Abstract
The essential function of penicillin-binding protein 2 (PBP2) in methicillin-susceptible Staphylococcus aureus RN4220 was clearly established by placing the pbp2 gene under control of the inducible Pspac promoter; the resulting bacteria were unable to grow in the absence of inducer. In contrast, the deficit in PBP2 caused by inhibition of transcription of the pbp2 gene did not block growth of a methicillin-resistant S. aureus strain expressing the extra penicillin-binding protein PBP2A, a protein of extraspecies origin that is central to the mechanism of methicillin resistance. Several lines of evidence indicate that the essential function of PBP2 that can be compensated for by PBP2A is the transpeptidase activity. This provides direct genetic evidence that PBP2A has transpeptidase activity.
The wide-spectrum resistance of methicillin-resistant Staphylococcus aureus (MRSA) to all β-lactam antibiotics has had a devastating impact on the chemotherapy of staphylococcal infections ever since the first appearance of MRSA in clinical specimens in the early 1960s. The key component of this resistance mechanism is an acquired penicillin-binding protein (PBP), PBP2A, which has unusually low affinity for all β-lactam antibiotics. The genetic determinant of PBP2A, the mecA gene, is not native to S. aureus but was imported from an as-yet-unidentified extraspecies source (2). Until recently, the accepted model of this resistance mechanism implied that in the presence of β-lactam antibiotics the only PBP that remains functional is the low-affinity PBP2A, since the other four staphylococcal PBPs had high enough affinities to penicillin type antibiotics to become rapidly and fully acylated and inactivated even at low drug concentrations that were several orders of magnitude below the concentration needed to inhibit the growth of many MRSA strains. In this model, PBP2A is a surrogate enzyme capable of taking over the normal functions of staphylococcal PBPs in cell wall biosynthesis.
However, some new observations require basic revision of this model. The structural determinant of staphylococcal PBP2 was shown to be an auxiliary gene; transposon inactivation of pbp2 resulted in a massive reduction in the level of resistance in an MRSA strain despite the fact that the bacteria continued to produce normal amounts of PBP2A (20). Recent experiments showed that expression of high-level resistance required the cooperative functioning of PBP2A and the penicillin-insensitive transglycosylase (TGase) domain of PBP2 (17).
These observations brought back into focus the possibility that native staphylococcal PBPs, primarily PBP2, are participants in the mechanism of β-lactam resistance. The purpose of the study described in this paper was to examine in more detail the role(s) PBP2 plays in staphylococcal wall synthesis in antibiotic-susceptible bacteria as well as resistant bacteria (i.e., both in the absence and in the presence of acquired PBP2A). To do this, pbp2 was put under control of the inducible promoter Pspac, which allowed testing of the essentiality of this gene in the backgrounds of methicillin-susceptible and -resistant strains of S. aureus.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are described in Table 1. S. aureus strains were grown in tryptic soy broth (TSB) (Difco Laboratories) with aeration at 37°C or on tryptic soy agar (TSA) (Difco Laboratories) plates at 37°C. Escherichia coli strains were grown in Luria-Bertani broth (Difco Laboratories) with aeration at 37°C.
TABLE 1.
Strains and plasmids used in this study
| Strain(s) or plasmida | Relevant characteristicsb | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus RN4220 | MSSA strain restriction negative | R. Novick |
| S. aureus COL | Homogeneous MRSA (MIC, 1,600 μg/ml) | RU collectionc |
| S. aureus RUSA4 | COL Tn551 mutant with insertion in mecA gene (MIC, 3 μg/ml) | 15 |
| RNpPBP2iII-1 and -5 | RN4220 with Pspac-pbp2 fusion in the chromosome, transformed with pMGPII | This study |
| COLpPBP2iII-1 and -5 | COL with Pspac-pbp2 fusion in the chromosome, transformed with pMGPII | This study |
| RUSA4pPBP2iC-2 and -3 | RUSA4 with Pspac-pbp2 fusion in the chromosome | This study |
| COLpPBP2iC-2 and -3 | COL with Pspac-pbp2 fusion in the chromosome | This study |
| Plasmids | ||
| pSP64 | E. coli plasmid, Apr | Promega |
| pSP64E | Integrational vector for S. aureus, Apr Emr | 18 |
| pSPT181c | Thermosensitive E. coli-S. aureus shuttle vector, Apr Tcr Cmr | S. Wu |
| pDH88 | Bacillus subtilis plasmid with IPTG-inducible Pspac promoter and lacI gene | 9 |
| pGC2 | E. coli-S. aureus shuttle vector, Apr Cmr | 24 |
| pMGPI | S. aureus integrational vector with IPTG-inducible Pspac promoter and lacI gene, Apr Emr | This study |
| pMGPII | S. aureus replicative plasmid containing lacI gene, Apr Cmr | This study |
| pPBP2i | pMGPI with pbp2 ribosome-binding site and first 171 codons fused to Pspac promoter, Apr Emr | This study |
| pPBP2iC | pSP64 with pbp2 ribosome-binding site and first 171 codons fused to Pspac promoter, Apr Cmr | This study |
The PBP2 constructs are underlined to indicate that the plasmids are integrated into the host cell's chromosome.
Apr, ampicillin resistant; Emr, erythromycin resistant; Cmr, chloramphenicol resistant.
RU, The Rockefeller University, Laboratory of Microbiology.
DNA methods.
Routine DNA manipulations were performed by using standard methods (1, 21). Restriction enzymes were purchased from New England Biolabs. Plasmid DNA was purified by using Wizard Plus SV Minipreps or Midipreps DNA purification systems (Promega). DNA sequencing was done at The Rockefeller University Protein/DNA Technology Center by the BigDye terminator cycle sequencing method with either a 3700 DNA analyzer for capillary electrophoresis or ABI Prism 377 DNA sequencers for slab gel electrophoresis.
Construction of pPBP2i.
Plasmid pDH88 was digested with EcoRI and BamHI. The digestion products were electrophoresed on a 1% low-melting-point agarose gel (Gibco-BRL Life Technologies), and the 1.6-kb fragment was cut from the gel and purified by using a QIAquick gel extraction kit. This fragment contained the IPTG (isopropyl-β-d-thiogalactoside)-inducible Pspac promoter followed by a polylinker and by the lacI gene under control of the constitutive Ppcn promoter. The 1.6-kb fragment was cloned into pSP64E, resulting in plasmid pMGPI.
A DNA fragment containing the ribosome-binding site and the first 171 codons of pbp2 was amplified by PCR by using PfuTurbo DNA polymerase (Stratagene) and primers PBP2pro20 (5′-TATCCCGGGAAAGTGAGGGACCGCGTATG-3′) and PBP2pro21 (5′-GGCATCGATGCTTCTTGAGCTTTACGTCC-3′). The following conditions were used: 94°C for 5 min; 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 1.5 min; and one final extension step of 72°C for 5 min. The PCR product was cleaned with a Wizard PCR Preps DNA purification system (Promega), digested with ClaI and AvaI, and fused with the inducible promoter present in pMGPI, which resulted in plasmid pPBP2i. The correct sequence of the pbp2 insert was confirmed.
Construction of S. aureus strains with inducible pbp2.
Plasmid pPBP2i was introduced into RN4220 by electroporation essentially as previously described (12). The transformation mixture was plated on TSA containing erythromycin (10 μg/ml) and different concentrations of IPTG (0, 0.05, 0.5, and 4 mM).
Plasmid pMGPII was constructed by cloning the BamHI/EcoRI 1.6-kb fragment from pDH88 containing the lacI gene into E. coli-S. aureus shuttle vector pGC2 and was electroporated into RN4220. Two transformants, RNpPBP2iII-1 and RNpPBP2iII-5, were chosen for further study.
Plasmids pPBP2i and pMGPII were sequentially transduced to MRSA strain COL by using phage 80α as previously described (16), except that 0.5 mM IPTG was added to the media used for preparation of the transducing lysate and for transduction. Transductants were selected with erythromycin (10 μg/ml) for pPBP2i and with chloramphenicol (20 μg/ml) for pMGPII. COLpPBP2iII-1 and COLpPBP2iII-5 were chosen for further study.
For construction of pPBP2iC, the EcoRI/BamHI fragment from pPBP2i containing the pbp2 fragment fused with the Pspac promoter was cloned into pSP64 (Promega). A 1.1-kb PCR fragment encoding the chloramphenicol resistance marker was amplified from pSPT181c by using primers 5′-CTGTCGACCAGTCATACCAATAAC-3′ and 5′-CATGTCGACGCTCAACGTCAATAAAGC-3′ and was cloned into the SalI site of pSP64.
Plasmid pPBP2iC was electroporated into RN4220 in the presence of IPTG and subsequently transduced to RUSA4 and to COL by using media supplemented with IPTG. Selection was done with 15 μg of chloramphenicol per ml.
Isolation of RNA and Northern blot hybridization.
Overnight cultures of COLpPBP2iII-1 and COLpPBP2iII-5 were grown in TSB supplemented with erythromycin (10 μg/ml) and chloramphenicol (10 μg/ml). Each strain was diluted 1:40 in two tubes of TSB supplemented with erythromycin (5 μg /ml) and chloramphenicol (5 μg /ml), and one of the preparations was also supplemented with 0.5 mM IPTG. After cells were grown to the mid-log phase (optical density at 620 nm [OD620], ∼0.7), they were pelleted and processed with an RNeasy mini kit (Qiagen) or with a FastRNA Blue isolation kit (Bio 101 Inc.) in combination with FastPrep FP120 (Bio 101 Savant) used according to the manufacturer's recommendations. RNA (5 μg) was electrophoresed through a 1.2% agarose–0.66 M formaldehyde gel in MOPS (morpholine propanesulfonic acid) running buffer (Sigma). Blotting of RNA onto Hybond N+ membranes (Amersham) was performed with Turboblotter alkaline transfer systems (Schleicher & Schuell). For detection of specific transcripts, a pbp2-specific DNA probe was constructed by PCR amplification with two primers, 5′-AGCTTGGCAATCAGTTAAGC-3′ and 5′-TCCCACCATAAAAGATGAAG-3′. The probe was labeled with a Ready To Go labeling kit (Pharmacia Biotech) by using [α-32P]dCTP (Amersham Life Sciences) and was hybridized under high-stringency conditions. The blots were subsequently washed and autoradiographed.
Depletion of PBP2.
Strains with pbp2 under control of IPTG-inducible promoter Pspac were tested on solid medium by using TSA plates containing the appropriate antibiotic(s) (10 μg of erythromycin per ml and 10 μg of chloramphenicol per ml for strains with pPBP2i and pMGPII or 10 μg of chloramphenicol per ml for strains with pPBP2iC) and supplemented or not supplemented with 0.5 mM IPTG.
Experiments in liquid culture were performed by using the following protocol. Strains were grown overnight at 37°C in TSB supplemented with 0.5 mM IPTG and the appropriate antibiotic(s) (see above). These starting cultures were then each diluted 1:100 in 50 ml of TSB containing 5 μg of erythromycin per ml and 5 μg of chloramphenicol per ml and supplemented with 0.5 mM IPTG, and they were allowed to grow until the OD620 was approximately 0.2. Each culture was centrifuged at room temperature, washed with TSB, centrifuged, resuspended in 50 ml of medium lacking IPTG, and divided into two portions, one of which was supplemented with 0.5 mM IPTG. The cultures were again incubated with agitation at 37°C, and OD620 was monitored.
To measure the growth rate of COLpPBP2iII-1 or COLpPBP2iII-5, an overnight culture was grown in TSB containing 10 μg of erythromycin per ml and 10 μg of chloramphenicol per ml and supplemented with 0.5 mM IPTG. This starting culture was washed and resuspended in TSB without IPTG to remove traces of the inducer before the culture was diluted 1:200 in TSB containing 5 μg of erythromycin per ml and 5 μg of chloramphenicol per ml and supplemented or not supplemented with 0.5 mM IPTG. Each culture was incubated with agitation at 37°C, and the OD620 was monitored.
Cell wall analysis.
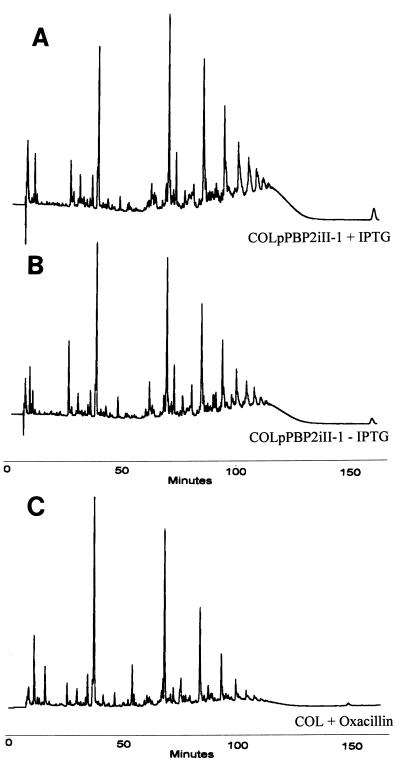
COL, COLpPBP2iII-1, and COLpPBP2iII-5 were grown overnight at 37°C in TSB supplemented with 10 μg of erythromycin per ml, 10 μg of chloramphenicol per ml, and 0.5 mM IPTG. Each starting culture was washed to remove traces of the inducer before it was diluted 1:5,000 in 500 ml of TSB containing 5 μg of erythromycin per ml and 5 μg of chloramphenicol per ml and supplemented or not supplemented with 0.5 mM IPTG. Cells were grown until the OD620 was approximately 0.3. Also included was a culture of strain COL grown in the presence of 1 μg of oxacillin per ml. Isolation of cell wall peptidoglycan and analysis of the family of enzymatically released muropeptides by reversed-phase high-pressure liquid chromatography (HPLC) were carried out essentially as previously described (3).
Electron microscopy.
COL was grown in TSB. COLpPBP2iII-5 was grown in TSB containing 5 μg of erythromycin per ml and 5 μg of chloramphenicol per ml and supplemented or not supplemented with 0.5 mM IPTG from a preinoculum that had been grown in the presence of the same antibiotics at concentrations of 10 μg/ml in medium that was supplemented or not supplemented with the inducer. When the OD620 reached 0.6, 5 ml of the culture was harvested by low-speed centrifugation and fixed with 1 ml of 2.5% glutaraldehyde. Electron microscopy was done at the Electron Microscopy Service of The Rockefeller University.
RESULTS
PBP2 is essential for growth of an MSSA strain but not for growth of an MRSA strain.
To determine if PBP2 is essential for survival of methicillin-susceptible S. aureus (MSSA) strain RN4220, we constructed a pbp2 conditional mutant in which pbp2 is regulated by the IPTG-inducible, LacI-repressible Pspac promoter (26). A fragment containing the ribosome-binding site and the first 171 codons of pbp2 was fused to the inducible promoter Pspac, creating plasmid pPBP2i, which was electroporated into RN4220. As a result of the integration of pPBP2i into the chromosome by a Campbell type of mechanism, a full-length copy of pbp2 was placed under control of Pspac, which is repressed by the product of the lacI gene that is also present in pPBP2i. Transformants were picked, and after correct insertion of the plasmid was confirmed by PCR, they were plated on selective plates in the presence or absence of IPTG. Supplementing the plates with 0.5 mM IPTG resulted in normal growth of the transformants, so this concentration was routinely used to propagate the mutants. In the absence of IPTG there was some growth in the area where there was heavy inoculum, where some colonies seemed to grow over a lawn of lysed cells. It has been found previously that in S. aureus tight regulation of genes under the control of Pspac can be obtained if lacI is present in a multiple-copy plasmid while a single copy of Pspac is present in the chromosome (11). Therefore, RN4220 with integrated pPBP2i was transformed with pMGPII, a plasmid encoding LacI, and two transformants, RNpPBP2iII-1 and RNpPBP2iII-5, were selected for further study. These transformants did not grow when they were plated on solid medium in the absence of IPTG, although the plates still had some colonies in the zone where there was heavy inoculum (Fig. 1a), indicating that PBP2 was essential for growth of RN4220.
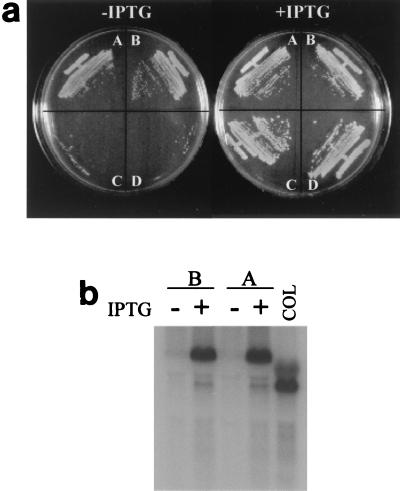
FIG. 1.
Effect of suppression of pbp2 transcription on growth of MRSA strain COL and MSSA strain RN4220. (a) Bacterial cultures grown on solid medium (TSA) supplemented or not supplemented with IPTG. Sector A, COLpPBP2iII-1; Sector B, COLpPBP2iII-5; Sector C, RNpPBP2iII-1; Sector D, RNpPBP2iII-5. (b) Northern blot hybridization with probe for pbp2. Lanes A, COLpPBP2iII-1 lanes B, COLpPBP2iII-5. Lane COL contained wild-type strain COL. RNA was prepared from cultures grown with or without 0.5 mM IPTG.
In order to test the essentiality of PBP2 in the background of an MRSA strain which contained the extra PBP2A, plasmids pPBP2i and pMGPII were sequentially transduced to MRSA strain COL. Two transductants, COLpPBP2iII-1 and COLpPBP2iII-5, were chosen for further study. When these transductants were plated in the presence and in the absence of IPTG, normal growth was observed on both plates (Fig. 1a), in sharp contrast to what occurred in the MSSA strain RN4220 background.
RNA was prepared from COLpPBP2iII-1 and COLpPBP2iII-5 grown in the presence and in the absence of IPTG. The amount of transcript produced in the presence of IPTG appeared to be similar to the amount of transcript produced from the two promoters of pbp2 in wild-type strain COL (19). However, transcription of pbp2 was severely suppressed in the absence of IPTG, confirming the deficit of PBP2 in the bacteria (Fig. 1b).
In order to rule out the possibility that survival of COLpPBP2iII strains depended on the presence of a small number of PBP2 molecules carried over from the inoculum (23), a single colony of COLpPBP2iII was streaked twice on medium without IPTG before it was plated on IPTG-free agar plates. Cells were still able to grow, and the promoter remained inducible.
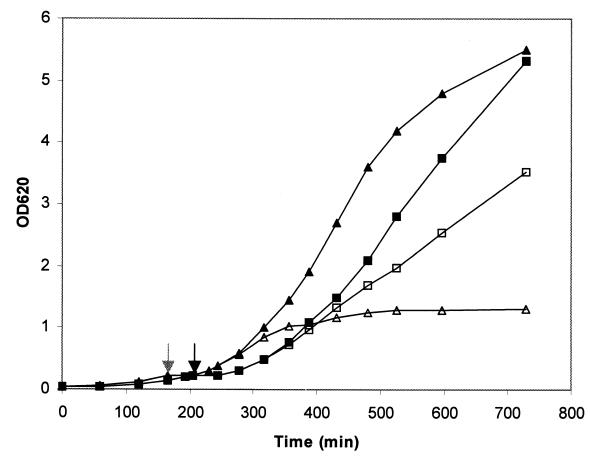
A study to confirm that PBP2 is essential in the background of drug-susceptible strain RN4220 but not in the background of drug-resistant strain COL was also done in liquid media. Strains RNpPBP2iII and COLpPBP2iII were grown in TSB containing 0.5 mM IPTG. When the OD620 reached 0.2, the cultures were washed with fresh medium lacking IPTG and divided into two portions, only one of which was supplemented with IPTG. After this the OD620 was monitored, and the growth curves indicated that removal of IPTG prevented growth of RNpPBP2iII-5 but only slowed growth of COLpPBP2iII-5 (Fig. 2). The presence of IPTG did not alter the growth rate of strain COL (data not shown).
FIG. 2.
Effect of suppression of pbp2 transcription on growth of MRSA strain COL and MSSA strain RN4220. Growth of the cultures in liquid medium was monitored by determining OD620. Symbols: ■ and □, COLpPBP2iII-5 grown with and without IPTG, respectively; ▴ and ▵, RNpPBP2iII-5 grown with and without IPTG, respectively. Cultures were grown until the OD620 was approximately 0.2. At that point (indicated by grey and black arrows for RNpPBP2iII-5 and COLpPBP2iII-5, respectively), the IPTG was removed by washing, the cells were resuspended in fresh medium without inducer, and each culture was divided into two portions, only one of which was supplemented with 0.5 mM IPTG. We continued to monitor the OD620 after this.
The growth rates of the COLpPBP2iII strains in the presence and in the absence of IPTG were measured, and the following results were obtained (each value is the average ± standard deviation based on three independent experiments): COLpPBP2iII-1 with IPTG, 0.75 ± 0.09 h−1; COLpPBP2iII-1 without IPTG, 0.60 ± 0.06 h−1; COLpPBP2iII-5 with IPTG, 0.74 ± 0.04 h−1; and COLpPBP2iII-5 without IPTG, 0.60 ± 0.06 h−1. These data showed that the deficit of PBP2, although not lethal, had a cost for the cells, as indicated by the reduced growth rates of the bacteria.
Replacement of the essential function of PBP2 by PBP2A.
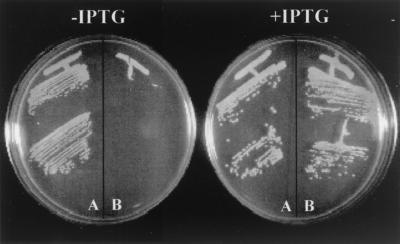
RN4220 and COL have unrelated genetic backgrounds. We assumed that the reason why PBP2 was essential in antibiotic-susceptible strain RN4220 but not in drug-resistant strain COL was the presence of PBP2A in the latter. In order to verify this, the experiments described above were repeated with strain RUSA4, an isogenic derivative of COL in which the mecA gene was inactivated by Tn551 insertion, which prevented expression of PBP2A (15). We constructed a RUSA4 strain with the pbp2 gene under control of the Pspac promoter by inserting plasmid pPBP2iC into its chromosome (plasmid pPBP2i could not be transduced to RUSA4 because its resistance marker, erm, is the same as the resistance marker present in Tn551). Transformants RUSA4pPBP2iC-2 and RUSA4pPBP2iC-3 were chosen for further study. Plasmid pPBP2iC was also inserted into the COL chromosome, and COLpPBP2iC-2 and COLpPBP2iC-3 were used for experiments. When these strains were plated on solid medium, RUSA4pPBP2iC was dependent on IPTG for growth, while COLpPBP2iC grew both in the presence and in the absence of IPTG (Fig. 3).
FIG. 3.
Effect of suppression of pbp2 transcription on growth of MRSA strain COL and its isogenic derivative RUSA4 with inactivated mecA gene. Bacterial cultures were grown on solid medium (TSA) supplemented or not supplemented with IPTG. Sector A, COLpPBP2iC-3 construct; Sector B, RUSA4pPBP2iC-3 construct.
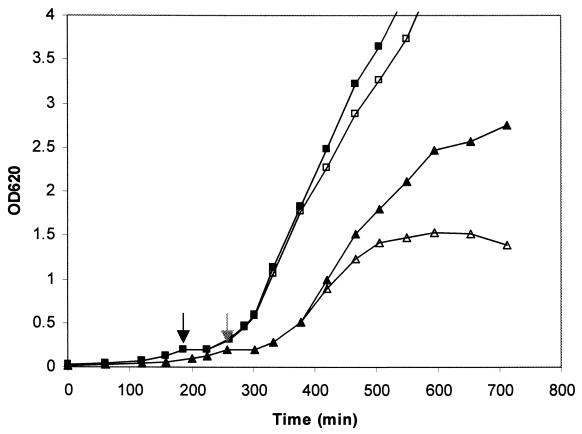
The constructs were also tested in liquid media. Cultures were grown in TSB containing IPTG, washed with fresh medium without IPTG, and divided into two portions, only one of which was supplemented with IPTG. Culture growth was monitored by determining OD620. A lack of IPTG prevented growth of the RUSA4pPBP2iC culture but only slowed the growth of the COLpPBP2iC culture (Fig. 4).
FIG. 4.
Effect of suppression of pbp2 transcription on growth of MRSA strain COL and its isogenic derivative RUSA4 with inactivated mecA gene. Growth of the cultures in liquid medium was monitored by determining the OD620. Symbols: ■ and □, COLpPBP2iC-2 grown with and without IPTG, respectively; ▴ and ▵, RUSA4pPBP2iC-2 grown with and without IPTG, respectively. Cultures were allowed to grow until the OD620 was approximately 0.2. At that point (indicated by grey and black arrows for RUSA4pPBP2iC-2 and COL pPBP2iC-2, respectively) the IPTG was removed by washing, the cells were resuspended in fresh medium without inducer, and each culture was divided into two portions, only one of which was supplemented with 0.5 mM IPTG. We continued to monitor the OD620 after this.
Cell wall composition and morphology of S. aureus constructs grown with and without PBP2.
Cell walls of COLpPBP2iII-1 and COLpPBP2iII-5 were purified from cultures grown in the presence and in the absence of IPTG. The cell walls of parental strain COL grown with and without IPTG were also analyzed to control for a possible effect of IPTG on cell wall composition; no evidence of such an effect was detected (data not shown). A comparison of the muropeptide compositions of strains with inducible pbp2 showed that the absence of PBP2 had only a minor, if any, effect on the muropeptide profile of the cell wall peptidoglycan (Fig. 5). Similar results were obtained for COLpPBP2iII-5 (data not shown).
FIG. 5.
Effect of PBP2 deficit on the muropeptide composition of the cell walls of MRSA strain COL. We determined the HPLC profiles of COLpPBP2iII-5 grown in the presence (A) and in the absence (B) of IPTG, as well as the HPLC profile of strain COL grown in the presence of 1 μg of oxacillin per ml (C). Muropeptides were purified and separated by HPLC as described in the text.
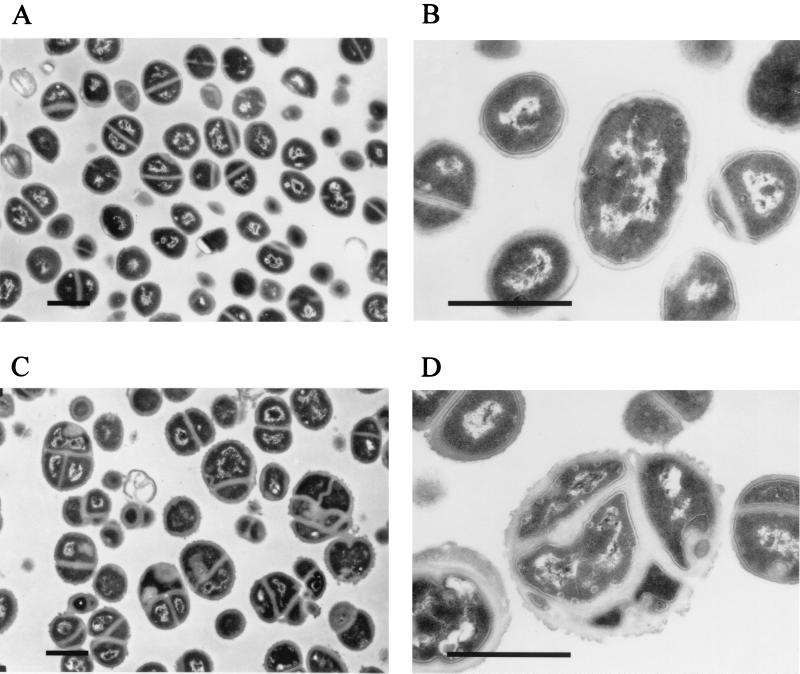
Replacement of PBP2 by PBP2A resulted in not only reduced growth rates but also some morphological abnormalities. Thin sections of cells of COL and COLpPBP2iII-5 grown in the presence and in the absence of IPTG were analyzed by electron microscopy (Fig. 6). Cells of COLpPBP2iII-5 grown with IPTG were morphologically indistinguishable from COL cells (data not shown). However, in the absence of the inducer, cells of COLpPBP2iII-5 exhibited premature septum formation or delayed division, which resulted in cells with more than one septum. When we scanned thin sections of 148 cells of strain COL, we did not find any bacteria with more than one septum (Fig. 6A and B), while about 14% of COLpPBP2iII-5 cells grown without IPTG (19 of 133 thin sections scanned) had two or more septa (Fig. 6C and D). In addition, an abnormal ruffled appearance of the cell wall occurred in COLpPBP2iII-5 grown without IPTG.
FIG. 6.
Morphological abnormalities in MRSA strain COL grown with a deficit of PBP2. Strain COL was grown in TSB (A and B), and construct COLpPBP2iII-5 was grown in medium without ITPG (C and D). Thin sections of bacteria were analyzed by electron microscopy. Bars = 1 μm.
DISCUSSION
The results described in this paper clarify several aspects of the function of staphylococcal PBP2, in both antibiotic-susceptible and antibiotic-resistant cells.
Contradictory reports have been published previously about the essentiality of PBP2 based on correlations between antibiotic affinities for different PBPs and antibacterial activities (5, 6, 8). It has also been suggested that PBP2 is not essential in MRSA strains (14) based on data obtained with a spontaneous mutant that was not genetically characterized.
Our observations clearly demonstrate that PBP2 is essential for growth of drug-susceptible strains of S. aureus; in a construct in which PBP2 of MSSA strain RN4220 was placed under control of IPTG-inducible promoter Pspac (an approach used previously to show the essentiality of murE in this organism [11]) bacterial growth depended on the presence of the IPTG inducer in both solid and liquid growth media.
When similar experiments were performed with drug-resistant strain COL and its isogenic transposon mutant RUSA4, in which the mecA gene was inactivated, strikingly different results were obtained. When pbp2 was made inducible in the background of strain COL expressing the mecA gene product PBP2A, growth of the bacteria did not depend on the presence of IPTG in the medium. When pbp2 was made inducible in the background of strain RUSA4, in which production of PBP2A is inhibited, bacterial growth occurred only in the presence of IPTG. These experiments clearly show that acquired drug resistance protein PBP2A can replace an essential function of native PBP2 in the absence of antibiotics in the surrounding medium.
PBP2 is a bifunctional protein that has both a transpeptidase (TPase) domain and a TGase domain (7, 13). The essential nature of this protein demonstrated in the experiments described above does not reveal which of these two activities is critical for survival and growth of antibiotic-susceptible S. aureus. A combination of the results of several experiments suggests that the critical function of PBP2 is the TPase function. This conclusion is based on the following observations. (i) In a recent study we showed that mutants with mutations in the TPase domain of PBP2 could not be obtained for MSSA while mutants with mutations in the TGase domain could be isolated, indicating that the TGase activity of PBP2 is not essential in the background of an MSSA strain (17). (ii) The experiments described in this paper demonstrate that suppression of production (transcription) of PBP2 is lethal in MSSA strains and also in an MRSA strain with inactivated PBP2A (strain RUSA4) but not in an MRSA strain with active PBP2A (strain COL). It follows that the essential activity of PBP2 replaced by PBP2A must be the TPase activity.
Our data also provide direct genetic evidence that the enzymatic activity of PBP2A is a TPase activity. Until now, the TPase activity for PBP2A has been assumed based on homology with other PBPs and current models of methicillin resistance in S. aureus (22). The exquisite dependence of antibiotic resistance on an intact TGase activity of PBP2 (17) also implies that PBP2A lacks such catalytic activity, which is in accordance with the current classification of this protein as a class B PBP with a TPase domain and a penicillin-insensitive second domain whose function is not known (7).
The muropeptide profiles of cell walls isolated from strain COLpPBP2iII grown in the presence and in the absence of IPTG (i.e., with and without PBP2) were very similar, if not identical, supporting the genetic evidence that the TPase function of PBP2 can be replaced by PBP2A. Under both conditions, HPLC analysis of enzymatic peptidoglycan hydrolysates revealed the multicomponent muropeptide profile with a high proportion of extensively cross-linked components typical of S. aureus cell walls. Such cell walls differ profoundly from the cell walls consisting of poorly cross-linked, monomer- and dimer-rich peptidoglycan produced by resistant strains of S. aureus grown in the presence of β-lactam antibiotics (Fig. 5), in which the TPase activity of PBP2A must play a major role (4). What factors determine the highly specific muropeptide composition of the staphylococcal peptidoglycan are not clear at present. Previously published data (25) clearly indicate that there is at least one additional PBP, namely, PBP4, that plays a major role in production of highly cross-linked staphylococcal peptidoglycan. It is likely that PBPs act in concert, i.e., in a manner possibly similar to that of the hypothetical multienzyme complex involved in biosynthesis of the cell wall of E. coli (10). If such complex exists in S. aureus, our results indicate that one of its characteristics is enough plasticity to enable integration of new components, such as PBP2A. However, the modest but frequent morphological abnormalities and slower growth rates detected for the staphylococci in which the deficit of PBP2 was compensated for by PBP2A suggest that integration of the foreign PBP2A into the cell wall synthetic apparatus may not be perfect.
ACKNOWLEDGMENTS
Mariana Pinho was supported by grant PRAXIS XXI/BD/9079/96. Sérgio Filipe was supported by grant PRAXIS XXI/BD/9071/96.
We thank Adriano Henriques and Shangwei Wu, who kindly provided plasmids pDH88 and pSPT181c, respectively.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1996. [Google Scholar]
- 2.Beck W D, Berger-Bachi B, Kayser F H. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J Bacteriol. 1986;165:373–378. doi: 10.1128/jb.165.2.373-378.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Jonge B L, Chang Y S, Gage D, Tomasz A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J Biol Chem. 1992;267:11248–11254. [PubMed] [Google Scholar]
- 4.de Jonge B L, Tomasz A. Abnormal peptidoglycan produced in a methicillin-resistant strain of Staphylococcus aureus grown in the presence of methicillin: functional role for penicillin-binding protein 2A in cell wall synthesis. Antimicrob Agents Chemother. 1993;37:342–346. doi: 10.1128/aac.37.2.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Georgopapadakou N H, Dix B A, Mauriz Y R. Possible physiological functions of penicillin-binding proteins in Staphylococcus aureus. Antimicrob Agents Chemother. 1986;29:333–336. doi: 10.1128/aac.29.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerogopapadakou N H, Liu F Y. Binding of beta-lactam antibiotics to penicillin-binding proteins of Staphylococcus aureus and Streptococcus faecalis: relation to antibacterial activity. Antimicrob Agents Chemother. 1980;18:834–836. doi: 10.1128/aac.18.5.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goffin C, Ghuysen J M. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayes M V, Curtis N A C, Wyke A W, Ward J B. Decreased affinity of a penicillin-binding protein for beta-lactam antibiotics in a clinical isolate of Staphylococcus aureus resistant to methicillin. FEMS Microbiol Lett. 1981;10:119–122. [Google Scholar]
- 9.Henner D J. Inducible expression of regulatory genes in Bacillus subtilis. Methods Enzymol. 1990;185:223–228. doi: 10.1016/0076-6879(90)85022-g. [DOI] [PubMed] [Google Scholar]
- 10.Holtje J V. A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology. 1996;142:1911–1918. doi: 10.1099/13500872-142-8-1911. [DOI] [PubMed] [Google Scholar]
- 11.Jana M, Luong T T, Komatsuzawa H, Shigeta M, Lee C Y. A method for demonstrating gene essentiality in Staphylococcus aureus. Plasmid. 2000;44:100–104. doi: 10.1006/plas.2000.1473. [DOI] [PubMed] [Google Scholar]
- 12.Kraemer G R, Iandolo J J. High-frequency transformation of Staphylococcus aureus by electroporation. Curr Microbiol. 1990;21:373–376. [Google Scholar]
- 13.Murakami K, Fujimura T, Doi M. Nucleotide sequence of the structural gene for the penicillin-binding protein 2 of Staphylococcus aureus and the presence of a homologous gene in other staphylococci. FEMS Microbiol Lett. 1994;117:131–136. doi: 10.1111/j.1574-6968.1994.tb06754.x. [DOI] [PubMed] [Google Scholar]
- 14.Murakami K, Nomura K, Doi M, Yoshida T. Increased susceptibility to cephamycin-type antibiotics of methicillin-resistant Staphylococcus aureus defective in penicillin-binding protein 2. Antimicrob Agents Chemother. 1987;31:1423–1425. doi: 10.1128/aac.31.9.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami K, Tomasz A. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J Bacteriol. 1989;171:874–879. doi: 10.1128/jb.171.2.874-879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oshida T, Tomasz A. Isolation and characterization of a Tn551-autolysis mutant of Staphylococcus aureus. J Bacteriol. 1992;174:4952–4959. doi: 10.1128/jb.174.15.4952-4959.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinho M G, de Lencastre H, Tomasz A. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc Natl Acad Sci USA. 2001;98:10886–10891. doi: 10.1073/pnas.191260798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinho M G, de Lencastre H, Tomasz A. Cloning, characterization, and inactivation of the gene pbpC, encoding penicillin-binding protein 3 of Staphylococcus aureus. J Bacteriol. 2000;182:1074–1079. doi: 10.1128/jb.182.4.1074-1079.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinho M G, de Lencastre H, Tomasz A. Transcriptional analysis of the Staphylococcus aureus penicillin binding protein 2 gene. J Bacteriol. 1998;180:6077–6081. doi: 10.1128/jb.180.23.6077-6081.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinho M G, Ludovice A M, Wu S, De Lencastre H. Massive reduction in methicillin resistance by transposon inactivation of the normal PBP2 in a methicillin-resistant strain of Staphylococcus aureus. Microb Drug Resist. 1997;3:409–413. doi: 10.1089/mdr.1997.3.409. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Song M D, Wachi M, Doi M, Ishino F, Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 1987;221:167–171. doi: 10.1016/0014-5793(87)80373-3. [DOI] [PubMed] [Google Scholar]
- 23.Williamson R, Tomasz A. Synthesis of penicillin-binding proteins in penicillin-treated Streptococcus pneumoniae. FEMS Microbiol Lett. 1984;22:301–305. [Google Scholar]
- 24.Wu S, de Lencastre H, Sali A, Tomasz A. A phosphoglucomutase-like gene essential for the optimal expression of methicillin resistance in Staphylococcus aureus: molecular cloning and DNA sequencing. Microb Drug Resist. 1996;2:277–286. doi: 10.1089/mdr.1996.2.277. [DOI] [PubMed] [Google Scholar]
- 25.Wyke A W, Ward J B, Hayes M V, Curtis N A. A role in vivo for penicillin-binding protein-4 of Staphylococcus aureus. Eur J Biochem. 1981;119:389–393. doi: 10.1111/j.1432-1033.1981.tb05620.x. [DOI] [PubMed] [Google Scholar]
- 26.Yansura D G, Henner D J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]