Abstract
Cell-cell signaling peptides (e.g., peptide hormones, neuropeptides) are among the largest class of cellular transmitters and regulate a variety of physiological processes. To identify and quantify the relative abundances of cell-cell signaling peptides in different physiological states, LC-MS-based peptidomics workflows are commonly utilized on freshly dissected tissue. In such animal experiments, the administration of general anesthetics is an important step for many research projects. However, acute anesthetic administration may rapidly change the measured abundance of transmitter molecules and metabolites, especially in the brain and endocrine system, which would confound experimental results. The aim of this study was to evaluate the effect of short-term (< 5 min) anesthetic administration on the measured abundance of cell-cell signaling peptides, as evaluated by a typical peptidomics workflow. To accomplish this goal, we compared endogenous peptide abundances in the rat pituitary following administration of 5% isoflurane, 200 mg/kg sodium pentobarbital, or no anesthetic administration. Label-free peptidomics analysis demonstrated that acute use of isoflurane changed the levels of a small number of peptides, primarily degradation products of the hormone somatotropin, but did not influence the levels of most other peptide hormones. Acute use of sodium pentobarbital had a negligible impact on the relative abundance of all measured peptides. Overall, our results suggest that anesthetics used in pituitary peptidomics studies do not dramatically confound observed results.
Keywords: peptidomics, anesthetics, mass spectrometry, pituitary, peptide hormones
Graphical Abstract
Introduction
Neuropeptides and peptide hormones represent a large class of cell-to-cell signaling molecules that modulate critical functions in the central nervous and endocrine systems. Because of the central role cell-cell signaling peptides play in biology, identifying and quantifying these molecules is of great interest to understand both normal physiology and disease. Recent developments in liquid chromatography-mass spectrometry (LC-MS)-based “peptidomics” methodologies have enabled the thorough characterization and comparison of cell-cell signaling peptides in a variety of tissues and physiological states.1–3 In contrast to antibody-based approaches (e.g., ELISA, Western blot), LC-MS-based peptidomics experiments do not require preselection of peptides of interest, allowing for non-targeted analysis of many endogenous peptide molecules simultaneously.
For peptidomics experiments involving animals, anesthetics may be implemented to facilitate specific experimental requirements. However, the use of anesthetics in peptidomics experiments is not standardized, and many protocols are adapted to meet experimental needs or conform with laboratory standards. Indeed, some peptidomics protocols call for use of volatile3–10 or injectable anesthetics,11–13 while others do not use any such anesthetic.14–21 General anesthetics have been reported to alter the activity and the level of both proteins and small molecule neurotransmitters in the central nervous system (CNS).22, 23 However, there is little data about the effects of these anesthetics on the full complement of cell-cell signaling peptides in brain and pituitary. Anesthetic use during a peptidomics experiment may alter the concentration or the enzymatic processing of peptides being measured, which may obscure changes resulting from experimental variables that are of real interest to researchers. Indeed, some prior studies suggest that common anesthetics like isoflurane, halothane, sevoflurane, or nitrous oxide can influence the levels of targeted cell-cell signaling peptides in the CNS, over either long-term or acute exposure.24–33 However, how anesthetics change the measured peptide abundances over the course of a typical peptidomics experiment, or what proportion of peptides may be affected, are unanswered questions.
Here, we performed label-free peptidomics analysis to evaluate the relative peptide levels from the rat pituitary under short-term (<5 min) administration of either isoflurane or sodium pentobarbital, two anesthetics commonly used in peptidomics studies prior to animal euthanasia. Our results demonstrate that anesthetic administration only minimally perturbs measured peptide abundances, suggesting that anesthetic use can be employed in peptidomics experimental designs without significant fear of confounding the results.
Results and Discussion
Analysis of peptide changes as a result of anesthetic administration
In this study, we sought to understand the influence of short-term anesthetic administration on peptide levels in the pituitary using label-free peptidomics (Figure 1).2 We chose to evaluate isoflurane and sodium pentobarbital because these anesthetics are commonly used in peptidomics studies.3, 4, 6, 12, 13 We chose to investigate the pituitary because it is a major producer of cell-cell signaling peptides that function in the hypothalamus-pituitary-adrenal (HPA) axis, and thus a common tissue analyzed for peptidomics experiments.13, 15, 17, 34–37 Prior studies using immunoassays have suggested that some peptide hormone levels in the pituitary may be altered by long-term (>1 hr) anesthetic administration,25, 26, 29 which make this an excellent tissue to examine for our study.
Figure 1.
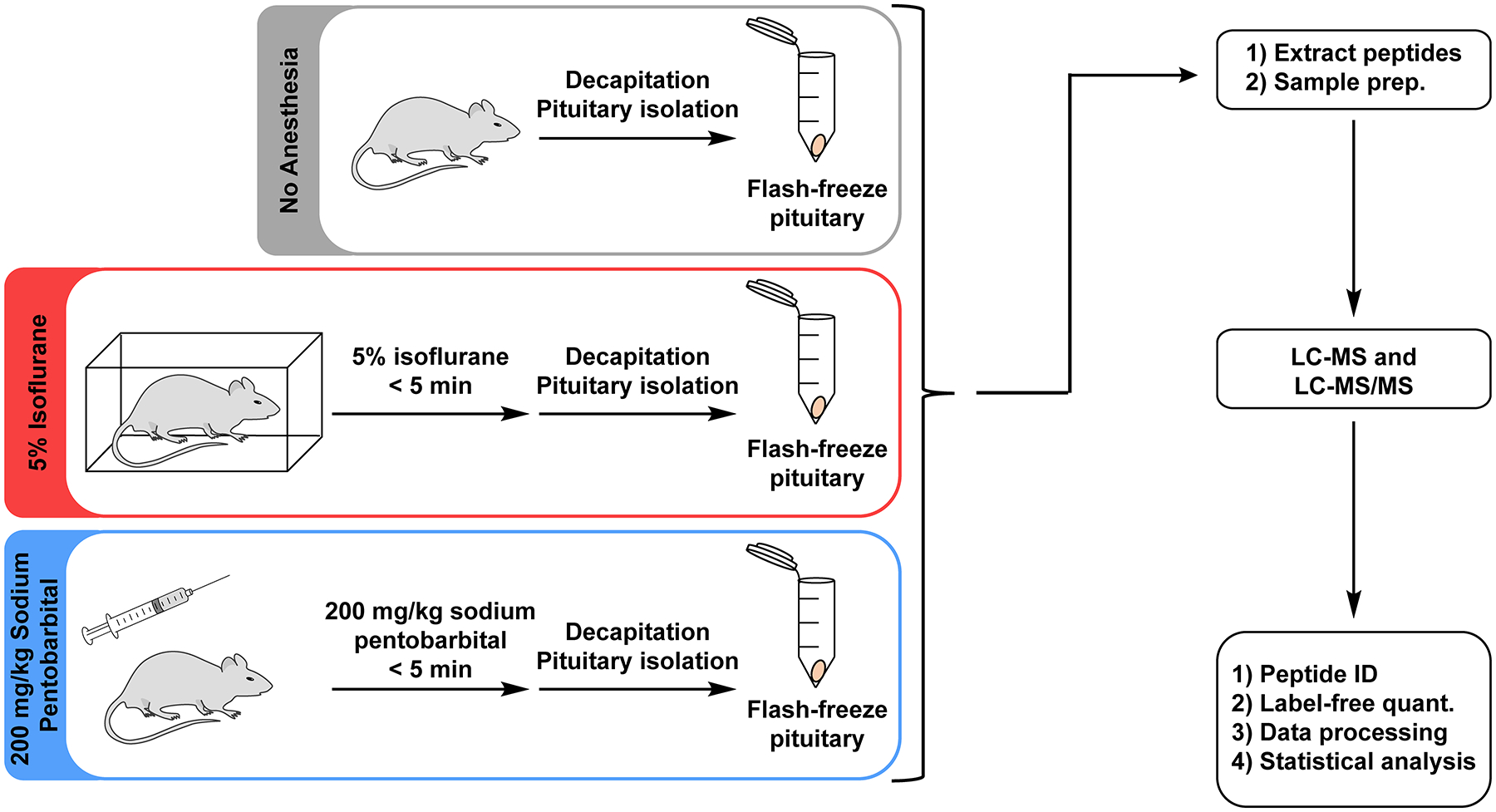
Overview of the experimental design. Rats were separated into “No Anesthesia”, “5% Isoflurane”, or “200 mg/kg Sodium Pentobarbital” groups. For each group, animals were subjected to the indicated condition prior to rapid decapitation and pituitary isolation, after which pituitaries were flash frozen. Peptides were then extracted from pituitary samples and analyzed using an LC-MS and LC-MS/MS peptidomics workflow.
Thirty-two Sprague-Dawley rats were assigned to one of three groups: “No Anesthesia” group, “5% Isoflurane” group, or “200 mg/kg Sodium Pentobarbital” group. Animals in the “No Anesthesia” group were sacrificed by rapid decapitation, while animals in the “5% Isoflurane” or “200 mg/kg Sodium Pentobarbital” groups were administered the indicated anesthetic, allowed time for the anesthetic to take effect (negative assessment by toe pinch), and then sacrificed by rapid decapitation. The total time from initiation of anesthetic to negative toe pinch was comparable in both anesthetic groups (166 ± 43 sec for “5% Isoflurane”, 180 ± 23 sec for “200 mg/kg Sodium Pentobarbital”, Figure S1a). Likewise, the total time from initiation of anesthetic to sacrifice was similar between both anesthetic groups (181 ± 43 sec for “5% Isoflurane”, 197 ± 26 sec for “200 mg/kg Sodium Pentobarbital”, Figure S1b). Following sacrifice, the pituitary was rapidly isolated and flash-frozen in liquid nitrogen. The average pituitary dissection times following decapitation were not statistically distinguishable between any of the three groups (195 ± 42 sec for “No Anesthesia”, 186 ± 24 sec for “5% Isoflurane”, 204 ± 36 sec for “200 mg/kg Sodium Pentobarbital”, Figure S2). The high similarity in dissections times is critical to ensure that any similarities/differences in peptide levels between groups are due to anesthetic treatment and not due to peptide degradation post-mortem by endogenous proteases.15, 18, 20
Peptides were extracted from pituitaries using a two-stage protocol consisting of homogenization in acidified acetone38, 39 followed by acidified water,34 removal of high-molecular-weight proteins using a centrifuge-based filtration device, and desalting via solid-phase extraction (SPE). The approximate total peptide concentration was determined by bicinchoninic acid (BCA) assay, and 200 ng of each peptide extract was analyzed by LC-MS and LC-MS/MS. Over 500 individual peptides were identified from all samples at the 1% false discovery rate (FDR) threshold. To facilitate downstream statistical analysis, data were filtered to remove peptides present in <50% of samples, absent from pooled quality control (QC) runs, or with <5 total MS2 identifications across runs, leaving a total of 164 peptides for statistical analysis (Table 1). As expected for the pituitary,15, 17, 34, 40 the majority of these peptides arose from the pro-opiomelanocortin (POMC) precursor protein (COLI_RAT). Peptides were also identified from other known pituitary proteins, such as somatotropin (also known as growth hormone), ProSAAS, proenkephalin-B (also known as prodynorphin), vasopressin-neurophysin 2-copeptin, and others. Data were then processed, imputed using k-nearest neighbors algorithm,41 and normalized using EigenMS (Figure S3).42, 43
Table 1.
Protein distribution of rat pituitary peptides analyzed in this study.
| Protein ID | Gene name | Protein name | Peptides |
|---|---|---|---|
| COLI_RAT | POMC | Pro-opiomelanocortin | 46 |
| SOMA_RAT | GH1 | Somatotropin | 25 |
| SCG1_RAT | CHGB | Secretogranin-1 | 16 |
| NEU2_RAT | AVP | Vasopressin-neurophysin 2-copeptin | 12 |
| HBB1_RAT/HBB2_RAT | HBB | Hemoglobin subunit beta-1/2 | 9 |
| HBA_RAT | HBA1 | Hemoglobin subunit alpha-1/2 | 6 |
| PCS1N_RAT | PCSK1N | ProSAAS | 6 |
| PDYN_RAT | PDYN | Proenkephalin-B | 5 |
| 7B2_RAT | SCG5 | Neuroendocrine protein 7B2 | 5 |
| CMGA_RAT | CHGA | Chromogranin-A | 4 |
| G3P_RAT | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 3 |
| SCG2_RAT | SCG2 | Secretogranin-2 | 3 |
| H4_RAT | H4C2 | Histone H4 | 3 |
| LSHB_RAT | LHB | Lutropin subunit beta | 2 |
| CBPE_RAT | CPE | Carboxypeptidase E | 2 |
| PGAM1_RAT | PGAM1 | Phosphoglycerate mutase 1 | 2 |
| SCG3_RAT | SCG3 | Secretogranin-3 | 2 |
| NEU1_RAT | OXT | Oxytocin-neurophysin 1 | 1 |
| PRL_RAT | PRL | Prolactin | 1 |
| ALBU_RAT | ALB | Albumin | 1 |
| MDHM_RAT | MDH2 | Malate dehydrogenase, mitochondrial | 1 |
| CLUS_RAT | CLU | Clusterin | 1 |
| NMU_RAT | NMU | Neuromedin-U | 1 |
| NEC2_RAT | PCSK2 | Neuroendocrine convertase 2 | 1 |
| MIF_RAT | MIF | Macrophage migration inhibitory factor | 1 |
| CX7A2_RAT | COX7A2 | Cytochrome c oxidase subunit 7A2, mitochondrial | 1 |
| COF1_RAT | CFL1 | Cofilin-1 | 1 |
| TPIS_RAT | TPI1 | Triosephosphate isomerase | 1 |
| UBB_RAT | UBB (and others) | Ubiquitin | 1 |
| GNAS3_RAT | GNAS | Neuroendocrine secretory protein 55 | 1 |
Principal component analysis (PCA) of the resulting data showed considerable overlap between the three experimental conditions (Figure 2), suggesting little difference between the three groups based on peptide profile when using an unsupervised multivariate analysis technique. There was also no separation between female and male rats in the PCA (Figure S4), indicating that we detected no major differences in peptide profile due to biological sex. Separation between groups could be visualized when the data were analyzed using partial least squares-discriminant analysis (PLS-DA, Figure S5). Peptides with the highest VIP scores in PLS-DA (See Supporting Document) include peptides from somatotropin, pro-opiomelanocortin, and secretogranin-1, indicating the abundance profiles of these peptides contribute to the separation using this supervised multivariate method. Perhaps not surprisingly, many of the peptides with the highest VIP scores from the PLS-DA were the peptides that also were found to significantly differ in our univariate approaches, as described below.
Figure 2.
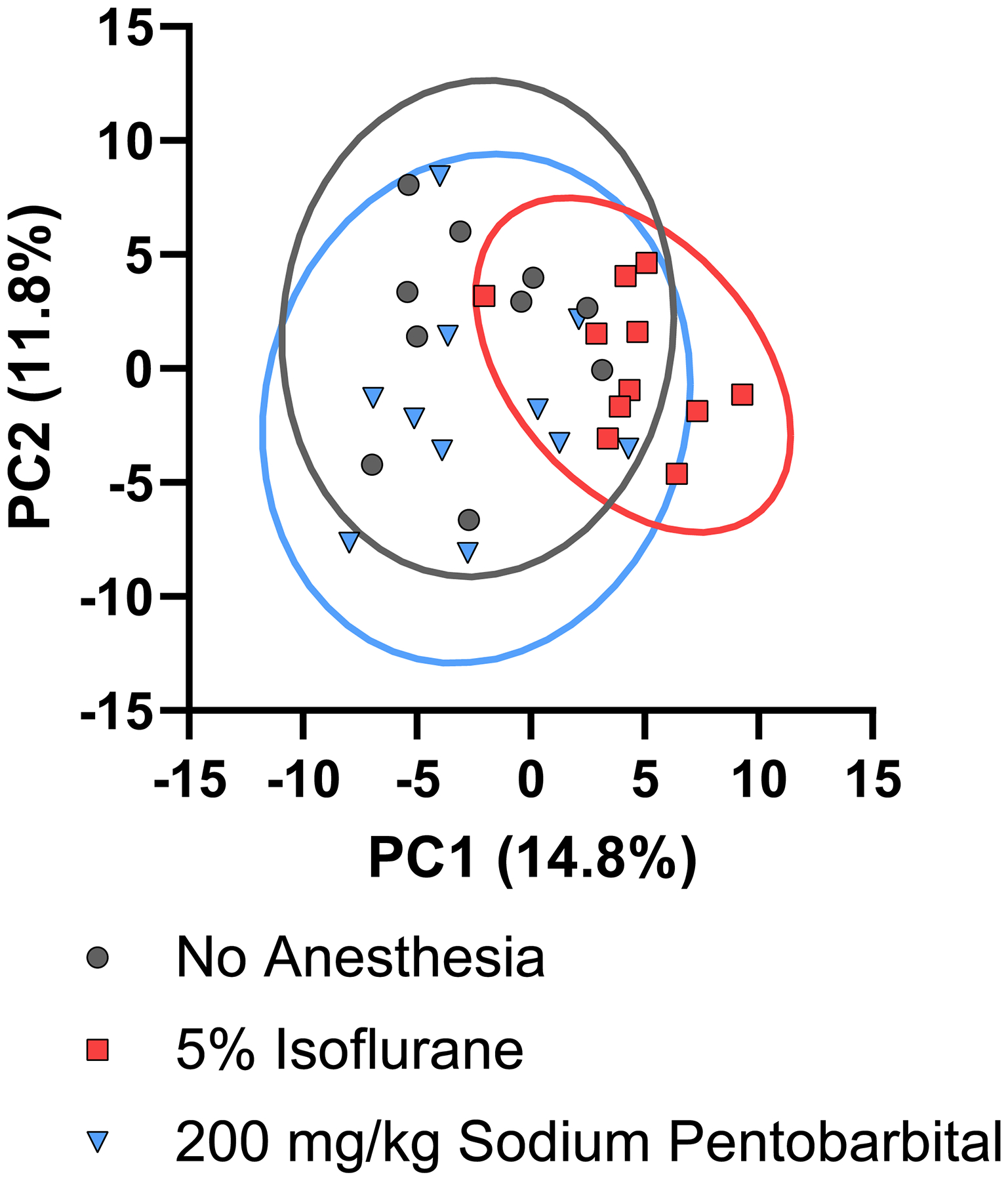
PCA scores plot for peptide abundance data after preprocessing and normalization by EigenMS shows no separation of the three experimental conditions based on peptide profile.
We next examined the abundances of individual peptides using univariate statistical analysis to identify if any peptide abundances were altered due to experimental conditions (Figure 3, Table 2, Figure S6). Overall, the abundances of most peptides in both the “5% Isoflurane” and “200 mg/kg Sodium Pentobarbital” conditions were similar to those in the “No Anesthesia” group, suggesting that neither anesthetic has a major impact on peptide profile. When comparing “5% Isoflurane” treatment to “No Anesthesia”, a total of 15 peptides out of 164 analyzed were found in significantly different abundances as a result of anesthesia treatment with log2 fold-change >0.6 or <−0.6 (Table 2). The majority of these peptides (11/15) were degradation products of the N-terminus of mature somatotropin (also known as growth hormone), a pituitary hormone which plays a major role in growth.44 Each of these somatotropin fragments were found to decrease as a result of 5% isoflurane administration ~2–5-fold. This result may indicate a change in somatotropin synthesis or its release from vesicle stores, both of which would lead to changes in the abundances in degradation products. Consistent with this observation, 15 min isoflurane administration has previously been shown to cause an increase in plasma somatotropin levels in humans, likely indicating increased release from the pituitary.45 Unfortunately, our peptidomics approach cannot detect the full-length somatotropin because this mature protein is relatively large (190 residues) and contains two disulfide bonds, making it difficult to sequence by tandem mass spectrometry without enzymatic digestion. As a result, it is not clear if the mature protein changes in abundance in the pituitary as a result of isoflurane administration similar to its degraded peptides. In contrast, two related peptides from secretogranin-1 (also known as chromogranin-B) increased as a result of 5% isoflurane administration, LLDEGHDPVHESPVDTA and LDEGHDPVHESPV, which were found to increase ~1.9- and ~2.3-fold, respectively. LLDEGHDPVHESPVDTA is a mature peptide hormone generated by the endogenous machinery responsible for the processing neuropeptides and peptide hormones46, 47, while LDEGHDPVHESPV is a partially degraded form of this peptide likely generated after cellular release by proteases in extracellular space. Secretogranin-1, along with other granin-related proteins, plays roles in the sorting and processing of prohormones into neuropeptides and peptide hormones in dense core vesicles.48 Secretogranin-1 also contains canonical basic cleavage sites which are hydrolyzed by prohormone processing machinery to generate smaller peptides, some of which may be biologically active.49–51 Little is known about the biological functions of LLDEGHDPVHESPVDTA, but this peptide was previously demonstrated to increase in the rat hippocampus as a result of acute stress52 and was found to decrease in the mouse hypothalamus as a result of chronic imipramine use.53 Thus, changes in the levels of this secretogranin-1 peptide may reflect changes in processing or release of these peptides due to stress resulting from isoflurane administration. Consistent with this hypothesis, isoflurane has previously been shown to increase blood/serum corticosterone levels in some studies.54, 55 One peptide each from glyceraldehyde-3-phosphate dehydrogenase and histone H4 were also found in slightly lower levels in the “5% Isoflurane” condition, although it is unclear if this observation has any biological relevance. Despite the changes described above, the majority of detected peptides, including the mature and truncated peptide hormones from the pro-opiomalanocortin, vasopressin-neurophysin 2-copeptin, proSAAS, and proenkephalin B precursors, did not show any significant changes as a result of 5% isoflurane administration relative to no anesthesia.
Figure 3.
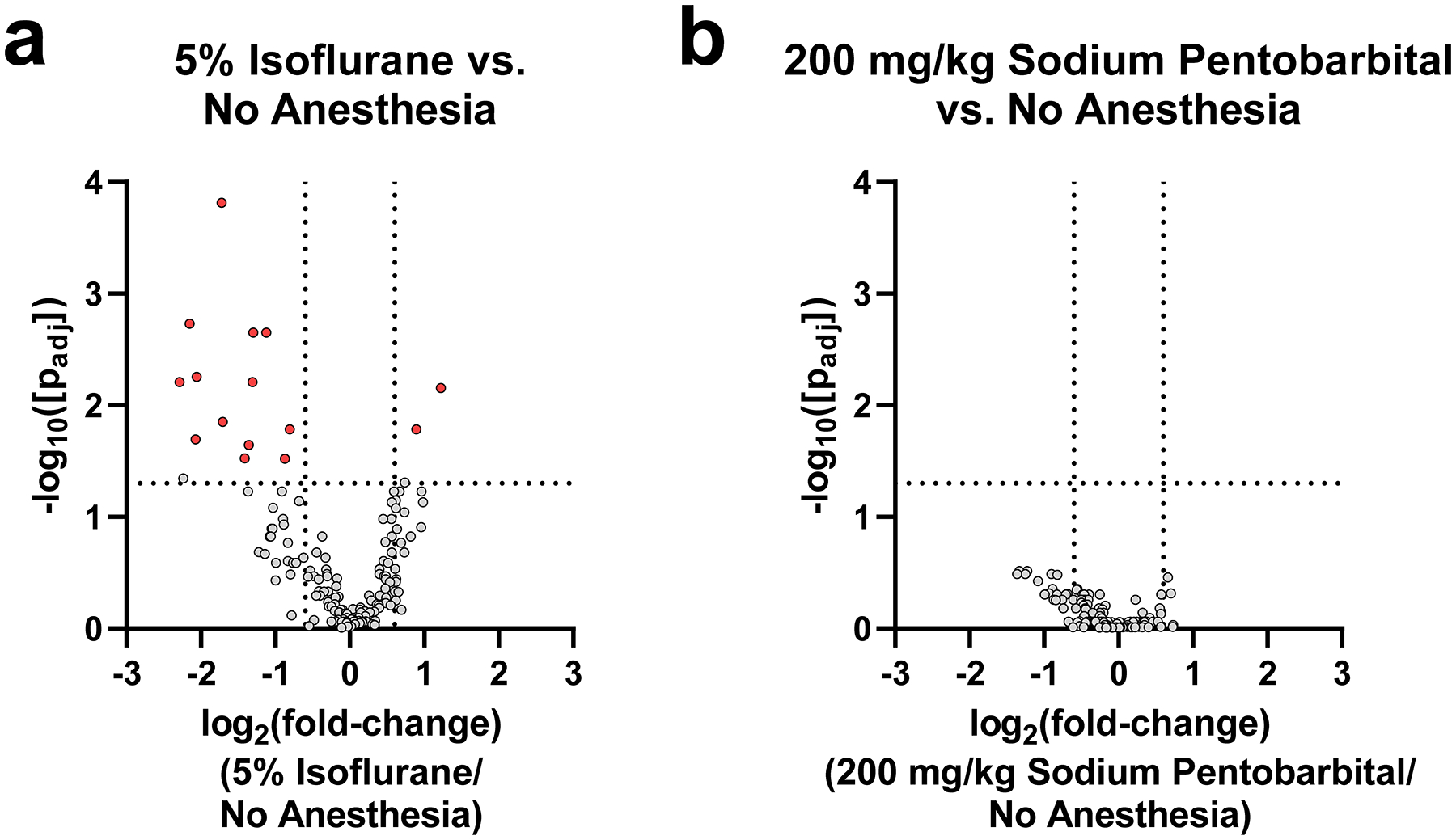
Volcano plots comparing pituitary peptide changes for (a) “5% Isoflurane” or (b) “200 mg/kg Sodium Pentobarbital” compared to “No Anesthesia” condition. Red data points indicate significantly changing peptides (passing both parametric and non-parametric tests) with a log2(fold-change) > 0.6 or < −0.6. The two vertical lines are demarcation points for the log2(fold-change) > 0.6 or < −0.6 between the groups. The horizontal line marks a corrected p-value of 0.05. Grey points above this horizontal line did not pass both the parametric and non-parametric tests.
Table 2.
Peptides found to significantly change as a result of “5% Isoflurane” administration relative to the “No Anesthesia” condition. A “fold-change” value >0 indicates the peptide was measured in higher abundance in the “5% Isoflurane” condition, while a “fold-change” value <0 indicates the peptide was measured in lower abundance in the “5% Isoflurane” condition.
| Peptide | Protein ID | Fold-change (log2) |
|---|---|---|
| LDEGHDPVHESPV | SCG1_RAT | 1.22 |
| LLDEGHDPVHESPVDTA | SCG1_RAT | 0.89 |
| VKVGVNGF | G3P_RAT | −0.81 |
| RTLYGFGG | H4_RAT | −0.87 |
| FPAMPLSSLFANAVLRAQHLHQ | SOMA_RAT | −1.12 |
| FPAMPLSSLFANAVLRAQHLH | SOMA_RAT | −1.30 |
| FPAMPLSSLF | SOMA_RAT | −1.31 |
| FPAMPLSSLFANAVLRAQHLHQLAAD | SOMA_RAT | −1.36 |
| FPAMPLSSLFANAVLRAQHLHQLA | SOMA_RAT | −1.41 |
| FPAMPLSSLFANAVLRA | SOMA_RAT | −1.71 |
| FPAMPLSSLFANAVLRAQH | SOMA_RAT | −1.72 |
| FPAMPLSSLFAN | SOMA_RAT | −2.06 |
| FPAMPLSSLFANA | SOMA_RAT | −2.07 |
| FPAMPLSSLFANAVLR | SOMA_RAT | −2.15 |
| FPAMPLSSLFA | SOMA_RAT | −2.29 |
When comparing “200 mg/kg Sodium Pentobarbital” treatment to “No Anesthesia”, no peptides were found to be significantly different by pairwise comparison. This result demonstrates that sodium pentobarbital injection has negligible impact on the measured abundance of detected peptides over the time period studied. When compared directly with the result of isoflurane administration described above, sodium pentobarbital appears to impact measured peptide levels to a lesser extent than isoflurane.
We also performed a pairwise comparison of the “200 mg/kg Sodium Pentobarbital” and “5% Isoflurane” conditions. Here, we identified 16 peptides that were significantly different between the two anesthetic treatments (Figure S7 and Table S1), including several peptides that were not identified in previous pairwise comparisons with the “No Anesthesia” group. Significant peptides found in higher abundance in the “5% Isoflurane” group relative to “200 mg/kg Sodium Pentobarbital” group include the C-terminal peptide from neuroendocrine protein 7B2 (also known as secretogranin-5), additional internal processed forms of secretogranin-1, and a truncated form of α-melanocyte-stimulating hormone (α-MSH) from pro-opiomalanocortin. Neuroendocrine protein 7B2 is known to generate an extended C-terminal peptide (SVNPYLQGKRLDNVVAKKSVPHFSEEEKEPE) that acts as a potent inhibitor of prohormone convertase 2 (PC2).56 Cleavage of this peptide at an internal dibasic site to generate the peptide seen in our experiments (SVPHFSEEEKEPE) by PC2 itself is a known method by which the full-length C-terminal peptide loses inhibitory activity.50, 51 Thus, the increase in 7B2 C-terminal peptide may indicate altered PC2 activity in the isoflurane condition, although there may be alternative explanations for this observation.
Discussion
Several prior studies have suggested that the administration of anesthetics can alter the levels of metabolites and signaling molecules in mammals. For example, short-term exposure (~2–3 min) to 100–150 mg/kg sodium pentobarbital in rats led to significant changes in the abundance of Hif1a and Tnfa mRNA and in the abundance of several metabolites and oxidative markers in serum and tissues.57 A separate study identified changes in serum glucose, cholesterol, and fatty acid levels upon 120 mg/kg pentobarbital exposure.58 Likewise, short-term exposure to isoflurane has been shown to alter plasma levels of a number of metabolites.59, 60 Some studies also suggest that neuropeptide/peptide hormone abundance can be affected by anesthetic treatment. For example, the neuropeptides somatostatin, substance P, and cholecystokinin were found to be altered in several brain regions within 30 min of ketamine injection, while the levels of thyrotropin-releasing hormone did not change during this same time period.61 Similarly, 15 min isoflurane administration has previously been shown to alter levels of Met-enkephalin and Leu-enkephalin in the brain.27 Changes in the concentration of cholecystokinin immunoreactivity in the rat brain were noted for anesthetics such as halothane (after 15 min of inhalation), chloral hydrate (30–120 min after injection), and pentobarbital (30–60 min after the injection).62 However, general anesthetics were not found to affect the release of cholecystokinin from isolated rat nerve terminals.63 Several studies have also demonstrated that the long-term use of anesthetics (hours-days) causes significant changes in neuropeptide transcription, abundance, or release.25, 26, 29–33
One important prior peptidomics study has examined the effects of general anesthetic conditions on neuropeptide levels.24 In this study, a mixture of isoflurane and nitrous oxide anesthesia was administered to tree shrews for 12 hours prior to euthanasia, peptide extraction, and LC-MS-based peptidomics analysis. The result of this study showed significant changes in several neuropeptides, most notably a >600% increase in somatostatin-14 abundances in the hypothalamus. However, the conditions of this experiment do not mimic a traditional peptidomics experiment, in which the animal would be subjected to the anesthetic for a much shorter period prior to euthanasia and would only receive isoflurane to induce anesthesia.
Our study differs from prior studies examining anesthetic effects on peptides in several important ways. First, many prior studies in this field measure transcript (mRNA) level changes resulting from anesthetic administration,57, 64, 65 rather than directly measuring the peptide/protein products. Although such mRNA measurements provide insight into changes in gene expression resulting from anesthetic administration, total peptide levels also depend highly on rates of translation, post-translational processing and modifications, cellular release, and signal degradation. Thus, final peptide levels often cannot be inferred from transcriptomic measurements, but must be directly measured, as we have done here. Second, because our study utilized a non-targeted LC-MS-based approach, we were able to monitor the changes in peptide modifications and degradation that would not be possible with traditional transcriptomic or antibody-based approaches. For example, the peptidomics approach easily allowed for evaluating the relative levels for multiple peptides from a single prohormone, including their individual degradation products, which would not be feasible using commonly used antibody-based approaches like radioimmunoassay.25–27, 29, 30, 54, 61, 62 Finally, we specifically examined peptide changes on a very short time-scale (<5 min total). This contrasts with many prior studies, which examine extended periods of anesthetic administration,24–26, 29–31, 65 or measure the changes at time points long after administration (≥30 min).54, 61, 64 Our study thus provides valuable insight into any rapid changes in pituitary peptides that may occur in a typical peptidomics study.
Changes in measured peptide levels resulting from anesthetic exposure could influence study conclusions, including for studies that measure target peptides to understand physiology or for diagnostic purposes. Additionally, there are also certain experimental protocols in which anesthetic use cannot be applied consistently to all treatment groups. For example, our group and others are interested in studying changes in the cell-cell signaling peptides of hibernating mammals, such as the thirteen-lined ground squirrel (Ictidomys tridecemlineatus).66 While anesthetic administration may be necessary for such wild-caught animals during the active season, the effectiveness of anesthetics is expected to change dramatically during the hibernation season when breathing, heart rate, and metabolism decrease dramatically. As a result, if anesthetic administration did significantly affect peptide levels, the animal’s inconsistent metabolism may lead to artificial changes that may be mistaken as related to natural physiology. Interestingly, some previous studies on mammalian hibernators used different anesthetic conditions on active animals versus torpid animals.67, 68 When embarking on such studies, it is important to understand the influence anesthetic treatment may have on levels of such molecules. However, it should be noted that other regions of the brain, particularly the hypothalamus, are also major producers of cell-cell signaling peptides. It will be of great interest for future efforts to examine the effect of anesthetics on neuropeptide levels in other brain regions using peptidomics approaches, and our study sets the stage for such research.
In conclusion, our results show that the use of short-term anesthetics (isoflurane or sodium pentobarbital) immediately prior to euthanasia only minimally perturbs the measured abundance of peptides in the rat pituitary. Although some differences were observed resulting from anesthetic administration, particularly for 5% isoflurane administration, these differences were limited to modest changes in a small number of peptides. Our results should be useful for future researchers in the area of peptidomics research, directly aiding in experimental design and interpretation of results.
Methods
Materials
LC-MS-grade solvents were used for all sample preparation prior to LC-MS analysis. Unless otherwise specified, all other reagents and solvents were purchased from ThermoFisher Scientific or MilliporeSigma.
Animals and tissue collection
All animal methods were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Nebraska (Project #2113).
Sprague Dawley rats, aged ~2 months, weight = 225 ± 24 g, were obtained from Charles River Laboratories (001CD). A total of 32 animals (17 females, 15 males) were assigned to one of three groups: “No Anesthesia” group (10 animals, 50% female/male), “5% Isoflurane” group (11 animals, 55% female/male), or “200 mg/kg Sodium Pentobarbital” group (11 animals, 55% female/male). Animal sacrifices were performed over the course of 4 consecutive days, with 2–3 animals from each group sacrificed per day. For animals in the “No Anesthesia” group, animals were sacrificed by rapid decapitation and the pituitary was immediately extracted, flash-frozen in liquid nitrogen, and stored on dry ice or −80 °C until processing. For animals in the “5% Isoflurane” group, animals were placed in a 3.2 L anesthesia chamber and administered 5% isoflurane in oxygen at 1.5 L/min. For animals in the “200 mg/kg Sodium Pentobarbital” group, animals were administered 200 mg/kg Fatal-Plus (stock concentration = 390 mg/mL, diluted to 39 mg/mL for animal injection) by intraperitoneal injection. For both anesthetic groups, animals were monitored for loss of righting reflex and a toe pinch test was used to confirm the loss of consciousness. Upon negative toe pinch, animals were sacrificed by rapid decapitation and the pituitary was immediately extracted, flash-frozen in liquid nitrogen, and stored on dry ice or −80 °C until processing.
Peptide extraction and sample preparation
Tissues were processed on the same day as isolation. For peptide extraction, frozen tissues were homogenized in 400 μL of ice-cold acidified acetone (40:6:1 acetone: water: con. HCl) using manual homogenization with a plastic pestle (Bel-Art) in microcentrifuge tubes, followed by sonication for 10 min and vortexing. Samples were then centrifuged at 15,000 × g, 10 min, 4 °C, and the supernatant was transferred to a clean tube and stored on ice. The remaining pellet was then subjected to the second stage of peptide extraction by adding 400 μL of acidified water (0.25% acetic acid in water). The tissue was again subjected to manual homogenization with a plastic pestle followed by sonication for 10 min and vortexing. Samples were centrifuged at 15,000 × g, 10 min, 4 °C, and the supernatant was combined with the acidified acetone extracts. The combined extracts were dried via a vacuum concentrator for at least 4 h to remove both water and acetone. Dried samples were redissolved in 300 μL of 5% acetonitrile (ACN)/water with 0.1% formic acid (FA) and centrifuged at 15,000 × g, 10 min, 4 °C to remove insoluble material. The resulting supernatant was passed through an Amicon Ultra-0.5 mL centrifugal filter unit with a 30 kDa molecular weight cutoff (Millipore Sigma, UFC503024), centrifuging at 14,000 × g for 15 min at 4 °C. The device was washed with 2 × 200 μL 5% ACN/water with 0.1% FA to ensure low molecular weight species passed through the filter. The resulting filtrate containing low molecular weight compounds was dried by a vacuum concentrator. The dried peptide extracts were then redissolved in 200 μL of 5% ACN/water with 0.1% FA and desalted via C18 SPE columns (Thermo Scientific, 889870). The resulting desalted peptide extracts were dried via vacuum concentration and stored at −20 °C until further analysis.
LC-MS and LC-MS/MS analysis
To prepare samples for LC-MS and LC-MS/MS analysis, dried and desalted peptide extracts were dissolved in 12 μL of 3% ACN/water with 0.1% FA, and the total protein concentration was estimated by BCA assay (Micro BCA Protein Assay Kit, Thermo Scientific, 23235). Each sample was diluted with 3% ACN/water with 0.1% FA to 33 ng/μL total protein. To account for batch effects, sample injection order was randomized, and injections of pooled quality control (QC) samples were included at regular intervals.
Prepared peptide extracts (6 μL of the above solution, ~200 ng total protein) were injected onto a Waters ACQUITY UPLC M-Class system coupled to a Waters Xevo G2-XS quadrupole time-of-flight mass spectrometer with Waters Z spray NanoLockSpray nano-electrospray ionization source. For nano-LC separations, the mobile phase Solvent A was composed of water and 0.1% FA, and Solvent B was composed of ACN and 0.1% FA. Samples were first loaded onto a Waters nanoEase M/Z symmetry C18 trap column (180 μm × 20 mm, product #186008821) for online desalting, using a loading solvent of 1% B and a flow rate of 5 μL/min for 10 min. Separations were then performed using a Waters nanoEase-C18 column (75 μm × 250 mm, product# 186008818) with a flow rate of 0.35 μL/min, column compartment temperature of 35 °C, and the autosampler compartment temperature of 8 °C. Peptides were separated using the following gradient: 0 – 40 min, 3 – 40% B; 40 – 44 min, 40 – 85% B; 44 – 48 min, 85% B; 48 – 50 min, 85 – 3% B; 50 – 60 min, 3% B. MS and MS/MS (collision-induced dissociation fragmentation) analysis was performed in positive ion mode with a mass range of 100–2000 m/z. For each MS scan, 5 precursor ions were selected for MS/MS using data-dependent acquisition based on a peak intensity threshold of 5000. The MS scan time was 0.5 s, and the MS/MS scan time was 1 s.
Data processing and statistics
The LC-MS data were processed using PEAKS Studio X Pro software (Bioinformatics Solutions Inc.)69, 70 with the rat SwissProt reference proteome from UniProt71 used as a database for peptide identifications. For peptide identifications, the parent mass error tolerance was set to 10 ppm, the fragment mass error tolerance was set to 0.1 Da, and the false discovery rate (FDR) threshold was set to 1%. The following PTMs were included as variable modifications: acetylation (N-terminus), amidation (C-terminus), oxidation (M), pyroglutamylation (E or Q), and phosphorylation (S, T, or Y). To calculate peptide peak areas, PEAKS Q Module for label-free quantification was used, and the PEAKS Q Module’s ID-transfer was used to reduce false missing values using a mass error tolerance of 20.0 ppm and a retention time shift tolerance of 0.5 minutes. All identified peptides and peak areas (sum of peak areas for each charge state) were normalized to total ion current (TIC) and exported from the PEAKS Q Module. Peptides were then manually inspected to identify any ambiguous identifications during the ID-transfer step for peptide peaks with similar m/z and retention times. Peptide peaks with ambiguous assignments or peptides with <5 total MS2 identifications were filtered out.
Data were batch corrected using the statTarget R package,72 where a peptide was considered present if it was measured in at least 50% of the samples per group, and was present in pooled QC samples (See note in Supporting Information). Peptides deemed absent were omitted after batch correction, leaving 164 peptides “present” for analysis. Signal imputation was performed using k-nearest neighbor (knn; k=10) method.41 Present peptides were then log2 transformed and normalized using EigenMS.42, 43 Data preprocessing was prepared using the R statistical software version 4.0.3.73
Differential peptides were assessed using parametric (t-test) and non-parametric (Wilcoxon rank-sum) statistical tests to account for the degree, direction, and difference between two groups. For both statistical tests, the Benjamini-Hochberg (BH) procedure was applied to correct for multiple hypothesis testing.74 Peptides were considered differential when they passed with a BH-corrected p-value less than 0.05 in both statistical tests. The fold change (FC) was calculated for each peptide to quantify the magnitude difference between the two groups. In this statistical analysis, three pairs of comparisons were performed: “5% Isoflurane” vs “No Anesthesia”, “200 mg/kg Sodium Pentobarbital” vs “No Anesthesia”,, and “200 mg/kg Sodium Pentobarbital” vs “5% Isoflurane”. Principal component analysis (PCA) was performed using R package pcaMethods75 and partial least squares-discriminant analysis (PLS-DA) was performed used R package ropls.76
The mass spectrometry peptidomics data have been deposited to the ProteomeXchange Consortium via the PRIDE77 partner repository with the dataset identifier PXD034821 and 10.6019/PXD034821.
Supplementary Material
Acknowledgments
This research was supported in part by the National Institutes of General Medical Sciences (R35 GM142784 to J.W.C.). We also acknowledge support from the Nebraska Center for Integrated Biomolecular Communication (NCIBC, National Institute of General Medical Sciences P20 GM113126). We are grateful for the assistance provided by the University of Nebraska-Lincoln Institutional Animal Care Program and the staff of Life Sciences Annex for oversight of animal protocols, training and experimental assistance, and additional resources. Finally, we thank the Nebraska Center for Mass Spectrometry, NCIBC Systems Biology Core Facility, and the Research Instrumentation Facility for providing instrument access.
Footnotes
Supporting Information
Supporting Note, Supporting Figures S1–S7, and Supporting Table S1, and Supporting Document.
The authors declare no competing financial interests.
References
- 1.Fricker LD, Lim J, Pan H, and Che FY (2006) Peptidomics: identification and quantification of endogenous peptides in neuroendocrine tissues, Mass Spectrom. Rev 25, 327–344. [DOI] [PubMed] [Google Scholar]
- 2.Romanova EV, and Sweedler JV (2015) Peptidomics for the discovery and characterization of neuropeptides and hormones, Trends Pharmacol. Sci 36, 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Secher A, Kelstrup CD, Conde-Frieboes KW, Pyke C, Raun K, Wulff BS, and Olsen JV (2016) Analytic framework for peptidomics applied to large-scale neuropeptide identification, Nat. Commun 7, 11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su J, Sandor K, Skold K, Hokfelt T, Svensson CI, and Kultima K (2014) Identification and quantification of neuropeptides in naive mouse spinal cord using mass spectrometry reveals [des-Ser1]-cerebellin as a novel modulator of nociception, J. Neurochem 130, 199–214. [DOI] [PubMed] [Google Scholar]
- 5.Dowell JA, Heyden WV, and Li L (2006) Rat neuropeptidomics by LC-MS/MS and MALDI-FTMS: Enhanced dissection and extraction techniques coupled with 2D RP-RP HPLC, J. Proteome Res 5, 3368–3375. [DOI] [PubMed] [Google Scholar]
- 6.Ye H, Wang J, Tian Z, Ma F, Dowell J, Bremer Q, Lu G, Baldo B, and Li L (2017) Quantitative mass spectrometry reveals food intake-induced neuropeptide level changes in rat brain: functional assessment of selected neuropeptides as feeding regulators, Mol. Cell. Proteomics 16, 1922–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sturm RM, Dowell JA, and Li L (2010) Rat brain neuropeptidomics: tissue collection, protease inhibition, neuropeptide extraction, and mass spectrometric analysis, Methods Mol. Biol 615, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saidi M, Kamali S, and Beaudry F (2019) Neuropeptidomics: Comparison of parallel reaction monitoring and data-independent acquisition for the analysis of neuropeptides using high-resolution mass spectrometry, Biomed. Chromatogr 33, e4523. [DOI] [PubMed] [Google Scholar]
- 9.Gervais JA, Otis C, Lussier B, Guillot M, Martel-Pelletier J, Pelletier JP, Beaudry F, and Troncy E (2019) Osteoarthritic pain model influences functional outcomes and spinal neuropeptidomics: A pilot study in female rats, Can. J. Vet. Res 83, 133–141. [PMC free article] [PubMed] [Google Scholar]
- 10.Ferland CE, Pailleux F, Vachon P, and Beaudry F (2011) Determination of specific neuropeptides modulation time course in a rat model of osteoarthritis pain by liquid chromatography ion trap mass spectrometry, Neuropeptides 45, 423–429. [DOI] [PubMed] [Google Scholar]
- 11.Anapindi KDB, Yang N, Romanova EV, Rubakhin SS, Tipton A, Dripps I, Sheets Z, Sweedler JV, and Pradhan AA (2019) PACAP and other neuropeptide targets link chronic migraine and opioid-induced hyperalgesia in mouse models, Mol. Cell. Proteomics 18, 2447–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowd SE, Mustroph ML, Romanova EV, Southey BR, Pinardo H, Rhodes JS, and Sweedler JV (2018) Exploring exercise- and context-induced peptide changes in mice by quantitative mass spectrometry, ACS Omega 3, 13817–13827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romanova EV, Rubakhin SS, Ossyra JR, Zombeck JA, Nosek MR, Sweedler JV, and Rhodes JS (2015) Differential peptidomics assessment of strain and age differences in mice in response to acute cocaine administration, J. Neurochem 135, 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonnard E, Burlet-Schiltz O, Francés B, Mazarguil H, Monsarrat B, Zajac JM, and Roussin A (2001) Identification of neuropeptide FF-related peptides in rodent spinal cord, Peptides 22, 1085–1092. [DOI] [PubMed] [Google Scholar]
- 15.Che FY, Lim J, Pan H, Biswas R, and Fricker LD (2005) Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary, Mol. Cell. Proteomics 4, 1391–1405. [DOI] [PubMed] [Google Scholar]
- 16.Che FY, Yuan Q, Kalinina E, and Fricker LD (2005) Peptidomics of Cpe fat/fat mouse hypothalamus: effect of food deprivation and exercise on peptide levels, J. Biol. Chem 280, 4451–4461. [DOI] [PubMed] [Google Scholar]
- 17.Che FY, and Fricker LD (2002) Quantitation of neuropeptides in Cpe(fat)/Cpe(fat) mice using differential isotopic tags and mass spectrometry, Anal. Chem 74, 3190–3198. [DOI] [PubMed] [Google Scholar]
- 18.Sköld K, Svensson M, Norrman M, Sjögren B, Svenningsson P, and Andrén PE (2007) The significance of biochemical and molecular sample integrity in brain proteomics and peptidomics: stathmin 2–20 and peptides as sample quality indicators, Proteomics 7, 4445–4456. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Che FY, Berezniuk I, Sonmez K, Toll L, and Fricker LD (2008) Peptidomics of Cpefat/fat mouse brain regions: implications for neuropeptide processing, J. Neurochem 107, 1596–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang N, Anapindi KDB, Romanova EV, Rubakhin SS, and Sweedler JV (2017) Improved identification and quantitation of mature endogenous peptides in the rodent hypothalamus using a rapid conductive sample heating system, Analyst 142, 4476–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JE, Atkins N Jr., Hatcher NG, Zamdborg L, Gillette MU, Sweedler JV, and Kelleher NL (2010) Endogenous peptide discovery of the rat circadian clock: a focused study of the suprachiasmatic nucleus by ultrahigh performance tandem mass spectrometry, Mol. Cell. Proteomics 9, 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller CP, Pum ME, Amato D, Schüttler J, Huston JP, and Silva MA (2011) The in vivo neurochemistry of the brain during general anesthesia, J. Neurochem 119, 419–446. [DOI] [PubMed] [Google Scholar]
- 23.Kalenka A, Gross B, Maurer MH, Thierse HJ, and Feldmann RE Jr. (2010) Isoflurane anesthesia elicits protein pattern changes in rat hippocampus, J. Neurosurg. Anesthesiol 22, 144–154. [DOI] [PubMed] [Google Scholar]
- 24.Fouillen L, Petruzziello F, Veit J, Bhattacharyya A, Kretz R, Rainer G, and Zhang X (2013) Neuropeptide alterations in the tree shrew hypothalamus during volatile anesthesia, J. Proteomics 80, 311–319. [DOI] [PubMed] [Google Scholar]
- 25.Karuri AR, Engelking LR, and Kumar MS (1998) Effects of halothane and methoxyflurane on the hypothalamic-pituitary-adrenal axis in rat, Brain Res. Bull 47, 205–209. [DOI] [PubMed] [Google Scholar]
- 26.Karuri AR, Agarwal RK, Engelking LR, and Kumar MS (1998) Effects of halothane and methoxyflurane on regional brain and spinal cord substance P-like and beta-endorphin-like immunoreactivities in the rat, Brain Res. Bull 45, 501–506. [DOI] [PubMed] [Google Scholar]
- 27.Chmielnicki Z, Was M, Kmieciak-Kołada K, Huzarska M, Spiewak Z, Pawłowski J, Kamiński M, Dyaczyńska-Herman A, and Herman ZS (1997) Influence of isoflurane on enkephalin levels and on some indicatory enzymes in the central nervous system of rabbits, Pol. J. Pharmacol 49, 97–106. [PubMed] [Google Scholar]
- 28.Finck AD, Samaniego E, and Ngai SH (1995) Nitrous oxide selectively releases Met5-enkephalin and Met5-enkephalin-Arg6-Phe7 into canine third ventricular cerebrospinal fluid, Anesth. Analg 80, 664–670. [DOI] [PubMed] [Google Scholar]
- 29.Agarwal RK, Court M, Chandna VK, Mohan A, Engelking LR, and Kumar AM (1994) Influence of halothane and methoxyflurane on regional brain and spinal cord concentrations of methionine-enkephalin in the rat, Brain Res. Bull 35, 273–277. [DOI] [PubMed] [Google Scholar]
- 30.Agarwal RK, Kugel G, Karuri A, Gwosdow AR, and Kumar MS (1996) Effect of low and high doses of nitrous oxide on preproenkephalin mRNA and its peptide methionine enkephalin levels in the hypothalamus, Brain Res. 730, 47–51. [DOI] [PubMed] [Google Scholar]
- 31.Kang W, Lu D, Yang X, Ma W, Chen X, Chen K, Xu X, Zhou X, Zhou L, and Feng X (2020) Sevoflurane induces hippocampal neuronal apoptosis by altering the level of neuropeptide Y in neonatal rats, Neurochem. Res 45, 1986–1996. [DOI] [PubMed] [Google Scholar]
- 32.Zuniga JR, Joseph SA, and Knigge KM (1987) The effects of nitrous oxide on the secretory activity of pro-opiomelanocortin peptides from basal hypothalamic cells attached to cytodex beads in a superfusion in vitro system, Brain Res. 420, 66–72. [DOI] [PubMed] [Google Scholar]
- 33.Kugel G, Zive M, Agarwal RK, Beumer JR, and Kumar AM (1991) Effect of nitrous oxide on the concentrations of opioid peptides, substance P, and LHRH in the brain and beta-endorphin in the pituitary, Anesth. Prog 38, 206–211. [PMC free article] [PubMed] [Google Scholar]
- 34.Che FY, Yan L, Li H, Mzhavia N, Devi LA, and Fricker LD (2001) Identification of peptides from brain and pituitary of Cpefat/Cpefat mice, Proc. Natl. Acad. Sci. U. S. A 98, 9971–9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin P, Bousquet-Moore D, Annangudi SP, Southey BR, Mains RE, Eipper BA, and Sweedler JV (2011) Probing the production of amidated peptides following genetic and dietary copper manipulations, PLoS One 6, e28679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fricker LD, Tashima AK, Fakira AK, Hochgeschwender U, Wetsel WC, and Devi LA (2021) Neuropeptidomic analysis of a genetically defined cell type in mouse brain and pituitary, Cell Chem. Biol 28, 105–112.e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeAtley KL, Colgrave ML, Cánovas A, Wijffels G, Ashley RL, Silver GA, Rincon G, Medrano JF, Islas-Trejo A, Fortes MRS, Reverter A, Porto-Neto L, Lehnert SA, and Thomas MG (2018) Neuropeptidome of the hypothalamus and pituitary gland of indicine × taurine heifers: evidence of differential neuropeptide processing in the pituitary gland before and after puberty, J. Proteome Res 17, 1852–1865. [DOI] [PubMed] [Google Scholar]
- 38.Livnat I, Tai HC, Jansson ET, Bai L, Romanova EV, Chen TT, Yu K, Chen SA, Zhang Y, Wang ZY, Liu DD, Weiss KR, Jing J, and Sweedler JV (2016) A D-amino acid-containing neuropeptide discovery funnel, Anal. Chem 88, 11868–11876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Checco JW, Zhang G, Yuan WD, Le ZW, Jing J, and Sweedler JV (2018) Aplysia allatotropin-related peptide and its newly identified D-amino acid-containing epimer both activate a receptor and a neuronal target, J. Biol. Chem 293, 16862–16873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cawley NX, Li Z, and Loh YP (2016) Biosynthesis, trafficking, and secretion of pro-opiomelanocortin-derived peptides, J. Mol. Endocrinol 56, T77–T97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cover T, and Hart P (1967) Nearest neighbor pattern classification, IEEE Trans. Inf. Theory, 21–27. [Google Scholar]
- 42.Karpievitch YV, Nikolic SB, Wilson R, Sharman JE, and Edwards LM (2014) Metabolomics data normalization with EigenMS, PLoS One 9, e116221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karpievitch YV, Taverner T, Adkins JN, Callister SJ, Anderson GA, Smith RD, and Dabney AR (2009) Normalization of peak intensities in bottom-up MS-based proteomics using singular value decomposition, Bioinformatics 25, 2573–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isaksson OG, Jansson JO, and Gause IA (1982) Growth hormone stimulates longitudinal bone growth directly, Science 216, 1237–1239. [DOI] [PubMed] [Google Scholar]
- 45.Oyama T, Latto P, and Holaday DA (1975) Effect of isoflurane anaesthesia and surgery on carbohydrate metabolism and plasma cortisol levels in man, Can. Anaesth. Soc. J 22, 696–702. [DOI] [PubMed] [Google Scholar]
- 46.Hook V, Lietz CB, Podvin S, Cajka T, and Fiehn O (2018) Diversity of neuropeptide cell-cell signaling molecules generated by proteolytic processing revealed by neuropeptidomics mass spectrometry, J. Am. Soc. Mass. Spectrom 29, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hook V, Funkelstein L, Lu D, Bark S, Wegrzyn J, and Hwang SR (2008) Proteases for processing proneuropeptides into peptide neurotransmitters and hormones, Annu. Rev. Pharmacol. Toxicol 48, 393–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Natori S, and Huttner WB (1996) Chromogranin B (secretogranin I) promotes sorting to the regulated secretory pathway of processing intermediates derived from a peptide hormone precursor, Proc. Natl. Acad. Sci. U. S. A 93, 4431–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Troger J, Theurl M, Kirchmair R, Pasqua T, Tota B, Angelone T, Cerra MC, Nowosielski Y, Mätzler R, Troger J, Gayen JR, Trudeau V, Corti A, and Helle KB (2017) Granin-derived peptides, Prog. Neurobiol 154, 37–61. [DOI] [PubMed] [Google Scholar]
- 50.Zhu X, Rouille Y, Lamango NS, Steiner DF, and Lindberg I (1996) Internal cleavage of the inhibitory 7B2 carboxyl-terminal peptide by PC2: a potential mechanism for its inactivation, Proc. Natl. Acad. Sci. U. S. A 93, 4919–4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang JR, and Lindberg I (2001) Inactivation of the 7B2 inhibitory CT peptide depends on a functional furin cleavage site, J. Neurochem 79, 437–444. [DOI] [PubMed] [Google Scholar]
- 52.Li W, Papilloud A, Lozano-Montes L, Zhao N, Ye X, Zhang X, Sandi C, and Rainer G (2018) Stress impacts the regulation neuropeptides in the rat hippocampus and prefrontal cortex, Proteomics 18, e1700408. [DOI] [PubMed] [Google Scholar]
- 53.Nilsson A, Stroth N, Zhang X, Qi H, Fälth M, Sköld K, Hoyer D, Andrén PE, and Svenningsson P (2012) Neuropeptidomics of mouse hypothalamus after imipramine treatment reveal somatostatin as a potential mediator of antidepressant effects, Neuropharmacology 62, 347–357. [DOI] [PubMed] [Google Scholar]
- 54.Jacobsen KR, Kalliokoski O, Teilmann AC, Hau J, and Abelson KS (2012) The effect of isoflurane anaesthesia and vasectomy on circulating corticosterone and ACTH in BALB/c mice, Gen. Comp. Endocrinol 179, 406–413. [DOI] [PubMed] [Google Scholar]
- 55.Teilmann AC, Kalliokoski O, Jacobsen KR, Hau J, and Abelson KS (2014) Impact of heparin and short term anesthesia on the quantification of cytokines in laboratory mouse plasma, Acta Vet. Scand 56, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muller L, and Lindberg I (1999) The cell biology of the prohormone convertases PC1 and PC2, Prog. Nucleic Acid Res. Mol. Biol 63, 69–108. [DOI] [PubMed] [Google Scholar]
- 57.Mohamed AS, Hosney M, Bassiony H, Hassanein SS, Soliman AM, Fahmy SR, and Gaafar K (2020) Sodium pentobarbital dosages for exsanguination affect biochemical, molecular and histological measurements in rats, Sci. Rep 10, 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pierozan P, Jernerén F, Ransome Y, and Karlsson O (2017) The choice of euthanasia method affects metabolic serum biomarkers, Basic Clin. Pharmacol. Toxicol 121, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deckardt K, Weber I, Kaspers U, Hellwig J, Tennekes H, and van Ravenzwaay B (2007) The effects of inhalation anaesthetics on common clinical pathology parameters in laboratory rats, Food Chem. Toxicol 45, 1709–1718. [DOI] [PubMed] [Google Scholar]
- 60.Arnold M, and Langhans W (2010) Effects of anesthesia and blood sampling techniques on plasma metabolites and corticosterone in the rat, Physiol. Behav 99, 592–598. [DOI] [PubMed] [Google Scholar]
- 61.Pongdhana K, Ogawa N, Hirose Y, Ono T, Kosaka F, and Mori A (1987) Effects of ketamine on the cholecystokinin, somatostatin, substance P, and thyrotropin releasing hormone in discrete regions of rat brain, Neurochem. Res 12, 73–77. [DOI] [PubMed] [Google Scholar]
- 62.Kushima Y, Takeda K, Oh-Hashi Y, Nakagawa T, and Kato T (1989) The effects of anesthetics on the concentrations of cholecystokinin octapeptide sulfate-like immunoreactivity in rat brain regions, Neuropeptides 14, 225–230. [DOI] [PubMed] [Google Scholar]
- 63.Pashkov VN, Westphalen RI, and Hemmings HC Jr. (2002) General anesthetics do not affect release of the neuropeptide cholecystokinin from isolated rat cortical nerve terminals, Anesthesiology 97, 1500–1506. [DOI] [PubMed] [Google Scholar]
- 64.Besnier E, Clavier T, Tonon MC, Pelletier G, Dureuil B, Castel H, and Compère V (2018) Anesthetic drugs modulate feeding behavior and hypothalamic expression of the POMC polypeptide precursor and the NPY neuropeptide, BMC Anesthesiol. 18, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Upton DH, Popovic K, Fulton R, and Kassiou M (2020) Anaesthetic-dependent changes in gene expression following acute and chronic exposure in the rodent brain, Sci. Rep 10, 9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andrews MT (2019) Molecular interactions underpinning the phenotype of hibernation in mammals, J. Exp. Biol 222. [DOI] [PubMed] [Google Scholar]
- 67.Su B, Wang X, Drew KL, Perry G, Smith MA, and Zhu X (2008) Physiological regulation of tau phosphorylation during hibernation, J. Neurochem 105, 2098–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gardi J, Nelson OL, Robbins CT, Szentirmai E, Kapas L, and Krueger JM (2011) Energy homeostasis regulatory peptides in hibernating grizzly bears, Gen. Comp. Endocrinol 172, 181–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma B, Zhang K, Hendrie C, Liang C, Li M, Doherty-Kirby A, and Lajoie G (2003) PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry, Rapid Commun. Mass Spectrom 17, 2337–2342. [DOI] [PubMed] [Google Scholar]
- 70.Zhang J, Xin L, Shan B, Chen W, Xie M, Yuen D, Zhang W, Zhang Z, Lajoie GA, and Ma B (2012) PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification, Mol. Cell. Proteomics 11, M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.UniProt Consortium . (2021) UniProt: the universal protein knowledgebase in 2021, Nucleic Acids Res. 49, D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luan H, Ji F, Chen Y, and Cai Z (2018) statTarget: A streamlined tool for signal drift correction and interpretations of quantitative mass spectrometry-based omics data, Anal. Chim. Acta 1036, 66–72. [DOI] [PubMed] [Google Scholar]
- 73.RStudioTeam. (2021) RStudio: Integrated Development for R, Boston, MA. [Google Scholar]
- 74.Benjamini Y, and Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing, J. R. Stat. Soc. Series B 57, 289–300. [Google Scholar]
- 75.Stacklies W, Redestig H, Scholz M, Walther D, and Selbig J (2007) pcaMethods--a bioconductor package providing PCA methods for incomplete data, Bioinformatics 23, 1164–1167. [DOI] [PubMed] [Google Scholar]
- 76.Thévenot EA, Roux A, Xu Y, Ezan E, and Junot C (2015) Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses, J. Proteome Res 14, 3322–3335. [DOI] [PubMed] [Google Scholar]
- 77.Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M, Walzer M, Wang S, Brazma A, and Vizcaíno JA (2022) The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences, Nucleic Acids Res. 50, D543–d552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


