Abstract
Amino acid-deprived rplK (previously known as relC) mutants of Escherichia coli cannot activate (p)ppGpp synthetase I (RelA) and consequently exhibit relaxed phenotypes. The rplK gene encodes ribosomal protein L11, suggesting that L11 is involved in regulating the activity of RelA. To investigate the role of L11 in the stringent response, a derivative of rplK encoding L11 lacking the N-terminal 36 amino acids (designated ′L11) was constructed. Bacteria overexpressing ′L11 exhibited a relaxed phenotype, and this was associated with an inhibition of RelA-dependent (p)ppGpp synthesis during amino acid deprivation. In contrast, bacteria overexpressing normal L11 exhibited a typical stringent response. The overexpressed ′L11 was incorporated into ribosomes and had no effect on the ribosome-binding activity of RelA. By several methods (yeast two-hybrid, affinity blotting, and copurification), no direct interaction was observed between the C-terminal ribosome-binding domain of RelA and L11. To determine whether the proline-rich helix of L11 was involved in RelA regulation, the Pro-22 residue was replaced with Leu by site-directed mutagenesis. The overexpression of the Leu-22 mutant derivative of L11 resulted in a relaxed phenotype. These results indicate that the proline-rich helix in the N terminus of L11 is involved in regulating the activity of RelA.
In Escherichia coli, amino acid deprivation results in a comprehensive reorganization of cellular metabolism known as the stringent response (see reference 7 for a recent review). The types of metabolic changes observed suggest that the stringent response is designed to promote the survival of the starved bacterium. One of the hallmarks of the stringent response is the rapid intracellular accumulation of guanosine 5′-triphosphate 3′diphosphate (pppGpp) and guanosine 3′,5′-bispyrophosphate (ppGpp). These nucleotides are collectively designated (p)ppGpp. They are believed to play a central role in signalling starvation and in mediating the various metabolic changes that constitute the stringent response.
In amino acid-deprived bacteria, the synthesis of (p)ppGpp is catalyzed by ppGpp synthetase I, the product of the relA gene (7). The synthesis of ppGpp during the stringent response is ribosome dependent, and RelA has been shown to be associated with the 50S subunit of the ribosome (22). The ppGpp synthetase activity is associated with a 455-residue N-terminal fragment of RelA (26). This fragment is constitutively active and is not ribosome associated. The C-terminal portion of RelA represents the regulatory domain which involves ribosome-binding and oligomerization (14). Amino acid deprivation restricts the levels of the corresponding aminoacyl-tRNA species. The in vitro studies of Haseltine et al. (15) indicate that the synthesis of (p)ppGpp is associated with the codon-specified binding of an uncharged tRNA species to the acceptor site on the ribosome and the resulting stall in polypeptide chain elongation. A conformational change during this event may activate RelA.
Mutations that confer relaxed phenotypes cause the inhibition of (p)ppGpp accumulation by amino acid-deprived E. coli. Such mutations map in either the relA (1) or the relC (11, 20) genes. The relC gene was later shown to be identical to rplK, the gene encoding ribosomal protein L11. This suggests that L11 plays a role in regulating the stringent response. The basis for the relaxed phenotype of rplK (relC) has not been determined. Here we report that the N-terminal domain of L11 is involved in the regulation of RelA activity. We show that the overexpression of a derivative of L11 lacking a 36-amino-acid N-terminal segment resulted in the inhibition of the RelA-dependent synthesis of (p)ppGpp and in the concomitant relaxation of the stringent response in amino acid-deprived relA+ bacteria. This N-terminal segment contains a proline-rich helix which is involved in regulating the elongation cycle of translation (8, 34). The Pro-22 residue of the proline-rich helix is shown, by site-directed mutagenesis, to be involved in RelA regulation.
MATERIALS AND METHODS
E. coli strains.
Strain W3110 was a prototrophic derivative of E. coli K-12 from our laboratory collection. Strain VC6216 was a derivative of W3110 which was lysogenized with phage λDE3 (obtained from Novagen). The λDE3 prophage contains the gene encoding phage T7 RNA polymerase under the control of the lac promoter. VC6216 was specifically constructed for the purpose of expressing genes controlled by the T7 promoter. Cells were routinely grown in a gyrotory shaker at 30°C in Luria broth (LB) (Difco), M9 minimal medium (19), or modified M53 low-phosphate medium (4).
General recombinant DNA techniques.
The procedures for plasmid and genomic DNA purification, restriction endonuclease digestion, DNA ligation, and PCR amplification were those described by Sambrook et al. (25). Restriction endonucleases and T4 DNA ligase were purchased from New England BioLabs Inc.
Cloning of the E. coli rplK and ′rplK genes.
The rplK gene was amplified by PCR from strain W3110 genomic DNA. The primers used for this purpose were 5′-GGGGGATCCTAATGGCTAAGAAAGTACAAGCCTA-3′ and 5′-TGCGTCGACTTTCTCGCGGATAACACGC-3′. They were designed to introduce BamHI and SalI restriction sites on the 5′ and the 3′ ends of rplK, respectively. The amplified PCR product was cloned into the BamHI-SalI-digested vector, pET30c(+) (Novagen), to generate the plasmid pXY51. Thus, in pXY51 the E. coli rplK gene was fused to His-tag and S-tag sequences at its 5′ terminus and was under the control of a phage T7 promoter.
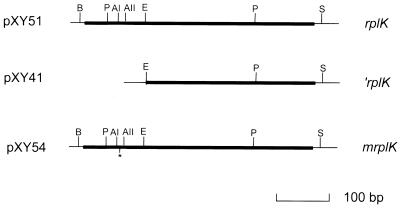
A derivative of rplK containing a deletion on its 5′ end was constructed by subcloning an EcoRI-SalI fragment from plasmid pXY51 into pET30a(+) (Novagen). The subcloned gene, designated ′rplK, encodes a derivative of the L11 protein which is missing the first 36 amino acids on its N-terminal end. This truncated protein is called ′L11. The plasmid carrying the ′rplK gene was designated pXY41. Figure 1 summarizes the relevant structural features of plasmids pXY51 and pXY41.
FIG. 1.
Relevant characteristics of plasmids pXY41, pXY51, and pXY54. Plasmid pXY51 is a derivative of pET30c(+) carrying the complete rplK gene, represented by the thick black line. The BamHI and SalI sites at 5′ and 3′ ends, respectively, were introduced through PCR primers. Plasmid pXY41 is a derivative of pET30a(+) carrying ′rplK, which contains a deletion extending from the 5′ terminus of rplK to the EcoRI site (nucleotide 109). Plasmid pXY54 is a derivative of pXY51 which contains a change of Pro-22 to Leu-22 (∗) created by site-directed mutagenesis. Abbreviations: AI, AvaI; AII, AvaII; B, BamHI; E, EcoRI; P, PstI; S, SalI.
Site-directed mutagenesis of rplK.
A derivative of rplK encoding a mutant L11 protein in which Pro-22 was changed to Leu-22 was constructed by site-directed mutagenesis. The sequence of the mutagenic primer was 5′-GGCTAACCCGAGTCTGCCAGTAGG-3′. PCR amplification of rplK was carried out using this primer and the 3′ end primer for the wild-type rplK gene. The PCR product was digested with AvaI and SalI. The AvaI-SalI fragment of the wild-type rplK gene in pXY51 was replaced by the mutant PCR product to generate plasmid pXY54 containing the mutant derivative of rplK designated mrplK (Fig. 1). The change of Pro-22 to Leu-22 was confirmed by an amino acid sequence determination of the purified mutant L11 protein performed by the Protein Chemistry Center of the University of Victoria.
Expression and purification of L11 and ′L11.
The methods for expression and purification of the L11 and ′L11 recombinant proteins were according to procedures described in the pET System Manual (Novagen). In summary, exponential-phase cultures of VC6216 carrying either pXY41 or pXY51 grown in LB at 30°C were induced by adding 0.5 mM isopropyl-β-d-galactoside (IPTG). After 3 h of incubation bacteria were harvested by centrifugation and broken by sonication. The recombinant His-tag L11 and His-tag ′L11 proteins in the crude extracts were purified by His-tag affinity chromatography (His-Bind Resin; Novagen).
Cloning of ′relA.
Plasmid pALS10 (30) contains the complete relA gene. A PstI-BamHI fragment from pALS10, encoding the C terminus of RelA, was subcloned from pALS10 into the vector pGEX-5X-1 (Pharmacia) to create plasmid pXY37. This C-terminal fragment, designated ′RelA, represents amino acid residues 455 to 743 of RelA. This region contains the ribosome-binding domain of RelA (14). The ′RelA fragment in plasmid pXY37 contains glutathione S-transferase (GST) fused to its N terminus.
Effect of L11 and L11 derivatives on the stringent response.
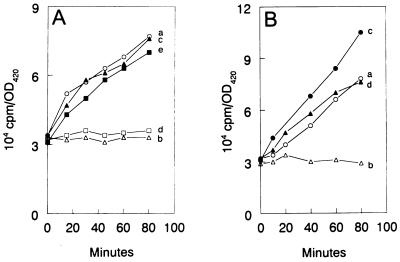
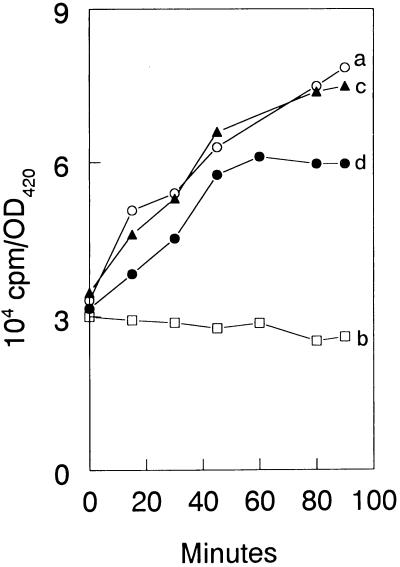
An exponential-phase culture of VC6216 carrying pXY41, pXY51, or pXY54 was grown in M9 minimal medium until it reached a density of about 2 × 108 cells per ml. The culture was divided into two portions, and 0.5 mM IPTG was added to one of these to induce the synthesis of L11, ′L11, or mL11. After 40 min, [5,6-3H]uracil (Amersham Corp.) was added to each of the two cultures at a final concentration of 1 μg per ml (0.5 μCi per ml). After an additional 20 min of incubation, each culture was further subdivided as indicated in Fig. 3 and 6, and portions were subjected to amino acid deprivation in the presence and absence of IPTG. From this point on the 100-μl samples were removed from the cultures at the indicated times, and the incorporation of [5,6-3H]uracil into cold trichloroacetic acid (TCA)-insoluble fractions was determined as described previously (17). Amino acid deprivation was achieved by the addition of either l-valine (13) or dl-serine hydroxamate (33) to cultures, each at 500 μg per ml. In addition to the amino acid-deprived cultures, an untreated control culture and a culture that was treated with IPTG alone were included in all experiments. In one experiment (see Fig. 3A), we relaxed the stringent response by treating an amino acid-deprived culture with 100 μg of chloramphenicol per ml (6).
FIG. 3.
(A) Effect of overexpression of wild-type L11 on the incorporation of [3H]uracil into cold TCA-insoluble fractions by E. coli strain VC6216 carrying the plasmid pXY51. Culture a (○) represents an untreated control. Culture b (▵) was amino acid-deprived with 500 μg of dl-serine hydroxamate per ml. Culture c (▴) was simultaneously amino acid deprived and treated with 100 μg of chloramphenicol per ml. Culture d (□) was amino acid deprived in the presence of 0.5 mM IPTG. Culture e (■) was treated with 0.5 mM IPTG. Where appropriate, IPTG was added to induce rplK 60 min prior to the start of amino acid deprivation at 0 min. (B) Effect of overexpression of ′L11 on the incorporation of [3H]uracil into stable RNA by E. coli strain VC6216 containing the plasmid pXY41. Culture a (○) was an untreated control. Culture b (▵) was amino acid deprived with 500 μg of dl-serine hydroxamate per ml. Culture c (●) was treated with 0.5 mM IPTG. Culture d (▴) was amino acid deprived in the presence of 0.5 mM IPTG. Where appropriate, IPTG was added to induce ′rplK 1 h prior to the start of amino acid deprivation at 0 min. OD420, optical density at 420 nm.
FIG. 6.
Effect of overexpression of L11Leu-22 on the incorporation of [3H]uracil into cold TCA-insoluble fractions by E. coli strain VC6216 carrying the plasmid pXY54. Culture a (○) was an untreated control. Culture d (●) was induced with 0.5 mM IPTG and then amino acid deprived by treatment with 500 μg of l-valine per ml. For comparison, cultures of VC6216 carrying plasmids pXY51 (b, □) and pXY41 (c, ▴), encoding L11 and ′L11, were induced with IPTG and were amino acid deprived like culture d. In all cases IPTG induction was initiated 60 min prior to the start of amino acid starvation at 0 min. OD420, optical density at 420 nm.
Synthesis of (p)ppGpp.
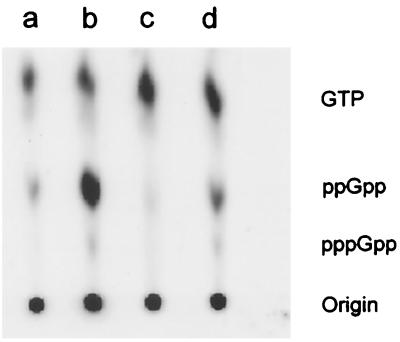
The synthesis of (p)ppGpp was measured in VC6216 carrying pXY41 grown in the modified M56LP medium of Bell (4). 32Pi (New England Nuclear; 40 μCi per ml) was added to an exponential-phase culture containing 2 × 108 cells per ml. One hour later the culture was divided into four portions as described in the legend to Fig. 4. One portion was a control that did not receive any further treatment for the duration of the experiment. Two of the portions received IPTG at 0.5 mM to induce the expression of ′L11. After one additional hour of incubation, the fourth portion and one of the IPTG-treated portions were subjected to amino acid deprivation by adding l-valine at 500 μg per ml. Ten minutes later, 200-μl samples were removed from each of the four cultures. Each sample was extracted with 20 μl of 11 M formic acid on ice for 30 min. The samples were centrifuged to remove cell debris, and 10-μl aliquots of the extracts were applied to a polyethyleneimine cellulose thin-layer chromatography plate (Aldrich Chemical Company Inc.). The chromatogram was developed in 1.5 M KH2PO4. The developed chromatogram was scanned on a Molecular Dynamics Storm PhosphorImager.
FIG. 4.
Effect of overexpression of ′L11 on ppGpp accumulation. Formic acid extracts of 32P-labeled strain VC6216 carrying plasmid pXY41 were fractionated by thin-layer chromatography. Lane a, no treatment; b, amino acid deprived; c, IPTG induction of ′L11; d, amino acid deprivation and IPTG induction of ′L11.
Ribosome preparation.
Ribosomes were prepared by the methods of Gentry and Cashel (12) and Homann and Nierhaus (16) with minor modifications. Briefly, bacteria were collected by centrifugation and washed once with Ribosome Buffer (10 mM Tris-HCl [pH 7.5], 14 mM MgCl, 10 mM K acetate, 1 mM dithiothreitol). The washed cells were resuspended in Ribosome Buffer and disrupted by sonication. Cell debris was removed by centrifugation at 20,000 × g for 30 min. The ribosomes were then recovered by centrifugation at 160,000 × g for 3 h. The ribosome pellets were resuspended in Ribosome Buffer and repurified in a two-step process. The crude ribosomes were first subjected to low-speed centrifugation (14,000 × g for 30 min), and the ribosomes in the supernatant from this step were pelleted by high-speed centrifugation (160,000 × g for 150 min). The concentration of ribosomes was estimated by measuring the absorbance of the preparation at 260 nm.
SDS-PAGE and Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed according to methods described by Ausubel et al. (2). To detect L11 or ′L11, Western blots were developed with polyclonal rabbit antibodies prepared against ′L11. Alternatively, a monoclonal antibody specific for S-tag (alkaline phosphatase-conjugated anti-S-tag antibody; Novagen) was used. A polyclonal rabbit antibody preparation specific for RelA was used for detection of ′RelA.
Yeast two-hybrid techniques.
The yeast two-hybrid technique was used in an attempt to determine whether L11 interacts with RelA. The technique was based on the Matchmaker Two-Hybrid System 3 (Clontech). The two cloning vectors, pGBKT7 and pGADT7, contain the GAL4 DNA-binding and activation domains, respectively. The BamHI-SalI fragment from pXY51 containing the complete rplK gene was subcloned into pGBKT7 and pGADT7 to yield plasmids pXY49 and pXY48, respectively. The source of the relA gene was plasmid pALS10 (30). We subcloned the PstI-BamHI fragment from pALS10 into pGBKT7 and pGADT7 to yield plasmids pXY62 and pXY63, respectively. As noted above, this fragment (′relA) encoded the C-terminal half of RelA, designated ′RelA, which contains the ribosome-binding domain of the enzyme (14). Plasmids pGBKT7-53 and pGADT7-T are positive controls encoding murine p53 fused to the GAL4 DNA-binding domain and simian virus 40 (SV40) T antigen fused to the GAL4 activation domain, respectively; these two proteins are known to interact with each other.
Saccharomyces cerevisiae (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS−GAL1TATAHIS3 GAL2UAS−GALITATA−ADE2 ura3::MELUAS−MEL1TATA−lacZ) was used as a host for the two-hybrid plasmids. Yeast were grown at 30°C in either YPD or SD synthetic medium as described in the Clontech User Manual for the Matchmaker System. In this system, a positive yeast two-hybrid was indicated by growth on SD medium lacking histidine, adenine, leucine, and tryptophan as well as by a high level of β-galactosidase activity from a reporter gene as measured by the method of Miller (19).
Attempt to demonstrate RelA-L11 interaction by affinity blotting.
The wild-type RelA protein was overexpressed in strain W3110 carrying plasmid pALS10 by induction with 1 mM IPTG for 2 h at 30°C. A crude extract prepared by sonication of these cells was fractionated by SDS-PAGE and transferred to a nitrocellulose membrane. The blot was incubated for 2 days at 4°C in a solution composed of 20 mM HEPES buffer at pH 7.9, 20% glycerol, 0.1 mM EDTA, 0.1 mM KCl, 10 mM MgCl, and 5% skim milk. The membrane was rinsed twice with HEPES buffer and incubated overnight at 4°C with purified L11 (from expression of pXY51). The blot was then probed with alkaline phosphatase-conjugated anti-S-tag antibody (Novagen) to detect any bound L11.
Attempt to demonstrate ′RelA-L11 interaction by affinity chromatography.
The GST-′RelA fusion protein was overexpressed in strain W3110 carrying plasmid pXY37 by induction with 0.1 mM IPTG for 2 h at 30°C. The GST-′RelA in a crude extract, prepared by sonication of these cells, was adsorbed to a Glutathione Sepharose 4B column (Pharmacia) according to a protocol provided by Pharmacia. A preparation of His-tag L11, purified as described above, was then loaded on the column. After the column was extensively washed with phosphate-buffered saline the adsorbed protein fraction was eluted with glutathione elution buffer (Pharmacia). The adsorbed fraction was analyzed in a Western blot developed with anti-S-tag antibody to detect L11.
Reproducibility of experiments.
All of the experiments described here were performed at least three times. The results in all cases were completely reproducible.
RESULTS
Expression and purification of the wild-type and truncated L11 protein.
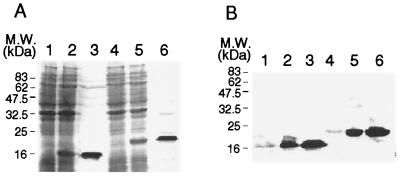
Plasmid pXY51 carries the complete rplK gene (Fig. 1). Figure 2A shows that the expression of rplK in pXY51 yielded a protein of approximately 21 kDa, which is consistent with the expected molecular size of L11 fused to His-tag and S-tag sequences (lanes 4 and 5). A truncated derivative of rplK, designated ′rplK, was cloned into plasmid pXY41 (Fig. 1). The ′rplK gene encoded a derivative of L11 containing a deletion of 36 amino acids at its N terminus. When ′rplK was expressed, a protein of 17 kDa was produced (Fig. 2A, lanes 1 and 2). Both ′L11 and L11 were readily purified by affinity chromatography (lanes 3 and 6, respectively). The identities of the purified ′L11 and L11 proteins were verified in Western blots developed with anti-S-tag antibodies (Fig. 2B, lanes 3 and 6, respectively).
FIG. 2.
Expression and purification of the L11 and ′L11 proteins. Panel A represents a Coomassie blue-stained SDS-PAGE gel, and panel B is a Western blot developed with anti-S-tag antibody. Lane 1, crude extract from VC6216/PXY41 uninduced cells; lane 2, crude extract from VC6216/PXY41 induced with 0.5 mM IPTG at 30°C for 2 h; lane 3, His-tag affinity chromatography-purified protein from induced VC6216/pXY41; lane 4, crude extract from VC6216/PXY51 uninduced cells; lane 5, crude extract from VC6216/PXY51 induced with 0.5 mM IPTG at 30°C for 2 h; lane 6, His-tag affinity chromatography-purified protein from induced VC6216/pXY51.
Effects of N-terminal deletion fragment of L11 protein on stringent response and accumulation of ppGpp.
Since a mutation in rplK (i.e., relC) is known to confer a relaxed phenotype, we tested the effects of overexpressing either L11 or ′L11 on the stringent response. Figure 3A shows the effect of the overproduction of L11 on the incorporation of [3H]uracil into cold TCA-insoluble fractions. Strain VC6216 carrying plasmid pXY51 exhibited a normal stringent response. Thus, amino acid deprivation inhibited [3H]uracil incorporation (compare curves a and b). Moreover, [3H]uracil incorporation was relaxed when the amino acid-deprived bacteria were treated with chloramphenicol (curve c). The addition of IPTG to induce the rplK gene on pXY51 prior to amino acid deprivation did not affect the stringent response (curve d). However, IPTG induction had a slight inhibitory effect on RNA synthesis in growing bacteria (curve e), and this probably reflects the inhibitory effect of IPTG induction on bacterial growth.
Figure 3B shows a similar experiment with strain VC6216 carrying plasmid pXY41. The stringent response in this strain was also normal, and [3H]uracil incorporation was inhibited during amino acid deprivation (compare curves a and b). When IPTG was added to a growing culture to induce ′rplK, [3H]uracil incorporation was actually stimulated (curve c). In contrast, the addition of IPTG to an amino acid-deprived culture resulted in the relaxation of [3H]uracil incorporation (curve d), indicating that the N-terminal domain of L11 was essential for normal stringent regulation. The intracellular levels of ppGpp under these various conditions were examined by thin-layer chromatography as shown in Fig. 4. As expected, a large amount of ppGpp, along with a small amount of pppGpp, accumulated within 10 min after the onset of amino acid deprivation (compare lanes a and b). The addition of IPTG to the growing culture actually caused a decrease in the basal level of ppGpp (compare lanes a and c). This may explain the stimulatory effect that IPTG had on [3H]uracil incorporation in growing bacteria (Fig. 3B, curve c). Finally, the treatment with IPTG significantly inhibited ppGpp accumulation by amino acid-deprived bacteria (compare lanes b and d). These results suggest that the overproduction of ′L11 prevents the activation of RelA during amino acid deprivation.
Incorporation of L11 and ′L11 into ribosomes.
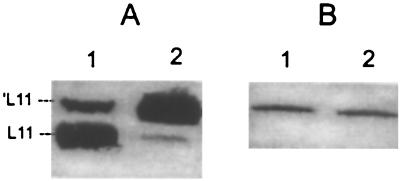
Ribosomes were purified from strain VC6216 carrying either pXY41 or pXY51. They were used to confirm that the recombinant L11 protein was incorporated into ribosomes and to determine whether the N-terminal deletion in ′L11 prevented this from happening. The ribosomal proteins from these preparations were analyzed in Western blots using anti-S-tag antibodies to detect the recombinant ribosomal proteins. Both L11 and ′L11 were readily detected (data not shown; see Fig. 5A). These results indicate that the N-terminal deletion in L11 did not affect its incorporation into ribosomes.
FIG. 5.
Effect of ′L11 on RelA-ribosome interaction. (A) Incorporation of ′L11 into ribosomes. Cultures of strain VC6216 carrying plasmid pXY41 were grown with (lane 2) or without (lane 1) 0.5 mM IPTG for 60 min. Equal amounts of ribosomes from these cultures were analyzed for ′L11 in a Western blot developed with anti-L11 antibody. (B) RelA-ribosome interaction. The same ribosome preparations were analyzed for RelA in a Western blot developed with anti-RelA antibody. Lane 1 represents ribosomes from untreated bacteria. Lane 2 represents ribosomes from IPTG-induced bacteria.
Effect of ′L11 on RelA-ribosome interaction.
The effect of ′L11 on the interaction of RelA with ribosomes was determined under the experimental conditions described in the legend to Fig. 3B. A culture of strain VC6216 carrying pXY41 was grown in M9 minimal medium and was divided into two portions, and one of the portions was treated with IPTG at 500 μM for 60 min. Ribosomes prepared from these cultures were then analyzed for ′L11 and RelA by Western blotting. As shown in Fig. 5A (lane 2), the overexpressed ′L11 was efficiently incorporated into ribosomes. Moreover, Fig. 5B shows that the ribosomes from the uninduced and the induced cultures had similar amounts of bound RelA, indicating that the presence of ′L11 had no significant effect on RelA-ribosome interaction. Therefore, the inhibition of ppGpp accumulation in amino acid-deprived bacteria overproducing ′L11 was not due to the inability of RelA to bind ribosomes.
Influence of proline-rich helix of L11 on RelA regulation.
The deletion in ′L11 removes a key region known as the proline-rich helix (8, 34). To determine whether this region was involved in RelA regulation, one residue in the proline-rich helix, Pro-22, was changed to Leu-22 by site-directed mutagenesis to create plasmid pXY54. The effect of overexpressing the resulting mutant protein, L11Leu-22, on the incorporation of [3H]uracil into cold TCA-insoluble material was tested as described in the legend to Fig. 6. Curve a represents an untreated control culture of VC6216 carrying pXY54. The other cultures shown were induced with 0.5 mM IPTG for 60 min and then subjected to isoleucine deprivation at 0 min. As already described, the stringent response was not affected when VC6216 with overexpressed levels of the wild-type L11 was amino acid deprived (curve b). In contrast, the overexpression of either ′L11 (curve c) or L11Leu-22 (curve d) resulted in the relaxation of the stringent response. These results indicate that Pro-22 plays an essential role in regulating the activity of RelA.
Attempts to demonstrate L11-RelA interaction.
Table 1 summarizes the results of a yeast two-hybrid analysis designed to test whether L11 interacts with ′RelA, the ribosome-binding domain of RelA. Transformant 1 was a negative control carrying the cloning vectors. Transformant 2 was a positive control carrying two-hybrid plasmids encoding two proteins, murine p53 and SV40 T antigen, that are known to interact with each other. Transformant 3 carried plasmids encoding ′RelA fused to the GAL4 DNA-binding domain and L11 fused to the GAL4 activation domain. This transformant clearly exhibited a negative two-hybrid test. As a check, the roles of the proteins were reversed in transformant 4. In this case, the L11 protein was fused to the GAL4 DNA-binding domain and the ′RelA protein was fused to the GAL4 activation domain. The results for this transformant were also negative. Therefore, L11 apparently does not bind to ′RelA.
TABLE 1.
Analysis of L11-RelA interaction in the yeast two-hybrid systema
| Transformant | GAL4 (DB) plasmid used | GAL4 (AD) plasmid used | Growth on SD selection medium | β-Galactosidase activity (Miller units)b |
|---|---|---|---|---|
| 1 | pGBKT7 (cloning vector) | pGADT7 (cloning vector) | − | 0.3 ± 0.1 |
| 2 | pGBKT7-53 (murine p53) | pGADT7-T (SV40 T antigen) | + | 18.8 ± 2.2 |
| 3 | pXY62 (′RelA) | pXY48 (L11) | − | 0.5 ± 0.4 |
| 4 | pXY49 (L11) | pXY63 (′RelA) | − | 0.6 ± 0.3 |
DB, DNA-binding; AD, activation domain.
Average of three independent determinations
Affinity blotting and protein affinity chromatography were two in vitro approaches used to test whether L11 interacts directly with RelA (data not shown). Both methods yielded negative results, indicating again that L11 and ′RelA do not bind to each other.
DISCUSSION
The removal of the C-terminal ribosome-binding domain of RelA results in a metabolically unstable constitutive ppGpp synthetase (26, 30), indicating that the ribosome plays an important role in regulating RelA-dependent (p)ppGpp synthesis. The existence of rplK (relC) mutants implicates ribosomal protein L11 in the regulation of RelA activity during the stringent response in E. coli (11, 20). In fact, L11 has, so far, been the only ribosomal protein demonstrated to be involved in the regulation of RelA. However, the basis for the relaxed phenotype of the rplK (relC) mutants has not been determined.
The involvement of L11 in the regulation of RelA activity is especially interesting in view of its important functional role in the bacterial ribosome. L11 is complexed to a 58-nucleotide segment (nucleotides 1051 to 1108) of 23S rRNA (34). The elongation factors, EF-Tu and EF-G, bind to overlapping sites on the ribosome (5, 23). The L11-23S rRNA complex is involved in regulating the activities of EF-Tu and EF-G in the elongation cycle of translation, and because the elongation factors are GTPases the L11-rRNA complex has been referred to as the GTPase-associated site (9). The complex is highly conserved in Bacteria and Archaea, and its crystal structure has recently been solved at 2.8- and 2.6-Å resolution by Conn et al. (8) and Wimberly et al. (34), respectively. The rRNA domain is precisely folded, and the crystal structure suggests that the primary role of L11 is to stabilize the tertiary structure of the rRNA (8). Two domains have been characterized in L11 (34). A C-terminal domain is tightly bound to the tertiary structure of RNA and is responsible for stabilizing its conformation. There are only limited contacts between the RNA and the N-terminal domain of L11. An N-terminal domain is involved in the cooperative binding of thiopeptide antibiotics such as thiostrepton, as discussed below.
We have shown here that bacteria exhibited a relaxed phenotype when ′L11 was overexpressed. On the other hand, the stringent response was unaffected when the complete L11 protein was overexpressed. Overexpressed ′L11 was incorporated into ribosomes and did not affect the binding of RelA. These results indicate that the 36-amino acid N terminus of L11 played an essential role in the activation of RelA during amino acid deprivation. In this regard, it is notable that the ribosomes from the original relC mutant were found to still bind RelA (11). Although the nucleotide sequence of the L11 mutation in this strain has not been reported, it is clear that it affects the activity of RelA, which, in an in vitro assay, was less than 10% of the activity exhibited by the wild type.
Results from both in vitro and in vivo experiments implicate an association between RelA and the thiostrepton-binding site on the ribosome (15, 27, 28, 31). For example, thiostrepton inhibits in vitro RelA-dependent (p)ppGpp synthesis (15, 31). High-affinity binding of thiostrepton to ribosomes requires the L11-rRNA complex (32, 35). Thiostrepton-binding involves nucleotides A1067 and A1095 of 23S rRNA (24) and the proline-rich helix of the L11 N-terminal domain (21, 32, 35). Our results further substantiate a relationship between RelA and the thiostrepton-binding site on L11. To prove that the L11 proline-rich helix was directly involved in regulating RelA, we created the L11Leu-22 derivative. We based the construction of this derivative on the observation that mutations in Pro-22 of L11 confer resistance to thiostrepton (21).
The mechanism by which L11 regulates RelA activity is still unknown. However, considerable progress has been made in elucidating the function of L11, and this information could be useful in formulating a working hypothesis. Based on the L11-23S rRNA crystal structure, Wimberly et al. (34) hypothesize that the N-terminal domain of L11 may represent a molecular switch that alternates between RNA-bound and RNA-free states during the ribosomal elongation cycle. This switching mechanism is proposed to be controlled by the binding of the elongation factors. The results of studies on the action of thiostrepton support this proposal (9, 21). It is noteworthy that, despite the apparent critical function of L11 in translation, E. coli mutants that completely lack this protein are viable (10, 29). To account for this, Wimberly et al. (34) suggest that the putative N-terminal switch may function by regulating the conformation or the accessibility of the RNA in the GTPase-associated site. These L11-deficient mutants may be useful in future studies on RelA regulation.
It has long been hypothesized that the activity of RelA is controlled by conformational changes in the ribosome. To date, we have been unable to demonstrate a direct interaction between L11 and RelA, suggesting that L11, through its N-terminal domain, indirectly controls RelA activation during the stringent response. Our data implicate the involvement of the proposed L11 N-terminal switch domain, and it is possible that this switch promotes the conformational change necessary to activate RelA during amino acid deprivation. Since there are no direct contacts between L11 and other ribosomal proteins (3), the putative L11 N-terminal switch may mediate RelA activation through 23S rRNA.
ACKNOWLEDGMENT
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
We dedicate this paper to the memory of our friend and colleague, Alistair T. Matheson, who passed away on 18 May 2001.
REFERENCES
- 1.Alfoldi L, Stent G S, Clowes R C. The chromosomal site for the RNA control (R.C.) locus in E. coli. J Mol Biol. 1962;5:348–355. doi: 10.1016/s0022-2836(62)80077-1. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Sediman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates & Wiley Interscience; 1994. [Google Scholar]
- 3.Ban N, Nissen P, Hansen J, Moore P B, Steitz T A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 4.Bell R M. Mutants of Escherichia coli defective in membrane phospholipid synthesis: macromolecular synthesis in an sn-glycerol 3-phosphate acyltransferase Km mutant. J Bacteriol. 1974;117:1065–1076. doi: 10.1128/jb.117.3.1065-1076.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabrer B, Vasquez D, Modolell J. Inhibition by elongation factor EF G of aminoacyl-tRNA binding to ribosomes. Proc Natl Acad Sci USA. 1972;69:733–736. doi: 10.1073/pnas.69.3.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cashel M. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid starved stringent strains. J Biol Chem. 1969;244:3133–3141. [PubMed] [Google Scholar]
- 7.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 8.Conn G L, Draper D E, Lattman F E, Gittis A G. Crystal structure of a conserved ribosomal protein-RNA complex. Science. 1999;284:1171–1174. doi: 10.1126/science.284.5417.1171. [DOI] [PubMed] [Google Scholar]
- 9.Cundliffe E. Involvement of specific portions of ribosomal RNA is defined ribosomal functions: a study utilizing antibiotics. In: Hardesty B, Kramer G, editors. Structure, function, and genetics of ribosomes. New York, N.Y: Springer-Verlag; 1986. pp. 586–604. [Google Scholar]
- 10.Dabbs E R. Selection for Escherichia coli mutants with proteins missing from the ribosome. J Bacteriol. 1979;140:734–737. doi: 10.1128/jb.140.2.734-737.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friesen J D, Fiil N P, Parker J M, Haseltine W A. A new relaxed mutant of Escherichia coli with an altered 50S ribosomal subunit. Proc Natl Acad Sci USA. 1974;71:3465–3469. doi: 10.1073/pnas.71.9.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gentry D R, Cashel M. Cellular localization of the Escherichia coli SpoT protein. J Bacteriol. 1995;177:3890–3893. doi: 10.1128/jb.177.13.3890-3893.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glover S W. Valine-resistant mutants of Escherichia coli K-12. Genet Res. 1962;3:448–460. [Google Scholar]
- 14.Gropp M, Strausz Y, Gross M, Glaser G. Regulation of Escherichia coli RelA requires oligomerization of the C-terminal domain. J Bacteriol. 2001;183:570–579. doi: 10.1128/JB.183.2.570-579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haseltine W A, Block R, Gilbert W, Weber K. MSI and MSII are made on ribosomes in an idling step of protein synthesis. Nature. 1972;238:381–384. doi: 10.1038/238381a0. [DOI] [PubMed] [Google Scholar]
- 16.Homann H E, Nierhaus K H. Ribosomal proteins. Protein compositions of biosynthetic precursors and artificial subparticles from ribosomal subunits in Escherichia coli K-12. Eur J Biochem. 1971;20:249–257. doi: 10.1111/j.1432-1033.1971.tb01388.x. [DOI] [PubMed] [Google Scholar]
- 17.Ishiguro E E, Ramey W D. Stringent control of peptidoglycan biosynthesis in Escherichia coli K-12. J Bacteriol. 1976;127:1119–1126. doi: 10.1128/jb.127.3.1119-1126.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lund E, Kjeldgaard N O. Metabolism of guanosine tetraphosphate in Escherichia coli. Eur J Biochem. 1972;28:316–326. doi: 10.1111/j.1432-1033.1972.tb01916.x. [DOI] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 20.Parker J, Watson R J, Friesen J D. A relaxed mutant with an altered ribosomal protein L11. Mol Gen Genet. 1976;144:111–114. doi: 10.1007/BF00277313. [DOI] [PubMed] [Google Scholar]
- 21.Porse B T, Leviev I, Mankin A S, Garrett R A. The antibiotic thiostrepton inhibits a functional transition within protein L11 at the ribosomal GTPase centre. J Mol Biol. 1998;276:391–404. doi: 10.1006/jmbi.1997.1541. [DOI] [PubMed] [Google Scholar]
- 22.Ramagopal S, Davis B D. Localization of the stringent protein of Escherichia coli on the 50S ribosomal subunit. Proc Natl Acad Sci USA. 1974;71:820–824. doi: 10.1073/pnas.71.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richman N, Bodley J W. Ribosomes cannot interact simultaneously with elongation factors EF Tu and EF G. Proc Natl Acad Sci USA. 1972;69:686–689. doi: 10.1073/pnas.69.3.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosendahl G, Douthwaite S. The antibiotics micrococcin and thiostrepton interact directly with 23S rRNA nucleotides 1067A and 1095A. Nucleic Acids Res. 1994;22:357–363. doi: 10.1093/nar/22.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Schreiber G, Metzger S, Aizenman E, Roza S, Cashel M, Glaser G. Overexpression of the relA gene in Escherichia coli. J Biol Chem. 1991;266:3760–3767. [PubMed] [Google Scholar]
- 27.Smith I, Paress P, Cabane K, Dubnau E. Genetics and physiology of the rel system of Bacillus subtilis. Mol Gen Genet. 1980;178:271–279. doi: 10.1007/BF00270472. [DOI] [PubMed] [Google Scholar]
- 28.Smith I, Paress P, Pestka S. Thiostrepton-resistant mutants exhibit relaxed synthesis of RNA. Proc Natl Acad Sci USA. 1978;75:5993–5997. doi: 10.1073/pnas.75.12.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stöffler G, Cundliffe E, Stöffler-Meilicke M, Dabbs E R. Mutants of Escherichia coli lacking ribosomal protein L11. J Biol Chem. 1980;255:10517–105122. [PubMed] [Google Scholar]
- 30.Svitil A L, Cashel M, Zyskind J W. Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J Biol Chem. 1993;268:2307–2311. [PubMed] [Google Scholar]
- 31.Sy J. Reversibility of the pyrophosphoryl transfer from ATP to GTP by Escherichia coli stringent factor. Proc Natl Acad Sci USA. 1974;71:3470–3473. doi: 10.1073/pnas.71.9.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson J, Cundliffe E, Stark M. Binding of thiostrepton to a complex of 23-S rRNA with ribosomal protein L11. Eur J Biochem. 1979;98:261–265. doi: 10.1111/j.1432-1033.1979.tb13184.x. [DOI] [PubMed] [Google Scholar]
- 33.Tosa T, Pizer L I. Effect of serine hydroxamate on the growth of Escherichia coli. J Bacteriol. 1971;106:966–971. doi: 10.1128/jb.106.3.966-971.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wimberly B T, Guymon R, McCutcheon J P, White S W, W. S, Ramakrishnan v. A detailed view of a ribosomal active site: the structure of the L11-RNA complex. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- 35.Xing Y, Draper D E. Cooperative interactions of RNA and thiostrepton antibiotic with two domains of ribosomal protein L11. Biochemistry. 1996;35:1581–1588. doi: 10.1021/bi952132o. [DOI] [PubMed] [Google Scholar]