Abstract
Clinical trials in patients with ER+ breast cancer with or without FGFR pathway somatic alterations have shown limited clinical benefit from treatment with FGFR tyrosine kinase inhibitors (TKIs) alone or in combination with endocrine therapy. This likely due to an inadequate predictive biomarker to select appropriate patients. In this study, we evaluated four anti-FGFR1 antibodies in breast cancer cell line and patient derived xenografts with FGFR1-amplification. We correlated D8E4 expression in 209 tumors from post-menopausal patients with stage I-III operable ER+ breast cancer with FGFR1 amplification status as determined by fluorescence in situ hybridization. FGFR1 amplification was identified in 10% of tumors (21/209), 80% of which exhibited membranous FGFR1 expression, however, only 50% of amplified cases showed strong, complete membranous staining (3+) based on established criteria to score HER2 by IHC. These findings suggest the combined evaluation of FGFR1 status by immunohistochemistry and FISH, may need to be incorporated in the selection of patients for trials with FGFR inhibitors.
Keywords: FGFR1, immunohistochemistry, biomarker, breast cancer
Introduction
Approximately 15% of patients with hormone receptor positive (HR+) breast cancer (BC) harbor amplification of the FGFR1 gene (8p11.23). FGFR1 amplification is associated with poor prognosis and resistance to endocrine therapy1 and CDK4/6 inhibition2. Early phase trials assessing the effect of multi-target and FGFR-selective tyrosine kinase inhibitors (TKIs) alone3–10 or in combination with endocrine therapy11,12 have shown limited clinical benefit in patients with BC harboring FGFR1 aberrations. This may be due to lack of an appropriately refined biomarker to predict patient benefit. Furthermore, data is inconsistent across studies with respect to the types of aberrations and the criteria used to establish FGFR1 amplification by in situ hybridization (FISH). However, evidence suggests that tumors with very high FGFR1 copy number (CN) and gene to centromere ratio are more likely to respond to FGFR inhibition9,13,14. High FGFR1 protein expression (defined as H-score >50) was also found to predict response9. However, in a recent study response was found to be unrelated to FGFR1 CN15. Furthermore, both FGFR1 amplified and FGFR1 non-amplified but overexpressing cells were sensitive to the addition of FGFR1 inhibition to antiestrogen administration and CDK4/6 inhibition in vitro16, suggesting that FGFR1 protein expression could also be considered in the prediction of response to FGFR1 inhibition.
Unlike HER2, the archetypical model for targeting amplified RTKs, where a direct correlation between HER2 amplification by FISH and increased expression of mRNA and protein has been demonstrated17, the literature regarding concordance among FGFR1 amplification and protein expression in breast cancer is less consistent. While FGFR1 gene and protein expression have been shown to have moderate to good concordance in breast cancer1,9,16,18–20, some studies show protein overexpression in non-amplified cases9,16,19,20, while others have found overexpression to be restricted to FGFR1 amplified cases1. Furthermore, methods and definitions for gene amplification and protein overexpression differ among these publications.
As reported, the analytical validity of a genomic biomarker must be first tested by evaluating the accuracy and consistency of the technique to detect the presence of protein21. In this study, we assessed analytical validation of four antibodies against FGFR1. We found a strong concordance between FGFR1 gene amplification by FISH and protein expression with D8E4 in a cohort of primary ER+ breast cancers and propose that the combined evaluation of FGFR1 status by immunohistochemistry and FISH, may be a better predictor of response to FGFR1 inhibition, than either marker alone.
Materials and Methods
Patients and Tumor samples.
DNA and RNA sequencing, FISH and IHC were performed on tissue from post-menopausal patients with stage I-III operable ER+/HER2− breast cancer enrolled in a clinical trial with the aromatase inhibitor letrozole administered for 10–21 days prior to surgery (NCT00651976). Tissue was obtained according to a protocol approved by the Vanderbilt-Ingram Cancer Center Institutional Review Board. TMAs were constructed utilizing two 1-mm punches of invasive carcinoma from patient’s surgical specimens post-letrozole administration.
Cell lines and patient derived xenografts (PDX).
MCF-7 (ATCC® HTB-22™) and MDA-MD-134-VI (ATCC® HTB-23™) human BC cells were obtained from the American Type Culture Collection (Manassas, VA, USA). Cell lines were authenticated by ATCC prior to purchase by the short tandem repeat (STR) method. All experiments were performed within 2 months after thawing early passage cells. Mycoplasma testing was conducted for each cell line before use. FGFR1 knock-out (MCF7FGFR1 KO) cells were created with siRNA as previously described22,23. TM00386 (Jackson Laboratory, Bar Harbor, ME, USA) PDX were established in ovariectomized SCID/beige mice and harvested.
Immunohistochemistry (IHC).
Formalin-fixed paraffin-embedded (FFPE) tissue sections were cut at 4 μm and deparaffinized. Antigen retrieval was performed with citrate buffer pH 6 or high pH buffer pH9. Endogenous peroxidases were blocked and protein block was applied. Sections were then incubated with the primary antibody overnight at 4°C. Visualization was achieved with the Envision (DAB) detection system (DAKO/Aligent, Glostrup, Denmark) and counterstained with hematoxylin. The following primary FGFR1 antibodies were used: D8E4 #9740 (Cell Signaling Technology, Danvers, MA, USA) at 1:200; Ab76464 (Abcam, Cambridge, UK) at 1:400; M2F12 sc-57132 (Santa Cruz Biotechnology, Dallas, TX, USA) at 1:400; and Ab10646 (Abcam) at 1:4000 (Figure 1A). Xenografts of MCF7FGFR1 KO and MDA-MB-134 cells were used as negative and positive controls, respectively. D8E4 was used to perform IHC on 3 TMAs containing 209 post-letrozole ER+ BC samples. In cases where very limited tumor (<200 cells) was present in the TMA, whole sections from the same tissue block used to construct the TMA were tested.
Figure 1: FGFR1 antibody validation.
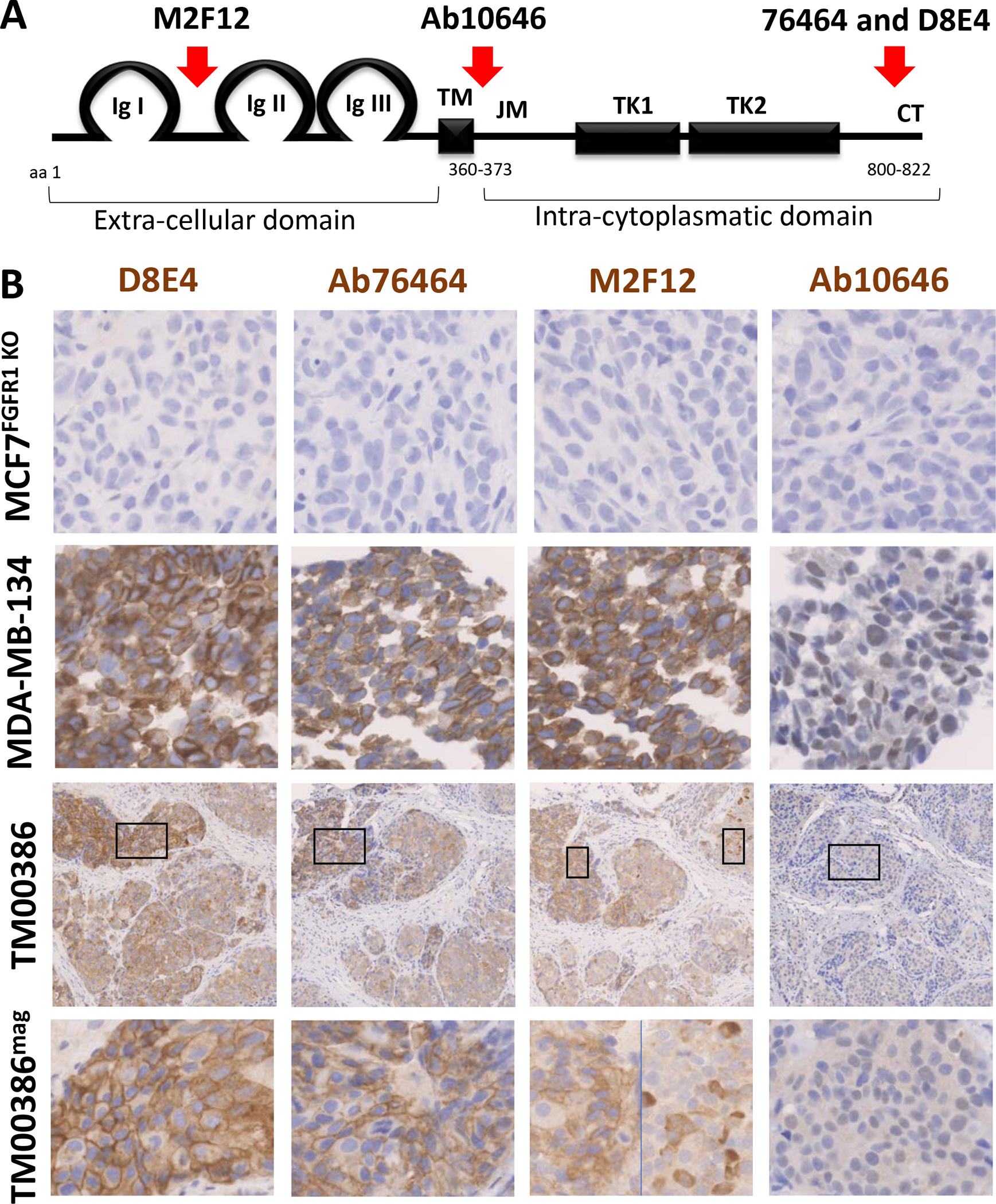
(A) FGFR1 protein structure, modified form the Atlas of Genetics and Cytogenetics in Oncology and Haematology30, showing binding sites for the four antibodies. Binding site data for each antibody were extracted from the manufacture’s data sheet. TM: transmembrane domain, JM: juxtamembrane domain, TK: tyrosine kinase subdomains, CT: C-terminal tail. (B) Representative FGFR1 IHC images of the validation for the four antibodies on negative control (FGFR1 KO cell line xenograft), positive control (FGFR1 amplified MDA-MB-134 cell line) and TM00386 (FGFR1 amplified pdx). Bottom images represent a higher magnification of the areas selected above.
Semiquantitative scoring was performed by a pathologist based on percentage of tumor cells demonstrating membranous and cytoplasmic staining (0–100%) and staining intensity (0–3+). H-scores were calculated using the following equation: [H-score = 3*(% of 3+ intensity cells) + 2*(% of 2+ intensity cells) + 1*(% of 1+ intensity cells)]24. Overexpression was defined as >10% intense complete membrane stain based on HER2 criteria.25
Subcellular fractionation assay:
Subcellular fractionation protocol was applied on frozen TM00386 chunks following the manufacturer’s instructions (Thermo Fisher Scientific, #87790). Derived proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked with 5% milk at room temperature for at least 1 h and then incubated overnight with primary antibodies at 4°C. Incubation with HRP-conjugated rabbit or mouse secondary antibodies was next performed for 1–2 h at room temperature. Protein bands were detected with an enhanced chemiluminescence substrate (Pierce ECL Western Blotting substrate, Thermo Fisher). The following primary antibodies were used: FGFR1 D8E4 #9740 (Cell Signaling Technology), FGFR1 Ab76464 (Abcam). FGFR1 Ab10646 (Abcam), α/β Tubulin #2148 (Cell Signaling Technology), AIF #5318 (Cell Signaling Technology), Lamin A/C #4777(Cell Signaling Technology).
Fluorescent in situ hybridization (FISH).
Deparaffinization, protease treatment and washes were performed as per standard protocols. Subsequently, 4-μm FFPE tissue sections were denatured in the presence of 10 μL of the probe for 6 min at 72°C and hybridized at 37°C overnight in StatSpin (Thermobrite, Abbott Molecular, Inc., Chicago, IL, USA) with SPEC FGFR1/CEN 8 Dual Color Probe catalog# ZTV-Z-2072 form ZytoVision (Bremerhaven, Germany). Post-hybridization saline-sodium citrate washes were performed at 72°C and the slides were then stained with DAPI before analysis. We performed FISH on 3 TMAs containing 209 post-letrozolesamples. In cases where very limited tumor (<100 cells) was present in the TMA, whole sections from the same tissue block used to make the TMA were tested. Tumor tissue was scanned at 20X magnification (BX60 Fluorescent microscope, Olympus, Tokyo, Japan) to identify appropriate regions for analysis. Images for cell counting were captured with a 100x oil immersion objective using Cytovision software (Leica, Wetzlar, Germany). At least 40 tumor cells per case were scored. Average gene/CEN ratio and gene copy number (CN) were calculated per case. Cases were considered to demonstrate amplification when FGFR1:CEP8 ratio ≥2.0 and FGFR1 CN >4 or FGFR1:CEP8 ratio was <2.0 and FGFR1 CN ≥6, based on 2018 HER2 guidelines25. High FGFR1 amplification (H-AMP) was defined as FGFR1:CEP8 ≥5 based on previous publications6,9. Low-level amplification was defined as FGFR1:CEP8 ratio between 2.0–4.0 or FGFR1:CEP8 ratio was <2.0 and FGFR1 CN ≥6.
Whole Exome Sequencing (WES).
DNA extraction was performed as previously described from frozen or FFPE tissue and matched normal/blood samples for 58 patients26 using the HiSeq 2000 v3 Sequencing-by-Synthesis method (Illumina, San Diego, CA, USA), and then analyzed using Real-Time Analysis (RTA) v.1.12.4.2. Exome sequence data processing was performed using established analytical pipelines at the Broad Institute (Seattle, WA, USA). Copy number ratios (CNR) were calculated for each captured target by dividing the tumor coverage by the median coverage obtained in a set of reference normal samples. Copy number alterations were detected by GISTIC analysis. Amplification was defined as log CNR≥2.
RNA sequencing.
H&E-stained sections were assessed for adequate tumor cellularity (>20%). RNA was extracted from 113 FFPE post letrozole treated samples using Promega Maxwell® (Madison, WI, USA) 16 LEV RNA FFPE Purification Kit and instrument, according to the manufacturer’s instructions. Total RNA was quantified, normalized, and used for library preparation. Amplification and sequencing were performed using HiSeq3000. Data were analyzed using the Broad Picard Pipeline (Seattle, WA, USA) which includes de-multiplexing and data aggregation. Transcripts were aligned, assembled and quantified using refseq transcripts as a guide with cufflinks, and normalized FPKMs generated using cuffnorm, following standard protocols.
Statistical Analysis.
Pearson correlation coefficient was assessed among tests. Student’s t-test or Fisher test was used for comparison of groups and pairs using Graphpad Prism 8 (San Diego, CA, USA). All tests were 2-tailed with p values <0.05 considered significant.
Results
FGFR1 antibody evaluation
In order to investigate FGFR1 expression intensity and localization in ER+ breast cancer, we first evaluated four commercially available antibodies targeting different epitopes of the FGFR1 protein (D8E4 and 76464: c-terminus, Ab106464: juxtamembrane domain, and M2F12: extracellular domain, Figure 1A). We used the ER+/FGFR1-amplified MDA-MB-134 cell line and an ER+/HER2−/FGFR1-amplified BC PDXs (TM00386) as positive controls (Supplementary Figure 1A and Supplementary Table 1). Xenograft of MCF7FGFR1 KO cells served as a negative control.
FGFR1 was expressed on most MDA-MB-134 cells (66–97%) with all antibodies. Strong membranous and mild to moderate cytoplasmic expression were observed with D8E4, Ab76464 and M2F12, while weak to moderate nuclear staining was seen for Ab10646 (Figure 1B). Absence of staining with all antibodies was observed in the negative control MCF7FGFR1 KO (Figure 1B). Specificity of the IHC technique was evidenced by the absence of protein observed on the FGFR1 knock-out cell lines with all antibodies, and the presence of protein matching the specific weight of FGFR1 on immunoblotting (Supplementary Figure 1B). Similar patterns, albeit less robust expression of each antibody, were seen in the PDX, with the greatest level of membranous expression observed with D8E4 (Figure 1B and Supplementary Table 1).
Given the finding of nuclear FGFR1 expression, also noted in previous publications22,23, was only observed with Ab10646, we performed subcellular fractionation of the PDX to further investigate FGFR1 cellular localization, which interestingly confirmed FGFR1 expression in the membranous, cytosolic and nuclear compartments with all antibodies (Supplementary Figure 1B). While nuclear FGFR1 has been linked to endocrine resistance, its activity was unaffected by FGFR-inhibition22. The biological relevance of cytoplasmic FGFR1, including isoforms or cleaved forms of FGFR1, is unknown. Hence, we report only membranous expression and selected D8E4 for further analysis of FGFR1 protein expression in patient tumor tissue.
FGFR1 overexpression and gene amplification in ER+ breast cancer
We subsequently performed immunohistochemistry for FGFR1 (D8E4) on 209 tumors from post-menopausal patients with stage I-III operable ER+ breast cancer treated with pre-surgical letrozole. In the absence of available tissue from patients treated with FGFR-inhibitors, we selected this cohort for study based on our prior work which found several patient’s endocrine therapy resistant tumors to harbor FGF alterations by NGS26. We identified a range of membranous expression in 9.6% of cases (20/209) which we scored utilizing clinical criteria for HER2 evaluation (3+, n = 9; 2+, n = 3; 1+, n =8) (Figure 2A). The percentage of cells staining in cases scored as 3+ ranged from 15 to 85%.
Figure 2: FGFR1 overexpression and gene amplification in ER+ breast cancer.
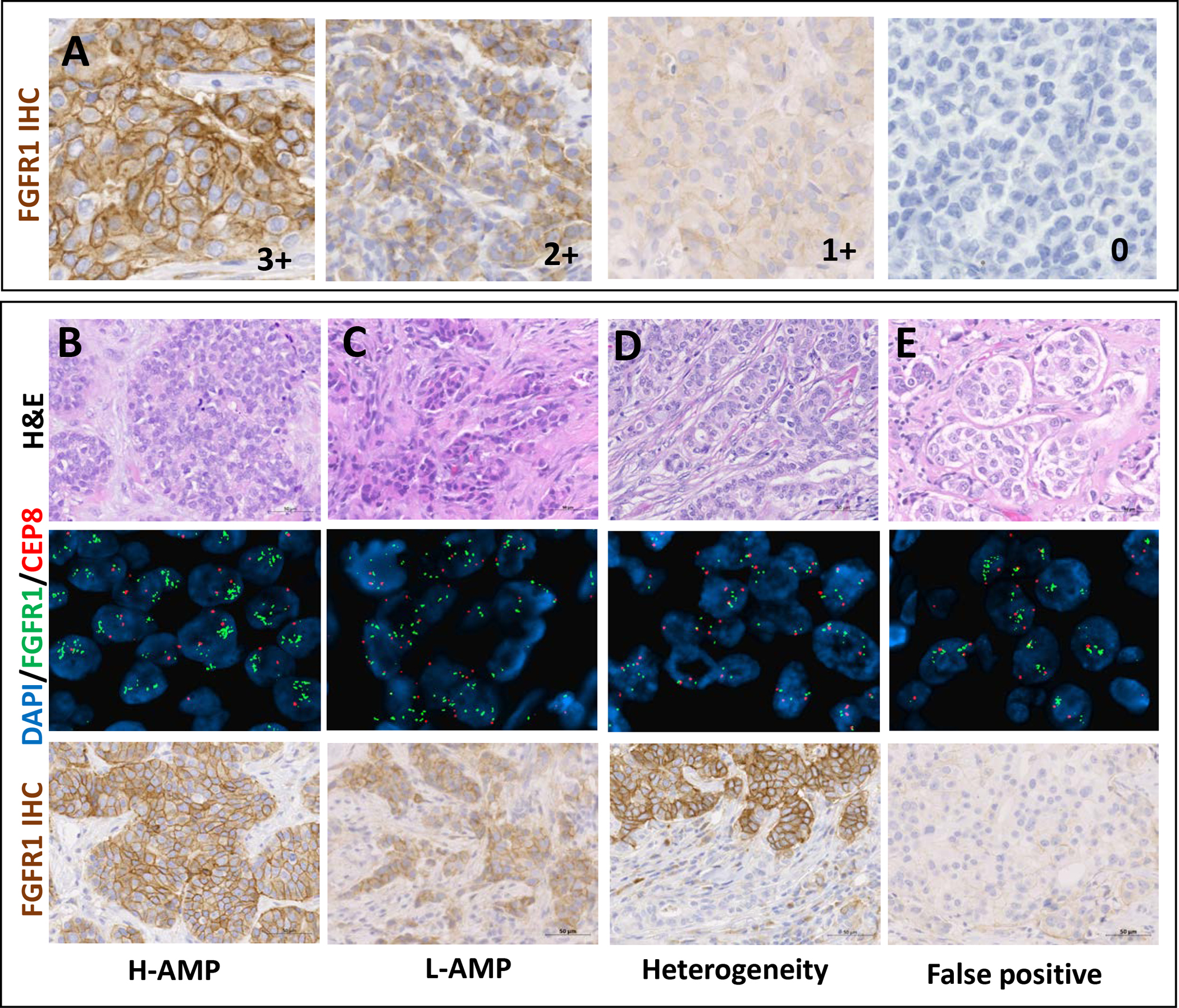
(A) Representative FGFR1 IHC images of membranous 3+, 2+, 1 and 0 (left to right) according to HER2 guidelines with D8E4 antibody. Scale bar 50μm. (B-E) Representative H&E (top), FGFR1 FISH (middle) and FGFR1 IHC (bottom) images of: (B) a highly amplified case with 3+ IHC expression, (C) a case showing low level FGFR1 amplification with 3+ protein expression, (D) a heterogeneously amplified case with corresponding zonal heterogenous protein expression (3+), and (E) a false positive for FISH FGFR1 amplification with discordant WES, RNA and protein expression (0+). Scale bar 50μm.
Next, we performed FGFR1 FISH on our cohort of 209 tumors. Twenty-one (10%) tumors were classified as amplified using previously proposed criteria (FGFR1:CEP8 ratio ≥2 and FGFR1 CN≥4 or FGFR1:CEP8 ratio <2 and FGFR1 CN≥6)25. We found high level FGFR1 amplification (H-AMP), defined as FGFR1:CEP8 ≥56,9, in 14 tumors (6.7%), representing 67% of FGFR1-amplified BC (Figure 2B). In 3 (14%) cases demonstrating low level FGFR1 amplification (Figure 2C), heterogeneous amplification was observed. In two cases the heterogeneity was localized and showed zonal FGFR1 overexpression by IHC (Figure 2D).
Membranous protein expression by IHC (H-score) and FGFR1 CN by FISH showed a strong correlation (r=0.76 p<0.0001, Figure 3A).
Figure 3: Only fifty percent of FGFR1 amplified tumors showed protein overexpression.
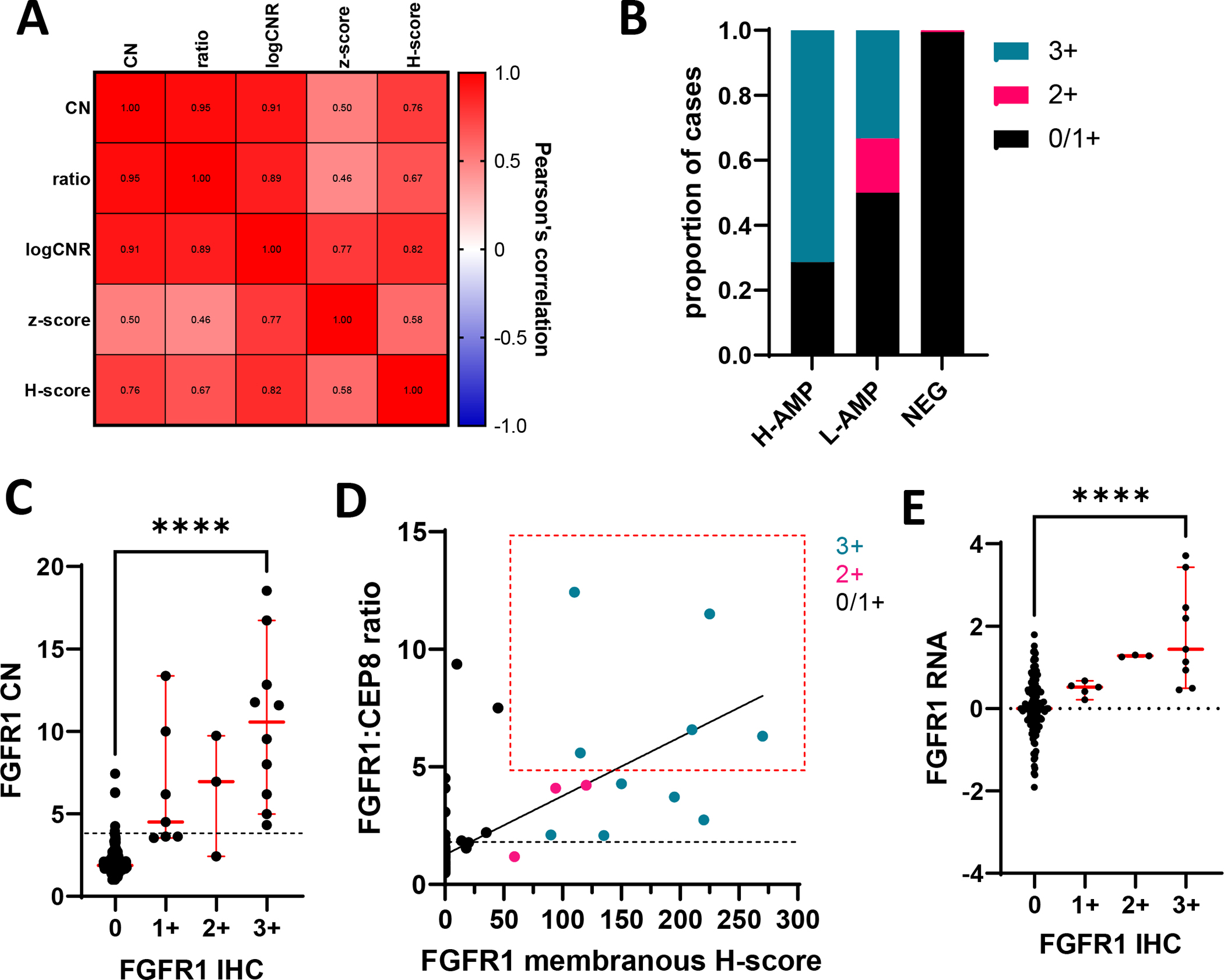
(A) Heatmap showing Pearson’s correlation between FGFR1 CN and ratio by FISH, logCNR by WES, ARN seq (z-score) and H-score by IHC. p<0.0001 for all correlations. (B) FGFR1 protein expression based on HER2 IHC criteria according to levels of amplification (C) FGFR1 CN according to protein expression graphed as values per case. Error bars represent mean and SD. p values represent the result of student t-test ****p<0.0001. Dotted line represents FGFR1 CN 4 (D) Correlation between FGFR1:CEP8 ratio and FGFR1 membranous H-score. Coloring represents protein expression based on HER2 IHC criteria. The red box highlights criteria that has identified patients that are more likely to respond to FGFR1 inhibition. Black dotted line represents FGFR1:CEP8 ratio 2 (E) RNA Z-score according to protein expression graphed as values per case. Error bars represent mean and SD. p values represent the result of student t-test ****p<0.0001.
We also performed RNA sequencing however adequate tumor was only available on 113 samples. Unlike that observed between protein and gene expression, correlation with mRNA was only moderate with both protein expression by IHC (H-score, r=0.58, p<0.0001) and FGFR1 CN by FISH (r=0.50 p<0.0001, Figure 3A).
FGFR1 over-expression and amplification correlate with pathologic characteristics
Clinicopathological characteristics of the tumors are shown in Table 2. Consistent with previous findings19,26, tumors exhibiting FGFR1 amplification and/or overexpression correlated with clinicopathologic features shown to correspond to resistance of anti-estrogen therapy, including higher combined histologic grade, proliferative rates, rates of lymph node metastasis at the time of primary diagnosis and high Ki67 index while on letrozole treatment according to the IMPACT study27,26.
Table 2:
Clinico-pathological characteristics of the 209 patients, comparing FGFR1 amplified and negative tumors.
| AMP | non-AMP | P value | Overexp | non-overexp | P value | |
|---|---|---|---|---|---|---|
| Number of cases | 20 | 188 | 10 | 199 | ||
| Mean age | 64 | 63 | 61 | 64 | ||
| Histological pattern (n=209) | ||||||
| No special type | 11 (52%) | 70 (37%) | 0.24 | 8 (80%) | 73 (37%) | 0.015 |
| Invasive lobular carcinoma | 3 (14%) | 32 (17%) | 1.00 | 0 | 35 (18%) | 1.00 |
| Tubular feature | 0 | 7 (4%) | 1.00 | 0 | 7 (3.5%) | 1.00 |
| Cribriform features | 0 | 6 (3%) | 1.00 | 0 | 6 (3%) | 1.00 |
| Lobular features | 2 (10%) | 40 (21%) | 0.38 | 2 (20%) | 40 (20%) | 1.00 |
| Mucin production | 1 (5%) | 7 (4%) | 0.54 | 1 (10%) | 7 (3.5%) | 0.33 |
| Invasive micropapillary features | 2 (10%) | 16 (9%) | 0.67 | 0 | 18 (9%) | 1.00 |
| Neuroendocrine features | 2 (10%) | 7 (4%) | 0.19 | 0 | 9 (4.5%) | 1.00 |
| Solid papillary pattern | 0 | 3 (2%) | 1.00 | 0 | 3 (1.5%) | 1.00 |
| Histological grade (n=208) | ||||||
| Low combined histologic grade | 1 (5%) | 67 (36%) | 0.003 | 1 (10%) | 67 (34%) | 0.29 |
| Intermediate combined histologic grade | 14 (67%) | 104 (56%) | 0.36 | 7 (70%) | 111 (56%) | 0.52 |
| High combined histologic grade | 6 (29%) | 16 (9%) | 0.013 | 2 (20%) | 20 (10%) | 0.29 |
| Proliferative rate (n=208) | ||||||
| Low proliferative rate | 11 (52%) | 161 (86%) | 0.017 | 5 (50%) | 166 (84%) | 0.018 |
| Intermediate proliferative rate | 8 (38%) | 19 (10%) | 0.001 | 4 (40%) | 23 (12%) | 0.028 |
| High proliferative rate | 2 (10%) | 7 (4%) | 0.19 | 1 (10%) | 8 (4%) | 0.37 |
| Mean tumor size (cm) | 2.22 | 2.05 | 0.60 | 2.22 | 2.06 | |
| Positive lymph nodes at dx (n=201) | 12 (57%) | 54 (30%) | 0.025 | 5 (55%) | 61 (32%) | 0.16 |
| Molecular surrogate (n=200) | ||||||
| Luminal A | 14 (67%) | 155 (87%) | 8 (80%) | 161 (85%) | ||
| Luminal B | 7 (33%) | 24 (13%) | 0.062 | 2 (20%) | 29 (15%) | 0.66 |
| Response to letrozole (n=177) | ||||||
| Sensitive | 7 (33%) | 100 (64%) | 0.009 | 4 (40%) | 103 (62%) | 0.12 |
| Intermediate | 4 (19%) | 31 (20%) | 1.00 | 3 (30%) | 32 (19%) | 0.46 |
| Resistant | 10 (48%) | 25 (16%) | 0.002 | 4 (30%) | 31 (19%) | 0.24 |
Tumors were designated as luminal A vs. B by a cut-off of Ki67 <14% as described 35. Response to letrozole was determined by Ki67 index in surgical sample, resected after 15 to 21 days of treatment with letrozole. The tertials were as follows: sensitive [Ki67 ln (natural log) ≤ 1.0; 0 to 2.7% Ki67+ tumor cells], intermediate (Ki67 ln = 1.1 to 1.9; 2.8 to 7.3%), or resistant (Ki67 ln ≥ 2.0; ≥7.4%) as previously published25. Statistically significant p values are shown in bold.
Amplification of other genes in the 8p11.23 loci may account for FGFR1 FISH false positive results
To evaluate the performance and sensitivity of the FGFR1 FISH assay, we compared the results with WES data previously obtained from tumor samples from 58 patients in our cohort26. We observed excellent correlation of FGFR1 CN (r=0.91 p<0.001) and FGFR1:CEP8 ratio (r=0.89 p<0.001) by FISH with log copy number ratio (CNR) and concordance with detection of amplification by WES (Figure 3A). However, one case demonstrating FGFR1 amplification by FISH was discordant with CNR, RNA expression and IHC results (Figure 2E). Interestingly, WES identified amplification of other genes in the 8p11.23 locus (ZNF703, WHSC1L1, LSM1, etc.) but not FGFR1 (log CNR of −0.074). Subsequently, we discovered that the FISH probe (catalog# ZTV-Z-2072, ZytoVision, Bremerhaven, Germany) localizes to the 8p11.23-p11.22 band but not specifically to the FGFR1 gene, which leads to a false positive result for FGFR1 amplification. This case was excluded from subsequent analyses.
Two other 2 discordant cases were considered amplified by FISH but classified as FGFR1 non-amplified by WES (CNR<2). Both cases demonstrated low level heterogenous FGFR1 amplification by FISH, and weak FGFR1 expression by IHC (Figure 2C). Neither case showed amplification of other genes at 8p11.23 by WES.
Only fifty percent of FGFR1 amplified tumors showed protein overexpression
Eighty percent of amplified cases showed some level of protein expression. When HER2 criteria25 are applied to define overexpression (>10% intense complete membranous staining) only 50% of amplified cases showed FGFR1 overexpression (Figure 3B). We observed no significant difference in staining intensity among low and high FGFR1 amplified tumors (data not shown). All cases classified as overexpressing (3+) also showed FGFR1 amplification (Figure 3B, C and D). Only overexpressing (3+) cases showed RNA expression above that observed for cases with null (0) protein expression (Figure 3E). Two tumors with low level amplification and one without gene amplification exhibited 2+ FGFR1 protein levels by IHC (Table 1). Forty-seven percent of non-amplified cases showed some protein expression, but 95% of protein expression in non-amplified cases was limited to the cytoplasmic compartment.
Table 1:
FGFR1 testing results for tumors demonstrating FGFR1 amplification and/or membranous expression.
| AVG FGFR1 copy # | AVG CEP8 copy # | Ratio FGFR1:CEP8 | FGFR1 FISH Results | AMP category | FGFR1 IHC |
|---|---|---|---|---|---|
| 18.53 | 1.49 | 12.43 | AMP | H-AMP | 3+ |
| 16.72 | 1.72 | 11.51 | AMP | H-AMP | 3+ |
| 12.83 | 2.52 | 6.58 | AMP | H-AMP | 3+ |
| 11.77 | 2.10 | 5.60 | AMP | H-AMP | 3+ |
| 11.58 | 1.83 | 6.31 | AMP | H-AMP | 3+ |
| 9.54 | 2.23 | 4.28 | AMP | L-AMP | 3+ |
| 6.19 | 1.86 | 3.72 | AMP | L-AMP | 3+ |
| 4.99 | 2.37 | 2.10 | AMP | L-AMP | 3+ |
| 4.32 | 2.33 | 2.08 | AMP | L-AMP | 3+ |
| 9.73 | 2.50 | 4.22 | AMP | L-AMP | 2+ |
| 6.95 | 1.70 | 4.09 | AMP | L-AMP | 2+ |
| 2.42 | 2.05 | 1.18 | NEG | NEG | 2+ |
| 13.36 | 1.43 | 9.37 | AMP | H-AMP | 1+ |
| 10.00 | 1.33 | 7.51 | AMP | H-AMP | 1+ |
| 8.00 | 2.92 | 2.74 | AMP | L-AMP | 1+ |
| 4.50 | 2.03 | 2.21 | AMP | L-AMP | 1+ |
| 6.19 | 3.52 | 1.78 | NEG | NEG | 1+ |
| 3.63 | 2.60 | 1.54 | NEG | NEG | 1+ |
| 3.62 | 2.25 | 1.85 | NEG | NEG | 1+ |
| 3.54 | 3.04 | 1.24 | NEG | NEG | 1+ |
| 7.43 | 2.00 | 4.52 | AMP | L-AMP | 0 |
| 6.30 | 1.61 | 4.10 | AMP | L-AMP | 0 |
| 6.27 | 2.85 | 4.07 | AMP | L-AMP | 0 |
| 4.25 | 1.63 | 3.08 | AMP | L-AMP | 0 |
Discussion
We found FGFR1 over-expression (3+) by IHC in 4.3% of cases (9/209), based on established criteria to score IHC for HER2. FGFR1 amplification by FISH was identified in 10% of tumors (21/209), 80% of which also exhibited membranous FGFR1 expression; however, only 50% showed strong, complete membranous staining (3+). While we report an excellent correlation between FGFR1 gene and protein expression, much can be learned from discordant cases. In clinical trials patient tumors with high level FGFR1 amplification responded better to FGFR inhibition9,14; however, not all amplified cases, or even all highly amplified cases, respond to treatment9,14. We observed that 40% of amplified cases showed low or no protein expression (1+/0, Figure 3B and Table 1), which suggests that some are not actually FGFR1-driven. In our study only cases defined as overexpressing FGFR1 based on established criteria to score HER2 by IHC, met both cut-offs predicting response to FGFR1 inhibition in clinical trials: FGFR1:CEP8 ≥59,13,14 and H-score >509 (Figure 3D). While cut offs tailored specifically to FGFR1 would need to be established, these findings suggest the combined evaluation of FGFR1 status by immunohistochemistry and FISH, may better predict response to FGFR1 inhibition, than either marker alone. Our results also suggest that several published trials evaluating clinical response to FGFR inhibitors which used only FISH to choose patients for inclusion may not have been selected adequately. This coupled with the lack of specificity of these therapeutic inhibitors has likely contributed to the limited clinical activity observed in these studies.
The amplicon containing FGFR1 is broad, with frequent co-amplification of other genes potentially contributing to carcinogenesis13. We describe an FGFR1 FISH false positive test result due to amplification of other genes in the 8p11.23 locus (ZNF703, WHSC1L1, LSM1, etc.) in the absence of FGFR1 amplification. This example highlights the importance of precise gene localization of FISH probes and well as the value of using two tests in combination, such as IHC and FISH, to more precisely identify patients with FGFR1-driven carcinomas. Although most tumors with 8p11.23 locus amplification included in the TCGA PanCancer Atlas data set28 demonstrated FGFR1 gene amplification, 19 cases with ZNF703 amplified ER+ BC are not associated with co-amplification of FGFR1. This means approximately 12% of tumors demonstrating FGFR1 amplification by FISH when using an amplicon-specific rather than FGFR1-specific probe would yield false positive results.
There are several limitations to our study. In the absence of available tissue from patients treated with FGF-inhibitors, we selected this cohort for study based on our prior work26 which found several patient’s tumors to harbor FGF alterations. FGFR1 inhibitors are currently only available to patients with locally advanced or metastatic breast cancer in the context of a clinical trial. Such patient would have already received at least one line of systemic treatment and long-term endocrine therapy. Therefore, we chose to study patients with early-stage breast cancer from a window study who received 10–20 days of presurgical letrozole as a surrogate for long term endocrine therapy. It is possible protein expression levels on metastatic samples after long-term drug treatment could differ from primary BC samples. However, obtaining metastatic tumor tissue from such patients is challenging as they may be in locations difficult to biopsy and/or contain scant tumor. Thus, even in the metastatic setting, primary diagnostic core biopsy tissue or surgical or samples must be used for biomarker evaluation. Availability of WES and RNA data on only a subset of patients is another limitation of the current study.
IHC is a low cost, reliable technique which is easily performed and incorporated into clinical trial protocols in combination with FGFR1 FISH. This work highlights the potential value of utilizing FISH and IHC for FGFR1 in combination as a biomarker to select patients for clinical trials with FGFR inhibitors. A clinical trial evaluating Fulvestrant, Palbociclib (CDK4/6 inhibitor) and Erdafitinib (JNJ-42756493, a pan-FGFR Tyrosine Kinase Inhibitor) in ER+/HER2−/FGFR-amplified metastatic breast cancer is currently underway at our institutions, which will allow testing of the predictive capacity of FGFR1 protein expression on relevant tissue. Since there is no current definition for FGFR1 overexpression as measured by IHC, a biologically relevant cut-off to define FGFR1 overexpression should be determined on standardized larger randomized controlled trial in conformance with REMARK guidelines29 and in conjunction with FGFR1 FISH results from the same specimen.
Supplementary Material
Supplementary Figure 1: FGFR1 antibody validation. (A) Representative FGFR1 FISH images of the MDA-MB-134 FGFR1 amplified cell lines used as positive control and the HR+HER2− FGFR1 amplified BC PDX model. (B) Subcellular protein fractionation assay was performed on TM00386PDX model, showing distribution of FGFR1 across cytosolic, membrane and soluble nuclear cellular compartments. Tubulin, AIF and Lamin represent markers of each cellular compartment, respectively.
Acknowledgements:
This study was funded by NIH Breast SPORE (Specialized Programs of Research Excellence) grant P50 CA098131. We thank the patients that participated on the clinical trial. We recognize Jennifer M. Giltnane, Katherine E. Hutchinson, Thomas P. Stricker and collaborators for the whole exome sequence data used in this manuscript that was acquired for their publication26. DNA and RNA sequencing were performed at VANTAGE Vanderbilt Technologies for Advanced Genomics core.
Footnotes
Conflicts of Interest and Source of Funding: This study was funded by NIH Breast SPORE (Specialized Programs of Research Excellence) grant P50 CA098131. Carlos L. Arteaga receives or has received research grants from Pfizer, Lilly, and Takeda; holds minor stock options in Provista; serves or has served in a scientific advisory role to Novartis, Merck, Lilly, Daiichi Sankyo, AstraZeneca, Immunomedics, Taiho Oncology, OrigiMed, Athenex, Arvinas, and Sanofi; and reports scientific advisory board remuneration from the Komen Foundation. The remaining authors declare no potential competing interests.
References
- 1.Turner N, Pearson A, Sharpe R, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer research. 2010;70(5):2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Formisano L, Lu Y, Servetto A, et al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nature communications. 2019;10(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soria J-C, DeBraud F, Bahleda R, et al. Phase I/IIa study evaluating the safety, efficacy, pharmacokinetics, and pharmacodynamics of lucitanib in advanced solid tumors. Annals of oncology. 2014;25(11):2244–2251. [DOI] [PubMed] [Google Scholar]
- 4.André F, Bachelot T, Campone M, et al. Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clinical cancer research. 2013:clincanres. 0190.2013. [DOI] [PubMed] [Google Scholar]
- 5.Nogova L, Sequist LV, Perez Garcia JM, et al. Evaluation of BGJ398, a fibroblast growth factor receptor 1–3 kinase inhibitor, in patients with advanced solid tumors harboring genetic alterations in fibroblast growth factor receptors: results of a global phase I, dose-escalation and dose-expansion study. Journal of Clinical Oncology. 2017;35(2):157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearson A, Smyth E, Babina IS, et al. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer discovery. 2016;6(8):838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smyth EC, Turner NC, Peckitt C, et al. Phase II multicenter proof of concept study of AZD4547 in FGFR amplified tumours. American Society of Clinical Oncology; 2015. [Google Scholar]
- 8.Chae YK, Hong F, Vaklavas C, et al. Phase II Study of AZD4547 in Patients With Tumors Harboring Aberrations in the FGFR Pathway: Results From the NCI-MATCH Trial (EAY131) Subprotocol W. Journal of Clinical Oncology. 2020;38(21):2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hui R, Pearson A, Cortes J, et al. Lucitanib for the treatment of HR+/HER2− metastatic breast cancer: Results from the multicohort phase II FINESSE study. Clinical Cancer Research. 2020;26(2):354–363. [DOI] [PubMed] [Google Scholar]
- 10.Voss MH, Hierro C, Heist RS, et al. A phase I, open-label, multicenter, dose-escalation study of the oral selective FGFR inhibitor Debio 1347 in patients with advanced solid tumors harboring FGFR gene alterations. Clinical Cancer Research. 2019;25(9):2699–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musolino A, Campone M, Neven P, et al. Phase II, randomized, placebo-controlled study of dovitinib in combination with fulvestrant in postmenopausal patients with HR+, HER2− breast cancer that had progressed during or after prior endocrine therapy. Breast Cancer Research. 2017;19(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seckl M, Badman PD, Liu X, et al. RADICAL trial: A phase Ib/IIa study to assess the safety and efficacy of AZD4547 in combination with either anastrozole or letrozole in ER positive breast cancer patients progressing on these aromatase inhibitors (AIs). American Society of Clinical Oncology; 2017. [Google Scholar]
- 13.Babina IS, Turner NCJNRC. Advances and challenges in targeting FGFR signalling in cancer. 2017;17(5):318. [DOI] [PubMed] [Google Scholar]
- 14.Pearson A, Smyth E, Babina IS, et al. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. 2016;6(8):838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coombes R, Badman P, Lozano-Kuehne J, et al. Results of the phase IIa RADICAL trial of the FGFR inhibitor AZD4547 in endocrine resistant breast cancer. Nature Communications. 2022;13(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mouron S, Manso L, Caleiras E, et al. FGFR1 amplification or overexpression and hormonal resistance in luminal breast cancer: rationale for a triple blockade of ER, CDK4/6, and FGFR1. Breast Cancer Research. 2021;23(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Press MF, Villalobos I, Santiago A, et al. Assessing the new American Society of Clinical Oncology/College of American Pathologists guidelines for HER2 testing by fluorescence in situ hybridization: Experience of an academic consultation practice. Archives of pathology & laboratory medicine. 2016;140(11):1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andre F, Job B, Dessen P, et al. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clinical cancer research. 2009;15(2):441–451. [DOI] [PubMed] [Google Scholar]
- 19.Tomiguchi M, Yamamoto Y, Yamamoto-Ibusuki M, et al. Fibroblast growth factor receptor-1 protein expression is associated with prognosis in estrogen receptor-positive/human epidermal growth factor receptor-2-negative primary breast cancer. 2016;107(4):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacquemier J, Adelaide J, Parc P, et al. Expression of the FGFR1 gene in human breast-carcinoma cells. 1994;59(3):373–378. [DOI] [PubMed] [Google Scholar]
- 21.Goossens N, Nakagawa S, Sun X, Hoshida YJTcr. Cancer biomarker discovery and validation. 2015;4(3):256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Formisano L, Stauffer KM, Young CD, et al. Association of FGFR1 with ERα maintains ligand-independent ER transcription and mediates resistance to estrogen deprivation in ER+ breast cancer. 2017;23(20):6138–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Servetto A, Kollipara R, Formisano L, et al. Nuclear FGFR1 regulates gene transcription and promotes antiestrogen resistance in ER+ breast cancer. Clinical Cancer Research. In review 2021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarty K Jr, Miller L, Cox E, Konrath J, McCarty K Sr. Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Archives of pathology & laboratory medicine. 1985;109(8):716. [PubMed] [Google Scholar]
- 25.Wolff AC, Hammond MEH, Allison KH, et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline focused update. Archives of pathology & laboratory medicine. 2018; [DOI] [PubMed] [Google Scholar]
- 26.Giltnane JM, Hutchinson KE, Stricker TP, et al. Genomic profiling of ER+ breast cancers after short-term estrogen suppression reveals alterations associated with endocrine resistance. Science translational medicine. 2017;9(402):eaai7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dowsett M, Smith IE, Ebbs SR, et al. Prognostic value of Ki67 expression after short-term presurgical endocrine therapy for primary breast cancer. Journal of the National Cancer Institute. 2007;99(2):167–170. [DOI] [PubMed] [Google Scholar]
- 28.Breast Invasive Carcinoma TCGA PanCancer Altas data set. https://www.cbioportal.org/
- 29.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast cancer research and treatment. 2006;100(2):229–235. [DOI] [PubMed] [Google Scholar]
- 30.Huret J-L. FGFR1 (fibroblast growth factor receptor 1). Atlas of Genetics and Cytogenetics in Oncology and Haematology. 2009; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: FGFR1 antibody validation. (A) Representative FGFR1 FISH images of the MDA-MB-134 FGFR1 amplified cell lines used as positive control and the HR+HER2− FGFR1 amplified BC PDX model. (B) Subcellular protein fractionation assay was performed on TM00386PDX model, showing distribution of FGFR1 across cytosolic, membrane and soluble nuclear cellular compartments. Tubulin, AIF and Lamin represent markers of each cellular compartment, respectively.


