Abstract
The polysaccharide chains of enterobacterial common antigen (ECA) are comprised of the trisaccharide repeat unit Fuc4NAc-ManNAcA-GlcNAc, where Fuc4NAc is 4-acetamido-4,6-dideoxy-d-galactose, ManNAcA is N-acetyl-d-mannosaminuronic acid, and GlcNAc is N-acetyl-d-glucosamine. Individual trisaccharide repeat units are assembled as undecaprenyl-linked intermediates in a sequence of reactions that culminate in the transfer of Fuc4NAc from TDP-Fuc4NAc to ManNAcA-GlcNAc-pyrophosphorylundecaprenol (lipid II) to yield Fuc4NAc-ManNAcA-GlcNAc-pyrophosphorylundecaprenol (lipid III), the donor of trisaccharide repeat units for ECA polysaccharide chain elongation. Most of the genes known to be involved in ECA assembly are located in the wec gene cluster located at ca. 85.4 min on the Escherichia coli chromosome. The available data suggest that the structural gene for the TDP-Fuc4NAc:lipid II Fuc4NAc transferase also resides in the wec gene cluster; however, the location of this gene has not been unequivocally defined. Previous characterization of the nucleotide sequence of the wec gene cluster in the region between o416 and wecG revealed that it contained three open reading frames: o74, o204, and o450. In contrast, the results of experiments described in the current investigation revealed that it contains only two open reading frames, o359 and o450. Mutants of E. coli possessing null mutations in o359 were unable to synthesize ECA, and they accumulated lipid II. In addition, the in vitro incorporation of [3H]FucNAc from TDP-[3H]Fuc4NAc into lipid II was not observed in reaction mixtures using cell extracts obtained from these mutants as a source of enzyme. The ECA-negative phenotype of these mutants was complemented by plasmid constructs containing the wild-type o359 allele, and Fuc4NAc transferase activity was demonstrated by using cell extracts obtained from the complemented mutants. Furthermore, partially purified o359 gene product, expressed as recombinant C-terminal His-tagged protein, was able to catalyze the in vitro transfer of [3H]Fuc4NAc from TDP-[3H]Fuc4NAc to lipid II. Our data support the conclusion that o359 of the wec gene cluster of E. coli is the structural gene for the TDP-Fuc4NAc:lipid II Fuc4NAc transferase involved in the synthesis ECA trisaccharide repeat units.
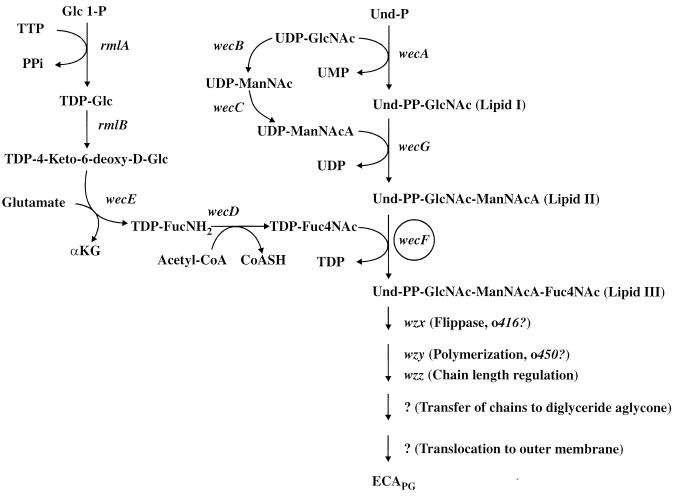
Enterobacterial common antigen (ECA) is a glycolipid found in the outer leaflet of the outer membrane of all gram-negative enteric bacteria (12, 15, 19, 27). The component sugars of the ECA polysaccharide are N-acetyl-d-glucosamine (GlcNAc), N-acetyl-d-mannosaminuronic acid (ManNAcA), and 4-acetamido-4,6-dideoxy-d-galactose (Fuc4NAc). These amino sugars are linked to one another to yield polysaccharide chains comprised of trisaccharide repeat units with the following structure: →3)-α-d-Fuc4NAc-(1→4)-β-d-ManNAcA-(1→4)-α-d-GlcNAc-(1→ (14, 16, 27). The linear trisaccharide repeat units of ECA are assembled as undecaprenyl-linked intermediates in a stepwise sequence of reactions that are initiated by the transfer of GlcNAc 1-P from UDP-GlcNAc to undecaprenylphosphate (Und-P) to yield Und-PP-GlcNAc (lipid I) (Fig. 1) (4, 26, 27). Subsequent steps involve the incorporation of ManNAcA and Fuc4NAc to yield ManNAcA-GlcNAc-PP-Und (lipid II) and Fuc4NAc-ManNAcA-GlcNAc-PP-Und (lipid III), respectively (3). Lipid III molecules are then presumably translocated across the cytoplasmic membrane to the periplasmic face of the membrane, where polysaccharide chain-elongation is believed to occur by a “block-polymerization” mechanism. Finally, Und-PP-linked ECA polysaccharide chains are transferred to an as-yet-unidentified glyceride acceptor to yield polysaccharide chains that are linked through the potential reducing terminal GlcNAc to diacylglycerol via phosphodiester linkage (ECAPG) (13, 25). Completed ECAPG molecules are subsequently translocated to the outer membrane by an unknown mechanism, where they are anchored in the outer leaflet by the hydrophobic portion of the phosphoglyceride aglycone (1, 29).
FIG. 1.
ECA biosynthetic pathway. The genetic determinants of the enzymes involved in the biosynthesis of ECA are indicated next to the reaction catalyzed by the respective enzyme. Abbreviations: Und-P, undecaprenylmonophosphate; Und-PP, undecaprenylpyrophosphate; GlcNAc, N-acetyl-d-glucosamine; ManNAcA, N-acetyl-d-mannosaminuronic acid; FucNH2, 4-amino-4,6-dideoxy-d-galactose; Fuc4NAc, 4-acetamido-4,6-dideoxy-d-galactose; Glc1-P, glucose-1-phosphate; PPi, inorganic pyrophosphate; αKG, α-ketoglutaric acid; acetyl-CoA, acetyl-coenzyme A; CoASH, coenzyme A; ECAPG, the phosphoglyceride-linked form of ECA.
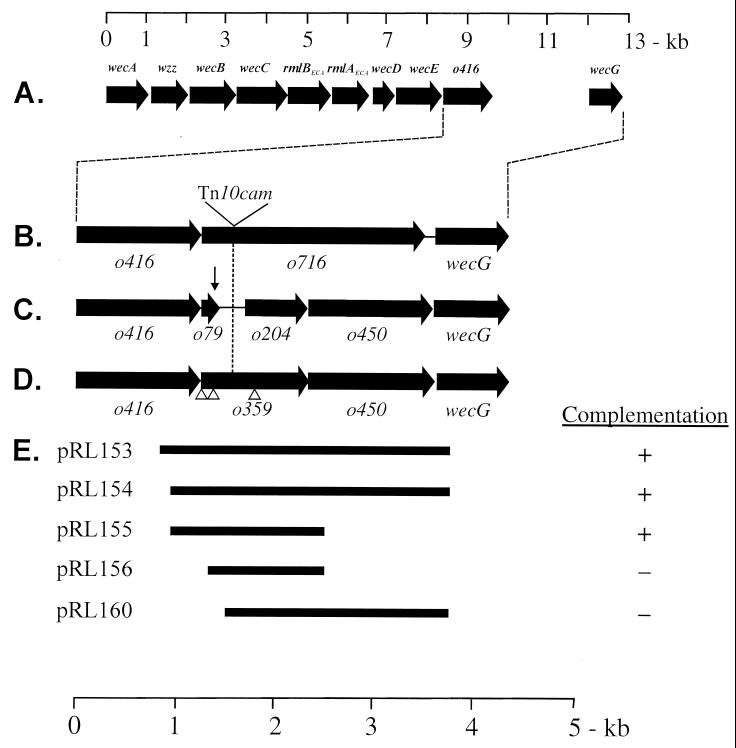
Most of the genes involved in the assembly of ECA are located in the wec gene cluster (formerly the rfe-rff gene cluster) located at ca. 85.4 min on the Escherichia coli chromosome (5, 12, 20, 27). Earlier studies on the genetics and biosynthesis of ECA in E. coli resulted in the isolation of a mutant that was unable to synthesize ECA due to a spontaneous mutation, termed rff-726 (21). Although these studies did not reveal the specific step of ECA synthesis affected by this mutation, rough-mapping studies indicated that it was located in the wec gene cluster near wecA (formerly rfe). Subsequent biochemical and genetic studies suggested that the rff-726 mutation resided in the structural gene for the TDP-Fuc4NAc:lipid II Fuc4NAc transferase (22). Although this gene was localized to the 3′ region of the wec gene cluster, its precise location was not determined. The complete nucleotide sequence of the E. coli wec gene cluster was later determined by Daniels et al. (9). These investigators defined a single open reading frame (ORF), o716, that was located between o416 and wecG in the 3′-terminal region of the wec cluster (Fig. 2B), and they tentatively identified o716 as the structural gene (wecF, formerly rffT) for the Fuc4NAc transfersase. This conclusion was supported by the results of experiments which demonstrated that the rff-726 mutation was complemented by a region of the wec cluster that included the putative wild-type o716 locus (21).
FIG. 2.
Identification of the genetic determinants located between o416 and wecG of the wec gene cluster. (A) wec gene cluster of E. coli K-12. ORFs are indicated by the solid arrows. The 13-kb scale above the gene cluster is provided for reference. The region between o416 and wecG, as defined by previous investigations, is presented below (B and C). (B) Identification of a single ORF, o716, between o416 and wecG, as determined by initial nucleotide sequencing experiments (9). (C) Revised characterization of the region between o416 and wecG, indicating the occurrence of ORFs o74, o204, and o450 (GenBank accession no. AE000455). (D) Identification of two ORFs, o359 and o450, in the region between o416 and wecG, as determined in the current study. The open triangles indicate the positions of potential translational start codons. The vertical arrow in panel C indicates the location of a frameshift resulting from the erroneous detection of two cytosines rather than a single cytosine immediately downstream of nucleotide 7617 of the wec cluster. The vertical dashed line indicates the position of the Tn10cam insertion in strain PND788. (E) Chromosomal insert fragments contained in the indicated plasmids, and the ability of these constructs to complement the ECA-negative phenotype of strain PND788 as determined by passive hemagglutination assay (26). The 5-kb scale at the bottom of the figure is presented to provide reference for the region circumscribed by o416 and wecG.
Morona et al. (24) noted that the hydropathy profile of the carboxy-terminal half of the putative protein encoded by o716 was strikingly similar to that of a wide variety of putative WzyOAg (Rfc, O-antigen polymerase) enzymes. These authors speculated that either the putative o716 gene product possessed both Fuc4NAc transferase and WzyECA activities or the nucleotide sequence including o716 was incorrect and this region contained structural genes for both the Fuc4NAc transferase and the WzyECA. A subsequent reexamination of the nucleotide sequence between o416 and wecG of the wec cluster (GenBank accession no. AE000455) indicated that it was incorrect. These experiments revealed that this region did not include an o716; rather, it was reported to contain three putative ORFs: o74, o204, and o450 (Fig. 2C). In addition, o450 was tentatively identified as the structural gene for the Fuc4NAc transferase. However, repeated examination of the nucleotide sequence of this region in our laboratory yielded results that were not in agreement with this conclusion. In contrast, our data revealed that the region between o416 and wecG contains only two ORFs, o359 and o450, and we report here the results of experiments which clearly demonstrate that o359 is the structural gene (wecF) for the TDP-Fuc4NAc:lipid II Fuc4NAc transferase involved in the synthesis of ECA.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. Where indicated, transductions were carried out by using P1 vir as described by Silhavy et al. (30). Cultures of E. coli were grown in Luria-Bertani (LB) broth (23) or on LB agar at 37°C. Cultures of Pseudomonas aeruginosa were grown at 32°C in a salts medium containing K2HPO4 (7 g/liter), KH2PO4 (3 g/liter), MgSO4 · 7H2O (0.1 g/liter), and (NH4)2SO4 (1.0 g/liter), as well as 3% glycerol as the sole source of carbon. The following antibiotics were added to media when appropriate at the indicated final concentrations: ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), and tetracycline (20 μg/ml).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genetic markers or characteristic(s) | Reference or source |
|---|---|---|
| E. coli K-12 | ||
| AB1133 | thr-1 λ(gpt-proA)66 leuB6 hisG4 argE3 thi-1 rfbD1 lacY1 ara-14 galK2 xyl-5 mtl-1 mgl-51 rpsL31 kdgK51 supE44 | CGSCa |
| PND788 | F−araD139 ΔlacU169 relA1 rpsL150 thi mot flb5301 deoC7 ptsF25 rbsR λR88[degP-lacZ] ompR::Tn10 wecF::cam | 8 |
| PND788F | PND788/pRL151; wecF::cam/C-terminal His6-tagged o359 fusion | This study |
| 21550 | Same as AB1133, but wecE::Tn10 | 22 |
| PR4144 | Same as 21550, but wecF::cam [PND788(P1) × 21550] | This study |
| P. aeruginosa 7700 | Wild type | ATCC |
| Plasmidsb | ||
| pBAD-TOPO | Cloning vector used to construct C-terminal histidine-tagged (His6) o359 gene product | Invitrogen |
| pCR2.1-TOPO | TA cloning vector | Invitrogen |
| pBlueScript IIKS+ | High-copy-number phagemid vector, used for expression of the C-terminal His6-tagged o359 gene product under control of the lac promoter | Stratagene |
| pRL150 | Fragment containing o359 (bp 7067 to 8519, E. coli GenBank accession no. AE000455) in pBAD-TOPO vector to yield C-terminal His6 tag | This study |
| pRL151 | PCR-amplified fragment of pRL150 containing o359 gene + C-terminal His6-tag subcloned into pBlueScript II KS(+) under control of the lac promoter | This study |
| pRL153 | PCR fragment containing the sequence from bp 7067 to 9948 (E. coli GenBank accession no. AE000455) in pCR 2.1-TOPO vector | This study |
| pRL154 | PCR fragment containing the sequence from bp 7160 to 9948 (E. coli GenBank accession no. AE000455) in pCR 2.1-TOPO vector | This study |
| pRL155 | PCR fragment containing the sequence from bp 7160 to 8683 (E. coli GenBank accession no. AE000455) in pCR 2.1-TOPO vector | This study |
| pRL156 | PCR fragment containing the sequence from bp 7463 to 8683 (E. coli GenBank accession no. AE000455) in pCR 2.1-TOPO vector | This study |
| pRL160 | PCR fragment containing the sequence from bp 7691 to 9948 (E. coli GenBank accession no. AE000455) in pCR 2.1 TOPO vector | This study |
E. coli Genetic Stock Center, M. Berlyn, Department of Biology, Yale University, New Haven, Conn.
PCR amplifications were carried out using genomic DNA from E. coli AB1133 as a template.
Radiochemicals, chemicals, and reagents.
d-[1-3H]glucose (15 Ci/mmol), UDP-N-acetyl-d-[6-3H](N)glucosamine (20.4 Ci/mmol), and UDP-N-acetyl-d-[U-14C]glucosamine (283 mCi/mmol) were purchased from New England Nuclear Corp. UDP-N-acetyl-d-[U-14C]mannosaminuronic acid (283 mCi/mmol) and unlabeled UDP-N-acetyl-d-mannosaminuronic acid were enzymatically synthesized from UDP-N-acetyl-d-[U-14C]glucosamine (283 mCi/mmol) and UDP-N-acetyl-d-glucosamine, respectively (4). d-[1-3H]glucose-6-phosphate was enzymatically synthesized from d-[1-3H]glucose (15 Ci/mmol) by using yeast hexokinase. TDP-d-[1-3H]glucose (TDP-[3H]glucose) was enzymatically synthesized from d-[1-3H]glucose-6-phosphate by using partially purified TDP-glucose pyrophosphorylase prepared from extracts of P. aeruginosa 7700 as described by Kornfeld and Glaser (11). TDP-4-acetamido-4,6-dideoxy-d-[1-3H]galactose (TDP-[3H]Fuc4NAc) and TDP-Fuc4NAc were synthesized from TDP-[3H]glucose and TDP-d-glucose, respectively, as described by Matsuhashi and Strominger (18). All other chemicals and reagents were purchased from standard commercial sources.
Plasmid constructions.
The plasmids used in this study are listed in Table 1. Primers used for PCR amplification of nucleotide sequences are listed in Table 2. PCR amplifications were carried out by using Taq polymerase (Sigma Chemicals) according to standard protocols. Plasmid pRL150 was constructed by PCR amplification of the DNA sequence from bp 7067 to 8519 (GenBank accession no. AE000455) by using primers F1 and F8 and genomic DNA from strain AB1133 as a template. The amplified sequence included o359 in addition to 375 bp immediately upstream of the translational start site, and it was cloned into the TA cloning site of the pBAD-TOPO vector (Invitrogen, Inc.). One of the resulting constructs contained the C-terminal nucleotide sequence of o359 in-frame with the polyhistidine (His6) encoding nucleotide sequence of the vector. Plasmid pRL151 was constructed by PCR amplification of the insert fragment of pRL150 containing the entire o359-C-terminal His6 tag fusion by using primers F1 and BAD1. The EcoRI- and BamHI-cleaved PCR product was subcloned into pBluescript II KS(+) (Stratagene) that was restricted by the same enzymes. Plasmids pRL153, pRL154, pRL155, pRL156, and pRL160 were constructed by PCR amplification of the DNA sequences from bp 7067 to 9948, 7160 to 9948, 7160 to 8683, 7463 to 8683, and 7691 to 9948 (GenBank accession no. AE000455) by using primer pairs F1-F7, F2-F7, F2-F6, F3-F6, and F4-F7 and genomic DNA from strain AB1133 as a template, respectively. The PCR products were cloned into the TA-cloning site of the pCR 2.1-TOPO cloning vector. T4 DNA ligase (Invitrogen) and restriction enzymes were used in accordance with the manufacturer's recommendations.
TABLE 2.
Nucleotide sequences of PCR primersa
| Primer (orientation) | Sequence | Restriction site created |
|---|---|---|
| F1 (forward) | 5′-GGAAGTGGTGAATTCGCTGA-3′ | EcoRI |
| F2 (forward) | 5′-GTTGTCGAATGAATTCACCGC-3′ | EcoRI |
| F3 (forward) | 5′-CTGGGATCGGAATTCCCTCAC-3′ | EcoRI |
| F4 (forward) | 5′-CGGTCAGTTGAATTCCACACTG-3′ | EcoRI |
| F6 (reverse) | 5′-GCTGGTCTGCAGGAAGCC-3′ | PstI |
| F7 (reverse) | 5′-GTGCTGCAGATCACGCCAA-3′ | PstI |
| F8 (reverse) | 5′-TGCGACCTCCCTGGCGGC-3′ | |
| BAD1 (reverse) | 5′-GATTTAATCTGGATCCGG-3′ | BamHI |
Nucleotide sequences for the indicated restriction sites are underlined. The nucleotide changes that were made in order to generate each of the respective restriction sites are indicated in boldface.
In vitro synthesis of lipid III.
Cell envelopes were prepared as previously described (4). Reaction mixtures for the in vitro synthesis of lipid III contained the following in a final volume of 55 μl: 50 mM Tris-HCl (pH 8.2), 30 mM MgCl2, 5 mM 2-mercaptoethanol, 0.14 μM TDP-[3H]Fuc4NAc (2.56 × 107 dpm/μmol), and cell envelope membranes (700 to 850 μg of protein). Partially purified native His6-tagged o359 gene product (6.75 μg of protein) and 0.24 μM UDP-[14C]ManNAcA (6.3 × 105 dpm/μmol) were added to reaction mixtures where indicated. Reactions were incubated at 37°C for 30 min and then terminated by the addition of 1.0 ml of chloroform-methanol (3:2, by volume). The radioactive products were extracted from reaction mixtures as described previously (3), and they were analyzed directly by ascending paper chromatography with SG-81 filter paper (Whatman, Inc.). Protein was determined by using the BCA Protein Assay Reagent Kit (Pierce) according to the instructions provided by the manufacturer.
In vivo assay for lipid II accumulation.
The incorporation of [3H]GlcNAc into lipid II was determined as previously described (28). Briefly, bacteria were grown with vigorous aeration at 37°C in 60 ml of LB medium supplemented with glucose (0.2%, final concentration) to an A600 of 0.4. The cells were then harvested by centrifugation, resuspended in fresh medium (6 ml), and incubated at 37°C with [3H]GlcNAc (75 μCi, 8.2 Ci/mmol) for 30 min. The labeled cells were then poured over crushed ice, harvested by centrifugation, and washed with cold 0.9% saline. The washed cells were then successively extracted with 95% ethanol (6 ml) and acetone (6 ml) and dried in vacuo. The dried cells were extracted with 1.5 ml of chloroform-methanol (3:2, by volume), and 75 μl of the resulting extract was analyzed by ascending paper chromatography on EDTA-treated SG-81 paper by using chloroform-methanol-water-concentrated ammonium hydroxide (88:48:10:1, by volume) as the developing solvent. The amount of radioactivity in the region of the chromatogram corresponding to lipid II was then determined as described below.
Partial purification of C-terminal histidine-tagged o359 gene product.
Strain PND788F was grown in 200 ml of LB-glucose medium at 37°C. IPTG (isopropyl-β-d-thiogalactopyranoside) was added at A600 = 0.2 to give a final concentration of 1 mM, and the culture was incubated overnight with vigorous aeration. The cells were harvested by centrifugation and lysed with 10 ml of B-PER (Pierce) according to the manufacturer's recommendation. The suspension was then briefly sonicated, and the insoluble material was then separated from the soluble fraction (B-PER extract) by centrifugation at 12,000 × g. The pellet was resuspended in 5 ml of a solution containing 8 M urea, 0.1 M NaH2PO4, and 0.01 M Tris-HCl (pH 8.0; final concentrations) and then incubated at room temperature for 60 min with gentle stirring. The urea-soluble fraction was next separated from cell debris by centrifugation at 12,000 × g for 20 min. The His-tagged o359 gene product present in the B-PER extract and the urea-soluble fraction were then partially purified by affinity chromatography by using nickel-nitrilotriacetic acid (Ni+-NTA) agarose resin (Qiagen, Inc.) under native and denaturing conditions, respectively. Briefly, for partial purification by using native conditions, imidazole was added to 4 ml of crude B-PER extract to give a final concentration of 10 mM. Then, 1 ml of a slurry containing 50% Ni+-NTA agarose resin was added, and the mixture was incubated with gentle shaking at 4°C for 60 min. The mixture was poured into a small column and, after the resin settled, the liquid fraction was allowed to flow out of the column. The resin was then washed twice with 4 ml of wash buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 20 mM imidazole). His-tagged protein was then eluted by four successive 0.5-ml washes of the resin with elution buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl, 250 mM imidazole), and fractions were analyzed by Western blot analyses by using mouse anti-Penta-His antibody (Qiagen, Inc.) and horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG) (Amersham). Fractions containing C-terminal His-tagged o359 were used for in vitro assay of Fuc4NAc transferase activity.
For partial purification of the C-terminal His-tagged o359 gene product using denaturing conditions, 4 ml of the urea-soluble extract was mixed with 1 ml of a slurry containing 50% Ni+-NTA agarose resin and incubated with gentle shaking for 60 min at room temperature. The mixture was poured into a small column and, after the resin settled, the liquid fraction was allowed to flow out of the column. The resin was then washed twice with 4 ml of buffer B (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-hydrochloride; pH 6.3). His-tagged protein was then eluted with four successive 0.5-ml washes of the resin with buffer C (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-hydrochloride; pH 5.9), followed by four successive 0.5-ml washes with buffer D (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-hydrochloride; pH 4.5). The fractions were analyzed by Western blot analyses by using mouse anti-Penta-His antibody (Qiagen) and horseradish peroxidase-conjugated anti-mouse IgG (Amersham). Increased yields of more highly purified urea-soluble His-tagged protein were obtained by combining sonicated B-PER extracts with the urea-soluble fraction, followed by batch purification by using a modification of the denaturing conditions as described above. Briefly, the material that eluted from the column with buffer C was concentrated by centrifugation by using a Microcon YM-30 centrifugal filtration device (Amicon Bioseparations) according to the manufacturer's recommendations and then reapplied to a new column containing Ni+-NTA agarose resin. The column was then eluted successively with buffers B and C as described above. The material that eluted after development of the column with buffer C was detected as a single band by Western blot analyses.
Cellular localization of C-terminal histidine-tagged o359 gene product.
The cellular localization of C-terminal His6-tagged o359 gene product was determined by using a modification of the microprocedure described by Carlone et al. (6). Strain PND788F was grown with vigorous aeration in 500 ml of LB broth at 37°C. IPTG was added at A600 = 0.2 to give a final concentration of 1 mM, and the culture was incubated overnight with vigorous aeration. The cells were then harvested by centrifugation, suspended in 5 ml of ice-cold 20 mM Tris-HCl–10 mM EDTA (pH 7.5) (Tris-EDTA) buffer, and disrupted by three passages through a French pressure cell (10,000 lb/in2, high ratio) at 0 to 4°C. Unbroken cells and cellular debris were removed by centrifugation at 20,000 × g for 20 min. Cell envelope membranes were isolated by centrifugation of the turbid supernatant solution at 200,000 × g for 2 h. The resulting pellet was resuspended in 1 ml of Tris-EDTA buffer, and the total cell envelope protein concentration was determined by using the BCA Protein Assay Reagent Kit (Pierce) according to the instructions provided by the manufacturer. The suspension was then diluted with Tris-EDTA buffer to give a protein concentration of 4.4 mg/ml, and 1/10 volume of a 10% (wt/vol) solution of Sarkosyl (N-lauroylsarcosine, sodium salt) (Sigma) was subsequently added with gentle mixing. The mixture was then incubated in an ice bath for 1 h, followed by centrifugation at 200,000 × g (2 h, 4°C). The supernatant solution was removed, and the pellet was resuspended in 0.5 ml of Tris-EDTA buffer and once again extracted with 1/10 volume of Sarkosyl, followed by centrifugation at 200,000 × g (2 h, 4°C) as described above. The quantity of protein in the Sarkosyl-insoluble fraction and pooled Sarkosyl-soluble fractions was determined. The proteins in these fractions were then precipitated by the addition of 8 volumes of cold acetone. The precipitates were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer.
Determination of the nucleotide sequence in the region between o416 and wecG
The nucleotide sequence of the wec gene cluster in the region between o416 and wecG (2,882 bp) contained in the cloned fragment of pRL153 was determined in both directions with BigDye Terminator Cycle Sequencing Ready Reaction kits (Applied Biosystems) and an ABI377 sequencer (Applied Biosystems). The data from these experiments were compared with the corresponding nucleotide sequence from E. coli (GenBank accession number AE000455). The nucleotide sequence of the same region of the chromosome of the mutant strain PND788 was also determined in order to confirm the position of the Tn10::cam insertion.
Passive hemagglutination assay.
The presence of ECA on the surface of bacterial strains was determined by passive hemagglutination assay with polyclonal rabbit anti-ECA antiserum as previously described (26).
Western blot analyses.
Western blot analyses were carried out as previously described (26), except that SDS-PAGE was conducted by using 10% polyacrylamide gels, and C-terminal His6-tagged WecF was detected by using mouse anti-tetra-His antibody (Qiagen), horseradish peroxidase-conjugated anti-mouse IgG (Amersham), and Western blot Chemiluminescence Reagent Plus (NEN Life Science Products).
Chromatographic and electrophoretic procedures.
Paper chromatography was carried out by using SG-81 paper (Whatman) that was prepared as previously described (28). Samples were spotted onto the paper, and the chromatogram was developed with a solvent mixture containing chloroform-methanol-water-concentrated ammonium hydroxide (88:48:10:1, by volume). Lanes of the air-dried chromatogram were cut into 1-cm sections, and the sections were soaked in 0.5 ml of 1.25% SDS for 6 to 8 h at 42°C. The samples were then analyzed for radioactivity by liquid scintillation counting.
Nucleotide sequence accession number.
The revised nucleotide sequence of the region of the wec gene cluster between o416 and wecG has been deposited in the GenBank database under accession number AF375882.
RESULTS AND DISCUSSION
Occurrence of ORFs o359 and o450 in the region between o416 and wecG of the wec gene cluster of E. coli.
The wec gene cluster of E. coli contains genes involved in the synthesis and assembly of ECA. Although the location and function of many of the genes in this cluster have been defined (2, 5, 17, 27), only the approximate location of the gene encoding the TDP-Fuc4NAc:lipid II Fuc4NAc transferase involved in the synthesis of the ECA trisaccharide repeat unit has been determined. The results of previous investigations support the conclusion that this gene is located in the region between o416 and wecG of the wec gene cluster (21). However, confusion regarding the nucleotide sequence of this region has hampered an unequivocal determination of the location of this gene. The nucleotide sequence between o416 and wecG of the wec gene cluster of E. coli has been reported to contain three putative ORFs: o74, o204, and o450 (GenBank accession no. AE000455). However, repeated determination of the nucleotide sequence of this region during the present study revealed only two putative ORFs, o359 and o450, whose transcriptional orientation is the same as that of the other genes in the wec gene cluster (Fig. 2D). These two ORFs overlap by 4 bp, and the overlapping sequence, 5′—o359—ATGA—o450—3′, includes the predicted translational start codon of o450 (underlined) and the predicted translational stop codon of o359 (boldface lettering). This organization suggests tight coupling at both the transcriptional and the translational levels. The previous assignments of o74 and o204 and the failure to detect o359 were found to be due to a frameshift stemming from the erroneous detection of two cytosines rather than a single cytosine immediately downstream of nucleotide 7617 of the nucleotide sequence of the wec cluster (Fig. 2C). However, with one exception, the predicted amino acid sequence of the o450 gene product as determined by nucleotide sequencing experiments carried out in this study revealed that it was essentially identical to that predicted by the published sequence. In this regard, the published sequence reported an aspartic acid residue at position 283, whereas the current study revealed a histidine residue at this position.
Complementation of mutants of E. coli defective in the synthesis of lipid III by o359 of the wec gene cluster.
Previous studies resulted in the isolation of E. coli PND788 which contains a Tn10cam insertion in the region of the chromosome originally identified as o716 of the wec gene cluster (8). These studies also demonstrated that mutant strain PND788 was unable to synthesize ECA and that it accumulated lipid II. A determination of the location of the Tn10cam insertion in the chromosome of strain PND788 revealed that it was inserted in codon 141 of the newly defined o359. This site corresponds to the insertion of Tn10cam between bp 7863 and 7864 as defined by the most recently published sequence of this region of the chromosome (GenBank accession no. AE000455).
ECA synthesis was rescued in transformants of PND788 containing either plasmid pRL153, pRL154, or pRL155, all of which contain o359 (Fig. 2E). Although the cloned fragment in plasmids pRL153 and pRL154 also contains o450 located immediately downstream of o359, the data clearly indicate that the observed complementation was not due to o450 since the cloned fragment in pRL155 contains o359 as the only complete ORF. This conclusion was supported by the observation that plasmid pRL160, which contains o450 as the only complete ORF, was unable to complement the ECA-negative phenotype of PND788. As previously stated, the inability of strain PND788 to synthesize ECA is accompanied by the accumulation of lipid II (8). However, lipid II accumulation was markedly reduced in strain PND788F, a derivative of PND788 that contains plasmid pRL151 which encodes the putative o359 gene product possessing a carboxy-terminal His6 tag (Table 3). Furthermore, the decreased accumulation of lipid II in strain PND788F was accompanied by the synthesis of ECA.
TABLE 3.
Complementation of the phenotype of strain PND788 (o359::cam) in a derivative expressing carboxy-terminal His6-tagged o359a
| Strain | Relevant properties | [3H]GlcNAc-lipid II recovered in extracts (dpm) | ECA synthesis |
|---|---|---|---|
| PND788 | o359::cam | 12,097 | − |
| PND788F | o359::cam/pRL151 (o359-His6) | 1,501 | + |
In vivo radiolabeling and extraction of [3H]GlcNAc-labeled lipid II are described in Materials and Methods. ECA synthesis was determined by the passive hemagglutination assay as described in Materials and Methods.
The nucleotide sequence of o359 contains three ATG codons: the putative translation initiation codon and two downstream ATG codons (codons 38 and 184) (Fig. 2D). However, translation does not appear to be initiated at either of the downstream codons since the ECA-negative phenotype of PND788 was not complemented by plasmid pRL156, which encodes a truncated o359 allele that includes codons 38 and 184 but which lacks the 5′ terminus from bp 1 to 121 (Fig. 2D and E). In addition, only the putative translational start codon is immediately preceded by a nucleotide sequence that conforms to the Shine-Dalgarno consensus sequence (data not shown). Taken together, the above data support the conclusion that o359 is indeed functional and that it is responsible for the observed complemention of the phenotype of mutant strain PND788.
The o359 gene product catalyzes the transfer of Fuc4NAc from TDP-Fuc4NAc to lipid II.
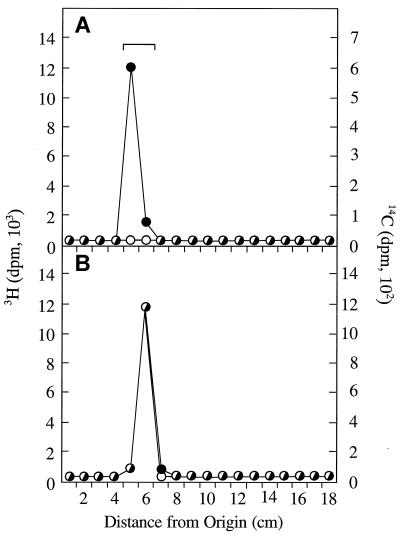
The accumulation of lipid II in strain PND788 suggested that o359 encoded the TDP-Fuc4NAc:lipid II Fuc4NAc transferase required for lipid III synthesis. However, the phenotype of this mutant is also compatible with the possibility that o359 is in some way required for TDP-Fuc4NAc synthesis. In order to distinguish between these possibilities, in vitro synthesis of lipid III was examined by utilizing cell envelopes prepared from E. coli PR4144, an o359::Tn10cam wecE::Tn10tet double mutant, as a source of enzymes and lipid acceptors in cell-free reaction mixtures. The null mutation in wecE precludes synthesis of TDP-Fuc4NH2, the immediate precursor of TDP-Fuc4NAc, thus resulting in the accumulation of lipid II as well as lesser amounts of lipid I (21). Incubation of cell envelopes obtained from strain PR4144 with UDP-[14C]ManNAcA and TDP-[3H]Fuc4NAc resulted in the incorporation of only [14C]ManNAcA into lipid II (Fig. 3A); thus, the incorporation of [14C]ManNAcA reflects the conversion of accumulated lipid I to lipid II. Similar results were obtained by using cell envelopes obtained from PND788 (data not shown). In contrast, the incorporation of both [14C]ManNAcA and [3H]Fuc4NAc into lipid III was observed in reaction mixtures containing cell envelopes obtained from transformants of strain PR4144 that contained plasmid pRL151 which encodes the putative o359 gene product containing a carboxy-terminal His6 tag (Fig. 3B). It is important to note that lipids II and III have the same chromatographic mobility under the assay conditions employed (3). Therefore, the single peak of radiolabeled material in Fig. 3B actually includes [14C]ManNAcA-labeled lipid II, as well as lipid III that is labeled with both [14C]ManNAcA and [3H]Fuc4NAc. These data are in agreement with the conclusion that o359 is not involved in TDP-Fuc4NAc synthesis but rather that it is the structural gene for the glycosyltransferase that catalyzes the transfer of Fuc4NAc from TDP-Fuc4NAc to lipid II to form lipid III.
FIG. 3.
In vitro transfer of [3H]Fuc4NAc from TDP-[3H]Fuc4NAc to lipid II present in cell envelopes of strains PR4144 and PR4144/pRL151. Cell envelope membranes (700 to 850 μg of protein) were incubated with UDP-[14C]ManNAcA and TDP-[3H]Fuc4NAc in the standard reaction mixture (55 μl) at 37°C for 30 min. Reactions were terminated by the addition of chloroform-methanol, and radioactive products were extracted from the reaction mixtures and subsequently analyzed by ascending paper chromatography by using SG-81 filter paper. Additional details are provided in Materials and Methods. (A) Cell envelopes of strain PR4144 (o359::Tn10cam wecE::Tn10tet). (B) Cell envelopes of strain PR4144/pRL151 (o359::Tn10cam wecE:: Tn10tet/C-terminal His6-tagged o359). Symbols: ●, carbon-14; ○, tritium. The chromatographic mobility of authentic lipid III is indicated by the bracket.
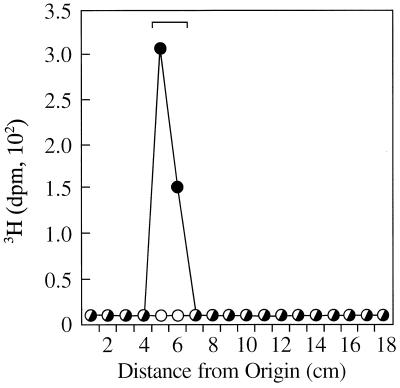
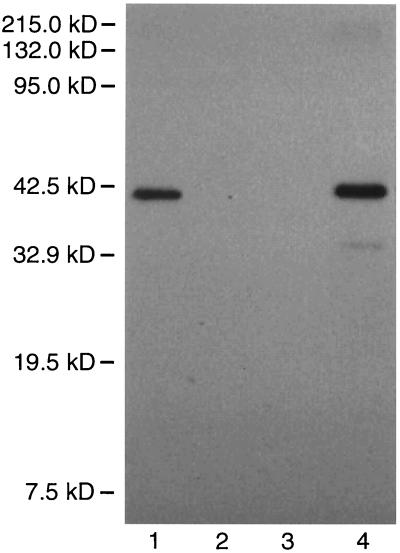
Additional evidence in support of the proposed function of o359 was obtained by analyses of partially purified C-terminal His6-tagged o359 gene product. Native His6-tagged protein was obtained by mild nonionic detergent extraction of strain PND788F cells after the induction of synthesis of the His-tagged protein with IPTG. The protein was then partially purified by affinity chromatography by using Ni+-NTA agarose resin. The in vitro transfer of [3H]Fuc4NAc from TDP-[3H]Fuc4NAc to endogenous lipid II present in the cell envelopes of strain PR4144 was observed in reaction mixtures containing affinity purified native His6-tagged protein (Fig. 4). In contrast, no [3H]Fuc4NAc-labeled product was observed in reaction mixtures lacking the affinity purified native His6-tagged protein. Furthermore, no Fuc4NAc-transferase activity was detected in column fractions obtained by chromatography of soluble B-PER extracts of PND788 and PND788/pBluescript II KS(+) when we used the same procedures for partial purification of the His6-tagged protein by Ni+-NTA agarose resin affinity chromatography (data not shown). Western blot analyses revealed that the affinity-purified native His6-tagged protein migrated as a 41.5-kDa component (Fig. 5). This observation is in excellent agreement with the predicted size of the His6-tagged o359 gene product since the calculated size of the wild-type protein is 39.5 kDa and the size of the additional C-terminal sequence of the His6-tagged protein, including the V5 epitope and polyhistidine region, is 2 kDa. The enzymatically active fraction eluted from the Ni+-NTA agarose resin was also considerably enriched for the 41.5-kDa component as determined by SDS-PAGE and Coomassie brilliant blue staining (data not shown). The above data strongly support the conclusion that o359 does indeed encode the TDP-Fuc4NAc:lipid II Fuc4NAc transferase involved in the synthesis of the ECA trisaccharide repeat unit and, accordingly, we propose that this gene locus be designated wecF.
FIG. 4.
In vitro transfer of [3H]Fuc4NAc from TDP-[3H]Fuc4NAc to endogenous lipid II present in cell envelopes of strain PR4144 catalyzed by partially purified C-terminal His6-tagged o359 gene product. Cell envelope membranes (700 to 850 μg of protein) prepared from strain PR4144 (o359::Tn10cam wecE::Tn10tet) were incubated with TDP-[3H]Fuc4NAc in standard reaction mixtures that also contained partially purified C-terminal His6-tagged o359 gene product (6.75 μg of protein) in a final volume of 55 μl. Experiments were also carried out with reaction mixtures (55 μl) that did not contain partially purified C-terminal His6-tagged o359 gene product. The reaction mixtures were incubated at 37°C for 30 min, and reactions were terminated by the addition of chloroform-methanol. Radioactive products were extracted from the reaction mixtures and subsequently analyzed by ascending paper chromatography with SG-81 filter paper. Additional details are provided in Materials and Methods. Symbols: ●, presence of His6-tagged o359 gene product; ○, absence of His6-tagged o359 gene product. The chromatographic mobility of authentic lipid III is indicated by the bracket.
FIG. 5.
Western blot analysis of partially purified C-terminal His6-tagged o359 gene product. Lane 1, native His6-tagged o359 gene product present in the soluble B-PER extract obtained from strain PND788F; lane 2, soluble B-PER extract obtained from strain PND788/pBluescript II KS(+) vector; lane 3, soluble B-PER extract obtained from strain PND788; lane 4, urea-soluble His6-tagged o359 gene product obtained from the insoluble fraction after B-PER extraction of strain PND788F. Details regarding the affinity purification and Western blot procedures are provided in Materials and Methods.
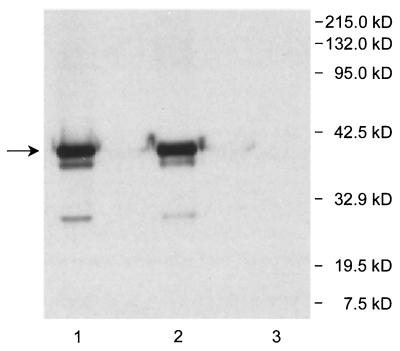
It is interesting that a significant amount of His6-tagged protein remained insoluble after extraction of cells with mild nonionic detergent; however, this material was rendered soluble by subsequent extraction with urea. In addition, Western blot analyses of this urea-soluble material revealed that it also migrated as a single 41.5-kDa component (Fig. 5), and it is likely that the urea-solubilized protein simply reflects inefficient detergent solubilization. In this regard, an examination of the hydropathy profile of the predicted wecF gene product suggests that it may contain a single transmembrane segment. These observations suggest that WecF is an integral membrane protein. In addition, C-terminal His6-tagged WecF protein was extracted from cell envelopes with the anionic detergent Sarkosyl, which preferentially solubilzes cytoplasmic membrane proteins (7, 10), and this protein was not detected in the Sarkosyl-insoluble fraction (Fig. 6). Accordingly, these data support the conclusion that WecF is indeed a component of the cytoplasmic membrane. However, details regarding the association of WecF with the cytoplasmic membrane and its membrane topology remain to be established.
FIG. 6.
Sarkosyl extraction of the C-terminal His6-tagged o359 gene product from cell envelopes. Cell envelopes obtained from strain PND788F were extracted with Sarkosyl, and the unextracted total cell envelope (lane 1) and the Sarkosyl-soluble (lane 2) and -insoluble (lane 3) fractions, were examined for the presence of the C-terminal His6-tagged o359 gene product by Western blot analysis by using mouse anti-Penta-His antibody. The arrow indicates the location of C-terminal His6-tagged o359 gene product that was partially purified by Ni+-NTA agarose resin affinity chromatography. Additional details are provided in Materials and Methods.
The available data suggest that ECA chain elongation occurs by a Wzy-mediated block-polymerization mechanism. However, the gene encoding the polymerase that catalyzes this reaction has not yet been identified. The wecF gene is located immediately upstream from o450; however, the function of o450 is not known. Previous observations suggest the possibility that o450 of the wec gene cluster is the structural gene for the polymerase involved in ECA polysaccharide chain elongation (24). This conclusion is supported by recent observations that mutants of E. coli K-12 containing a null mutation in o450 are unable to synthesize ECA and that they accumulate lipid III (unpublished results). However, biochemical data that directly demonstrate the ability of the o450 gene product to catalyze polymerization of ECA trisaccharide repeat units have not yet been obtained.
ACKNOWLEDGMENT
This work was supported by an NIGMS grant (GM52882) to P.D.R.
REFERENCES
- 1.Acker G, Knapp W, Wartenberg K, Mayer H. Localization of enterobacterial common antigen in Yersinia enterocolitica by immunoferritin technique. J Bacteriol. 1981;147:602–611. doi: 10.1128/jb.147.2.602-611.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barr K, Klena J, Rick P D. The modality of enterobacterial common antigen polysaccharide chain lengths is regulated by o349 of the wec gene cluster of Escherichia coli K-12. J Bacteriol. 1999;181:6564–6568. doi: 10.1128/jb.181.20.6564-6568.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr K, Nunes-Edwards P, Rick P D. In vitro synthesis of a lipid-linked trisaccharide involved in synthesis of enterobacterial common antigen. J Bacteriol. 1989;171:1326–1332. doi: 10.1128/jb.171.3.1326-1332.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr K, Rick P D. Biosynthesis of enterobacterial common antigen in Escherichia coli. In vitro synthesis of lipid-linked intermediates. J Biol Chem. 1987;262:7142–7150. [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Carlone G M, Thomas M L, Rumschlag H S, Sottnek F O. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J Clin Microbiol. 1986;24:330–332. doi: 10.1128/jcm.24.3.330-332.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chopra I, Shales S W. Comparison of the polypeptide composition of Escherichia coli outer membranes prepared by two methods. J Bacteriol. 1980;144:425–427. doi: 10.1128/jb.144.1.425-427.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danese P N, Oliver G R, Barr K, Bowman G D, Rick P D, Silhavy T J. Accumulation of the enterobacterial common antigen lipid II biosynthetic intermediate stimulates degP transcription in Escherichia coli. J Bacteriol. 1998;180:5875–5884. doi: 10.1128/jb.180.22.5875-5884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels D L, Plunkett III G, Burland V, Blattner F R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992;257:771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- 10.Filip C, Fletcher G, Wulff J L, Earhart C F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kornfeld S, Glaser L. The enzymatic synthesis of thymidine-linked sugars. I. Thymidine diphosphate glucose. J Biol Chem. 1961;236:1791–1794. [PubMed] [Google Scholar]
- 12.Kuhn H-M, Meier-Dieter U, Mayer H. ECA the enterobacterial common antigen. FEMS Microbiol Rev. 1988;54:195–222. doi: 10.1111/j.1574-6968.1988.tb02743.x. [DOI] [PubMed] [Google Scholar]
- 13.Kuhn H-M, Neter E, Mayer H. Modification of the lipid moiety of the enterobacterial common antigen by the “Pseudomonas” factor. Infect Immun. 1983;40:696–700. doi: 10.1128/iai.40.2.696-700.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lugowski C, Romanowska E, Kenne L, Lindberg B. Identification of a trisaccharide repeating unit in the enterobacterial common antigen. Carbohydr Res. 1983;118:173–181. [Google Scholar]
- 15.Mäkelä P H, Mayer H. Enterobacterial common antigen. Bacteriol Rev. 1976;40:591–632. doi: 10.1128/br.40.3.591-632.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Männel D, Mayer H. Isolation and chemical characterization of the entero-bacterial common antigen. Eur J Biochem. 1978;86:361–370. doi: 10.1111/j.1432-1033.1978.tb12318.x. [DOI] [PubMed] [Google Scholar]
- 17.Marolda C L, Valvano M A. Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J Bacteriol. 1995;126:579–586. doi: 10.1128/jb.177.19.5539-5546.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuhashi M, Strominger J L. Thymidine diphosphate 4-acetamido-4,6-dideoxyhexoses. I. Enzymatic synthesis by strains of Escherichia coli. J Biol Chem. 1964;239:2454–2463. [PubMed] [Google Scholar]
- 19.Mayer H, Schmidt G. Chemistry and biology of the enterobacterial common antigen (ECA) Curr Top Microbiol Immunol. 1979;85:99–153. doi: 10.1007/978-3-642-67322-1_3. [DOI] [PubMed] [Google Scholar]
- 20.Meier U, Mayer H. Genetic location of genes encoding enterobacterial common antigen. J Bacteriol. 1985;163:756–762. doi: 10.1128/jb.163.2.756-762.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meier-Dieter U, Barr K, Starman R, Hatch L, Rick P D. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. Molecular cloning of the rfe-rff gene cluster. J Biol Chem. 1992;267:746–753. [PubMed] [Google Scholar]
- 22.Meier-Dieter U, Starman R, Barr K, Mayer H, Rick P D. Biosynthesis of enterobacterial common antigen in Escherichia coli. Biochemical characterization of Tn10 insertion mutants defective in enterobacterial common antigen synthesis. J Biol Chem. 1990;265:13490–13497. [PubMed] [Google Scholar]
- 23.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 24.Morona R, Mavris M, Fallarino A, Manning P A. Characterization of the rfc region of Shigella flexneri. J Bacteriol. 1994;176:733–747. doi: 10.1128/jb.176.3.733-747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rick P D, Hubbard G L, Kitaoka M, Nagaki H, Kinoshita T, Dowd S, Simplaceanu V, Ho C. Characterization of the lipid-carrier involved in the synthesis of enterobacterial common antigen (ECA) and identification of a novel phosphoglyceride in a mutant of Salmonella typhimurium defective in ECA synthesis. Glycobiology. 1998;8:557–567. doi: 10.1093/glycob/8.6.557. [DOI] [PubMed] [Google Scholar]
- 26.Rick P D, Mayer H, Neumeyer B A, Wolski S, Bitter-Suermann D. Biosynthesis of enterobacterial common antigen. J Bacteriol. 1985;162:494–503. doi: 10.1128/jb.162.2.494-503.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rick P D, Silver R P. Enterobacterial common antigen and capsular polysaccharides. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 104–122. [Google Scholar]
- 28.Rick P D, Wolski S, Barr K, Ward S, Ramsay-Sharer L. Accumulation of a lipid-linked intermediate in enterobacterial common antigen synthesis in mutants lacking dTDP-glucose pyrophosphorylase. J Bacteriol. 1988;170:4008–4014. doi: 10.1128/jb.170.9.4008-4014.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rinno J, Golecki R, Mayer H. Localization of enterobacterial common antigen: immunogenic and nonimmunogenic enterobacterial antigen-containing Escherichia coli. J Bacteriol. 1980;141:814–821. doi: 10.1128/jb.141.2.814-821.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. p. 107. [Google Scholar]