Abstract
Outer membrane lipoproteins of Escherichia coli are released from the inner membrane upon the formation of a complex with a periplasmic chaperone, LolA, followed by localization to the outer membrane. In vitro biochemical analyses revealed that the localization of lipoproteins to the outer membrane generally requires an outer membrane lipoprotein, LolB, and occurs via transient formation of a LolB-lipoprotein complex. On the other hand, a mutant carrying the chromosomal lolB gene under the control of the lac promoter-operator grew normally in the absence of LolB induction if the mutant did not possess the major outer membrane lipoprotein Lpp, suggesting that LolB is only important for the localization of Lpp in vivo. To examine the in vivo function of LolB, we constructed a chromosomal lolB null mutant harboring a temperature-sensitive helper plasmid carrying the lolB gene. At a nonpermissive temperature, depletion of the LolB protein due to loss of the lolB gene caused cessation of growth and a decrease in the number of viable cells irrespective of the presence or absence of Lpp. LolB-depleted cells accumulated the LolA-lipoprotein complex in the periplasm and the mature form of lipoproteins in the inner membrane. Taken together, these results indicate that LolB is the first example of an essential lipoprotein for E. coli and that its depletion inhibits the upstream reactions of lipoprotein trafficking.
More than 80 species of lipoproteins, lipid-modified proteins, are predicted to exist in the Escherichia coli cell envelope. Each lipoprotein is synthesized in the cytoplasm as a precursor with a signal peptide and then translocated across the cytoplasmic (inner) membrane by protein translocation machinery (5, 14, 18). Lipid modification and processing to the mature lipoprotein take place on the outer surface of the inner membrane. Lipoprotein-specific signal peptidase cleaves the signal peptide after the cysteine residue present at the N terminus of the mature domain has been modified with diglyceride (7, 14). Further fatty acylation of the cysteine residue then takes place to complete the processing. The mature lipoprotein thus formed is then localized to either the inner or the outer membrane, depending on the amino acid residue immediately after the N-terminal cysteine (14). Aspartate at this position functions as an inner membrane retention signal, whereas other amino acid residues target lipoproteins to the outer membrane (22). It was recently reported that, in addition to aspartate, some other residues at the +2 position also function as an inner membrane retention signal, although these residues are rarely found in native lipoproteins (15).
We found that five Lol proteins are involved in the sorting signal-dependent localization of lipoproteins (12, 13, 17, 19, 21, 24). An ABC (ATP binding cassette) transporter comprising LolC, LolD, and LolE releases outer membrane-directed lipoproteins from the inner membrane in an ATP-dependent manner, leading to the formation of a LolA-lipoprotein complex (12, 19, 21). LolA, as a periplasmic chaperone, then transports lipoproteins from the inner to the outer membrane (12, 17, 24). Lipoproteins with the inner membrane sorting signal remain in the inner membrane even in the presence of LolA and the ABC transporter (12, 19, 24). Upon interaction of the LolA-lipoprotein complex with an outer membrane lipoprotein, LolB, lipoproteins are transferred from LolA to LolB and then incorporated into the outer membrane (13, 24).
We previously constructed a mutant in which the chromosomal lolB gene is under the control of the lac promoter-operator (13) and prepared the outer membrane from this mutant to show that in vitro outer membrane localization of lipoproteins generally requires LolB (13, 24). A mutant possessing the major outer membrane lipoprotein Lpp could not grow in the absence of the lac inducer (13). However, we later found that a mutant lacking Lpp grew normally in the absence of the inducer, as if LolB were dispensable for the in vivo localization of lipoproteins except Lpp. Therefore, the significance of the in vivo function of LolB, especially for the outer membrane localization of lipoproteins other than Lpp, remains unclear. To address this issue, we constructed a conditional lolB null mutant in which the kan gene replaced the chromosomal lolB gene and the intact lolB gene was supplied by a helper plasmid carrying a temperature-sensitive replicon. We show here that depletion of LolB results in a decrease in viable cells, irrespective of the presence or absence of Lpp, and the accumulation of a LolA-lipoprotein complex in the periplasm as a localization intermediate.
MATERIALS AND METHODS
Bacteria and plasmids.
The E. coli K-12 strains and plasmids used in this study are listed in Table 1.
TABLE 1.
E. coli K-12 strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Reference |
|---|---|---|
| Strains | ||
| FS1576 | supE44 hsdR thi-1 thr-1 leuB6 lacY1 tonA21 recD1009 | 16 |
| JE5506 | pps his proA argE thi gal lac xyl mtl tsx | 8 |
| JE5505 | JE5506 lpp | 8 |
| SM704 | JE5505 lac-lolB | 13 |
| JC10240 | Hfr thi-300 recA56 srl::Tn10 relA1 ilv-318 spoT1 thi rpsE2300 | 4 |
| KT1 | FS1576 ΔlolB::kan/pKT021 | This study |
| KT2 | recA56 srl::Tn10 conjugant of KT1 (donor, JC10240) | This study |
| KT3 | ΔlolB::kan transductant of JE5505 harboring pKT021 (donor, KT1) | This study |
| KT4 | ΔlolB::kan transductant of JE5506 harboring pKT021 (donor, KT1) | This study |
| KT5 | recA56 srl::Tn10 conjugant of KT3 | This study |
| KT6 | recA56 srl::Tn10 conjugant of KT4 | This study |
| Plasmids | ||
| pEL3 | bla, temperature-sensitive replicon | 1 |
| pSY343 | kan str | 23 |
| pMAN997 | bla, temperature-sensitive replicon, multicloning sites | This study |
| pKT020 | bla, lolB on pMAN651 replaced by kan | This study |
| pKT021 | bla, cloned gene, lolB, temperature-sensitive replicon | This study |
| pMAN651 | bla, cloned gene, lolB ychB | 13 |
| pTAN20 | cat; cloned gene, arabinose promoter-controlled pal | 24 |
Media and chemicals.
L broth was used as a standard medium. Labeling experiments were carried out with M63 minimal medium supplemented with all of the amino acids, except methionine and cysteine, at 20 μg/ml each. When required, ampicillin, kanamycin, tetracycline, and chloramphenicol were added at concentrations of 50, 25, 25, and 25 μg/ml, respectively. Restriction endonucleases, T4 DNA ligase, and a PCR kit were purchased from Takara Shuzo Co. Tran35S-label (a mixture of 70% [35S]methionine and 20% [35S]cysteine, 1,000 Ci/mmol) was obtained from ICN. Antibodies were raised against the respective purified proteins in rabbits.
Construction of plasmids.
A DNA fragment containing the kan gene and the truncated str gene was amplified by PCR with pSY343 (23) as a template and a pair of primers (5′-ACTCGCTAGCAAGCTTCACGCTGCCGCAAG-3′ and 5′-CGTACTCGAGGTGGGTGGTGAGCAGCTCGC-3′) and then digested with NheI and XhoI. The 1.7-kb DNA fragment thus obtained was ligated with an 8.4-kb NheI-SalI DNA fragment of pMAN651 (13) to construct pKT020. In this plasmid, the kan gene replaced the region encompassing the Shine-Dalgarno sequence for lolB through the C-terminal coding region of lolB. The lolB gene forms an operon with downstream ychB, which was recently reported to be essential for E. coli (3, 6). In order to circumvent the negative polar effect of the lolB deletion on ychB expression, pKT020 carried the truncated str gene, which was fused in frame to the C-terminal coding region of lolB.
To construct pMAN997 carrying the bla gene, multicloning sites, and a temperature-sensitive replicon, a 0.8-kb ScaI-BglII fragment obtained from pSP72 (Promega) was ligated with a 3.2-kb ScaI-BamHI fragment of pEL3 (1). A 1.7-kb BglII-EcoRI fragment containing the entire lolB gene was inserted into the BamHI-EcoRI sites of pMAN997 to construct a helper plasmid, pKT021.
Construction of ΔlolB::kan strains.
Construction of a lolB null allele mutant harboring the helper plasmid was carried out at 30°C. A 7.4-kb SacI-XbaI DNA fragment containing the ΔlolB::kan gene was isolated from pKT020 and transformed into a recD strain, FS1576, harboring pKT021. One of the kanamycin-resistant transformants, KT1, was mated with JC10240 (recA) to construct a recA mutant derivative, KT2. The ΔlolB::kan allele of KT1 was introduced into JE5505 and JE5506 harboring pKT021 by P1 transduction, followed by conjugation to introduce recA. All recA strains were scored for UV sensitivity. Disruption of the chromosomal lolB gene and the organization of the downstream region were confirmed by PCR with the recA mutant transconjugants KT5 and KT6.
Immunoprecipitation, sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and Western blot analyses.
Immunoprecipitation was carried out as previously described (11). SDS-PAGE for Lpp was performed as previously described (9). Other proteins were analyzed on 12.5% polyacrylamide gels as described by Laemmli (10). Proteins labeled with Tran35S-label were analyzed by SDS-PAGE, followed by fluorography with Enlightning (NEN Life Science Products, Inc). Western blot analyses were carried out as previously described (11).
Preparation of the periplasm fraction.
Cells were converted into spheroplasts in the presence of 0.5 mM EDTA and lysozyme at 60 μg/ml as previously described (12) and then centrifuged at 16,000 × g for 2 min. The supernatant was centrifuged again at 100,000 × g for 30 min to remove insoluble materials. The supernatant thus obtained was used as the periplasm fraction.
Separation of the inner and outer membranes.
Spheroplasts prepared as described above were sonicated and centrifuged at 100,000 × g for 30 min. The pellets were fractionated by 25 to 55% (wt/wt) sucrose density gradient centrifugation to separate the inner and outer membranes as previously described (12).
RESULTS
Effect of LolB depletion on the growth of a lac-lolB mutant.
We previously constructed a mutant in which the chromosomal lolB gene was under the control of the lac promoter-operator. We then revealed with the outer membrane prepared from this mutant that the in vitro outer membrane incorporation of lipoproteins, Lpp, Pal, NlpB, and LolB per se, requires LolB (13, 24). Moreover, we showed that LolB depletion causes severe growth inhibition with a concomitant decrease in the number of viable cells (13). Strikingly, however, we found that such severe growth defects were only observed when the mutant expressed Lpp. The lac-lolB mutant lacking Lpp continued to grow normally, and lipoproteins were localized to the outer membrane even after LolB had become hardly detectable in the total membrane fraction prepared from uninduced cells (data not shown). Although the previous in vitro experiments revealed that the LolB function is generally important for the outer membrane incorporation of lipoproteins (13, 24), these results suggested that LolB is dispensable in the absence of Lpp. However, since the number of Lpp molecules in a single cell is several orders of magnitude higher than those of other lipoproteins (7), it seemed possible that a trace amount of LolB expressed in the absence of an inducer is sufficient for the in vivo localization of other lipoproteins. To address this, we constructed a mutant carrying a conditional lolB null allele.
Construction of a conditional lolB null mutant harboring a helper plasmid.
The lolB gene was first replaced with the kan gene derived from pSY343 to construct pKT020. A 7.4-kb DNA fragment of pKT020 was transformed into recD strain FS1576. Double-crossover transformants possessing the ΔlolB::kan gene were readily isolated at 30°C only when the lolB gene was provided by helper plasmid pKT021. The ΔlolB::kan gene was next introduced into isogenic strains JE5505 (lpp) and JE5506 (lpp+), harboring pKT021, by P1 transduction. The resulting transductants, KT3 (ΔlolB::kan lpp/pKT021) and KT4 (ΔlolB::kan lpp+/pKT021), were expected to lose the lolB gene at 42°C since pKT021 carries a temperature-sensitive replication origin. To prevent the integration of the helper plasmid into the chromosome through homologous recombination, the recA allele was further introduced into KT3 and KT4 by conjugation.
KT5 (ΔlolB::kan lpp recA/pKT021) and KT6 (ΔlolB::kan lpp+ recA/pKT021) thus constructed were grown at 30°C in the absence of ampicillin and then transferred to 42°C. Helper plasmid pKT021 became undetectable on ethidium bromide staining after 7 to 8 generations of growth at 42°C, suggesting that both KT5 and KT6 had lost the functional lolB gene due to the lack of plasmid replication. Since the copy number of the original vector plasmid, pSC101, was 5 to 7 (2), the average copy number of pKT021 in a single cell was expected to be less than 0.03 after 8 generations at 42°C, provided that no replication of the helper plasmid occurred at a nonpermissive temperature.
LolB is an essential lipoprotein, irrespective of the presence or absence of Lpp.
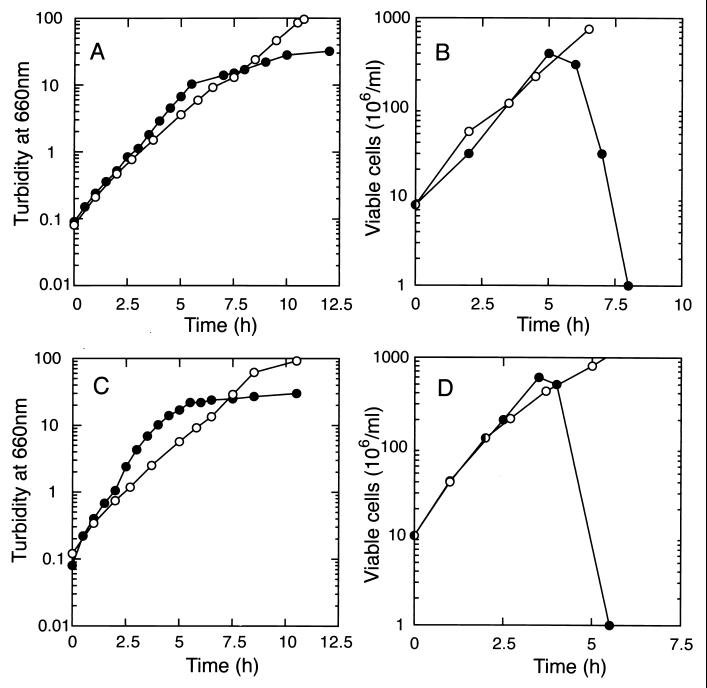
The effect of Lpp on growth in the absence of the functional lolB gene was examined (Fig. 1). The cessation of growth observed after 6 to 7 generations of growth at 42°C was more complete with KT6 than with KT5 (Fig. 1A and C). The number of viable cells started to decrease dramatically for both mutants before the culture turbidity stopped increasing. The decrease in viable cells occurred earlier and more drastically for KT6 than for KT5 (Fig. 1B and D). These results indicate that the lolB gene is essential, irrespective of the presence or absence of Lpp, although the defect is observed earlier in the presence of Lpp. The mutants grown at 30 and 42°C were indistinguishable in morphology and the same as the parental strains.
FIG. 1.
Effect of Lpp on the growth of the ΔlolB::kan mutant. KT5 (A and B) and KT6 (C and D) were grown at 30°C (open circles) or 42°C (closed circles) for the indicated times by inoculating portions of the cultures into fresh medium. The culture turbidity at 660 nm was monitored and plotted after correction for the culture dilution (A and C). The number of viable cells (B and D) during growth at 30°C and 42°C was determined at the specified times by plating aliquots of the cultures onto L broth containing kanamycin at 25 μg/ml, followed by overnight incubation at 30°C.
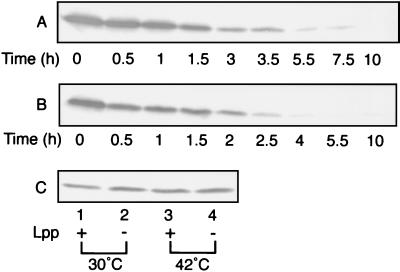
The amounts of LolB in KT5 and KT6 grown at 42°C were determined by immunoblotting with anti-LolB antibodies (Fig. 2A and B). The LolB protein became undetectable in both KT5 and KT6 after 8 to 10 generations at 42°C, whereas LolB in parental strains JE5505 and JE5506 was stable at 42°C (Fig. 2C). The LolB depletion did not affect the level of LolA (data not shown).
FIG. 2.
Depletion of LolB in the ΔlolB::kan mutant at 42°C. KT5 (A) and KT6 (B) grown at 30°C on L broth containing kanamycin at 25 μg/ml were transferred to 42°C and then cultured for the indicated times. Aliquots of the cultures were withdrawn and treated with Laemmli sample buffer at 100°C for 5 min. Cellular proteins (100 μg) were analyzed by SDS-PAGE and immunoblotting with anti-LolB antibodies. As controls (C), JE5505 (lanes 2 and 4) and JE5506 (lanes 1 and 3) grown for 10 h at 30°C (lanes 1 and 2) and 42°C (lanes 3 and 4) were also analyzed.
Periplasmic accumulation of the LolA-lipoprotein complex in LolB-depleted cells.
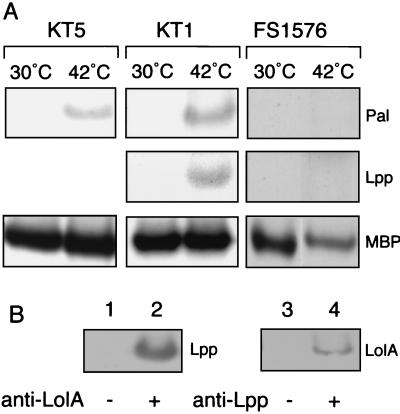
The effect of LolB depletion on lipoprotein localization was examined in vivo. Cultures of KT1, KT5, and FS1576 grown at permissive and nonpermissive temperatures were labeled with Tran35S-label for 30 s. The periplasm fractions of these cells were immunoprecipitated with anti-Pal, anti-Lpp, or anti-maltose binding protein (MBP) antibodies and then analyzed by SDS-PAGE and fluorography. When grown at 42°C, both mutants accumulated Pal in the periplasm (Fig. 3A). Lpp was also found in the periplasm of KT1 grown at 42°C. In contrast, neither the mutant grown at 30°C nor the wild-type cells grown at 42°C accumulated these lipoproteins in their periplasm (Fig. 3A). The periplasmic MBP was found in the periplasm of all strains. When the periplasm fraction of KT1 was subjected to immunoprecipitation with anti-LolA antibodies, Lpp was coimmunoprecipitated (Fig. 3B, lane 2). Moreover, on immunoprecipitation with anti-Lpp antibodies, LolA was coprecipitated (Fig. 3B, lane 4). These results indicate that accumulated Lpp in the periplasm of KT1 grown at 42°C exists as a soluble complex with LolA.
FIG. 3.
Accumulation of Pal and Lpp in the periplasm of LolB-depleted cells. (A) KT5, KT1, and FS1576 were grown on M63 minimal medium supplemented with 0.2% maltose at 30°C or 42°C and then labeled with Tran35S-label for 30 s. The labeling at 42°C was started immediately before growth arrest occurred and stopped by the addition of cold methionine and cysteine on ice. Periplasm fractions were prepared from these cells as described in Materials and Methods and then subjected to immunoprecipitation with antibodies raised against Pal, Lpp, and MBP. The precipitates were analyzed by SDS-PAGE and fluorography. (B) The periplasm fraction prepared from KT1 cells grown on L broth at 42°C was immunoprecipitated with nonimmune (lanes 1 and 3), anti-LolA (lane 2), and anti-Lpp (lane 4) antibodies and then analyzed by SDS-PAGE and immunoblotting with anti-Lpp (lanes 1 and 2) and anti-LolA (lanes 3 and 4) antibodies.
LolB depletion causes mislocalization of Pal in the inner membrane.
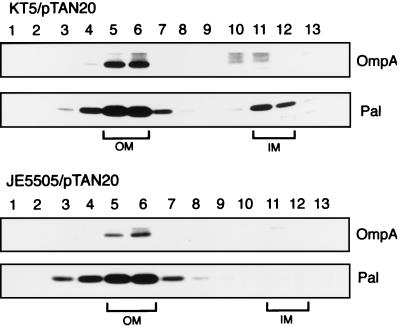
We expressed and radiolabeled Pal in strain KT5 and parental strain JE5505 at 42°C. Envelope fractions prepared from these cells were fractionated into inner and outer membranes by sucrose density gradient centrifugation (Fig. 4). A portion of Pal was found as a mature form in the inner membrane of the LolB-depleted KT5 cells, whereas Pal was localized exclusively to the outer membrane of JE5505 (Fig. 4). LolB depletion did not affect the outer membrane localization of OmpA. Taken together, these results indicate that LolB depletion causes accumulation of the LolA-lipoprotein complex in the periplasm, which inhibits the catalytic cycle of LolA, thereby causing accumulation of the mature lipoprotein in the inner membrane.
FIG. 4.
Accumulation of Pal in the inner membrane of LolB-depleted cells. KT5/pTAN20 and JE5505/pTAN20 were grown at 42°C and then labeled with Tran35S-label as described in the legend to Fig. 3. Envelope fractions were prepared from the labeled cells and subjected to sucrose density gradient centrifugation, followed by fractionation into 13 fractions from the bottom to the top of the gradient. Each fraction was analyzed by SDS-PAGE and fluorography. OmpA and Pal were identified on the gel by their molecular masses and migration positions. OM and IM represent the outer and inner membrane fractions, respectively.
DISCUSSION
In contrast to the lac-lolB mutant (13), LolB depletion was lethal for the lolB null mutant constructed in this study, irrespective of the presence or absence of Lpp. Although LolB was undetectable in both mutants after several generations of growth under LolB depletion conditions, the significance of LolB depletion seems to differ between these two mutants. In the lac-lolB mutant, a basal level of LolB is likely to be expressed in most mutant cells and to be sufficient for the localization of all lipoproteins except Lpp. On the other hand, most lolB null mutant cells are expected to have lost the helper plasmid carrying lolB after several generations of growth at a nonpermissive temperature. Therefore, the depletion of LolB should be more complete for the lolB null mutant than for the lac-lolB mutant. The results presented here indicate that LolB is an essential lipoprotein and critically important for the in vivo localization of not only Lpp but also other lipoproteins to the outer membrane. Previous observations and the results presented here also indicate that in vivo outer membrane localization is very efficient, so the basal level of LolB is sufficient to support the localization of all E. coli lipoproteins except Lpp. However, the basal level of LolB was undetectable with conventional anti-LolB antibodies, even when a large amount of membranes was examined.
LolB depletion caused accumulation of the LolA-lipoprotein complex in the periplasm and mature forms of lipoproteins in the inner membrane. Judging from the amino-terminal sequences of more than 80 putative lipoproteins, most lipoproteins are assumed to be localized to the outer membrane. Although not examined in this study, it seems likely, therefore, that various lipoproteins are also accumulated in the periplasm and the inner membrane in the absence of LolB, causing cessation of growth and a decrease in the number of viable cells. We previously showed that the mislocalization of Lpp to the inner membrane inhibits the growth of cells (20). The results presented here indicate that, in addition to Lpp and LolB (13, 20), there may be other lipoproteins whose mislocalization is toxic to E. coli. Alternatively, mislocalization of various lipoproteins may perturb the integrity of the cell surface structure and thus be lethal for E. coli. We speculate that the lipoprotein family contains physiologically important cell surface proteins. The correct localization of lipoproteins depends exclusively on the Lol system in E. coli.
ACKNOWLEDGMENTS
We thank Rika Ishihara for technical assistance and secretarial support.
This work was supported by grants to H.T. from CREST of the Japan Science and Technology Corporation and from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Armstrong K A, Acosta R, Ledner E, Machida Y, Pancotto M, McCormick M, Ohtsubo H, Ohtsubo E. A 37 × 103 molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J Mol Biol. 1984;175:331–348. doi: 10.1016/0022-2836(84)90352-8. [DOI] [PubMed] [Google Scholar]
- 2.Cabello F, Timmis K, Cohen S N. Replication control in a composite plasmid constructed by in vitro linkage of two distinct replicons. Nature. 1976;259:285–290. doi: 10.1038/259285a0. [DOI] [PubMed] [Google Scholar]
- 3.Campos N, Rodriguez-Concepcion M, Sauret-Gueto S, Gallego F, Lois L M, Boronat A. Escherichia coli engineered to synthesize isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate: a novel system for the genetic analysis of the 2-C-methyl-d-erythritol 4-phosphate pathway for isoprenoid biosynthesis. Biochem J. 2001;353:59–67. [PMC free article] [PubMed] [Google Scholar]
- 4.Csonka L N, Clark A J. Construction of an Hfr strain useful for transferring recA mutations between Escherichia coli strains. J Bacteriol. 1980;143:529–530. doi: 10.1128/jb.143.1.529-530.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duong F, Wickner W. Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freiberg C, Wieland B, Spaltmann F, Ehlert K, Brotz H, Labischinski H. Identification of novel essential Escherichia coli genes conserved among pathogenic bacteria. J Mol Microbiol Biotechnol. 2001;3:483–489. [PubMed] [Google Scholar]
- 7.Hayashi S, Wu H C. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 8.Hirota Y, Suzuki H, Nishimura Y, Yasuda S. On the process of cellular division in Escherichia coli: a mutant of E. coli lacking a murein-lipoprotein. Proc Natl Acad Sci USA. 1977;74:1417–1420. doi: 10.1073/pnas.74.4.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussain M, Ichihara S, Mizushima S. Accumulation of glyceride-containing precursor of the outer membrane lipoprotein in the cytoplasmic membrane of Escherichia coli treated with globomycin. J Biol Chem. 1980;255:3707–3712. [PubMed] [Google Scholar]
- 10.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Matsuyama S, Fujita Y, Mizushima S. SecD is involved in the release of translocated secretory proteins from the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:265–270. doi: 10.1002/j.1460-2075.1993.tb05652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuyama S, Tajima T, Tokuda H. A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane. EMBO J. 1995;14:3365–3372. doi: 10.1002/j.1460-2075.1995.tb07342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuyama S, Yokota N, Tokuda H. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 1997;16:6947–6955. doi: 10.1093/emboj/16.23.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seydel A, Gounon P, Pugsley A P. Testing the '+2 rule' for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol Microbiol. 1999;34:810–821. doi: 10.1046/j.1365-2958.1999.01647.x. [DOI] [PubMed] [Google Scholar]
- 16.Stahl F W, Kobayashi I, Thaler D, Stahl M M. Direction of travel of RecBC recombinase through bacteriophage lambda DNA. Genetics. 1986;113:215–227. doi: 10.1093/genetics/113.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tajima T, Yokota N, Matsuyama S, Tokuda H. Genetic analyses of the in vivo function of LolA, a periplasmic chaperone involved in the outer membrane localization of Escherichia coli lipoproteins. FEBS Lett. 1998;439:51–54. doi: 10.1016/s0014-5793(98)01334-9. [DOI] [PubMed] [Google Scholar]
- 18.Tokuda H. Biochemical characterization of the presecretory protein translocation machinery of Escherichia coli. FEBS Lett. 1994;346:65–68. doi: 10.1016/0014-5793(94)00317-3. [DOI] [PubMed] [Google Scholar]
- 19.Yakushi T, Masuda K, Narita S, Matsuyama S, Tokuda H. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat Cell Biol. 2000;2:212–218. doi: 10.1038/35008635. [DOI] [PubMed] [Google Scholar]
- 20.Yakushi T, Tajima T, Matsuyama S, Tokuda H. Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. J Bacteriol. 1997;179:2857–2862. doi: 10.1128/jb.179.9.2857-2862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yakushi T, Yokota N, Matsuyama S, Tokuda H. LolA-dependent release of a lipid-modified protein from the inner membrane of Escherichia coli requires nucleoside triphosphate. J Biol Chem. 1998;273:32576–32581. doi: 10.1074/jbc.273.49.32576. [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi K, Yu F, Inouye M. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell. 1988;53:423–432. doi: 10.1016/0092-8674(88)90162-6. [DOI] [PubMed] [Google Scholar]
- 23.Yasuda S, Takagi T. Overproduction of Escherichia coli replication proteins by the use of runaway-replication plasmids. J Bacteriol. 1983;154:1153–1161. doi: 10.1128/jb.154.3.1153-1161.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokota N, Kuroda T, Matsuyama S, Tokuda H. Characterization of the LolA-LolB system as the general lipoprotein localization mechanism of Escherichia coli. J Biol Chem. 1999;274:30995–30999. doi: 10.1074/jbc.274.43.30995. [DOI] [PubMed] [Google Scholar]