Abstract
Besides Helicobacter pylori, a Gram-negative bacterium that may cause gastric disorders in humans, non-Helicobacter pylori helicobacters (NHPH) may also colonize the stomach of humans and animals. In pigs, H. suis can induce gastritis and may play a role in gastric ulcer disease, possibly in association with Fusobacterium gastrosuis. In the present study, gastric samples from 71 slaughtered pigs and 14 hunted free range wild boars were tested for the presence of DNA of F. gastrosuis and gastric Helicobacter species associated with pigs, dogs cats and humans, using species-specific PCR assays, followed by sequencing of the amplicon. These gastric samples were also histopathologically evaluated. Almost all the pigs presented gastritis (95.8%). Helicobacter spp. were detected in 78.9% and F. gastrosuis in 35.2% of the animals. H. suis was the most frequently identified Helicobacter species (57.7% of the animals), followed by a H. pylori-like species (50.7%) and less often H. salomonis and H. felis (each in 2.8% of the animals). H. suis was most often detected in the glandular (distal) part of the stomach (pars oesophagea 9.9%, oxyntic mucosa 35.2%, antral mucosa 40.8%), while the H. pylori-like species was mainly found in the non-glandular (proximal) part of the stomach (pars oesophagea 39.4%, oxyntic mucosa 14.1%, antral mucosa 4.2%). The great majority of wild boars were also affected with gastritis (71.4%) and Helicobacter spp. and F. gastrosuis were detected in 64.3% and 42.9% of the animals, respectively. H. bizzozeronii and H. salomonis were the most frequently detected Helicobacter species, while a H. pylori-like species and H. suis were only occasionally identified. These findings suggest that these microorganisms can colonize the stomach of both porcine species and may be associated with gastric pathology. This should, however, be confirmed through bacterial isolation. This is the first description of the presence of F. gastrosuis DNA in the stomach of wild boars and a H. pylori-like species in the pars oesophagea of the porcine stomach.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13567-022-01101-5.
Keywords: Sus scrofa, stomach, gastritis, gastric pathology, One Health
Introduction
Helicobacter species are Gram-negative, spiral-shaped motile bacteria that colonize the gastrointestinal tract of humans and a wide range of animals [1–4]. Studies have been done over the years to investigate their association with gastrointestinal and extra-gastrointestinal diseases [5]. In humans, Helicobacter pylori (H. pylori) is the most common gastric pathogen, with an estimated prevalence of 44.3% worldwide [6]. Other spiral-shaped non-Helicobacter pylori helicobacters (NHPH) have also been associated with gastric diseases in humans. These gastric helicobacters can be responsible for development of gastritis, gastroduodenal ulcers, gastric adenocarcinoma and mucosa-associated lymphoid tissue (MALT) lymphoma [2, 7, 8].
Zoonotic NHPH such as H. suis, H. felis, H. bizzozeronii, H. salomonis, and H. heilmannii, with an estimated prevalence ranging from 0.2 to 6% in symptomatic humans, naturally colonize the stomach of dogs, cats and pigs [6, 9].
Helicobacter suis is found mainly in the fundic and pyloric gland zone of the pig stomach [10]. This bacterium presents tropism for the gastric acid-producing parietal cells [11]. Its prevalence appears to be very low prior to weaning, but increases rapidly thereafter, being very high at slaughter age (77%) and in adults (>90%) [10–12]. H. suis infection causes gastritis, decreased daily weight gain, and may play a role in induction of gastric ulcers possibly in association with Fusobacterium gastrosuis (F. gastrosuis), further affecting animal production and welfare [13–15].
Fusobacteria are Gram-negative, non-sporing, strict anaerobic bacilli. They tend to form long filamentous rods, described as fusiform, spindle-shaped short with rounded ends [16]. There are 27 recognized species, including F. gastrosuis [17]. Fusobacteria can be involved in a wide range of mixed infections. Depending on the host species they can play different roles: in humans they can be associated with gingivitis, dental plaque formation, dental and periodontal abscesses and liver abscesses; while in pigs, they have been associated with lameness, facial skin necrosis and gastric ulceration [17].
Fusobacterium gastrosuis was primarily identified in the pars oesophagea of the pig stomach [17]. This anatomical region may be affected by ulceration, with a prevalence of up to 93% [18]. Gastric ulceration can lead to discomfort, decrease in daily weight gain, and sudden dead [17]. It is known that gastric ulceration is multifactorial and multiple agents can be involved such as H. suis and F. gastrosuis [17].
Several Helicobacter spp. have been described to have a zoonotic potential and therefore close contact between humans, domestic animals, and wild animals deserves more attention [19–23]. Although reservoirs of wild and domestic animals can be considered important sources of emerging infectious diseases, it is the human impact on ecological systems that determines the level of risk at the human/animal interface upon the occurrence of emerging zoonotic diseases [22, 23]. From an eco-epidemiological perspective, wild boars have an important role in spreading several pathogens [24–26].
The aim of this study is to determine the presence of gastric helicobacters and F. gastrosuis in a group of pigs and wild boars and describe the most relevant gastric histopathological alterations associated.
Materials and methods
Sample collection
Pig gastric tissues samples were collected from a total of 71 domestic pigs slaughtered at two slaughterhouses in the center of Portugal: one in Coimbra (n = 46, from 3 different herds) and another in Santarém (n = 25, from 2 different herds). All sampled animals were older than 6 months according to the slaughter animal registries.
Wild boar gastric tissue samples were collected from 14 hunted animals during two national campaigns, one in the north and other in the center of Portugal (Vila Real and Coimbra districts, respectively). Based on tooth evaluation [26], all sampled animals were older than 9 months.
From each animal, two samples were collected from the three different anatomical gastric regions: pars oesophagea, antrum and fundus, using a sterile disposable Kruuse® biopsy punch of 8 mm per site. After collection, from each region one sample was fixed in 10% phosphate-buffered formalin for histopathology and the other sample was stored at −20 °C for further DNA extraction and molecular analysis.
Gastric tissue sampling was performed within one hour and 4 h after slaughter, in pigs and wild boars respectively. The animals were not slaughtered, euthanized, or hunted to carry out this study, and the fresh gastric tissue specimens were purchased and obtained as sub-products derived from the normal meat inspection activity occurring in the slaughterhouses or during the national campaigns. Additionally, any of these actions were performed solely for research purposes and the researchers had no influence in the selection and execution of the slaughters nor in the national hunting campaign’s inspection procedures. Ethical approval was obtained from the i3S Animal Welfare and Ethics Review Body (ref. 2021-4).
Sample evaluation
Histological examination
After fixation, gastric tissues were routinely processed, paraffin-embedded and stained with hematoxylin and eosin (HE) for histopathology. The severity of gastritis was scored according to the human Updated Sydney System [14, 27] classification, with some modifications [13]. The same criteria were applied to the pars oesophagea, as described by Yamasaki et al., although this gastric region is not included within the human Updated Sydney System [28, 29]. Diffuse infiltration with inflammatory cells and the presence of lymphoid aggregates and lymphoid follicles in the mucosa and submucosa were also evaluated [13, 14, 27, 28] (Table 1).
Table 1.
Histological parameters used to establish a scoring system based on the Updated Sydney System and De Witte et al. and Gastritis overall scoring system [13]
| Parameter | Scoring | |
|---|---|---|
| Inflammatory cells (lymphocytes, plasm cells, neutrophils, eosinophils) | Absent | 0 |
| Mild infiltration (<5 cells per HPF) | 1 | |
| Moderate infiltration (≥5–20 cells per HPF) | 2 | |
| Severe infiltration (≥20 cells per HPF) | 3 | |
| Lymphoid follicles in the superficial and deep mucosa | No aggregates of lymphoid cells | 0 |
| Presence of one aggregate of lymphoid cells with minimal organization into a follicular structure | 1 | |
| Presence of at least one large follicle measuring at least 300 µm in diameter and/or more than one small aggregate of lymphoid cells per histological section | 2 | |
| At least two large lymphoid follicles measuring at least 300 µm in diameter per histological section and/or deformation of the mucosal caused by large lymphoid follicles | 3 | |
| Gastritis overall scoring system | ||
| Score | ||
| 0 | Normal | |
| 1 | Mild gastritis | |
| 2 | Moderate gastritis | |
| ≥ 3 | Severe gastritis | |
HPF: high power field, i.e., 400× total magnification.
A semi quantitative estimation of the overall inflammatory degree for each gastric area (pars oesophagea, oxyntic and antral mucosa) was calculated using a composite score which consisted of adding the partial values previously obtained [13, 27]. Additionally, each gastric section was also microscopically evaluated for the presence of hyperplasia, erosion, ulceration, and fibrosis.
Finally, the overall inflammatory score was correlated with the presence of H. pylori, H. suis, H. felis, H. salomonis, H. bizzozeronii, H. heilmannii, H. ailurogastricus and F. gastrosuis using Pearson Correlation coefficient.
DNA extraction, PCR conditions, and sequencing
DNA was extracted from the frozen tissue samples, using EXTRACTME® DNA tissue Kit (BLIRT, Poland) according to the manufacturer’s instructions.
All the samples were tested for the presence of H. pylori, H. suis, H. salomonis, H. bizzozeronii, H. felis, H. heilmannii, H. ailurogastricus and F. gastrosuis DNA through conventional PCR, according to previously described protocols (Additional file 1) [2, 18, 30, 31]. As can be seen in Additional file 1: Table S1, for detection of H. pylori DNA, two different PCR tests were used, one targeting the ureAB gene and the other one targeting the glmM gene.
Aliquots of each PCR product were electrophoresed on 1.5% agarose gel stained with Xpert Green Safe DNA gel stain (GRISP, Porto, Portugal) and examined for the presence of specific fragment under UV light. DNA fragment size was compared with the standard molecular weight, 100 bp DNA ladder (GRISP, Porto, Portugal) and the molecular weight of the positive controls (Additional file 1). For negative control, distilled water was used.
To exclude false-positive samples, the amplicons from each positive sample were sequenced. Bidirectional sequencing was performed using Sanger method at the genomics core facility of the Institute of Molecular Pathology and Immunology of the University of Porto. Sequence editing and multiple alignments were performed using MegaX Molecular Evolutionary Genetic Analysis version 10.1.8. The sequences obtained were subject to the basic local alignment search tool (BLAST) using the non-redundant nucleotide database.
Statistical analysis
Statistical analysis was performed using SAS®9.4 (SAS Institute Inc., 2019. Copyright® 2019 SAS Institute Inc., Cary, NC, USA). Correlations between different variables (erosion, ulceration, hyperplasia, fibrosis and presence of Helicobacter spp. and or F. gastrosuis) were investigated using Pearson correlation coefficient, r results of 0 meaning no correlation, r < 0.3 low degree of correlation, r ≥ 0.3 to r < 0.5 moderate degree of correlation, and r ≥ 0.5 to 1 high degree of correlation. Differences were considered statistically significant at p ≤ 0.05.
Differences between the different variables (erosion, ulceration, hyperplasia, fibrosis and presence of Helicobacter spp. and or F. gastrosuis) of the pars oesophagea, oxyntic mucosa, and antral mucosa were investigated using non-parametric Kruskal–Wallis, Chi-Square test. A p-value ≤ 0.05 was considered to be significant.
Results
Pigs
A total of 426 gastric samples were collected from 71 animals: 213 for histopathology and 213 for molecular evaluation.
Histopathology
Only 192 of the 213 available samples for histopathology were microscopically evaluated and classified, since 21 samples were in poor preservation conditions impairing histological examination. Thus 57 samples from pars oesophagea, 69 from the oxyntic mucosa, and 66 from the antral mucosa were analyzed.
Normal histological features were only observed in 3/71 (4.2%) pigs. Signs of inflammation were diagnosed in 95.8% of pigs. Indeed, 52 out of 57 samples (91.2%) of pars oesophagea, 60 out of 69 samples (86.9%) of oxyntic mucosa and all the samples of antral mucosa analyzed (100%) exhibited gastritis (Figure 1). Among samples with gastritis, hyperplasia was present in 28/52 (53.8%) of pars oesophagea and fibrosis in 49/60 (81.6%) of oxyntic and in 44/66 (66.6%) of antral mucosa (Table 2).
Figure 1.
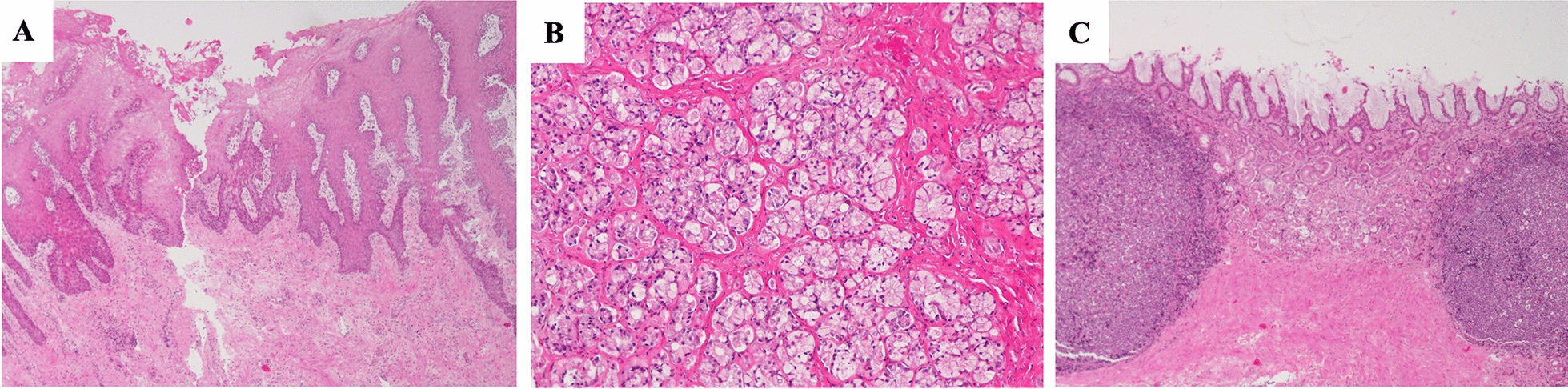
Main histopathological features observed in the different regions of pigs’ stomach. A Erosion and irregular and papillary hyperplasia of the lining epithelium of pars oesophagea. HE, ×40; B Mild inflammation of oxyntic mucosa. HE, ×100. C Severe inflammation and deformation of the antral mucosal caused by large lymphoid follicles and fibrosis. HE, ×40.
Table 2.
Results of the histopathology of the different gastric regions of pigs’ stomach
| Pars oesophagea | Oxyntic mucosa | Antral mucosa | ||||
|---|---|---|---|---|---|---|
| Number of samples per gastric zone | N = 57 | N = 69 | N = 66 | |||
| n/N | % | n/N | % | n/N | % | |
| Normal mucosa | 5/57 | 8.8 | 9/69 | 13.0 | 0/66 | 0.0 |
| Score 0 | ||||||
| Fibrosis | 0/5 | 0.0 | 6/9 | 66.7 | 0/0 | 0.0 |
| Erosion | 1/5 | 20.0 | 0/9 | 0.0 | 0/0 | 0.0 |
| Ulceration | 0/5 | 0.0 | 0/9 | 0.0 | 0/0 | 0.0 |
| Hyperplasia | 3/5 | 60.0 | 0/9 | 0.0 | 0/0 | 0.0 |
| Mild gastritis | 16/57 | 28.1 | 28/69 | 40.6 | 2/66 | 3.0 |
| Score 1 | ||||||
| Fibrosis | 2/16 | 12.5 | 23/28 | 82.1 | 1/2 | 50.0 |
| Erosion | 4/16 | 25.0 | 1/28 | 3.6 | 1/2 | 50.0 |
| Ulceration | 2/16 | 12.5 | 0/28 | 0.0 | 0/2 | 0.0 |
| Hyperplasia | 8/16 | 50.0 | 0/28 | 0.0 | 0/2 | 0.0 |
| Moderate gastritis | 9/57 | 15.8 | 13/69 | 18.8 | 3/66 | 4.5 |
| Score 2 | ||||||
| Fibrosis | 0/9 | 0.0 | 12/13 | 92.3 | 3/3 | 100.0 |
| Erosion | 0/9 | 0.0 | 2/13 | 15.4 | 0/3 | 0.0 |
| Ulceration | 0/9 | 0.0 | 0/13 | 0.0 | 0/3 | 0.0 |
| Hyperplasia | 6/9 | 66.7 | 0/13 | 0.0 | 0/3 | 0.0 |
| Severe gastritis | 27/57 | 47.4 | 19/69 | 27.5 | 61/66 | 92.4 |
| Score ≥ 3 | ||||||
| Fibrosis | 0/27 | 0.0 | 14/19 | 73.7 | 40/61 | 65.6 |
| Erosion | 5/27 | 18.5 | 1/19 | 5.3 | 12/61 | 19.7 |
| Ulceration | 1/27 | 3.7 | 0/19 | 0.0 | 21/61 | 34.4 |
| Hyperplasia | 14/27 | 51.9 | 0/19 | 0 | 2/61 | 3.3 |
Remarkably, almost all samples of antral mucosa presented severe inflammation (61/66 or 92.4%) and amongst these, fibrosis was also a common finding (40/61 or 65.6%) (Table 2). When comparing the three different gastric zones, there were statistically significant differences for the presence of erosion, ulceration, hyperplasia, and fibrosis (p ≤ 0.05) (Additional file 2).
Presence of Helicobacter spp. and F. gastrosuis DNA through PCR analysis
In ten pigs, no DNA amplification was achieved, neither for Helicobacter spp. nor for F. gastrosuis.
Out of all animals (n = 71), Helicobacter spp. were detected in 56 stomachs (78.9%) with H. suis being the most frequently identified species (41/71 or 57.7%), followed by a H. pylori-like species (36/71 or 50.7%) and less often H. salomonis and H. felis (2/71 or 2.8% each).
Regarding the H. pylori PCR results, 36/71 samples amplified H. pylori DNA with homologies ranging from 96.3 to 100% (Additional file 3) using the ureAB gene primers but no amplification was achieved when using the glmM gene primers. Therefore, these cases were reclassified as H. pylori-like.
Positive results for F. gastrosuis were obtained in 25 out of the 71 pigs (35.2%), although F. gastrosuis was mainly found in association with helicobacters (20/71 or 28.2%) rather than alone (5/71 or 7.0%). Indeed, in 27/71 of the positive samples (38.0%), only one species was identified, while mixed infections with two or more distinct bacteria were detected in 34/71 (47.9%) (Table 3). The most frequent bacterial combination was H. pylori-like + H. suis (12/71 or 16.9%) and H. pylori-like + H. suis + F. gastrosuis (10/71 or 14.1%) (Table 3).
Table 3.
Helicobacter spp. and F. gastrosuis DNA PCR positive samples per pigs’ stomach
| Specific PCR-positive results | n/N | % |
|---|---|---|
| Overall identification | ||
| Helicobacter spp. and/or F. gastrosuis | 61/71 | 85.9 |
| Helicobacter spp. | 56/71 | 78.9 |
| H. suis | 41/71 | 57.7 |
| H. pylori-like | 36/71 | 50.7 |
| H. felis | 2/71 | 2.8 |
| H. salomonis | 2/71 | 2.8 |
| F. gastrosuis | 25/71 | 35.3 |
| Single bacteria | ||
| H. suis | 14/71 | 19.7 |
| H. pylori-like | 7/71 | 9.9 |
| F. gastrosuis | 5/71 | 7.0 |
| H. felis | 1/71 | 1.4 |
| Multiple bacteria | ||
| H. pylori-like + H. suis | 12/71 | 16.9 |
| H. pylori-like + F. gastrosuis | 5/71 | 7.0 |
| H. suis + F. gastrosuis | 4/71 | 5.6 |
| H. pylori-like + H. salomonis | 1/71 | 1.4 |
| H. suis + H. felis | 1/71 | 1.4 |
| H. pylori-like + H. suis + H. salomonis + F. gastrosuis | 1/71 | 1.4 |
| H. pylori-like + H. suis + F. gastrosuis | 10/71 | 14.1 |
Bacteria were differently distributed throughout the porcine gastric compartments (Table 4). H. pylori-like DNA was mostly detected in the pars oesophagea (28/71, 39.4%), while H. suis was most frequently identified in the oxyntic (25/71, 35.2%) and antral mucosa (29/71, 40.8%). There was also a statistically highly significant moderate degree of correlation regarding the presence of H. pylori-like + H. suis DNA in the pars oesophagea, as well as a statistically significant low degree of correlation between the presence of H. pylori-like and H. suis in the pars oesophagea (r = 0.29 p ≤ 0.05) and oxyntic mucosa (r = 0.25, p ≤ 0.05) (Table 4).
Table 4.
Helicobacter spp. and F. gastrosuis detected through PCR in the different regions of pigs’ stomach
| Pars oesophagea | r | p | Oxyntic mucosa | r | p | Antral mucosa | r | p | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of samples | N = 71 | N = 71 | N = 71 | |||||||||
| n/N | % | n/N | % | n/N | % | |||||||
| Single bacteria | ||||||||||||
| H. pylori-like | 28/71 | 39.4 | 10/71 | 14.1 | 3/71 | 4.2 | ||||||
| H. suis | 7/71 | 9.9 | 25/71 | 35.2 | 29/71 | 40.8 | ||||||
| H. felis | 0/71 | 0.0 | 2/71 | 2.8 | 0/71 | 0.0 | ||||||
| H. salomonis | 0/71 | 0.0 | 0/71 | 0.0 | 2/71 | 2.8 | ||||||
| F. gastrosuis | 12/71 | 16.9 | 4/71 | 5.6 | 10/71 | 7.6 | ||||||
| Multiple bacteria | ||||||||||||
| F. gastrosuis + H. pylori-like | 8/71 | 11.3 | 0.29 | 0.03 | 1/71 | 1.4 | 0.25 | 0.04 | 1/71 | 1.4 | 0.11 | 0.38 |
| H. pylori-like + H. suis | 6/71 | 8.5 | 0.36 | < 0.01 | 1/71 | 1.4 | −0.07 | 0.56 | 2/71 | 2.8 | −0.05 | 0.71 |
| F. gastrosuis + H. suis | 0/71 | 0.0 | −0.04 | 0.79 | 2/71 | 2.8 | 0.20 | 0.09 | 4/71 | 5.6 | −0.03 | 0.79 |
| H. suis + H. felis | 0/71 | 0.0 | – | – | 1/71 | 1.4 | 0.05 | 0.69 | 0/71 | 0.0 | – | – |
| F. gastrosuis + H. pylori-like + H. suis | 1/71 | 1.4 | – | – | 1/71 | 1.4 | – | – | 0/71 | 0.0 | – | – |
r: Pearson correlation coefficient.
p: p-value ≤ 0.05 was considered to be significant.
Regarding the correlation between the presence of both bacteria and the respective histological findings, a statistically highly significant moderate correlation was observed between the presence of H. felis and erosion in the oxyntic mucosa (r = 0.33, p ≤ 0.01); a statistically significant low degree of correlation between the presence of H. pylori-like and erosion (r = 0.23, p ≤ 0.05) in the antral mucosa as well as a statistically significant correlation between the presence of F. gastrosuis and ulceration of the antral mucosa (r = 0.31, p ≤ 0.05) (Table 5).
Table 5.
Correlation between the PCR-positive results obtained in the different stomach regions and the gastric histological findings in pigs
| Erosion | Ulceration | Hyperplasia | Fibrosis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n/N | 10/57 | r | p | 3/57 | r | p | 31/57 | r | p | 2/57 | r | p | |
| Pars oesophagea | |||||||||||||
| H. pylori-like | 28/57 | 1/10 | −0.11 | 0.40 | 1/3 | −0.03 | 0.81 | 14/31 | 0.04 | 0.64 | 2/2 | 0.20 | 0.13 |
| H. suis | 7/57 | 1/10 | −0.03 | 0.83 | 0 | −0.08 | 0.57 | 4/31 | 0.04 | 0.77 | 0 | −0.07 | 0.63 |
| H. felis | 0/57 | 0 | – | – | 0 | – | – | 0 | – | – | 0 | – | – |
| F. gastrosuis | 12/57 | 2/10 | −0.04 | 0.74 | 0 | −0.12 | 0.39 | 7/31 | −0.01 | 0.93 | 0 | –0.10 | 0.46 |
| n/N | 4/69 | r | p | 0 | r | p | 0 | r | p | 4 | r | p | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Oxyntic mucosa | |||||||||||||
| H. pylori-like | 10/69 | 0 | −0.10 | 0.40 | 0 | – | – | 0 | – | – | 0 | −0.21 | 0.08 |
| H. suis | 25/69 | 2/4 | 0.07 | 0.56 | 0 | – | – | 0 | – | – | 2/4 | 0.0 | 0.16 |
| H. felis | 2/69 | 1/4 | 0.33 | < 0.01 | 0 | – | – | 0 | – | – | 1/4 | 0.15 | 0.19 |
| F. gastrosuis | 4/69 | 1/4 | 0.20 | 0.09 | 0 | – | – | 0 | – | – | 1/4 | 0.08 | 0.51 |
| n/N | 13/66 | r | p | 21/66 | r | p | 2/66 | r | p | 44/66 | r | p | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antral mucosa | |||||||||||||
| H. suis | 29/66 | 5/13 | −0.02 | 0.85 | 10/21 | 0.13 | 0.31 | 1/2 | −0.04 | 0.78 | 16/44 | 0.02 | 0.86 |
| H. pylori-like | 3/66 | 2/13 | 0.23 | 0.02 | 0 | −0.01 | 0.31 | 0 | −0.04 | 0.77 | 1/44 | −0.02 | 0.86 |
| H. felis | 0/66 | 0 | – | – | 0 | – | – | 0 | – | – | 0 | – | – |
| F. gastrosuis | 10/66 | 2/13 | −0.11 | 0.36 | 4/21 | 0.31 | 0.011 | 1/2 | 0.08 | 0.50 | 3/44 | −0.20 | 0.11 |
r: Pearson correlation coefficient.
p: p-value ≤ 0.05 was considered to be significant.
Wild boars
A total of 84 samples were collected from 14 animals: 42 samples for histopathological examination and 42 samples for molecular evaluation.
Histopathology
Only 30 of the 42 available samples for histopathology were microscopically evaluated and classified, since 12 were in poor preservation conditions impairing histological examination (10 of the pars oesophagea, 10 of the oxyntic mucosa and 10 of the antral mucosa).
Normal histological features were not identified in wild boars. Microscopically, gastritis was diagnosed in 8 out of 10 samples of pars oesophagea (80.0%), 4 out of 10 samples of oxyntic mucosa (40.0%), and 9 out of 10 samples of antral mucosa (90.0%). In most representative cases of pars oesophagea the inflammation was mild (4/10 or 40.0%), whereas in the antral mucosa the inflammation was severe (7/10 or 70.0%) (Figure 2).
Figure 2.
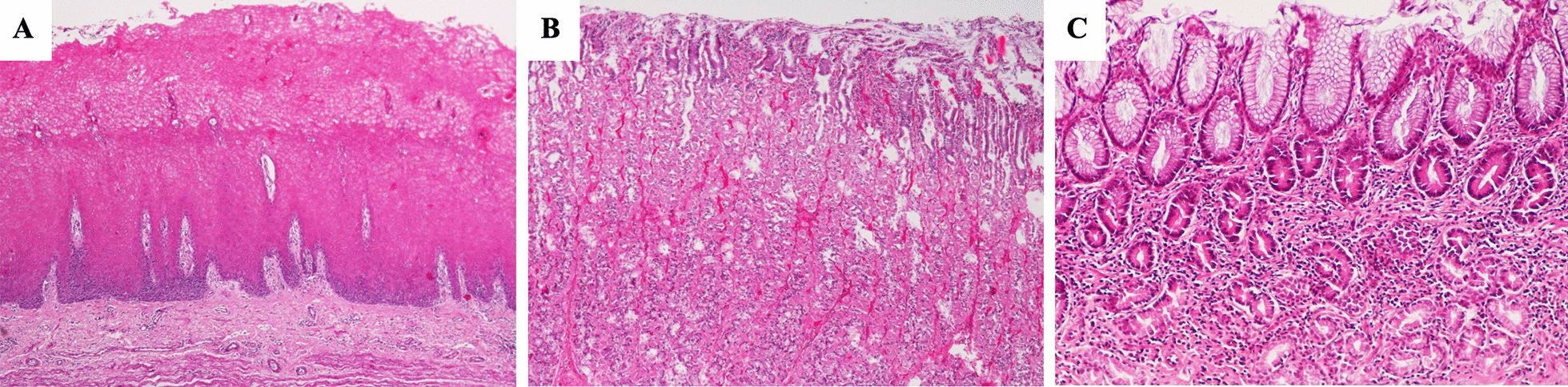
Main histopathological features observed in the different regions of wild boars’ stomach. A Hyperkeratosis and hyperplasia of the lining epithelium of pars oesophagea. HE, ×40; B Mild inflammation of oxyntic mucosa. HE, ×40 and C Moderate and diffuse inflammatory infiltration of antral mucosa. HE, ×100.
Regarding the cases diagnosed with gastritis of pars oesophagea (8/10), 3 out of 8 were associated with erosion (37.5%) and 2 out of 8 with hyperplasia (25.0%) (Table 6).
Table 6.
Results of the histopathology of the different gastric regions of wild boars’ stomach
| Pars oesophagea | Oxyntic mucosa | Antral mucosa | ||||
|---|---|---|---|---|---|---|
| Number of samples per gastric zone | N = 10 | N = 10 | N = 10 | |||
| n/N | % | n/N | % | n/N | % | |
| Normal mucosa | 2/10 | 20.0 | 6/10 | 60.0 | 1/10 | 10.0 |
| Score 0 | ||||||
| Fibrosis | 0/2 | 0.0 | 0/6 | 0.0 | 0/1 | 0.0 |
| Erosion | 1/2 | 50.0 | 0/6 | 0.0 | 0/1 | 0.0 |
| Ulceration | 0/2 | 0.0 | 0/6 | 0.0 | 0/1 | 0.0 |
| Hyperplasia | 0/2 | 0.0 | 0/6 | 0.0 | 0/1 | 0.0 |
| Mild gastritis | 4/10 | 40.0 | 1/10 | 10.0 | 1/10 | 10.0 |
| Score 1 | ||||||
| Fibrosis | 0/4 | 0.0 | 0/1 | 0.0 | 0/1 | 0.0 |
| Erosion | 2/4 | 50.0 | 0/1 | 0.0 | 0/1 | 0.0 |
| Ulceration | 0/4 | 0.0 | 0/1 | 0.0 | 0/1 | 0.0 |
| Hyperplasia | 1/4 | 25.0 | 0/1 | 0.0 | 0/1 | 0.0 |
| Moderate gastritis | 2/10 | 20.0 | 2/10 | 20.0 | 1/10 | 10.0 |
| Score 2 | ||||||
| Fibrosis | 0/2 | 0.0 | 0/2 | 0.0 | 0/1 | 0.0 |
| Erosion | 0/2 | 0.0 | 0/2 | 0.0 | 0/1 | 0.0 |
| Ulceration | 0/2 | 0.0 | 0/2 | 0.0 | 0/1 | 0.0 |
| Hyperplasia | 0/2 | 0.0 | 1/2 | 50.0 | 0/1 | 0.0 |
| Severe gastritis | 2/10 | 20.0 | 1/10 | 10.0 | 7/10 | 70.0 |
| Score ≥ 3 | ||||||
| Fibrosis | 0/2 | 0.0 | 0/1 | 0.0 | 0/7 | 0.0 |
| Erosion | 1/2 | 25.0 | 0/1 | 0.0 | 0/7 | 0.0 |
| Ulceration | 0/2 | 0.0 | 0/1 | 0.0 | 0/7 | 0.0 |
| Hyperplasia | 1/2 | 25.0 | 0/1 | 0.0 | 0/7 | 0.0 |
Helicobacter spp. and F. gastrosuis identification through PCR analysis
In 3 out of 14 wild boars, no DNA amplification was achieved, neither for Helicobacter spp. nor for F. gastrosuis. Out of all animals, Helicobacter spp. were detected in 9 sampled stomachs (78.6%) with both H. bizzozeronii and H. salomonis being the most frequently identified species (4/11 or 36.4%), while a H. pylori-like species (2/11 or 18.2%) and H. suis (1/11 or 9.1%) were less often detected (Table 7).
Table 7.
Helicobacter spp. and F. gastrosuis DNA PCR positive samples per wild boars’ stomach
| Specific PCR-positive results | n/N | % |
|---|---|---|
| Overall identification | ||
| Helicobacter spp. + F. gastrosuis | 11/14 | 78.6 |
| Helicobacter spp. | 9/14 | 64.3 |
| H. bizzozeronii | 4/14 | 28.6 |
| H. salomonis | 4/14 | 28.6 |
| H. pylori-like | 2/14 | 14.3 |
| H. suis | 1/14 | 7.1 |
| F. gastrosuis | 6/14 | 42.9 |
| Single bacteria | ||
| F. gastrosuis | 2/14 | 14.3 |
| H. salomonis | 2/14 | 14.3 |
| H. pylori-like | 0/14 | 0.0 |
| H. suis | 1/14 | 7.1 |
| H. bizzozeronii | 1/14 | 7.1 |
| Multiple bacteria | ||
| H. bizzozeronii + F. gastrosuis | 2/14 | 14.3 |
| H. pylori + H. salomonis | 1/14 | 7.1 |
| H. pylori-like + F. gastrosuis | 1/14 | 7.1 |
| H. bizzozeronii + H. salomonis + F. gastrosuis | 1/14 | 7.1 |
Regarding the H. pylori PCR results, 2/14 samples amplified H. pylori DNA with homologies of 100% (Additional file 2: Table S2) using the ureAB gene primers but no amplification was achieved when using the glmM gene primers. Therefore, these cases were reclassified as H. pylori-like.
Positive results for F. gastrosuis were obtained in 6 out of the 14 wild boars (42.9%), although it was mainly found in association with helicobacters (4/11 or 36.4%) rather than alone (2/11 or 18.2%). Indeed, in 6/14 of the positive sampled stomachs (42.9.0%), only one species was identified while mixed infections with two or more distinct bacteria were detected in 5/14 (35.7%) (Table 7). The most frequent bacterial combination was H. salomonis + F. gastrosuis (2/14 or 14.3%) (Table 7).
Bacteria were differently distributed throughout the different regions of wild boars’ stomach (Table 8).
Table 8.
Specific Helicobacter spp. and F. gastrosuis detected through PCR in the different regions of wild boars’ stomach
| Pars oesophagea | r | p | Oxyntic mucosa | r | p | Antral mucosa | r | p | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of samples | N = 14 | N = 14 | N = 14 | |||||||||
| n/N | % | n/N | % | n/N | % | |||||||
| Single bacteria | ||||||||||||
| H. pylori-like | 0/14 | 0.0 | 1/14 | 7.1 | 0/14 | 0.0 | ||||||
| H. suis | 0/14 | 0.0 | 1/14 | 7.1 | 0/14 | 0.0 | ||||||
| H. bizzozeronii | 1/14 | 7.1 | 1/14 | 7.1 | 0/14 | 0.0 | ||||||
| H. salomonis | 2/14 | 14.3 | 0/14 | 0.0 | 3/14 | 21.4 | ||||||
| F. gastrosuis | 2/14 | 14.3 | 4/14 | 28.6 | 5/14 | 28.5 | ||||||
| Multiple bacteria | ||||||||||||
| H. pylori-like + F. gastrosuis | 0/14 | 0.0 | – | – | 1/14 | 7.1 | – | – | 0/14 | 0.0 | – | – |
| H. bizzozeronii + F. gastrosuis | 1/14 | 7.1 | 0.22 | 0.55 | 1/14 | 7.1 | – | – | 0/14 | 0.0 | – | – |
| H. bizzozeronii + H. salomonis + F. gastrosuis | 0 | 0 | – | – | 0 | 0.0 | – | – | 1/14 | 7.1 | 0.67 | 0.04 |
r: Pearson correlation coefficient.
p: p-value ≤ 0.05 was considered to be significant.
Regarding the identification of DNA of only one bacteria per sample site, the pars oesophagea samples presented 2/14 (14.3%) DNA for F. gastrosuis, 2/14 (14.3%) DNA for H. salomonis and 1/14 (7.1%) DNA for H. bizzozeronii. The oxyntic mucosa had 2/14 (13.3%) of the samples with F. gastrosuis DNA, 1/14 (7.1%) with H. pylori-like DNA, 1/14 (7.1%) with H. suis DNA and 1/14 (7.1%) with H. bizzozeronii DNA. The antral mucosa had 4/14 (28.5%) of the samples with F. gastrosuis DNA followed by H. salomonis DNA in 3/14 (21.4%) of the samples (Table 8).
Regarding the antral mucosa, there was a statistically significant high degree of correlation between the presence of H. salomonis and H. bizzozeronii (r = 0.67, p ≤ 0.05) (Table 8).
Concerning the correlation between the presence of Helicobacter spp., F. gastrosuis DNA and different histological findings, there was a highly statistically significant correlation between the presence of H. bizzozeronii and hyperplasia in the pars oesophagea (Table 9).
Table 9.
Correlation between the PCR-positive results obtained in the different stomach regions and the gastric histological findings in wild boars
| Erosion | Ulceration | Hyperplasia | Fibrosis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n/N | 4/10 | r | p | 0/10 | r | p | 2/10 | r | p | 0/10 | r | p | |
| Pars oesophagea | |||||||||||||
| H. pylori-like | 0/10 | 0 | – | – | 0 | – | – | 0 | – | – | 0 | – | – |
| H. suis | 0/10 | 0 | – | – | 0 | – | – | 0 | – | – | 0 | – | – |
| H. bizzozeronii | 1/10 | 0 | – | – | 0 | – | – | 2/2 | 0.94 | < 0.01 | 0 | – | – |
| H. salomonis | 0/10 | 0 | – | – | 0 | – | – | 0 | – | – | 0 | – | – |
| F. gastrosuis | 4/10 | 2/4 | 0.36 | 0.31 | 0 | – | – | 1/2 | 0.03 | 0.93 | 0 | – | – |
| n/N | 0/10 | r | p | 0/10 | r | p | 1/10 | r | p | 0 | r | p | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Oxyntic mucosa | |||||||||||||
| H. pylori-like | 2/10 | 0 | – | – | 0 | – | – | 0 | – | – | 0 | – | – |
| H. suis | 1/10 | 0 | – | – | 0 | – | – | 0 | – | – | 0 | – | – |
| H. bizzozeronii | 0/10 | 0 | – | – | 0 | – | – | 0 | – | – | 0 | – | – |
| H. salomonis | 0/10 | 0 | – | – | 0 | – | – | 0 | – | – | 0 | – | – |
| F. gastrosuis | 5/10 | 0 | – | – | 0 | – | – | 0 | – | – | 0 | – | – |
| n/N | 0/10 | r | p | 0/10 | r | p | 0/10 | r | p | 0 | r | p | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antral mucosa | |||||||||||||
| H. pylori-like | 0/10 | 0 | – | – | 0 | – | – | 0 | – | – | 0 | – | – |
| H. suis | 0/10 | 0 | – | – | 0 | – | – | 0 | – | – | 0 | – | – |
| H. bizzozeronii | 2/10 | 0 | – | – | 0 | – | – | 0 | – | – | – | – | |
| H. salomonis | 5/10 | 0 | – | – | 0 | – | – | 0 | – | – | 0 | – | – |
| F. gastrosuis | 6/10 | 0 | – | – | 0 | – | – | 0 | – | – | 0 | – | – |
r: Pearson correlation coefficient.
p: p-value ≤ 0.05 was considered to be significant.
Discussion
It is known that the presence of helicobacters may be associated with gastric disease in humans and animals. F. gastrosuis has been described to possibly play a role in gastric ulceration in pigs [17, 18].
This study reports a high occurrence of gastritis in pigs and wild boars, 95.8% and 100% respectively, as was previously described by Hessing et al., Robertson et al., De Witte et al. [18, 32, 33].
Regarding the pig samples that presented gastritis: 51.9% of the pars oesophagea tissues were positive for one or more Helicobacter spp. and F. gastrosuis and, specifically 23.1% were positive for a H. pylori-like species and 11.5% for a H. pylori-like species in combination with F. gastrosuis (Additional file 4). In the oxyntic mucosa samples, 50.0% were positive for Helicobacter spp. and F. gastrosuis but, in contrast, the species most often detected was H. suis. In addition, positive cases for F. gastrosuis were always accompanied by a Helicobacter species (Additional file 4). Among the porcine antral gastritis cases, 57.6% were positive for Helicobacter spp. and F. gastrosuis, while 36.4% were H. suis positive, 7.6% F. gastrosuis positive and 6.1% F. gastrosuis plus H. suis positive (Additional file 4).
In both pigs and wild boars, F. gastrosuis was detected in association with Helicobacter species which corroborates their potential synergy to induce gastric pathology (Additional files 4 and 5).
Particularly in pigs, F. gastrosuis may have a synergetic role with H. suis in gastric ulceration as described by De Witte et al. [18]. De Bruyne et al. described a correlation between H. suis infection and the development of gastritis [14]. H. suis can affect the acid gastric secretion by altering the number and/or function of parietal D and G cells, as well as interfere with the sonic hedgehog (Shh) signaling pathway that regulates the gastric acid secretion and is involved in the gastric organogenesis, glandular differentiation and gastric homeostasis. This may lead to gastroesophageal ulceration as well as affect the gastric microbiota since the presence of H. suis alters the gastric environment which may promote the proliferation of other microorganisms such as F. gastrosuis in the non-glandular zone leading to gastritis and ulceration [10, 11, 34].
In pig samples, gastric ulceration and erosion were higher in the antral mucosa samples positive for H. suis and F. gastrosuis and a statistically significant correlation for the presence of F. gastrosuis with ulceration (r = 0.31, p ≤ 0.05), and H. pylori-like with erosion (r = 0.27, p ≤ 0.05) was demonstrated. The presence of H. suis and F. gastrosuis in pigs was previously reported and it was hypothesized that bacterial interaction can lead to gastric lesions and ulceration of the pars oesopahea [10, 18].
Compared to pigs, wild boars had a lower percentage of a H. pylori-like species and H. suis in the analyzed samples. However, both had close percentages of samples positive for F. gastrosuis (35.2% in pigs vs 42.9% in wild boars). Previous studies have reported the presence of Helicobacter spp. in wild boars [26, 35, 36], but this remains the first description of F. gastrosuis DNA detection and its possible relation with gastric erosion and ulceration in this species, as it has been described in pigs [10, 17, 18].
DNA of zoonotic Helicobacter spp. were detected in the pigs’ and wild boars’ stomach samples screened in this study, so the close contact between wildlife, domestic animals, and humans should be a concern for the transmission of bacteria with zoonotic potential that raises awareness in a One health perspective [4, 5, 19, 21, 24–26].
In the present study, DNA from H. felis, H. bizzozeronii and H. salomonis, which naturally colonize the stomach of dogs and cats, was detected in the stomach of pigs and/or wild boars. It is not clear whether these species are really able to colonize the stomach of these animals or whether our results are due to, for instance, environmental contamination.
Two PCR assays were used for detection of H. pylori DNA, one targeting the ureAB gene and the other one targeting the glmM gene. Although a number of studies have been published diagnosing H. pylori infection based on a ureAB gene PCR only [19, 37, 38], some reports indeed claim that H. pylori identification should include these two different target genes amplification [39–41]. Lu et al. compared five PCR methods for detection of H. pylori DNA of 24 culture-positive samples obtained from 50 human gastric samples, including PCRs targeting the ureA and glmM gene. These authors considered the glmM gene PCR to be the most appropriate method for detection of H. pylori organisms in clinical samples [42]. After analyzing 290 human gastric samples, Elnosh et al. concluded that a glmM gene based PCR showed higher sensitivity and specificity than a ureA gene based PCR [43]. In another investigation, only 10 out of the 50 human gastric samples analyzed turned out positive using the glmM gene whereas 25 were PCR-positive using the ureA gene [44].
It is remarkable that more than 50% of the pigs were positive in the ureAB PCR, while none of the samples were positive in the glmM PCR. Sequencing of the amplicons obtained in the ureAB PCR, revealed high homologies with H. pylori ureAB ranging from 96.3 to 100%. Natural colonization of the stomach of pigs with H. pylori has not been described before. We hypothesize that a H. pylori-like species, with a similar ureAB gene as H. pylori, was detected. This organism seems to preferably colonize the proximal, non-glandular part of the stomach whereas H. suis mainly colonizes the distal part. In any case, our results should be confirmed by isolation and identification of this H. pylori-like species from the stomach of pigs.
Krakowka et al. described a Helicobacter species present in the stomach of pigs, which had a curve-shaped morphology similar to that of H. pylori [45]. Unfortunately, no taxonomic studies or genome sequences have been published for this Helicobacter species and its exact identity is unclear [10]. Comparison with our results is therefore not possible.
This study reports the presence of a H. pylori-like species, gastric NHPH, and F. gastrosuis DNA in the stomach of pigs and wild boars and the putative gastric histopathological alterations associated. The results suggest that pigs and wild boars may act as reservoirs for these bacteria.
Further research should be carried out, including studies with a larger sampled population of wild boars, to assess the real prevalence of these bacteria in this animal species and to better understand their possible role in the development of gastric pathology.
Supplementary Information
Additional file 1. Primer sequences used for detection of Helicobacter spp. and F. gastrosuis and thermocycling conditions.
Additional file 2. Nonparametric Kruskal–Wallis, Chi Square test applied to the three gastric zones comparing the different variables in pigs.
Additional file 3. BLAST identity percentage interval of the different Helicobacter spp. and F. gastrosuis sequences obtained.
Additional file 4. Number of Helicobacter spp. and F. gastrosuis DNA positive samples associated with gastritis score per pig gastric zone.
Additional file 5. Number of Helicobacter spp. and F. gastrosuis DNA positive samples associated with gastritis score per wild boar’s gastric zone.
Acknowledgements
The participation of Teresa Letra Mateus was supported by the projects UIDB/CVT/00772/2020 and LA/P/0059/2020 funded by the Portuguese Foundation for Science and Technology (FCT).
Authors' contributions
Conceptualization: FCN, TLM, FH and IA; Methodology: FCN, TLM, FH and IA; formal analysis: FCN, NC, TLM, ET, FH and IA; investigation: FCN, ST, ET, SB, AR; resources: FCN, TLM and IA; data curation: FCN, TLM, FH and IA; writing—original draft preparation: FCN, TLM and IA; writing—review and editing: FCN, ST, TLM, AR, FG, ET, FH and IA; supervision: TLM, IA, FH. All authors read and approved the final manuscript.
Funding
This research was funded by FCT-the Portuguese Foundation for Science and Technology, MCTES-Ministry of Science, Technology and Higher Education, FSE- European Social Fund and UE-European Union (SFRH/BD/147761/2019).
Declarations
Ethics approval and consent to participate
This study was approved by i3S Animal Welfare and Ethics Review Body (ref. 2021-4).
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Freddy Haesebrouck and Irina Amorim shared senior authorship
References
- 1.Choi YK, Han JH, Joo HS. Identification of novel Helicobacter species in pig stomachs by PCR and partial sequencing. J Clin Microbiol. 2001;39:3311–3315. doi: 10.1128/JCM.39.9.3311-3315.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahadori A, De Witte C, Agin M, De Bruyckere S, Smet A, Tümgör G, Gökmen T, Haesebrouck F, Köksal F. Presence of gastric Helicobacter species in children suffering from gastric disorders in Southern Turkey. Helicobacter. 2018;23:e12511. doi: 10.1111/hel.12511. [DOI] [PubMed] [Google Scholar]
- 3.Haesebrouck F, Pasmans F, Flahou B, Chiers K, Baele M, Meyns T, Decostere A, Ducatelle R. Gastric helicobacters in domestic animals and nonhuman primates and their significance for human health. Clin Microbiol Rev. 2009;22:202–223. doi: 10.1128/CMR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cortez Nunes F, Letra Mateus T, Teixeira S, Barradas PF, Gärtner F, Haesebrouck F, Amorim I. Molecular detection of human pathogenic gastric Helicobacter species in wild rabbits (Oryctolagus cuniculus) and wild quails (Coturnix coturnix) Zoonotic Dis. 2021;1:42–50. doi: 10.3390/zoonoticdis1010005. [DOI] [Google Scholar]
- 5.Mladenova-Hristova I, Grekova O, Patel A. Zoonotic potential of Helicobacter spp. J Microbiol Immunol Infect. 2017;50:265–269. doi: 10.1016/j.jmii.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Berlamont H, De Witte C, De Bruyckere S, Fox JG, Backert S, Smet A, Boyen F, Haesebrouck F. Differentiation of gastric Helicobacter species using MALDI-TOF mass spectrometry. Pathogens. 2021;10:366. doi: 10.3390/pathogens10030366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopo I, Libânio D, Pita I, Dinis-Ribeiro M, Pimentel-Nunes P. Helicobacter pylori antibiotic resistance in Portugal: systematic review and meta-analysis. Helicobacter. 2018;23:e12493. doi: 10.1111/hel.12493. [DOI] [PubMed] [Google Scholar]
- 8.Guevara B, Cogdill AG. Helicobacter pylori: a review of current diagnostic and management strategies. Dig Dis Sci. 2020;65:1917–1931. doi: 10.1007/s10620-020-06193-7. [DOI] [PubMed] [Google Scholar]
- 9.Taillieu E, Chiers K, Amorim I, Gärtner F, Maes D, Van Steenkiste C, Haesebrouck F. Gastric Helicobacter species associated with dogs, cats and pigs: significance for public and animal health. Vet Res. 2022;53:42. doi: 10.1186/s13567-022-01059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Witte C, Ducatelle R, Haesebrouck F. The role of infectious agents in the development of porcine gastric ulceration. Vet J. 2018;236:56–61. doi: 10.1016/j.tvjl.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Zhang G, Ducatelle R, Mihi B, Smet A, Flahou B, Haesebrouck F. Helicobacter suis affects the health and function of porcine gastric parietal cells. Vet Res. 2016;47:101. doi: 10.1186/s13567-016-0386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellemans A, Chiers K, De Bock M, Decostere A, Haesebrouck F, Ducatelle R, Maes D. Prevalence of ‘Candidatus Helicobacter suis’ in pigs of different ages. Vet Rec. 2007;16:189–192. doi: 10.1136/vr.161.6.189. [DOI] [PubMed] [Google Scholar]
- 13.De Witte C, Devriendt B, Flahou B, Bosschem I, Ducatelle R, Smet A, Haesebrouck F. Helicobacter suis induces changes in gastric inflammation and acid secretion markers in pigs of different ages. Vet Res. 2017;48:34. doi: 10.1186/s13567-017-0441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Bruyne E, Flahou B, Chiers K, Meyns T, Kumar S, Vermoote M, Pasmans F, Millet S, Dewulf J, Haesebrouck F, Ducatelle R. An experimental Helicobacter suis infection causes gastritis and reduced daily weight gain in pigs. Vet Microbiol. 2012;160:449–454. doi: 10.1016/j.vetmic.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 15.Baele M, Decostere A, Vandamme P, Ceelen L, Hellemans A, Mast J, Chiers K, Ducatelle R, Haesebrouck F. Isolation and characterization of Helicobacter suis sp. nov. from pig stomachs. Int J Syst Evol Microbiol. 2008;58:1350–1358. doi: 10.1099/ijs.0.65133-0. [DOI] [PubMed] [Google Scholar]
- 16.Booth SJ (2014) Fusobacterium infections. In: Reference module in biomedical sciences. Elsevier
- 17.De Witte C, Flahou B, Ducatelle R, Smet A, De Bruyne E, Cnockaert M, Taminiau B, Daube G, Vandamme P, Haesebrouck F. Detection, isolation and characterization of Fusobacterium gastrosuis sp. nov. colonizing the stomach of pigs. Syst Appl Microbiol. 2017;40:42–50. doi: 10.1016/j.syapm.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 18.De Witte C, Demeyere K, De Bruyckere S, Taminiau B, Daube G, Ducatelle R, Meyer E, Haesebrouck F. Characterization of the non-glandular gastric region microbiota in Helicobacter suis-infected versus non-infected pigs identifies a potential role for Fusobacterium gastrosuis in gastric ulceration. Vet Res. 2019;50:39. doi: 10.1186/s13567-019-0656-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubota-Aizawa S, Matsubara Y, Kanemoto H, Mimuro H, Uchida K, Chambers J, Tsuboi M, Ohno K, Fukushima K, Kato N, Yotsuyanagi H, Tsujimoto H. Transmission of Helicobacter pylori between a human and two dogs: a case report. Helicobacter. 2021;26:e12798. doi: 10.1111/hel.12798. [DOI] [PubMed] [Google Scholar]
- 20.De Cooman L, Houf K, Smet A, Flahou B, Ducatelle R, De Bruyne E, Pasmans F, Haesebrouck F. Presence of Helicobacter suis on pork carcasses. Int J Food Microbiol. 2014;187:73–76. doi: 10.1016/j.ijfoodmicro.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 21.Joosten M, Flahou B, Meyns T, Smet A, Arts J, De Cooman L, Pasmans F, Ducatelle R, Haesebrouck F. Case report: Helicobacter suis infection in a pig veterinarian. Helicobacter. 2013;18:392–396. doi: 10.1111/hel.12054. [DOI] [PubMed] [Google Scholar]
- 22.Flahou B, Rossi M, Bakker J, Langermans JA, Heuvelman E, Solnick JV, Martin ME, O'Rourke J, Ngoan LD, Hoa NX, Nakamura M, Overby A, Matsui H, Ota H, Matsumoto T, Foss DL, Kopta LA, Omotosho O, Franciosini MP, Casagrande Proietti P, Guo A, Liu H, Borilova G, Bracarense AP, Lindén SK, De Bruyckere S, Zhang G, De Witte C, Smet A, Pasmans F, Ducatelle R, Corander J, Haesebrouck F. Evidence for a primate origin of zoonotic Helicobacter suis colonizing domesticated pigs. ISME J. 2018;12:77–86. doi: 10.1038/ismej.2017.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sleeman JM, Richgels KLD, White CL, Stephen C. Integration of wildlife and environmental health into a One Health approach. Rev Sci Tech. 2019;38:91–102. doi: 10.20506/rst.38.1.2944. [DOI] [PubMed] [Google Scholar]
- 24.Meng XJ, Lindsay DS, Sriranganathan N. Wild boars as sources for infectious diseases in livestock and humans. Philos Trans R Soc Lond B Biol Sci. 2009;364:2697–2707. doi: 10.1098/rstb.2009.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meier R, Ryser-Degiorgis M. Wild boar and infectious diseases: evaluation of the current risk to human and domestic animal health in Switzerland: a review. Schweiz Arch Tierheilkd. 2018;160:443–460. doi: 10.17236/sat00168. [DOI] [PubMed] [Google Scholar]
- 26.Cortez Nunes F, Letra Mateus T, Teixeira S, Barradas P, de Witte C, Haesebrouck F, Amorim I, Gärtner F. Presence of Helicobacter pylori and H. suis DNA in free-range wild boars. Animals. 2021;11:1269. doi: 10.3390/ani11051269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. the updated Sydney system. International workshop on the histopathology of gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 28.Yamasaki L, Boselli-Grotti CC, Alfieri AA, Silva EO, Oliveira RL, Camargo PL, Bracarense APFRL. Histological findings in swine pars esophagea and its Helicobacter spp. relationship. Braz J Vet Anim Sci. 2009;61:553–560. [Google Scholar]
- 29.Queiroz DM, Rocha GA, Mendes EN, De Moura SB, De Oliveira AM, Miranda D. Association between Helicobacter and gastric ulcer disease of the pars esophagea in swine. Gastroenterology. 1996;111:19–27. doi: 10.1053/gast.1996.v111.pm8698198. [DOI] [PubMed] [Google Scholar]
- 30.Amorim I, Smet A, Alves O, Teixeira S, Saraiva AL, Taulescu M, Reis C, Haesebrouck F, Gärtner F. Presence and significance of Helicobacter spp. in the gastric mucosa of Portuguese dogs. Gut Pathog. 2015;7:12. doi: 10.1186/s13099-015-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flahou B, Deun KV, Pasmans F, Smet A, Volf J, Rychlik I, Ducatelle R, Haesebrouck F. The local immune response of mice after Helicobacter suis infection: strain differences and distinction with Helicobacter pylori. Vet Res. 2012;43:75. doi: 10.1186/1297-9716-43-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hessing MJ, Geudeke MJ, Scheepens CJ, Tielen MJ, Schouten WG, Wiepkema PR. Mucosal lesions in the pars esophagus in swine: prevalence and the effect of stress. Tijdschr Diergeneeskd. 1992;117:445–450. [PubMed] [Google Scholar]
- 33.Robertson ID, Accioly JM, Moore KM, Driesen SJ, Pethick DW, Hampson DJ. Risk factors for gastric ulcers in Australian pigs at slaughter. Prev Vet Med. 2002;53:293–303. doi: 10.1016/S0167-5877(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 34.Sapierzyński R, Fabisiak M, Kizerwetter-Swida M, Cywińska A. Effect of Helicobacter sp. infection on the number of antral gastric endocrine cells in swine. Pol J Vet Sci. 2007;10:65–70. [PubMed] [Google Scholar]
- 35.Zanoni RG, Piva S, Florio D, Bassi P, Mion D, Cnockaert M, Luchetti A, Vandamme P. Helicobacter apri sp. nov., isolated from wild boars. Int J Syst Evol Microbiol. 2016;66:2876–2882. doi: 10.1099/ijsem.0.001071. [DOI] [PubMed] [Google Scholar]
- 36.Fabisiak M, Sapierzyński R, Salamaszyńska-Guz A, Kizerwetter-Swida M. The first description of gastric Helicobacter in free-ranging wild boar (Sus scrofa) from Poland. Pol J Vet Sci. 2010;13:171–174. [PubMed] [Google Scholar]
- 37.Shafaie S, Kaboosi H, PeyraviiGhadikolaii F. Prevalence of non Helicobacter pylori gastric Helicobacters in Iranian dyspeptic patients. BMC Gastroenterol. 2020;20:190. doi: 10.1186/s12876-020-01331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matos R, Taillieu E, De Bruyckere S, De Witte C, Rêma A, Santos-Sousa H, Nogueiro J, Reis CA, Carneiro F, Haesebrouck F, Amorim I, Gärtner F. Presence of Helicobacter species in gastric mucosa of human patients and outcome of helicobacter eradication treatment. J Pers Med. 2022;12:181. doi: 10.3390/jpm12020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabbagh P, Mohammadnia-Afrouzi M, Javanian M, Babazadeh A, Koppolu V, Vasigala VR, Nouri HR, Ebrahimpour S. Diagnostic methods for Helicobacter pylori infection: ideals, options, and limitations. Eur J Clin Microbiol Infect Dis. 2019;38:55–66. doi: 10.1007/s10096-018-3414-4. [DOI] [PubMed] [Google Scholar]
- 40.Wang YK, Kuo FC, Liu CJ, Wu MC, Shih HY, Wang SS, Wu JY, Kuo CH, Huang YK, Wu DC. Diagnosis of Helicobacter pylori infection: current options and developments. World J Gastroenterol. 2015;21:11221–11235. doi: 10.3748/wjg.v21.i40.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gastli N, Allain M, Lamarque D, Abitbol V, Billoët A, Collobert G, Coriat R, Terris B, Kalach N, Raymond J. Diagnosis of Helicobacter pylori infection in a routine testing workflow: effect of bacterial load and virulence factors. J Clin Med. 2021;10:2755. doi: 10.3390/jcm10132755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu J-J, Perng C-L, Shyu R-Y, Chen C-H, Lou Q, Chong SKF, Lee C-H. Comparison of five PCR methods for detection of Helicobacter pylori DNA in gastric tissues. J Clin Microbiol. 1999;37:772–774. doi: 10.1128/JCM.37.3.772-774.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elnosh M, Altayb H, Hamedelnil Y, Elshareef W, Abugrain A, Osman E, Albasha A, Abdelhamid A, Moglad E, AbdAlla A, Ismail A. Comparison of invasive histological and molecular methods in the diagnosis of Helicobacter pylori from gastric biopsies of Sudanese patients: a cross-sectional study. F1000Res. 2022;11:113. doi: 10.12688/f1000research.75873.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wahab H, Khan T, Ahmad I, Jan A, Younas M, Shah H, AbdEl-Salam NM, Ayaz S, Ullah R, Wasim MA. Detection of H. pylori By PCR method using UreA and UreC gene in gastric biopsy sample. J Pure Appl Microbiol. 2015;9:2165–2174. [Google Scholar]
- 45.Krakowka S, Ringler SS, Flores J, Kearns RJ, Eaton KA, Ellis JA. Isolation and preliminary characterization of a novel Helicobacter species from swine. Am J Vet Res. 2005;66:938–944. doi: 10.2460/ajvr.2005.66.938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Primer sequences used for detection of Helicobacter spp. and F. gastrosuis and thermocycling conditions.
Additional file 2. Nonparametric Kruskal–Wallis, Chi Square test applied to the three gastric zones comparing the different variables in pigs.
Additional file 3. BLAST identity percentage interval of the different Helicobacter spp. and F. gastrosuis sequences obtained.
Additional file 4. Number of Helicobacter spp. and F. gastrosuis DNA positive samples associated with gastritis score per pig gastric zone.
Additional file 5. Number of Helicobacter spp. and F. gastrosuis DNA positive samples associated with gastritis score per wild boar’s gastric zone.


