Abstract
Background:
Cystatin SN (CST1) and cystatin SA (CST2) are cysteine protease inhibitors that protect against allergen, viral, and bacterial proteases. Cystatins are overexpressed in the setting of allergic rhinitis and chronic rhinosinusitis with nasal polyps (CRSwNP); however, their role in promoting type 2 inflammation remains poorly characterized.
Objective:
The purpose of this study was to use integrated poly-omics and a murine exposure model to explore the link between cystatin overexpression in CRSwNP and type 2 inflammation.
Methods:
In this institutional review board– and institutional animal care and use committee–approved study, we compared tissue, exosome, and mucus CST1 and CST2 between CRSwNP and controls (n = 10 per group) by using matched whole exome sequencing, transcriptomic, proteomic, posttranslational modification, histologic, functional, and bioinformatic analyses. C57/BL6 mice were dosed with 3.9 μg/mL of CST1 or PBS intranasally for 5 to 18 days in the presence or absence of epithelial ABCB1a knockdown. Inflammatory cytokines were quantified by using Quansys multiplex assays or ELISAs.
Results:
Of the 1305 proteins quantified, CST1 and CST2 were among the most overexpressed protease inhibitors in tissue, exosome, and mucus samples; they were localized to the epithelial layer. Multiple posttranslational modifications were identified in the polyp tissue. Exosomal CST1 and CST2 were strongly and significantly correlated with eosinophils and Lund-Mackay scores. Murine type 2 cytokine secretion and TH2 cell infiltration increased in a time-dependent manner following CST1 exposure and was abrogated by epithelial knockdown of ABCB1a, a regulator of epithelial cytokine secretion.
Conclusion:
CST1 is a potent upstream initiator of epithelial-derived type 2 inflammation in CRSwNP. Therapeutic strategies targeting CST activity and its associated posttranslational modifications deserve further interrogation.
Keywords: Epithelium, sinonasal mucus, cystatin SA, cystatin SN, P-glycoprotein, proteomics, transcriptome, exome, posttranslational modification
Chronic rhinosinusitis (CRS) affects more than 30 million Americans, resulting in $6.9 to $9.9 billion in annual health care expenditures1,2 and $12.8 billion in productivity costs.3 The subset of patients with CRS with nasal polyps (CRSwNP) has an estimated prevalence between 2% to 5%4–6 and represents the most challenging population with respect to disease severity, recurrence after surgery, and lack of effective therapies.5–7 Current state-of-the-art treatments for CRSwNP have targeted the TH2 cytokine cascade,8–11 as type 2 skewing has been shown to be a dominant feature in the majority of cases.12 Despite the recent success of targeted biologics in CRSwNP, the persistence of non-responders and relapsing nature of the disease following treatment cessation13 suggests that the search for upstream drivers of type 2 inflammation remains salient.
Current descriptions of the immune pathways underpinning type 2 CRS fail to implicate which, if any, environmental pathogens may be driving the aberrant inflammation and epithelial barrier dysfunction. Multiple hypotheses have been put forth, including bacteria (eg, Staphylococcus aureus14 and biofilms15), fungus,16 and allergens17; however, none has been definitively linked to the development of nasal polyps, and more specifically, their eradication has not been convincingly shown to reverse the disease process.
Proteomic investigations by our group have previously identified cystatins as being among the most sensitive and specific predictors of CRSwNP.18 These findings have since been validated by multiple independent laboratories.19–21 Cystatins constitute a large group of evolutionarily related proteins acting as protease inhibitors on papain-like proteases belonging to the enzyme family C1. The type 2 cystatins C, D, E/M, F, S, SN, and SA are characterized by 2 conserved disulfide bridges and the presence of a signal peptide for extracellular targeting. Cystatin SN (CST1) and cystatin SA (CST2) are expressed at the nasal epithelial surface; their purpose is to control cysteine proteases, which are widely expressed proteolytic enzymes within allergens,22 viruses, and bacteria and have established roles in inflammatory tissue remodeling.23,24 Cystatins have also been shown to have direct immunomodulatory,25 antimicrobial,25,26 and antiviral properties.27
Prior studies have shown that CST1 expression is induced following exposure to type 2 cytokines, including TSLP and IL-33, and can reciprocally stimulate TSLP expression in combination with double-stranded DNA and IL-4, thereby forming a proinflammatory feedback loop.19 Although intriguing, these results fail to resolve the “chicken or egg” question as to which event precedes which. The epithelial localization of CST1, coupled with its role in protecting against diverse type 2 CRS–associated environmental pathogens, led us to hypothesize that CST1 functions as an upstream driver of epithelial-derived type 2 inflammation rather than a consequence thereof. The goal of this study was to therefore test this hypothesis by (1) using poly-omics to characterize the expression of cystatins in nasal polyps, (2) determining the time-dependent impact of recombinant CST1 exposure on type 2 cytokine secretion in a murine model, and (3) abrogating epithelial inflammation by using ABCB1a knockdown to dissect the impact of CST1 on type 2 cytokine secretion.
METHODS
Patient tissue and mucus sampling
Tissue and mucus sampling were approved by the Massachusetts Eye and Ear Infirmary institutional review board. Matched tissue and mucus exosomal samples were taken from patients who were undergoing sinonasal surgery and had not been exposed to antibiotics or any topical/systemic steroids for at least 4 weeks. In accordance with the inclusion criteria, the study population consisted of patients diagnosed with CRSwNP (as specified by the international consensus statement on allergy and rhinology Rhinosinusitis 202128 criteria) and healthy patients (ie, controls) undergoing surgery for noninflammatory disease. The exclusion criteria included ciliary dysfunction, autoimmune disease, cystic fibrosis, and immunodeficiency. Among the controls, additional exclusion criteria included the presence of environmental allergy or asthma so as to avoid potentially confounding comorbidities. Mucus samples were taken before tissue sampling by placing compressed polyvinyl alcohol sponges (Medtronic, Minneapolis, Minn) against the middle meatus for 5 minutes, taking care to not abrade the mucosa or contaminate the sponge with blood. Tissue and exosomes were sampled from 10 and 20 patients per group, respectively.
Exosome purification technique from whole mucus
The exosome purification procedure was adapted from the ultracentrifugation technique described by Théry et al29 and previously reported by our group.30 Mucus samples were extracted from the polyvinyl alcohol sponges by centrifugation (1500 g at 4°C for 30 minutes). The mucus was then diluted in 150 μL of 1× PBS (Life Technologies, Carlsbad, Calif) with protease inhibitor cocktail (1:100, Sigma, St Louis, Mo). Cellular debris was pelleted by centrifugation for 45 minutes at 12,000 g at 4°C. The supernatant was then suspended in 4.5 mL of PBS in polypropylene tubes (Thinwall, 5.0 mL, 13 × 51 mm, Beckman Coulter, Indianapolis, Ind) and ultracentrifuged for 2 hours at 110,000 g, at 4°C. The supernatant was collected, and the pellet was resuspended in 4.5 mL of 1× PBS. The suspension was filtered through a 0.22-μm filter (Fisher Scientific, Pittsburgh, Pa) and collected in a fresh ultracentrifuge tube. The filtered suspension was then centrifuged for 70 minutes at 110,000 g at 4°C. The supernatant was collected, and the pellet was resuspended in 175 μL of PBS M-per with protease inhibitor for further proteomic analysis.
Western blots
Western blots were performed in an independent group of CRSwNP and control (n = 6 per group) tissue- and mucus-derived exosomal samples to validate the proteomic results. Homogenization (T25 Basic, IKA Labortechnik, Staufen, Germany) of 30 to 40 mg of each tissue sample was accomplished in 1.0 mL of lysis buffer (T-PER Mammalian Protein Extraction Reagent [Thermo Scientific, Bonn, Germany] and protease inhibitor cocktail Complete1 [Roche, Mannheim, Germany]). The samples were then incubated at 4°C for 2 hours and centrifuged at 16,000 g for 1 hour. The total protein concentration of the supernatant was determined with bicinchoninic acid assays (Thermo Fisher Scientific, Bonn, Germany). A quantity of 50 μg of lysed tissue protein was used for each patient. Exosomes were isolated as already described; 2 μg of lysed exosomal protein was used for each patient. After denaturation at 90°C for 5 minutes with SDS loading buffer including mercaptoethanol, lysates were applied on 8% to 15% SDS-PAGE and transferred to nitrocellulose membranes (Protran-BA-83, Schleicher & Schuell, Keene, NH). Glyceraldehyde-3–phosphate dehydrogenase (GAPDH) (rabbit mAb against GAPDH, clone D16H11, New England Biolabs GmbH, Frankfurt, Germany) staining served as a control. The primary detection antibodies were mouse anti-CST1, clone 213139 (R&D Systems, Abingdon, UK) for CST1 and CST2 antibody (OTI1D6), TrueMAB Mouse Monoclonal (Thermo Fisher Scientific, Bonn, Germany) for CST2, followed by the secondary antibody (peroxidase-labeled anti-mouse/rabbit IgG, F(ab’)2 antibody (KPL, Gaithersburg, Md). The blot was incubated with SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific), and the signals were imaged using ChemStudio PLUS (Analytik Jena, Jena, Germany). Quantification of band intensity was performed using VisionWorks 8.2 (Analytik Jena).
Immunohistochemistry
Paraffin-embedded, formalin-fixed tissue samples were sectioned, deparaffinized, and rehydrated before staining. Antigen retrieval was performed by using proteinase K (Thermo Fisher Scientific, Waltham, Mass) for 5 minutes at room temperature. After being washed twice, the tissue was blocked with BLOXALL endogenous peroxidase and alkaline phosphatase blocking solution (Vector Laboratories, Burlingame, Calif) for 10 minutes at room temperature in a humidified chamber. The slides were washed once and then blocked with horse serum (2.5% normal horse serum blocking solution, Vector Laboratories) for 20 minutes at room temperature in a humidified chamber. The blocking solution was removed, and the primary antibody (1:7500 anti–cystatin SA antibody or 1:3000 anti–cystatin SN antibody, Abcam, Cambridge, Mass) in 1% BSA/PBS was incubated on the slides overnight at 4°C. The secondary antibody, from the ImmPRES Horseradish Peroxidase Anti-Mouse IgG (Peroxidase) Polymer Detection Kit (Vector Laboratories), was applied for 30 minutes at room temperature. After being rinsed 3 times, the slides were developed with ImmPACT 3,3’-diaminobenzidine peroxidase (horseradish peroxidase) substrate (Vector Laboratories) for 1 minute at room temperature, after which the process was stopped with double-distilled H2O for 5 minutes. The slides were counterstained with hematoxylin, dehydrated with xylenes, and coverslipped with Permount mounting medium.
Proteomic analysis
The SOMAscan proteomic analysis (SomaLogic, Boulder, Colo) on nasal tissue and isolated exosomes was run by using the SOMAscan Assay Cells and Tissue Kit, 1.3k (SomaLogic 900-00009) following the recommended protocol from the manufacturer. For nasal tissue extracts and mucus, 2.4 μg of protein from each sample was run. With exosomes isolated from nasal mucus, 1.7 μg per sampleof proteinwas used in the SOMAscan assay. Three kit-provided controls and 1 no-protein buffer control per plate were run in parallel with the samples. Median normalization and calibration of the SOMAscan data was performed according to the standard quality control protocols at SomaLogic.
DNA and RNA extraction
DNA and RNA extraction are described in the Methods section of the Supplementary Material (see the Online Repository at www.jacionline.org).
Transcriptome analysis
The transcriptome analysis is described in the Methods section of the Supplementary Material.
PTMs
The posttranslational modification (PTM) analysis is described in the Methods section of the Supplementary Material.
Murine model
All the animal procedures were approved by the Northeastern University institutional animal care and use committee. Male and female C57BL/6 mice were purchased from Charles River Laboratories (Wilmington, Mass), kept in specific pathogen–free conditions, and used at 6 to 8 weeks of age.
Liposome preparation and characterization
The cationic liposome formulations were adapted from Pawar et al.31 First, stock solutions of 1,2-dioleoyl-3-trimethylammonium-propane chloride salt (Avanti Lipids, Alabaster, Ala), a cationic lipid cholesterol (stabilizer, Avanti Lipids), and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (Avanti Lipids), a neutral lipid, were prepared individually in chloroform. Cationic lipid films were prepared by combining the lipids in a 5:3:5 molar ratio, after which the chloroform solvent was evaporated using a Rotovap and dried overnight.
Small interfering RNA (siRNA) for murine ABCB1a (synthesized by Sigma Aldrich, HPLC-purified) and a universal negative control (MISSION siRNA Universal Negative Control 1, Sigma Aldrich) were used. The knockdown approach was selected over pharmacologic inhibition to more efficiently abrogate ABCB1a activity throughout the course of the experiments. The ABCB1a sequence was adapted from Matsui et al32; it had the sense sequence 5’-AAU GUU GUC UGG ACA AGC ACU dTdT-3’ and the antisense sequence 5’[Phos]AGU GCU UGU CCA GAC AAC AUU dTdT-3’. siRNA was diluted to 1 mg/mL in 1 mL 5% dextrose (dextrose [D-glucose], anhydrous, Fisher Scientific) prepared with UltraPure distilled water (Invitrogen). This solution was added to the liposome film and vortexed for 2 minutes. The solution was then subjected to 5 freeze-thaw cycles as follows: ice for 2 minutes, vortexing for 30 seconds, water bath at 37°C for 2 minutes, and vortexing for 30 seconds. The liposomal preparation was then probe-sonicated twice for 8 minutes on ice. The liposomes were then centrifuged twice in a 100-kDa filter (Amicon Ultra-15; Sigma Aldrich) at 14,000 rpm for 15 minutes at 4°C. The resulting volume of solution was then brought back to 0.25 mL with 5% dextrose. A Zetasizer (Nano-ZS90, Malvern Instruments, Inc, Westborough, Mass) was used to characterize the hydrodynamic diameter, polydispersity index, and surface charge (zeta potential) of the liposomes. The encapsulation efficiency was indirectly measured by using the Quant-iT RiboGreen RNA Assay Kit (Invitrogen, Thermo Fisher) according to the manufacturer’s instructions.
In vivo dosing
To induce sinonasal inflammation, mice were anesthetized and treated intranasally with 3.9 μg/mL of CST1 (Recombinant Human Cystatin SN Protein, CF, R&D Systems) in 20 μL of PBS, a concentration taken from Fukuoka et al.33 A 7.8-μg/mL dose was initially tested but was poorly tolerated. Animals were dosed daily for 5, 10, or 18 days for the cytokine experiments and 18 days for all other experiments. The groups receiving liposomal siRNA were dosed with 0.1 or 1 mg/kg on days 11 to 15, for a total of 0.5 or 5 mg/kg. Liposomes were dosed in 20 μL of 5% dextrose intranasally.
Tissue harvest
At 24 hours after the last administration of CST1, the mice were humanely killed and their tissue was collected. Septal tissue was harvested by decapitating the mouse after it had been killed and removing the lower jaw, front teeth, and skin. Then, to remove the septal tissue using a razor blade (Grainger Razor Blades, Thermo Fisher Scientific), the skull was cut along the suture line of the nasal bones. Fine scissors were used to isolate the septal tissue as previously described.34
Protein extraction and analysis
Protein was extracted from the mouse tissue by using CelLytic MT (Sigma Aldrich). The tissue and CelLytic MT solution were homogenized for 60 seconds and then incubated at 4°C for 15 minutes. Tissue debris was pelleted by centrifugation at 17,000 g at 4°C for 10 minutes, and the supernatant was frozen at −80°C until analysis. The lysate supernatants were then analyzed by using a Custom Mouse Cytokine Q-Plex Array for the detection of IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-10, IL-12, IL-13, IL-17, MCP1, IFN-γ, TNF-α, MIP-1a, GM-CSF, RANTES, and Eotaxin as well as ELISA kits for an ABCB1b (P-glycoprotein; USCN Life Sciences, Inc, Wuhan, China) and ABCB1a (MyBiosource, Inc, San Diego, Calif). All values were normalized to the total protein concentration within the same sample by using a Micro BCA Protein Assay Kit (Pierce, Rockford, Ill).
RNA extraction and analysis
The harvested murine sinonasal tissue was placed in RNAlater (Thermo Fisher) and frozen at −80°C until RNA isolation. RNA was isolated by using TRIzol reagent (Invitrogen) followed by the Qiagen QIAshredder and RNeasy Mini kits (Qiagen, Hilden, Germany), according to the manufacturer’s instructions for tissue samples. Total RNA was quantified by using the nanodrop, and 2 μg was used for the cDNA kit (High-Capacity cDNA Reverse Transcription Kit, Thermo Fisher). Mouse TaqMan primers for ABCB1a (Mm00440761_m1), ABCB1b (Mm00440736_m1), and the housekeeping gene (peptidylpropyl isomerase B, Mm00478295_m1) and TaqMan Gene Expression Master Mix (Thermo Fisher) were used for the quantitative PCR procedure. The samples were analyzed by using a LightCycler 480 PCR system (Roche Diagnostics).
Flow cytometry
The mucosal tissue from 4 mice was pooled was digested into a single-cell suspension. The tissue was placed in 6 mL of dissociation buffer consisting of RPMI 1640 medium (Invitrogen, ThermoFisher, Bedford, Mass), penicillin/streptomycin (Invitrogen), 0.4 mg/mL of DNase I (Sigma Aldrich, 2×), and 50 μg/mL of Liberase TL (Roche Diagnostics). The tissue was digested for 45 minutes at 37°C with rotation, and the digestion was stopped with 1200 μL of FBS (Corning Life Sciences, Bedford, Mass). The resulting solution was then filtered and centrifuged to obtain a cell pellet that was resuspended in PBS.
A Nexcelom Cellaca MX High-throughput Automated System (Nexcelom Bioscience, Lawrence, Mass) was used for counting of all processed suspensions before staining. For intracellular cytokine staining, suspension cells were each aliquoted for an unstimulated and a PMA/ionomycin-stimulated sample by using the eBioscience Cell Stimulation Cocktail (plus protein transport inhibitor [500×]) (ThermoFisher) and the eBioscience Protein Transport Inhibitor Cocktail (500×) (ThermoFisher) following the manufacturer’s recommendations. Both unstimulated and stimulated aliquots were incubated in RPMI-1640 medium containing 10% FBS (vol/vol) for 4 hours at 37°C in a 5% CO2 atmosphere.
Cells were then harvested, washed, and prepared for staining. Cells were stained with Fixable Viability Dye (eBioscience Fixable Viability Dye eFluor 780, ThermoFisher), mouse CD45 (fluorescein isothiocyanate, clone 11-0451-82, ThermoFisher), mouse CD3 (phycoerythrin–cyanine 7, clone 17A2, BioLegend, San Diego, Calif), mouse CD4 (peridinin-chlorophyll-protein–cyanine 5.5, clone GK1.5, BioLegend), mouse IFN-γ (phycoerythrin, clone XMG1.2, BioLegend), and mouse IL-4 (BV 421, clone 11B11, BioLegend). Fluorescence minus 1 controls were prepared from spleen for each cell marker and used for gating. Fixed cells were finally transferred to a 96-well plate in a final volume of 300 μL of BD stain buffer and kept at 4°C until they were acquired at a rate of 100 μL per minute on an Attune NxT flow cytometry system (ThermoFisher, 4 lasers). Compensation controls were prepared from Ultra-Comp eBeads Plus Compensation Beads (Invitrogen, ThermoFisher) and single-cell suspensions. Raw data were captured as .fcs files and analyzed in FlowJo software (version 10) for gating immune cell populations. GraphPad Prism (version 5.02) was used for graphical representations of data.
Statistical analysis
Protein expression profiling was performed by using the SomaLogic SOMAscan platform35 for 1305 proteins, including 12 controls. All samples passed the established technical quality control criteria according to the standard quality control protocols at SomaLogic. Raw data (abundance signal values) were normalized by using the rank invariant set method.36 Briefly, proteins that did not exhibit large differences in their signal rank between 2 samples were labeled as the rank invariant set, and a lowest smoothing line was fitted through their expression values to define the normalization equation.
Similarity between samples in the same phenotypic group was assessed by calculating the Pearson correlation coefficient between each pair of samples in the group. Differential protein expression was calculated by using the Student t test followed by the Benjamini-Hochberg procedure37 for multiple hypotheses testing correction. A false discovery rate (FDR) value less than 0.05 was considered statistically significant.
Transcriptome assembly and gene expression quantification were performed by using StringTie (version 1.3.3b). Ballgown (version 2.0.0) was then invoked to perform differential gene expression analysis between phenotypic groups. An FDR cutoff of 0.05 was used to select significantly differentially expressed genes, which were then correlated to the protein expression by using a Spearman correlation analysis. Principal component analysis (PCA) was carried out with log2 (fragments per kilobase of transcript per million mapped reads + 1) of expressed genes by using the function pca() in R (version 3.2.2) package FactoMineR (version 1.31.4), which identified sample P9 as an outlier that was excluded from downstream analysis.
Clustering of samples and/or proteins was performed by using the unweighted pair group method with arithmetic mean method with Pearson correlation as the distance measure.38 The expression data matrix was row-normalized before the application of average linkage clustering. A linear regression analysis was performed for the correlation between tissue and exosomes. R2 = 1 was defined as the maximum correlation. For analysis of the relationship between tissue, mucus, and exosomal samples, hierarchic cluster analysis and Pearson correlation analyses were performed by using Morpheus software (https://software.broadinstitute.org/morpheus/) and data were log2-transformed in the analysis. Two-tailed t tests were used to compare individual groups.
All cytokine and RNA analyses were performed in technical duplicates. Statistical significance was determined by using the Student t test or repeated 2-way t tests, followed by the Benjamini-Hochberg procedure for multiple hypotheses testing correction.37 An FDR value less than 0.05 was considered statistically significant. Values falling outside 1.5 times the interquartile range (IQR) of their respective data sets were considered outliers and indiscriminately excluded from analysis, except for the flow cytometry data. Results were considered significant when a P value of .05 or less was obtained.
RESULTS
Proteomic and transcriptomic expression analyses
Among the entire 1,305 protein SOMAscan array of biologically relevant immune and inflammatory proteins, CST1 and CST2 were the seventh and second most overexpressed mucosal proteins between the samples from patients with CRSwNP (mean ± SD 5 52,214 ± 23,857 relative fluorescence units [RFU] and 67,820 ± 52,932 RFU, respectively) and the controls (mean ± SD = 7,735 ± 14,377 RFU and 4,427 ± 10,345 RFU, respectively) (fold change [FC] = 6.75 [P = 6.70 × 10−5] and 15.31 [P = 1.72 × 10−4], respectively). A similar pattern was seen in matched exosome and mucus samples. Among the 44 protease and protease inhibitors present within the human nose, CST1 and CST2 demonstrated the greatest positive mRNA FC between the samples from patients with CRSwNP and the controls (FC = 73.11 [P = .002] and 18.37 [P = .02], respectively) among samples matched to the protein analysis (Fig 1 and Table I).
FIG 1.
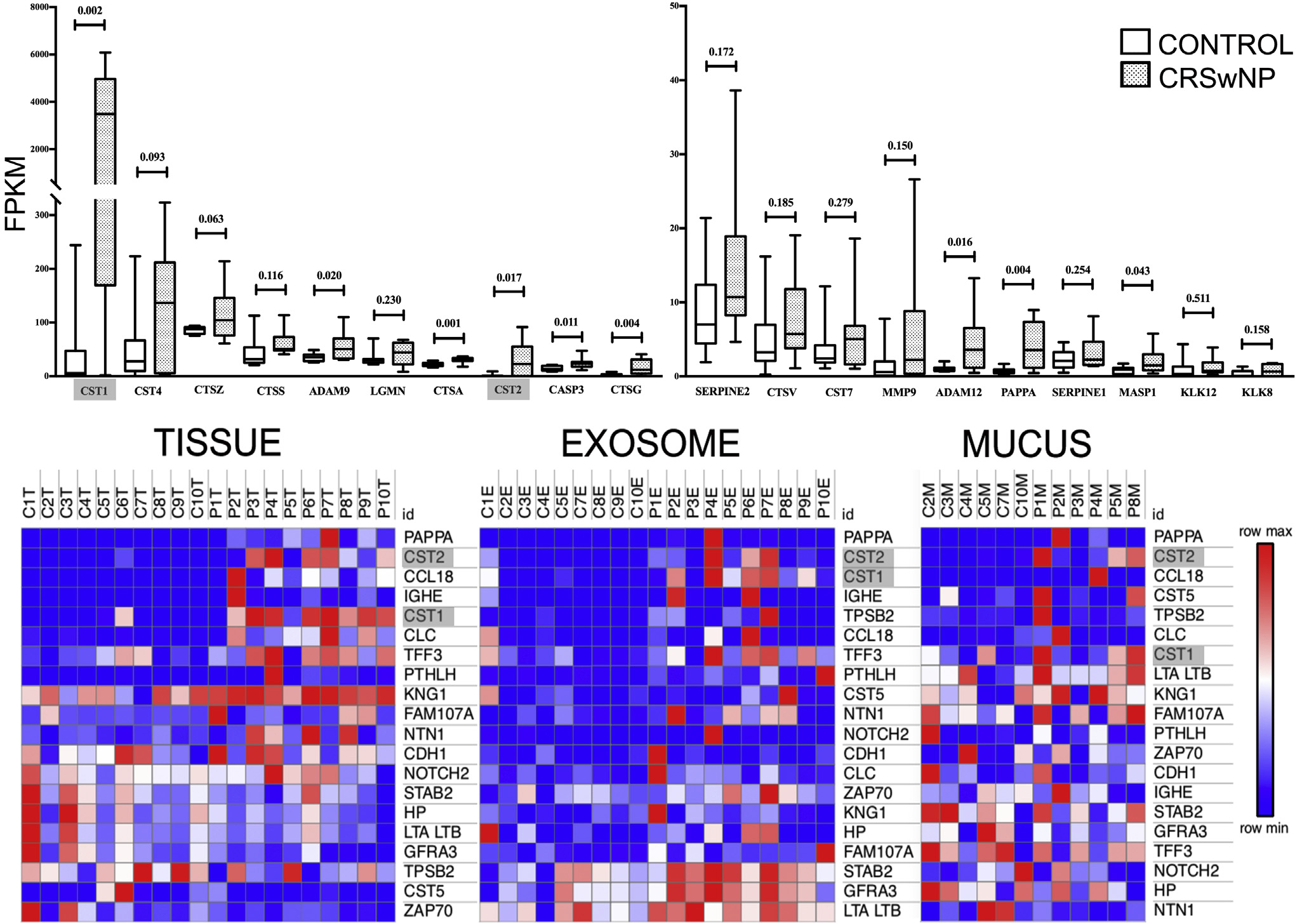
Proteomics data on matched clinical samples. Top row, Box and whisker plots of tissue mRNA expression (by fragments per kilobase of transcript per million mapped reads [FPKM]) of the top 20 protease and protease inhibitors included in the proteomic array. Note that CST1 has both the highest expression in CRSwNP and the greatest FC relative to the control (73.1; CST2 FC = 18.4) among the proteins examined. Bottom row, Heatmaps of top 20 proteins by FC. Individual boxes represent protein concentration by RFU) within matched tissue, exosome, and mucus samples among the entire 1305-protein SOMAscan array. Note that CST1 and CST2 are among the most overexpressed proteins across all CRSwNP sample sources. Data represent 10 control patients (C) and 10 patients with CRSwNP (P).
TABLE I.
CST1 and CST2 expression among matched tissue mRNA, exosome, and mucus samples
| Control |
CRSwNP |
|||||
|---|---|---|---|---|---|---|
| Cystatin | Mean | SD | Mean | SD | FC | P value |
|
| ||||||
| CST1 | ||||||
| mRNA (FPKM) | 38.38 | 75.13 | 2,844.39 | 2,470.62 | 73.11 | <.0001 |
| Exosome (RFU) | 28,503 | 39,401 | 109,146 | 68,733 | 3.82 | .003 |
| Mucus (RFU) | 33,126 | 27,545 | 41,646 | 34,468 | 1.6 | .23 |
| CST2 | ||||||
| mRNA (FPKM) | 1.51 | 3.17 | 29.17 | 33.02 | 18.37 | .016 |
| Exosome (RFU) | 7,102 | 13,395 | 48,559 | 43,260 | 6.8 | <.001 |
| Mucus (RFU) | 11,666 | 9,808 | 66,269 | 80,094 | 7.3 | .04 |
FPKM, Fragments per kilobase of transcript per million mapped reads.
Western blot confirmation and histopathologic localization of CST1 and CST2 expression
Among the tissue samples, protein expression by Western blot harmonized with the SOMAscan data. The GAPDH-normalized CST1 and CST2 concentrations within CRSwNP polyp tissue (median = 7714.0 [IQR = 3866.0–9853.0] and 2317.0 [IQR = 756.5–4065.0], respectively) were significantly greater than those within CRSwNP inferior turbinate tissue (median = 212.3 [IQR = 150.0–501.5 (P = .002)] and 103.4 [IQR = 50.6–479.8 (P = .002)], respectively) and the control tissue (median = 80.7 [IQR = 12.6–276.0 (P = .002)] and 0.0 [0.0 (P = .02)], respectively). A similar pattern was seen among exosome samples (Fig 2, A). With use of immunohistochemistry, this expression was found to be localized to the epithelial cytoplasm in CRSwNP tissue. No significant staining was seen within the stroma or epithelial glands (Fig 2, B).
FIG 2.
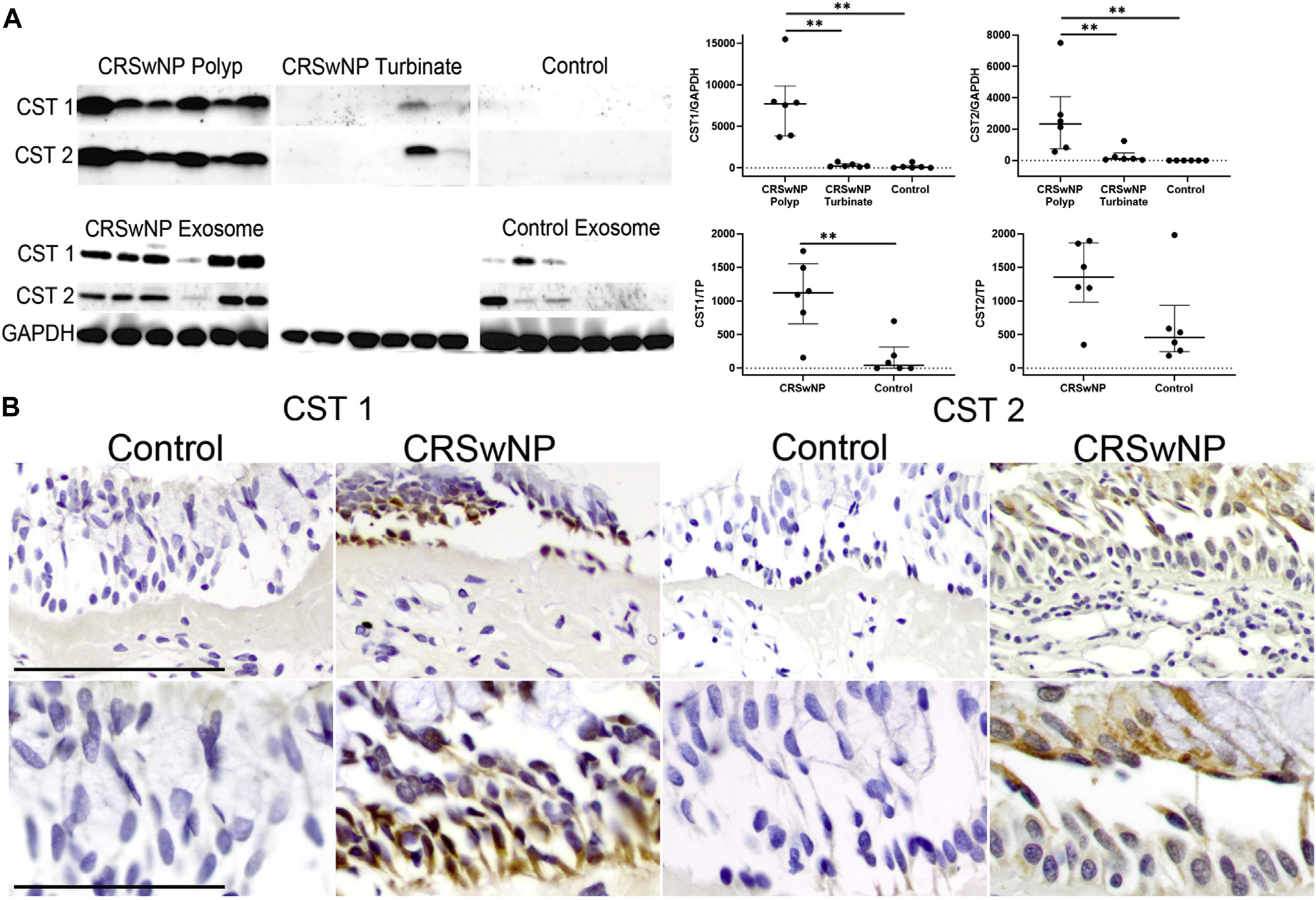
CST1 is present in the epithelium of the control patients and patients with CRSwNP. A, Images and semiquantitative scatter dot plots of Western blots confirming the same expression pattern of CST1 and CST2 in tissue from patients with CRSwNP and exosomes as seen in the SOMAscan readouts (**P < .01). Data represent 6 control patients and 6 patients with CRSwNP (median [IQR]). B, Immunohistochemistry demonstrating differential expression and localization of CST1 and CST2 to the epithelial cytoplasm in the samples from patients with CRSwNP (top row bar = 5 μm; bottom row bar = 2.5 μm). TP, Total protein.
Exosomal CST1 and CST2 are correlated with tissue expression and clinical disease severity
Both exosomal CST1 and CST2 exhibited a strong and significant correlation with matched tissue protein (r = 0.62 [P = .005] and r = 0.68 [P = .001], respectively) and mRNA expression levels (r = 0.59 [P = .008] and r = 0.80 [P < .001], respectively). Similarly, exosomal CST1 and CST2 were strongly and significantly correlated with tissue eosinophil per hpf and Lund-Mackay scores. Exosomal CST1 was significantly correlated with total and domain 1 22-Item Sino-Nasal Outcome Test scores, whereas CST2 was correlated only with domain 1 scores. Mucus CST1 and CST2 protein levels tended to be poorly correlated with other sample sources (eg, exosomal protein, tissue protein, and tissue mRNA), as well as with clinical disease parameters (Fig 3).
FIG 3.
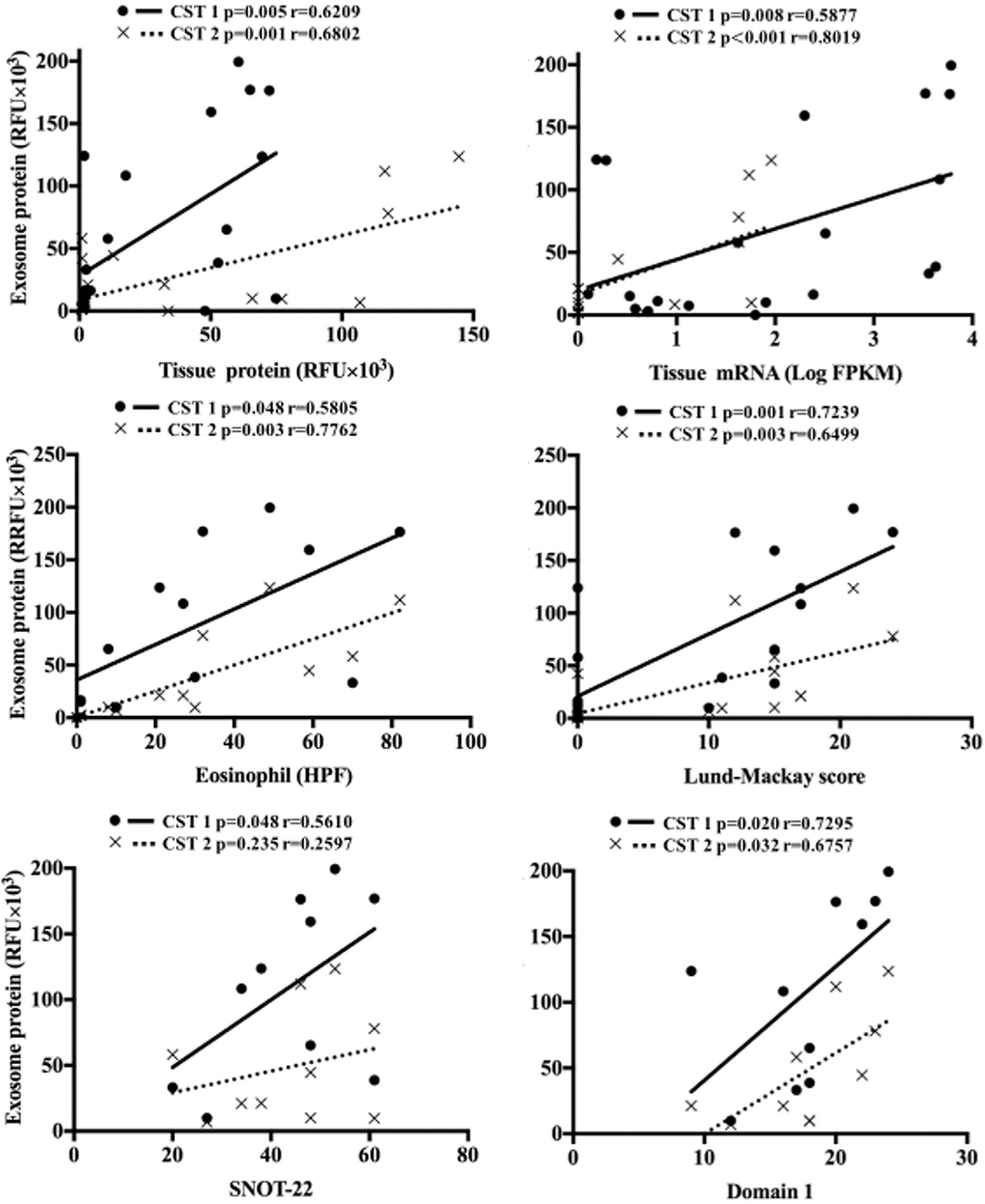
Elevated CST1 and CST2 levels are correlated with clinical measures. A, Scatterplots detailing positive Pearson correlations between exosomal CST1 and CST2 protein with tissue CST1 and CST2 protein, tissue CST1 and CST2 mRNA, and clinical parameters. Data represent 10 patients with CRSwNP.
Exosomal CST1 and CST2 are associated with allergy and AERD
We next examined the association between exosomal CST expression in the setting of comorbidities associated with increased type 2 inflammation, including seasonal allergic rhinitis and aspirin-exacerbated respiratory disease (AERD). None of the control patients (n = 20) had allergy or AERD. In the 20 patients with CRSwNP, 3 had a history of allergy or AERD, with 1 patient endorsing a history of both. Patients with CRSwNP with allergy demonstrated a nonsignificant trend toward increased exosomal CST1 and CST2 expression (mean ± SEM = 80,266 ± 55,527 and 28,001 ± 17,988 RFU, respectively) relative to CRSwNP without allergy (mean ± SEM = 39,992 ± 8,528 [P = .18] and 11,872 ± 5,981 RFU [P = .19], respectively). A similar trend was seen with respect to AERD (see Fig E1 in the Online Repository at www.jacionline.org).
CST1 and CST2 exome analysis
Among the patients with CRSwNP, 3 variants were identified in the promoter region of the CST1 transcript, as compared with 18 variants identified in the transcript region. Similarly, 4 variants were identified in the promoter region for CST2, as compared with 16 variants identified in the transcript region. None of the variants in the CST1 or CST2 transcript region were of high or moderate impact according to functional consequence checking as described in the variant prioritization section (see Table E1 in the Online Repository at www.jacionline.org).
CST1 and CST2 are associated with multiple PTMs in CRSwNP
Among the CST1 proteins, 8 PTMs were identified at an incidence of more than 50% among the CRSwNP group with no corresponding modifications in the control group. These included deamidation at N36 (75%), N40 (50%), N56 (57%), N100 (88%), and Q116 (60%); acetylation at K114 (57%) and K115 (100%); and methylation at R46 (50%). Among the CST2 proteins, 6 PTMs were identified at an incidence of more than 50% in the polyp group with no corresponding modifications in the control group. These included deamidation at Q24 (100%), Q76 (100%), and N82 (100%); acetylation at K114 (50%); and methylation at R28 (100%) and K96 (100%) (Fig 4). No significant PTMs were identified in the control tissue.
FIG 4.

PTMs of CST1 and CST2. Amino acid sequences of CST1 and CST2 proteins with PTMs, which are present in 50% or more of polyp tissue samples but absent in the controls.
CST1 induces type 2 inflammation in murine sinonasal mucosa
In light of the findings that CSTs are both upregulated in CRS and associated with increased type 2 inflammation, we next developed a murine model to determine whether prolonged CST exposure was capable of independently inducing mucosal inflammation within an otherwise healthy background. We elected to focus on CST1 given its relative abundance over CST2. Exposure to CST1 preferentially induced cytokine production among type 2–associated cytokines (Fig 5, A), particularly after 18 days of CST1 treatment. This effect was most pronounced among the epithelial-derived cytokines (IL-6 [mean ± SEM = 128.7% ± 3.0% compared with baseline (P = .003)]) and TH 2 cytokines (IL-4 [mean ± SEM = 130.1% ± 1.7% (P = .012)], IL-5 [mean ± SEM = 155.1% ± 2.8% (P = .0093)], and IL-13 [mean ± SEM = 138% ± 2.5% (P = .0145)]) when compared with baseline. CST1 exposure also increased eotaxin (mean ± SEM = 171.2% ± 9.3% [P = .045]) and P-gp (mean ± SEM =182.1% ± 2.5% [P = .0035]) protein expression in the sinonasal tissue when compared with baseline, which is also indicative of a type 2 response. The most significant cytokine responses occurred after 18 days of CST1 treatment. TH2 cell infiltration was characterized by using flow cytometry. The CST1 treatment increased TH2 cell infiltration in the murine nasal tissue (median [IQR] = 344.4% [75.5%-980.4% (P = .12)]) when compared with the untreated baseline control (Fig 5, B and see Fig E2 in the Online Repository at www.jacionline.org). This is consistent with the increase in production of TH2 cytokines.
FIG 5.
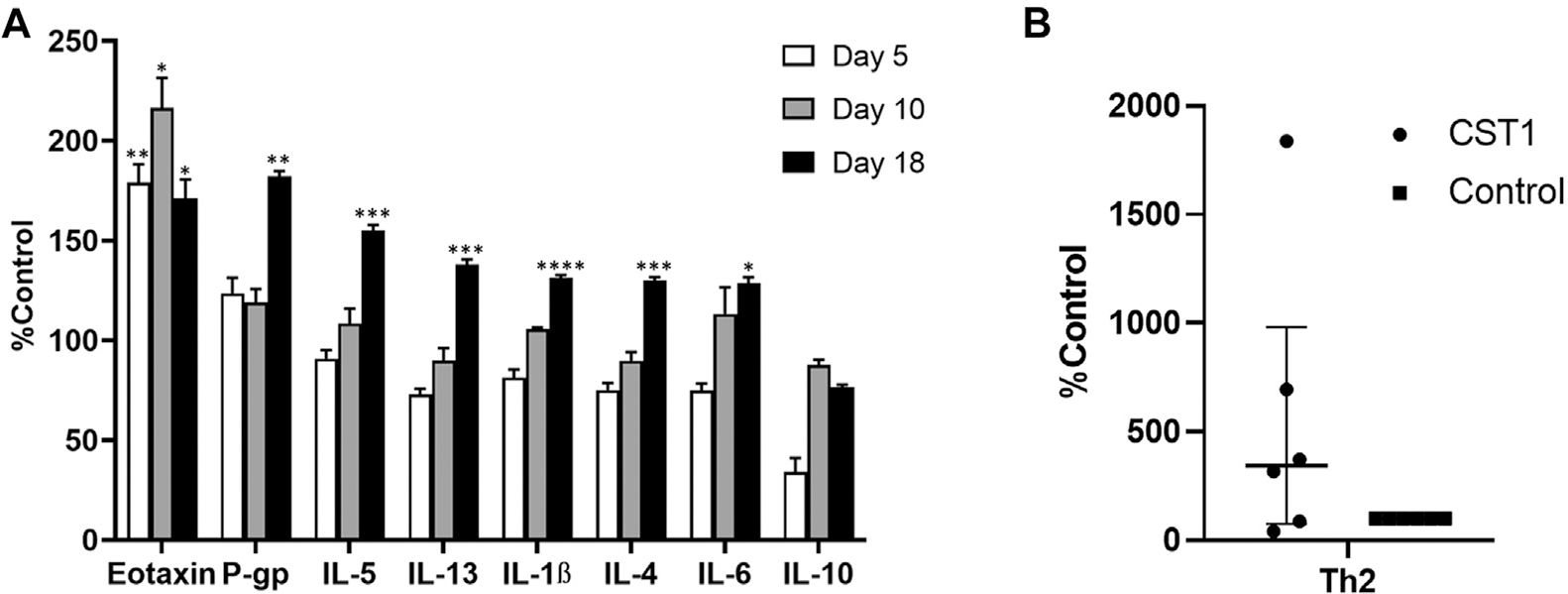
CST1-induced TH2 cytokine production in mouse septonasal tissue. A, Bar graph of cytokine production in mouse septonasal tissue, in which CST1 increased production of TH2 cytokines. Data represent 4 mice per group, mean s ± SEMs. *P < .05; **P < .01; ***P < .001; ****P < .0001. B, Bar graph of TH2 cell infiltration by flow cytometry. Data represent 6 independent experiments with 4 pooled mice per group expressed as medians (IQRs).
ABCB1a siRNA–loaded liposomes knock down TH2 cytokines in a CST1 murine model
After establishing the recombinant CST1 (rCST1) mouse model, we sought to determine whether type 2 inflammation was being orchestrated by the epithelial cells themselves. To achieve this, we utilized knockdown of ABCB1a, which encodes P-glycoprotein, a membrane-bound ATP-dependent pump capable of promoting epithelial type 2 cytokine secretion in a dose-dependent manner.39 ABCB1a siRNA–loaded liposomes significantly knocked down ABCB1a expression at both 1 mg/kg (mean ± SEM = 67.3% ± 16.3% knockdown [P = .01] compared with the CST1 control) and 0.1 mg/kg (mean ± SEM = 56.6% ± 6.6% knockdown [P = .003] compared with the CST1 control) (Fig 6, A).
FIG 6.
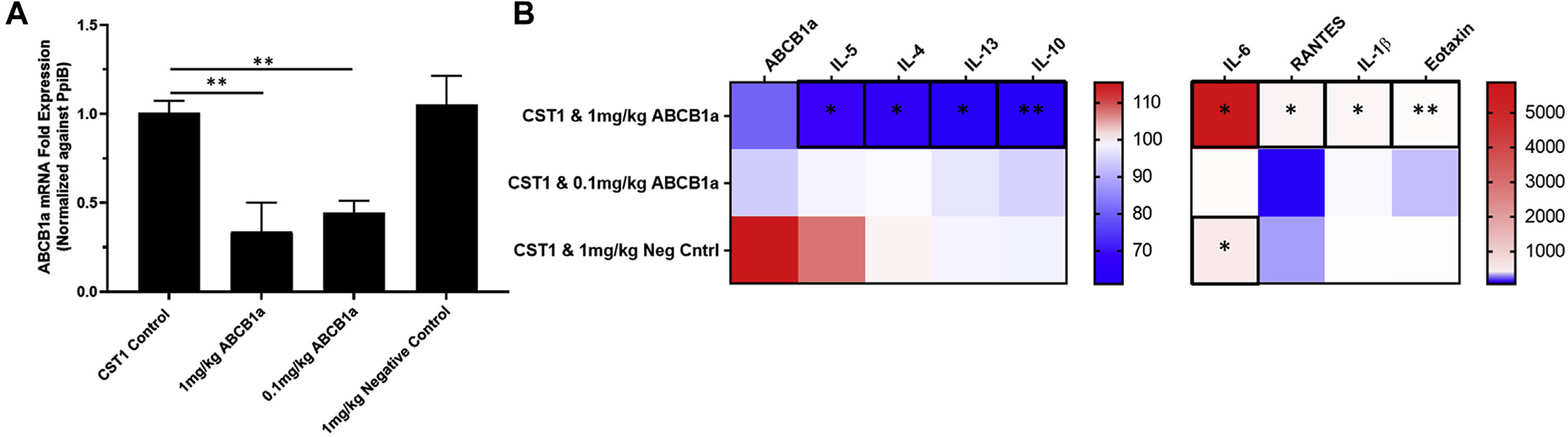
ABCB1a siRNA–encapsulated liposomes knocked down target mRNA and TH2 cytokine expression. A, Bar graph of mRNA expression of ABCB1a mRNA after ABCB1a siRNA–loaded liposome exposure. Data represent 4 mice per group, means ± SEMs. *P < .05; **P < .01; ***P < .001, ****P < .0001. B, Heatmap of cytokine and protein production in mouse septal tissue expressed as percentage of baseline of CST1 control mice. Data represent 4 mice per group, means ± SEMs. * P < .05; **P < .01.
After validating success using our knockdown approach, we next quantified the effect of the siRNA-loaded liposomes on target and cytokine protein levels. Epithelial ABCB1a knockdown reduced CST1-induced type 2 cytokine expression in a dose-dependent manner, particularly in the 1-mg/kg group: IL-4 (66.4% ± 4.4% [P = .03]), IL-5 (68.9% ± 4.8% [P = .04]), IL-10 (61.0% ± 3.4% [P = .004]), and IL-13 (67.9% ± 4.9% [P =.063]) when compared with baseline CST1 (Fig 6, B).
DISCUSSION
Integrated poly-omics refers to a biologic analysis in which multiple omics data sets may be combined through bioinformatics approaches to identify biomarkers and unravel the complex geno-pheno-enviro–type relationships even with low sample numbers.40,41 Our analysis confirmed that both CST1 and CST2 are profoundly overexpressed in CRSwNP at both the protein and mRNA levels, are localized to the epithelial surface, and may be sampled from tissue, whole mucus, and mucus-derived exosomes. Although these results confirm those of prior studies20 focusing largely on CST1, we elected to also study CST2, as it shares a conserved “cystatin fold” formed by a 5- stranded antiparallel β-pleated sheet wrapped around a 5-turn α-helix,42 suggesting similar functional effects.
In our next set of studies, we examined the relationship between CST1 and CST2 expression and several measures of CRS disease severity. We found that exosomal CST was significantly correlated with both objective and subjective measures of CRSwNP disease severity, including eosinophils per hpf, radiographic inflammation, and domain 1 22-Item Sino-Nasal Outcome Test scores. These results are generally consistent with those of Wu et al43 who showed that CST1 was an independent predictor of uncontrolled CRSwNP disease status over 2 years.
In light of the CST overexpression evident in our data set and confirmed by others, we next studied several primary mechanisms to help explain this observation. Our genetic analysis failed to reveal any transcript variants with moderate impact. Thus, if any disease-causing differences in the exome exist, they would have to be due to regulatory variants in the promoter regions that were not adequately covered in our whole exome approach. Interestingly, we did find several PTMs that were present only in the CRSwNP group. These modifications could, in theory, lead to a reduction in CST function with compensatory overexpression. However, the variability in PTMs across all patients with CRSwNP suggests that this concept requires further future study.
To further explore CST as a driver of CRSwNP, we next examined the ability of rCST1 to induce type 2 inflammation in a murine chronic exposure model. Our findings demonstrated that chronic CST exposure is capable of producing significant type 2 inflammation in a time-dependent manner in otherwise healthy nasal epithelium. The elaboration of canonic TH2 cell–derived cytokines such as IL-4, IL-5, and IL-13 in the mouse is consistent with the evidence of TH2 cell infiltration by flow cytometry. This result also harmonizes with those of Yan et al,20 who showed that incubation of dispersed nasal polyp cells with rCST1 could induce IL-5 expression. Furthermore, the upregulation of P-gp, a member of the ABCB1 family, is consistent with prior findings by our laboratory that P-gp is an epithelial driver of type 2 inflammation,44,45 as well as an independent and clinically validated therapeutic target.46
Having confirmed that CST exposure alone can induce type 2 inflammation, we sought to confirm whether we could localize this effect to the level of the epithelium. To achieve this, we elected to knock down murine P-gp by developing and validating a murine ABCB1a siRNA–loaded cationic liposomal formulation. After confirming a dose-dependent knockdown of P-gp mRNA, we next coadministered the liposomal siRNA and CST1 to the mouse. These studies verified a dose-dependent abrogation of CST1-induced type 2 inflammation, thereby confirming the mechanism of CST1 as a promotor of epithelial-derived type 2 inflammation.
There are several limitations to our current study design that bear noting. First, although our murine model confirms that CST1 can independently induce type 2 inflammation, correlative clinical studies remain necessary to confirm this mechanism in patients with CRS. Additionally, although our -omics studies confirmed the overexpression patterns of CST1 and CST2 previously reported in the literature, the overall population studied was limited, and thus the potential for selection bias remains.
CST1 and CST2 are profoundly and focally overexpressed within nasal polyp epithelium, secreted in exosomes, and correlated with CRSwNP disease severity. Chronic recombinant CST1 exposure is capable of inducing a type 2 inflammatory response in healthy murine mucosa in a time-dependent manner along with concomitant P-gp overexpression. Knockdown of the immunomodulator P-gp at the level of the epithelium results in abrogation of the downstream type 2 inflammation. Taken together, these data suggest that CST1 is capable of inducing epithelial-derived type 2 inflammation. However, the degree to which CST1 serves as an initiating event in CRS will continue to require further clinical interrogation, as will the potential array of allergen-, bacterial-, and viral-derived proteases capable of triggering cystatin upregulation.
We would like to acknowledge Towia Libermann and Justin Li for their technical expertise, data analysis, and mentoring.
Supplementary Material
Key messages.
CST1 and CST 2 are among the most highly overexpressed proteins in both nasal polyp epithelium and mucus-derived exosomes in CRSwNP relative to the control among a proteomic array of more than 1300 proteins.
CST1 and CST2 levels are strongly correlated with subjective and objective measures of CRSwNP disease severity.
Recombinant CST1 induces type 2 epithelial cytokine secretion and TH2 cell infiltration in healthy mouse sinonasal mucosa.
Acknowledgments
Supported by the National Institutes of Health/National Institute of Neurological Disorders and Stroke (grant 1R01NS108968-01 [to B.S.B.]).
Abbreviations used
- AERD:
Aspirin-exacerbated respiratory disease
- CRS:
Chronic rhinosinusitis
- CRSwNP:
Chronic rhinosinusitis with nasal polyps
- CST1:
Cystatin SN
- CST2:
Cystatin SA
- FC:
Fold change
- FDR:
False discovery rate
- GAPDH:
Glyceraldehyde-3–phosphate dehydrogenase
- IQR:
Interquartile range
- PTM:
Posttranslational modification
- rCST1:
Recombinant cystatin SN
- RFU:
Relative fluorescence unit
- siRNA:
Small interfering RNA
Footnotes
Disclosure of potential conflict of interest: B. S. Bleier has received grants from the MEEI Curing Kids Fund, Cook Medical, Medtronics; has consultant arrangements with Olympus, Medtronics, 3D Matrix, Third Wave Therapeutics, Bear-ENT, and Karl Storz; has provided expert testimony on ear, nose, and throat–related cases; has a patent for P-gp and cystatin inhibition for chronic rhinosinusitis and receives royalties from this patent; and has stock and/or stock options in Interscope, Inquis Medical, and Diceros Therapeutics. The rest of the authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.Wallace DV, Dykewicz MS, Bernstein DI, Blessing-Moore J, Cox L, Khan DA, et al. The diagnosis and management of rhinitis: an updated practice parameter. J Allergy Clin Immunol 2008;122:S1–84. [DOI] [PubMed] [Google Scholar]
- 2.Smith KA, Orlandi RR, Rudmik L. Cost of adult chronic rhinosinusitis: a systematic review. Laryngoscope 2015;1–10. [DOI] [PubMed] [Google Scholar]
- 3.Rudmik L, Smith TL, Schlosser RJ, Hwang PH, Mace JC, Soler ZM. Productivity costs in patients with refractory chronic rhinosinusitis. Laryngoscope 2014;124: 2007–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giannoni C, Stewart M, Alford E. Intracranial complications of sinusitis. Laryngoscope 1997;107:863–7. [DOI] [PubMed] [Google Scholar]
- 5.Hedman J, Kaprio J, Poussa T, Nieminen MM. Prevalence of asthma, aspirin intolerance, nasal polyposis and chronic obstructive pulmonary disease in a population-based study. Int J Epidemiol 1999;28:717–22. [DOI] [PubMed] [Google Scholar]
- 6.Klossek JM, Neukirch F, Pribil C, Jankowski R, Serrano E, Chanal I, et al. Prevalence of nasal polyposis in France: a cross-sectional, case-control study. Allergy 2005;60:233–7. [DOI] [PubMed] [Google Scholar]
- 7.Johansson L, Akerlund A, Holmberg K, Meleén I, Bende M. Prevalence of nasal polyps in adults: the Skövde population-based study. Ann Otol Rhinol Laryngol 2003;112:625–9. [DOI] [PubMed] [Google Scholar]
- 8.Gevaert P, Lang-Loidolt D, Lackner A, Stammberger H, Staudinger H, Van Zele T, et al. Nasal IL-5 levels determine the response to anti-IL-5 treatment in patients with nasal polyps. J Allergy Clin Immunol 2006;118:1133–41. [DOI] [PubMed] [Google Scholar]
- 9.Gevaert P, Van Bruaene N, Cattaert T, Van Steen K, Van Zele T, Acke F, et al. Mepolizumab, a humanized anti-IL-5 mAb, as a treatment option for severe nasal polyposis. J Allergy Clin Immunol 2011;128:989–95.e1–8. [DOI] [PubMed] [Google Scholar]
- 10.Wenzel S, Ford L, Pearlman D, Spector S, Sher L, Skobieranda F, et al. Dupilumab in persistent asthma with elevated eosinophil levels. N Engl J Med 2013;368:2455–66. [DOI] [PubMed] [Google Scholar]
- 11.Bachert C, Mannent L, Naclerio RM, Mullol J, Ferguson BJ, Gevaert P, et al. Effect of subcutaneous dupilumab on nasal polyp burden in patients with chronic sinusitis and nasal polyposis: a randomized clinical trial. JAMA 2016;315:469–79. [DOI] [PubMed] [Google Scholar]
- 12.Tomas sen P, Vandeplas G, Van Zele T, Cardell L, Arebro J, Olze H, et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol 2016; 137:1449–56.e4. [DOI] [PubMed] [Google Scholar]
- 13.Bachert C, Han JK, Desrosiers M, Hellings PW, Amin N, Lee SE, et al. Efficacy and safety of dupilumab in patients with severe chronic rhinosinusitis with nasal polyps (LIBERTY NP SINUS-24 and LIBERTY NP SINUS-52): results from two multicentre, randomised, double-blind, placebo-controlled, parallel-group phase 3 trials. Lancet 2019;394:1638–50. [DOI] [PubMed] [Google Scholar]
- 14.Tantilipikorn P, Bunnag C, Nan Z, Bachert C. Staphylococcus aureus superantigens and their role in eosinophilic nasal polyp disease. Asian Pac J Allergy Immunol 2012;30:171–6. [PubMed] [Google Scholar]
- 15.Zhang Z, Kofonow JM, Finkelman BS, Doghramji L, Chiu AG, Kennedy DW, et al. Clinical factors associated with bacterial biofilm formation in chronic rhinosinusitis. Otolaryngol Head Neck Surg 2011;144:457–62. [DOI] [PubMed] [Google Scholar]
- 16.Ponikau JU, Sherris DA, Kephart GM, Adolphson C, Kita H. The role of ubiquitous airborne fungi in chronic rhinosinusitis. Clin Rev Allergy Immunol 2006; 30:187–94. [DOI] [PubMed] [Google Scholar]
- 17.Tan BK, Zirkle W, Chandra RK, Lin D, Conley DB, Peters AT, et al. Atopic profile of patients failing medical therapy for chronic rhinosinusitis. Int Forum Allergy Rhinol 2011;1:88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mueller SK, Nocera AL, Dillon ST, Gu X, Wendler O, Otu HH, et al. Noninvasive exosomal proteomic biosignatures, including cystatin SN, peroxiredoxin-5, and glycoprotein VI, accurately predict chronic rhinosinusitis with nasal polyps. Int Forum Allergy Rhinol 2019;9:177–86. [DOI] [PubMed] [Google Scholar]
- 19.Kato Y, Takabayashi T, Sakashita M, Imoto Y, Tokunaga T, Ninomiya T, et al. Expression and functional analysis of CST1 in intractable nasal polyps. Am J Respir Cell Mol Biol 2018;59:448–57. [DOI] [PubMed] [Google Scholar]
- 20.Yan B, Lou H, Wang Y, Li Y, Meng Y, Qi S, et al. Epithelium-derived cystatin SN enhances eosinophil activation and infiltration through IL-5 in patients with chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol 2019;144:455–69. [DOI] [PubMed] [Google Scholar]
- 21.Ordovas-Montanes J, Dwyer DF, Nyquist SK, Buchheit KM, Vukovic M, Deb C, et al. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature 2018;560:649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sokol CL, Barton GM, Farr AG, Medzhitov R. A mechanism for the initiation of allergen-induced T helper type 2 responses. Nat Immunol 2008;9:310–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imoto Y, Tokunaga T, Matsumoto Y, Hamada Y, Ono M, Yamada T, et al. Cystatin SN upregulation in patients with seasonal allergic rhinitis. PLoS One 2013;8:e67057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bobek LA, Levine MJ. Cystatins–inhibitors of cysteine proteinases. Crit Rev Oral Biol Med 1992;3:307–32. [DOI] [PubMed] [Google Scholar]
- 25.Fábiáan TK, Hermann P, Beck A, Fejérdy P, Fábián G. Salivary defense proteins: their network and role in innate and acquired oral immunity. Int J Mol Sci 2012; 13:4295–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganeshnarayan K, Velliyagounder K, Furgang D, Fine DH. Human salivary cystatin SA exhibits antimicrobial effect against Aggregatibacter actinomycetemcomitans. J Periodontal Res 2012;47:661–73. [DOI] [PubMed] [Google Scholar]
- 27.Dickinson DP. Salivary (SD-type) cystatins: over one billion years in the making–but to what purpose? Crit Rev Oral Biol Med 2002;13:485–508. [DOI] [PubMed] [Google Scholar]
- 28.Orlandi RR, Kingdom TT, Smith TL, Bleier B, DeConde A, Luong AU, et al. International consensus statement on allergy and rhinology: rhinosinusitis 2021. Int Forum Allergy Rhinol 2021;11:213–739. [DOI] [PubMed] [Google Scholar]
- 29.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 2006;Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 30.Nocera AL, Miyake MM, Seifert P, Han X, Bleier BS. Exosomes mediate interepithelial transfer of functional P-glycoprotein in chronic rhinosinusitis with nasal polyps. Laryngoscope 2017;127:E295–300. [DOI] [PubMed] [Google Scholar]
- 31.Pawar GN, Parayath NN, Nocera AL, Bleier BS, Amiji MM. Direct CNS delivery of proteins using thermosensitive liposome-in-gel carrier by heterotopic mucosal engrafting. PLoS One 2018;13:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsui Y, Kobayashi N, Nishikawa M, Takakura Y. Sequence-specific suppression of mdrla/lb expression in mice via RNA interference. Pharm Res 2005; 22:2091–8. [DOI] [PubMed] [Google Scholar]
- 33.Fukuoka A, Matsushita K, Morikawa T, Adachi T, Yasuda K, Kiyonari H, et al. Human cystatin SN is an endogenous protease inhibitor that prevents allergic rhinitis. J Allergy Clin Immunol 2019;143:1153–62.e12. [DOI] [PubMed] [Google Scholar]
- 34.Tharakan A, Dobzanski A, London NR, Khalil SM, Surya N, Lane AP, et al. Characterization of a novel, papain-inducible murine model of eosinophilic rhinosinusitis. Int Forum Allergy Rhinol 2018;00:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 2010;5:e15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol 2001;2: RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamini YHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 1995;57:289–300. [Google Scholar]
- 38.Sneath P, Sokal R. Numerical Taxonomy: The principles and practice of numerical classification. San Francisco, CA: WH Free Company; 1973:573. [Google Scholar]
- 39.Bleier BS, Nocera AL, Iqbal H, Hoang JD, Alvarez U, Feldman RE, et al. P-glycoprotein promotes epithelial T helper 2-associated cytokine secretion in chronic sinusitis with nasal polyps. Int Forum Allergy Rhinol 2014;4:488–94. [DOI] [PubMed] [Google Scholar]
- 40.Vilanova C, Porcar M. Are multi-omics enough? Nat Microbiol 2016; 1:16101. [DOI] [PubMed] [Google Scholar]
- 41.Bersanelli M, Mosca E, Remondini D, Giampieri E, Sala C, Castellani G, et al. Methods for the integration of multi-omics data: mathematical aspects. BMC Bioinformatics 2016;17(suppl 2): 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kopitar-Jerala N. The role of cystatins in cells of the immune system. FEBS Lett 2006;580:6295–301. [DOI] [PubMed] [Google Scholar]
- 43.Wu D, Yan B, Wang Y, Wang C, Zhang L. Prognostic and pharmacologic value of cy statin SN for chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol 2021;148:450–60. [DOI] [PubMed] [Google Scholar]
- 44.Bleier BS, Kocharyan A, Singleton A, Han X. Verapamil modulates interleukin-5 and interleukin-6 secretion in organotypic human sinonasal polyp explants. Int Forum Allergy Rhinol 2015;5:10–3. [DOI] [PubMed] [Google Scholar]
- 45.Nocera AL, Meurer AT, Miyake MM, Sadow PM, Han X, Bleier BS. Secreted P-glycoprotein is a noninvasive biomarker of chronic rhinosinusitis. Laryngoscope 2017;127:E1–4. [DOI] [PubMed] [Google Scholar]
- 46.Miyake MMM, Nocera A, Levesque P, Guo R, Finn CA, Goldfarb J, et al. Double-blind placebo-controlled randomized clinical trial of verapamil for chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol 2017;140:271–3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


