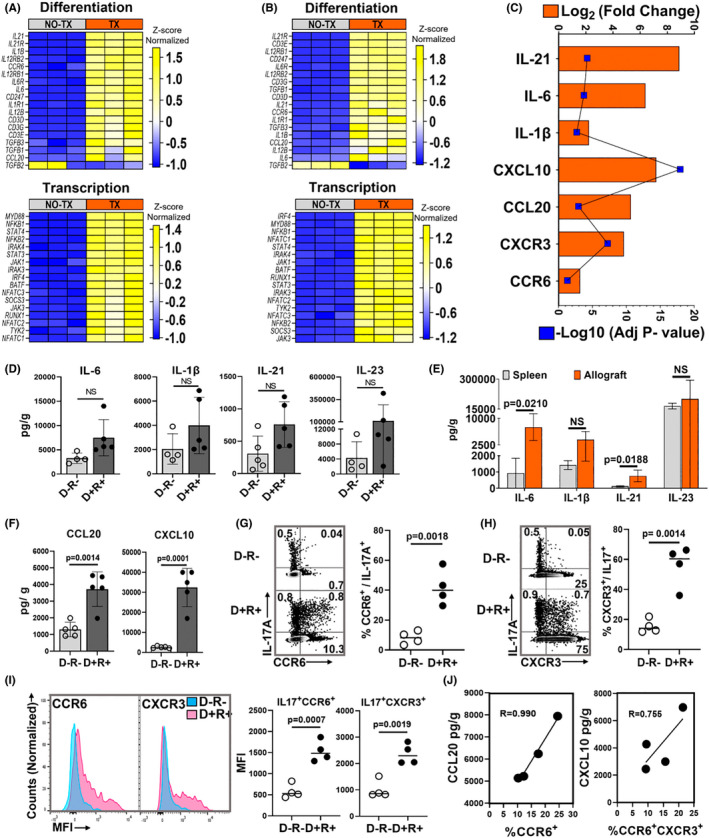
MCMV‐infected allograft microenvironment favors Th17 cell recruitment. (A–C) Gene expression profiles by RNA‐seq. (A) Heat map shows differentially expressed genes encoding molecules required for Th17 cell differentiation and transcription factors between D+ allografts (TX) and infected non‐transplant kidneys (NO‐TX) and (B) differentially expressed Th17 cell‐related genes between D− allografts (TX) and uninfected non‐transplant kidneys (NO‐TX). Each column represents a single sample whereas rows represent intensities of gene expression. Hierarchical clustering of the genes was performed based on the average column z‐score, highest (top) to lowest (bottom). (C) Transcripts for Th17 cell differentiating cytokines and recruiting chemokines are upregulated in D+ transplants compared to MCMV‐infected native kidneys. (D) Comparison of intragraft Th17 cell differentiating cytokine quantities between D−R− and D+R+ transplants. (E) Quantities of Th17 cell differentiating cytokines in D+R+ spleens and allografts. (F) Comparison of Th17 cell recruiting chemokine quantities in D−R− and D+R+ allografts. (G,H) Representative flow plots and frequencies of CCR6+ and CXCR3+ Th17 cells in D−R− and D+R+ allografts. (I) Mean fluorescence intensity (MFI) of CCR6 and CXCR3 expression for Th17 cells from D−R− (blue) and D+R+ (pink) allografts. (J) Correlation between intragraft chemokines and receptors expressed by Th17 cells from D+R+ transplants. All data are represented as mean ± standard deviation (SD) and are analyzed by two‐sided Student's t‐test or Pearson correlation. NS, not significant (p > .05)
As the basis for differential Th17 cell infiltration was not determined by transcriptional profiling, Th17 cell‐associated cytokine and chemokine protein levels were compared in D− and D+ allografts. Th17 cell differentiating cytokines, IL‐6 and IL‐21, trended higher in D+ allografts (p = .0664 and p = .0538) but did not reach statistical significance (Figure 3D). These cytokines were significantly higher in D+ allografts than spleens (Figure 3E). Th17 cell‐recruiting chemokines, CCL20 and CXCL10, were higher in D+ than D− allografts (Figure 3F) and correlated with frequencies of graft‐infiltrating Th17 cells expressing CCR6 and CXCR3 (Figure 3G–J; Figure S4A). These findings support that CCL20 and CXCL10 are differentially expressed in MCMV‐infected allografts and correlate with CCR6+ and CCR6+CXCR3+ Th17 cell infiltrates.
3.4. Ischemia reperfusion injury and MCMV infection each contribute to intragraft Th17 cell recruitment
Ischemia–reperfusion injury (IRI) alone can generate trafficking signals for CD4+ T cells into allografts and can also induce MCMV reactivation from latently infected transplant organs. 32 , 61 , 62 To assess the contribution of IRI without alloimmune responses, syngeneic transplants were performed using D− (IRI alone) and D+ (IRI + MCMV) kidneys (Figure 4A). CCL20, CXCL10, and Th17 cells were detected in syngeneic D− transplants (Figure 4B,C), confirming that IRI alone can induce Th17 cell recruitment. Syngeneic D+ grafts had higher quantities of CCL20, CXCL10, and Th1/17 cells than syngeneic D− grafts (Figure 4B,D), indicating that MCMV reactivation after IRI increases chemokine‐mediated Th17 cell recruitment in the absence of allogeneic signals. IRI, MCMV infection, and allogeneic transplantation each contribute to Th17 cell recruitment but is highest after D+ allogeneic transplantation (Figure 4E).
FIGURE 4.
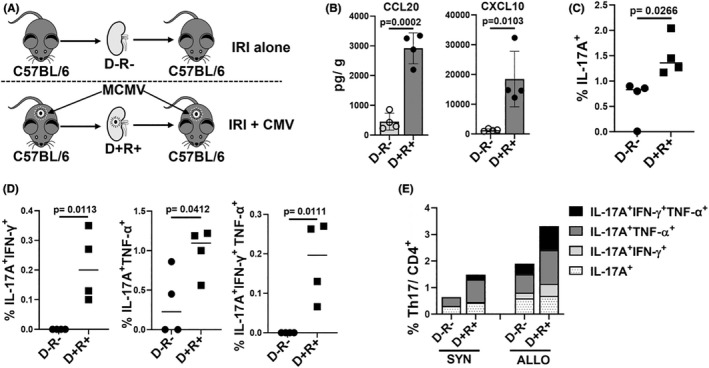
Ischemia reperfusion injury, MCMV infection, and allogeneic transplantation each contribute to intragraft Th17 cell infiltration. (A) Syngeneic transplantation was performed using D−R− and D+R+ grafts to evaluate role of ischemia–reperfusion injury (IRI) without alloimmune responses. (B) CCL20 and CXCL10 quantities in D−R− and D+R+ syngeneic transplants. (C) Frequencies of Th17 infiltrates in syngeneic D−R− and D+R+ grafts. (D) Intragraft Th17 cells co‐expressing IFN‐γ and/or TNF‐α in D−R− and D+R+ grafts. (E) Percentage and proportions of single or multiple cytokines expressing Th17 cells in syngeneic and allogeneic grafts. For (A–D), data are represented as mean ± standard deviation (SD) and are analyzed by two‐sided Student's t‐test. For (E), mean values are shown.
3.5. Th17 cells correlate with graft‐infiltrating Th1 cells and reduced Tregs
Signals promoting development of Th17 cells can suppress Th1 and regulatory T (Treg) subsets. 63 , 64 However, in IPA analysis, the Th1/Th2 activation pathway was the second most highly upregulated canonical pathway (Table S5), consistent with acute T cell–mediated rejection. 65 , 66 , 67 D+ allografts showed higher expression of 64 genes encoding proteins and transcription factors involved in the Th1 pathway, including IFN‐γ (Figure 5A), with similar findings in D− allografts (Figure S4). Th1 cell frequencies were higher in D+ than D− allografts and correlated directly with Th17 cells (Figure 5B). D+ allografts had higher Th17:Th1 ratios than spleens (Figure 5C,D). Conversely, Tregs (Figure S1) were lower in D+ than D− allografts and spleens (Figure 5E–H). These results demonstrate that Th17 cells correlate directly with Th1 cell infiltrates and inversely with Treg frequencies in D+R+ transplants.
FIGURE 5.
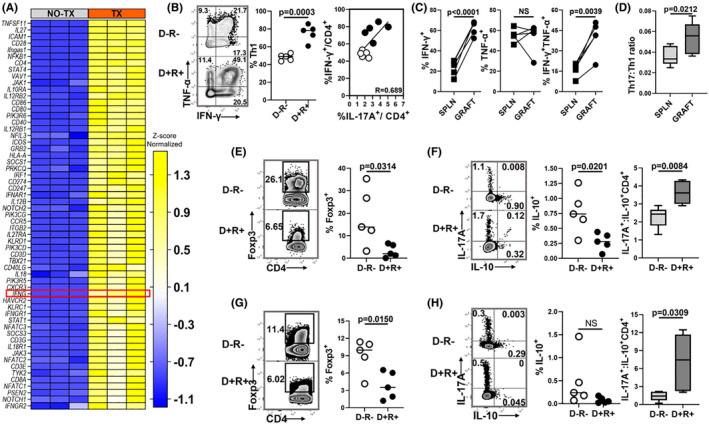
MCMV infection is associated with increased Th1 cells and decreased Tregs. (A) Heat map shows differential expression of transcripts involved with Th1 cell activation in D+ allografts (TX) compared to non‐transplant MCMV‐infected kidneys (NO TX). Hierarchical clustering of the genes was performed based on the average column z‐score, highest to lowest. (B) Representative flow plots and frequencies of Th1 cells in D−R− and D+ R+ allografts. Intragraft frequencies of Th17 cells were correlated with Th1 cells in D−R− and D+ R+ transplants. (C) Frequencies of IFN‐γ and/or TNF‐α expressing Th1 cells in D+R+ spleens and allografts. (D) Ratio of Th17:Th1 cell infiltrates in D+R+ spleens and allografts. (E,F) Representative flow plots and frequencies of (E) Foxp3+ Tregs and (F) IL‐10 expressing Tregs in allografts. (F) Box plot shows ratio of Th17:Treg cells (IL‐17A+: IL‐10+ CD4+). (G,H) Representative flow plots and frequencies of (G) Foxp3+ Tregs and (H) IL‐10 expressing Tregs in spleens. (H) Box plot shows ratio of Th17:Treg cells in spleens. All data are represented as mean ± standard deviation (SD) and are analyzed by two‐sided Student's t‐test or Pearson correlation. NS, not significant (p > .05)
3.6. IL‐17A inhibition reduces neutrophils and Th1 cells, increases Tregs, and ameliorates allograft injury
IL‐17A mobilizes neutrophils to sites of inflammation via CXCL1 and CXCL5. 68 , 69 , 70 Intragraft CXCL1 was significantly higher in D+ allografts (Figure 6A) and correlated with intragraft Th17 cell frequencies (Figure 6B), but CXCL5 did not (Figure 6A). To disrupt IL‐17A signaling, D+R+ recipients were treated with either αIL‐17A or isotype antibodies (Figure 6C). At posttransplant day 7, αIL‐17A treated animals had significantly reduced frequencies of Th17 cells and neutrophils in allografts compared to isotype‐treated mice (Figure 6D,E; Figure S4B), but no effect upon neutrophil frequencies in spleens and blood (Figure 6E). αIL‐17A treated grafts also had significantly increased Foxp3+ Treg frequencies and decreased Th17:Tregs ratio (Figure 6F,G). Unexpectedly, total Th1 cell infiltrates were lower in αIL‐17A treated grafts, with selective decrease in total (PMA+) but not MCMV antigen‐specific (peptide+) Th1 cells, whereas splenic MCMV‐specific and total Th1 cell frequencies were unchanged (Figure 6H–J). At posttransplant day 14, αIL‐17A treated allografts had significantly lower damage scores (18.0 ± 1.7) compared to isotype treated (21.3 ± 0.6) and untreated grafts (20.7 ± 0.6) (p = .0224) (Figure 6K,L). Viral loads were similar in allografts from αIL‐17A, isotype, and untreated groups (Figure 6M). These data show that αIL‐17A treatment reduces neutrophils and total Th1 cells, increases Tregs, and ameliorates allograft damage without increasing viral replication.
FIGURE 6.
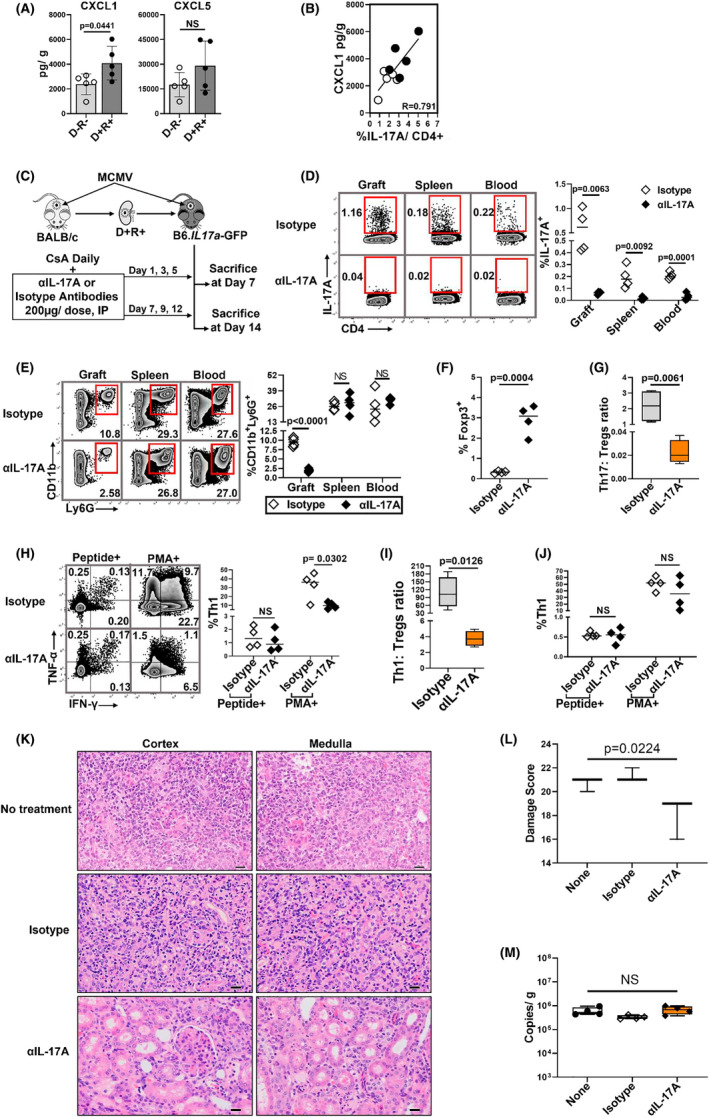
IL‐17A inhibition modulates neutrophil, Th1/Treg cell infiltrates and reduces allograft injury. (A) IL‐17A induced neutrophil chemoattractants, CXCL1 and CXCL5, were compared for D−R− and D+R+ allografts. (B) Correlation between intragraft CXCL1 and Th17 cell infiltrates. (C) Study design. D + R+ recipients were treated with αIL‐17A antibodies or isotype control antibodies at 200 μg/dose, intraperitoneally (IP), on indicated days post‐transplantation and sacrificed at day 7 or 14. (D–J) Day 7 post‐transplantation. (D) Representative flow plots and frequencies of Th17 cells in allograft, spleen, and peripheral blood of recipient mice from αIL‐17A treated and isotype control groups. (E) Representative flow plots and frequencies of CD11b+Ly6G+ neutrophils in organs and blood of αIL‐17A and isotype treated transplant recipients. (F) Frequencies of Foxp3+ Tregs in allografts of αIL‐17A and isotype treated mice. (G) Intragraft Th17:Tregs ratio between the groups. (H) Representative flow plots and frequencies of MCMV‐specific and PMA+ intragraft Th1 cell infiltrates between the groups. (I) Ratio of Th1:Tregs in allografts. (J) Frequencies of MCMV‐specific and PMA+ Th1 cells in spleens. (K) Day 14 posttransplant, hematoxylin and eosin (H&E) stained allografts from D+R+ No treatment (none), isotype control and αIL‐17A treated groups (40×). Scale bar 20 μm. (L) Histopathology was scored in blinded fashion using grading scale as shown in Table S3. (M) Viral loads in allografts. All data are represented as mean ± standard deviation (SD) and are analyzed by two‐sided Student's t‐test, Pearson correlation and ANOVA. NS, not significant (p > .05)
3.7. Th17 cell‐associated allograft damage does not require Treg and Th1 cells
The αIL‐17A treatment could modulate histopathologic graft injury due to direct inhibition of IL‐17A, or indirectly by increasing Tregs that in turn suppress Th1 cell‐mediated graft damage. To distinguish these mechanisms, Tregs were depleted independent of IL‐17A signaling using αCD25 antibodies (Figure 7A). 71 At day 7, Foxp3+ Treg frequencies were significantly lower in grafts, spleens, and blood of αCD25 treated recipients compared to isotype controls (Figure 7B), whereas total IL‐17A+ Th17 cell frequencies were similar between the groups (Figure 7C). Unexpectedly, αCD25 treatment also reduced Th1/17 cells and Th1 cells in grafts and spleens (Figure 7C–F). This result was contrary to our prediction that Treg depletion would increase Th1 cell frequencies; however, these data separated the effect of Th17 cells from that of Treg/Th1 cells in allograft injury. Despite reduced Tregs and Th1 cells, histopathology scores were similar between αCD25 and isotype ‐treated groups in the presence of Th17 cells (Figure 7G,H). These results support that αIL‐17A modulates allograft injury by inhibiting IL‐17A directly, without requiring the downstream effect of IL‐17A upon Tregs or Th1 cell activity.
FIGURE 7.
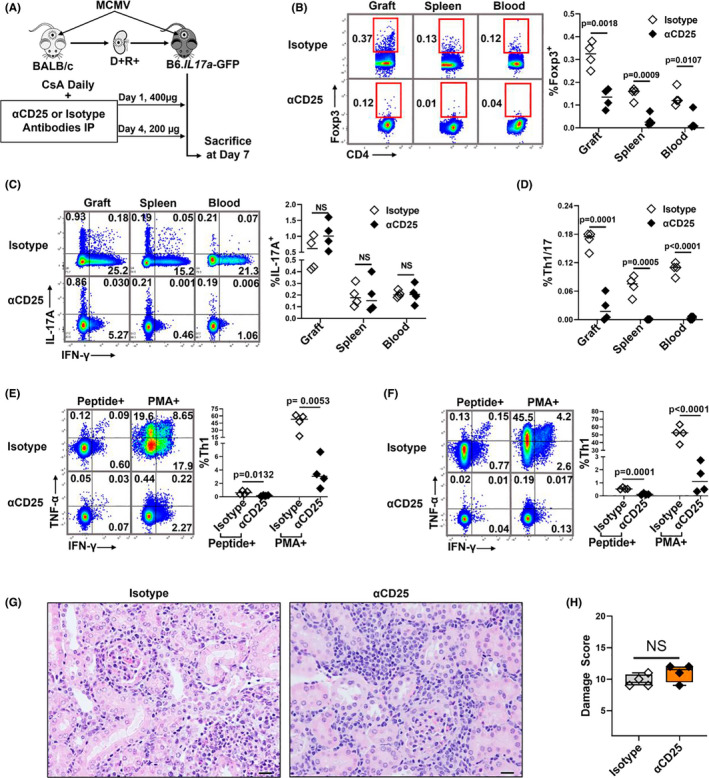
Depletion of Tregs and Th1 cells does not change allograft damage in the presence of Th17 cells. (A) D+R+ recipients were treated with αCD25 antibodies or isotype control antibodies at days 1 (400 μg) and 4 (200 μg) post‐transplantation and sacrificed at day 7. (B) Representative flow plots and summary data showing frequencies of Foxp3+ Tregs between αCD25‐treated mice and isotype controls. (C,D) Representative flow plots and summary data showing the frequencies of Th17 and Th1/17 cells between the groups. (E,F) Representative flow plots and summary data showing the frequencies of total (PMA+) and MCMV‐specific (Peptide+) Th1 cells in allografts (E) and spleens (F) between the groups. (G) H&E‐stained allografts from αCD25 treated and control groups (40×). Scale bar 20 μm. (H) Damage score. All data are represented as mean ± standard deviation (SD) and are analyzed by two‐sided Student's t‐test and ANOVA. NS, not significant (p > .05)
3.8. HCMV DNAemia is associated with elevated serum IL‐17A quantities during acute rejection in clinical renal transplantation
To determine if HCMV DNAemia is associated with IL‐17A in clinical renal transplantation, we retrospectively analyzed samples collected from a prospectively enrolled cohort of renal transplant patients (Figure S6). Of 211 subjects, 35 had biopsy proven acute rejection (AR) in the first‐year posttransplant, whereas 173 had no acute rejection (NR). AR cases with a first episode of acute cellular or mixed rejection and blood samples with sufficient RNA quality for analysis (N = 24) were matched with NR controls (N = 29) by age and sex (Table S6). The AR group had significantly higher serum IL‐17A quantities at the time of rejection compared to the NR group (Figure 8A), which remained significantly elevated even after the AR episode. The AR group also had higher blood RORyt and lower FOXP3 mRNA and IL‐10 cytokine quantities than the NR group, shown as higher IL‐17A:IL‐10 ratio (Figure 8B–F). Together, these findings are consistent with published reports showing elevated Th17 cell signatures during clinical AR. 37
FIGURE 8.
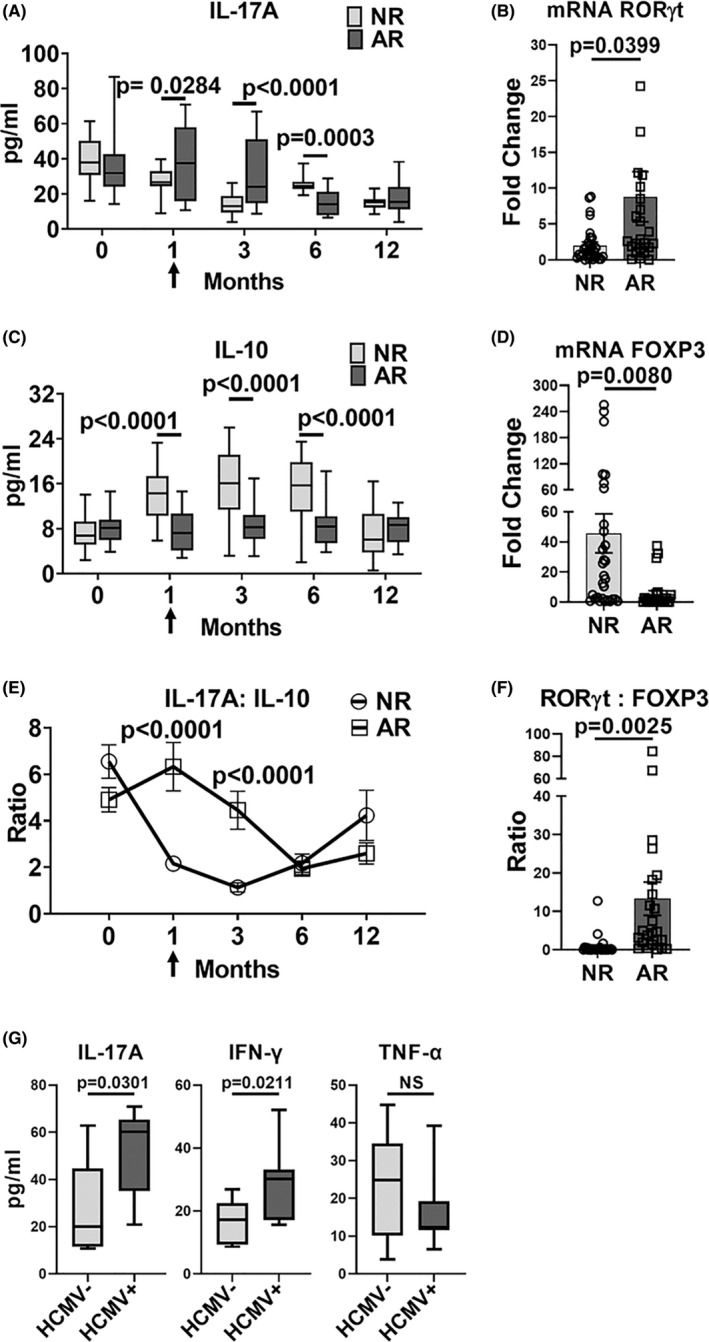
CMV DNAemia is associated with elevated serum IL‐17A and IFN‐γ in kidney transplant patients with acute rejection. Clinical renal transplant recipients with acute rejection (AR, N = 24) were compared to those with no rejection (NR, N = 29) over the first 12 months posttransplant. Blood was analyzed at pretransplant (0), 1‐, 3‐, 6‐ and 12‐month posttransplant and at the time of AR (arrow). (A,C,E) Serum cytokine levels were measured at indicated posttransplant timepoints. (B,D) Expression of transcription factors were quantified in whole blood by RT‐PCR at the time of AR. (F) RORγt:FOXP3 mRNA ratio was compared at the time of rejection for AR and NR groups. (G) HCMV DNAemia in the AR group was determined by quantitative DNA PCR. Serum cytokine quantities were compared for patients with (HCMV+) and without (HCMV−) HCMV DNAemia. All data are represented as mean ± SEM and are analyzed by two‐sided Student's t‐test. NS, not significant (p > .05)
We next examined HCMV DNAemia among 20 AR patients and 17 NR patients for whom blood was available for HCMV PCR testing. In the AR group, 40% (8/20) had HCMV DNAemia at the time of AR, compared to 6% (1/17) of the NR group (p = .0159) (Table S7), consistent with HCMV reactivation during AR. Within the AR group, serum IL‐17A and IFN‐γ were significantly higher among patients with HCMV DNAemia (HCMV+) compared to those without HCMV DNAemia (HCMV‐), but no difference was observed for TNF‐α (Figure 8G). This cytokine profile strikingly resembles that observed in MCMV+ allografts in the murine model (Figure 1D) and supports that HCMV reactivation is associated with elevated IL‐17A and IFN‐γ during clinical acute rejection.
4. DISCUSSION
In prior work, MCMV infection promoted intragraft Th17 cell infiltration, and treatment with αIL‐6 antibodies reduced Th17 cell infiltrates and ameliorated MCMV‐induced histopathologic damage. 30 In this study, the phenotype and antigen specificity of MCMV‐induced, graft‐infiltrating Th17 cells and MCMV‐induced microenvironmental cues promoting Th17 cell recruitment were identified. Intragraft MCMV‐induced Th17 cells co‐express Th1 cytokines that differ according to antigen specificity, with MCMV‐specific Th1/17 cells co‐expressing IFN‐γ, and viral antigen‐independent Th1/17 cells co‐expressing TNF‐α with or without IFN‐γ. MCMV‐specific Th17 cells comprise only a small minority of total graft‐infiltrating Th17 cells; instead, CCL20 and CXCL10 in MCMV‐infected allografts may favor recruitment of MCMV antigen‐independent CCR6+ CXCR3+ Th17 cells. MCMV‐infected syngeneic grafts also express CCL20 and CXCL10, indicating that IRI‐induced MCMV reactivation can alter the graft microenvironment in the absence of allogeneic stimuli. In contrast, MCMV‐infected native kidneys are transcriptionally indistinguishable from uninfected native kidneys, indicating that MCMV infection alone does not promote Th17 cell‐recruiting signals. MCMV‐induced Th17 cells are associated with neutrophils, Th1 cells, and reduced Tregs, promoting the pro‐inflammatory milieu of acute rejection. Together, these findings indicate that MCMV infection exacerbates recruitment of Th17 cells to allografts not only via expression of viral antigens, but also by altering the chemokine microenvironment.
IRI contributes to Th17 cell trafficking to ischemic kidneys. 72 , 73 , 74 STAT‐3 deficient mice are protected from renal IRI, corroborating the role of Th17 cells in IRI‐induced inflammation. 75 Our study confirms the previous observation that IRI induces intrarenal Th17 infiltration, but further shows that CMV infection enhances IRI‐induced Th17 cell recruitment. Renal cells secrete a wide range of cytokines after IRI, including IL‐6, IL‐1β, and TNF‐α, which can promote CMV reactivation in vitro and after syngeneic and allogeneic transplantation in animal models. 32 , 53 , 62 , 76 , 77 , 78 , 79 , 80 , 81 , 82 In vitro, CMV infection induces expression of IL‐6, IL‐8, and TNF‐α from monocytes, TGF‐β1 from renal tubular epithelial cells, and IL‐8, CXCL11, RANTES, IL‐1β, IL‐6, fractalkine (CX3CL1), and CXCL1 from endothelial cells, and in vivo induces RANTES, MCP‐1, MIP‐1α, and CXCL10 expression after allogeneic renal transplantation. 29 , 83 , 84 , 85 , 86 Among renal transplant patients, higher plasma levels of IL‐8, MIP‐1α, and MCP‐1 are associated with active CMV infection and are reduced by ganciclovir treatment. 87 In this work, MCMV‐infected allografts expressed CCL20 and CXCL10, expanding the list of chemokines induced by CMV infection in vivo to include those recruiting Th17 cells. MCMV‐induced Th17 cells then exacerbate Th1 and neutrophil activity via IL‐17A and CXCL1, revealing a previously undescribed pathway by which CMV promotes Th1 and myeloid cell infiltration.
In this model, MCMV infection promotes the development of Th1/17 cells, which have been described in inflammatory diseases including murine experimental autoimmune encephalitis (EAE) and colitis, and among patients with Candida albicans infection, systemic lupus erythematosus, and Crohn's disease. 88 , 89 , 90 , 91 , 92 MCMV‐specific Th1/17 cells infiltrate D + R+ allografts and co‐express the antiviral cytokine, IFN‐γ, whereas MCMV‐induced, antigen‐independent Th1/17 cells co‐express both TNF‐α and IFN‐γ. To our knowledge, this is the first description of MCMV‐specific and cytokine‐induced Th1/17 cells arising during a pathologic inflammatory state. CMV can cause pathological inflammation in numerous organs other than allografts, including CMV pneumonitis, hepatitis, colitis, retinitis, and encephalitis. 16 , 17 , 93 , 94 Although Th17 cells have not been assessed in these CMV end‐organ diseases, Th17 cells can infiltrate these same organs during other infectious and inflammatory diseases, including RSV and COVID19 pneumonia, hepatitis due to HBV, HCV, and murine hepatitis virus, inflammatory bowel disease (IBD) colitis, EAE, and encephalitis due to Theiler's murine encephalomyelitis virus. 46 , 47 , 48 , 49 , 50 , 90 , 95 , 96 , 97 , 98 , 99 , 100 It is thus possible that Th17 cell‐associated inflammation in other CMV end‐organ diseases could be targeted with adjunctive anti‐inflammatory therapies in addition to antiviral drugs.
Th17 cells correlate directly with Th1 cells in MCMV‐infected allografts. Inhibition of IL‐17A reduces Th1 cells and increases Tregs, consistent with the known reciprocal relationship between Th17 and Treg cells. 101 To determine if αIL‐17A ameliorates graft injury indirectly through Treg/Th1 cells, αCD25 antibodies were used to deplete Tregs, in whose absence Th1 cells were predicted to increase and exacerbate allograft damage. However, αCD25 reduced not only Tregs but also unexpectedly reduced Th1 cell infiltrates, possibly due to a direct effect of αCD25 upon Th1 cells. CD25, the IL‐2 receptor‐α subunit, is expressed by activated T cells. 102 , 103 , 104 Studies have shown that the PC61 clone of αCD25 not only depletes Tregs but also induces defective IFN‐γ production from CD4+ T cells in murine organs 105 and abolishes STAT5 signaling in CD4+ effector T cells, impairing STAT5‐induced differentiation of IFN‐γ producing Th1 effector cells. 106 , 107 , 108 In our experimental system, αCD25 depleted both Tregs and Th1 cells, but not Th17 cells, distinguishing the function of Th17 cells independent of Treg/Th1 cells. Reduction of both Tregs and Th1 cells by αCD25 treatment failed to alter graft injury in the presence of Th17 cells, indicating that αIL‐17A modulates graft damage by directly inhibiting IL‐17A and not via indirect effects upon Treg/Th1 subsets. In addition, αIL‐17A treatment did not reduce frequencies of CMV‐specific Th1 cells or increase intragraft viral loads, indicating that αIL‐17A treatment may ameliorate allograft damage without impairing protective antiviral immunity. Consistent with this interpretation, CMV is not a known complication of clinically utilized αIL‐17A monoclonal antibodies such as secukinumab. 109
Clinical studies have previously shown that Th17 cells are associated with acute rejection and late allograft dysfunction in populations with high HCMV seroprevalence, 34 , 37 but HCMV DNAemia was not assessed as a clinical variable in those studies. Our work shows that HCMV DNAemia is associated with higher serum IL‐17A and IFN‐γ, but not TNF‐α, a cytokine profile that strongly resembles that observed in the MCMV transplant model. Further studies among HCMV seropositive renal transplant patients may determine the extent to which the epidemiologic association between HCMV infection and allograft dysfunction is mediated by Th17 cells, and whether Th17 cell activity is promoted by subclinical HCMV DNAemia in the absence of AR.
There are limitations to this study. Due to the technical complexity of murine microvascular renal transplant surgery, most cohorts were analyzed at day 7 to permit direct comparison of results between experiments. Consequently, the temporal relationship between MCMV‐induced chemokine expression, Th17 cell recruitment, and Th1 cell activation were not well defined. Similarly, gene expression profiling of transplant tissues did not identify differential expression of genes encoding Th17 cell‐related transcriptional pathways between D+ and D− allografts, possibly due to insufficient sensitivity of tissue profiling to distinguish differential expression of low‐abundance transcripts in Th17 cells, or due to inability of transcriptional analysis at a single timepoint to capture kinetic differences in transcriptional signaling preceding protein expression. Despite the lack of observed differences in transcriptional signaling, functional T cell and protein expression analyses showed significant differences in Th17 cells and associated chemokines in MCMV‐infected allografts. We also did not assess developmental plasticity between Th17 and Th1 cell subsets during the course of acute rejection, and it is possible that transdifferentiation between Th17 and Th1 cells could contribute to the Th1/17 cytokine co‐expression profiles observed in these studies. In addition, these studies only analyzed antiviral T cell responses during acute T cell–mediated rejection in MCMV R+ recipients. MCMV‐induced Th17 cell frequencies and phenotypes were not examined in allogeneic models with less fulminant rejection, or in D+R− transplants with primary MCMV infection. Finally, although the renal transplant population evaluated in this study has nearly universal HCMV seropositivity, HCMV D/R serostatus was not confirmed, so it is possible that patients with HCMV D−R− serostatus had no viral detection. However, this serostatus is probably rare in this population and, if present, would most likely be distributed similarly among the groups with and without AR.
In summary, MCMV infection induces recruitment of Th1/17 cells into infected renal allografts, which arise in response to viral antigens and chemokine‐induced pathways and are associated with neutrophils, Th1 cells, and reduced Tregs. Among renal transplant recipients, Th1/17 cytokines are associated with HCMV reactivation. These findings identify several pathways by which CMV exacerbates inflammation during acute rejection and raises the possibility that inhibition of CMV‐induced Th17 cell effector cytokines might ameliorate organ inflammation without impairing protective antiviral Th1 cell responses. In a larger context, CMV‐induced Th1/17 cells might also contribute to pathological inflammation during CMV end‐organ disease, which could be targeted with anti‐cytokine treatments as an adjunct to antiviral therapy.
DISCLOSURE
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Supporting information
Appendix S1 Supplementary Information
ACKNOWLEDGMENTS
We are grateful to the renal transplant patients for their participation in this study. We thank Sonya Maher, Jessica Ghere, Irina Kaptsan, and Kaitlyn Flint for providing technical assistance. This research was supported by NIH R01AI101138 (MS), the Abigail Wexner Research Institute at Nationwide Children's Hospital (MS), and the Intramural Research Grant of the Post Graduate Institute of Medical Education and Research (MM and RWM). The authors declare no competing financial interests.
Dhital R, Anand S, Graber B, et al.. Murine cytomegalovirus promotes renal allograft inflammation via Th1/17 cells and IL‐17A . Am J Transplant. 2022;22:2306‐2322. doi: 10.1111/ajt.17116
DATA AVAILABILITY STATEMENT
Data are available in the main text or Supporting Information. RNA sequencing data are deposited in the NCBI Gene Expression Omnibus 110 (accession #GSE179788). A pre‐print version of this manuscript is available at https://www.biorxiv.org/content/10.1101/2021.08.05.455061v1. 111 However, this manuscript differs from the pre‐print version by showing that αIL‐17A modulates allograft damage by direct inhibition of IL‐17A and not through indirect effects on Treg/Th1 cell infiltrates, and that αIL‐17A treatment does not increase viral replication. Therefore, this manuscript differs from the pre‐print in content and interpretation.
REFERENCES
- 1. Das B, Kaur G, Basu S. Seroprevalence of cytomegalovirus antibodies among blood donors and multitransfused recipients–a study from North India. Transfus Apher Sci. 2014;50(3):438‐442. doi: 10.1016/j.transci.2014.02.022 [DOI] [PubMed] [Google Scholar]
- 2. Lachmann R, Loenenbach A, Waterboer T, et al. Cytomegalovirus (CMV) seroprevalence in the adult population of Germany. PLoS One. 2018;13(7):e0200267. doi: 10.1371/journal.pone.0200267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Warnecke JM, Pollmann M, Borchardt‐Loholter V, et al. Seroprevalences of antibodies against torch infectious pathogens in women of childbearing age residing in Brazil, Mexico, Germany, Poland, Turkey and China. Epidemiol Infect. 2020;148:e271. doi: 10.1017/S0950268820002629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bate SL, Dollard SC, Cannon MJ. Cytomegalovirus seroprevalence in the United States: the national health and nutrition examination surveys, 1988‐2004. Clin Infect Dis. 2010;50(11):1439‐1447. doi: 10.1086/652438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Myerson D, Hackman RC, Nelson JA, Ward DC, McDougall JK. Widespread presence of histologically occult cytomegalovirus. Hum Pathol. 1984;15(5):430‐439. doi: 10.1016/s0046-8177(84)80076-3 [DOI] [PubMed] [Google Scholar]
- 6. Hendrix RM, Wagenaar M, Slobbe RL, Bruggeman CA. Widespread presence of cytomegalovirus DNA in tissues of healthy trauma victims. J Clin Pathol. 1997;50(1):59‐63. doi: 10.1136/jcp.50.1.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klenerman P, Oxenius A. T cell responses to cytomegalovirus. Nat Rev Immunol. 2016;16(6):367‐377. doi: 10.1038/nri.2016.38 [DOI] [PubMed] [Google Scholar]
- 8. Sester M, Sester U, Gärtner B, et al. Levels of virus‐specific CD4 T cells correlate with cytomegalovirus control and predict virus‐induced disease after renal transplantation. Transplantation. 2001;71(9):1287‐1294. doi: 10.1097/00007890-200105150-00018 [DOI] [PubMed] [Google Scholar]
- 9. Walter EA, Greenberg PD, Gilbert MJ, et al. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T‐cell clones from the donor. N Engl J Med. 1995;333(16):1038‐1044. doi: 10.1056/nejm199510193331603 [DOI] [PubMed] [Google Scholar]
- 10. Reddehase MJ, Mutter W, Münch K, Bühring HJ, Koszinowski UH. CD8‐positive T lymphocytes specific for murine cytomegalovirus immediate‐early antigens mediate protective immunity. J Virol. 1987;61(10):3102‐3108. doi: 10.1128/jvi.61.10.3102-3108.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dunn HS, Haney DJ, Ghanekar SA, Stepick‐Biek P, Lewis DB, Maecker HT. Dynamics of CD4 and CD8 T cell responses to cytomegalovirus in healthy human donors. J Infect Dis. 2002;186(1):15‐22. doi: 10.1086/341079 [DOI] [PubMed] [Google Scholar]
- 12. Komanduri KV, Donahoe SM, Moretto WJ, et al. Direct measurement of CD4+ and CD8+ T‐cell responses to CMV in HIV‐1‐infected subjects. Virology. 2001;279(2):459‐470. doi: 10.1006/viro.2000.0697 [DOI] [PubMed] [Google Scholar]
- 13. Gamadia LE, Rentenaar RJ, van Lier RA, ten Berge IJ. Properties of CD4(+) T cells in human cytomegalovirus infection. Hum Immunol. 2004;65(5):486‐492. doi: 10.1016/j.humimm.2004.02.020 [DOI] [PubMed] [Google Scholar]
- 14. Jeitziner SM, Walton SM, Torti N, Oxenius A. Adoptive transfer of cytomegalovirus‐specific effector CD4+ T cells provides antiviral protection from murine CMV infection. Eur J Immunol. 2013;43(11):2886‐2895. doi: 10.1002/eji.201343690 [DOI] [PubMed] [Google Scholar]
- 15. Rentenaar RJ, Gamadia LE, van DerHoek N, et al. Development of virus‐specific CD4(+) T cells during primary cytomegalovirus infection. J Clin Invest. 2000;105(4):541‐548. doi: 10.1172/JCI8229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Razonable RR, Humar A. Cytomegalovirus in solid organ transplant recipients‐guidelines of the American society of transplantation infectious diseases community of practice. Clin Transplant. 2019;33(9):e13512. doi: 10.1111/ctr.13512 [DOI] [PubMed] [Google Scholar]
- 17. Ljungman P, Reusser P, de la Camara R, et al. Management of CMV infections: recommendations from the infectious diseases working party of the ebmt. Bone Marrow Transplant. 2004;33(11):1075‐1081. doi: 10.1038/sj.bmt.1704505 [DOI] [PubMed] [Google Scholar]
- 18. Freeman RB Jr. The 'indirect' effects of cytomegalovirus infection. Am J Transplant. 2009;9(11):2453‐2458. doi: 10.1111/j.1600-6143.2009.02824.x [DOI] [PubMed] [Google Scholar]
- 19. Humar A, Gillingham KJ, Payne WD, Dunn DL, Sutherland DE, Matas AJ. Association between cytomegalovirus disease and chronic rejection in kidney transplant recipients. Transplantation. 1999;68(12):1879‐1883. [DOI] [PubMed] [Google Scholar]
- 20. Tong CY, Bakran A, Peiris JS, Muir P, Herrington CS. The association of viral infection and chronic allograft nephropathy with graft dysfunction after renal transplantation. Transplantation. 2002;74(4):576‐578. doi: 10.1097/00007890-200208270-00026 [DOI] [PubMed] [Google Scholar]
- 21. Toupance O, Bouedjoro‐Camus MC, Carquin J, et al. Cytomegalovirus‐related disease and risk of acute rejection in renal transplant recipients: a cohort study with case‐control analyses. Transpl Int. 2000;13(6):413‐419. doi: 10.1007/s001470050723 [DOI] [PubMed] [Google Scholar]
- 22. Gerstenkorn C, Balupuri S, Mohamed MA, et al. The impact of cytomegalovirus serology for 7‐year graft survival in cadaveric kidney transplantation‐‐the Newcastle experience. Transpl Int. 2000;13(Suppl 1):S372‐S374. doi: 10.1007/s001470050364 [DOI] [PubMed] [Google Scholar]
- 23. Gerstenkorn C, Robertson H, Bell A, Shenton B, Talbot D. CMV infection as a contributory factor for renal allograft injury and loss. Transplant Proc. 2001;33(4):2461‐2462. doi: 10.1016/s0041-1345(01)02047-4 [DOI] [PubMed] [Google Scholar]
- 24. Leeaphorn N, Garg N, Thamcharoen N, Khankin EV, Cardarelli F, Pavlakis M. Cytomegalovirus mismatch still negatively affects patient and graft survival in the era of routine prophylactic and preemptive therapy: a paired kidney analysis. Am J Transplant. 2019;19(2):573‐584. doi: 10.1111/ajt.15183 [DOI] [PubMed] [Google Scholar]
- 25. Kliem V, Fricke L, Wollbrink T, Burg M, Radermacher J, Rohde F. Improvement in long‐term renal graft survival due to cmv prophylaxis with oral ganciclovir: results of a randomized clinical trial. Am J Transplant. 2008;8(5):975‐983. doi: 10.1111/j.1600-6143.2007.02133.x [DOI] [PubMed] [Google Scholar]
- 26. Mavrakanas TA, Fournier MA, Clairoux S, et al. Neutropenia in kidney and liver transplant recipients: risk factors and outcomes. Clin Transplant. 2017;31(e13058):10. doi: 10.1111/ctr.13058 [DOI] [PubMed] [Google Scholar]
- 27. Rha B, Redden D, Benfield M, Lakeman F, Whitley RJ, Shimamura M. Correlation and clinical utility of pp65 antigenemia and quantitative polymerase chain reaction assays for detection of cytomegalovirus in pediatric renal transplant patients. Pediatr Transplant. 2012;16(6):627‐637. doi: 10.1111/j.1399-3046.2012.01741.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lautenschlager I, Soots A, Krogerus L, et al. Cmv increases inflammation and accelerates chronic rejection in rat kidney allografts. Transplant Proc. 1997;29(1):802‐803. doi: 10.1016/S0041-1345(96)00109-1 [DOI] [PubMed] [Google Scholar]
- 29. Soule JL, Streblow DN, Andoh TF, Kreklywich CN, Orloff SL. Cytomegalovirus accelerates chronic allograft nephropathy in a rat renal transplant model with associated provocative chemokine profiles. Transplant Proc. 2006;38(10):3214‐3220. doi: 10.1016/j.transproceed.2006.10.187 [DOI] [PubMed] [Google Scholar]
- 30. Li M, Boddeda SR, Chen B, et al. NK cell and Th17 responses are differentially induced in murine cytomegalovirus infected renal allografts and vary according to recipient virus dose and strain. Am J Transplant. 2018;18(11):2647‐2662. doi: 10.1111/ajt.14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shimamura M, Seleme MC, Guo L, et al. Ganciclovir prophylaxis improves late murine cytomegalovirus‐induced renal allograft damage. Transplantation. 2013;95(1):48‐53. doi: 10.1097/TP.0b013e3182782efc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saunders U, Li M, Boddeda SR, et al. Murine cytomegalovirus‐induced complement‐fixing antibodies deposit in murine renal allografts during acute rejection. Transplantation. 2020;105(8):12. doi: 10.1097/tp.0000000000003548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crispim JC, Grespan R, Martelli‐Palomino G, et al. Interleukin‐17 and kidney allograft outcome. Transplant Proc. 2009;41(5):1562‐1564. doi: 10.1016/j.transproceed.2009.01.092 [DOI] [PubMed] [Google Scholar]
- 34. Chung BH, Kim KW, Kim BM, Doh KC, Cho ML, Yang CW. Increase of Th17 cell phenotype in kidney transplant recipients with chronic allograft dysfunction. PLoS One. 2015;10(12):e0145258. doi: 10.1371/journal.pone.0145258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fabrega E, Lopez‐Hoyos M, San Segundo D, Casafont F, Pons‐Romero F. Changes in the serum levels of interleukin‐17/interleukin‐23 during acute rejection in liver transplantation. Liver Transpl. 2009;15(6):629‐633. doi: 10.1002/lt.21724 [DOI] [PubMed] [Google Scholar]
- 36. Fan H, Li L‐X, Han D‐D, Kou J‐T, Li P, He Q. Increase of peripheral Th17 lymphocytes during acute cellular rejection in liver transplant recipients. Hepatobiliary Pancreat Dis Int. 2012;11(6):606‐611. doi: 10.1016/s1499-3872(12)60231-8 [DOI] [PubMed] [Google Scholar]
- 37. Chung BH, Oh HJ, Piao SG, et al. Higher infiltration by Th17 cells compared with regulatory T cells is associated with severe acute T‐cell‐mediated graft rejection. Exp Mol Med. 2011;43(11):630‐637. doi: 10.3858/emm.2011.43.11.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gorbacheva V, Fan R, Li X, Valujskikh A. Interleukin‐17 promotes early allograft inflammation. Am J Pathol. 2010;177(3):1265‐1273. doi: 10.2353/ajpath.2010.091106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Strehlau J, Pavlakis M, Lipman M, et al. Quantitative detection of immune activation transcripts as a diagnostic tool in kidney transplantation. Proc Natl Acad Sci U S A. 1997;94(2):695‐700. doi: 10.1073/pnas.94.2.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Woltman AM, de Fijter JW, van der Kooij SW, et al. Mip‐3alpha/CCL20 in renal transplantation and its possible involvement as dendritic cell chemoattractant in allograft rejection. Am J Transplant. 2005;5(9):2114‐2125. doi: 10.1111/j.1600-6143.2005.00997.x [DOI] [PubMed] [Google Scholar]
- 41. Yapici U, Kers J, Bemelman FJ, et al. Interleukin‐17 positive cells accumulate in renal allografts during acute rejection and are independent predictors of worse graft outcome. Transpl Int. 2011;24(10):1008‐1017. doi: 10.1111/j.1432-2277.2011.01302.x [DOI] [PubMed] [Google Scholar]
- 42. Yuan X, Paez‐Cortez J, Schmitt‐Knosalla I, et al. A novel role of CD4 Th17 cells in mediating cardiac allograft rejection and vasculopathy. J Exp Med. 2008;205(13):3133‐3144. doi: 10.1084/jem.20081937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Khader SA, Gaffen SL, Kolls JK. Th17 cells at the crossroads of innate and adaptive immunity against infectious diseases at the mucosa. Mucosal Immunol. 2009;2(5):403‐411. doi: 10.1038/mi.2009.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Khader SA, Gopal R. IL‐17 in protective immunity to intracellular pathogens. Virulence. 2010;1(5):423‐427. doi: 10.4161/viru.1.5.12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mukherjee S, Lindell DM, Berlin AA, et al. IL‐17‐induced pulmonary pathogenesis during respiratory viral infection and exacerbation of allergic disease. Am J Pathol. 2011;179(1):248‐258. doi: 10.1016/j.ajpath.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paquissi FC. Immunity and fibrogenesis: the role of Th17/IL‐17 axis in hbv and hcv‐induced chronic hepatitis and progression to cirrhosis. Front Immunol. 2017;8:1195. doi: 10.3389/fimmu.2017.01195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bera MM, Lu B, Martin TR, et al. Th17 cytokines are critical for respiratory syncytial virus‐associated airway hyperreponsiveness through regulation by complement c3a and tachykinins. J Immunol. 2011;187(8):4245‐4255. doi: 10.4049/jimmunol.1101789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stoppelenburg AJ, de Roock S, Hennus MP, Bont L, Boes M. Elevated Th17 response in infants undergoing respiratory viral infection. Am J Pathol. 2014;184(5):1274‐1279. doi: 10.1016/j.ajpath.2014.01.033 [DOI] [PubMed] [Google Scholar]
- 49. Yang W, Ding X, Deng J, et al. Interferon‐gamma negatively regulates Th17‐mediated immunopathology during mouse hepatitis virus infection. J Mol Med (Berl). 2011;89(4):399‐409. doi: 10.1007/s00109-010-0711-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hou W, Kang HS, Kim BS. Th17 cells enhance viral persistence and inhibit T cell cytotoxicity in a model of chronic virus infection. J Exp Med. 2009;206(2):313‐328. doi: 10.1084/jem.20082030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Arens R, Wang P, Sidney J, et al. Cutting edge: murine cytomegalovirus induces a polyfunctional CD4 T cell response. J Immunol. 2008;180(10):6472‐6476. doi: 10.4049/jimmunol.180.10.6472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yue FY, Merchant A, Kovacs CM, Loutfy M, Persad D, Ostrowski MA. Virus‐specific interleukin‐17‐producing CD4+ T cells are detectable in early human immunodeficiency virus type 1 infection. J Virol. 2008;82(13):6767‐6771. doi: 10.1128/JVI.02550-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hummel M, Zhang Z, Yan S, et al. Allogeneic transplantation induces expression of cytomegalovirus immediate‐early genes in vivo: a model for reactivation from latency. J Virol. 2001;75(10):4814‐4822. doi: 10.1128/JVI.75.10.4814-4822.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schlub TE, Sun JC, Walton SM, et al. Comparing the kinetics of NK cells, CD4, and CD8 T cells in murine cytomegalovirus infection. J Immunol. 2011;187(3):1385‐1392. doi: 10.4049/jimmunol.1100416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shimamura M, Saunders U, Rha B, et al. Ganciclovir transiently attenuates murine cytomegalovirus‐associated renal allograft inflammation. Transplantation. 2011;92(7):759‐766. doi: 10.1097/TP.0b013e31822c6e89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Loupy A, Haas M, Roufosse C, et al. The banff 2019 kidney meeting report (i): updates on and clarification of criteria for T cell‐ and antibody‐mediated rejection. Am J Transplant. 2020;20(9):2318‐2331. doi: 10.1111/ajt.15898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roufosse C, Simmonds N, Clahsen‐van Groningen M, et al. A 2018 reference guide to the banff classification of renal allograft pathology. Transplantation. 2018;102(11):1795‐1814. doi: 10.1097/TP.0000000000002366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dhital R, Minz M, Minz RW, et al. Flow cytometric quantitation of T‐cell subsets in peripheral blood of renal allograft recipients. Indian J Transpl. 2016;10(4). doi: 10.1016/j.ijt.2016.09.031 [DOI] [Google Scholar]
- 59. Dhital R, Minz M, Minz RW, et al. Or19 quantification of peripheral b‐cell subsets in acute allograft rejection in recipients with renal transplantation. Hum Immunol. 2016;77. doi: 10.1016/j.humimm.2016.07.031 [DOI] [Google Scholar]
- 60. Minz M, Dhital R, Jha V, Minz RW, Sharma A. Mrna expression of baff and april receptors increases in acute rejection in kidney transplant recipients. Am J Transplant. 2016;16(Suppl 3). [Google Scholar]
- 61. Takada M, Nadeau KC, Shaw GD, Marquette KA, Tilney NL. The cytokine‐adhesion molecule cascade in ischemia/reperfusion injury of the rat kidney. Inhibition by a soluble p‐selectin ligand. J Clin Invest. 1997;99(11):2682‐2690. doi: 10.1172/jci119457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang Z, Qiu L, Yan S, et al. A clinically relevant murine model unmasks a "two‐hit" mechanism for reactivation and dissemination of cytomegalovirus after kidney transplant. Am J Transplant. 2019;19(9):2421‐2433. doi: 10.1111/ajt.15376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Harrington LE, Hatton RD, Mangan PR, et al. Interleukin 17‐producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6(11):1123‐1132. doi: 10.1038/ni1254 [DOI] [PubMed] [Google Scholar]
- 64. Yang XO, Nurieva R, Martinez GJ, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29(1):44‐56. doi: 10.1016/j.immuni.2008.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. D'Elios MM, Josien R, Manghetti M, et al. Predominant th1 cell infiltration in acute rejection episodes of human kidney grafts. Kidney Int. 1997;51(6):1876‐1884. doi: 10.1038/ki.1997.256 [DOI] [PubMed] [Google Scholar]
- 66. Obata F, Yoshida K, Ohkubo M, et al. Contribution of CD4+ and CD8+ T cells and interferon‐gamma to the progress of chronic rejection of kidney allografts: the th1 response mediates both acute and chronic rejection. Transpl Immunol. 2005;14(1):21‐25. doi: 10.1016/j.trim.2004.11.003 [DOI] [PubMed] [Google Scholar]
- 67. Sadeghi M, Daniel V, Weimer R, Wiesel M, Hergesell O, Opelz G. Pre‐transplant th1 and post‐transplant th2 cytokine patterns are associated with early acute rejection in renal transplant recipients. Clin Transplant. 2003;17(2):151‐157. doi: 10.1034/j.1399-0012.2003.00037.x [DOI] [PubMed] [Google Scholar]
- 68. Scozzi D, Ibrahim M, Menna C, Krupnick AS, Kreisel D, Gelman AE. The role of neutrophils in transplanted organs. Am J Transplant. 2017;17(2):328‐335. doi: 10.1111/ajt.13940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sawant KV, Poluri KM, Dutta AK, et al. Chemokine cxcl1 mediated neutrophil recruitment: role of glycosaminoglycan interactions. Sci Rep. 2016;6:33123. doi: 10.1038/srep33123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Disteldorf EM, Krebs CF, Paust HJ, et al. Cxcl5 drives neutrophil recruitment in th17‐mediated gn. J Am Soc Nephrol. 2015;26(1):55‐66. doi: 10.1681/ASN.2013101061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Setiady YY, Coccia JA, Park PU. In vivo depletion of cd4+foxp3+ treg cells by the pc61 anti‐cd25 monoclonal antibody is mediated by fcgammariii+ phagocytes. Eur J Immunol. 2010;40(3):780‐786. doi: 10.1002/eji.200939613 [DOI] [PubMed] [Google Scholar]
- 72. Guo L, Lee HH, Noriega ML, Paust HJ, Zahner G, Thaiss F. Lymphocyte‐specific deletion of ikk2 or nemo mediates an increase in intrarenal Th17 cells and accelerates renal damage in an ischemia‐reperfusion injury mouse model. Am J Physiol Renal Physiol. 2016;311(5):F1005‐F1014. doi: 10.1152/ajprenal.00242.2016 [DOI] [PubMed] [Google Scholar]
- 73. Mehrotra P, Collett JA, McKinney SD, Stevens J, Ivancic CM, Basile DP. IL‐17 mediates neutrophil infiltration and renal fibrosis following recovery from ischemia reperfusion: compensatory role of natural killer cells in athymic rats. Am J Physiol Renal Physiol. 2017;312(3):F385‐F397. doi: 10.1152/ajprenal.00462.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mehrotra P, Patel JB, Ivancic CM, Collett JA, Basile DP. Th‐17 cell activation in response to high salt following acute kidney injury is associated with progressive fibrosis and attenuated by at‐1r antagonism. Kidney Int. 2015;88(4):776‐784. doi: 10.1038/ki.2015.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lee JW, Bae E, Kwon SH, et al. Transcriptional modulation of the T helper 17/interleukin 17 axis ameliorates renal ischemia‐reperfusion injury. Nephrol Dial Transplant. 2019;34(9):1481‐1498. doi: 10.1093/ndt/gfy370 [DOI] [PubMed] [Google Scholar]
- 76. Hargett D, Shenk TE. Experimental human cytomegalovirus latency in CD14+ monocytes. Proc Natl Acad Sci U S A. 2010;107(46):20039‐20044. doi: 10.1073/pnas.1014509107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. O'Connor CM, Murphy EA. A myeloid progenitor cell line capable of supporting human cytomegalovirus latency and reactivation, resulting in infectious progeny. J Virol. 2012;86(18):9854‐9865. doi: 10.1128/JVI.01278-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Reeves MB, Compton T. Inhibition of inflammatory interleukin‐6 activity via extracellular signal‐regulated kinase‐mitogen‐activated protein kinase signaling antagonizes human cytomegalovirus reactivation from dendritic cells. J Virol. 2011;85(23):12750‐12758. doi: 10.1128/JVI.05878-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. de Vries DK, Lindeman JH, Tsikas D, et al. Early renal ischemia‐reperfusion injury in humans is dominated by IL‐6 release from the allograft. Am J Transplant. 2009;9(7):1574‐1584. doi: 10.1111/j.1600-6143.2009.02675.x [DOI] [PubMed] [Google Scholar]
- 80. Donnahoo KK, Shames BD, Harken AH, Meldrum DR. Review article: the role of tumor necrosis factor in renal ischemia‐reperfusion injury. J Urol. 1999;162(1):196‐203. doi: 10.1097/00005392-199907000-00068 [DOI] [PubMed] [Google Scholar]
- 81. Prieto‐Moure B, Lloris‐Carsí JM, Belda‐Antolí M, Toledo‐Pereyra LH, Cejalvo‐Lapeña D. Allopurinol protective effect of renal ischemia by downregulating TNF‐α, IL‐1β, and IL‐6 response. J Invest Surg. 2017;30(3):143‐151. doi: 10.1080/08941939.2016.1230658 [DOI] [PubMed] [Google Scholar]
- 82. Forte E, Zhang Z, Thorp EB, Hummel M. Cytomegalovirus latency and reactivation: an intricate interplay with the host immune response. Front Cell Infect Microbiol. 2020;10:130. doi: 10.3389/fcimb.2020.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Alcendor DJ, Charest AM, Zhu WQ, Vigil HE, Knobel SM. Infection and upregulation of proinflammatory cytokines in human brain vascular pericytes by human cytomegalovirus. J Neuroinflammation. 2012;9:95. doi: 10.1186/1742-2094-9-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bolovan‐Fritts CA, Trout RN, Spector SA. Human cytomegalovirus‐specific CD4+−T‐cell cytokine response induces fractalkine in endothelial cells. J Virol. 2004;78(23):13173‐13181. doi: 10.1128/JVI.78.23.13173-13181.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shimamura M, Murphy‐Ullrich JE, Britt WJ. Human cytomegalovirus induces TGF‐beta1 activation in renal tubular epithelial cells after epithelial‐to‐mesenchymal transition. PLoS Pathog. 2010;6(11):e1001170. doi: 10.1371/journal.ppat.1001170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Grundy JE, Lawson KM, MacCormac LP, Fletcher JM, Yong KL. Cytomegalovirus‐infected endothelial cells recruit neutrophils by the secretion of c‐x‐c chemokines and transmit virus by direct neutrophil‐endothelial cell contact and during neutrophil transendothelial migration. J Infect Dis. 1998;177(6):1465‐1474. doi: 10.1086/515300 [DOI] [PubMed] [Google Scholar]
- 87. Nordøy I, Müller F, Nordal KP, Rollag H, Aukrust P, Frøland SS. Chemokines and soluble adhesion molecules in renal transplant recipients with cytomegalovirus infection. Clin Exp Immunol. 2000;120(2):333‐337. doi: 10.1046/j.1365-2249.2000.01221.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Annunziato F, Cosmi L, Santarlasci V, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204(8):1849‐1861. doi: 10.1084/jem.20070663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Duhen T, Campbell D. Human CCR6+CXCR3+ “Th1/17” cells are polyfunctional and can develop from Th17 cells under the influence of IL‐1β and IL‐12 (p4122). J Immunol 2013;190(1 Suppl.):191.197. [Google Scholar]
- 90. Harbour SN, Maynard CL, Zindl CL, Schoeb TR, Weaver CT. Th17 cells give rise to th1 cells that are required for the pathogenesis of colitis. Proc Natl Acad Sci U S A. 2015;112(22):7061‐7066. doi: 10.1073/pnas.1415675112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zielinski CE, Mele F, Aschenbrenner D, et al. Pathogen‐induced human Th17 cells produce IFN‐gamma or IL‐10 and are regulated by IL‐1beta. Nature. 2012;484(7395):514‐518. doi: 10.1038/nature10957 [DOI] [PubMed] [Google Scholar]
- 92. Tsanaktsi A, Solomou EE, Liossis SC. Th1/17 cells, a subset of Th17 cells, are expanded in patients with active systemic lupus erythematosus. Clin Immunol. 2018;195:101‐106. doi: 10.1016/j.clim.2018.08.005 [DOI] [PubMed] [Google Scholar]
- 93. Kadambari S, Whittaker E, Lyall H. Postnatally acquired cytomegalovirus infection in extremely premature infants: how best to manage? Arch Dis Child Fetal Neonatal ed. 2020;105(3):334‐339. doi: 10.1136/archdischild-2019-317650 [DOI] [PubMed] [Google Scholar]
- 94. Kaplan JE, Benson C, Holmes KK, Brooks JT, Pau A, Masur H. Guidelines for prevention and treatment of opportunistic infections in HIV‐infected adults and adolescents: recommendations from CDC, the national institutes of health, and the HIV medicine association of the infectious diseases society of america. MMWR Recomm Rep 2009;58(Rr‐4):1–207; quiz CE201‐204. [PubMed] [Google Scholar]
- 95. Grifka‐Walk HM, Lalor SJ, Segal BM. Highly polarized Th17 cells induce EAE via a T‐bet independent mechanism. Eur J Immunol. 2013;43(11):2824‐2831. doi: 10.1002/eji.201343723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Rowan AG, Fletcher JM, Ryan EJ, et al. Hepatitis C virus‐specific Th17 cells are suppressed by virus‐induced TGF‐beta. J Immunol. 2008;181(7):4485‐4494. doi: 10.4049/jimmunol.181.7.4485 [DOI] [PubMed] [Google Scholar]
- 97. Zhang JY, Zhang Z, Lin F, et al. Interleukin‐17‐producing CD4(+) T cells increase with severity of liver damage in patients with chronic hepatitis B. Hepatology. 2010;51(1):81‐91. doi: 10.1002/hep.23273 [DOI] [PubMed] [Google Scholar]
- 98. Schultheiss C, Paschold L, Simnica D, et al. Next‐generation sequencing of T and B cell receptor repertoires from COVID‐19 patients showed signatures associated with severity of disease. Immunity. 2020;53(2):442‐455. doi: 10.1016/j.immuni.2020.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420‐422. doi: 10.1016/s2213-2600(20)30076-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhao Y, Kilian C, Turner JE, et al. Clonal expansion and activation of tissue‐resident memory‐like Th17 cells expressing GM‐CSF in the lungs of severe COVID‐19 patients. Sci Immunol. 2021;6(56). doi: 10.1126/sciimmunol.abf6692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Mucida D, Park Y, Kim G, et al. Reciprocal Th17 and regulatory t cell differentiation mediated by retinoic acid. Science. 2007;317(5835):256‐260. doi: 10.1126/science.1145697 [DOI] [PubMed] [Google Scholar]
- 102. Dooms H, Kahn E, Knoechel B, Abbas AK. IL‐2 induces a competitive survival advantage in T lymphocytes. J Immunol. 2004;172(10):5973‐5979. doi: 10.4049/jimmunol.172.10.5973 [DOI] [PubMed] [Google Scholar]
- 103. Jelley‐Gibbs DM, Lepak NM, Yen M, Swain SL. Two distinct stages in the transition from naive CD4 T cells to effectors, early antigen‐dependent and late cytokine‐driven expansion and differentiation. J Immunol. 2000;165(9):5017‐5026. doi: 10.4049/jimmunol.165.9.5017 [DOI] [PubMed] [Google Scholar]
- 104. Malek TR, Yu A, Vincek V, Scibelli P, Kong L. CD4 regulatory T cells prevent lethal autoimmunity in IL‐2rbeta‐deficient mice. Implications for the nonredundant function of IL‐2. Immunity. 2002;17(2):167‐178. doi: 10.1016/s1074-7613(02)00367-9 [DOI] [PubMed] [Google Scholar]
- 105. Couper KN, Lanthier PA, Perona‐Wright G, et al. Anti‐CD25 antibody‐mediated depletion of effector T cell populations enhances susceptibility of mice to acute but not chronic Toxoplasma gondii infection. J Immunol. 2009;182(7):3985‐3994. doi: 10.4049/jimmunol.0803053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Shi M, Lin TH, Appell KC, Berg LJ. Janus‐kinase‐3‐dependent signals induce chromatin remodeling at the IFNG locus during T helper 1 cell differentiation. Immunity. 2008;28(6):763‐773. doi: 10.1016/j.immuni.2008.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Solomon I, Amann M, Goubier A, et al. CD25‐Treg‐depleting antibodies preserving IL‐2 signaling on effector T cells enhance effector activation and antitumor immunity. Nat Cancer. 2020;1(12):1153‐1166. doi: 10.1038/s43018-020-00133-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. van Besouw NM, Balk AH, van Vliet M, et al. Anti‐CD25 therapy impairs donor‐specific Th1 and Th2 cytokine‐producing peripheral blood cells after clinical heart transplantation. Transplant Proc. 2002;34(7):2942‐2943. doi: 10.1016/s0041-1345(02)03498-x [DOI] [PubMed] [Google Scholar]
- 109. Deodhar A, Mease PJ, McInnes IB, et al. Long‐term safety of secukinumab in patients with moderate‐to‐severe plaque psoriasis, psoriatic arthritis, and ankylosing spondylitis: integrated pooled clinical trial and post‐marketing surveillance data. Arthritis Res Ther. 2019;21(1):111. doi: 10.1186/s13075-019-1882-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207‐210. doi: 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Dhital R, Anand S, Zeng Q, et al. Th1/17 cells infiltrate murine cytomegalovirus‐infected renal allografts via virus‐induced CCL20 and promote Th1 cells through IL‐17a. bioRxiv 2021:2021.2008.2005.455061. doi: 10.1101/2021.08.05.455061 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supplementary Information
Data Availability Statement
Data are available in the main text or Supporting Information. RNA sequencing data are deposited in the NCBI Gene Expression Omnibus 110 (accession #GSE179788). A pre‐print version of this manuscript is available at https://www.biorxiv.org/content/10.1101/2021.08.05.455061v1. 111 However, this manuscript differs from the pre‐print version by showing that αIL‐17A modulates allograft damage by direct inhibition of IL‐17A and not through indirect effects on Treg/Th1 cell infiltrates, and that αIL‐17A treatment does not increase viral replication. Therefore, this manuscript differs from the pre‐print in content and interpretation.


